Abstract
The UL69 gene product of human cytomegalovirus belongs to a family of regulatory proteins conserved among all herpesviruses that have in part been characterized as posttranscriptional transactivators participating in the nuclear export of RNA. Recent experiments suggested that pUL69 also acts as a posttranscriptional activator since it was demonstrated that nucleocytoplasmic shuttling via a CRM1-independent nuclear export signal is a prerequisite for its stimulatory effect on gene expression. Based on these findings we initiated studies to investigate the role of pUL69 in mRNA export and demonstrate that pUL69 efficiently promotes the cytoplasmic accumulation of unspliced RNA. Furthermore, we show that this pUL69 activity is linked to the cellular mRNA export machinery by direct protein interaction with the highly related DEXD/H-box RNA helicases UAP56 and URH49. Particularly, we identified a 12-amino-acid domain within the N terminus of pUL69 which is required for binding to UAP56 and URH49, and we could demonstrate that UAP56 interaction and nucleocytoplasmic shuttling are both prerequisites for pUL69-mediated mRNA export. Thus, we identified a novel cellular target which provides a herpesviral regulatory protein with access to a conserved cellular transport system in order to promote nuclear export of unspliced RNA.
A defining feature of eukaryotic cells is their division into nucleoplasm and cytoplasm. This segregation requires specific mechanisms for the continuous transport of large numbers of macromolecules between the two compartments. One essential class of macromolecules that are produced in the nucleus, yet are used primarily in the cytoplasm, are mRNAs. Eukaryotic pre-mRNAs require extensive nuclear processing after synthesis in the nucleus, including addition of the 5′ cap, splicing, and polyadenylation. Following these processing events mRNAs are efficiently transported to the cytoplasm where they direct protein synthesis. This transport occurs through the nuclear pore complex and is mediated by shuttling RNA transport factors and the corresponding transport receptors (reviewed in reference 59).
Viruses that replicate in the nucleus have to make extensive use of, or even modify, these transport mechanisms in order to optimize the cellular environment for efficient viral multiplication. In this regard the analysis of nuclear RNA export pathways accessed by different primate retroviruses led to the identification of CRM1 as an important member of the karyopherin/exportin family of nucleocytoplasmic transport receptors and of TAP as a key nuclear mRNA export factor in metazoan cells (reviewed in reference 9). Human immunodeficiency virus type 1 (HIV-1), which belongs to the complex retroviruses, encodes the sequence-specific mRNA export factor Rev, which acts as an adaptor between Rev-responsive element (RRE)-containing HIV-1 mRNAs and the cellular CRM1 nuclear export receptor (12, 14). However, it is now clear that CRM1 not only mediates the nuclear export of retroviral RNAs but is also responsible for the nuclear export of proteins containing a leucine-rich nuclear export signal (NES) and of some noncoding cellular RNAs like U snRNAs or rRNAs (9). TAP was first identified as the cellular factor that interacts with the constitutive transport element (CTE) present in RNAs from type D retroviruses, and TAP promotes the nuclear export of CTE-containing transcripts (25). Later on, it was demonstrated that TAP interacts with p15 and that this heterodimer functions as the major mRNA transport receptor in metazoan cells that facilitates mRNA export to the cytoplasm via direct interaction with the nuclear pore (26, 53).
Although TAP is able to interact directly with CTE-containing viral mRNAs, additional factors are needed to bridge the interaction between TAP-p15 and metazoan mRNAs (33). Among these are members of the evolutionarily conserved REF protein family (alternatively termed Aly and in Saccharomyces cerevisiae termed Yra1). REF proteins shuttle between the nucleus and the cytoplasm and bind directly to both mRNAs and TAP, thereby facilitating the association of the export receptor TAP with cellular mRNAs (47, 55). Nuclear export of mRNA is highly coupled to other processes in gene expression including transcription and splicing. A key factor that brings the transcription, splicing, and nuclear export machineries into close functional context is the RNA helicase UAP56 and its yeast homolog Sub2. These proteins belong to the DEXD/H-box family of RNA helicases (also referred to as DEAD-box proteins) (46). Although UAP56 has been implicated in splicing (13), this protein plays an essential role in mRNA export (37, 56). It has been reported that UAP56/Sub2 (i) interacts with the polymerase II-dependent transcription elongation machinery to form the TREX complex that links transcription to mRNA export (57) and (ii) interacts with the splicing machinery to form the exon junction complex that links splicing to mRNA export (45). Current models assume that UAP56 is recruited either cotranscriptionally or splicing-coupled to both intron-containing and intron-free pre-mRNAs and, then, UAP56 itself recruits REF and hence indirectly the TAP-p15 export receptor. After loading of REF onto the mRNA, UAP56 must be released from REF to allow binding of TAP, because binding of UAP56 and that of TAP to REF are mutually exclusive (9, 45, 59). Interestingly, a protein termed URH49 that is 90% identical to UAP56 has recently been identified in mammalian cells, and it is thought that the two proteins have similar or redundant functions in mammalian mRNA biogenesis (44).
While most metazoan mRNAs are derived from spliced genes, Herpesviridae are dependent on the nuclear export of intronless mRNAs. Since these mRNAs are unable to recruit mRNA export factors via splicing, human herpesviruses have evolved regulatory proteins to promote the nuclear export of unspliced RNAs (reviewed in reference 51). The most intensively studied member of this group is the multifunctional regulatory protein ICP27 of herpes simplex virus type 1 (HSV-1) (52). ICP27 is a nucleocytoplasmic shuttling protein that functions mainly on the posttranscriptional level. With regard to RNA export it has been demonstrated that ICP27 recruits the cellular mRNA export factors REF and TAP to intronless viral mRNA, thus allowing these RNAs to access the cellular mRNA nuclear export pathway (6, 27, 50). Proteins with homology to ICP27 are present in all human herpesviruses, suggesting a conserved function of these polypeptides. Consistently, similar results have recently been reported for the homologous proteins EB2 of Epstein-Barr virus (EBV) (11, 19) or ORF57 of Kaposi's sarcoma-associated herpesvirus (39). The ICP27 homolog of human cytomegalovirus (HCMV) is the pleiotropic transactivator pUL69. The overall amino acid identity between the 744-amino-acid (aa) protein pUL69 and the 512-aa ICP27 is approximately 24%. However, the C-terminal 160 amino acids of ICP27, which are known to be of functional importance, show a higher conservation with several positionally conserved amino acids (52). This amino acid sequence with higher conservation corresponds to a central domain within pUL69. The betaherpesviral proteins differ from their homologous proteins by a unique C-terminal domain that is not contained within the alpha- or gammaherpesvirus members of this protein family (60). An initial functional characterization of pUL69 revealed several differences from ICP27. For instance, in contrast to ICP27, pUL69 is expressed as an early/late gene during viral replication and is incorporated into viral particles (61, 62). Recently, however, evidence has been presented to suggest that pUL69 also acts at the posttranscriptional level and shares properties with mRNA export factors. In particular, it has been demonstrated that (i) pUL69 shuttles between the nucleus and the cytoplasm, (ii) pUL69 nuclear export is not dependent on CRM1 activity, and (iii) nucleocytoplasmic shuttling is crucial for pUL69-mediated activation of gene expression (34).
Here, we extend these findings and report that pUL69 functions as a viral nuclear RNA export factor that promotes the nuclear export of unspliced reporter mRNAs. Most importantly, our results indicate that pUL69 gains access to the cellular mRNA export pathway via an interaction with the cellular DEXD/H-box RNA helicases UAP56 and URH49. This constitutes a novel mechanism for how viruses target the cellular machinery for mRNA export.
MATERIALS AND METHODS
Plasmids.
The chloramphenicol acetyltransferase (CAT) reporter plasmids pDM128/CMV/RRE and pDM128/CMV/RxRE and the HIV-1 Rev (pcREV), the HTLV-1 Rex (pcREX), and the EBV EB2 (pSVSM) expression plasmids were described elsewhere and were obtained from J. Hauber (Hamburg, Germany) and M. Marschall (Erlangen, Germany) (7, 17). The eukaryotic expression plasmids coding for pUL69, pIE1, and the UL69 mutants aa92-744, aa1-521, PP598/99AA, PP602/03AA, QQ607/08AA, GE613/14AA, E617/18A, and ED619/20AA were described previously, as was the prokaryotic expression plasmid for glutathione S-transferase-IE1 (GST-IE1) (1, 34, 61). UL69 92-744 NLS was cloned by inserting the simian virus 40 (SV40) T antigen (TAg) nuclear localization signal (NLS) sequence in frame with the 5′ end of the UL69 aa92-744 cDNA. The FLAG-UL69-expressing vector F-UL69 was created by inserting a XhoI/BamHI PCR fragment of UL69 into plasmid FLAG-pcDNA3 (20). Site-directed mutagenesis within F-UL69 was performed using the QuickChange site-directed mutagenesis kit as instructed by the manufacturer (Stratagene). The resulting plasmid was termed UL69 mUAP. The bait plasmid pHM300 for the Saccharomyces cerevisiae two-hybrid screen and the yeast plasmids expressing UL69 deletion mutants aa315-744 and aa92-744 fused in frame to the GAL4-BD were described elsewhere (60). Additional UL69 N- or C-terminal deletion mutants fused to GAL4-BD (see Fig. 3B) were constructed by inserting EcoRI/BamHI PCR fragments of UL69 with the corresponding residues into the pGBT9 vector (Clontech). Plasmids GST-UAP56 and FLAG-UAP56 (F-UAP56) were a kind gift of K. Nagata (42); plasmid pCFN-β-Gal was provided by M. Dobbelstein (49). Full-length UAP56 fused to the GAL4-AD (AD-UAP56) was constructed by inserting a UAP56 PCR fragment into pGAD424 (Clontech) via BamHI/PstI. Full-length URH49 cDNA was PCR amplified from a human cDNA (RZPD clone ID IMAGp958H155Q2 [32]) and inserted into pGAD424 via EcoRI/SalI, resulting in plasmid GAL4-AD-URH49. The yeast plasmids expressing the UAP56 or URH49 deletion mutants aa151-428 and aa150-427 fused in frame to the GAL4-BD were constructed by inserting BamHI/PstI PCR fragments of UAP56 or URH49 with the corresponding residues into the pGBT9 vector (Clontech). Plasmid FLAG-URH49 (F-URH49) was generated by insertion of the respective full-length cDNA into the ClaI and XhoI sites of FLAG-pcDNA3 (20). Eukaryotic expression plasmids encoding myc-UAP56 or the N-terminal UAP56 deletion mutant myc-UAP56 aa140-428 (ΔN-UAP56) were generated by inserting the respective BamHI/XhoI PCR fragments of UAP56 into plasmid myc-pcDNA3 (20); the eukaryotic myc-URH49- and myc-URH49 aa168-427 (ΔN-URH)-expressing plasmids were constructed likewise, albeit with insertion of the corresponding URH49 PCR fragments via EcoRI/XbaI restriction sites into myc-pcDNA3.
