Abstract
Polyadenylation in eukaryotes is conventionally associated with increased nuclear export, translation, and stability of mRNAs. In contrast, recent studies suggest that the Trf4 and Trf5 proteins, members of a widespread family of noncanonical poly(A) polymerases, share an essential function in Saccharomyces cerevisiae that involves polyadenylation of nuclear RNAs as part of a pathway of exosome-mediated RNA turnover. Substrates for this pathway include aberrantly modified tRNAs and precursors of snoRNAs and rRNAs. Here we show that Cid14 is a Trf4/5 functional homolog in the distantly related fission yeast Schizosaccharomyces pombe. Unlike trf4 trf5 double mutants, cells lacking Cid14 are viable, though they suffer an increased frequency of chromosome missegregation. The Cid14 protein is constitutively nucleolar and is required for normal nucleolar structure. A minor population of polyadenylated rRNAs was identified. These RNAs accumulated in an exosome mutant, and their presence was largely dependent on Cid14, in line with a role for Cid14 in rRNA degradation. Surprisingly, both fully processed 25S rRNA and rRNA processing intermediates appear to be channeled into this pathway. Our data suggest that additional substrates may include the mRNAs of genes involved in meiotic regulation. Polyadenylation-assisted nuclear RNA turnover is therefore likely to be a common eukaryotic mechanism affecting diverse biological processes.
In eukaryotic cells, most genes are transcribed into precursor messenger RNAs that have to undergo several processing events before maturation, including addition of a cap structure to the 5′ terminus, removal of introns by splicing, and 3′ cleavage and polyadenylation. A key player in the polyadenylation process is the nuclear poly(A) polymerase (PAP), part of a large cleavage and polyadenylation complex that becomes activated during transcription by RNA polymerase II. The poly(A) tail is a hallmark of mRNA and is necessary both for subsequent transport into the cytoplasm and efficient translation. Shortening of the poly(A) tail in the cytoplasm can trigger translational repression and acts as a signal for rapid decay of transcripts (50).
In addition to the canonical nuclear enzyme, eukaryotic cells contain several unconventional PAPs. In a study of the fission yeast Schizosaccharomyces pombe we previously identified the cid1 gene family, which encodes a distinct group of PAPs conserved from yeast to humans (46). In Caenorhabditis elegans, the Cid1-related protein GLD-2 was characterized as a cytoplasmic PAP (42), and Xenopus laevis GLD-2 was subsequently found to be required for CPEB-mediated cytoplasmic polyadenylation during early development (3). Kwak et al. (20) recently described five human orthologs of the Cid1 protein, named Hs1 to Hs5. Among these, Hs4 is responsible for the synthesis of long poly(A) tails in human mitochondria (37). Sequence comparisons identify six members of the S. pombe cid1 gene family (cid1, cid11, cid12, cid13, cid14, and cid16) (46). Cid1 (for caffeine-induced death suppressor), the first member of this family to be described, was identified through its ability when overexpressed to confer resistance specifically to the combination of hydroxyurea and caffeine, which overrides the replication checkpoint (44). Cid13 was identified independently based on its ability, when overexpressed, to rescue the hydroxyurea sensitivity of checkpoint rad mutants (29, 31). In addition to its related role in checkpoint control, Cid12 is required for faithful chromosome segregation and RNA interference-mediated heterochromatin assembly at centromeres (24, 46). Purified recombinant Cid1 catalyzed polyadenylation in vitro (29), as did Cid13-myc immunoprecipitated from S. pombe (31). Both Cid1 and Cid13 are cytoplasmic proteins, indicating that their function is likely to be distinct from that of the nuclear PAP. It has been proposed that Cid13 acts to extend the poly(A) tails of ribonucleotide reductase mRNA to counteract deadenylation and enhance protein synthesis (31).
In Saccharomyces cerevisiae the Cid1-related protein Trf4 (which has a redundant homolog, Trf5) was reported to have DNA polymerase activity in vitro (47). Subsequently, immunoprecipitates of Trf4-hemagglutinin (HA) from S. cerevisiae were found to have PAP, but not DNA polymerase, activity (31). However, unlike most of the Cid proteins, Trf4 and Trf5 are nuclear (12) and are proposed to function in a nuclear RNA surveillance system, whereby the Trf4-mediated oligoadenylation of RNAs targets them for rapid degradation by the exosome. This resembles the role of polyadenylation in bacterial RNA turnover (15, 21, 40, 51).
In this study, we describe the characterization of fission yeast Cid14, a functional homologue of S. cerevisiae Trf4/5. We present evidence that Cid14 is required for the polyadenylation of rRNAs as a prelude to their degradation by the exosome. Polyadenylation mediated by Cid1-related proteins therefore appears to have different consequences and to affect diverse functions, depending on the subcellular localization and identity of the target RNA.
MATERIALS AND METHODS
Fission yeast strains and methods.
Conditions for growth, maintenance, and genetic manipulation of fission yeast were as described previously (23). A complete list of the strains used in this study is given in Table 1. Except where stated otherwise, strains were grown at 30°C in YE5S or EMM2 medium (23) with appropriate supplements. Meiosis and sporulation were induced by plating onto malt extract agar, and tetrad dissection was performed with an MSM micromanipulator (Singer Instruments, Watchet, United Kingdom). Cell concentration was determined with a Sysmex F-800 cell counter (TOA Medical Electronic, Kobe, Japan).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| 972 | h− | Lab stock |
| SW0962 | h−cid14::ura4 ura4 | This study |
| SW0139 | h−cid14::LEU2 leu1 ura4 | This study |
| SW0269 | h−top1::LEU2 leu1 ura4 | 41 |
| SW0130 | h−cid14::ura4 top1::LEU2 leu1 ura4 | This study |
| SW0155 | h−cid14-GFP::kanrleu1 ura4 | This study |
| SW0295 | h−nda3-KM311 cid14-GFP::kanrleu1 ura4 | This study |
| SW0106 | h−cid14-HA::kanrleu1 ura4 | This study |
| SW0161 | h−cdc25-22 cid14-HA::kanr | This study |
| SW0292 | h−ndc80-GFP::kanrleu1 ura4 | 49 |
| SW0305 | h+nda3-KM311 ndc80-GFP::kanrleu1 ura4 his2 | 45 |
| SW0563 | h+cid14::ura4 nda3-KM311 ndc80-GFP::kanrleu1 ura4 his2 | This study |
| SW0646 | h−cid12::ura4 ura4 | 46 |
| SW0554 | h+cid12::ura4 nda3-KM311 ndc80-GFP::kanrleu1 ura4 his2 | This study |
| SW0240 | h−gar2-GFP::kanrleu1 ura4 | 34 |
| SW0131 | h+cid14::ura4 gar2-GFP::kanrleu1 ura4 | This study |
| SW0231 | h−bub1::LEU2 ade6 leu1 ura4 his1 | 4 |
| SW0262 | h−mad2::ura4 ade6 leu1 ura4 | 10 |
| SW1092 | h−dis3-54 | 17 |
| SW1195 | h+dis3-54 cid14::ura4 ura4 | This study |
| SW0428 | h−pat1-114 | Lab stock |
| CY950a | MATatrf4ts top1::LEU2 ura3 ade2 his3 leu2 trp1 | 7 |
S. cerevisiae strain.
Minichromosome loss rates were measured as described previously (1). Briefly, cells from ade+ colonies were plated on yeast extract (YE) plates and incubated at 30°C for 5 days. The colonies with red sectors covering at least one-half of the colony were counted. The rate of chromosome loss per division is the number of these half-sectored colonies divided by the total number of white colonies plus half-sectored colonies.
Plasmids and site-directed mutagenesis.
