Abstract
Telomerase maintains cell viability and chromosomal stability through the addition of telomere repeats to chromosome ends. The reactivation of telomerase through the upregulation of TERT, the telomerase protein subunit, is an important step during cancer development, yet TERT protein function and regulation remain incompletely understood. Despite its close sequence similarity to human TERT (hTERT), we find that mouse TERT (mTERT) does not immortalize primary human fibroblasts. Here we exploit these differences in activity to understand TERT protein function by creating chimeric mouse-human TERT proteins. Through the analysis of these chimeric TERT proteins, we find that sequences in the human carboxy-terminal domain are critical for telomere maintenance in human fibroblasts. The substitution of the human carboxy-terminal sequences into the mouse TERT protein is sufficient to confer immortalization and maintenance of telomere length and function. Strikingly, we find that hTERT protein accumulates to markedly higher levels than does mTERT protein and that the sequences governing this difference in protein regulation also reside in the carboxy-terminal domain. These elevated protein levels, which are characteristic of hTERT, are necessary but not sufficient for telomere maintenance because stabilized mTERT mutants cannot immortalize human cells. Thus, the TERT carboxy terminus contains sequences that regulate TERT protein levels and determinants that are required for productive action on telomere ends.
Telomeres are protein-DNA structures that cap the ends of linear chromosomes (11). These guanine-rich nucleotide repeats are bound by a large protein complex that protects chromosomal ends from recombination and prevents telomere ends from activating checkpoint responses (21). Telomeres shorten with cell proliferation and with aging, and this shortening is countered by telomerase, a reverse transcriptase that synthesizes telomere repeats (17). Telomerase is expressed in a restricted pattern in mammalian tissues, predominantly in stem cells and progenitor cells, and is upregulated in approximately 90% of human cancers (24, 36, 37, 47, 53). Telomerase activity is regulated at the level of expression of TERT, the protein catalytic subunit, which together with TERC, the telomerase RNA component, comprises the catalytic core of the enzyme (10, 26, 47, 54, 65).
Telomeres shorten with cell division because of the end replication problem, which is the inability of DNA polymerase to complete synthesis of the lagging strand. Consequently, telomeres shorten in settings of insufficient telomerase activity, including in primary human cells in culture and in human tissues with aging in vivo (28, 30, 41). Telomere shortening has profound effects on cell proliferation and survival. After 60 to 80 population doublings (pd), shortened telomeres in human fibroblasts trigger a DNA damage signal that causes replicative senescence (20), an arrest that is enforced by the activation of the tumor suppressor proteins p53 and Rb (27, 62). The inactivation of both p53 and Rb prevents the replicative senescence response; however the continued cell division that is associated with this extended lifespan results in further telomere loss and the initiation of a second response, which is termed crisis. In cells that have reached crisis, a subset of telomeres become critically short or uncapped; that is, they can no longer protect the chromosome end from recombination, resulting in end-to-end chromosome fusions (18). Each chromosome fusion harbors two centromeres that can attach to opposite spindle poles during mitosis, resulting in chromosome breakage and elevated rates of apoptosis in cell cultures in crisis. Ectopic expression of human TERT (hTERT) in primary human cells reconstitutes telomerase activity and prevents senescence and crisis responses by adding telomere repeats de novo, thereby stabilizing chromosome ends (13, 19, 68). In addition to synthesizing telomere repeats, telomerase has been shown to protect telomere ends from becoming dysfunctional by contributing to the functional cap at the chromosome terminus (12, 46, 68).
The catalytic action of telomerase on telomere ends is tightly regulated, in part through protein-protein interactions. In Saccharomyces cerevisiae, the catalytic core of telomerase interacts with Est1p, which in turn binds Cdc13p, a critical telomere end-binding protein. Like the TERT and TERC homologues Est2p and TLC1, Est1p and Cdc13p are both required for telomere maintenance and cell viability (43, 44). Est1p and Cdc13p may function either by recruiting telomerase to the telomere end (23) or by activating telomerase after it has become telomere associated (64). In humans, telomerase associates with the Est1p homologue Est1A, which can facilitate telomerase-mediated telomere lengthening (59, 63). However, our understanding of the molecular events in human cells that are necessary for telomerase recruitment and activation remains incomplete.
Additional insights into the function of telomerase have come from structure-function studies that characterized the TERT sequences that are required for enzymatic catalysis, TERT-TERC association, and telomerase multimerization in vitro. These studies have revealed four general domains that are conserved in TERT proteins: two N-terminal regions, a central reverse transcriptase (RT) domain, and a C-terminal region (34). The RT domain contains seven motifs with similarity to viral RTs, and these conserved sequences are required for telomerase enzymatic activity in vitro (29, 42, 54). In the N terminus, regions II, III, and T comprise a domain that is critical for binding TERC and this domain is therefore required for enzymatic function in vitro (5, 6, 38, 49, 55, 67). The most amino-terminal portion of the protein, region I, is also important for enzymatic activity as well as telomere length regulation (9, 25, 33, 51). In both yeast and human cells, telomerase functions as a dimer comprised of two TERT molecules and two TERC molecules (57, 66). In humans, dimerization is in part mediated by TERT protein, through interaction between N-terminal regions II and III and amino acids immediately carboxy terminal to motif D in the RT domain (1, 8, 50). In addition to these interactions between TERT proteins, the P3 pseudoknot region of human TERC participates functionally in telomerase dimer formation (45, 60).
The C-terminal domain also contains sequences conserved among TERT proteins from divergent species (7, 32, 56). In S. cerevisiae, the C-terminal domain modulates telomerase processivity, although TERT mutants in which this domain is entirely deleted retain enzymatic function and the corresponding mutant yeast strains remain viable (31, 56). In contrast, C-terminal deletions in hTERT abrogate telomerase activity, indicating that this region is necessary for catalytic function (5, 8). Similarly, the deletion or mutation of conserved residues in the C-terminal domain of hTERT impairs telomerase processivity (32). The fact that TERT enzymatic function in vitro is sensitive to both N- and C-terminal deletion analysis has complicated efforts to understand TERT function in vivo through the identification of protein domains that control immortalization or telomere maintenance.
One approach circumvented these limitations and identified domains in TERT that are required for immortalization of primary human cells through scanning mutagenesis, replacing groups of six amino acids with the sequence NAAIRS throughout the amino- and carboxy-terminal regions whole protein. These efforts led to the identification of two regions, one in the N-terminal domain and one in the C-terminal domain, which are required for cellular immortalization but preserve enzymatic activity in vitro (2, 7). These DAT (dissociates activities of telomerase) domains were proposed to be required for the recruitment of telomerase to telomeric chromatin (3, 4). Other analyses of these TERT mutants found that the DAT mutants showed reduced processivity, principally on oligonucleotide primers matching the natural telomere sequence, indicating that the DAT regions contribute to interactions between TERT and its telomere substrate (40, 52). These data suggest that the DAT mutants may fail to immortalize human cells because of impaired interaction with telomere DNA sequences or with telomeric chromatin. Consistent with a potential role for the C terminus in facilitating telomerase action on telomere ends, the addition of heterologous amino acid sequences to the hTERT C terminus prevented immortalization or impaired telomere maintenance in human cells (19, 68). Despite this progress, the protein domains involved in recruiting telomerase to telomere ends or in posttranslational regulation of TERT remain incompletely understood.
To identify sequences that are required for TERT protein regulation and for TERT protein function in vivo, we have compared the activities of human and mouse TERT (mTERT) orthologues in human cells. We find that mouse TERT cannot immortalize primary human cells, despite efficient reconstitution of telomerase enzymatic activity. We exploit this important difference in function to identify sequences that are critical for telomerase action in immortalization and in telomere maintenance in vivo by creating chimeric proteins between mTERT and hTERT. Unlike conventional mutagenesis, which may alter protein structure in some cases, this approach takes advantage of evolutionary conservation throughout the TERT open reading frame to introduce subtle amino acid substitutions at positions divergent between the two species and is therefore more likely to preserve overall domain architecture. This strategy identified the C terminus as a critical regulatory domain controlling telomere maintenance, immortalization, and steady-state protein levels in human cells.
MATERIALS AND METHODS
Cell culture.
Human foreskin fibroblasts BJ cells (ATCC) and BJ cells transduced with the simian virus 40 (SV40) early region (BJT), including both large T and small t antigens (gift of R. Weinberg), were cultured in a 4:1 mixture of Dulbecco's modified Eagle's medium (Invitrogen) and medium 199 (Invitrogen) with 15% fetal bovine serum (Invitrogen) and 1% penicillin-streptomycin at 37°C under 5% CO2. Phoenix A cells (gift of G. Nolan) were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and 1% penicillin-streptomycin at 37°C under 5% CO2. For serial passage experiments, transduced BJT cells were counted and plated every 3 to 4 days. Population doublings were calculated using the formula log2(number cells harvested/number cells plated). Population doublings were plotted against time (days). For metaphase analysis, cells treated with Colcemid and metaphase chromosomes were prepared on glass slides as previously described (16).
Construction of retroviruses and TERT mutants.
