Abstract
The normal expression of human β globin is critically dependent upon the constitutively high stability of its encoding mRNA. Unlike with α-globin mRNA, the specific cis-acting determinants and trans-acting factors that participate in stabilizing β-globin mRNA are poorly described. The current work uses a linker-scanning strategy to identify a previously unknown determinant of mRNA stability within the β-globin 3′ untranslated region (3′UTR). The new determinant is positioned on an mRNA half-stem opposite a pyrimidine-rich sequence targeted by αCP/hnRNP-E, a factor that plays a critical role in stabilizing human α-globin mRNA. Mutations within the new determinant destabilize β-globin mRNA in intact cells while also ablating its 3′UTR-specific interaction with the polyfunctional RNA-binding factor nucleolin. We speculate that 3′UTR-bound nucleolin enhances mRNA stability by optimizing αCP access to its functional binding site. This model is favored by in vitro evidence that αCP binding is enhanced both by cis-acting stem-destabilizing mutations and by the trans-acting effects of supplemental nucleolin. These studies suggest a mechanism for β-globin mRNA stability that is related to, but distinct from, the mechanism that stabilizes human α-globin mRNA.
Erythroid cells accumulate hemoglobin through a process that is critically dependent upon the high stabilities of mRNAs that encode their constituent α- and β-globin subunits (10, 64). In vivo analyses estimate a half-life for human α-globin mRNA of between 24 and 60 h (47, 62, 63, 74), while similar studies with cultured NIH 3T3 and murine erythroleukemia (MEL) cells (2, 36, 42), primary mouse hematopoietic cells (4), and human erythroid progenitors (62, 63) suggest a half-life value for human β-globin mRNA that exceeds 16 to 20 h. Globin mRNAs survive, and continue to translate at high levels, for as long as a week following nuclear condensation and extrusion in transcriptionally silent erythroid progenitor cells. As might be anticipated, mutations that impair the normal stabilities of globin mRNAs can severely impact the levels of their encoded proteins. For example, an mRNA-destabilizing mutation reduces the expression of αConstant Spring to less than 2% of normal levels (45, 51, 80), resulting in a clinically important form of thalassemia characterized by a substantial imbalance in α- and β-globin chain accumulation (10, 64).
The cis-acting determinants and trans-acting factors that participate in regulating α-globin mRNA stability have recently been identified, and the relevant molecular mechanisms have been described in detail. Mutational analyses carried out with cultured cells (80, 81) and with animal models (52, 66) clearly demonstrate the importance of the 3′ untranslated region (3′UTR) to the constitutively high stability of α-globin mRNA (45). Other studies have mapped this characteristic to a phylogenically conserved, 16-nucleotide (nt) C/U-rich element in this region (39, 75, 76). The cis-acting pyrimidine-rich element (PRE) assembles an mRNP “α-complex” that comprises a member of the αCP/hnRNP-E family of mRNA-binding proteins (37, 39, 48, 75) and possibly one or more additional trans-acting factors (17, 38, 77). The α-complex may slow α-globin mRNA decay by enhancing the binding of poly(A)-binding protein to the poly(A) tail (77, 79). The α-complex may also prevent the access of an erythroid-cell-specific endoribonuclease to the α-PRE (61, 78, 79), mimicking mechanisms through which several nonglobin mRNAs evade endonucleolytic cleavage (5, 6, 8).
Unlike with α-globin mRNA, neither the cis elements nor the trans-acting factors that specify the constitutively high stability of human β-globin mRNA have been fully described. Although several hundred mutations are known to affect β-globin gene expression, few offer any insight into the position of a specific β-globin mRNA stability-enhancing region or its likely mechanism (10). Common mutations that encode premature translation termination codons or adversely affect processing of β-globin pre-mRNA, though accelerating its degradation, utilize mRNA-indifferent decay pathways (10, 26, 49) and consequently do not illuminate the putative β-globin mRNA-restricted mechanism(s) that defines its high baseline stability.
The search for a discrete β-globin mRNA stability element has focused largely on its 3′UTR, since sequences dictating the stabilities of mRNAs encoding α globin (80, 81), transferrin receptor (40), histones (57), and a variety of cytokines and lymphokines (70) are commonly positioned in this ribosome-free region. This expectation has been fulfilled by studies of chimeric fos-β-globin mRNA in cultured cells (36) and full-length β-globin mRNA in MEL cells and in whole-animal models (65, 84). The latter studies were particularly informative, demonstrating that β-globin mRNA can be destabilized by engineered mutations that permit ribosomes to read past its native translational terminal codon and through the entire 3′UTR, disrupting hypothetical mRNP structures positioned in that region. In contrast, β-globin expression is minimally impacted by naturally occurring and synthetic frameshift mutations that restrict ribosomal readthrough to the proximal one-third of the 3′UTR (11, 20, 25, 65), suggesting that critical determinants of mRNA stability are likely to be positioned further downstream. Scattered, naturally occurring single-nucleotide substitutions and small deletions in the terminal portion of the 3′UTR do not materially affect β-globin mRNA stability (3, 12, 34) and are consequently of little help in mapping important cis-acting stability elements. Recent interest has focused on a 14-nt pyrimidine-rich element in the β-globin 3′UTR (the β-PRE) because of its obvious similarity to the α-PRE stability determinant. Deletion or purine substitution of the β-PRE reduces the stability of human β-globin mRNA in transgenic mice by approximately one-half (84), indicating the importance of this structure to β-globin gene expression.
Although the mechanism through which the β-PRE confers stability to the β-globin mRNA is not known, several observations suggest that it, like the α-PRE, may be functionally targeted by αCP. First, the human α- and β-globin genes evolved from a single ancestral globin gene (28, 29), raising the possibility of a primitive, evolutionally conserved mechanism for stabilizing their mRNAs. Second, αCP is known to play an important role in stabilizing several nonglobin mRNAs that contain pyrimidine-rich 3′UTR motifs [e.g., 15-lipoxygenase (55), tyrosine hydroxylase (18), and α1(I) collagen (73)] and may participate in stabilizing other globin mRNAs (66). Third, electrophoretic mobility studies demonstrate that the β-globin 3′UTR assembles an mRNP structure in vitro that may contain αCP (84). Formal attempts to directly link αCP to β-globin mRNA stability, however, have been unsuccessful, suggesting that the constitutive stability of β-globin mRNA in vivo requires mRNA structural conformations or additional trans-acting factors that are poorly recapitulated in vitro. Consequently, the posttranscriptional mechanisms that maintain β-globin mRNA levels in vivo remain poorly understood, despite their obvious relevance to common disorders of globin gene expression.
The current article provides new insights into the structural basis for β-globin mRNA stability, suggesting an underlying mechanism that differs in several fundamental respects from the mechanism dictating the stability of human α-globin mRNA. A previously unrecognized cis determinant of β-globin mRNA stability is identified by assessing the survival of derivative mRNAs containing defined site-specific mutations in vivo. Independent experiments in vitro, in cultured cells, and in primary human erythroid progenitors identify nucleolin, a ubiquitous, structurally heterogeneous, polyfunctional RNA-binding protein, as a cytoplasmic factor that binds to the β-globin 3′UTR in a sequence-specific manner. A link between nucleolin binding and mRNA stability is provided by subsequent in vitro and in vivo analyses demonstrating that functional mutations within the stability determinant also interfere with nucleolin binding. These data, and previous experimental evidence favoring an mRNA-stabilizing role for αCP, are accommodated by a model in which nucleolin facilitates the access of αCP to its functional β-globin 3′UTR target site. We demonstrate a key aspect of this model by showing that disruption of a high-order structure within the β-globin 3′UTR facilitates αCP binding in vitro. These studies suggest a mechanism for β-globin mRNA stability that is related to, but distinct from, the mechanism that stabilizes human α-globin mRNA.
MATERIALS AND METHODS
Cell culture.
HeLa cells expressing the tetracycline-regulated transactivator (tTA) fusion protein (BD Biosciences) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum in a humidified 5% CO2 environment. Suspension MEL cells were cultured under similar conditions, while human K562 cells were grown in Iscove's modified Dulbecco's medium containing 4 mM glutamine and 1.5 g/liter sodium bicarbonate and supplemented with 10% fetal bovine serum. Cells (∼5 × 105) were transfected with 5 μg supercoiled DNA using Superfect reagent as recommended by the manufacturer (QIAGEN). Doxycycline was added to a final concentration of 1 μg/ml when required.
Gene cloning.
pTRE-βWT was constructed from a 3.3-kb fragment of human genomic DNA containing the intact β-globin gene and contiguous 3′ flanking region, inserted into the SacII-ClaI polylinker site of pTRE2 (BD Biosciences). Linker-scanning mutations were introduced into the human β-globin gene by a splice overlap extension-PCR method (65, 84) using paired, complementary 30-nt primers containing the desired HindIII mutation (5′AAGCTT3′). The resulting mutated 904-bp cDNAs were then substituted for the cognate EcoRI-EcoNI fragment of pTRE-βWT. Chemically competent DH5α Escherichia coli cells were transformed (Invitrogen), mini-prep DNA was prepared from individual colonies (QIAGEN), and the structures of the variant β-globin genes were subsequently validated by HindIII digestion and by automated dideoxy sequencing. pTRE-βARE104 and pTRE-βARE130 were constructed by introducing a 59-bp A/U-rich mRNA instability element (70) into the HindIII sites of pTRE-βH104 and pTRE-βH130, respectively.