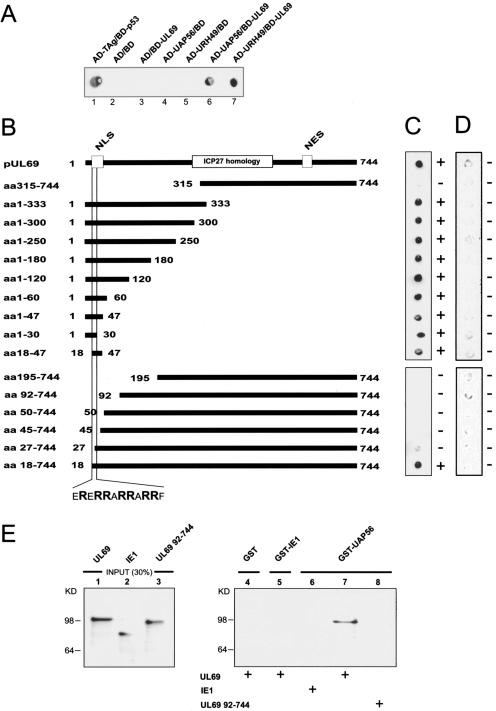
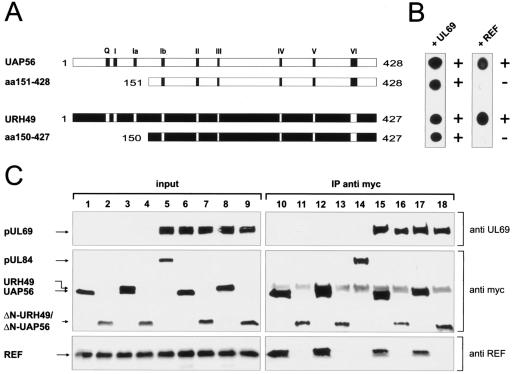
FIG. 3.
Interaction between pUL69 and cellular DEXD/H-box proteins UAP56 and URH49. (A) Yeast two-hybrid system: qualitative analysis of interactions between pUL69 and the cellular DEXD/H-box proteins UAP56 and URH49 as determined in filter lift experiments after staining for β-Gal activity. Yeast cells were transformed with two separate expression plasmids, one of which encoded either BD-UL69 or the GAL4 DNA binding domain alone (BD). The second plasmid encoded either UAP56 or URH49 as a fusion with the GAL4 activation domain (AD-UAP56 and AD-URH49) or the GAL4 activation domain alone (AD). The association of murine p53 (BD-p53) and SV40 TAg (AD-TAg) served as a positive control. (B) Mapping of the UAP56/URH49 interaction domain within pUL69. N- and C-terminal deletion mutants of pUL69 generated as in-frame fusions to the GAL4 DNA binding domain are indicated. (C) Yeast cells were transfected with a combination of vectors encoding the pUL69 deletion mutants (see panel B) and UAP56 fused to the GAL4 activation domain. Yeast colonies, selected for the presence of both plasmids, were analyzed for the expression of β-Gal by filter lift assays. (D) Qualitative analysis of the respective interaction between the various pUL69 fragments and the GAL4 activation domain alone as determined in filter lift experiments. (E) pUL69 interacts physically with UAP56 in GST pull-down experiments. In vitro-translated 35S-labeled pUL69, IE1, and an N-terminally deleted pUL69 mutant lacking the UAP56 interaction domain (UL69 aa92-744) were used for pull-down assays. Lanes 1 to 3, input protein; 30% of the amount used in the pull-down analysis is shown. Lanes 4 to 8, proteins are shown that were recovered after GST pull-down analysis. GST (lane 4), GST-IE1 (lane 5), or GST-UAP56 (lanes 6 to 8) was incubated with in vitro-translated proteins as indicated. After extensive washing, the bound proteins were resolved by SDS-PAGE, and an autoradiograph of the gel is shown.
Infection and transfection of cells and CAT RNA export assays.
Primary human foreskin fibroblasts (HFFs) and HeLa and HEK 293T cells were cultured as described previously (20, 61). HFFs were infected with HCMV (strain AD169) at a multiplicity of infection of 1 to 2 PFU per cell. HeLa and 293T cells were transfected via the calcium phosphate coprecipitation procedure as described earlier (20, 61). CAT reporter assays were performed essentially as described in reference 11. To quantify CAT protein expression, a CAT enzyme-linked immunosorbent assay was used (Roche Molecular Biochemicals). After transfection, half of the cells were used for CAT assays according to the manufacturer's instructions, and the other half was used to monitor protein expression by Western blotting. Each transfection was performed in triplicate and was repeated at least three times. If inhibition of CRM1-dependent protein export was required, cells were treated with 2.5 ng/ml leptomycin B (LMB) (kindly provided by M. Yoshida, Tokyo, Japan) for 6 hours before cell extracts were prepared.
RNA isolation and RNase protection assay.
Total cellular RNA was harvested 48 h after transfection according to the method in reference 8. To isolate cytoplasmic RNA, cells were lysed in 50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 5 mM MgCl2, 0.5% Nonidet P-40, 40 units of RNasin (Roche), 1 mM dithiothreitol on ice for 30 seconds. After centrifugation, the supernatant was incubated with 20% sodium dodecyl sulfate (SDS) and 25 mg/ml proteinase K at 37°C for 15 min and then extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1) and once with chloroform-isoamyl alcohol (24:1) followed by ethanol precipitation of the RNA. The pellet was resuspended in 100 mM Tris-HCl, pH 7.5, 100 mM MgCl2 and treated with 10 U of RNase-free DNase (Roche) at 37°C for 15 min followed by an additional round of proteinase K treatment, phenol extraction, and ethanol precipitation. The antisense riboprobe was prepared using plasmid pHM2119 containing a 655-bp SalI-EcoRI fragment of pDM128/CMV/RRE downstream of the T7 promoter of the pBluescript vector (Stratagene). Preparation of the riboprobe and RNase protection analysis were performed as described previously (54).
Yeast two-hybrid screen and mapping.
The GAL4-based yeast two-hybrid screening procedure to identify pUL69-interacting proteins has been described previously (60). For mapping of the pUL69 interaction domain using the yeast two-hybrid system, the respective UL69 deletion mutants were transformed together with the interactor plasmids into yeast strain Y153 and tested as described in reference 35.
Purification of GST fusion proteins and pull-down assays.
The purification of GST fusion proteins was described in detail elsewhere (30). For pull-down assays, 5 to 30 μl of glutathione-Sepharose-bound proteins were preincubated for 10 min in 200 μl of ELB+ buffer (250 mM NaCl, 50 mM HEPES, pH 7.0, 0.1% NP-40, 0.5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol, 10 μg/ml RNase A) containing bovine serum albumin to a final concentration of 1 mg/ml. After addition of 1 to 5 μl of the indicated in vitro-translated, radiolabeled protein which had been generated by using the TNT system (Promega), the beads loaded with GST fusion proteins were gently mixed for 2 h at 4°C. The beads were then washed five times in 1 ml ELB+ buffer, pelleted, and boiled in SDS sample buffer. Finally, the bound proteins were resolved using SDS-polyacrylamide gels. The gels were fixed, incubated in Amplify (Amersham Life Science) for 30 min, dried, and subjected to autoradiography.
Western blotting and immunoprecipitation analysis.