Plasmid pCB427 (TRF4 URA3 CEN) was kindly provided by M. F. Christman (7). Plasmids for overexpression of cid1 and cid14 under the control of the GAL1 promoter were constructed by PCR amplification of the cid1+ and cid14+ open reading frames from S. pombe genomic DNA using the primer pairs CGCGGATCCGCGAATATGAACATTTCTTCTGCAC and ATAGTTTAGCGGCCGCATTCTTATTCACTCAGAATTGTCACCATC and CGCGGATCCGCGGAGATGGGTAAAAAAAGCGTGTC and ATAGTTTAGCGGCCGCATTCTTATCTAAAAACGTTTGCGTATTTTTTTCGC, respectively, followed by digestion with BamHI and NotI and subsequent ligation into BamHI- and NotI-cleaved pYES2 (Invitrogen), generating pYES2cid1 and pYES2cid14. pREP1cid14 was constructed by PCR amplification of the cid14 open reading frame from S. pombe genomic DNA using the primer pair CID14F (TTTGGATCCCATGGGTAAAAAAAGCGTGTCATTTAACCGC) and CID14R (TTTGGATCCCTAAAAACGTTTGCGTATTTTTTTCGCTCG), followed by digestion with BamHI and subsequent ligation into BamHI-cleaved pREP1. PCR using primers CID14R and DADAF (CCAAATTATATTTACCTACTTCAGCTCTTGCTCTTGTAATAATATCACCTGAG) and primers CID14F and DADAR (CTCAGGTGATATTATTACAAGAGCAAGAGCTGAAGTAGGTAAATATAATTTGG) was used to generate the cid14 open reading frame in two fragments overlapping by 53 bp, with the region of overlap spanning codons 298 and 300, which were altered in the primer sequence to encode alanine rather than the aspartate residues specified by the wild-type gene at these positions. The resulting fragments were mixed and used in a secondary PCR with primers CID12F and CID12R. After digestion with BamHI, the final product was ligated into pREP1 to generate pREP1cid14DADA. All plasmid constructions were confirmed by complete sequencing of the inserts using an ABI sequencer and ABI PRISM dRhodamine reagents (Applera UK).
Gene disruption.
The one-step disruption method was used following PCR-mediated generation of the entire ura4+ gene flanked by 80-bp segments from the 5′ and 3′ regions of cid14+ using oligonucleotides TGGTTCAATTAGTCATATATTGATTTTTATAAGAATTCTACAAGATAGGTATAGAAAACCATTCTATCCATGTCTTAGAGAAATCCCACTGGCTATATGT and ACATT TTAACATATGCTCACGATGTTGACGAACCCCTTCATCAACTGATAAT ATGGTACTTAGGATACTAGGGAAATCTTAATTCTAAATGCCTTCTG AC. Construction of the chromosomally HA- and green fluorescent protein (GFP)-tagged cid14 strains (cid14-HA and cid14-GFP) was accomplished by an analogous method using primers CGGAAGACGAAGTACCTATTATTGAGGACACCACTGCTTCAGATGAAGAATCTCGAGCGAAAAAAATACGCAAACGTTTTCGGATCCCCGGGTTAATTAA and AAAATTACTACAGATCCATAGTGAGTTTAAGGTGTTGATTAATTTATTACTGGTATTCGAGTTT ACTGGCTAAATCGTAAGAATTCGAGCTCGTTTAAAC. The cid14::LEU2 allele was generated by a secondary one-step disruption of the cid14::ura4+ allele using primers as described previously (46).
Antibodies and immunoblotting.
Cell extracts were prepared by trichloroacetic acid precipitation following glass bead disruption (6). Immunoblotting was performed essentially as described elsewhere (2). The mouse anti-influenza virus HA monoclonal antibody HA-11 (Babco, Berkeley, CA) was used for detection of HA-tagged Cid14 proteins. Cdc2 was detected using the mouse monoclonal antibody Y100 (generated by J. Gannon and kindly provided by H. Yamano). Horseradish peroxidase-conjugated anti-mouse antibodies (Sigma, Poole, United Kingdom) and enhanced chemiluminescence (ECL; Amersham) were used to detect bound antibody.
Gel filtration chromatography.
Chromatographic separation of S. pombe lysate from a 100-ml culture of the cid14-HA strain in midexponential growth was carried out using a Superose-6 column attached to an fast protein liquid chromatography workstation (Amersham Pharmacia) as previously described (45). The eluate was collected in 0.5-ml fractions, and these were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting. The column was calibrated using the standards dextran blue (∼2,000 kDa), thyroglobulin (669 kDa), apoferritin (443 kDa), β-amylase (200 kDa), bovine serum albumin (66 kDa), and carbonic anhydrase (29 kDa).
Microscopy.
Visualization of GFP in living cells, embedded in 0.6% low-melting-point agarose after staining with Hoechst 33342 (5 μg/ml), was performed at room temperature as previously described (45). Images were acquired using a Zeiss Axioplan 2 microscope equipped with a Planapochromat 100× objective, an Axiocam cooled charge-coupled device camera, and Axiovision software (Carl Zeiss, Welwyn Garden City, United Kingdom) and were assembled using Adobe PhotoShop.
RNA isolation and quantitative PCR.
Total RNA was isolated by hot-phenol extraction and purified using RNeasy (QIAGEN). Total S. pombe RNA (0.5 μg) was reverse transcribed (SuperScript; Invitrogen) using an oligo(dT) primer according to the manufacturer's instructions. Quantitative PCR was performed using SYBR green PCR Master Mix (Applera UK) and primer pairs specific for ste11 (ATACCTGGAGAAGCGACGAA and GAATGAGCGGCGAACTCATA), mei2 (TATGGACGAGAACCCTGCTT and GACGTGCATTATTGGGAGCA), and cdc2 (normalizer gene; CCAGCTAGTGAACGGTGTAA and TCCGCAAAGGGACACCAAAT). Samples were run in triplicate on the Applied Biosystems 7300 real-time PCR system (Applera UK). Relative quantities of transcript were determined using the 2−ΔΔCt formula, where Ct is defined as the cycle at which fluorescence is determined to be statistically above background, ΔCt is the difference in Ct of the gene of interest and Ct of the normalizer gene, and ΔΔCt is the difference in ΔCt of the strain tested and ΔCt of the control cells.
PCR poly(A) test.
Polyadenylation tests were performed according to the rapid amplification of cDNA ends-polyadenylation technique (RACE-PAT) method as described previously (32) using a reverse transcription-PCR (RT-PCR) kit (SuperScript; Invitrogen) with oligo(dT) anchor primer AT (GCGAGCTCCGCGGCCGCGT12) and a primer specific for 25S rRNA (25F1: GCGGCTGCCTGACAAGGCAATGCCGCGGAG), 5.8S rRNA (5.8F1: TCAGCAACGGATCTCTTGGCTCTCGCATCG), or 5S rRNA (5F1: GTTCCCGTCCGATCACTGCAGTTAAGCGTC). PCR products were cloned (TOPO TA cloning; Invitrogen) and sequenced using an ABI sequencer and ABI PRISM dRhodamine reagents (Applera UK). For detection of mature rRNA, primer pairs 25F2 (GAAGATGGGCGATGGTTGATGAAACGGAAGTG) and 25R1 (ACAAATCTTGGGAACAAAGGCTTAATCTCAGCAG), 5.8F1 and 5.8R1 (AATGACACTCAAACAGGCATGCCTTTGGTAG), and 5F1 and 5R1 (AGCCTACAGCACCCCGGATTCCCATGTTGT) were used for 25S, 5.8S, and 5S rRNA, respectively.