RNase protection analysis.
Cellular RNAs prepared from cultured cells using TRIzol reagent (Gibco-BRL) were analyzed as described previously (66, 84). 32P-labeled β-globin and β-actin probes were prepared by in vitro transcription of DNA templates using SP6 RNA polymerase (Ambion). The 287-nt β-globin probe protects a 199-nt sequence of human β-globin mRNA exon II, while the 313-nt β-actin probe protects a 160-nt exonic fragment of human β-actin mRNA (84). Band intensities were quantitated from PhosphorImager files using ImageQuant software (Amersham Biosciences).
RT-PCR+1 analysis (65).
Purified RNAs (∼500 ng) were reverse transcribed and thermally amplified using Superscript one-step reagents under conditions recommended by the manufacturer (Invitrogen) and then amplified for 40 cycles using exon II (5′ACCTGGACAACCTCAAGG3′) and exon III (5′TTTTTTTTTTGCAATGAAAATAAATG3′) primers that generate a 355-bp cDNA product encompassing the full β-globin 3′UTR. Reaction mixtures were subsequently augmented with 100 μmol of a nested 32P-labeled exon II primer (5′CCACACTGAGTGAGCTGC3′) and 0.5 μl Platinum Taq (Invitrogen) and product DNA amplified for one additional cycle. This method generates 328-nt 32P-labeled homodimeric DNAs that fully digest with HindIII to generate 32P-labeled products between 189 and 285 bp in length.
Proteomics.
Analyses were carried out by the University of Pennsylvania Proteomics Facility. Tryptic digests were resolved on a Voyager DE Pro (Applied Biosystems), and protein identities were deduced from MS-Fit (University of California) analysis of peptide fragments using the NCBInr database. Time-of-flight (TOF)-TOF analysis was carried out using a 4700 proteomics analyzer (Applied Biosystems) equipped with Global Proteomics Server analytical software.
Cytosolic extract.
Extracts were prepared as previously described (19, 21). Briefly, phosphate-buffered saline (PBS)-washed cells were incubated for 20 min at 4°C in RNA immunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1 mM Na3VO4, 1 mM NaF, and 1× protease inhibitor cocktail [BD Biosciences]). The lysate was centrifuged at 13,000 × g for 15 min, and the supernatant was collected and stored at −80°C. For cross-linking studies, in vitro-transcribed, 32P-labeled RNAs were incubated with cytoplasmic extract and exposed to UV light (3,000 mJ/cm2) for 5 min.
Fluorescence-activated cell sorter (FACS) analysis.
A protocol for all animal work was approved by the Institutional Animal Care and Use Committee at the University of Pennsylvania School of Medicine. EDTA-anticoagulated whole blood was stained with thiazole orange as directed by the manufacturer (Sigma) (31). Erythroid cells were identified by their characteristic forward- and side-scatter properties using a FACSVantage cell sorter equipped with Digital Vantage options (Becton-Dickinson). Thiazole orange-staining cells (reticulocytes) were collected, excluding a small population of hyper-staining nucleated erythroid progenitor cells.
Affinity enrichment studies.
Custom 5′-terminal biotinylated single-stranded DNAs (ssDNAs) were purchased from Integrated DNA Technologies (Coralville, IA). Molar equivalents of each ssDNA (3 pmol) were incubated for 1 h at 4°C in PBS (pH 7.2) along with 100 μl of preequilibrated ImmunoPure immobilized avidin agarose beads (Pierce Biotechnology). The pelleted beads were washed four times with PBS, incubated at 4°C for 1 h with 1 ml cytoplasmic extract, and then washed five times with PBS. Bound proteins were eluted with loading buffer and resolved on precast 4 to 12% gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels as recommended by the manufacturer (Invitrogen). A parental ssDNA corresponding to the β-globin 3′UTR stem-loop structure (5′ATTTCTATTAAAGGTTCCTTTGTTCCCTAAGTCCAACTACTAAACTGGGGGATATTATGAAGGGCCTTGAGCATC3′) was modified by the deletion of an internal 18-nt sequence (5′GGGGGATATTATGAAGGG3′) and by the substitution of an unrelated 18-nt sequence (5′ATGCCGTAATGCCGTAAT3′) or a sequence encompassing the β-PRE (5′TTCCTTTGTTCCCTAAGT3′) at the same site.
Western blotting.
Antibodies purchased from Santa Cruz Biotechnology included mouse monoclonal anti-human nucleolin (MS-3), rabbit polyclonal anti-human nucleolin (H-250), goat polyclonal anti-human HDAC-2 (C-19), rabbit polyclonal anti-human tumor necrosis factor alpha, and goat polyclonal anti-human hnRNP-E1 (T-18). Rabbit polyclonal anti-human actin antibodies were purchased from Sigma (A-2066). Protein samples in loading buffer were denatured at 100°C for 5 min, resolved on a precast 4 to 12% gradient SDS-PAGE gel, and transferred to a nitrocellulose membrane using an XCell II blot module according to the manufacturer's instructions (Invitrogen). Blots were blocked for 1 h at room temperature in PBS containing 0.1% Tween 20, supplemented with 3% dried milk, and then incubated for an additional hour following antibody addition. Membranes washed with the Tween 20-PBS mixture were subsequently incubated for 1 h with a horseradish peroxidase-conjugated secondary antibody (Amersham Biosciences) and analyzed using a chemiluminescence method (ECL kit; Amersham).
RNA immunoprecipitation.
HeLa cell extracts were prepared as previously described (15, 16). PBS-washed erythrocytes were isolated from EDTA-anticoagulated whole blood by fractionation over a Histopaque 1.077/1.119 bilayer cushion (Sigma). Extracts prepared in RIPA buffer (1 ml) were precleared with 60 μl protein A-agarose beads (Invitrogen) and then incubated at 4°C for 3 h with nucleolin H-250 antibodies. Fresh protein A-agarose beads (60 μl) were then added, and the incubation continued for another 2 h. Immunoprecipitates were washed three times in RIPA buffer, and bound RNAs were collected by TRIzol extraction and ethanol precipitation for subsequent analysis. Control 18S pre-RNAs were RT-PCR amplified using oligomers 5′GTTCGTGCGACGTGTGGCGTGG3′ and 5′CAGACCCGCGACGCTTCTTCGT3′, producing a 501-bp cDNA fragment (1).
Preparation of recombinant αCP and purification of nucleolin.
A glutathione S-transferase-αCP1 fusion protein was purified from DH5α cells transfected with pEGX-6P-αCP1 (kind gift of M. Kiledjian, Rutgers University); the glutathione S-transferase domain was subsequently cleaved with PreScission proteinase (Pharmacia Biotech). Human nucleolin was affinity enriched from HeLa and/or K562 cell extract using an agarose-immobilized 2′-O-methyl RNA sequence (5′UAUUAAAGGUUCCUUUGUUCCCUAAGUCCAAC3′). A related method was used to prepare nucleolin-depleted extract.
RESULTS
Validation of a method for analyzing the stability of β-globin mRNA in intact cells.
To facilitate our studies of β-globin mRNA stability, we developed a system in which a single defined gene can be transcriptionally silenced in intact, translationally competent cells. This approach permits mRNA decay to be assessed in vivo using a transcriptional chase approach that does not compromise cell viability (41, 82). The method requires cells that constitutively express a tTA fusion protein that activates genes linked to a recombinant hybrid tetracycline response element (TRE). tTA activity is rapidly and efficiently inhibited in the presence of tetracycline or doxycycline (Dox), which does not affect the expression of other, constitutively expressed eukaryotic genes. Consequently, the stabilities of mRNAs encoded by TRE-linked genes can be estimated by assessing their rate of disappearance from Dox-treated cells.
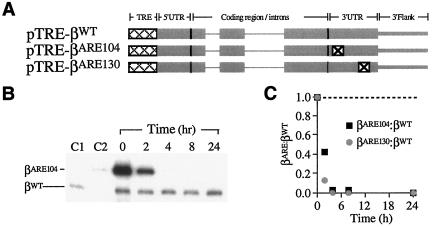
The proposed use of tTA-expressing HeLa cells was tested by assessing the fate of mRNAs carrying a known mRNA-destabilizing determinant, the 3′UTR A/U-rich element (ARE) derived from human granulocyte-macrophage colony-stimulating factor mRNA (70) (Fig. 1A). TRE-linked β-globin genes were constructed to contain either the native 3′UTR (pTRE-βWT) (84) or 3′UTRs engineered to contain single-copy ARE inserts (pTRE-βARE104 and pTRE-βARE130). pTRE-βWT was cotransfected into HeLatTA cells with either pTRE-βARE104 or pTRE-βARE130, and the levels of their encoded mRNAs were established at defined intervals following Dox exposure. Unlike with βWT mRNA, the level of each βARE mRNA fell rapidly (Fig. 1B and C), confirming the utility of the tTA-TRE system for differentiating unstable and stable mRNAs in intact, cultured cells.
FIG. 1.