For Western blot analysis, transfected cells were lysed, diluted in SDS-Laemmli buffer, and boiled at 94°C for 5 min (20). Samples were electrophoresed by SDS-polyacrylamide gel electrophoresis (PAGE) on 8 to 12.5% polyacrylamide gels, and the proteins were transferred onto nitrocellulose membranes. Western blotting and chemiluminescence detection were performed according to the manufacturer's protocol (ECL Western detection kit; Amersham Pharmacia Biotech). Coimmunoprecipitation analysis was performed as described previously (35). Briefly, transfected 293T cells or HCMV-infected (72 h postinfection) and mock-infected HFFs were lysed in 800 μl of NP-40 lysis buffer (50 mM Tris-HCl, pH 8.0; 150 mM NaCl; 5 mM EDTA; 0.5% NP-40; 1 mM phenylmethylsulfonyl fluoride; 1 mg each of aprotinin, leupeptin, and pepstatin per ml) and incubated for 20 min at 4°C. After centrifugation, the supernatant was incubated with the appropriate antibody coupled to protein A-Sepharose beads for 1 h at 4°C. The Sepharose beads were collected and washed five times in NP-40 lysis buffer. Antigen-antibody complexes were recovered by boiling the mixture in SDS sample buffer and analyzed by Western blotting. For RNase A treatment, 100 μg RNase A was added to the cell extracts and preincubated at room temperature 15 min before immunoprecipitation.
Antibodies, indirect immunofluorescence analysis, and heterokaryon assays.
The polyclonal antiserum used to detect pUL69 was described previously (61), the polyclonal antisera against REF (KJ58) and TAP (αTAP) were obtained from E. Izaurralde (Heidelberg, Germany), and the polyclonal anti-UAP56 antibody (αUAP56) was provided by Michael Green (HHMI, Worcester, MA) (13). The anti-FLAG monoclonal antibody M2 was purchased from INTEGRA Bioscience (Fernwald, Germany), and the anti-β-galactosidase (β-Gal) antibody was obtained from Roche (Mannheim, Germany). Anti-mouse and anti-rabbit horseradish peroxidase- or fluorescein isothiocyanate- and tetramethyl rhodamine isothiocyanate-conjugated secondary antibodies were obtained from Dianova (Hamburg, Germany). Indirect immunofluorescence analysis and interspecies heterokaryon analyses were carried out exactly as described previously (34). Images were analyzed using a Zeiss Axioplan-2 microscope and recorded with a cooled Spot color digital camera (Diagnostic Instruments, Sterling Heights, MI). The Meta-Imaging series and Adobe Photoshop package (Universal Imaging Corp., Brandywine, PA; Adobe Systems Incorporated) were used for processing.
RESULTS
The UL69 protein promotes the nuclear export of inefficiently spliced mRNA.
Human herpesviruses encode posttranscriptional activators that are believed to up-regulate viral replication by facilitating viral mRNA export. We have reported previously that the HCMV transactivator pUL69 is a nucleocytoplasmic shuttling protein that contains a novel type of CRM1-independent nuclear export signal. Initial functional experiments revealed a correlation between nucleocytoplasmic shuttling and pUL69-mediated transactivation, indicating that pUL69 might function as an mRNA export factor (34). Consequently, we initiated studies to investigate the role of pUL69 in mRNA export. For this, we took advantage of a functional reporter assay which has been developed by Hope and colleagues to monitor RNA export in vivo (22). This assay is based on the pDM128/CMV reporter plasmid (Fig. 1A) and has been used successfully to (i) study mRNA export activity of the HIV-1 Rev protein (58), (ii) quantify TAP-mediated stimulation of RNA nuclear export (4), and (iii) identify novel factors with mRNA export activity (11, 39). pDM128/CMV/RRE harbors the CAT coding sequence and the RRE target sequence of the HIV-1 mRNA export factor Rev inserted into an intron, which is flanked by HIV-1 splice sites (23, 41) (Fig. 1A). Since HIV-1 splice sites are inefficiently recognized in primate cells, unspliced pre-mRNA expressed from this plasmid is retained in the nucleus of transfected cells, yielding only minimal levels of CAT protein expression (23, 41). However, cotransfection of the pDM128/CMV/RRE reporter with plasmids expressing nucleocytoplasmic shuttling proteins that promote nuclear export of the inefficiently spliced pre-mRNA leads to an increase in CAT expression. HIV-1 Rev and EBV EB2 both have been shown previously to increase the nuclear export of unspliced mRNA generated from the pDM128/CMV/RRE reporter construct. Whereas in the case of Rev the cis-acting RNA element RRE was found to be necessary for activity, EB2-mediated nuclear export of unspliced RNA was shown to be RRE independent (11, 19, 22, 40).
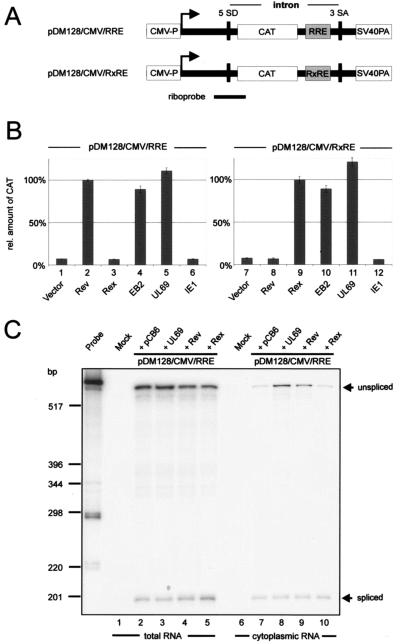
FIG. 1.
pUL69 mediates the nuclear export of unspliced CAT mRNA. (A) Schematic view of the pDM128/CMV reporter plasmids showing locations of the splice donor (SD) and splice acceptor (SA) sites that flank the CAT reporter gene, the Rev (RRE) or Rex (RXRE) response element, and the SV40 late poly(A) signal. The localization of the antisense riboprobe used in the RNase protection assay is indicated. (B) HeLa cells were transfected with the respective pDM128/CMV reporter constructs and with plasmids expressing HIV Rev, human T-cell leukemia virus Rex, EBV EB2, HCMV UL69, and HCMV IE1 as indicated. CAT protein expression was measured in cell lysates 48 h after transfection using a CAT enzyme-linked immunosorbent assay. The relative amounts of CAT protein expressed were given as percentages of the amount obtained for the Rev positive control. One hundred percent represents an 8.2-fold increase of CAT protein in comparison to basal levels. (C) pUL69- and Rev-mediated cytoplasmic accumulation of CAT mRNA was monitored by RNase protection analysis of total (lanes 1 to 5) or cytoplasmic (lanes 6 to 10) RNA from 293T cells transfected with pDM128/CMV/RRE. In lanes 3, 4, 8, and 9, plasmids encoding pUL69 or Rev were cotransfected. As a control, the reporter plasmid was cotransfected either with empty vector (pCB6, lanes 2 and 7) or with a vector encoding Rex (lanes 5 and 10).
In order to test whether the HCMV UL69 protein is active in this reporter assay, plasmids expressing HIV-1 Rev, HTLV-1 Rex, EBV EB2, HCMV pUL69, and HCMV IE1-p72 (IE1) were transfected into HeLa cells together with the pDM128/CMV/RRE reporter construct, followed by quantification of CAT protein expression (Fig. 1B, lanes 1 to 6). As expected, Rev and EB2 expression induced the cytoplasmic accumulation of unspliced RNA, and therefore, a strong increase in the amount of CAT protein was detected (Fig. 1B, lanes 2 and 4). Conversely, the sequence-specific mRNA export factor Rex, encoded by the human T-cell leukemia virus type 1, and the HCMV transactivator IE1 had no effect on the level of CAT protein expression compared to the value obtained with the reporter plasmid alone (Fig. 1B, compare lanes 1, 3, and 6). When a plasmid expressing pUL69 was cotransfected with pDM128/CMV/RRE, the amount of CAT protein significantly increased and reached a level similar to or even higher than that in the presence of the Rev or EB2 positive controls (Fig. 1B, lane 5). In order to exclude the possibility that CAT induction upon pUL69 expression was dependent on the RRE, we repeated the transfection experiments using the reporter plasmid pDM128/CMV/RxRE, in which the HIV-1 RRE had been replaced by the cis-acting RXRE response element specific for the mRNA export factor Rex (Fig. 1A and B, lanes 7 to 12) (17, 21). As expected, Rev did not induce CAT expression in pDM128/CMV/RxRE-transfected cells, whereas now a strong induction of CAT could be observed in response to Rex (Fig. 1B, lanes 8 and 9). However, CAT induction upon pUL69 expression was similar in value to that for the RRE-bearing reporter plasmid (Fig. 1B, lane 11), indicating that pUL69-mediated transactivation in the pDM128/CMV reporter system is RRE independent.
To confirm that the stimulation of CAT protein expression observed in the reporter assay reflects an increased export of unspliced CAT RNA, we performed an RNase protection assay with either total (Fig. 1C, lanes 1 to 5) or cytoplasmic (Fig. 1C, lanes 6 to 10) RNA fractions derived from mock-transfected cells or cells cotransfected with the pDM128/CMV/RRE reporter and Rev-, pUL69-, or Rex-expressing vectors. In order to determine levels of spliced and unspliced CAT RNAs, we used a riboprobe spanning the 5′ splice site of pDM128/CMV/RRE as depicted in Fig. 1A. When total cellular RNA was analyzed, we consistently detected comparable amounts of spliced and unspliced CAT RNA in all samples, indicating that the total amount of CAT RNA is not altered in the presence of pUL69 or Rev (Fig. 1C, lanes 1 to 5). This suggests that the effect of pUL69 on CAT expression is not due to increased transcription. In RNase protection experiments with cytoplasmic RNA, unspliced CAT RNA was detected only at low levels after cotransfection with pDM128/CMV/RRE and an empty control vector or a vector encoding HTLV-1 Rex (Fig. 1C, lanes 6 and 10). However, there was a marked increase in the relative amount of unspliced cytoplasmic CAT RNA after cotransfection of pDM128/CMV/RRE with expression plasmids for pUL69 or the positive control Rev (Fig. 1C, lanes 8 and 9). This increase in the levels of unspliced RNA in the cytoplasm correlated very well with the increase in CAT expression observed in Fig. 1B. Thus, the results obtained by quantitative RNA analysis are consistent with the assumption that pUL69 functions as an RNA export factor that promotes CAT expression by allowing the unspliced transcripts to enter the cytoplasm.
pUL69-mediated nuclear export of unspliced RNA requires nucleocytoplasmic shuttling but is independent of the CRM1 export receptor.