EcoRI fragments comprising the open reading frame of either hTERT or mTERT were cloned into the EcoRI site of the pLPC retroviral plasmid (gift of S. Lowe). HMM was created by PCR of the 5′ region of hTERT with the primers 5′-GAATTCAGATCTCCGCGATGCCGCGCG-3′ and 5′-GAATTCAGATCTCCTCACGCAGACGGTGC-3′. This PCR product was digested with HindIII/BamHI and ligated to a 3′ BamHI/EcoRI-digested fragment of mTERT and a HindIII/EcoRI-digested vector. MHH was engineered by ligating a 3′ hTERT PCR product (primers 5′-GAATTCAGATCTTGGCCAAGTTCCTGC-3′ and 5′-GAATTCAGATCTTCA GTCCAGGATGGTCTTGAAGTC-3′) with a 5′ mTERT HindIII/BamHI fragment and a HindIII/EcoRI-digested vector. HMH and MMH were created by replacing a 3′ XbaI/EcoRI mTERT fragment from HMM and mTERT, respectively, with a 3′ hTERT PCR fragment (primers 5′-GGAATTCTAGACTTGCAGGTGAACAGCCTC-3′ and 5′-ATGGAATTCAGTCCAGGATGGTCTTGAAG-3′) flanked by XbaI/EcoRI sites. MHM and HHM were engineered by replacing the 3′ ApaLI/EcoRI fragment of hTERT in MHH and hTERT, respectively, with an ApaLI/RI PCR fragment of mTERT (primers 5′-GAATTCGTGCACCAATATATACAAGATCTTCC-3′ and 5′-ATGGAATTCTTAGTCCAAAATGGTCTGAAAGTC-3′). mTERT-hC2 and hTERT-mC1 were created by using a chimeric primer (5′-AGCCGCACATTGGCTCTGCTACCAAGCATTCCTGCTCAAGCTG-3′) with the reverse primer from MMH to create a long 3′ primer for a second PCR with the forward primer from HHM. This chimeric PCR product was digested with ApaLI/RI and used to replace the 3′ end of HHM or mTERT, respectively, to create hTERT-mC1 and mTERT-hC2. hTERT-mC2 was created by replacing a BsmI/RI fragment of hTERT with an mTERT PCR fragment (primer 5′-GAATTCAAGCATTCCTGCTCAAGCTG-3′ and the reverse primer from HHM) flanked by these same restriction sites. mTERT-hC1 was created by PCR as follows. Primer 5′-GAATTCATGCATGTGTGCTGCAGCTC-3′ and the reverse primer from HHM were used with hTERT-mC2 as a template. This PCR product was digested with NsiI/RI and ligated to an mTERT fragment cut with HindIII/NsiI and vector HindIII/RI. mTERT-h2-4, -3-4, and -4 used 5′ primers 5′-CCTTTGACCAGCGTGTTAGGAAGAACCCCACATTTTTCC-3′, 5′-GCTATGCTATCCTGAAGGTCAAGAATGCAGGGATGTCGCTGGG-3′, and 5′-GGAATGACACTAAAGGCCTCTGGCCCTCTGCCCTCCG-3′, respectively, with the reverse primer from HHM with hTERT-mC2 as a template to create chimeric PCR products. These chimeric PCR products were subsequently used as the 3′ primer for PCR on mTERT with the 5′ primer from HHM. Each product was digested with NsiI/EcoRI, ligated to HindIII/NsiI-digested mTERT, and vector digested with HindIII/EcoRI. All regions generated by PCR were verified by DNA sequencing.
Retroviral transductions.
Phoenix A cells (gift of G. Nolan) were plated at 1.5 × 106 to 2 × 106 cells per 10-cm dish and transfected the following day with 15 μg of plasmid DNA by calcium phosphate precipitation. Forty-eight hours after transfection, Phoenix A supernatants were used to infect BJ or BJT cells that were plated at 7 × 105 to 8 × 105 cells per 10-cm dish on the previous day. Three serial infections were performed at 8- to 12-h intervals. Transduced cells were selected by using 1 to 2 μg/ml puromycin beginning 24 h after the last infection.
Telomerase repeat amplification protocol (TRAP).
TRAP was performed according to standard protocols (TRAPeze; Chemicon).
Telomere length analysis by Southern blotting.
Genomic DNA was isolated from BJT cells by using lysis buffer (100 mM Tris [pH 8.5], 5 mM EDTA, 0.2% sodium dodecyl sulfate [SDS], 200 mM NaCl) and 10 μg/ml proteinase K overnight at 55°C. Genomic DNA was precipitated with isopropanol, washed with 70% ethanol, and resuspended in Tris-EDTA buffer at 55°C overnight. DNA (10 μg) was digested with RsaI/HinfI overnight. Two micrograms of digested DNA was fractionated on a 0.8% agarose gel and transferred overnight to Hybond-N membrane (Amersham) in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The ultraviolet light-cross-linked membrane was blocked by prehybridization and then hybridized with a 32P-end-labeled (TTAGGG)4 oligonucleotide by using Rapid Hyb (Amersham Biosciences) at 42°C for 1 h. Membranes were washed once in 5× SSC and 0.1% SDS at room temperature for 20 min and twice in 1× SSC and 0.1% SDS at 42°C for 15 min and then exposed to film. Telomere signal intensities were quantitated by scanning the autoradiographs and using ImageJ software and the following equation: mean telomere length = Σ(intensityi)/Σ(intensityi/kbi) (28).
RNA analysis by Northern blotting.
RNA was isolated from cells by using Trizol (Invitrogen). Five micrograms of RNA was resolved on a 1.2% formaldehyde agarose gel and then transferred to Hybond-N membrane (Amersham) in 10× SSC overnight. After cross-linking and prehybridization, the membrane was incubated in Ultrahyb at 42°C overnight with an appropriate cDNA fragment labeled by random priming with [α-32P]-ATP. Membranes were washed twice with 2× SSC and 0.1% SDS for 5 min at 42°C and twice with 0.1× SSC and 0.1% SDS for 15 min at 42°C.
Immunofluorescence.
BJ cells were plated to be subconfluent on coverslips in six-well plates. The next day, phosphate-buffered saline (PBS)-washed coverslips were permeabilized at room temperature with 0.2% Triton X-100 in PBS for 10 min and then washed three times with 2 ml PBS. Primary antibody (PAP; Sigma) was diluted 1:400 in PBS-1% bovine serum albumin (BSA) and incubated with the coverslips in a humidity chamber at 37°C for 50 min. Coverslips were washed three times with 2 ml PBS before incubation with Cy3-conjugated anti-rabbit antibody (Jackson ImmunoResearch) at 1:500 dilution in PBS-1% BSA plus DAPI (4′,6′-diamidino-2-phenylindole) at 37°C for 50 min in a humidity chamber. Coverslips were washed in PBS and then mounted on slides with polyvinyl alcohol (Mowiol), sealed, and photographed by fluorescence microscopy.
Western blotting.
All antibodies were diluted in blocking buffer (1× TBST [Tris-buffered saline-Tween] and 5% nonfat milk). Staphylococcus aureus protein A (SPA)-tagged TERTs were visualized using PAP (Sigma) at 1:1,000 dilution for 1 to 3 h at room temperature. Anti-α-tubulin antibodies (Sigma) were diluted 1:20,000 and incubated with membranes for 1 h at room temperature, followed by a 1-h incubation with horseradish peroxidase-conjugated anti-mouse antibody (Jackson ImmunoResearch) at 1:20,000. Anti-FLAG Western analyses were performed with M2 antibody (Sigma) at 1:400 for 1 h at room temperature, followed by horseradish peroxidase-conjugated anti-mouse antibody at 1:5,000 for 1 h at room temperature.
Coimmunoprecipitation.
293T cells were transiently transfected with FLAG-TERT-FLAG and/or CMV-hTERC or CMV-U64 constructs (gifts of K. Collins) by calcium phosphate precipitation. Cells were lysed in hypotonic lysis buffer (20 mM HEPES, 2 mM MgCl2, 0.2 mM EGTA, 10% glycerol, 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride), incubated on ice 5 min, and snap-frozen/thawed twice. NaCl was added to 400 mM final concentration in two steps. Lysates were cleared by centrifugation, diluted 1:2 in hypotonic lysis buffer plus 0.2% NP-40, and cleared again. M2 beads (Sigma) were blocked in wash buffer (20 mM HEPES, 2 mM MgCl2, 0.2 mM EGTA, 200 mM NaCl, 0.1% NP-40, 10% glycerol, 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride) and blocking reagents (100 ng/ml yeast tRNA, 100 ng/ml yeast RNA, 100 ng/ml glycogen, 200 ng/ml BSA, 200 ng/ml casein) for 1 h at 4°C. Immunoprecipitations were carried out with blocked M2 beads at 4°C for 1 h. Beads were washed with 1 ml 3× wash buffer and then divided in half (48). For RNA analysis, beads were boiled in 2× RNA loading buffer and the RNA was resolved on a 7 M urea-5% acrylamide gel and transferred to Hybond-N (Amersham). The blot was hybridized with 32P-labeled hTERC probe. Membranes were washed twice for 5 min in 2× SSC and 0.1% SDS at 42°C and once for 15 min in 0.1× SSC and 0.1% SDS at 42o and then exposed to film overnight. Beads also were analyzed for protein by Western blotting using M2 antibody (Sigma) as denoted above.
Telomere length analysis by quantitative fluorescence in situ hybridrization (Q-FISH).
Metaphase chromosomes were prepared from BJT cells by treatment with Colcemid (0.1 μg/ml), followed by hypotonic KCl and fixation in cold methanol-acetic acid. Chromosomes were hybridized with a Cy3-conjugated (CCCTAA)3 peptide-nucleic acid probe as described previously (39). Briefly, chromosomes were spread on glass slides and dried overnight. Slides were fixed with 4% formaldehyde in PBS for 2 min, treated with pepsin (Sigma) at 1 mg/ml for 10 min at 37°C, and hybridized with the telomere peptide-nucleic acid probe. After hybridization for 2 h at room temperature, the slides were washed and stained with DAPI at 1.5 μg/ml in antifade solution (VectaShield; Vector Laboratories Inc.). Cy3 and DAPI digital images were captured with a SPOT RT 2.3.0 charge-coupled device camera (Diagnostic Instruments) on an Eclipse E800 microscope (Nikon) using SPOT Advanced software. Telomere intensities were analyzed using TFL-Telo V.2.1 (gift of P. Lansdorp). At least 11 metaphases (>2,000 telomeres) per sample were analyzed to determine the distribution of telomere lengths in each sample. Telomere length distributions between samples were statistically compared by the two-tailed Wilcoxon rank sum test.
RESULTS
mTERT cannot substitute for hTERT in immortalization of human fibroblasts.