RESULTS
S. pombe cid14+ is a functional homologue of S. cerevisiae TRF4.
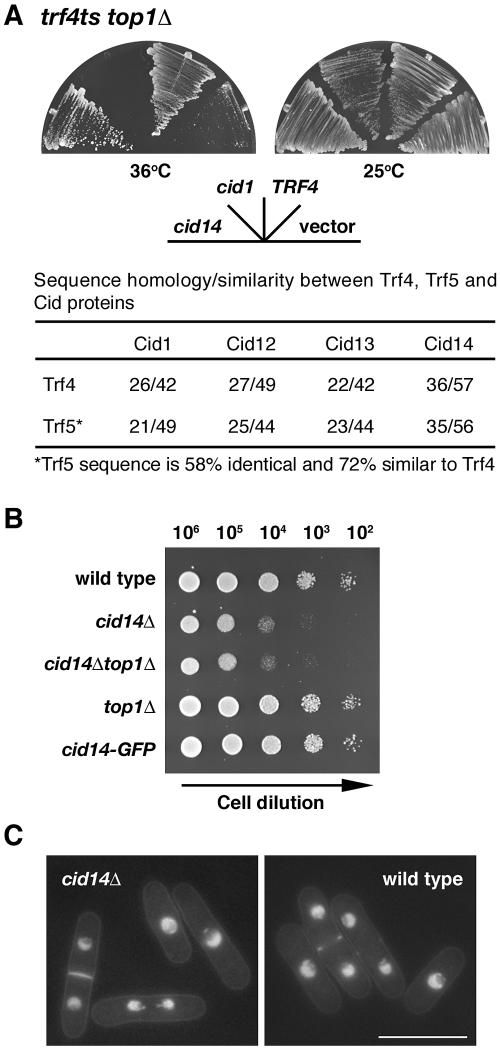
Fission yeast contains six cid1-like genes, whereas budding yeast has just two, TRF4 and TRF5, which share an essential function as trf4 trf5 double mutants are inviable. We set out to determine which cid gene performs a TRF4/5-like function in S. pombe. Sequence comparisons identified Cid14 as the closest relative of Trf4/5 (Fig. 1A). We therefore asked whether or not cid14+ could rescue the phenotype of trf4 mutants. In S. cerevisiae, TRF4 and TRF5 were identified through mutations that are synthetically lethal with a mutation in DNA topoisomerase I (7). To determine if cid14+ could complement this growth defect, we transformed a trf4ts top1Δ double mutant with cid14+ under the control of a galactose-inducible promoter. As shown in Fig. 1A, cid14+ complemented the temperature-sensitive growth phenotype of the trf4ts top1Δ double mutant. The double mutant containing vector alone grew at 25°C but was unable to grow at 36°C. Expression of TRF4 or cid14+ but not cid1+ was able to support growth of the double mutant at both temperatures. We conclude that the S. pombe cid14+ and S. cerevisiae TRF4 genes are functionally homologous. Multiple sequence comparisons suggest that TRF4 and TRF5 were generated by a relatively recent gene duplication event, so it is likely that the single cid14 gene performs the TRF4/5-like function in S. pombe. Consistent with this hypothesis, no synthetic lethality was observed in cid14Δ mutants in combination with deletion of any other cid genes (data not shown).
FIG. 1.
S. pombe cid14 is a functional homologue of S. cerevisiae TRF4 and is required for normal cell growth. (A) trf4ts top1Δ mutants transformed with plasmids carrying full-length cid1, cid14, and TRF4, as indicated, under the control of the GAL1 promoter or “empty” vector were streaked onto synthetic defined plates containing galactose. Plates were photographed after 3 days of incubation at 25°C or the nonpermissive temperature of 36°C. (B) Tenfold serial dilutions of the indicated strains spanning the range from 106 to 102 cells, as indicated, were spotted onto YE-agar plates and photographed after 2 days of incubation at 30°C. (C) Micrographs of Hoechst 33342-stained living wild-type and cid14Δ cells. Bar: 10 μm.
S. pombe cid14 is not essential for mitotic growth.
The one-step gene disruption method was used in a ura4−/ura4− diploid S. pombe strain to replace one copy of the entire cid14 open reading frame with the ura4+ selectable marker. After induction of meiosis and sporulation, microdissection of tetrads showed that cid14::ura4+ alleles segregated 2:2, indicating that, in contrast to its S. cerevisiae orthologue pair TRF4/5, S. pombe cid14+ is not essential for mitotic growth despite the slow-growth phenotype (Fig. 1B). This phenotype was most marked in cells growing in liquid minimal medium in which cid14Δ cells grew with a doubling time of approximately 9 h, compared with 2.5 h for a cid14+ strain. In addition, unlike trf4 and trf5 mutants, cid14 deletion mutants remained fully viable on mutation of top1, which encodes the fission yeast topoisomerase I (41). Microscopic examination showed that cid14Δ cells retained an essentially normal morphology as judged by cell staining to reveal distribution of DNA, septa, and actin (Fig. 1C and data not shown).
Deletion of cid14 leads to an increased chromosome segregation failure rate but not precocious sister chromatid separation during metaphase arrest.
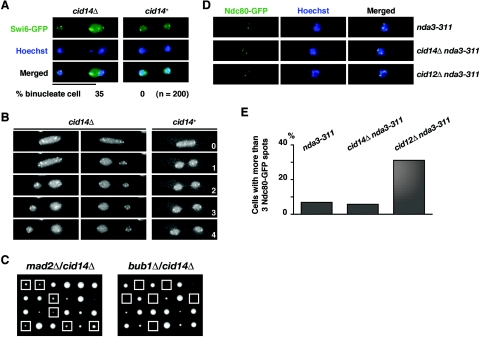
In S. cerevisiae Trf4 was reported to be required for the establishment of sister chromatid cohesion during S phase to ensure faithful chromosome segregation at mitosis (47). We therefore asked whether or not Cid14 is required for faithful chromosome segregation. To address this point, wild-type and cid14Δ strains containing an integrated GFP-tagged swi6 gene were constructed. GFP-Swi6 is localized in the nucleoplasm, giving a background of faint nuclear signal and several brightly fluorescent spots corresponding to the heterochromatic centromeres and telomeres (28). As shown in Fig. 2A and B, anaphase chromosome segregation in living cells as monitored by GFP-Swi6 fluorescence appeared normal in the cid14+ background. In contrast, abnormal anaphase progression (segregation of two unequal masses of GFP-Swi6) was observed in 35% of binucleate cid14Δ cells. Consistent with these apparent chromosome segregation defects, cid14Δ mutants had a moderate increase in the rate of minichromosome loss. Mean loss rates were 0.14% and 0.04% per division for cid14Δ and wild-type strains, respectively (average of three independent clones), using strains containing the nonessential ade6-M216-marked Ch16 minichromosome derivative of chromosome 3 (25). Despite the chromosome segregation defects described, cid14Δ mutants are viable, indicating that the spindle checkpoint might be required for the survival of these cells. Indeed, as shown in Fig. 2C, deletion of bub1 (4) but not mad2 (10) is lethal in cid14Δ cells, indicating that a subset of the spindle checkpoint factors are required for the survival of cid14Δ cells.
FIG. 2.
Deletion of cid14 leads to an increased chromosome segregation failure rate but not to precocious sister chromatid separation during metaphase arrest. (A) Merged images of fluorescence micrographs showing GFP-Swi6 and DNA (Hoechst 33342) localization in living cells. Bar: 10 μm. Percentages of cells with lagging chromosomes are indicated. (B) Visualization of unequal segregation of GFP-Swi6 signal in cid14Δ cells. Individual GFP-swi6 (cid14+) and GFP-swi6 cid14Δ cells were observed as for panel A, over a 4-min period, with images collected every minute. (C) Six tetrads derived from diploid strains h+/h− mad2::ura4+/mad2+ cid14::LEU2/cid14+ (left) and bub1::ura4+/bub1+ cid14::LEU2/cid14+ (right) were microdissected onto YE-agar, and the resulting colonies were photographed after 5 days of growth at 30°C. The genotypes of the segregants were determined by replica plating. Boxes indicate the position of cid14 double mutants with mad2 and bub1, respectively. (D) Merged images of fluorescence micrographs showing Ndc80-GFP localization in nda3-KM311, cid14Δ nda3-KM311, and cid12Δ nda3-KM311 cells in EMM medium after incubation at 20°C for 8 h. Fluorescence micrographs of living cells were acquired after staining with Hoechst 33342, revealing green fluorescence (Ndc80-GFP) and DNA (Hoechst). (E) Percentages of cells with more than three Ndc80-GFP spots, indicative of precocious sister chromatid separation, in each strain are shown (n = 200).