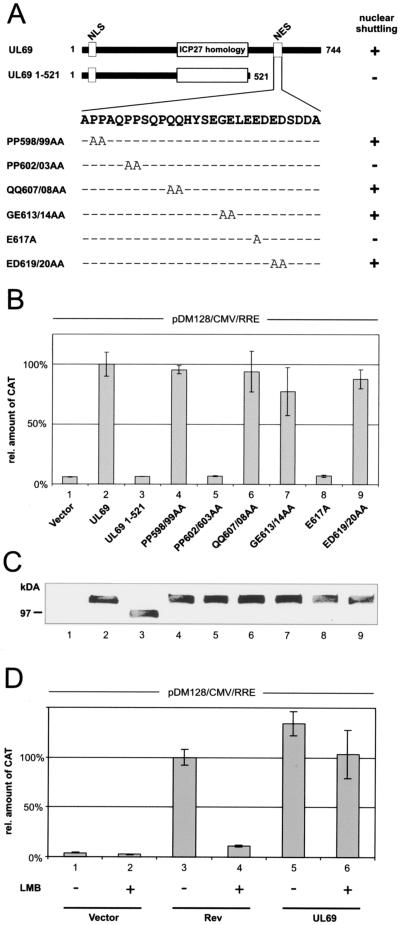
Since we could demonstrate that pUL69 promotes the nuclear export of unspliced mRNA, we next queried the role of nucleocytoplasmic shuttling for pUL69-mediated RNA export. For this, a pUL69 deletion mutant lacking the C-terminally located NES (UL69 1-521) and a series of pUL69 alanine replacement mutants carrying point mutations within the NES were tested for their capacity to export RNA, using the pDM128/CMV/RRE reporter system (Fig. 2). It has previously been reported that the UL69 deletion mutant UL69 1-521 and the UL69 point mutants PP602/03AA and E617A show a nuclear localization but are nonshuttling proteins (34). Vectors expressing the pUL69 mutants were cotransfected into HeLa cells with the pDM128/CMV/RRE reporter plasmid. All shuttling-competent pUL69 mutants very efficiently increased CAT protein levels (Fig. 2B, lanes 2, 4, 6, 7, and 9), whereas the shuttling-deficient mutants failed to do so (Fig. 2B, lanes 3, 5, and 8). This effect was not due to instability of the respective mutant polypeptides, since all tested proteins were expressed at comparable levels (Fig. 2C). Thus, these results revealed a perfect correlation between shuttling and pUL69 RNA export capability, strongly indicating that nucleocytoplasmic shuttling is crucial for pUL69-dependent nuclear RNA export stimulation.
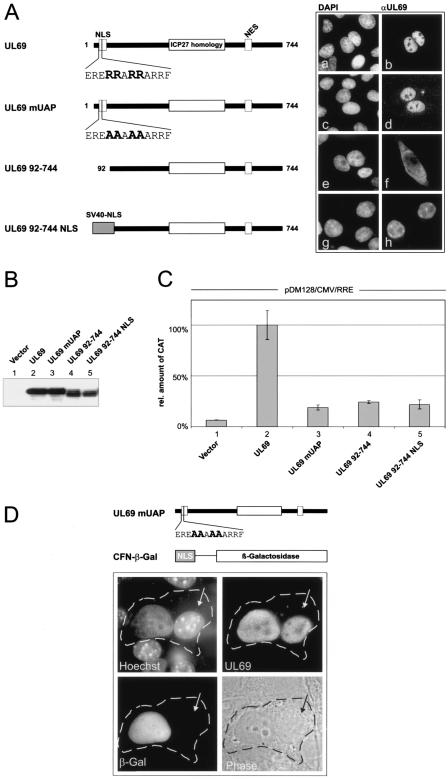
FIG. 2.
pUL69-mediated RNA export requires nucleocytoplasmic shuttling but is CRM1 independent. (A) Schematic representation of the wt UL69 protein, a C-terminal deletion mutant lacking the NES (UL69 1-521), and a series of pUL69 alanine replacement mutants carrying point mutations within the NES. The region of highest homology to the HSV-1 ICP27 and the location of the nuclear import signal (NLS) and the NES are depicted for pUL69. + or − indicates the nucleocytoplasmic shuttling activity of each pUL69 mutant as determined by heterokaryon analysis (34). (B) CAT protein expression was measured in HeLa cell lysates cotransfected with pDM128/CMV/RRE and plasmids encoding the proteins depicted in panel A. The values are given as percentages of the amount of CAT protein detected with wt UL69; 100% represents a 10.4-fold increase of CAT protein in comparison to basal levels. (C) Western blot analysis of HeLa cell extracts after transfection of the pUL69 mutants indicated in panel A using a polyclonal antiserum specific for pUL69. (D) Relative CAT protein amount expressed in HeLa cells transfected with pDM128/CMV/RRE and plasmids expressing Rev or pUL69 as indicated. As indicated by a + sign cells were washed with fresh medium supplemented with 2.5 ng/ml LMB at 12 h posttransfection and incubated for a further 6 h. At this point cell extracts were made and CAT protein was quantified. The results are expressed as the amount of CAT relative to the amount of CAT protein expressed in the presence of Rev.
Next, we were interested in the nuclear export pathway which is targeted by pUL69 in order to facilitate nuclear RNA export. Previous studies had demonstrated that sequence-specific RNA binding proteins encoded by complex retroviruses (e.g., HIV-1 Rev) recruit the cellular nuclear export receptor CRM1 to incompletely spliced viral mRNAs (3, 14). In contrast, posttranscriptional transactivators encoded by specific herpesviruses have been described to facilitate RNA export independently of CRM1 by accessing the cellular mRNA export factor REF (18, 27, 39). To address the question whether the HCMV UL69 protein promotes cytoplasmic accumulation of unspliced RNA via a CRM1-dependent or -independent pathway we tested whether pUL69-associated nuclear export of RNA is resistant to LMB, a specific inhibitor of the CRM1 transport receptor (63). For this, HeLa cells were cotransfected with the pDM128/CMV/RRE reporter and HIV-1 Rev or HCMV pUL69 expression vectors followed by incubation in the presence or absence of LMB. The presence of LMB effectively inhibited Rev-dependent RNA export, confirming that LMB treatment of cells was able to block the CRM1-dependent export pathway (Fig. 2D, compare lanes 3 and 4). However, LMB had only minor effects on pUL69 activity (Fig. 2D, compare lanes 5 and 6), indicating that pUL69 induces the nuclear export of intronless RNA by targeting a CRM1-independent transport pathway. Importantly, this result is consistent with our previous findings that defined pUL69 as a CRM1-independent nucleocytoplasmic shuttling protein (34).
pUL69 interacts with the cellular mRNA export factors URH49 and UAP56 in vitro and in vivo.
To identify potential interacting partners of pUL69, we screened a human cDNA library derived from B lymphocytes by yeast two-hybrid analysis and selected the putative DEXD/H-box RNA helicase URH49 (also termed DDX39) as a potential pUL69 binding partner (31, 44, 60). Since URH49 shares more than 90% amino acid identity with the multifunctional intranuclear RNA helicase UAP56, we further analyzed both URH49 and UAP56 for their ability to bind pUL69 in a yeast two-hybrid assay. As shown in Fig. 3A, we could observe an interaction of both URH49 and UAP56 with pUL69 in yeast cells. These interactions between pUL69 and the two DEXD/H-box proteins were considered specific, since the results of control transformations argued against a nonspecific activation of the reporter genes in this system (Fig. 3A, lanes 2 to 5). As outlined in the introduction, UAP56 has been defined as a key player in cellular mRNA export that acts upstream of the adaptor protein REF and is thought to couple intranuclear steps in mRNA biogenesis with mRNA export. Recent work provided evidence that the highly related DEXD/H-box protein URH49 carries out similar or redundant functions in mammalian mRNA biogenesis (44).
The identification of protein-protein interactions between pUL69 and UAP56 or URH49 raised the question of the responsible interaction motif within pUL69. To uncover the region of pUL69 required for binding to UAP56, we constructed a series of N- or C-terminal pUL69 deletion mutants fused to the GAL4 DNA binding domain (Fig. 3B). These mutants were analyzed in the yeast two-hybrid assay for their interaction with UAP56 fused to the GAL4 activation domain (Fig. 3B and C). The regions encompassing aa 1 to 30 and 18 to 47, which contain the NLS of pUL69 (34), were both required and sufficient for interaction with UAP56. By cotransformation of the UL69 mutants with the empty GAL4 activation domain vector, it was excluded that the pUL69 fusions with the GAL4 DNA binding domain were able to activate the reporter genes in yeast in the absence of UAP56 (Fig. 3B and D). The expression of pUL69 mutants that lack an interaction with UAP56 has been verified by filter lift assays after transformation of these plasmids into a yeast strain that expresses the known pUL69 binding protein hSPT6 (60) (data not shown). Thus, we could identify a short peptide sequence comprising 12 aa within the N terminus of pUL69 as a binding motif for UAP56 (Fig. 3B and C). No differences were observed when we assayed binding of pUL69 to either UAP56 or URH49 (data not shown), indicating that the two proteins bind to an identical domain within pUL69.