Unstable and stable variant β-globin mRNAs can be distinguished in intact cells. (A) Structures of conditionally expressed reporter genes encoding variant β-globin mRNAs. pTRE-βWT contains the full-length human β-globin gene, including native intronic, exonic, and 3′-flanking sequences (thin, thick, and intermediate gray lines, respectively), downstream of a Tet-conditional TRE promoter (dotted cross-hatching). pTRE-βARE104 and pTRE-βARE130 are identical to pTRE-βWT except for a 59-bp ARE instability element (⊠) at either of two 3′UTR positions. (B) Variant βARE104 mRNA is unstable in cultured cells. The levels of βWT and βARE104 mRNAs in transiently transfected HeLatTA cells were assessed by RT-PCR+1 at defined intervals following Dox exposure. The intensities of the βWT bands were balanced by adjusting sample loading. C1 and C2 contain RNA from cells transfected singly with pTRE-βWT and pTRE-βARE104, respectively. (C) ARE-mediated destabilization of β-globin mRNA in cultured cells. The autoradiograph in panel B was analyzed by PhosphorImager densitometry and the βARE104/βWT ratio plotted (black squares). Results from a parallel study of βARE130 are also plotted (gray circles). The dashed line indicates the βARE/βWT ratio that would be observed if the two mRNAs were equally stable.
Human β-globin mRNA is destabilized by either of two adjacent site-specific 3′UTR mutations.
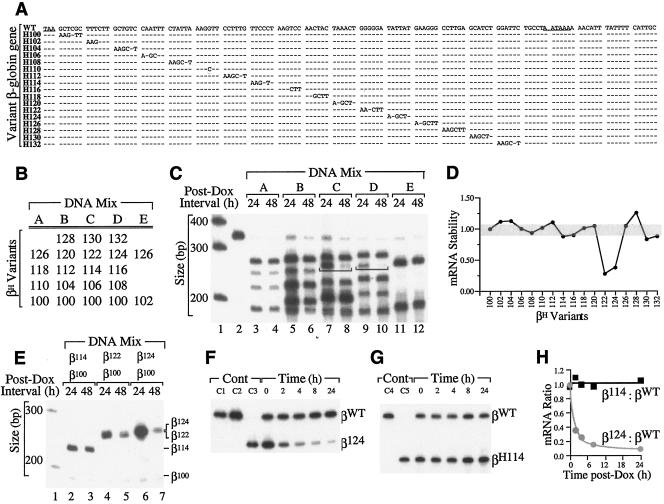
To map critical cis determinants of β-globin mRNA stability, 17 full-length β-globin genes were constructed, each containing a hexanucleotide substitution at a unique 3′UTR position (Fig. 2A). Collectively, the mutations saturate 102 nt of the 107-nt sequence of β-globin 3′UTR between the native TAA translational termination codon and the AATAAA polyadenylation signal. HeLatTA cells were cotransfected with DNA mixes comprising different combinations of TRE-linked, variant βH-globin genes, including one (βH100) that was arbitrarily selected as an internal control (Fig. 2B). The level of each variant βH mRNA, relative to that of βH100 mRNA, was subsequently determined by RT-PCR+1 following 24- and 48-hour exposures to Dox (65). Two of the variant βH mRNAs containing hexanucleotide substitutions at 3′UTR positions 122 and 124 displayed levels that fell four- to fivefold faster than those of other variant βH mRNAs (Fig. 2C and D). These results were confirmed in a duplicate analysis utilizing a different post-Dox interval (not shown) and in related experiments in which genes encoding unstable variant βH122 and βH124 mRNAs and stable variant βH114 mRNA were separately transfected into HeLatTA cells along with internal control pTRE-βH100 (Fig. 2E). Formal mRNA stability studies were subsequently carried out using Dox-exposed HeLatTA cells that had been cotransfected with TRE-linked genes encoding βWT and either βH114 or βH124 mRNA (Fig. 2F to H). By comparison to the level of βWT mRNA, that of βH124 mRNA fell rapidly (Fig. 2F and H), while that of control βH114 mRNA remained stable (Fig. 2G and H). The combined results of screening and formal mRNA stability analyses confirm the importance of the 12-nt H122/H124 sequence to the intrinsically high stability of β-globin mRNA.
FIG. 2.
Two adjacent hexanucleotide mutations destabilize β-globin mRNA in intact cultured cells. (A) Structures of variant β-globin genes. The 3′UTR of the wild-type β-globin gene (WT) is illustrated, with the TAA termination codon and AATAAA polyadenylation signal underlined. Each variant β-globin gene (designated H100, H102, and H104, etc.) contains a site-specific AAGCTT hexanucleotide substitution encoding a HindIII recognition site. Dashes indicate identity with the WT sequence. (B) Composition of DNA mixes used for mRNA stability studies in cultured cells. Mixes A to D each contain four or five variant TRE-linked βH-globin genes, including one (βH100) whose mRNA is used as a normalization control in subsequent analyses. Mix E contains a control variant βH126 gene for the same purpose. (C) Relative stabilities of variant β-globin mRNAs following transcriptional silencing of their encoding genes. HeLatTA cells transfected with DNA mixes A to E were exposed to Dox, and total RNA was recovered from aliquots following an additional 24 or 48 h of culture. RT-PCR+1-amplified products were restricted with HindIII to generate differently sized DNA fragments whose quantities correspond to the levels of individual variant βH mRNAs in the original sample. Brackets emphasize the rapid interval decline in βH122 mRNA (lanes 7 and 8) and βH124 mRNA (lanes 9 and 10), relative to levels of other variant βH mRNAs. Lanes 1 and 2 contain 32P-labeled size markers and the undigested PCR product from mix A, respectively. (D) Relative stabilities of variant βH mRNAs. The stabilities of individual variant βH mRNAs are plotted. Stability is defined as [(βH)48/(βH)24]/[(βH100)48/(βH100)24], with the stability of βH100 arbitrarily assigned unit value (subscript values represent the post-Dox intervals in hours). (E) Accelerated decay of variant βH mRNAs in intact cultured cells. The stabilities of mRNAs encoded by variant βH114, βH122, and βH124 genes (top) were established singly, relative to that of internal control βH100 mRNA, as described for panel C. The positions of individual HindIII-restricted RT-PCR+1 products are indicated to the right. Lane 1 contains a DNA size marker. (F and G) Formal decay analyses of βH124 and control βH114 mRNAs. Mixes containing pTRE-βWT and either pTRE-βH124 (F) or pTRE-βH114 (G) were transfected into HeLatTA cells, and relative mRNA levels were established by RT-PCR+1 at defined intervals following Dox exposure. Controls (Cont) include undigested βWT (C1), HindIII-digested βWT (C2), HindIII-digested βH124 (C3), undigested βH114 (C4), and HindIII-digested βH114 (C5). (H) Relative stabilities of βH124 and control βH114 mRNAs. Band intensities were established from the autoradiographs in panels F and G by PhosphorImager densitometry. Levels of βH124 and βH114 mRNAs, relative to levels of coexpressed βWT mRNA and normalized to the corresponding ratio at time zero, are plotted in gray and black, respectively.
Nucleolin binds to the β-globin 3′UTR in intact cultured cells and primary erythroid cells.
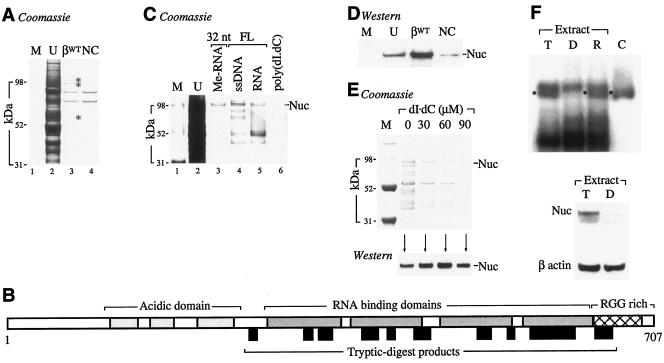
The stabilities of many mRNAs, including those encoding α globin (75), α1(I) collagen (73), tyrosine hydroxylase (18), histone (57), and the transferrin receptor (53), require the assembly of defined mRNP effector complexes on specific determinants within their 3′UTRs. To identify candidate trans-acting factors that might functionally interact with the β-globin 3′UTR, agarose-immobilized ssDNAs corresponding to the βWT 3′UTR and to negative-control poly(dI · dC) were separately incubated with cytoplasmic extract prepared from cultured human erythroid K562 cells. Three bands that displayed relative specificities for the βWT 3′UTR were subsequently excised and subjected to matrix-assisted laser desorption ionization (MALDI)-TOF analysis (Fig. 3A). The ∼100-kDa band was unambiguously identified as nucleolin from 14 tryptic peptide fragments representing 22% coverage (molecular weight search, 1.469 × 104) (Fig. 3B); the identities of the remaining two bands could not be established with certainty. Companion experiments indicated that nucleolin binds equally well to related full-length and truncated agarose-immobilized RNAs and 2′-O-methylated RNAs, respectively (Fig. 3C). These results were corroborated by parallel TOF-TOF analyses of affinity-enriched erythroid MEL cell extract that also unequivocally identified nucleolin (data not shown). This dual preliminary identification was subsequently confirmed by Western blot analysis of affinity-enriched proteins using a polyclonal nucleolin antibody (Fig. 3D). Nucleolin appears to bind to the β-globin 3′UTR in a sequence-specific manner, as increasing quantities of an unrelated soluble competitor ssDNA effectively compete background proteins from an agarose-immobilized ssDNA β-globin 3′UTR ligand but do not affect nucleolin binding (Fig. 3E). In addition, UV-cross-linked nucleolin-β-3′UTR mRNPs assemble in K562 cytoplasmic extract but not in extracts that are affinity depleted of nucleolin, confirming that nucleolin also binds to βWT RNA (Fig. 3F, lanes T and D, respectively). These results document the sequence-specific binding of nucleolin to the β-globin mRNA 3′UTR in vitro and suggest that this interaction may subserve a critical function in vivo.