To test whether the observed chromosome segregation defects are due to defects in sister chromatid cohesion, we monitored sister centromere separation by the use of a strain expressing a GFP-tagged version of the kinetochore component Ndc80 (49). Ndc80-GFP appears as a single fluorescent spot in interphase cells, while in anaphase the clustered kinetochores are seen as two spots, one in each daughter nucleus. When this strain was arrested in metaphase by inactivation of nda3 (11), a mutation affecting β-tubulin, most cells (63%) still showed a single Ndc80-GFP spot, indicating that the sister centromeres were clustered. The remaining cells had three well-defined masses of condensed chromatin, presumably corresponding to the three S. pombe chromosomes, with Ndc80-GFP associated with each (Fig. 2D). This pattern of Ndc80-GFP distribution was seen in nda3-KM311 cid14Δ cells after shifting to the restrictive temperature to induce arrest at metaphase (Fig. 2E). In contrast, cells displaying separated Ndc80-GFP signals (more than three spots) were dramatically increased in a cid12Δ mutant defective in centromeric cohesion mediated by the RNA interference machinery (24). Thus, unlike S. cerevisiae trf4 mutants, S. pombe cid14Δ cells showed no indication of premature sister centromere separation during metaphase arrest.
Characterization of Cid14 protein in S. pombe.
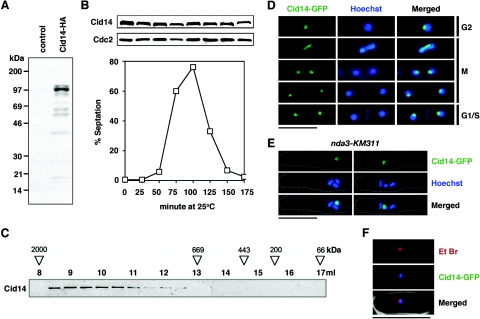
Targeted recombination was used to add an HA epitope tag sequence to the 3′ end of the cid14 open reading frame in its normal chromosomal context, generating the cid14-HA strain. Western blot analysis showed that the Cid14 protein had an aberrant mobility when analyzed by SDS-PAGE. While the calculated molecular mass of the protein is 78 kDa, the Cid14 protein isolated from S. pombe cells migrated as a predominant band of approximately 100 kDa (Fig. 3A). Next, we examined the expression of the Cid14 protein during the cell cycle. For this, cdc25-22 cid14-HA cells, released synchronously after cell cycle arrest at the restrictive temperature, were collected every 25 min and their lysates were subjected to immunoblotting. Cell cycle synchrony and position were monitored by determining the septation index. As shown in Fig. 3B, the Cid14 protein level remains constant with no obvious mobility shift through the cell cycle. Recently, S. cerevisiae Trf4 was reported to function in a complex with Mtr4 and Air1/2 (21, 40, 51). To determine whether or not Cid14 also functions as part of a higher-order complex, we used gel filtration chromatography to examine the native size of S. pombe Cid14 in a cell lysate prepared from the cid14-HA strain. As shown in Fig. 3C, the cellular Cid14 protein was found to be almost exclusively included in a large complex with an apparent size between 669 and 2,000 kDa. This result indicates that Cid14, like Trf4, forms part of a large complex in vivo.
FIG. 3.
S. pombe Cid14 localizes to the nucleolus throughout the cell cycle. (A) Whole-cell protein extracts from wild-type (control) and cid14-HA strains were prepared by trichloroacetic acid precipitation following glass bead disruption. The extracts were separated by SDS-PAGE and subjected to immunoblotting using anti-HA antibodies. (B) Cells expressing Cid14-HA were synchronously released from the G2 block imposed by a temperature-sensitive cdc25-22 mutation. Samples taken at the times indicated were used to score percentages of septated cells and subjected to immunoblotting using anti-HA (Cid14) or anti-Cdc2 (loading control) antibodies as indicated. (C) The size of soluble Cid14-HA was estimated by gel filtration chromatography. Samples of each 0.5-ml Superose-6 column fraction were processed for immunoblotting with an anti-HA antibody. The elution volume (ml) of each fraction is shown above the blots. The positions at which dextran blue (∼2,000 kDa), thyroglobulin (669 kDa), apoferritin (443 kDa), β-amylase (200 kDa), and bovine serum albumin (66 kDa) migrated are indicated. (D and E) Merged images of fluorescence micrographs showing Cid14-GFP and DNA (Hoechst 33342) localization in (D) living wild-type cells grown at 30°C and (E) nda3-KM311 mutants after incubation at 20°C for 8 h. (F) Merged images of fluorescence micrographs showing Cid14-GFP and nucleolar localization (Et Br, ethidium bromide) in living cells grown at 30°C. Bar: 10 μm.
S. pombe Cid14 is a nuclear protein enriched in the nucleolus.
To monitor the localization of Cid14 in living cells, the one-step gene replacement method was used to generate a strain (cid14-GFP) encoding a C-terminally GFP-tagged version of Cid14. The GFP-tagged protein appeared to be functional, as judged by the normal growth of the cid14-GFP strain compared with the cid14Δ strain (Fig. 1B). Examination of living cid14-GFP cells by fluorescence microscopy showed that Cid14-GFP (and, by inference, Cid14) was predominantly localized to the nucleus and was concentrated in a condensed spot in the nonchromosomal domain (Fig. 3D), previously assigned as the nucleolus (39). This phenomenon was further explored by examination of Cid14-GFP localization in an nda3-KM311 mutant in which chromosomes condense after incubation at the restrictive temperature of 20°C for 8 h. As shown in Fig. 3E, the intense fluorescence of Cid14-GFP signal was associated with the smallest chromosome, consistent with the fact that the shortest S. pombe chromosome (III) contains the rRNA gene clusters. In line with these results, Cid14 colocalized with the region of intense ethidium bromide staining (Fig. 3F), which distinguishes the nucleolus from the rest of the nucleus (38). The nucleolar concentration of Cid14-GFP was observed in cells from all stages of the cell cycle (Fig. 3D).
Aberrant nucleolar structures in cid14 mutants.
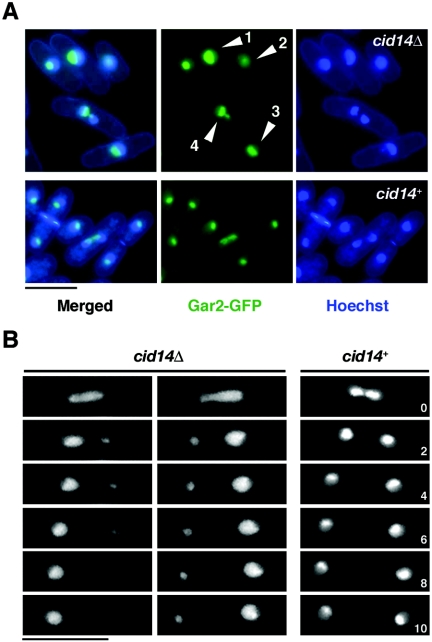
Next, we asked whether or not Cid14 deficiency might affect nucleolar architecture. Nucleolar structure was visualized in living cells using a protein that was a fusion between GFP and the nucleolar protein Gar2 (34). In wild-type cells, Gar2-GFP occupied roughly one-half of the nucleus, in a discrete region distinct from the bulk chromosomal DNA, against a background of fainter nuclear signal (Fig. 4A, lower panels). In contrast, a more diffuse Gar2-GFP fluorescence signal was observed in the majority of cid14Δ cells (examples 1 to 3) and, upon entry into mitosis, these cells often failed to segregate Gar2-GFP properly (example 4). This phenomenon was further explored by time-lapse microscopy. As shown in Fig. 4B, in contrast to wild-type cells, in which Gar2-GFP separated equally into the daughter cells at mitosis, abnormal anaphase progression (segregation of two unequal masses of Gar2-GFP) frequently occurred in cid14Δ cells. Taken together, these results suggest that Cid14 might play a role in the organization of the nucleolus to ensure faithful segregation during mitosis.