To confirm that the observed interaction of pUL69 with UAP56 can also be detected in an independent experimental approach, GST pull-down analyses were performed using a GST-UAP56 fusion protein purified from Escherichia coli (Fig. 3E). Wild-type (wt) pUL69, an N-terminally truncated pUL69 mutant lacking amino acid residues 1 to 91 (UL69 92-744), and the viral protein IE1 were produced by in vitro transcription/translation in the presence of [35S]methionine (Fig. 3E, lanes 1 to 3). The radiolabeled polypeptides were then incubated with the bacterially expressed GST-UAP56 fusion protein or with GST alone and GST-IE1 that served as negative controls. Since UAP56 and also pUL69 are likely to be RNA binding proteins, RNase A was added to all reaction mixtures in order to avoid nonspecific bridging through RNA. As shown in Fig. 3E, lanes 6 and 7, pUL69 was able to interact strongly with the GST-UAP56 fusion protein whereas IE1 did not. No pUL69 interaction was observed with GST alone or GST-IE1 (Fig. 3E, lanes 4 and 5), arguing against a nonspecific protein-protein interaction. Furthermore, consistent with the data obtained in yeast experiments, a pUL69 fragment lacking aa 1 to 91 failed to bind to GST-UAP56. Taken together, these data indicate that pUL69 can interact directly with UAP56 in an in vivo and in an in vitro binding assay and show that an N-terminal region of pUL69 which includes the UL69 NLS is necessary for this interaction.
pUL69 is associated with UAP56- and URH49-containing protein complexes in mammalian cells.
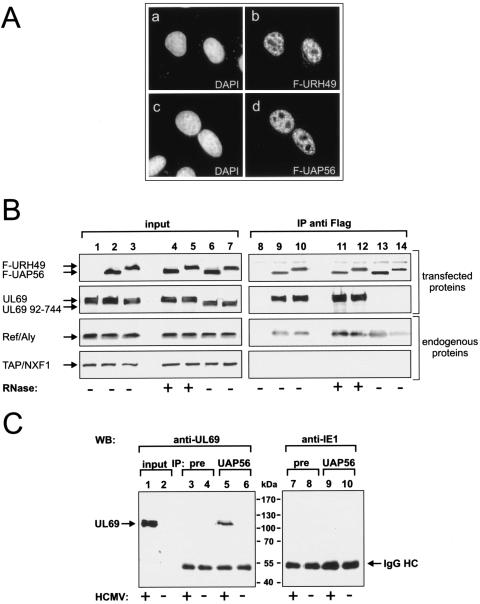
Although the yeast two-hybrid screen and the in vitro interaction experiments suggested that pUL69 associates with UAP56- or URH49-containing protein complexes, we sought to confirm this within the context of a mammalian cell. In order to be able to immunoprecipitate the two highly related DEXD/H-box RNA helicases from mammalian cell extracts, we constructed eucaryotic vectors, expressing the indicated proteins in fusion with the FLAG epitope. Since very little is known about the URH49 protein expression pattern, we first examined the intracellular localization of F-URH49 in HeLa cells. As shown in Fig. 4A, F-URH49 is localized predominantly in the nucleoplasm, is excluded from nucleoli, and shows a pattern similar to that observed for F-UAP56 (42). Next, we investigated the expression of F-UAP56 and F-URH49 in HEK 293T cells by Western blotting with an anti-FLAG antibody (Fig. 4B, lanes 2 to 7). Interestingly, although UAP56 and URH49 are 90% identical to each other and both proteins have a predicted molecular mass of 49 kDa, F-URH49 showed a significantly reduced mobility in SDS-PAGE in comparison to F-UAP56 (Fig. 4B, compare lanes 2 and 3). To determine whether pUL69 can be found in UAP56- or URH49-containing complexes, cotransfection experiments in HEK 293T cells were performed using expression vectors for F-UAP56 or F-URH49 and pUL69 (Fig. 4B, lanes 1 to 7), followed by immunoprecipitation with an anti-FLAG antibody. Coimmunoprecipitated proteins were resolved by SDS-PAGE and analyzed by Western blotting with several different antibodies (Fig. 4B, lanes 8 to 14). As shown in Fig. 4B, lanes 9 and 10, pUL69 was coimmunoprecipitated efficiently with F-UAP56 and F-URH49. This precipitation was specific, since no signal was present when pUL69 was expressed alone (Fig. 4B, compare lanes 1 and 8) or when a nonspecific control antibody was used for precipitation (not shown). Since pUL69 promotes nuclear export of a reporter RNA and both UAP56 and URH49 have been implicated in mRNA transport, we also carried out identical immunoprecipitation experiments after digestion with RNase A. As shown in Fig. 4B, lanes 11 and 12, RNase digestion did not affect binding of pUL69 to F-UAP56 or F-URH49, indicating that these interactions do not occur through RNA bridging. In addition, an N-terminal pUL69 deletion mutant (UL69 92-744) lacking the putative UAP56 binding domain could not be coprecipitated with either of the two DEXD/H-box proteins (Fig. 4B, compare lanes 6 and 7 and lanes 13 and 14), further confirming that the very N-terminal region of pUL69 is required for interaction with UAP56 and URH49. As outlined in the introduction, current models assume that mRNA-associated UAP56 recruits REF and hence indirectly the TAP-p15 export receptor to mRNPs marked for nuclear export. However, after loading of REF onto the mRNA, UAP56 must be released from REF to allow binding of TAP, because binding of UAP56 and that of TAP to REF are mutually exclusive (56). Due to these findings, we tested for the presence of REF and TAP proteins in UAP56 or URH49 protein complexes containing pUL69. This revealed that immunoprecipitation of UAP56 or URH49 brought down cell-endogenous REF, but not TAP (Fig. 4B, endogenous proteins). Thus, our binding assays are in agreement with previous observations and we concluded that in transfected cells pUL69 is able to interact with UAP56 and URH49 in mRNP complexes that also contain REF.
FIG. 4.
pUL69 is associated with nuclear UAP56- or URH49-containing complexes. (A) URH49 is mainly localized in the nucleoplasm of HeLa cells and shows a pattern similar to that observed for UAP56. FLAG-URH49 (a and b)- or FLAG-UAP56 (c and d)-expressing plasmids were transfected into HeLa cells which were subsequently immunostained using an anti-FLAG antibody. DAPI, 4′,6′-diamidino-2-phenylindole. (B) pUL69 is associated with UAP56 or URH49 and REF. HEK 293T cells were cotransfected with plasmids encoding wt UL69 (lanes 1 to 5 and 8 to 12) or an N-terminal UL69 deletion mutant (UL69 92-744, lanes 6 and 7 and lanes 13 and 14) and FLAG-tagged proteins UAP56 (lanes 2, 4, 6, 9, 11, and 13) or URH49 (lanes 3, 5, 7, 10, 12, and 14). The amount of protein in the input was analyzed by Western blotting (input, lanes 1 to 7). Immunoprecipitation was performed using anti-FLAG antibody M2 (IP anti-FLAG, lanes 8 to 14). Coimmunoprecipitated proteins were visualized by Western blotting using either anti-FLAG, anti-UL69, anti-REF, or anti-TAP antibodies to detect the indicated proteins. For RNase treatment RNase A was added to the indicated extracts as described in Materials and Methods. (C) pUL69 forms a complex with UAP56 in HCMV-infected cells. HFF cells were mock infected (lanes 2, 4, 6, 8, and 10) or infected with HCMV AD169 (lanes 1, 3, 5, 7, and 9), and immunoprecipitation was performed using a polyclonal anti-UAP56 antibody (lanes 5, 6, 9, and 10) or a nonspecific preimmune serum (lanes 3, 4, 7, and 8). The amount of UL69 protein in the input (input; lanes 1 and 2) and the coimmunoprecipitated proteins (IP; lanes 3 to 10) were visualized by Western blotting. Lanes 1 to 6, Western blotting was performed with anti-UL69 monoclonal antibody; lanes 7 to 10, Western blotting was performed with anti-IE1 monoclonal antibody. IgG HC, immunoglobulin G heavy chain.
In light of these results we next asked whether pUL69 can also interact with UAP56 in HCMV-infected cells. To do so, we performed anti-UAP56 immunoprecipitations using extracts of HCMV-infected or mock-infected primary HFF cells followed by Western blotting analysis. As shown in Fig. 4C, lanes 5 and 6, a signal of approximately 110 kDa, corresponding to pUL69, could be detected with lysates from HCMV-infected fibroblasts but not with lysates from mock-infected cells. No pUL69 was detectable either when preimmune serum was used for precipitation or when an IE1-specific antibody was used for Western blot analysis (Fig. 4C, lanes 3 and 4 and lanes 7 to 10). Thus, pUL69 is also able to interact with UAP56 in the context of an HCMV-infected cell.
pUL69 does not rescue REF recruitment to a REF-binding-deficient UAP56 or URH49 mutant.