FIG. 3.
Identification of a cytoplasmic factor that exhibits binding specificity for the βWT 3′UTR. (A) Affinity enrichment of candidate β-globin 3′UTR-binding factors. Agarose-immobilized ssDNAs corresponding to the 132-nt full-length β-globin 3′UTR (βWT) or to a poly(dI · dC) negative control (NC) were incubated with K562 cytoplasmic extract, and adherent factors were resolved by SDS-PAGE. Three bands were analyzed by MALDI-TOF (asterisks). Lanes M and U contain protein size markers and unfractionated extract, respectively. (B) Identification of nucleolin as a β-globin 3′UTR-binding factor. A diagram illustrates key structural features of full-length human nucleolin, including amino-terminal acidic domains (light shading), RNA-binding domains (dark shading), and a carboxy-terminal, RGG-rich domain (crosshatched). The sizes and positions of tryptic-digest fragments, identified by MALDI-TOF analysis of affinity-enriched K562 cell extract, are indicated as black boxes below the diagram. (C) Nucleolin (Nuc) binds liganded ssDNAs and RNAs corresponding to the β-globin 3′UTR. K562 extract was affinity enriched using a 32-nt ligand corresponding to the H122/H124 site (32 nt) or ligands comprising the full-length (FL) β-globin 3′UTR. Ligands comprised ssDNA, in vitro-transcribed RNA (RNA), or 2′-O-methyl RNA (Me-RNA). Poly(dI · dC) was assessed in parallel as a negative control. Lanes M and U contain protein size markers and unfractionated extract, respectively. (D) Immunological confirmation of nucleolin as a β-globin 3′UTR-binding factor. Affinity-enriched lysate from panel A was analyzed by Western transfer analysis using nucleolin antibody MS-3. Lane U contains unfractionated extract analyzed in parallel as a migration control. (E) Sequence-specific binding of nucleolin to the β-globin 3′UTR. Agarose-immobilized ssDNAs corresponding to the βWT 3′UTR were incubated with MEL cytoplasmic extract in the presence of defined quantities of competitor poly(dI · dC). Adherent proteins were resolved on a Coomassie blue-stained SDS-polyacrylamide gel (top) and subjected to Western blot analysis using nucleolin antibody MS-3 (bottom). (F) Nucleolin binds to the 3′UTR of β-globin mRNA. In vitro-transcribed, 32P-labeled RNAs corresponding to the βWT 3′UTR were incubated with total (lane T) or nucleolin-depleted (lane D) K562 extract and cross-linked with UV light, and mRNPs were resolved on a nondenaturing acrylamide gel. RNAs incubated in reconstituted lysate (lane R) and with affinity-purified nucleolin (lane C) were analyzed in parallel as controls. Bands corresponding to nucleolin-β-3′UTR mRNPs are indicated (black spots). (Bottom) The efficiency of nucleolin depletion was assessed by Western blot analysis of reagent extracts using nucleolin antibodies (bottom). The stripped blot was rehybridized with a β-actin antibody to control for variations in sample loading.
Nucleolin localizes to the cytoplasm of intact erythroid progenitor cells.
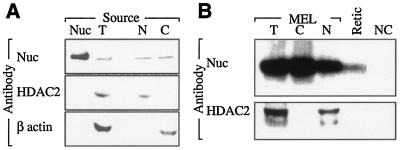
Although nucleolin has been identified in the cytoplasm of nonerythroid cells (7, 68), its presence in erythroid cytoplasm has never been formally established. Two methodologically independent approaches were used to demonstrate that nucleolin can be found in the cytoplasm of erythroid cells representing temporally distinct stages of terminal differentiation. Nucleolin was easily detected by Western analysis of cytoplasm prepared from murine erythroid MEL cells (Fig. 4A) and was also identified in extract prepared from FACS-sorted murine reticulocytes (Fig. 4B). These results confirm that nucleolin is abundant in erythroid cytoplasm, permitting consideration of its potential role in stabilizing the relatively pure population of globin mRNAs that also populate these cells.
FIG. 4.
Nucleolin is present in the cytoplasms of differentiating erythroid cells. (A) Nucleated erythroid progenitors contain cytoplasmic nucleolin. Western blot analysis was performed on total (T), nuclear (N), and cytoplasmic (C) extracts prepared from MEL cells using nucleolin (Nuc) antibody. The blot was stripped and rehybridized with antibodies directed against nucleus- and cytoplasm-specific histone deacetylase-2 (HDAC-2) and β actin, respectively. Affinity-purified nucleolin was analyzed in parallel as a positive control. (B) Anucleate erythroid progenitors (reticulocytes) contain cytoplasmic nucleolin. Hemolysate prepared from FACS-sorted murine reticulocytes (Retic) was analyzed by Western transfer analysis using nucleolin antibody. Total, cytoplasmic, and nuclear extracts prepared from MEL cells were analyzed in parallel as positive controls, and recombinant αCP was run as a negative control (NC). The blot was stripped and rehybridized with HDAC-2 antibody to confirm the absence of contaminating nucleoplasm in the Retic sample.
Nucleolin binds human β-globin mRNA in both cultured cells and primary human erythroid progenitors.
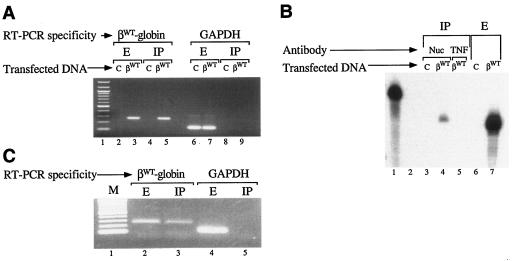
The demonstration that nucleolin binds to ssDNA and RNA corresponding to the β-globin 3′UTR in vitro predicted its capacity to interact with full-length β-globin mRNA transcripts in vivo in intact cells. This hypothesis was subsequently tested using an RNA-immunoprecipitation (RIP) method (15, 16). Human β-globin mRNA was detected in cell extract as well as in a nucleolin immunoprecipitate prepared from cells transfected with pTRE-βWT (Fig. 5A, lanes 3 and 5) but not in fractions prepared from cells transfected with an empty pTRE control vector (lanes 2 and 4). The specificity of the nucleolin-globin mRNA interaction was indicated by control experiments in which constitutively expressed GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA was observed in cell extract (Fig. 5A, lanes 6 and 7) but not in the nucleolin immunoprecipitate (Fig. 5A, lanes 8 and 9). Human β-globin mRNA was not identified in immunoprecipitate prepared with an unrelated antibody (Fig. 5B, compare lanes 4 and 5), demonstrating that the results do not arise from artifactual binding of β-globin mRNA to immunoglobulin. The likely physiological importance of the interaction between nucleolin and the β-globin mRNA was indicated by RIP analyses of lysate prepared from density-fractionated human erythroid progenitors. Both β-globin mRNA and control GAPDH mRNA were observed in the unfractionated lysate (Fig. 5C, lanes 2 and 4), while β-globin mRNA, but not GAPDH mRNA, was detected in immunoprecipitate prepared using nucleolin antibody (Fig. 5C, compare lanes 3 and 5). These experiments confirm that β-globin mRNA and nucleolin interact with high mutual specificity in intact cultured cells as well as in primary human erythrocytes.
FIG. 5.
Nucleolin binds to β-globin mRNA in intact cells. (A, B) Specificity of nucleolin-β-globin mRNA interaction in vivo. (A) HeLatTA cells were transfected with pTRE-βWT (βWT) or with an empty pTRE vector control (C). Total RNA recovered from cell extract (E) or nucleolin immunoprecipitate (IP) was RT-PCR amplified using βWT sequence-specific oligomers, generating a 261-bp product (lanes 2 to 5), or with GAPDH mRNA-specific oligomers, producing a 116-bp product (lanes 6 to 9). Lane 1 contains a 100-bp DNA ladder. (B) Total RNA was recovered from immunoprecipitate (lanes 3 to 5) or extract (lanes 6 and 7) prepared from cells transfected with pTRE-βWT (βWT) or with the empty pTRE vector control (C). Immunoprecipitates were prepared using nucleolin- or tumor necrosis factor-specific antibodies (Nuc or TNF, respectively). RNAs were analyzed by RNase protection using in vitro-transcribed, 32P-labeled RNA probes (84). Intact and RNase-digested 32P-labeled probes were run in lanes 1 and 2, respectively. (C) Nucleolin binds β-globin mRNA in intact human erythroid cells. Purified RNA prepared from the extract or nucleolin immunoprecipitate of density-fractionated human erythroid cells was RT-PCR amplified using human β-globin- and GAPDH-specific oligomers. M, DNA size markers.