FIG. 4.
Aberrant nucleolar structures in the cid14Δ mutants. (A) Merged images of fluorescence micrographs showing Gar2-GFP and DNA (Hoechst 33342) localization in living cells. Arrowheads indicate cells with aberrant nucleolar structures. (B) Visualization of unequal segregation of Gar2-GFP signal in cid14Δ cells. Individual gar2-GFP (cid14+) and gar2-GFP cid14Δ cells were observed as in panel A, over a 10-min period, with images collected every 2 min. Bar: 10 μm.
Cid14-dependent polyadenylated rRNAs are accumulated in a dis3-54 mutant.
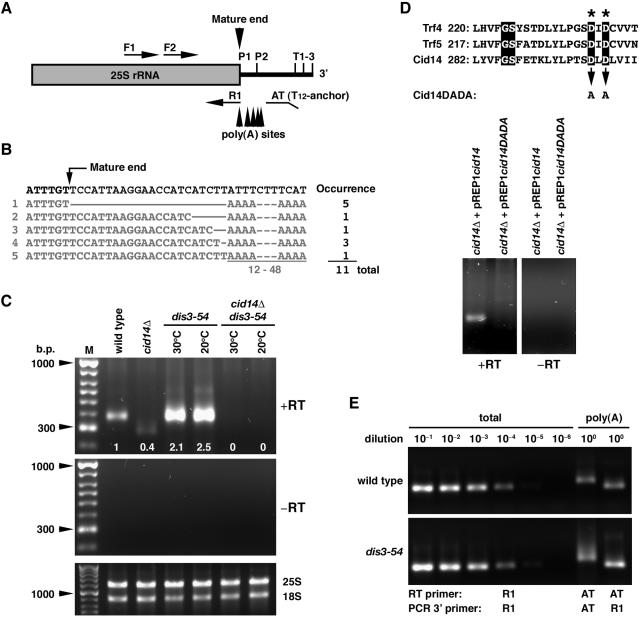
We reasoned that, given its cellular localization and requirement in the maintenance of nucleolar structure, Cid14 might have a role in rRNA metabolism through its predicted polyadenylation activity. In contrast to mRNA, rRNAs are not generally considered to be polyadenylated because they lack recognizable polyadenylation signals and are not synthesized by RNA polymerase II. However, recent data seriously undermine this notion by demonstrating that, in S. cerevisiae, some rRNAs can indeed be polyadenylated (9, 19). To explore whether Cid14 polyadenylates rRNA, we examined the 3′ end of the 25S rRNA using a RACE-PAT assay (32). As shown in Fig. 5C, distinct products of lengths consistent with polyadenylation in the region of the mature 25S rRNA 3′ end were observed in wild-type cells that were not detected in the “no-RT” control. The PCR-amplified products were cloned and sequenced. Each of 11 randomly selected clones contained a poly(A) sequence 12 to 48 nucleotides long immediately downstream of the mature 3′ end of the 25S rRNA, which is generated in several steps by posttranscriptional processing during rRNA biogenesis (Fig. 5B). There is no templated poly(A) tract in this region, suggesting that the poly(A) detected in this assay was not due to nonspecific priming. We conclude that rRNA can contain 3′ poly(A) tails in S. pombe.
FIG. 5.
Cid14-dependent polyadenylated 25S rRNAs are accumulated in a dis3-54 mutant. (A) Schematic diagram of the primers for RT-PCR. Alternative processing cleavage sites (P1 and P2) and transcription termination sites (T1-3) are indicated. (B) The 3′-terminal sequences of cloned reverse transcription products. (C) Total RNA from cultures of indicated strains grown at 30°C or the nonpermissive temperature of 20°C for dis3-54 mutants was reverse transcribed using the anchored oligo(dT) primer AT and then PCR amplified with AT and the F1 primer specific for 25S rRNA. RT-PCR products were separated on a 1% agarose gel, and the relative signals were quantified as indicated. A control using total RNA and a non-reverse-transcribed negative control were included. (D) Total RNA from cultures of cid14Δ strains containing the indicated plasmids (with or without mutation in the nucleotide transferase motif: GS X10 DXD of Cid14) and grown in the presence of 15 μΜ thiamine was subjected to RACE-PAT assay as for panel C. RT-PCR products were separated on a 1% agarose gel. A non-reverse-transcribed negative control was included. (E) Reverse transcripts were made with the R1 or AT primer and RNA from wild-type and dis3-54 strains grown at 30°C. Serial dilutions of R1 transcripts (for total 25S rRNA) were PCR amplified with F2 and R1 and compared with AT transcripts (polyadenylated 25S rRNA) amplified with primer pair F2 and AT or F2 and R1.
To estimate the relative levels of polyadenylated 25S rRNA, the levels of these products were compared with a series of RT-PCR assays performed on diluted samples of the same RNAs (Fig. 5E). This analysis showed that the polyadenylated 25S rRNA product is approximately 0.02% of total 25S rRNA in wild-type cells. Consistent with a role for the exosome in rRNA processing and degradation, the polyadenylated 25S rRNA products were accumulated in a dis3/RPR44 exosome mutant and increased further after 8 h of incubation at the nonpermissive temperature of 20°C (Fig. 5C). The amounts of mature 18S and 25S rRNAs were very similar in all cell types examined (Fig. 5C and E), indicating that cleavage steps were not adversely affected by the mutations. These results are consistent with a model in which a polyadenylation-assisted degradation mechanism is responsible for the degradation of aberrant RNA transcripts. Furthermore, these polyadenylated 25S rRNA products were not detectable in the cid14Δ and cid14Δ dis3-54 strains, suggesting that Cid14 plays a role in adenylating the rRNAs. In line with its predicted polyadenylation activity, the polyadenylated 25S rRNA products were found to be dependent on the conserved aspartate residues 298 and 230 in the nucleotide transferase motif (GS X10 DXD) of Cid14 that are known to be essential for the catalytic activity of this protein family (Fig. 5D). When expressed in the cid14Δ strain from the nmt1 promoter in the plasmid pREP1cid14DADA in the presence of thiamine, this mutant form of Cid14, unlike the wild-type protein, was unable to restore the polyadenylated 25S rRNA species in these cells.
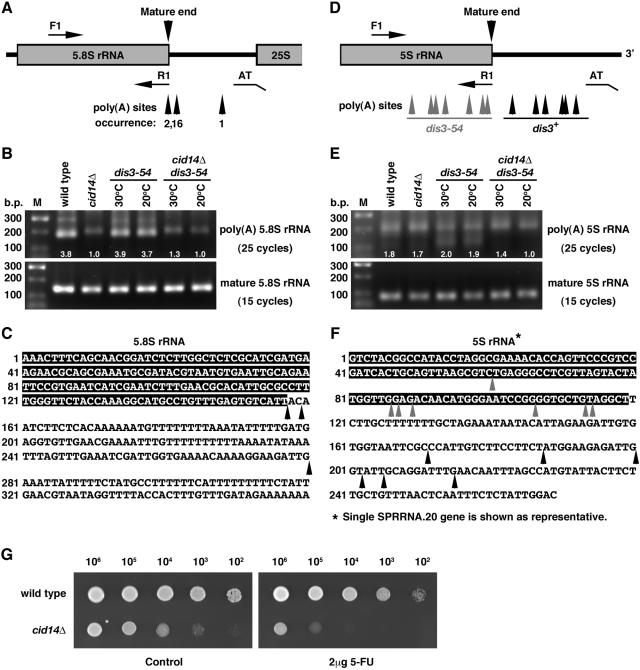
Next, we asked whether or not the other three types of rRNA might be polyadenylated and accumulate in a dis3-54 mutant. As shown in Fig. 6A, similar results were observed in the RACE-PAT assay of 5.8S RNA. Sequencing analysis confirmed Cid14-dependent polyadenylation of 5.8S RNAs, as these products were dramatically reduced in the cid14Δ and cid14Δ dis3-54 strains. The residue products presented in cid14Δ cells were due to nonspecific priming as confirmed by sequencing of 17 randomly selected clones (data not shown). No obvious difference was observed in the amount of these polyadenylated 5.8S rRNA products in dis3-54 mutants compared with the wild-type strain. Although it has been reported that 18S RNAs are polyadenylated to a certain level (19), we were not able to detect any polyadenylated 18S rRNA products using an approach similar to that described above (data not shown).