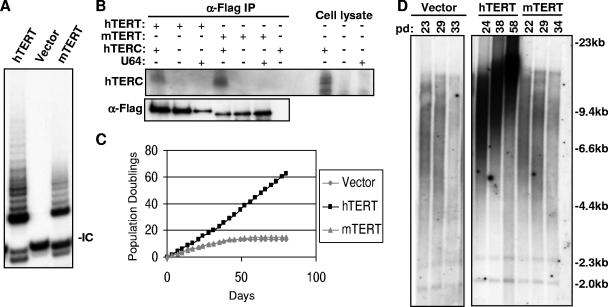
To compare the activities of mTERT and hTERT, we assessed their ability to prevent the critical telomere shortening that is characteristic of human cells in crisis. A subset of telomeres becomes uncapped during crisis, resulting in chromosomal end-to-end fusions and apoptosis. We therefore studied human BJ fibroblasts that were stably expressing the SV40 early region, which abrogates the replicative senescence response, allowing telomeres to become critically short at crisis with continued cell division. The expression of TERT alleles in this cellular context is therefore a test of their ability to synthesize and stabilize the shortest telomeres in the population. Enforced expression of hTERT reconstituted telomerase activity as measured by the TRAP (Fig. 1A). Telomerase activity was detected neither in untransduced BJT cells nor in BJT cells that were infected with the retroviral vector alone (Fig. 1A). The transduction of BJT cells with mTERT retrovirus reproducibly resulted in telomerase activity by TRAP, although the levels of TRAP activity were consistently lower than that in hTERT-transduced cells (Fig. 1A and data not shown). These data suggest that mTERT can assemble with human TERC (hTERC) to allow the formation of enzymatically active telomerase ribonucleoprotein complexes. To assess directly the ability of mTERT to interact with hTERC, coimmunoprecipitation experiments were performed in 293T cells that were transfected with expression plasmids for hTERC and either FLAG-tagged hTERT or FLAG-tagged mTERT. TERT protein in lysates from transfected cells was immunoprecipitated by using anti-FLAG antibody agarose. FLAG antibody efficiently pulled down the tagged TERT proteins. Northern blotting on the affinity-purified complexes showed that hTERC was associated with both hTERT and mTERT at comparable levels (Fig. 1B). Together, these data show that mTERT efficiently interacts with hTERC in human cells, giving rise to an enzymatically active chimeric telomerase RNP, consistent with previous findings (15).
FIG. 1.
mTERT cannot immortalize primary human fibroblasts. (A) Expression of mTERT in primary BJT cells reconstitutes telomerase activity by TRAP assay. Note the telomerase elongation products in cells expressing hTERT and mTERT but not with empty vector alone. IC, internal control for PCR. (B) mTERT interacts with hTERC in transfected human 293T cells. Left, α-FLAG immunoprecipitates (IP) were analyzed for hTERC RNA by Northern blotting (top) and for TERT protein by α-FLAG Western blotting (bottom). Right, total cellular RNA from transfected 293T cells analyzed by Northern blotting. U64, small nucleolar RNA used as a negative control. (C) mTERT is unable to immortalize primary human fibroblasts during serial passage of hTERT-, mTERT-, or empty vector-BJT fibroblasts. (D) mTERT does not lengthen telomeres. Telomere restriction fragment (TRF) Southern blot analysis of genomic DNA from hTERT-, mTERT-, or empty vector-BJT cells over multiple population doublings is shown.
To determine whether reconstitution of telomerase with mTERT is sufficient to immortalize human fibroblasts, the proliferative capacity of mTERT-BJT cells was compared to that of hTERT-BJT cells through serial passage. Cells were counted and seeded every 3.5 days for up to 60 population doublings (80 days). Cells that were transduced with the empty retroviral vector entered crisis after approximately 10 population doublings, whereas hTERT-BJT cells showed a dramatically increased lifespan, proliferating in an unimpeded fashion for at least 60 population doublings (Fig. 1C). Despite the efficient reconstitution of telomerase enzymatic activity, mTERT did not extend the replicative lifespan of BJT cells (Fig. 1C). Both vector-BJT and mTERT-BJT cultures showed characteristic apoptotic morphology (data not shown) as well as elevated rates of chromosomal end-to-end fusions (Table 1). In contrast, metaphase chromosomes from hTERT-transduced cells showed no chromosomal end-to-end fusions, indicating that telomeres remain protected in the hTERT cultures (P was 0.001 for hTERT versus mTERT by t test) (Table 1). Experiments with BJ fibroblasts and IMR90 fibroblasts lacking expression of the SV40 early region similarly showed that mTERT cannot avert replicative senescence of primary human cells (data not shown). These results show that, unlike hTERT, mTERT cannot prevent chromosomal end-to-end fusions or extend the proliferative lifespan of human cells.
TABLE 1.
Telomere analysis of stably transduced BJT cellsa
| TERT type | Metaphasesb | Fusionsc | Fusions/metaphase | Mean telomere length at crisisd | Mean telomere length at pd150d |
|---|---|---|---|---|---|
| Vector | 12 | 8 | 0.67 | 5.68 ± 0.44 | NA |
| hTERT | 21 | 0 | 0 | NA | 10.80 ± 0.56 |
| mTERT | 21 | 15 | 0.71 | 5.75 ± 0.23 | NA |
| HMM | 15 | 7 | 0.47 | 5.88 ± 1.11 | NA |
| MHH | 17 | 8 | 0.47 | ND | ND |
| HMH | 23 | 1 | 0.04 | NA | 3.86 ± 0.21 |
| MMH | 29 | 5 | 0.17 | NA | 3.94 ± 0.22 |
| HHM | 12 | 7 | 0.58 | 5.5 ± 0.69 | NA |
| mTERT-hC1 | 6 | 8 | 1.33 | ND | ND |
| hTERT-mC2 | 32 | 4 | 0.13 | NA | 4.56 ± 0.21 |
NA, not applicable; ND, not determined.
Results are shown as the total number of metaphases analyzed.
Results are shown as the number of fusions detected.
Results are shown as mean ± standard error.
The appearance of end-to-end fusions in mTERT cultures suggests that, despite efficient reconstitution of telomerase enzymatic activity, mTERT cannot synthesize telomere repeats in human cells. To determine whether mTERT can extend human telomeres, telomere lengths were measured by Southern blot analysis. Telomeres increased in length in BJT cells that were transduced with hTERT with advancing cell divisions, while cells that were transduced with vector alone showed telomere shortening (Fig. 1D). Telomeres in mTERT-BJT cells shortened progressively and were indistinguishable from vector-only controls, with mean telomere lengths at crisis of 5.8 kb and 5.7 kb, respectively (Fig. 1D and Table 1). Together, these data indicate that despite strong sequence similarity to hTERT and reconstitution of enzymatically active telomerase complexes in human cells, mTERT cannot effectively extend human telomeres and fails to immortalize human fibroblasts.
The N terminus of hTERT cannot confer upon mTERT the ability to immortalize human cells.
The observation that mTERT reconstitutes enzymatic activity but cannot extend the lifespan of human cells creates a unique opportunity to understand the protein domains and sequences that account for this important difference in function. The similarity between the two proteins allows for the exchange of homologous regions that should conserve the overall structure of TERT while allowing for the investigation of the effect of infrequent but functionally important amino acid differences. To study the effects of these sequence differences on TERT function, we created chimeric TERT proteins by exchanging mouse and human sequences in three main functional domains spanning the TERT open reading frame: (i) the amino-terminal domain containing the essential domain I and TERC-binding domains II and III, (ii) the reverse transcriptase domain, including the T motif and the conserved RT motifs, and (iii) the carboxy-terminal region (Fig. 2A). Chimeric cDNAs were expressed from retroviruses to characterize their function in primary human cells.
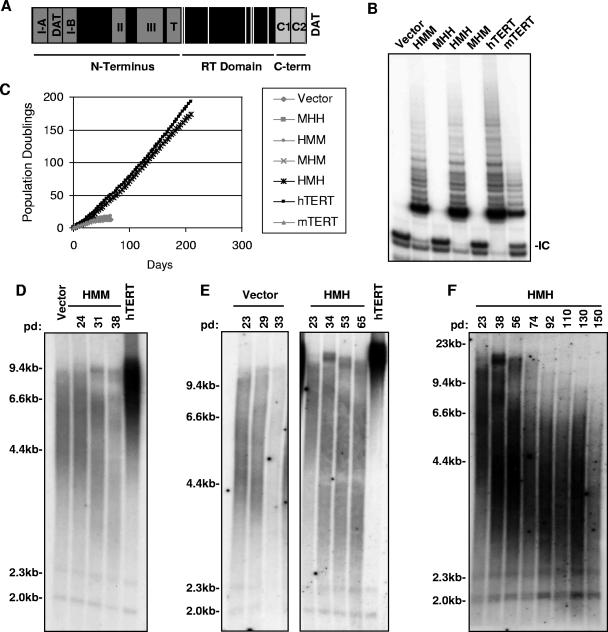
FIG. 2.
Analysis of the N-terminal domain and RT domain in immortalization and telomere maintenance. (A) Schematic diagram of TERT delineating the three major domains: N terminus, RT domain, and the C terminus (C-term). Functional regions I, II, III, T, N-DAT, and C-DAT are indicated in dark gray. Conserved RT motifs 1, 2, A, B′, C, D, and E are designated by white lines. C1 and C2 regions that are defined in this paper are shown in light gray. (B) TRAP analysis and (C) serial passage of BJT cells that were transduced with retroviruses expressing HMM, MHH, HMH, MHM, hTERT, mTERT, or vector alone (IC, internal control). (D) Telomeres shorten in HMM-BJT cells. TRF analysis of HMM-BJT cells over multiple population doublings (vector-BJT and hTERT-BJT are shown for comparison). (E and F) Telomeres shorten, but then stabilize in HMH-expressing cells. TRF analysis of HMH-BJT and empty vector-BJT over multiple population doublings by TRF Southern blot analysis (hTERT, hTERT-BJT sample for comparison) is shown.