An mRNA-destabilizing mutation in the β-globin 3′UTR reduces nucleolin binding in vitro and in vivo.
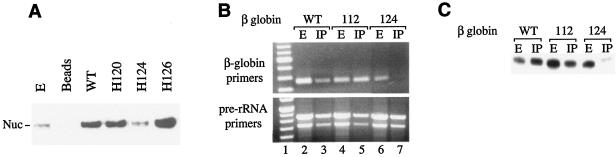
The proposed functional linkage between nucleolin binding and β-globin mRNA stability was subsequently investigated by assessing the affinity of nucleolin for variant βH-globin mRNAs containing destabilizing and control nondestabilizing 3′UTR hexanucleotide linker-scanning substitutions. The affinity of purified nucleolin for ssDNAs corresponding to the β-globin 3′UTR was substantially reduced by the mRNA-destabilizing H124 mutation but not by flanking mutations at position H120 or H126 that had had no discernible effect on β-globin mRNA stability in earlier in vivo studies (Fig. 6A). The adverse effect of the H124 mutation on nucleolin binding was also demonstrated in vivo using RIP analyses of HeLatTA cells expressing βWT, βH112, and βH124 mRNAs (Fig. 6B). Each mRNA was easily detected in the cell extract (Fig. 6B, lanes 2, 4, and 6), while only the stable βWT and βH112 mRNAs—but not the unstable βH124 mRNA—were present in the nucleolin immunoprecipitate (Fig. 6B, lanes 3, 5, and 7). Pre-rRNA, which is known to bind nucleolin strongly (1), was observed in all samples, confirming the quality of the mRNAs and controlling for other aspects of the experimental method. These results were corroborated by parallel analyses of βWT, βH112, and βH124 mRNAs using an independent RNase protection approach (Fig. 6C) and confirmed in repeat analyses (data not shown). Consequently, the native sequence targeted by the H124 mutation appears to function both as a determinant of β-globin mRNA stability and as a binding site for nucleolin, providing a critical link between these two processes.
FIG. 6.
Differential binding of nucleolin to mRNA-stabilizing and -destabilizing 3′UTR determinants. (A) β-Globin mRNA-destabilizing linker-scanning mutations reduce nucleolin binding in vitro. Agarose-immobilized, 59-nt ssDNAs corresponding to the proposed 3′UTR nucleolin-binding region of β-globin mRNA were incubated in cytoplasmic extract, and adherent proteins were assessed by Western transfer analysis using nucleolin antibody. The wild-type sequence (WT) as well as sequences containing destabilizing (H124) and nondestabilizing (H120 and H126) HindIII mutations were assessed. Unfractionated extract (E) and extract adhering to unliganded agarose beads were run in the first two lanes as controls. (B, C) Full-length, unstable βH124 mRNA binds nucleolin poorly in vivo in intact, cultured cells. Unfractionated cell extract or nucleolin immunoprecipitate (IP) was prepared from cultured cells transfected with genes encoding βWT, βH112, and βH124 mRNAs. (B) Recovered RNAs were RT-PCR amplified using primers specific to β-globin mRNA (top) or to internal control pre-rRNA (bottom). The reaction products were resolved on an ethidium bromide-stained, nondenaturing polyacrylamide gel. Lane 1 contains a 100-bp DNA ladder. (C) Recovered RNAs were assessed by RNase protection using an in vitro-transcribed, 32P-labeled β-globin RNA probe.
A model for β-globin mRNA stability.
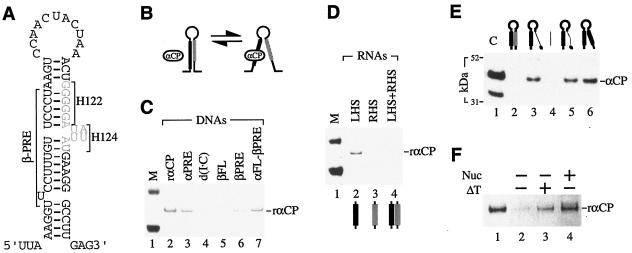
Although the β-PRE appears to be a determinant of β-globin mRNA stability in vivo (84), its anticipated role as a target for αCP binding has been difficult to recapitulate in vitro. We propose a model for β-globin mRNA stability that incorporates the findings presented here and, in addition, accounts for previous experimental evidence that indirectly implicates αCP in this process. In this model, the β-globin 3′UTR has the potential to assume a highly stable stem-loop structure that incorporates the β-PRE and nucleolin-binding sites into its left and right half-stems, respectively (Fig. 7A). If secondary structure were to inhibit the access of αCP to the β-PRE-binding site, then any process that weakens the stem structure would be predicted to facilitate αCP binding (Fig. 7B).
FIG. 7.
Model for regulated β-globin mRNA stability. (A) A secondary structure exists within the β-globin 3′UTR. A stable stem-loop structure within the β-globin 3′UTR is predicted by the Zuker algorithm using default parameters (50, 87). The positions of the β-PRE and the two previously identified mRNA-destabilizing hexanucleotide mutations (H122 and H124) (gray) are indicated. (B) Predicted effect of the secondary structure on αCP binding. The access of αCP to its functional β-PRE-binding site (black) is favored by the relaxation of a native β-globin mRNA stem-loop motif. The positioning of a binding site for nucleolin on the opposite (right) half-stem suggests a role for nucleolin in shaping the high-order 3′UTR structure. (C) RNA context-dependent binding of αCP to the β-PRE. ssDNA ligand-bound rαCP was resolved by Coomassie blue staining after SDS-PAGE. Agarose-immobilized ligands (top), including the α-PRE and β-PRE (lanes 3 and 6), the full-length β-3′UTR (lane 5), a full-length α-globin 3′UTR in which the β-PRE is substituted for the α-PRE (lane 7), and a negative-control poly(dI · dC) (lane 4), are identified. Lanes 1 and 2 contain protein standards (M) and rαCP, respectively. (D) αCP binding to the β-PRE is inhibited by its participation in a stable stem structure. Agarose-immobilized 2′-O-methylated RNAs corresponding to the predicted left and right half-stems (LHS and RHS, respectively) of the 3′UTR structure (32 nt each) were incubated with rαCP either singly (lanes 2 and 3) or in combination (lane 4), and adherent αCP was resolved by Coomassie blue staining of SDS-PAGE gels. The LHS (black) and RHS (gray) contain the β-PRE and the H122/H124 nucleolin-binding sites, respectively. M, protein size markers. (E) Mutations that disrupt the 3′UTR secondary structure enhance αCP binding to β-globin mRNA. Agarose-immobilized ssDNAs were incubated with HeLa cell extract, and adherent factor was analyzed by Western blot analysis using αCP antibody. The predicted structures of individual ssDNAs are schematically illustrated (top). The β-PRE and proposed nucleolin-binding sites are represented as thick black and gray lines. Right-half-stem modifications include the deletion of a native 18-nt sequence (broken thin black line) (lane 5), the substitution of an unrelated 18-nt sequence (thin gray line) (lane 3), and the substitution of a stem-destabilizing 18-nt region containing the β-PRE (lane 6). The unrelated stem-destabilizing sequence was analyzed as a control (lane 4). Lane 1 contains recombinant αCP as a migration control (C). See Materials and Methods for details of each ssDNA sequence. (F) Nucleolin (Nuc) enhances αCP binding to the β-globin 3′UTR in vitro. Agarose-immobilized ssDNAs corresponding to the β-globin 3′UTR were incubated with rαCP following no pretreatment (lane 2), heat denaturation at 95°C for 5 min (ΔT) (lane 3), or preincubation with affinity-purified nucleolin (lane 4). Ligand-bound rαCP was analyzed by SDS-PAGE. Lane 1 contains rαCP as a migration control.
The possibility that native secondary structure inhibits αCP binding was tested in three independent affinity-binding studies. Results from the first study suggest that αCP access to the β-PRE is highly dependent upon its mRNA context: recombinant αCP (rαCP) binds poorly to an ssDNA corresponding to the full-length β-3′UTR (Fig. 7C, lane 5), while binding avidly to ssDNAs corresponding to the β-PRE either in isolation (Fig. 7C, lane 6) or when inserted into a different 3′UTR (Fig. 7C, lane 7). In a second study, baseline interaction of rαCP with the left-half-stem β-PRE was ablated by its preincubation with an ssDNA corresponding to the right half-stem (Fig. 7D). A third study demonstrated that αCP binds poorly to the intact 3′UTR stem-loop structure (Fig. 7E, lane 2) while, in agreement with our predictions, binding strongly to 3′UTRs that contain stem-destabilizing substitutions (Fig. 7E, lanes 3 and 6) or deletions (Fig. 7E, lane 5). The results of all three experiments are consistent with a model in which native structure within the β-globin 3′UTR must be remodeled as a precondition for αCP interaction with the β-PRE.