FIG. 6.
Analysis of the polyadenylation status of 5.8S and 5S rRNA in different genetic backgrounds. (A and D) Schematic diagram of the primers for RT-PCR. (B and E) Total RNA from cultures of indicated strains grown at 30°C or the nonpermissive temperature of 20°C for dis3-54 mutants was reverse-transcribed with the oligo(dT) anchor primer AT and then PCR amplified with AT and forward primers F1 specific for 5.8S and 5S rRNA, respectively. For detection of mature rRNA, reverse transcripts were made with the R1 primer specific for 5.8S and 5S rRNA and then PCR amplified with pair F1 and R1. RT-PCR products were separated on a 1.5% agarose gel, and the relative signals were quantified as indicated. (C and F) Genomic sequences of 5.8S and 5S genes. Arrowheads indicate polyadenylation sites identified. (G) Tenfold serial dilutions of wild-type and cid14Δ strains spanning the range from 106 to 102 cells, as indicated, were spotted onto YE-agar containing 2 μg/ml 5-FU or no drug (control). Plates were photographed after 3 days of incubation at 30°C.
In contrast to the genes for the large rRNAs, the 5S gene sequences are dispersed within the genome and are transcribed by RNA polymerase III independently of other rRNA genes. As shown in Fig. 6E, RACE-PAT assays identified two forms of polyadenylated 5S rRNA products. A broad band of longer transcripts with poly(A) tails downstream of the mature 3′ end of 5S rRNA was observed in all strains examined. Interestingly, deletion of cid14 had no influence on the amount of these transcripts, in sharp contrast to what we observed for the 5.8S and 25S genes. In addition, a band of shorter products which contained fragments of 5S RNAs with poly(A) tails (gray arrowheads in Fig. 6F) were detected only in the dis3-54 cells. The dependence of these products on dis3 mutation implied that these molecules were destined for exosome-mediated degradation. Consistent with a role of Cid14 in polyadenylation-assisted degradation, the polyadenylated 5S-related RNAs were not detectable in the cid14Δ dis3-54 strain.
In line with these results, cid14Δ cells were hypersensitive to 5-fluorouracil (5-FU; Fig. 6G), a chemical which interferes with the degradation of rRNA precursors (9).
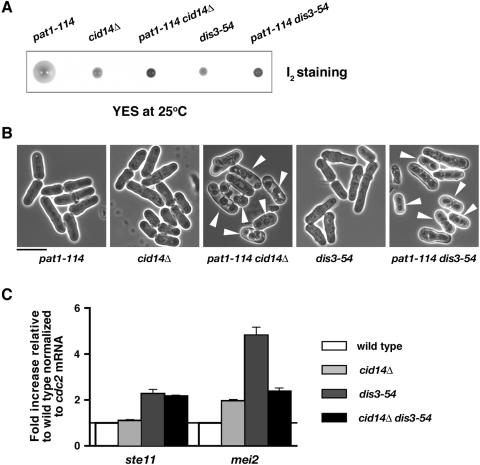
Deletion of cid14 leads to stimulation of meiosis in pat1-114 cells.
In C. elegans, the Cid1-related protein GLD-2 together with its RNA binding partner GLD-3 controls various aspects of germ line development, including the mitosis/meiosis decision (42), while in Xenopus xGLD-2 is required for CPEB-mediated polyadenylation-induced translation during oocyte maturation (3). We were therefore interested to see whether or not Cid14 plays a role in meiosis in fission yeast. To test this, we examined the genetic interaction between cid14 and pat1, which encodes a general inhibitor of sexual differentiation (22). As shown in Fig. 7A and B, whereas the pat1-114 single mutants enter meiosis at 30°C, the pat1-114 cid14Δ mutants initiated meiosis at the normally permissive temperature of 25°C. Similarly, pat1-114 dis3-54 mutants underwent meiosis at the lower temperature. These results suggest that Cid14 together with the exosome might play a role in meiosis, closely interacting with the Pat1 kinase.
FIG. 7.
Deletion of cid14 leads to stimulation of meiosis in pat1-114 cells. (A) Iodine staining of colonies derived from spores of diploid h+/h− cid14::ura4+/cid14+ pat1-114/pat1+ and h+/h− dis3-54/dis3+ pat1-114/pat1+ strains after 5 days of growth on YE-agar at 25°C. Iodine stains cells that have undergone meiosis and sporulation dark brown. (B) Micrographs of the strains indicated from panel A. Arrowheads indicate cells that have undergone meiosis and sporulation. Bar: 10 μm. (C) Total RNA from cultures of the indicated strains grown at 30°C was reverse transcribed using an oligo(dT) primer. The cDNA was amplified using quantitative PCR and SYBR green in triplicate with primers specific for the indicated genes. RNA amounts normalized to cdc2 mRNA were expressed relative to the wild type.
In S. pombe, the switch from mitosis to meiosis is controlled by the Pat1 kinase-Mei2 system. Mei2, an RNA-binding protein, is essential for the initiation of premeiotic DNA synthesis and meiosis I (48). Transcription of mei2 is activated by nutrient deprivation and a concomitant decrease in the intracellular cyclic AMP level and is directly regulated by Ste11, which is a transcription factor of the high-mobility-group family (36). During the normal mitotic cycle, Pat1 suppresses meiotic entry via transcriptional repression through Ste11, in addition to the direct inhibition of Mei2 by phosphorylation (18). To understand the role of Cid14 in meiotic control, we measured the levels of ste11 and mei2 mRNAs in cid14Δ cells as well as in dis3-54 mutants grown at the permissive temperature 30°C. As shown in Fig. 7C, deletion of cid14, or dis3 mutation, led to accumulation of mei2 transcripts as determined by quantitative PCR. Mutation of dis3 also affected the stell mRNA level. The elevated levels of mei2 mRNA could account for the stimulation of meiosis in pat1-114 cid14Δ and pat1-114 dis3-54 cells.
DISCUSSION
The Cid1 family of nucleotidyltransferases is widespread in eukaryotes (20). Members of this family (Cid1 and Cid13 in fission yeast and GLD-2 in C. elegans and vertebrates) have been shown to use RNA substrates and define a cytoplasmic family of poly(A) polymerases (29, 31, 42). In contrast, the S. cerevisiae Cid1-related protein Trf4 was identified as a component of a nuclear polyadenylation complex and is required for polyadenylation and degradation of rRNA and snoRNA precursors that are characterized as exosome substrates (21, 51). In this study, we have identified Cid14 as the functional homologue of S. cerevisiae Trf4/5. We have provided evidence suggesting that in S. pombe subpopulations at least three of the four rRNAs contain 3′ poly(A) tails and, at some level, Cid14 appears responsible for the polyadenylation of these rRNAs. It has been proposed that polyadenylation by the Trf4 complex stimulates RNA degradation by the exosome. Consistent with this hypothesis, the Cid14-dependent polyadenylated rRNAs were accumulated in a dis3 mutant defective in a component of the exosome. In addition, our results also revealed a fraction of the polyadenylated rRNA species that are not dependent on Cid14 (Fig. 6E). In the absence of Cid14, an alternative pathway must be responsible for the polyadenylation of these rRNAs; low levels of polyadenylated 25S rRNA products with alternative polyadenylation sites were observed in cid14Δ mutants (data not shown). It has been reported that polyadenylation of several rRNA species in a rrp6Δ background depends on the nuclear poly(A) polymerase (19, 51). Whether or not Pla1, the canonical S. pombe poly(A) polymerase, is responsible for the polyadenylation of these transcripts remains to be determined. However, the significance of polyadenylation of these transcripts is unclear; it does not appear to stimulate their degradation by the exosome, as the mutation of dis3 has no influence on the amount of these transcripts. In line with these results, cells that lack a functional cid14+ gene grow slowly and are hypersensitive to 5-FU (Fig. 6G).