To analyze the importance of sequence differences in the N terminus, this region was exchanged between mouse and human TERT to yield the chimeric proteins HMM and MHH. The nomenclature of these constructs is designed to show the identity, whether mouse or human, of the N terminus, RT domain, and the C terminus, based on position. Stable retroviral expression of the reciprocal mutants HMM and MHH in BJT cells showed that HMM-BJT cells exhibited telomerase activity similar to that of cells expressing wild-type hTERT, whereas telomerase activity was not detected in MHH-BJT cells (Fig. 2B). The lack of telomerase activity in the MHH mutant was unexpected because mTERT efficiently interacted with hTERC and because the human N-terminal domain could be readily substituted in mTERT to yield an active telomerase complex. To determine whether this mutant demonstrates enzymatic function in vitro, MHH protein was expressed in rabbit reticulocyte lysates in the presence of hTERC RNA. When equal amounts of mTERT and MHH proteins were used to program TRAP reactions, MHH showed a significantly reduced activity compared to that of wild-type mTERT, thus explaining the absence of enzymatic activity in transduced cells (see Fig. S1 in the supplemental material). Although the MHH mutant exhibited reduced enzymatic activity, the HMM mutant was fully active, allowing us to determine whether the human N terminus could confer the ability to immortalize human cells.
To assess the importance of the N-terminal region in immortalization, HMM- and MHH-transduced BJT cells were cultured for approximately 20 population doublings (60 days). Cells that were transduced with empty vector entered crisis by population doubling 13 (day 50) as did MHH-BJT cells, which was expected for an enzymatically impaired mutant (Fig. 2C). Despite robust enzymatic activity by TRAP, HMM protein did not extend the replicative lifespan of BJT cells. HMM-BJT cells entered crisis by population doubling 18 (day 60) (Fig. 2C). Metaphase preparations demonstrated chromosomal end-to-end fusions for both MHH-BJT and HMM-BJT (0.47 fusions per metaphase for each mutant) (P was 0.004 for MHH and 0.008 for HMM versus hTERT by t test) (Table 1), and both cultures showed apoptotic morphologies (data not shown). Telomere Southern blot analysis demonstrated equivalent telomere shortening for MHH, HMM, and vector-transduced cells (mean telomere lengths at crisis were 5.9 kb and 5.7 kb for HMM and vector, respectively) (Fig. 2D, Table 1, and data not shown). These data indicate that, despite the strong telomerase activity exhibited by the HMM mutant, the N-terminal domain of human TERT did not allow telomere maintenance or extension of replicative lifespan when combined with the RT and C-terminal domains of mTERT.
The mouse RT domain substitutes effectively for the human RT domain in immortalization and telomere maintenance.
To determine whether the sequence differences that allow immortalization by hTERT but not by mTERT lie in the critical RT domain, mouse and human RT domains were exchanged to yield HMH and MHM proteins. The presence of the mouse RT domain in HMH allowed telomerase activity in BJT cells that was comparable to that of hTERT (Fig. 2B). Moreover, when expressed in BJT cells through retroviral transduction, the HMH mutant dramatically extended the replicative lifespan for at least 170 population doublings (over 200 days), an activity that was indistinguishable from hTERT (Fig. 2C). In contrast, empty vector controls and mTERT-transduced cells entered crisis within 15 population doublings (day 50) (Fig. 2C). Consistent with the ability of the HMH mutant to immortalize cells, chromosomal end-to-end fusions were rare in HMH cultures (0.043 fusions per metaphase) (P was 0.001 for HMH versus mTERT by t test) (Table 1). To assess the ability of HMH to maintain telomere length, telomere Southern blot analyses were performed on DNA that was isolated during the serial passage of HMH-BJT cells. Interestingly, telomeres in the bulk population of HMH-BJT cells initially shortened, despite the fact that the HMH mutant prevented telomere uncapping and supported continued cell proliferation (Fig. 2E). Upon continued cell passage, it became evident that after 90 population doublings, mean telomere length stabilized at approximately 3.9 kb (Fig. 2F and Table 1). These data show that the mouse RT domain, in the context of the N-terminal and C-terminal domains from hTERT, is sufficient to maintain telomeres, prevent telomere uncapping, and immortalize BJT cells.
Analysis of the reciprocal mutant MHM revealed an absence of telomerase activity in transduced cells, which was reminiscent of the MHH protein (Fig. 2B). When expressed in vitro in rabbit reticulocyte lysates with hTERC RNA, MHM also showed a markedly reduced specific activity compared to that of mTERT (see Fig. S1 in the supplemental material). Thus, both the MHH and the MHM mutants exhibit a defect in enzymatic activity, likely specific to the juxtaposition of the mouse N terminus and human RT domain. As anticipated for a mutant with such a functional defect, MHM-transduced BJT cells entered crisis simultaneously with empty vector controls, after 12 population doublings (by day 50) (Fig. 2C). In addition, chromosomal end-to-end fusions were elevated just as in empty vector controls, consistent with an absence of telomere maintenance in these cultures (data not shown). Together, these data show that amino acid differences in neither the N-terminal domain nor the RT domain explain the inability of mTERT to maintain telomeres and prevent crisis in primary human cells.
The C-terminal domain of TERT controls immortalization and telomere maintenance.
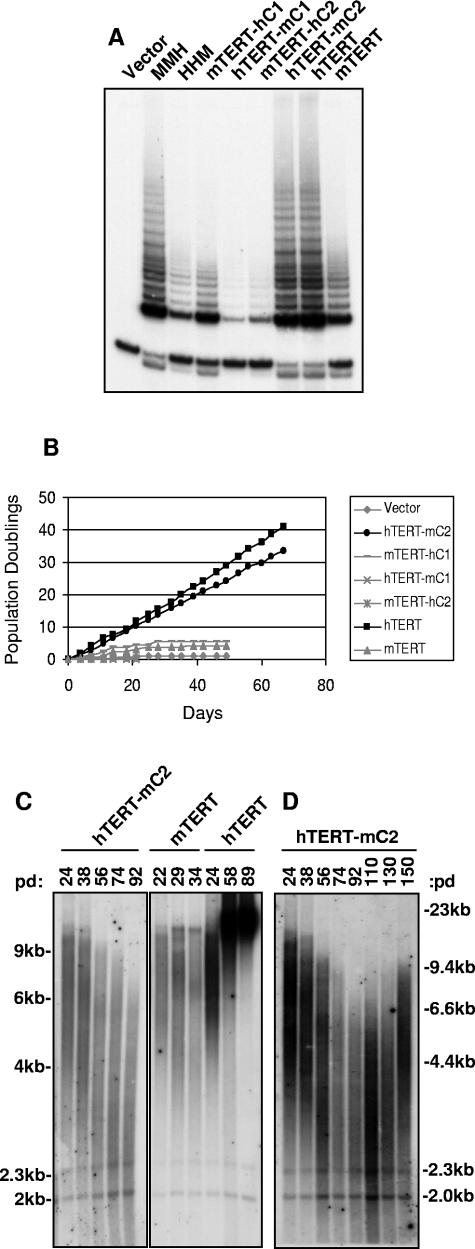
The preceding data demonstrate that the substitution of the N-terminal sequences of hTERT into the mTERT protein does not allow telomere maintenance in human cells and that the mouse RT domain can effectively substitute for the human RT sequences in telomere maintenance and immortalization. These results show that the inability of mTERT to maintain telomeres in human cells is not due to sequence variation within these domains. To determine whether amino acid variation in the C terminus accounts for the differences in telomere maintenance between mTERT and hTERT, we exchanged carboxy-terminal domains between the two orthologues, creating HHM and MMH TERT mutants. Retroviruses expressing HHM and MMH proteins were used to infect BJT cells, and cell protein extracts were collected to analyze telomerase activity. Interestingly, the transfer of the 120-amino-acid C-terminal domain reversed the relative TRAP activity of the mutants such that MMH exhibited significantly higher activity than HHM. The enzymatic activity of HHM was comparable to that of mTERT, whereas the activity of MMH was elevated to that of hTERT (Fig. 3A).
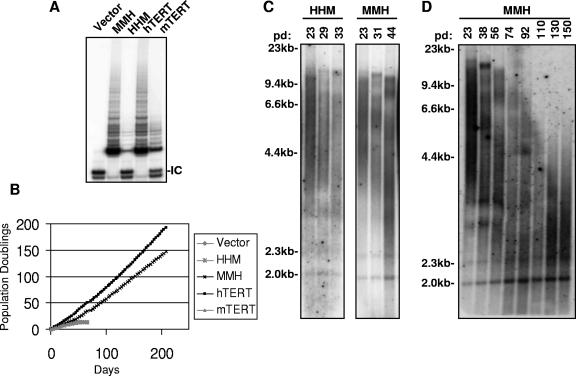
FIG. 3.
C terminus controls enzymatic activity and immortalization. (A) C terminus of hTERT dictates activity level. TRAP analysis of BJT cells that were transduced with retroviruses expressing HHM, MMH, hTERT, and mTERT (IC, internal control). (B) Serial passage of cells in panel A shows that the C terminus of hTERT is necessary and sufficient for immortalization. (C and D) Telomeres shorten in HHM-BJT cells, leading to crisis. Telomeres shorten and then stabilize in immortally proliferating MMH-BJT cells for up to 150 population doublings by TRF Southern blot analysis.
To assess the effect of exchanging C-terminal domains on cellular immortalization, the proliferative capacity of MMH- and HHM-transduced BJT cultures was studied through serial passage. Cells expressing HHM showed a finite lifespan that was limited by crisis (population doubling 14, day 60) in a fashion similar to that of mTERT-transduced cells and empty vector controls (Fig. 3B). These results indicate that the inability of mTERT to immortalize human cells is due to sequence divergence in the C terminus. Indeed, MMH-transduced cells showed a dramatically extended lifespan comparable to that of the cells expressing hTERT. MMH-BJT cultures demonstrated continued proliferation for over 140 population doublings (200 days) (Fig. 3B). Cytogenetic analysis showed that MMH protein, but not the HHM mutant, efficiently preserved telomere function. MMH-transduced cultures showed only 0.17 chromosomal fusions per metaphase, a rate of fusion similar to that of hTERT (P was 0.003 for MMH versus mTERT by t test). In contrast, cells expressing HHM exhibited a significantly elevated prevalence of chromosomal fusions (0.58 fusions per metaphase) (P was 0.012 for HHM versus hTERT by t test) (Table 1). These data show that divergent sequences in the C-terminal domain underlie the differential activity of mouse and human orthologues in immortalization and telomere maintenance of human cells.