The potential role that nucleolin may play in remodeling the 3′UTR stem-loop structure in vivo was investigated by assessing the binding of rαCP to agarose-immobilized β-globin 3′UTRs in vitro under different conditions. The poor baseline affinity of rαCP for the naked probe is significantly enhanced by preincubating the β-globin 3′UTR with affinity-purified nucleolin (Fig. 7F, compare lanes 2 and 4). Although this result does not favor any specific mechanism, the possibility that nucleolin facilitates αCP binding through its effect on mRNA structure is suggested by the observation that αCP binding is also enhanced, in the absence of nucleolin, by prior heat denaturation of the agarose-immobilized β-3′UTR ligand (Fig. 7F, lane 3). In the aggregate, the results of these in vitro analyses are consistent with the assembly of a stable structure within the β-globin 3′UTR that inhibits αCP binding and suggest that nucleolin facilitates αCP access through interaction with this structure.
DISCUSSION
Although the high stability of human β-globin mRNA has been repeatedly demonstrated (2, 4, 36, 42, 62, 63) and the importance of this characteristic to normal β-globin expression firmly established (10, 64), the relevant molecular mechanisms have yet to be defined. The current article provides original evidence that this process is likely to require cis determinants and trans-acting factors that have not previously been appreciated. We initially designed and validated an important method for assessing the stability of an mRNA in vivo in intact cultured cells without affecting the expression or function of other cellular mRNAs (Fig. 1). Using this approach, we identified a defined 3′UTR region that is critical to normal β-globin mRNA stability (Fig. 2), thus linking this important functional characteristic to a discrete, previously unrecognized structural determinant. This evidence does not rule out the possibility that other cis elements may participate in this process, since the experimental method is likely to overlook extensive and/or highly degenerate structural determinants. This technical limitation may explain why the β-PRE element, whose importance has previously been implicated in vitro and in vivo (84), was not functionally identified. Nevertheless, the critical nature of the H122-H124 region to β-globin mRNA stability is clear.
Among three candidate 3′UTR-binding factors, nucleolin appears most likely to play a central role in stabilizing β-globin mRNA in vivo. Nucleolin displays a relative specificity for ssDNAs corresponding to the β-globin 3′UTR in vitro (Fig. 3) and interacts with full-length β-globin mRNA both in intact cultured cells and in primary human erythroid progenitors (Fig. 5). Moreover, binding is ablated in vivo by mRNA-destabilizing mutations but preserved in β-globin mRNAs carrying control nondestabilizing mutations, firmly linking nucleolin binding to its proposed mRNA-stabilizing function (Fig. 6).
Although it is tempting to speculate that nucleolin directly stabilizes β-globin mRNA, we think it more likely that this highly abundant factor facilitates functional interaction of other, known globin mRNA-stabilizing factors, such as αCP. Our structural analyses are consistent with this possibility; nucleolin binds to the right half-stem of a stable 3′UTR stem-loop structure, directly opposite to the β-PRE (Fig. 7A). Among several mechanistic possibilities, we favor one in which nucleolin binding is required to relax a stem-loop structure that is predicted to interfere with αCP binding (Fig. 7B). In vitro studies show enhanced αCP binding to 3′UTRs in which the stem-loop structure is disrupted (Fig. 7C to E), consistent with the proposed mechanism. A specific role for nucleolin in this process is suggested by the demonstration that αCP binding to the β-globin 3′UTR can be enhanced either by heat denaturation or by preincubation with immunopurified nucleolin (Fig. 7F). Though potentially informative, these studies do not resolve the final common pathway through which the combined activities of nucleolin and αCP effect mRNA stabilization, including potential effects on β-globin mRNA polyadenylation and/or translational efficiency. Nevertheless, we find the proposed model to be particularly attractive because it accommodates both our current data and evidence from previous studies favoring a critical role for αCP in stabilizing the β-globin mRNA.
The apparent role that nucleolin plays in stabilizing β-globin mRNA is consistent with its participation in a wide range of molecular processes. In the nucleus, nucleolin is associated with ribosome biogenesis (9, 35), chromatin remodeling (23), immunoglobulin isotype switching (30), telomere formatting (33), and posttranscriptional processing of nascent mRNAs (33). In the cytoplasm, nucleolin binds to the 5′ and 3′UTRs of specific mRNAs, enhancing both their stabilities and their translational efficiencies (15, 56, 58, 69, 71, 72, 85, 86). This functional diversity reflects both the complexity of the nucleolin core structure and the heterogeneity of isoforms that it can assume. The core structure, which comprises acidic and glycine-rich domains as well as four RNA-binding domains (RBDs) (43), is extensively modified by targeted proteolysis (14, 24), phosphorylation (13, 54, 59, 60, 67), ADP ribosylation (44), and methylation (46), resulting in combinatorial structural complexity that may form the basis for its observed functional heterogeneity.
The four centrally positioned RBDs of nucleolin mediate its interaction with RNA both in the nucleus (32, 33) and in the cytoplasm (15, 56, 58, 69, 71, 72, 85, 86). These domains, which are structurally similar to RBDs in protein factors that regulate the stabilities and translational efficiencies of other mRNAs (22), appear to subserve a parallel spectrum of functions in nucleolin. Nucleolin has been reported to stabilize mRNAs encoding amyloid precursor protein (85, 86), renin (58, 72), CD154 (71), and Bcl-2 (56, 69) by binding to structurally distinct cis elements within their 3′UTRs. The heterogeneity in its posttranslational modification may account for nucleolin's equally heterogeneous mRNA-binding specificities. The nucleolin-binding sites of interleukin 2 and amyloid precursor protein mRNAs, which share a common 5′CUCUCUUUA3′ target sequence (15, 85, 86), differ from the A/U-rich nucleolin-binding site in the 3′UTR of Bcl-2 mRNA (56, 69) and from the 5′UCCCGA3′ motif mediating its binding to rRNA (1, 27). Nucleolin may also bind to motifs corresponding to splice acceptor sequences (5′UUAGG3′) (33) and to G-quartet and other related nonlinear, thermodynamically favorable nucleic acid structures that are not predicted by common mRNA-folding algorithms (83). The β-globin mRNA nucleolin-binding determinant that we describe (Fig. 2) is dissimilar to each of these linear elements, possibly reflecting interaction with a subset of nucleolin structural isoforms that carry specific phosphoryl, ADP-ribosyl, or methyl modifications.
The wide variety of molecular processes that require nucleolin suggests that it may serve a general role—as a molecular scaffold or perhaps a substrate-remodeling factor—acting in concert with other proteins that provide the required functional specificity. We suggest that a specific nucleolin-β-globin mRNP may have to assemble before αCP can bind, and subsequently stabilize, the full-length β-globin mRNA. This hypothesis may explain the difficulties that we and others have encountered in attempting to demonstrate bimolecular interaction between the β-globin 3′UTR and αCP in vitro, despite the clear importance of the proposed αCP-binding site to human transgenic β-globin mRNA stability observed in vivo in mice (84). The potential importance of cooperative interactions between these two protein factors is additionally indicated by analyses of renin mRNA, which, like β-globin mRNA, displays nucleolin-dependent stability and binds both nucleolin and αCP to the same 3′UTR region (58, 72). These aspects of nucleolin function merit close scrutiny as they may provide opportunities for novel therapeutic approaches to the treatment of common thalassemias and hemoglobinopathies.
Acknowledgments
We thank Lorelle Bradley for technical assistance and Stephen Liebhaber for critical reading of the manuscript.
This work was supported in part by NIH grants HL-R01-061399 and HL-U54-070596.
REFERENCES
- 1.Allain, F. H.-T., P. Bouvet, T. Dieckmann, and J. Feigon. 2000. Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J. 19:6870-6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aviv, H., Z. Voloch, R. Bastos, and S. Levy. 1976. Biosynthesis and stability of globin mRNA in cultured erythroleukemic Friend cells. Cell 8:495-503. [DOI] [PubMed] [Google Scholar]
- 3.Basak, A., A. Ozer, B. Kirdar, and N. Akar. 1993. A novel 13 bp deletion in the 3′UTR of the β-globin gene causes β-thalassemia in a Turkish patient. Hemoglobin 17:551-555. [DOI] [PubMed] [Google Scholar]
- 4.Bastos, R., Z. Volloch, and H. Aviv. 1977. Messenger RNA population analysis during erythroid differentiation: a kinetic approach. J. Mol. Biol. 110:191-203. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, P. L., D. J. Herrick, R. D. Prokipcak, and J. Ross. 1992. Control of c-myc mRNA half-life in vitro by a protein capable of binding to a coding region stability determinant. Genes Dev. 6:642-654. [DOI] [PubMed] [Google Scholar]
- 6.Binder, R., J. A. Horowitz, J. P. Basilion, D. M. Koeller, R. D. Klausner, and J. B. Harford. 1994. Evidence that the pathway of transferrin receptor mRNA degradation involves an endonucleolytic cleavage within the 3′UTR and does not involve poly(A) tail shortening. EMBO J. 13:1969-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borer, R. A., C. F. Lehner, H. M. Eppenberger, and E. A. Nigg. 1989. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 56:379-390. [DOI] [PubMed] [Google Scholar]
- 8.Brown, B. D., I. D. Zipkin, and R. M. Harland. 1993. Sequence-specific endonucleolytic cleavage and protection of mRNA in Xenopus and Drosophila. Genes Dev. 7:1620-1631. [DOI] [PubMed] [Google Scholar]
- 9.Bugler, B., M. Caizergues-Ferrer, G. Bouche, H. Bourbon, and F. Amalric. 1982. Detection and localization of a class of proteins immunologically related to a 100-kDa nucleolar protein. Eur. J. Biochem. 128:475-480. [DOI] [PubMed] [Google Scholar]
- 10.Bunn, H. F., and B. G. Forger. 1986. Hemoglobin: molecular, genetic, and clinical aspects. W. B. Saunders, Philadelphia, Pa.