After having shown that pUL69 forms a complex with both UAP56 or URH49 and REF in eukaryotic cells, we went on and asked whether the presence of pUL69 in mRNP complexes leads to an enhanced recruitment of REF, as shown for ICP27 of HSV-1 (6, 27). To address this question, we took advantage of the observation that different domains within UAP56 and URH49 are required for binding to pUL69 or REF. In detail, N-terminal UAP56 or URH49 deletion mutants lacking ∼150 amino acids failed to interact with REF in the yeast two-hybrid assay; however, binding to pUL69 was unimpaired (Fig. 5A). Next, we validated these findings in human cells. To do so, we transiently expressed UAP56 and URH49 or N-terminally truncated UAP56 or URH49 deletion mutants, each with a myc tag at its N terminus, in 293T cells either in the absence (Fig. 5C, lanes 1 to 4 and 10 to 13) or in the presence (Fig. 5C, lanes 5 to 9 and 14 to 18) of pUL69. Then, immunoprecipitations were performed with an anti-myc antibody followed by Western blotting with anti-UL69, anti-REF, or anti-myc antibodies. As expected, endogenous REF protein efficiently coprecipitated with full-length UAP56 and full-length URH49 but not with an N-terminally truncated UAP56 or URH49 deletion mutant (Fig. 5C, compare lanes 10, 12, 15, and 17 and lanes 11, 13, 16, and 18). In contrast, pUL69 was found to associate with both full-length UAP56/URH49 and the two REF-binding-deficient deletion mutants (Fig. 5C, lanes 15 to 18). Most interestingly, although pUL69 is coprecipitated with REF-binding-deficient UAP56/URH49 mutants, REF assembly into UAP56- or URH49-containing protein complexes was not rescued (Fig. 5C, lanes 16 and 18; compare pUL69 and REF). Thus, these results strongly suggest (i) that pUL69 interacts in a REF-independent manner with UAP56 or URH49, (ii) that there is no direct interaction between pUL69 and REF in UAP56- or URH49-containing complexes, and (iii) that binding of pUL69 to primarily UAP56 does not result in an increased recruitment of REF.
FIG. 5.
pUL69 does not rescue REF recruitment to a REF-binding-deficient UAP56 or URH49 mutant. (A) Schematic diagram illustrating N-terminal deletion mutants of UAP56 and URH49 generated as in-frame fusions to the GAL4 DNA binding domain. Conserved motifs of the DEXD/H-box family are indicated. (B) Proteins indicated in panel A were analyzed for their ability to bind GAL4 activation domain fusions of pUL69 or REF in a yeast two-hybrid assay. The experiments were performed as described in the legend to Fig. 3. (C) 293T cells were transfected with plasmids encoding myc-tagged UAP56 (lanes 1, 6, 10, and 15), myc-tagged URH49 (lanes 3, 8, 12, and 17), myc-tagged UAP56 mutant ΔN-UAP56 (lanes 2, 7, 11, and 16), myc-tagged URH49 mutant ΔN-URH49 (lanes 4, 9, 13, and 18), or a myc-UL84-expressing control plasmid (lanes 5 and 14) either in the absence (lanes 1 to 4 and 10 to 13) or in the presence (lanes 5 to 9 and 14 to 18) of a pUL69-expressing vector. The amount of protein in the input was analyzed by Western blotting (input, lanes 1 to 9). Immunoprecipitation was performed using anti-myc antibody (IP anti-myc, lanes 10 to 18). Coimmunoprecipitated proteins were visualized by Western blotting using either anti-UL69, anti-myc, or anti-REF antibodies to detect the indicated proteins. Cells cotransfected with expression plasmids encoding pUL69 and myc-tagged pUL84 (lanes 5 and 14) served as a negative control.
UAP56 binding is crucial for pUL69-mediated nuclear RNA export.
The DEXD/H-box RNA helicase UAP56 has been implicated in nuclear mRNA export as well as in RNA splicing (10). To determine whether the interaction with UAP56 is necessary for pUL69-mediated stimulation of nuclear RNA export, we investigated the RNA export activity of pUL69 mutants incapable of binding to UAP56 (Fig. 6). Mutant UL69 mUAP carries alanine substitutions within the UAP56 binding domain whereas the N-terminal deletion mutant UL69 92-744 lacks the entire UAP56 interaction domain. However, since we have shown previously that the NLS of pUL69 is also located in the N-terminal region of the protein (34), we fused the NLS of the SV40 T antigen to UL69 92-744 in order to reconstitute the nuclear localization of this mutant (Fig. 6A). Next, comparable expression levels of the UL69 mutants were verified by Western blotting (Fig. 6B) and the nuclear RNA export activities of the proteins were assayed by transient transfection into HeLa cells along with the pDM128/CMV/RRE reporter plasmid. As shown in Fig. 6C, lanes 3 to 5, mutation or deletion of the UAP56 binding domain within pUL69 eliminates not only its UAP56 binding (Fig. 4B, lanes 13 and 14; data not shown for UL69 mUAP) but also its nuclear RNA export activity. This strongly suggests that binding to UAP56 is crucial for pUL69-mediated stimulation of RNA export.
FIG. 6.
Binding to UAP56 is crucial for pUL69-mediated RNA export. (A) Schematic diagram illustrating FLAG-tagged UL69 proteins with either an intact or a mutated/deleted UAP56 binding domain. The subcellular localization of each mutant was analyzed via indirect immunofluorescence analysis. DAPI, 4′,6′-diamidino-2-phenylindole. (B) Expression of wild-type and mutant UL69 proteins after transfection of HeLa cells as detected by Western blotting. (C) Relative amount of CAT protein expressed in HeLa cells cotransfected with pDM128/CMV/RRE and plasmids expressing the proteins indicated in panel A. (D) Interspecies heterokaryon analysis to detect nucleocytoplasmic shuttling of a UL69 protein with mutated UAP56 binding site. Upper part: schematic representation of mutant UL69 mUAP and an NLS/β-Gal fusion protein (CFN-β-Gal) that was used as an internal nonshuttling control in the interspecies heterokaryon analysis. Lower part: immunodetection of UL69 protein and β-Gal in a heterokaryon consisting of HeLa and NIH 3T3 cells. Human and murine nuclei were differentiated via counterstaining with Hoechst 33258 dye (Hoechst panel; arrows indicate the mouse nucleus). Phase, phase-contrast image of the heterokaryon; the cytoplasmic edge is highlighted by a broken line.
Since the RNA export function of the UL69 protein correlated with its ability to shuttle between the nucleus and the cytoplasm (Fig. 2B), we finally examined the nucleocytoplasmic shuttling activity of the mutated UL69 variants using an interspecies heterokaryon analysis. HeLa cells were cotransfected with an expression plasmid for mutant UL69 mUAP and the CFN-β-Gal control plasmid that encodes a nucleus-restricted β-galactosidase (Fig. 6D). After heterokaryon formation with murine NIH 3T3 cells, immunostaining was performed to detect both the UL69 mutant and the control protein β-Gal. Murine nuclei were identified by counterstaining with Hoechst 33258 dye, yielding a characteristic punctate heterochromatin staining pattern. Two hours after heterokaryon formation, only UL69 mUAP was found to be present in both human and murine nuclei (UL69), whereas NLS-β-Gal was detected exclusively in the human nuclei. Identical results were obtained when UL69 92-744 NLS was used in this assay (not shown). Thus, UL69 mUAP and UL69 92-744 NLS were selectively transported from the transfected HeLa cell nucleus into the cytoplasm and subsequently into the murine nucleus. These data indicate that the UAP56-binding-deficient UL69 mutants constitute bona fide nucleocytoplasmic shuttling proteins as does the wild-type UL69 protein. Taken together, we concluded from this set of experiments that binding to UAP56 is crucial for pUL69-mediated stimulation of mRNA export.
DISCUSSION
Recently, it has become apparent that some homologous proteins of the transactivator pUL69 within the subfamilies of human alpha- and gammaherpesviruses, including HSV-1 ICP27, EBV EB2, and Kaposi's sarcoma-associated herpesvirus ORF57, are implicated in RNA processing and transport (27, 39). The experiments described here provide distinct lines of evidence supporting the identification of pUL69 of the betaherpesvirus HCMV as an additional important herpesviral regulatory protein that promotes the cytoplasmic accumulation of unspliced RNA. Importantly, although it appears that all pUL69 counterparts in herpesviruses share this conserved function, we identified significant differences in how pUL69 mediates RNA export.
Utilizing a reporter system widely used and designed to identify factors with mRNA export potential, we demonstrate that expression of pUL69 in human cells bypasses nuclear retention and promotes the nuclear export of unspliced reporter RNAs that are otherwise exported inefficiently. Interestingly, our characterization of this RNA export revealed that shuttling of pUL69 is linked to the export of unspliced RNAs, suggesting coexport of pUL69 with RNAs to the cytoplasm. We have previously reported that pUL69 shuttles between the nucleus and the cytoplasm in a CRM1-independent manner (34). Thus, it was tempting to speculate that, unlike with HIV-1 Rev, pUL69 nuclear export of RNA was also not dependent on the CRM1 pathway. Consistent with this idea, our results showed that pUL69-mediated nuclear RNA export is not sensitive to LMB, a drug that specifically binds to and inactivates CRM1 (29).