To investigate directly the role of the C-terminal domain in telomere length regulation, telomere Southern blot analyses were performed by using genomic DNA that was isolated from cells during serial passage. Mean telomere length decreased progressively in HHM-BJT cells (Fig. 3C) similarly to that in cells expressing empty vector or wild-type mTERT (Fig. 1D and 2E). Interestingly, the mean telomere length in the population of HHM-BJT cells at crisis remained long (5.5 kb). In fact, all the cultures that failed to immortalize, including those transfected with HHM, HMM, wild-type mTERT, or empty vector, entered crisis with similar mean telomere lengths (5.5 kb, 5.9 kb, 5.8 kb, and 5.7 kb, respectively). Despite the apparently ample telomere reserve, all these cultures showed similarly elevated rates of chromosomal fusions, which is clear evidence of telomere dysfunction (Table 1). In comparison, telomeres shortened in immortally proliferating MMH-BJT cells for as long as 110 population doublings and were then maintained at a stable length of approximately 3.9 kb (Fig. 3C and D; Table 1). The mean telomere length of proliferating MMH-BJT cells was significantly shorter than that of HHM-BJT cells in crisis, indicating that the MMH mutant may effectively maintain the shortest telomeres in the population.
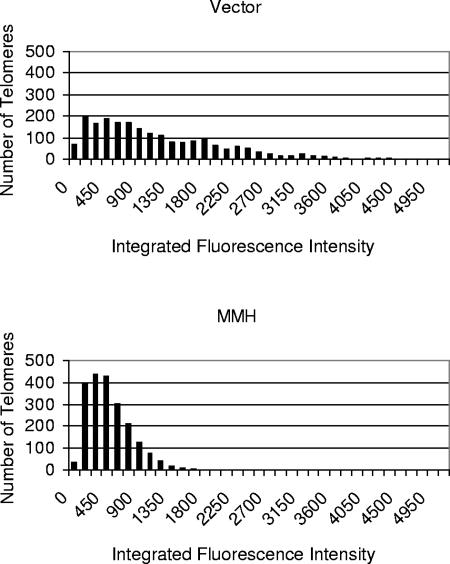
To investigate whether the MMH mutant immortalizes human fibroblasts by synthesizing telomere repeats on the shortest telomeres, we performed Q-FISH, which allows quantitation of the telomere signal at each chromosome end (69). In cultures that had been transduced with empty vector and were therefore approaching crisis (pd 25), telomeres showed a broad distribution with a median telomere signal of 827 fluorescence units. In contrast, telomeres were markedly shorter in MMH-BJT cells analyzed after a larger number of population doublings (pd 150), showing a median telomere signal of 359 fluorescence units (P was <0.0001 by two-tailed Wilcoxon rank sum tests, vector versus MMH) (Fig. 4). Although telomeres were, on average, shorter in MMH cells, the number of chromosomal ends with telomere signals below the level of detection was greater in vector-transduced fibroblasts (6.2 signal-free ends [SFEs] per metaphase for vector versus 3.0 SFEs per metaphase for MMH cells, P was 0.0046). This elevated frequency of SFEs in vector cells is consistent with the observation that vector-transduced cells enter crisis, whereas MMH cells continue proliferating in immortal fashion. Together, these data show that the human C-terminal sequences in MMH allow telomere maintenance at a short set point and that these short telomeres remain capped, as evidenced by the continued proliferation of the culture and absence of chromosomal fusions. In addition to maintaining these short telomeres through telomere synthesis, it is also possible that the presence of telomerase enhances telomere stability in this setting by contributing to the functional cap at chromosome ends (12).
FIG. 4.
Telomeres are maintained at a short length and within a narrow range in MMH-expressing cells. Q-FISH analysis of MMH-BJT and vector-BJT cells. MMH cells at advanced passage (pd 150) demonstrate a significantly shorter mean telomere length than those for empty vector controls studied immediately before crisis (pd 25) (median fluorescence value was 359 for MMH versus 827 for the empty vector) (P was <0.0001 by the two-tailed Wilcoxon rank sum test).
An important role for the C1 domain in immortalization.
To understand which residues in the C terminus control immortalization, we compared the sequences in the TERT C-terminal domain among vertebrates, including mice and humans. Sequence alignment shows similarity throughout the length of the C-terminal domain, with blocks of strong conservation separated by smaller regions of divergence (see Fig. S2 in the supplemental material). Based on this analysis, we divided the C terminus into two regions such that divergent residues between the mouse and human orthologues fall equally into each region, C1 or C2 (Fig. 2A; see Fig. S2 in the supplemental material). Mutants comprising the open reading frame of mTERT with a substitution of the C1 domain from hTERT (mTERT-hC1) and the open reading frame of hTERT with the C1 domain from mTERT (hTERT-mC1) were expressed in BJT cells by retroviral transduction. The substitution of the C1 domain reversed the telomerase activity levels of these mutants; mTERT-hC1 showed significantly increased TRAP activity compared to that of hTERT-mC1 (Fig. 5A). Thus, the ability of the C-terminal domain to alter telomerase activity, as seen in the HHM and MMH mutants (Fig. 3A), maps to the C1 domain.
FIG. 5.
Human C1 is crucial for extended lifespan and telomere maintenance. (A) TRAP analysis of BJT cells transduced with retroviruses expressing MMH, HHM, mTERT-hC1, hTERT-mC1, mTERT-hC2, hTERT-mC2, hTERT, or mTERT. (B) C1 domain of hTERT is required, but not sufficient for extended lifespan. Serial passage of cells in panel A is shown. (C and D) Telomeres shorten then stabilize in immortal hTERT-mC2-BJT cells over 150 population doublings by TRF Southern blot analysis (mTERT-BJT and hTERT-BJT shown for comparison).
To analyze the ability of hTERT-mC1 and mTERT-hC1 to extend cellular lifespan, BJT cells that were transduced with retroviruses expressing these alleles were serially passaged for approximately 50 days. Neither hTERT-mC1 nor mTERT-hC1 could extend lifespan, and both cultures entered crisis by population doubling 5 (day 30); this behavior was similar to that of the vector-only control (Fig. 5B). Consistent with their inability to immortalize BJT cells, both hTERT-mC1- and mTERT-hC1-transduced cells showed an increase in chromosome end-to-end fusions by cytogenetic analysis, with mTERT-hC1-transduced cells demonstrating 1.3 fusions per metaphase (Table 1 and data not shown). These data suggest that the C1 domain harbors important sequences for telomere maintenance and immortalization but that the human C1 sequences are insufficient to allow immortalization in the context of mTERT. The fact that MMH can immortalize cells, whereas mTERT-hC1 cannot, indicates that there are additional residues in the C2 region that are necessary for a fully functional C-terminal domain to facilitate telomere maintenance in human cells.
Exchanging the C2 sequences between mTERT and hTERT further supported a critical role for the C1 domain in immortalization. Substitution of the C2 regions did not alter telomerase activity. Telomerase activity of mTERT-hC2 was similar to that of mTERT, and activity of hTERT-mC2 was unchanged compared to that of hTERT (Fig. 5A). Interestingly, hTERT-mC2 was capable of immortalizing BJT cells for at least 140 population doublings (200 days), whereas mTERT-hC2-BJT cells entered crisis similarly to controls (less than 5 population doublings, 30 days) (Fig. 5B and data not shown). Consistent with their extended proliferative lifespan, hTERT-mC2-BJT cells showed only 0.075 end-to-end fusions per metaphase, a number similar to that of hTERT-transduced cells in cytogenetic analysis (P was 0.001 for hTERT-mC2 versus mTERT by t test) (Table 1). Telomere measurements by Southern blot analysis showed that telomeres shortened progressively in hTERT-mC2-BJT cultures and then stabilized at approximately 4.6 kb (Fig. 5C and D; Table 1); this was similar to telomere dynamics in immortal HMH-BJT cells (Fig. 2E) and MMH-BJT cells (Fig. 3D). The stabilization of telomere length was not due to upregulation of the endogenous hTERT gene because allele-specific RT-PCR showed that endogenous TERT cDNA was undetectable in all retrovirally transduced cultures analyzed (see Fig. S3 in the supplemental material). Taken together, these data show that the sequences in the C terminus that allow immortalization in human cells reside predominantly in the C1 region, although additional residues in the C2 region are likely important in facilitating immortalization in the MMH mutant (Table 2).
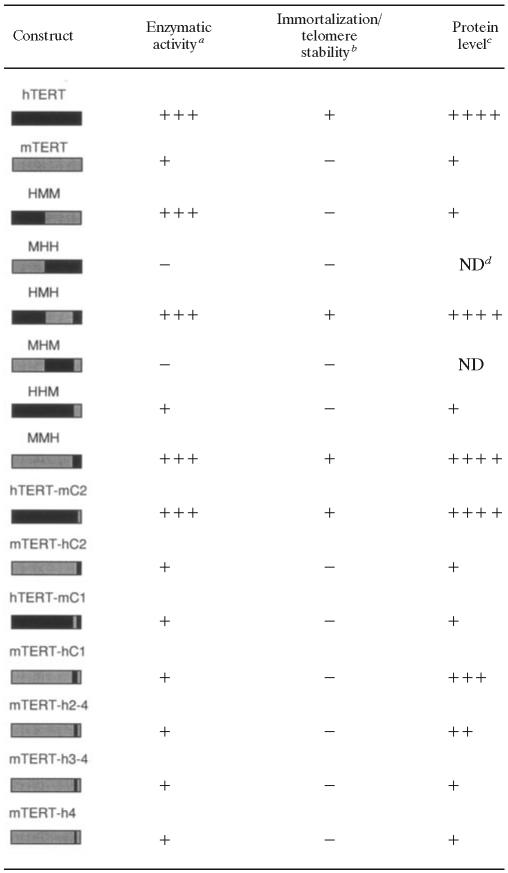
TABLE 2.
Summary of activities of TERT mutants
Enzymatic activity was determined by TRAP.
Immortalization of BJT cells and telomere stability were determined by metaphase analysis. +, allowed immortalization and telomere maintenance; −, cells entered crisis, and chromosomes showed end-to-end fusions.
Protein analysis was determined by Western blotting.
ND, not determined.
The TERT C terminus controls steady-state protein levels.