- 11.Bunn, H. F., G. J. Schmidt, D. N. Haney, and R. G. Dluhy. 1975. Hemoglobin Cranston, an unstable variant having an elongated beta chain due to nonhomologous crossover between two normal beta chain genes. Proc. Natl. Acad. Sci. USA 72:3609-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai, S. P., B. Eng, W. H. Francombe, N. F. Olivieri, A. G. Kendall, J. S. Waye, and D. H. Chui. 1992. Two novel beta-thalassemia mutations in the 5′ and 3′ noncoding regions of the beta-globin gene. Blood 79:1342-1346. [PubMed] [Google Scholar]
- 13.Caizergues-Ferrer, M., P. Mariottini, C. Curie, B. Lapeyre, N. Gas, F. Amalric, and F. Amaldi. 1989. Nucleolin from Xenopus laevis: cDNA cloning and expression during development. Genes Dev. 3:324-333. [DOI] [PubMed] [Google Scholar]
- 14.Chen, C. M., S. Y. Chiang, and N. H. Yeh. 1991. Increased stability of nucleolin in proliferating cells by inhibition of its self-cleaving activity. J. Biol. Chem. 266:7754-7758. [PubMed] [Google Scholar]
- 15.Chen, C.-Y., R. Gherzi, J. S. Andersen, G. Gaietta, K. Jurchott, H.-D. Royer, M. Mann, and M. Karin. 2000. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 14:1236-1248. [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, C. -Y., F. Del Gatto-Konczak, Z. Wu, and M. Karin. 1998. Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science 280:1945-1949. [DOI] [PubMed] [Google Scholar]
- 17.Chkheidze, A. N., D. L. Lyakhov, A. V. Makeyev, J. Morales, J. Kong, and S. A. Liebhaber. 1999. Assembly of the α-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′ untranslated region determinant and poly(C) binding protein αCP. Mol. Cell. Biol. 19:4572-4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Czyzyk-Krzeska, M. F., and J. E. Beresh. 1996. Characterization of the hypoxia-inducible protein binding site within the pyrimidine-rich tract in the 3′-untranslated region of the tyrosine hydroxylase mRNA. J. Biol. Chem. 271:3293-3299. [DOI] [PubMed] [Google Scholar]
- 19.Dalton, T. P., D. Bittel, and G. K. Andrews. 1997. Reversible activation of mouse metal response element-binding transcription factor 1 DNA binding involves zinc interaction with the zinc finger domain. Mol. Cell. Biol. 17:2781-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delanoe-Garin, J., Y. Blouquit, N. Arous, J. Kister, C. Poyart, M. L. North, J. Bardakdjian, C. Lacombe, J. Rosa, and F. Galacteros. 1988. Hemoglobin Saverne: a new variant with elongated beta chains: structural and functional properties. Hemoglobin 12:337-352. [DOI] [PubMed] [Google Scholar]
- 21.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dreyfuss, G., V. N. Kim, and N. Kataoka. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3:195-205. [DOI] [PubMed] [Google Scholar]
- 23.Erard, M., F. Lakhdar-Ghazal, and F. Amalric. 1990. Repeat peptide motifs which contain beta-turns and modulate DNA condensation into chromatin. Eur. J. Biochem. 191:19-26. [DOI] [PubMed] [Google Scholar]
- 24.Fang, S. H., and N. H. Yeh. 1993. The self-cleaving activity of nucleolin determines its molecular dynamics in relation to cell proliferation. Exp. Cell Res. 208:48-53. [DOI] [PubMed] [Google Scholar]
- 25.Flatz, G., J. L. Kinderlerer, J. V. Kilmartin, and H. Lehmann. 1971. Haemoglobin Tak: a variant with additional residues at the end of the beta-chains. Lancet i:732-733. [DOI] [PubMed] [Google Scholar]
- 26.Frischmeyer, P. A., and H. C. Dietz. 1999. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 8:1893-1900. [DOI] [PubMed] [Google Scholar]
- 27.Ghisolfi-Nieto, L., G. Joseph, F. Puvion-Dutilluel, F. Amalric, and P. Bouvet. 1996. Nucleolin is a sequence-specific RNA binding protein: characterization of targets on pre-ribosomal RNA. J. Mol. Biol. 260:34-53. [DOI] [PubMed] [Google Scholar]
- 28.Goodman, M. 1981. Decoding the pattern of protein evolution. Prog. Biophys. Mol. Biol. 37:105-164. [DOI] [PubMed] [Google Scholar]
- 29.Goodman, M., M. L. Weiss, and J. Czelusniak. 1982. Molecular evolution above the species level: branching patterns, rates, and mechanisms. Syst. Zool. 31:376-399. [Google Scholar]
- 30.Hanakahi, L. A., L. A. Dempsey, M.-J. Li, and N. Maizels. 1997. Nucleolin is one component of the B-cell specific transcription factor and switch region binding protein, LR-1. Proc. Natl. Acad. Sci. USA 94:3605-3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He, Z., and J. E. Russell. 2004. Anti-sickling effects of an endogenous human alpha-like globin. Nat. Med. 10:365-367. [DOI] [PubMed] [Google Scholar]
- 32.Herrera, A. H., and M. O. Olson. 1986. Association of protein C23 with rapidly labeled nucleolar RNA. Biochemistry 25:6258-6264. [DOI] [PubMed] [Google Scholar]
- 33.Ishikawa, F., M. J. Matunis, G. Dreyfuss, and T. R. Cech. 1993. Nuclear proteins that bind to the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol. Cell. Biol. 13:4301-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jankovic, L., A. J. Dimovsky, P. Kollia, M. Karageorga, D. Loukopoulos, and T. H. J. Huisman. 1991. A C----G mutation at nt position 6 3′ to the terminating codon may be the cause of a silent beta-thalassemia. Int. J. Hematol. 54:289-293. [PubMed] [Google Scholar]
- 35.Jordan, G. 1987. At the heart of the nucleolus. Nature 329:489-490. [DOI] [PubMed] [Google Scholar]
- 36.Kabnick, K. S., and D. E. Housman. 1988. Determinants that contribute to cytoplasmic stability of human c-fos and β-globin mRNAs are located at several sites in each mRNA. Mol. Cell. Biol. 8:3244-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiledjian, M., N. Day, and P. Trifillis. 1999. Purification and RNA binding properties of the polycytidylate-binding proteins αCP1 and αCP2. Methods 17:84-91. [DOI] [PubMed] [Google Scholar]
- 38.Kiledjian, M., C. T. DeMaria, G. Brewer, and K. Novick. 1997. Identification of AUF1 (heterogeneous nuclear ribonucleoprotein D) as a component of the α-globin mRNA stability complex. Mol. Cell. Biol. 17:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiledjian, M., X. Wang, and S. A. Liebhaber. 1995. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. EMBO J. 14:4357-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klausner, R. D., T. A. Rouault, and J. B. Harford. 1993. Regulating the fate of mRNA: the control of cellular iron metabolism. Cell 72:19-28. [DOI] [PubMed] [Google Scholar]
- 41.Koeller, D. M., J. A. Horowitz, J. L. Casey, R. D. Klausner, and J. B. Harford. 1991. Translation and the stability of mRNAs encoding the transferrin receptor and c-fos. Proc. Natl. Acad. Sci. USA 88:7778-7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krowczynska, A., R. Yenofsky, and F. Brawerman. 1985. Regulation of messenger RNA stability in mouse erythroleukemia cells. J. Mol. Biol. 181:231-239. [DOI] [PubMed] [Google Scholar]
- 43.Lapeyre, B., H. Bourbon, and F. Amalric. 1987. Nucleolin, the major nucleolar protein of eukaryotic cells: an unusual protein structure revealed by the nucleotide sequence. Proc. Natl. Acad. Sci. USA 84:1472-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leitinger, N., and J. Wesierska-Gadek. 1993. ADP-ribosylation of nucleolar proteins in HeLa tumor cells. J. Cell. Biochem. 52:153-158. [DOI] [PubMed] [Google Scholar]
- 45.Liebhaber, S. A., and Y. W. Kan. 1983. Alpha thalassemia caused by an unstable alpha-globin mutant. J. Clin. Investig. 71:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lischwe, M. A., K. D. Roberts, L. C. Yeoman, and H. Busch. 1982. Nucleolar specific acidic phosphoprotein C23 is highly methylated. J. Biol. Chem. 257:14600-14602. [PubMed] [Google Scholar]
- 47.Lodish, H. F., and B. Small. 1976. Different lifetimes of reticulocyte messenger RNA. Cell 7:59-65. [DOI] [PubMed] [Google Scholar]
- 48.Makeyev, A. V., A. N. Chkheidze, and S. A. Liebhaber. 1999. A set of highly conserved RNA-binding proteins, αCP-1 and αCP-2, implicated in mRNA stabilization, are coexpressed from an intronless gene and its intron-containing paralog. J. Biol. Chem. 274:24849-24857. [DOI] [PubMed] [Google Scholar]
- 49.Maquat, L. E. 1995. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA 1:453-465. [PMC free article] [PubMed] [Google Scholar]