If not targeting the CRM1 pathway, how does pUL69 promote the nuclear export of unspliced RNA? Recently, it turned out that some members of the ICP27 family of herpesviral proteins directly bind to specific intronless mRNAs and direct these RNAs to the TAP-dependent cellular mRNA export pathway (reviewed in reference 51). Consistently, we could demonstrate that pUL69 is also able to interact directly with RNA, although, similar to its homologs, no apparent specificity for defined RNA sequences could be detected up to now (Z. Toth et al., unpublished data). However, whereas for ICP27, EB2, and ORF57 a direct protein interaction with the adaptor protein REF has been described elsewhere (19, 27, 39), our findings suggest that pUL69 gains access to the mRNA export pathway via its physical interaction with the highly related DEXD/H-box RNA helicases UAP56 and/or URH49 initially described by Fleckner et al. (13) and Pryor et al. (44). In this context it is of note that, although initial data suggest that the highly related proteins URH49 and UAP56 are likely to carry out similar functions in mammalian mRNA biogenesis, details of the biological function of URH49 are unknown so far (44). Interestingly, recent publications reported a differential up-regulation of URH49 expression during HIV-1 infection and a requirement of DEAD box RNA helicase DDX3 for HIV-1 Rev-RRE export function (28, 64). This suggests that RNA helicases of the DEAD box family may be of general importance for viral infections.
The conclusion that UAP56 is involved in pUL69-mediated nuclear RNA export is based on several results. First, we have demonstrated that both UAP56 and URH49 associate with pUL69 in yeast two-hybrid assays, as well as in in vitro binding and in coimmunoprecipitation experiments. These interactions were not bridged by RNA since we could observe coimmunoprecipitation of the respective proteins after RNase treatment of cell extracts. Second, the domain required for UAP56 or URH49 binding has been narrowed down to pUL69 amino acids 18 to 30 and we verified that pUL69 mutants, lacking this domain or carrying amino acid exchanges within this domain, failed to bind the DEXD/H-box proteins in vitro and in vivo. It should be noted that the UAP56 binding sequence of pUL69 is not conserved within other members of the ICP27 family. Third, functional characterization of these protein-protein interactions demonstrated that pUL69 mutants incapable of binding to UAP56/URH49 lost their stimulatory effect on nuclear RNA export. Interestingly, nucleocytoplasmic shuttling of UAP56-binding-deficient mutants was not impaired, which is consistent with the fact that the previously defined CRM1-independent nuclear export signal of pUL69 is clearly distinct from the 12-amino-acid domain required for UAP56 binding. Thus, nucleocytoplasmic shuttling per se appears to be insufficient for pUL69 to export unspliced RNA; however, an interaction with UAP56 or URH49 is additionally required. This scenario is not without precedent since the REF binding site within ICP27 also differs from sequences required for nuclear export (5). Thus, the nuclear export pathway accessed by the pUL69 NES still remains to be defined, but our experiments unambiguously demonstrate a requirement of UAP56 binding for pUL69 RNA export activity.
In coimmunoprecipitation experiments we detected the adaptor protein REF in RNase-insensitive protein complexes that contained UAP56 or URH49 together with pUL69. This observation is consistent with the assumption that pUL69 acts to stimulate nuclear export of intronless viral RNAs by accessing the same pathway proposed for nuclear export of spliced cellular mRNAs via its association with UAP56 and/or REF. Interestingly, however, a closer look at the composition of pUL69-containing protein complexes using REF-binding-deficient mutants of UAP56 or URH49 did not reveal that the interaction of pUL69 with UAP56 or URH49 leads to an increased recruitment of REF to UAP56/URH49-containing complexes; furthermore, the result of this experiment argues against a direct protein-protein interaction between REF and pUL69 (Fig. 5). Thus, it is tempting to speculate that, in contrast to ICP27, which is thought to recruit REF, pUL69 targets the cellular mRNA export machinery at a locus that is upstream of the point at which ICP27 gains access to it.
UAP56 was defined as a factor which couples transcription and splicing to cellular mRNA export, thus acting at multiple steps during mRNA biogenesis (10). The importance of this protein is also emphasized by the recent demonstration that UAP56 homologs in yeast, Drosophila melanogaster, and Caenorhabditis elegans are essential for mRNA export and viability (15, 38, 56). It has been shown that UAP56 associates with the THO proteins of transcription elongation factors, thus forming the TREX (transcription/export) complex. TREX is thought to have a dual role both in the control of transcription elongation and in coupling transcription elongation with mRNA export (48, 57). Interestingly, we have reported previously an interaction between pUL69 and a transcription elongation factor termed hSPT6 (60). One might speculate that pUL69, via its interaction with hSPT6, is recruited to elongating RNA polymerase II. This could then result in an enhanced cotranscriptional loading of UAP56 onto nascent mRNPs, thus ultimately stimulating the nuclear export of intronless RNAs. This scenario would be consistent with the finding that different domains of pUL69 are required for an interaction with either hSPT6 or UAP56 (60). Such a mechanism of enhanced cotranscriptional loading of an essential mRNA export factor could account for an involvement of pUL69 in pleiotropic up-regulation of cellular and viral gene expression as observed for HCMV-infected cells at late stages of the replication cycle.
Studies using an HCMV mutant virus with a deletion of the UL69 coding region showed that the lack of pUL69 led to a substantially diminished level of several viral late transcripts (16). At present, it cannot be distinguished whether this is a direct or an indirect consequence of the lack of pUL69. As an indirect effect, viral transcripts that are dependent on pUL69 for efficient nuclear export might be misleadingly retained in the nucleus with the consequence that these unexported mRNAs are rapidly degraded by the nuclear exosome complex (24). This idea fits with observations that have recently been published for an HSV mutant lacking ICP27 or an EB2-deleted EBV (2, 43). Alternatively, the nuclear export of specific viral early RNAs might also be affected, thus leading to a general delay of the replication cycle which would then in turn result in reduced transcription of viral late genes. However, we cannot exclude that pUL69 may also exert a direct effect on transcription elongation of specific viral late RNAs which could be mediated via its interaction with hSPT6. A detailed comparison of nuclear and cytoplasmic transcript levels as well as a quantification of the transcriptional elongation rate of viral late genes in the absence or presence of pUL69 will be required in order to differentiate between these possibilities. In addition to its modulatory influence on gene expression, it was reported that pUL69 is also able to induce a G1 cell cycle arrest in both transfected and infected cells (16, 36). Although several publications report that either depletion or overexpression of UAP56 leads to a deregulation of mRNA export, subsequently eliciting a growth arrest, our data do not allow us to conclude that the UAP56-UL69 interaction contributes to the cell cycle arrest induced by pUL69 (15, 37, 38, 56). Further experiments will be required to clarify this.
In summary, our current work identifies a novel cellular target which provides a herpesvirally encoded regulatory protein with access to a conserved cellular transport system in order to promote export of unspliced RNA. In this regard, future studies on the activities of pUL69 may help to provide further insights into the mechanisms by which viruses are able to optimize the coupling of transcription with nuclear RNA export in order to enhance their gene expression.
Acknowledgments
We thank Minoru Yoshida for his generous gift of LMB and Joachim Hauber, Michael Dobbelstein, Michael Green, Kyosuke Nagata, and Elisa Izaurralde for providing plasmids or antibodies.
This work was supported by the DFG (SFB473), the IZKF Erlangen, and the Wilhelm Sander Stiftung.