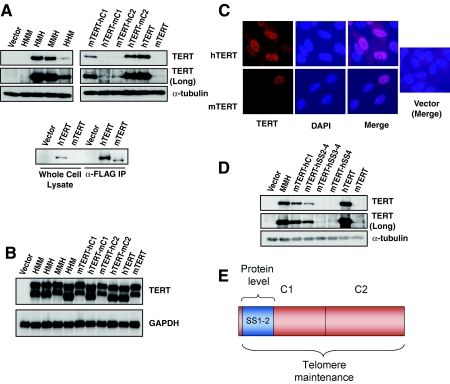
To understand the regulation of our TERT mutants at the protein level, each mutant was tagged with an immunoglobulin-binding repeat from SPA, facilitating sensitive and quantitative detection by Western blotting (14). SPA-tagged TERT proteins were expressed in BJ cells via retroviral transduction, and total cell protein extracts were analyzed by Western blotting. Surprisingly, whereas SPA-hTERT was readily detected by Western blotting, SPA-mTERT protein was expressed at much lower levels (Fig. 6A). This difference in protein level was also seen with FLAG-hTERT and FLAG-mTERT, indicating that the lower levels of mTERT protein are independent of the specific tag used. In this case, immunoprecipitation with anti-FLAG antibody agarose followed by α-FLAG Western blotting was required to detect FLAG-mTERT protein (Fig. 6A). To determine whether the difference in steady-state mTERT and hTERT protein levels could be due to variation in mRNA levels, RNA was collected from these cells for Northern blot analysis. Using a probe to the SPA tag sequences, we found no difference in the levels of retroviral transcripts for mTERT and hTERT (Fig. 6B). Furthermore, indirect immunofluorescence revealed that both hTERT and mTERT proteins were predominantly expressed in the nucleus, therefore the different activities of the TERT orthologues is unlikely to be due to altered compartment of expression (Fig. 6C). These findings indicate that expressing mTERT and hTERT from the same retroviral promoter results in identical mRNA transcript levels but a pronounced difference in steady-state protein levels.
FIG. 6.
hTERT accumulates to markedly higher protein levels than those of mTERT and these differences in protein level are controlled by SS1-2 sequences within the C1 domain. (A) Sequences regulating differences in mTERT and hTERT protein levels localize to the C1 domain. Western analysis detecting SPA-tagged TERT proteins HMM, HMH, MMH, HHM, mTERT-hC1, hTERT-mC1, mTERT-hC2, hTERT-mC2, hTERT, and mTERT (long, extended exposure time; α-tubulin, loading control) is shown. Differences in protein levels between mTERT and hTERT were also seen with FLAG-tagged TERTs (bottom panel). Protein analysis of FLAG-TERTs in total cell lysates (left) and α-FLAG immunoprecipitates (IP) (right) by FLAG Western blotting. (B) Differences in steady-state protein levels are not due to variation in RNA expression. Data shown are results of Northern blot analysis of SPA-TERT cells in panel A (TERT, SPA probe; GAPDH, loading control). (C) Nuclear localization of SPA-TERT in BJ cells. Indirect immunofluorescence analysis on vector-, SPA-hTERT-, and SPA-mTERT-transduced BJ cells (DAPI, nuclear counterstain; merge, DAPI and TERT signals combined). (D) Human SS1-2 sequences control steady-state protein levels. Western blot analysis of SPA-tagged chimeric proteins mTERT-hSS2-4, mTERT-hSS3-4 and mTERT-hSS4 (vector, MMH, mTERT-hC1, hTERT, and mTERT for comparison; long, extended exposure time; α-tubulin, loading control) is shown. (E) Model of TERT C-terminal domain. Sequences required for telomere maintenance are encoded in both C1 and C2 regions (red). Amino acids controlling protein accumulation are embedded within the C1 region (SS1-2, blue). These residues controlling protein level are separable from the larger domain regulating telomere maintenance.
Given this unexpected and marked dissociation between transcript and protein levels, we analyzed each of our tagged TERT mutants in order to map a domain that was controlling TERT protein level. Each SPA-tagged TERT construct was stably expressed in BJ cells via retroviral transduction. SPA-HMM was expressed at low levels, similar to those of SPA-mTERT, indicating that when transferred to mTERT, the N-terminal domain of hTERT does not lead to increased protein levels (Fig. 6A). SPA-HMH accumulated to levels similar to those of SPA-hTERT, suggesting that sequences in the RT domain are not responsible for the low levels of mTERT protein (Fig. 6A). To determine whether the remaining domain, the C terminus, dictates protein levels, we analyzed SPA-HHM and SPA-MMH. SPA-HHM protein was expressed at mTERT-like levels, whereas SPA-MMH accumulated to levels similar to those of hTERT (Fig. 6A). Similar results were obtained in transformed human embryonic kidney cells (293T) and in mouse embryonic fibroblasts, indicating that the difference in protein levels is intrinsic to the mTERT and hTERT proteins and not dependent on cell type (data not shown). These results show that the marked difference in steady-state protein levels between mouse and human TERT orthologues is due to sequence variation in the C-terminal domain.
Sequences within the SS1-2 regions of C1 dictate TERT protein levels.
To further delineate the region in the C terminus of hTERT that controls protein levels, we analyzed SPA-tagged versions of the C1 and C2 region mutants. An exchange of the C1 region between mTERT and hTERT dramatically altered protein levels. Protein levels of hTERT-mC1 were significantly reduced, and conversely, mTERT-hC1 protein levels were markedly increased (Fig. 6A). In contrast, substitution of the C2 sequences had no effect on protein levels. The levels of neither hTERT-mC2 nor mTERT-hC2 were affected by the exchange of the C2 regions (Fig. 6A). The results of Northern blot analysis of mRNA transcripts from BJ cells that were stably transfected with each retroviral construct showed that each chimeric cDNA was expressed equally at the mRNA level, supporting the conclusion that the different protein levels are intrinsic properties of each mutant protein (Fig. 6B). These data demonstrate that sequences in the C1 region control steady-state TERT protein levels in both species.
To delineate the sequences responsible for increased steady-state protein levels, the C1 domain was divided into four regions, denoted SS1 to -4, each approximately 14 amino acids in length (see Fig. S2 in the supplemental material). New chimeric proteins were constructed in which the human steady-state (hSS) sequences were substituted for the endogenous mouse sequences in mTERT, yielding mTERT-hSS2-4, mTERT-hSS3-4, and mTERT-hSS4 chimeric proteins. SPA-tagged mTERT-hSS2-4 demonstrated markedly increased protein levels compared to those of mTERT but slightly reduced levels compared to those of mTERT-hC1, which contains the entire C1 region from hTERT. However, replacing the human SS2 sequence with the mouse SS2 sequence resulted in a dramatic reduction in protein levels, comparable to those of wild-type mTERT (Fig. 6D, mTERT-hSS3-4 and mTERT-hSS4 lanes). Together, these data identify the SS1 and SS2 sequences within the C1 domain as those controlling TERT protein level.
All of the mutations that reduced hTERT protein levels abrogated immortalization, suggesting that these elevated levels are required for telomere maintenance. Interestingly, both the mTERT-hC1 mutant and the mTERT-hSS2-4 mutant were expressed at markedly higher levels than mTERT, and yet neither immortalized human fibroblasts or prevented telomere uncapping (Fig. 5, Table 1, and data not shown). These data indicate that the protein level determinants and the sequences allowing immortalization are separable. Furthermore, these data show that elevated TERT protein levels may be necessary but are not sufficient for immortalization. In addition to controlling protein level, the C-terminal domain serves a critical and species-specific function in enabling an assembled, enzymatically active telomerase complex to productively act on or to protect telomere ends.
DISCUSSION
Although the protein sequences that are required for TERT's enzymatic function have been well delineated, less is known regarding protein domains that mediate telomere maintenance in human cells. Through comparison of mouse and human TERT orthologues, we have identified the C-terminal domain (a region whose function is poorly understood) as a complex regulatory domain that is controlling telomere maintenance and TERT protein levels. We find that the sequence determinants that are regulating protein levels are separable from the larger domain that is required for telomere maintenance and stability (Table 2). These data identify the C terminus as a critical regulatory domain controlling TERT action in vivo and TERT posttranscriptional regulation.
Functional analysis of chimeric mouse-human TERT proteins revealed that the inability of mTERT to synthesize telomere repeats in human cells is not due to defects in the N terminus or RT domains, which comprise the TERC-binding region and the active site of the enzyme, respectively. The MMH mutant effectively immortalized human fibroblasts, indicating that the mouse N terminus and RT domains function appropriately in human cells. Furthermore, these results show that the N-DAT domain in mTERT, which is 82% similar to the human DAT region, functions properly in human cells (2). The N-DAT domain has been proposed to mediate telomerase access to telomeric chromatin (3, 4) or to contribute to interactions between telomerase and telomere DNA repeats (40, 52). Instead, our data identify the C terminus as a critical regulatory domain controlling the ability of TERT to stabilize telomeres in cells and to extend replicative lifespan.
We found that sequences in the human C-terminal domain are both necessary and sufficient for telomere maintenance. The substitution of the human C terminus for the mouse sequences in mTERT resulted in immortalization (MMH mutant); conversely, replacing the human C terminus with the mouse domain prevented immortalization (HHM mutant) (Fig. 3B). The C1 sequences of hTERT are absolutely essential for immortalization of human cells because the substitution of the C1 region from mTERT into hTERT prevented immortalization. However, the human C1 sequences were not sufficient to allow telomere maintenance in the context of mTERT protein (mTERT-hC1) (Fig. 5B). Instead, both human C1 and C2 regions were required in the mTERT protein (MMH). The stricter requirement for both human C1 and C2 regions in mTERT likely reflects subtle cooperative effects between the C-terminal domain and more N-terminal sequences that render the hTERT protein more tolerant of substitution with the mouse C2 sequences. Together, these results highlight the importance of both the C1 and C2 regions within the C terminus in allowing immortalization of human fibroblasts.