- 50.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 51.Milner, P. F., J. B. Clegg, and D. J. Weatherall. 1971. Haemoglobin H disease due to a unique haemoglobin variant with an elongated α-chain. Lancet i:729-732. [DOI] [PubMed] [Google Scholar]
- 52.Morales, J., J. E. Russell, and S. A. Liebhaber. 1997. Destabilization of human alpha-globin mRNA by translation anti-termination is controlled during erythroid terminal differentiation and paralleled by phased shortening of the poly(A) tail. J. Biol. Chem. 272:6607-6613. [DOI] [PubMed] [Google Scholar]
- 53.Mullner, E. W., B. Neupert, and L. C. Kuhn. 1989. A specific mRNA binding factor regulates the iron-dependent stability of cytoplasmic transferrin receptor mRNA. Cell 58:373-382. [DOI] [PubMed] [Google Scholar]
- 54.Olson, M. O., E. G. Ezrailson, K. Guetzow, and H. Busch. 1975. Localization and phosphorylation of nuclear, nucleolar, and extranucleolar nonhistone proteins of Novikoff hepatoma ascites cells. J. Mol. Biol. 97:611-619. [DOI] [PubMed] [Google Scholar]
- 55.Ostareck-Lederer, A., D. H. Ostareck, N. Standart, and B. J. Thiele. 1994. Translation of 15-lipoxygenase mRNA is inhibited by a protein that binds to a repeated sequence in the 3′ untranslated region. EMBO J. 13:1476-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Otake, Y., T. K. Sengupta, S. Bandyopadhyay, E. K. Spicer, and D. J. Fernandes. 2005. Retinoid-induced apoptosis in HL-60 cells is associated with nucleolin down-regulation and destabilization of Bcl-2 mRNA. Mol. Pharmacol. 67:319-326. [DOI] [PubMed] [Google Scholar]
- 57.Pandey, N. B., and W. F. Marzluff. 1987. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol. Cell. Biol. 7:4557-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Persson, P. B., A. Skalweit, R. Mrowka, and B.-J. Thiele. 2003. Control of renin synthesis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 285:R491-R497. [DOI] [PubMed] [Google Scholar]
- 59.Peter, M., J. Nakagawa, M. Doree, J. C. Labbe, and E. A. Nigg. 1990. Identification of major nucleolar proteins as candidate mitotic substrates of cdc2 kinase. Cell 60:791-801. [DOI] [PubMed] [Google Scholar]
- 60.Rao, S. V., M. D. Mamrack, and M. O. Olson. 1982. Localization of phosphorylated highly acidic regions in the NH2-terminal half of nucleolar protein C23. J. Biol. Chem. 257:15035-15041. [PubMed] [Google Scholar]
- 61.Rodgers, N. D., Z. Wang, and M. Kiledjian. 2002. Characterization and purification of a mammalian endoribonuclease specific for the alpha-globin mRNA. J. Biol. Chem. 277:2597-2604. [DOI] [PubMed] [Google Scholar]
- 62.Ross, J., and A. Pizarro. 1983. Human beta and delta globin messenger RNAs turn over at different rates. J. Mol. Biol. 167:607-617. [DOI] [PubMed] [Google Scholar]
- 63.Ross, J., and T. D. Sullivan. 1985. Half-lives of beta and gamma globin messenger RNAs and of protein synthetic capacity in cultured human reticulocytes. Blood 66:1149-1154. [PubMed] [Google Scholar]
- 64.Russell, J. E., and S. A. Liebhaber. 1993. Molecular genetics of thalassemia, p. 283-353. In R. S. Verma (ed.), Advances in genome biology, vol. 2. JAI Press, Greenwich, Conn. [Google Scholar]
- 65.Russell, J. E., and S. A. Liebhaber. 1996. The stability of human beta-globin mRNA is dependent on structural determinants positioned within its 3′ untranslated region. Blood 87:5314-5323. [PubMed] [Google Scholar]
- 66.Russell, J. E., J. Morales, A. Makeyev, and S. A. Liebhaber. 1998. Sequence divergence in the 3′ untranslated regions of human ζ- and α-globin mRNAs mediates a difference in their stabilities and contributes to efficient ζ-to-α gene developmental switching. Mol. Cell. Biol. 18:2173-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schneider, H. R., G. U. Reichert, and O. G. Issinger. 1986. Enhanced casein kinase II activity during mouse embryogenesis. Eur. J. Biochem. 161:733-738. [DOI] [PubMed] [Google Scholar]
- 68.Semenkovich, C. F., R. E. Ostlund, M. O. Olson, and J. W. Yang. 1990. A protein partially expressed on the surface of HepG2 cells that binds lipoproteins specifically is nucleolin. Biochemistry 29:9708-9713. [DOI] [PubMed] [Google Scholar]
- 69.Sengupta, T. K., S. Bandyopadhyay, D. J. Fernandes, and E. K. Spicer. 2004. Identification of nucleolin as an AU-rich element binding protein involved in bcl-2 mRNA stabilization. J. Biol. Chem. 279:10855-10863. [DOI] [PubMed] [Google Scholar]
- 70.Shaw, G., and R. Kamen. 1986. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46:659-667. [DOI] [PubMed] [Google Scholar]
- 71.Singh, K., J. Laughlin, P. A. Kosinski, and L. R. Covey. 2004. Nucleolin is a second component of the CD154 mRNA stability complex that regulates mRNA turnover in activated T cells. J. Immunol. 173:976-985. [DOI] [PubMed] [Google Scholar]
- 72.Skalweit, A., A. Doller, A. Huth, T. Kahne, P. B. Persson, and B. J. Thiele. 2003. Posttranscriptional control of renin synthesis: identification of proteins interacting with renin mRNA 3′-untranslated region. Circ. Res. 92:419-427. [DOI] [PubMed] [Google Scholar]
- 73.Stefanovic, B., C. Hellerbrand, M. Holcik, M. A. Briendl, S. A. Liebhaber, and D. A. Brenner. 1997. Posttranscriptional regulation of collagen α1(I) mRNA in hepatic stellate cells. Mol. Cell. Biol. 17:5201-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Volloch, V., and D. Housman. 1981. Stability of globin mRNA in terminally differentiating murine erythroleukemia cells. Cell 23:509-514. [DOI] [PubMed] [Google Scholar]
- 75.Wang, X., M. Kiledjian, I. M. Weiss, and S. A. Liebhaber. 1995. Detection and characterization of a 3′ untranslated region ribonucleoprotein complex associated with human α-globin mRNA stability. Mol. Cell. Biol. 15:1769-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang, X., and S. A. Liebhaber. 1996. Complementary change in cis determinants and trans factors in the evolution of an mRNP stability complex. EMBO J. 15:5040-5051. [PMC free article] [PubMed] [Google Scholar]
- 77.Wang, Z., N. Day, P. Trifillis, and M. Kiledjian. 1999. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell. Biol. 19:4552-4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang, Z., and M. Kiledjian. 2000. Identification of an erythroid-enriched endoribonuclease activity involved in specific mRNA cleavage. EMBO J. 19:295-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang, Z., and M. Kiledjian. 2000. The poly(A)-binding protein and an mRNA stability protein jointly regulate an endoribonuclease activity. Mol. Cell. Biol. 20:6334-6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weiss, I. M., and S. A. Liebhaber. 1994. Erythroid cell-specific determinants of α-globin mRNA stability. Mol. Cell. Biol. 14:8123-8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weiss, I. M., and S. A. Liebhaber. 1995. Erythroid cell-specific mRNA stability elements in the α2-globin 3′ nontranslated region. Mol. Cell. Biol. 15:2457-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wisdom, R., and W. Lee. 1991. The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev. 5:232-243. [DOI] [PubMed] [Google Scholar]
- 83.Xu, X., F. Hamhouyia, S. D. Thomas, T. J. Burke, A. C. Girvan, W. G. McGregor, J. O. Trent, D. M. Miller, and P. J. Bates. 2001. Inhibition of DNA replication and induction of S phase cell cycle arrest by G-rich oligonucleotides. J. Biol. Chem. 276:43221-43230. [DOI] [PubMed] [Google Scholar]
- 84.Yu, J., and J. E. Russell. 2001. Structural and functional analysis of an mRNP complex that mediates the high stability of human β-globin mRNA. Mol. Cell. Biol. 21:5879-5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zaidi, S. H. E., R. Denman, and J. S. Malter. 1994. Multiple proteins interact at a unique cis-element in the 3′-untranslated region of amyloid precursor protein mRNA. J. Biol. Chem. 269:24000-24006. [PubMed] [Google Scholar]
- 86.Zaidi, S. H. E., and J. Malter. 1995. Nucleolin and heterogeneous nuclear ribonucleoprotein C proteins specifically interact with the 3′-untranslated region of amyloid protein precursor mRNA. J. Biol. Chem. 270:17292-17298. [DOI] [PubMed] [Google Scholar]
- 87.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]