Consistent with its role in rRNA metabolism, S. pombe Cid14 is a nucleolar protein and is required for maintenance of nucleolar structure (Fig. 3 and 4). Similarly, Dis3 protein (and, by inference the exosome) is enriched in the nucleolus (17). Intriguingly, polyadenylation of 5S rRNAs, transcribed by RNA polymerase III outside the nucleolus, is also dependent on Cid14 to a degree (Fig. 6). This could be due to the function of nucleolus in the processing of rRNAs and their subsequent assembly into preribosomes. The fact that a Trf4-dependent polyadenylation-assisted degradation mechanism is also responsible for the degradation of RNA polymerase II transcripts (51) broadens the implication of these results. Possible involvement of the nucleolus in mRNA export has been proposed, based on observations made in yeasts and higher eukaryotic cells (5, 8, 13, 33). These results suggest that the nucleolus might function as a site of general RNA quality control, although it is still unclear how the cell determines which transcripts should be degraded.
Chromosome segregation defects were another obvious feature of cid14-deficient cells (Fig. 2). In S. cerevisiae Trf4 was reported to be required for the establishment of sister chromatid cohesion during S phase to ensure faithful chromosome segregation at mitosis (47). However, unlike S. cerevisiae Trf4 mutants, S. pombe cid14Δ cells showed no indication of premature sister centromere separation during metaphase arrest (Fig. 2D and E). Given the function of Cid14 in rRNA metabolism, ribosome biogenesis and translation defects might have a direct effect on some aspect of chromosome structure or kinetochore function. Alternatively, the impact of a loss of Cid14 function in mitosis may be through impaired expression of genes required for faithful chromosome segregation. Intriguingly, in S. pombe, the dis3+ gene was first identified through a screen for mutants defective in mitotic chromosome separation (17, 26, 27). The same screen identified dis2+, which encodes a serine-threonine protein phosphatase identified as a component of the pre-mRNA cleavage and polyadenylation complex (30). Similar defects have been reported for pfs2 mutants, which are defective in another component of the pre-mRNA cleavage and polyadenylation complex (43). The characterization of S. pombe mutants defective in the 5′-3′ exoribonuclease Dhp1 suggested an additional connection between RNA processing and chromosome segregation (35). It will be interesting to determine whether or not chromosome segregation defects are a general feature of mutants defective in RNA processing.
Our results also revealed a possible function of Cid14 in the control of meiosis. In C. elegans, the Cid1-related protein GLD-2 promotes entry into meiosis from the mitotic cell cycle (16). In contrast, deletion of cid14 leads to stimulation of meiosis in pat1-114 cells (Fig. 7). This effect could be due to the deprivation of nutrients as a consequence of defective ribosome biogenesis. However, pat1-144 cells arrested at G1 rather than entering meiosis after deprivation of nutrients by nitrogen starvation at the permissive temperature 25°C (data not shown). These results suggest that Cid14 together with the exosome might have a more direct role in meiotic control. Additional connections between RNA processing and meiosis were suggested by the identification of S. pombe pac1+, which encodes an RNase III-like RNase, as a multicopy suppressor of the uncontrolled meiosis driven by pat1 deficiency (14). The pac1 gene product probably inhibits mating and meiosis by degrading mRNAs required for sexual development. The accumulation of mRNAs required for meiosis in exosome mutants suggests that these transcripts might be constantly degraded by the exosome in mitotic cells to prevent ectopic meiosis. Experiments are under way to explore this possibility.
Cid1-like proteins control a variety of functions in S. pombe, C. elegans, and vertebrates. The identification of Cid14 as the functional homologue of S. cerevisiae Trf4/5 indicates the generality of a polyadenylation-assisted degradation mechanism, which is likely to be conserved in higher eukaryotes. Interestingly, in contrast to the situation in other organisms, no cytoplasmic function of Trf4/5 has been described. Studying the way in which Cid1-like proteins function in lower eukaryotes is expected to provide greater understanding of the biological functions of polyadenylation in eukaryotic cells in general.
Acknowledgments
We thank Michael F. Christman, Jean-Paul Javerzat, Nancy C. Walworth, Mitsuhiro Yanagida, and Masayuki Yamamoto for yeast strains; Hiro Yamano for Cdc2 antibody; and Stephen Kearsey for useful discussions and comments on the manuscript.
This work was supported by Cancer Research UK and The Wellcome Trust (Research Career Development Fellowship to Shao-Win Wang).
REFERENCES
- 1.Allshire, R. C., E. R. Nimmo, K. Ekwall, J. P. Javerzat, and G. Cranston. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218-233. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley and Son, Inc., New York, N.Y.
- 3.Barnard, D. C., K. Ryan, J. L. Manley, and J. D. Richter. 2004. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119:641-651. [DOI] [PubMed] [Google Scholar]
- 4.Bernard, P., K. Hardwick, and J. P. Javerzat. 1998. Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J. Cell Biol. 143:1775-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bond, V. C., and B. Wold. 1993. Nucleolar localization of myc transcripts. Mol. Cell. Biol. 13:3221-3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caspari, T., M. Dahlen, G. Kanter-Smoler, H. D. Lindsay, K. Hofmann, K. Papadimitriou, P. Sunnerhagen, and A. M. Carr. 2000. Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9. Mol. Cell. Biol. 20:1254-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castano, I. B., S. Heath-Pagliuso, B. U. Sadoff, D. J. Fitzhugh, and M. F. Christman. 1996. A novel family of TRF (DNA topoisomerase I-related function) genes required for proper nuclear segregation. Nucleic Acids Res. 24:2404-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deak, I., E. Sidebottom, and H. Harris. 1972. Further experiments on the role of the nucleolus in the expression of structural genes. J. Cell Sci. 11:379-391. [DOI] [PubMed] [Google Scholar]
- 9.Fang, F., J. Hoskins, and J. S. Butler. 2004. 5-Fluorouracil enhances exosome-dependent accumulation of polyadenylated rRNAs. Mol. Cell. Biol. 24:10766-10776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He, X., T. E. Patterson, and S. Sazer. 1997. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA 94:7965-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiraoka, Y., T. Toda, and M. Yanagida. 1984. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell 39:349-358. [DOI] [PubMed] [Google Scholar]
- 12.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 13.Ideue, T., A. K. Azad, J. Yoshida, T. Matsusaka, M. Yanagida, Y. Ohshima, and T. Tani. 2004. The nucleolus is involved in mRNA export from the nucleus in fission yeast. J. Cell Sci. 117:2887-2895. [DOI] [PubMed] [Google Scholar]
- 14.Iino, Y., A. Sugimoto, and M. Yamamoto. 1991. S. pombe pac1+, whose overexpression inhibits sexual development, encodes a ribonuclease III-like RNase. EMBO J. 10:221-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadaba, S., A. Krueger, T. Trice, A. M. Krecic, A. G. Hinnebusch, and J. Anderson. 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 18:1227-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadyk, L. C., and J. Kimble. 1998. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 125:1803-1813. [DOI] [PubMed] [Google Scholar]