Surprisingly, we found that hTERT protein accumulates to a markedly elevated level compared to that of mTERT protein and that this difference is attributed entirely to the C-terminal domain. Within this domain, the C1 sequences are responsible for regulating TERT protein accumulation because exchanging these sequences between mTERT and hTERT reversed their relative levels. A detailed mutational analysis of the C1 region localized the region that regulated TERT protein level to the SS1 and SS2 sequences (Fig. 6A and D). The elevated protein levels that are characteristic of hTERT are likely necessary for immortalization because replacing the C1 region in hTERT with mTERT abrogated telomere maintenance and immortalization. Importantly, the substitution of the stabilizing residues, including either the entire C1 domain or SS2-4, from hTERT into mTERT dramatically increased protein levels but did not result in immortalization, indicating that elevated TERT protein levels are not sufficient to prevent crisis. These results clearly show that the sequences that control protein accumulation and telomere maintenance in the C terminus are separable. We therefore propose that sequences that regulate TERT protein level are embedded in a larger domain that is required for immortalization and telomere maintenance (Fig. 6E).
How does this larger C-terminal domain so profoundly influence TERT's ability to immortalize human cells? To address this question, we analyzed telomere lengths in MMH-expressing cells by Q-FISH. Telomeres in MMH cells at population doubling 150 were maintained at a very short set point and in a narrow range (Fig. 4). The fact that the vast majority of chromosome ends showed detectable telomere repeats, coupled with the stabilization of telomere lengths seen by Southern blot analysis, indicates that the MMH mutant synthesizes telomeres repeats. These data show that the C terminus facilitates immortalization in part by enabling TERT to synthesize telomere repeats in vivo. However, telomeres were undetectable at a subpopulation of chromosome ends (signal-free ends). Metaphases from MMH cells exhibited 3.0 SFEs per metaphase at pd 150, yet these cultures never entered crisis and chromosomal fusions remained suppressed. We cannot determine precisely the length of the SFE telomeres; however, based upon the known sensitivity of Q-FISH, we estimate that these telomeres are less than 150 nucleotides. Although the more abundant SFEs in the empty vector-transduced cells resulted in crisis and chromosomal fusions, the SFEs in the MMH cultures remained stable. One possibility for the stability of the SFEs and other short telomeres in the MMH cultures is that these telomeres, while short, retain sufficient repeats to allow T-loop formation and interaction with the shelterin protein complex (22). However, another nonmutually exclusive possibility is that telomerase is physically capping or enhancing telomere stability through direct association with the telomere, as seen in experiments with both yeast (58, 61) and human cells (46, 68). Together, these data support a model in which the TERT C terminus is required to enable telomerase to productively act on and/or stabilize telomere ends.
The TERT C terminus may facilitate immortalization by direct interaction with other molecules that are required for productive telomere engagement. The inability of the mouse C terminus to facilitate telomere maintenance in human cells could therefore reflect evolutionary divergence in both the C-terminal domain and its molecular targets such that these important interactions are maintained in each species. The compensatory evolutionary changes in TERT's molecular targets might therefore prevent the mouse domains from functioning fully in human cells. In the future, the identification of the molecules that regulate TERT and that mediate its ability to act on telomere ends in vivo will allow testing of this hypothesis and will further our understanding of TERT function in normal progenitor cells and in human cancer.
These findings provide insight into results from earlier studies showing that the addition of heterologous amino acids to the C terminus dominantly interfered with immortalization and telomere synthesis (19, 68). These heterologous C-terminal amino acids likely alter the function of the C-terminal domain in telomere maintenance or in regulating TERT protein levels. A systematic mutational analysis of the C terminus of hTERT showed that most of the domain, including the C1 region, was essential for enzymatic activity in vitro. This requirement for the C1 sequences in enzymatic function explains why the C1 region was not identified as a domain that was specifically required for telomere maintenance in that mutational study (7). That study identified the final six amino acids of the C terminus of hTERT as a C-DAT domain, one required for immortalization in vivo but not for TRAP activity in vitro (7). However, this C-DAT region does not account for the different activities of the mouse and human C-terminal domains for at least two reasons. First, the mouse sequence in the C-DAT region (FQTILD) differs from the human sequence (FKTILD) at only the second position, which was shown to be tolerant of single-amino-acid substitutions in the immortalization assay. Second, the principal sequences in the hTERT C terminus that confer telomere maintenance reside in the C1 region, which is nonoverlapping with the C-DAT sequences. Therefore, the C-terminal sequences identified here comprise a novel domain in TERT that is required for telomere maintenance and immortalization in human cells.
Our results showing that the SS1-2 sequences embedded within the C terminus control TERT protein accumulation suggest the possibility that regulation of TERT protein level is coupled to telomere synthesis. Because mRNA levels for all our TERT mutants were indistinguishable, the SS1-2 region must specify protein levels either by affecting the rate of translation or by altering the rate of protein degradation. Consistent with a role for the C-terminal domain in protein degradation, the C terminus of hTERT was recently shown to bind E3 ubiquitin-ligase MKRN1. MKRN1 ubiquitinated hTERT in vitro, and the overexpression of MKRN1 in human cancer cells reduced telomerase activity and led to telomere shortening (35). In the future, it will be important to determine which pathways regulate endogenous TERT protein in both mouse and human cells and to identify the molecular targets of the C terminus that mediate telomere synthesis and stability.
Supplementary Material
Acknowledgments
We thank J. Shay for the hTERT cDNA, R. Weinberg for the BJT fibroblasts, K. Collins for the hTERC expression plasmids, and P. Lansdorp for the TFL-Telo V.2.1 software. We thank L. Attardi, J. Sage, and members of the Artandi laboratory for critical reading of the manuscript.
E.J.M. was supported by PHS training grant CA09302 from the NCI. J.C. is supported by a Samsung Fellowship. A.S.V. was supported by Medical Scientist Training Program grant GM07365. This work was supported by NIH RO1 CA111691, CA109088, KO8 CA082176, the Rita Allen Foundation and the V Foundation.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Arai, K., K. Masutomi, S. Khurts, S. Kaneko, K. Kobayashi, and S. Murakami. 2002. Two independent regions of human telomerase reverse transcriptase are important for its oligomerization and telomerase activity. J. Biol. Chem. 277:8538-8544. [DOI] [PubMed] [Google Scholar]
- 2.Armbruster, B. N., S. S. Banik, C. Guo, A. C. Smith, and C. M. Counter. 2001. N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol. Cell. Biol. 21:7775-7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armbruster, B. N., K. T. Etheridge, D. Broccoli, and C. M. Counter. 2003. Putative telomere-recruiting domain in the catalytic subunit of human telomerase. Mol. Cell. Biol. 23:3237-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armbruster, B. N., C. M. Linardic, T. Veldman, N. P. Bansal, D. L. Downie, and C. M. Counter. 2004. Rescue of an hTERT mutant defective in telomere elongation by fusion with hPot1. Mol. Cell. Biol. 24:3552-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachand, F., and C. Autexier. 2001. Functional regions of human telomerase reverse transcriptase and human telomerase RNA required for telomerase activity and RNA-protein interactions. Mol. Cell. Biol. 21:1888-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachand, F., I. Triki, and C. Autexier. 2001. Human telomerase RNA-protein interactions. Nucleic Acids Res. 29:3385-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banik, S. S., C. Guo, A. C. Smith, S. S. Margolis, D. A. Richardson, C. A. Tirado, and C. M. Counter. 2002. C-terminal regions of the human telomerase catalytic subunit essential for in vivo enzyme activity. Mol. Cell. Biol. 22:6234-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beattie, T. L., W. Zhou, M. O. Robinson, and L. Harrington. 2001. Functional multimerization of the human telomerase reverse transcriptase. Mol. Cell. Biol. 21:6151-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beattie, T. L., W. Zhou, M. O. Robinson, and L. Harrington. 2000. Polymerization defects within human telomerase are distinct from telomerase RNA and TEP1 binding. Mol. Biol. Cell 11:3329-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beattie, T. L., W. Zhou, M. O. Robinson, and L. Harrington. 1998. Reconstitution of human telomerase activity in vitro. Curr. Biol. 8:177-180. [DOI] [PubMed] [Google Scholar]
- 11.Blackburn, E. H. 2001. Switching and signaling at the telomere. Cell 106:661-673. [DOI] [PubMed] [Google Scholar]
- 12.Blackburn, E. H. 2000. Telomere states and cell fates. Nature 408:53-56. [DOI] [PubMed] [Google Scholar]
- 13.Bodnar, A. G., M. Ouellette, M. Frolkis, S. E. Holt, C. P. Chiu, G. B. Morin, C. B. Harley, J. W. Shay, S. Lichsteiner, and W. E. Wright. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279:349-352. [DOI] [PubMed] [Google Scholar]
- 14.Borggrefe, T., R. Davis, A. Bareket-Samish, and R. D. Kornberg. 2001. Quantitation of the RNA polymerase II transcription machinery in yeast. J. Biol. Chem. 276:47150-47153. [DOI] [PubMed] [Google Scholar]
- 15.Chen, J. L., and C. W. Greider. 2003. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J. 22:304-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chin, L., S. E. Artandi, Q. Shen, A. Tam, S. L. Lee, G. J. Gottlieb, C. W. Greider, and R. A. DePinho. 1999. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell 97:527-538. [DOI] [PubMed] [Google Scholar]
- 17.Collins, K., and J. R. Mitchell. 2002. Telomerase in the human organism. Oncogene 21:564-579. [DOI] [PubMed] [Google Scholar]