REFERENCES
- 1.Arlt, H., D. Lang, S. Gebert, and T. Stamminger. 1994. Identification of binding sites for the 86-kilodalton IE2 protein of human cytomegalovirus within an IE2-responsive viral early promoter. J. Virol. 68:4117-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batisse, J., E. Manet, J. Middeldorp, A. Sergeant, and H. Gruffat. 2005. Epstein-Barr virus mRNA export factor EB2 is essential for intranuclear capsid assembly and production of gp350. J. Virol. 79:14102-14111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogerd, H. P., A. Echarri, T. M. Ross, and B. R. Cullen. 1998. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J. Virol. 72:8627-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun, I. C., A. Herold, M. Rode, E. Conti, and E. Izaurralde. 2001. Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J. Biol. Chem. 276:20536-20543. [DOI] [PubMed] [Google Scholar]
- 5.Chen, I. H., L. Li, L. Silva, and R. M. Sandri-Goldin. 2005. ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export. J. Virol. 79:3949-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, I. H., K. S. Sciabica, and R. M. Sandri-Goldin. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 76:12877-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chevallier-Greco, A., E. Manet, P. Chavrier, C. Mosnier, J. Daillie, and A. Sergeant. 1986. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 5:3243-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 9.Cullen, B. R. 2003. Nuclear mRNA export: insights from virology. Trends Biochem. Sci. 28:419-424. [DOI] [PubMed] [Google Scholar]
- 10.Erkmann, J. A., and U. Kutay. 2004. Nuclear export of mRNA: from the site of transcription to the cytoplasm. Exp. Cell Res. 296:12-20. [DOI] [PubMed] [Google Scholar]
- 11.Farjot, G., M. Buisson, M. D. Dodon, L. Gazzolo, A. Sergeant, and I. Mikaelian. 2000. Epstein-Barr virus EB2 protein exports unspliced RNA via a Crm-1-independent pathway. J. Virol. 74:6068-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer, U., J. Huber, W. C. Boelens, I. W. Mattaj, and R. Luhrmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475-483. [DOI] [PubMed] [Google Scholar]
- 13.Fleckner, J., M. Zhang, J. Valcarcel, and M. R. Green. 1997. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 11:1864-1872. [DOI] [PubMed] [Google Scholar]
- 14.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 15.Gatfield, D., H. Le Hir, C. Schmitt, I. C. Braun, T. Kocher, M. Wilm, and E. Izaurralde. 2001. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 11:1716-1721. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi, M. L., C. Blankenship, and T. Shenk. 2000. Human cytomegalovirus UL69 protein is required for efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc. Natl. Acad. Sci. USA 97:2692-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heger, P., O. Rosorius, C. Koch, G. Casari, R. Grassmann, and J. Hauber. 1998. Multimer formation is not essential for nuclear export of human T-cell leukemia virus type 1 Rex trans-activator protein. J. Virol. 72:8659-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiriart, E., L. Bardouillet, E. Manet, H. Gruffat, F. Penin, R. Montserret, G. Farjot, and A. Sergeant. 2003. A region of the Epstein-Barr virus (EBV) mRNA export factor EB2 containing an arginine-rich motif mediates direct binding to RNA. J. Biol. Chem. 278:37790-37798. [DOI] [PubMed] [Google Scholar]
- 19.Hiriart, E., G. Farjot, H. Gruffat, M. V. Nguyen, A. Sergeant, and E. Manet. 2003. A novel nuclear export signal and a REF interaction domain both promote mRNA export by the Epstein-Barr virus EB2 protein. J. Biol. Chem. 278:335-342. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann, H., S. Floss, and T. Stamminger. 2000. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol. 74:2510-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hope, T. J., B. L. Bond, D. McDonald, N. P. Klein, and T. G. Parslow. 1991. Effector domains of human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex are functionally interchangeable and share an essential peptide motif. J. Virol. 65:6001-6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hope, T. J., X. J. Huang, D. McDonald, and T. G. Parslow. 1990. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc. Natl. Acad. Sci. USA 87:7787-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, X. J., T. J. Hope, B. L. Bond, D. McDonald, K. Grahl, and T. G. Parslow. 1991. Minimal Rev-response element for type 1 human immunodeficiency virus. J. Virol. 65:2131-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen, T. H., K. Dower, D. Libri, and M. Rosbash. 2003. Early formation of mRNP: license for export or quality control? Mol. Cell 11:1129-1138. [DOI] [PubMed] [Google Scholar]
- 25.Kang, Y., and B. R. Cullen. 1999. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 13:1126-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katahira, J., K. Strasser, A. Podtelejnikov, M. Mann, J. U. Jung, and E. Hurt. 1999. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 18:2593-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koffa, M. D., J. B. Clements, E. Izaurralde, S. Wadd, S. A. Wilson, I. W. Mattaj, and S. Kuersten. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 20:5769-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishnan, V., and S. L. Zeichner. 2004. Alterations in the expression of DEAD-box and other RNA binding proteins during HIV-1 replication. Retrovirology 1:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kudo, N., N. Matsumori, H. Taoka, D. Fujiwara, E. P. Schreiner, B. Wolff, M. Yoshida, and S. Horinouchi. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96:9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang, D., S. Gebert, H. Arlt, and T. Stamminger. 1995. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J. Virol. 69:6030-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leaw, C. L., E. C. Ren, and M. L. Choong. 2004. Hcc-1 is a novel component of the nuclear matrix with growth inhibitory function. Cell Mol. Life Sci. 61:2264-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lennon, G., C. Auffray, M. Polymeropoulos, and M. B. Soares. 1996. The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics 33:151-152. [DOI] [PubMed] [Google Scholar]
- 33.Liker, E., E. Fernandez, E. Izaurralde, and E. Conti. 2000. The structure of the mRNA export factor TAP reveals a cis arrangement of a non-canonical RNP domain and an LRR domain. EMBO J. 19:5587-5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lischka, P., O. Rosorius, E. Trommer, and T. Stamminger. 2001. A novel transferable nuclear export signal mediates CRM1-independent nucleocytoplasmic shuttling of the human cytomegalovirus transactivator protein pUL69. EMBO J. 20:7271-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lischka, P., G. Sorg, M. Kann, M. Winkler, and T. Stamminger. 2003. A nonconventional nuclear localization signal within the UL84 protein of human cytomegalovirus mediates nuclear import via the importin alpha/beta pathway. J. Virol. 77:3734-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu, M., and T. Shenk. 1999. Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J. Virol. 73:676-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo, M. L., Z. Zhou, K. Magni, C. Christoforides, J. Rappsilber, M. Mann, and R. Reed. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413:644-647. [DOI] [PubMed] [Google Scholar]
- 38.MacMorris, M., C. Brocker, and T. Blumenthal. 2003. UAP56 levels affect viability and mRNA export in Caenorhabditis elegans. RNA 9:847-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malik, P., D. J. Blackbourn, and J. B. Clements. 2004. The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 279:33001-33011. [DOI] [PubMed] [Google Scholar]
- 40.Malim, M. H., L. S. Tiley, D. F. McCarn, J. R. Rusche, J. Hauber, and B. R. Cullen. 1990. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell 60:675-683. [DOI] [PubMed] [Google Scholar]
- 41.McDonald, D., T. J. Hope, and T. G. Parslow. 1992. Posttranscriptional regulation by the human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex proteins through a heterologous RNA binding site. J. Virol. 66:7232-7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Momose, F., C. F. Basler, R. E. O'Neill, A. Iwamatsu, P. Palese, and K. Nagata. 2001. Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J. Virol. 75:1899-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pearson, A., D. M. Knipe, and D. M. Coen. 2004. ICP27 selectively regulates the cytoplasmic localization of a subset of viral transcripts in herpes simplex virus type 1-infected cells. J. Virol. 78:23-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pryor, A., L. Tung, Z. Yang, F. Kapadia, T. H. Chang, and L. F. Johnson. 2004. Growth-regulated expression and G0-specific turnover of the mRNA that encodes URH49, a mammalian DExH/D box protein that is highly related to the mRNA export protein UAP56. Nucleic Acids Res. 32:1857-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reed, R., and E. Hurt. 2002. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108:523-531. [DOI] [PubMed] [Google Scholar]
- 46.Rocak, S., and P. Linder. 2004. DEAD-box proteins: the driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 5:232-241. [DOI] [PubMed] [Google Scholar]
- 47.Rodrigues, J. P., M. Rode, D. Gatfield, B. Blencowe, M. Carmo-Fonseca, and E. Izaurralde. 2001. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. USA 98:1030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rondon, A. G., S. Jimeno, M. Garcia-Rubio, and A. Aguilera. 2003. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J. Biol. Chem. 278:39037-39043. [DOI] [PubMed] [Google Scholar]
- 49.Roth, J., and M. Dobbelstein. 1997. Export of hepatitis B virus RNA on a Rev-like pathway: inhibition by the regenerating liver inhibitory factor IκBα. J. Virol. 71:8933-8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sandri-Goldin, R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandri-Goldin, R. M. 2004. Viral regulation of mRNA export. J. Virol. 78:4389-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandri-Goldin, R. M., and G. E. Mendoza. 1992. A herpesvirus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes Dev. 6:848-863. [DOI] [PubMed] [Google Scholar]
- 53.Segref, A., K. Sharma, V. Doye, A. Hellwig, J. Huber, R. Luhrmann, and E. Hurt. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16:3256-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stamminger, T., H. Fickenscher, and B. Fleckenstein. 1990. Cell type-specific induction of the major immediate early enhancer of human cytomegalovirus by cyclic AMP. J. Gen. Virol. 71:105-113. [DOI] [PubMed] [Google Scholar]
- 55.Strasser, K., and E. Hurt. 2000. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 19:410-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strasser, K., and E. Hurt. 2001. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413:648-652. [DOI] [PubMed] [Google Scholar]
- 57.Strasser, K., S. Masuda, P. Mason, J. Pfannstiel, M. Oppizzi, S. Rodriguez-Navarro, A. G. Rondon, A. Aguilera, K. Struhl, R. Reed, and E. Hurt. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417:304-308. [DOI] [PubMed] [Google Scholar]
- 58.Thomas, S. L., M. Oft, H. Jaksche, G. Casari, P. Heger, M. Dobrovnik, D. Bevec, and J. Hauber. 1998. Functional analysis of the human immunodeficiency virus type 1 Rev protein oligomerization interface. J. Virol. 72:2935-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vinciguerra, P., and F. Stutz. 2004. mRNA export: an assembly line from genes to nuclear pores. Curr. Opin. Cell Biol. 16:285-292. [DOI] [PubMed] [Google Scholar]
- 60.Winkler, M., S. T. aus Dem, and T. Stamminger. 2000. Functional interaction between pleiotropic transactivator pUL69 of human cytomegalovirus and the human homolog of yeast chromatin regulatory protein SPT6. J. Virol. 74:8053-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winkler, M., S. A. Rice, and T. Stamminger. 1994. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J. Virol. 68:3943-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Winkler, M., and T. Stamminger. 1996. A specific subform of the human cytomegalovirus transactivator protein pUL69 is contained within the tegument of virus particles. J. Virol. 70:8984-8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolff, B., J. J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4:139-147. [DOI] [PubMed] [Google Scholar]
- 64.Yedavalli, V. S., C. Neuveut, Y. H. Chi, L. Kleiman, and K. T. Jeang. 2004. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119:381-392. [DOI] [PubMed] [Google Scholar]