- 17.Kinoshita, N., M. Goebl, and M. Yanagida. 1991. The fission yeast dis3+ gene encodes a 110-kilodalton essential protein implicated in mitotic control. Mol. Cell. Biol. 11:5839-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitamura, K., S. Katayama, S. Dhut, M. Sato, Y. Watanabe, M. Yamamoto, and T. Toda. 2001. Phosphorylation of Mei2 and Ste11 by Pat1 kinase inhibits sexual differentiation via ubiquitin proteolysis and 14-3-3 protein in fission yeast. Dev. Cell 1:389-399. [DOI] [PubMed] [Google Scholar]
- 19.Kuai, L., F. Fang, J. S. Butler, and F. Sherman. 2004. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101:8581-8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwak, J. E., L. Wang, S. Ballantyne, J. Kimble, and M. Wickens. 2004. Mammalian GLD-2 homologs are poly(A) polymerases. Proc. Natl. Acad. Sci. USA 101:4407-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaCava, J., J. Houseley, C. Saveanu, E. Petfalski, E. Thompson, A. Jacquier, and D. Tollervey. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121:713-724. [DOI] [PubMed] [Google Scholar]
- 22.McLeod, M., and D. Beach. 1988. A specific inhibitor of the ran1+ protein kinase regulates entry into meiosis in Schizosaccharomyces pombe. Nature 332:509-514. [DOI] [PubMed] [Google Scholar]
- 23.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 24.Motamedi, M. R., A. Verdel, S. U. Colmenares, S. A. Gerber, S. P. Gygi, and D. Moazed. 2004. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119:789-802. [DOI] [PubMed] [Google Scholar]
- 25.Niwa, O., T. Matsumoto, Y. Chikashige, and M. Yanagida. 1989. Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J. 8:3045-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noguchi, E., N. Hayashi, Y. Azuma, T. Seki, M. Nakamura, N. Nakashima, M. Yanagida, X. He, U. Mueller, S. Sazer, and T. Nishimoto. 1996. Dis3, implicated in mitotic control, binds directly to Ran and enhances the GEF activity of RCC1. EMBO J. 15:5595-5605. [PMC free article] [PubMed] [Google Scholar]
- 27.Ohkura, H., Y. Adachi, N. Kinoshita, O. Niwa, T. Toda, and M. Yanagida. 1988. Cold-sensitive and caffeine-supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. EMBO J. 7:1465-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pidoux, A. L., S. Uzawa, P. E. Perry, W. Z. Cande, and R. C. Allshire. 2000. Live analysis of lagging chromosomes during anaphase and their effect on spindle elongation rate in fission yeast. J. Cell Sci. 113(Pt. 23):4177-4191. [DOI] [PubMed] [Google Scholar]
- 29.Read, R. L., R. G. Martinho, S. W. Wang, A. M. Carr, and C. J. Norbury. 2002. Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proc. Natl. Acad. Sci. USA 99:12079-12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roguev, A., A. Shevchenko, D. Schaft, H. Thomas, and A. F. Stewart. 2004. A comparative analysis of an orthologous proteomic environment in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Mol. Cell. Proteomics 3:125-132. [DOI] [PubMed] [Google Scholar]
- 31.Saitoh, S., A. Chabes, W. H. McDonald, L. Thelander, J. R. Yates, and P. Russell. 2002. Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell 109:563-573. [DOI] [PubMed] [Google Scholar]
- 32.Salles, F. J., W. G. Richards, and S. Strickland. 1999. Assaying the polyadenylation state of mRNAs. Methods 17:38-45. [DOI] [PubMed] [Google Scholar]
- 33.Schneiter, R., T. Kadowaki, and A. M. Tartakoff. 1995. mRNA transport in yeast: time to reinvestigate the functions of the nucleolus. Mol. Biol. Cell 6:357-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimada, T., A. Yamashita, and M. Yamamoto. 2003. The fission yeast meiotic regulator Mei2p forms a dot structure in the horse-tail nucleus in association with the sme2 locus on chromosome II. Mol. Biol. Cell 14:2461-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shobuike, T., K. Tatebayashi, T. Tani, S. Sugano, and H. Ikeda. 2001. The dhp1+ gene, encoding a putative nuclear 5′→3′ exoribonuclease, is required for proper chromosome segregation in fission yeast. Nucleic Acids Res. 29:1326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugimoto, A., Y. Iino, T. Maeda, Y. Watanabe, and M. Yamamoto. 1991. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev. 5:1990-1999. [DOI] [PubMed] [Google Scholar]
- 37.Tomecki, R., A. Dmochowska, K. Gewartowski, A. Dziembowski, and P. P. Stepien. 2004. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res. 32:6001-6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Umesono, K., Y. Hiraoka, T. Toda, and M. Yanagida. 1983. Visualization of chromosomes in mitotically arrested cells of the fission yeast Schizosaccharomyces pombe. Curr. Genet. 7:123-128. [DOI] [PubMed] [Google Scholar]
- 39.Uzawa, S., and M. Yanagida. 1992. Visualization of centromeric and nucleolar DNA in fission yeast by fluorescence in situ hybridization. J. Cell Sci. 101(Pt. 2):267-275. [DOI] [PubMed] [Google Scholar]
- 40.Vanacova, S., J. Wolf, G. Martin, D. Blank, S. Dettwiler, A. Friedlein, H. Langen, G. Keith, and W. Keller. 19. April 2005, posting date. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 3:e189. [Online.] http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371/journal/pbio.0030189. [DOI] [PMC free article] [PubMed]
- 41.Wan, S., H. Capasso, and N. C. Walworth. 1999. The topoisomerase I poison camptothecin generates a Chk1-dependent DNA damage checkpoint signal in fission yeast. Yeast 15:821-828. [DOI] [PubMed] [Google Scholar]
- 42.Wang, L., C. R. Eckmann, L. C. Kadyk, M. Wickens, and J. Kimble. 2002. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419:312-316. [DOI] [PubMed] [Google Scholar]
- 43.Wang, S. W., K. Asakawa, T. Z. Win, T. Toda, and C. J. Norbury. 2005. Inactivation of the pre-mRNA cleavage and polyadenylation factor Pfs2 in fission yeast causes lethal cell cycle defects. Mol. Cell. Biol. 25:2288-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, S. W., C. Norbury, A. L. Harris, and T. Toda. 1999. Caffeine can override the S-M checkpoint in fission yeast. J. Cell Sci. 112(Pt. 6):927-937. [DOI] [PubMed] [Google Scholar]
- 45.Wang, S. W., R. L. Read, and C. J. Norbury. 2002. Fission yeast Pds5 is required for accurate chromosome segregation and for survival after DNA damage or metaphase arrest. J. Cell Sci. 115:587-598. [DOI] [PubMed] [Google Scholar]
- 46.Wang, S. W., T. Toda, R. MacCallum, A. L. Harris, and C. Norbury. 2000. Cid1, a fission yeast protein required for S-M checkpoint control when DNA polymerase delta or epsilon is inactivated. Mol. Cell. Biol. 20:3234-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, Z., I. B. Castano, A. De Las Penas, C. Adams, and M. F. Christman. 2000. Pol kappa: a DNA polymerase required for sister chromatid cohesion. Science 289:774-779. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe, Y., and M. Yamamoto. 1994. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell 78:487-498. [DOI] [PubMed] [Google Scholar]
- 49.Wigge, P. A., and J. V. Kilmartin. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152:349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilusz, C. J., M. Wormington, and S. W. Peltz. 2001. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2:237-246. [DOI] [PubMed] [Google Scholar]
- 51.Wyers, F., M. Rougemaille, G. Badis, J. C. Rousselle, M. E. Dufour, J. Boulay, B. Regnault, F. Devaux, A. Namane, B. Seraphin, D. Libri, and A. Jacquier. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121:725-737. [DOI] [PubMed] [Google Scholar]