- 18.Counter, C. M., A. A. Avilion, C. E. LeFeuvre, N. G. Stewart, C. W. Greider, C. B. Harley, and S. Bacchetti. 1992. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 11:1921-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Counter, C. M., W. C. Hahn, W. Wei, S. D. Caddle, R. L. Beijersbergen, P. M. Lansdorp, J. M. Sedivy, and R. A. Weinberg. 1998. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc. Natl. Acad. Sci. USA 95:14723-14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.d'Adda di Fagagna, F., P. M. Reaper, L. Clay-Farrace, H. Fiegler, P. Carr, T. Von Zglinicki, G. Saretzki, N. P. Carter, and S. P. Jackson. 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature 426:194-198. [DOI] [PubMed] [Google Scholar]
- 21.de Lange, T. 2002. Protection of mammalian telomeres. Oncogene 21:532-540. [DOI] [PubMed] [Google Scholar]
- 22.de Lange, T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19:2100-2110. [DOI] [PubMed] [Google Scholar]
- 23.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286:117-120. [DOI] [PubMed] [Google Scholar]
- 24.Forsyth, N. R., W. E. Wright, and J. W. Shay. 2002. Telomerase and differentiation in multicellular organisms: turn it off, turn it on, and turn it off again. Differentiation 69:188-197. [DOI] [PubMed] [Google Scholar]
- 25.Friedman, K. L., and T. R. Cech. 1999. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 13:2863-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greider, C. W., and E. H. Blackburn. 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337:331-337. [DOI] [PubMed] [Google Scholar]
- 27.Hara, E., H. Tsurui, A. Shinozaki, S. Nakada, and K. Oda. 1991. Cooperative effect of antisense-Rb and antisense-p53 oligomers on the extension of life span in human diploid fibroblasts, TIG-1. Biochem. Biophys. Res. Commun. 179:528-534. [DOI] [PubMed] [Google Scholar]
- 28.Harley, C. B., A. B. Futcher, and C. W. Greider. 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345:458-460. [DOI] [PubMed] [Google Scholar]
- 29.Harrington, L. 1997. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 11:3109-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hastie, N. D., M. Dempster, M. G. Dunlop, A. M. Thompson, D. K. Green, and R. C. Allshire. 1990. Telomere reduction in human colorectal carcinoma and with ageing. Nature 346:866-868. [DOI] [PubMed] [Google Scholar]
- 31.Hossain, S., S. Singh, and N. F. Lue. 2002. Functional analysis of the C-terminal extension of telomerase reverse transcriptase. A putative “thumb” domain. J. Biol. Chem. 277:36174-36180. [DOI] [PubMed] [Google Scholar]
- 32.Huard, S., T. J. Moriarty, and C. Autexier. 2003. The C terminus of the human telomerase reverse transcriptase is a determinant of enzyme processivity. Nucleic Acids Res. 31:4059-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji, H., M. H. Platts, L. M. Dharamsi, and K. L. Friedman. 2005. Regulation of telomere length by an N-terminal region of the yeast telomerase reverse transcriptase. Mol. Cell. Biol. 25:9103-9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelleher, C., M. T. Teixeira, K. Forstemann, and J. Lingner. 2002. Telomerase: biochemical considerations for enzyme and substrate. Trends Biochem. Sci. 27:572-579. [DOI] [PubMed] [Google Scholar]
- 35.Kim, J. H., S. M. Park, M. R. Kang, S. Y. Oh, T. H. Lee, M. T. Muller, and I. K. Chung. 2005. Ubiquitin ligase MKRN1 modulates telomere length homeostasis through a proteolysis of hTERT. Genes Dev. 19:776-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, N. W., M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, P. L. Ho, G. M. Coviello, W. E. Wright, S. L. Weinrich, and J. W. Shay. 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 266:2011-2015. [DOI] [PubMed] [Google Scholar]
- 37.Kolquist, K. A., L. W. Ellisen, C. M. Counter, M. Meyerson, L. K. Tan, R. A. Weinberg, D. A. Haber, and W. L. Gerald. 1998. Expression of TERT in early premalignant lesions and a subset of cells in normal tissues. Nat. Genet. 19:182-186. [DOI] [PubMed] [Google Scholar]
- 38.Lai, C. K., J. R. Mitchell, and K. Collins. 2001. RNA binding domain of telomerase reverse transcriptase. Mol. Cell. Biol. 21:990-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lansdorp, P. M., N. P. Verwoerd, F. M. van de Rijke, V. Dragowska, M. T. Little, R. W. Dirks, A. K. Raap, and H. J. Tanke. 1996. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 5:685-691. [DOI] [PubMed] [Google Scholar]
- 40.Lee, S. R., J. M. Wong, and K. Collins. 2003. Human telomerase reverse transcriptase motifs required for elongation of a telomeric substrate. J. Biol. Chem. 278:52531-52536. [DOI] [PubMed] [Google Scholar]
- 41.Lindsey, J., N. I. McGill, L. A. Lindsey, D. K. Green, and H. J. Cooke. 1991. In vivo loss of telomeric repeats with age in humans. Mutat. Res. 256:45-48. [DOI] [PubMed] [Google Scholar]
- 42.Lingner, J., T. R. Hughes, A. Shevchenko, M. Mann, V. Lundblad, and T. R. Cech. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276:561-567. [DOI] [PubMed] [Google Scholar]
- 43.Lundblad, V. 2003. Telomere replication: an Est fest. Curr. Biol. 13:R439-R441. [DOI] [PubMed] [Google Scholar]
- 44.Lundblad, V., and J. W. Szostak. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57:633-643. [DOI] [PubMed] [Google Scholar]
- 45.Ly, H., L. Xu, M. A. Rivera, T. G. Parslow, and E. H. Blackburn. 2003. A role for a novel ‘trans-pseudoknot’ RNA-RNA interaction in the functional dimerization of human telomerase. Genes Dev. 17:1078-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masutomi, K., E. Y. Yu, S. Khurts, I. Ben-Porath, J. L. Currier, G. B. Metz, M. W. Brooks, S. Kaneko, S. Murakami, J. A. DeCaprio, R. A. Weinberg, S. A. Stewart, and W. C. Hahn. 2003. Telomerase maintains telomere structure in normal human cells. Cell 114:241-253. [DOI] [PubMed] [Google Scholar]
- 47.Meyerson, M., C. M. Counter, E. N. Eaton, L. W. Ellisen, P. Steiner, S. D. Caddle, L. Ziaugra, R. L. Beijersbergen, M. J. Davidoff, Q. Liu, S. Bacchetti, D. A. Haber, and R. A. Weinberg. 1997. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 90:785-795. [DOI] [PubMed] [Google Scholar]
- 48.Mitchell, J. R., J. Cheng, and K. Collins. 1999. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol. 19:567-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitchell, J. R., and K. Collins. 2000. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol. Cell 6:361-371. [DOI] [PubMed] [Google Scholar]
- 50.Moriarty, T. J., S. Huard, S. Dupuis, and C. Autexier. 2002. Functional multimerization of human telomerase requires an RNA interaction domain in the N terminus of the catalytic subunit. Mol. Cell. Biol. 22:1253-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moriarty, T. J., D. T. Marie-Egyptienne, and C. Autexier. 2004. Functional organization of repeat addition processivity and DNA synthesis determinants in the human telomerase multimer. Mol. Cell. Biol. 24:3720-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moriarty, T. J., R. J. Ward, M. A. Taboski, and C. Autexier. 2005. An anchor site-type defect in human telomerase that disrupts telomere length maintenance and cellular immortalization. Mol. Biol. Cell 16:3152-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morrison, S. J., K. R. Prowse, P. Ho, and I. L. Weissman. 1996. Telomerase activity in hematopoietic cells is associated with self-renewal potential. Immunity 5:207-216. [DOI] [PubMed] [Google Scholar]
- 54.Nakamura, T. M., G. B. Morin, K. B. Chapman, S. L. Weinrich, W. H. Andrews, J. Lingner, C. B. Harley, and T. R. Cech. 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277:955-959. [DOI] [PubMed] [Google Scholar]
- 55.O'Connor, C. M., C. K. Lai, and K. Collins. 2005. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J. Biol. Chem. 280:17533-17539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng, Y., I. S. Mian, and N. F. Lue. 2001. Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol. Cell 7:1201-1211. [DOI] [PubMed] [Google Scholar]
- 57.Prescott, J., and E. H. Blackburn. 1997. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 11:2790-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prescott, J., and E. H. Blackburn. 1997. Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev. 11:528-540. [DOI] [PubMed] [Google Scholar]
- 59.Reichenbach, P., M. Hoss, C. M. Azzalin, M. Nabholz, P. Bucher, and J. Lingner. 2003. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr. Biol. 13:568-574. [DOI] [PubMed] [Google Scholar]
- 60.Rivera, M. A., and E. H. Blackburn. 2004. Processive utilization of the human telomerase template: lack of a requirement for template switching. J. Biol. Chem. 279:53770-53781. [DOI] [PubMed] [Google Scholar]
- 61.Roy, J., T. B. Fulton, and E. H. Blackburn. 1998. Specific telomerase RNA residues distant from the template are essential for telomerase function. Genes Dev. 12:3286-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shay, J. W., O. M. Pereira-Smith, and W. E. Wright. 1991. A role for both RB and p53 in the regulation of human cellular senescence. Exp. Cell Res. 196:33-39. [DOI] [PubMed] [Google Scholar]
- 63.Snow, B. E., N. Erdmann, J. Cruickshank, H. Goldman, R. M. Gill, M. O. Robinson, and L. Harrington. 2003. Functional conservation of the telomerase protein Est1p in humans. Curr. Biol. 13:698-704. [DOI] [PubMed] [Google Scholar]
- 64.Taggart, A. K., S. C. Teng, and V. A. Zakian. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297:1023-1026. [DOI] [PubMed] [Google Scholar]
- 65.Weinrich, S. L., R. Pruzan, L. Ma, M. Ouellette, V. M. Tesmer, S. E. Holt, A. G. Bodnar, S. Lichsteiner, N. W. Kim, J. B. Tragner, R. D. Taylor, R. Carlos, W. H. Andrews, W. E. Wright, J. W. Shay, C. B. Harley, and G. B. Morin. 1997. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 17:498-502. [DOI] [PubMed] [Google Scholar]
- 66.Wenz, C., B. Enenkel, M. Amacker, C. Kelleher, K. Damm, and J. Lingner. 2001. Human telomerase contains two cooperating telomerase RNA molecules. EMBO J. 20:3526-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xia, J., Y. Peng, I. S. Mian, and N. F. Lue. 2000. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol. Cell. Biol. 20:5196-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu, J., H. Wang, J. M. Bishop, and E. H. Blackburn. 1999. Telomerase extends the lifespan of virus-transformed human cells without net telomere lengthening. Proc. Natl. Acad. Sci. USA 96:3723-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zijlmans, J. M., U. M. Martens, S. S. Poon, A. K. Raap, H. J. Tanke, R. K. Ward, and P. M. Lansdorp. 1997. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl. Acad. Sci. USA 94:7423-7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.