Abstract
The mitogen-activated protein kinase (MAPK) p38/MAPK-activated protein kinase 2 (MK2) signaling pathway plays an important role in the posttranscriptional regulation of tumor necrosis factor (TNF), which is dependent on the adenine/uridine-rich element (ARE) in the 3′ untranslated region of TNF mRNA. After lipopolysaccharide (LPS) stimulation, MK2-deficient macrophages show a 90% reduction in TNF production compared to the wild type. Tristetraprolin (TTP), a protein induced by LPS, binds ARE and destabilizes TNF mRNA. Accordingly, macrophages lacking TTP produce large amounts of TNF. Here, we generated MK2/TTP double knockout mice and show that, after LPS stimulation, bone marrow-derived macrophages produce TNF mRNA and protein levels comparable to those of TTP knockout cells, indicating that in the regulation of TNF biosynthesis TTP is genetically downstream of MK2. In addition, we show that MK2 is essential for the stabilization of TTP mRNA, and phosphorylation by MK2 leads to increased TTP protein stability but reduced ARE affinity. These data suggest that MK2 inhibits the mRNA destabilizing activity of TTP and, in parallel, codegradation of TTP together, with the target mRNA resulting in increased cellular levels of TTP.
The p38 mitogen-activated protein kinase (MAPK) pathway is activated by stress and inflammatory stimuli. One of the major and best understood functions of this module is posttranscriptional regulation of mRNAs containing adenylate/uridylate-rich elements (AREs) in their 3′ untranslated region (3′UTR) (13, 14, 30, 31). AREs that contain the pentamer AUUUA are wide spread, and a computational estimation suggests that as much as 8% of human genes contain functional AREs, making a total of 2,800 to 9,600 ARE-containing transcripts (1, 2). Experimentally, the p38 pathway has been implicated in the regulation of mRNA stability of cyclooxygenase 2, tumor necrosis factor alpha (TNF-α), interleukin-3 (IL-3), IL-6, IL-8, macrophage inhibitory protein 1α, granulocyte-macrophage colony-stimulating factor (GM-CSF), vascular endothelial growth factor, urokinase-type plasminogen activator, and inducible NO synthase (13-15, 30, 31).
MAPK-activated protein kinase 2 (MK2) is one of several downstream targets of p38. The generation and analysis of MK2-deficient mice demonstrated that MK2 is the major kinase downstream of p38 responsible for posttranscriptional regulation of cytokine biosynthesis. Spleen cells of MK2-deficient mice produced levels of TNF, IL-6, and gamma interferon that were 10 to 20% of those of wild type (WT) cells after lipopolysaccharide (LPS) stimulation (20). The effect of MK2 on TNF biosynthesis is directly dependent on the ARE in the 3′UTR of TNF mRNA (28).
The zinc finger protein tristetraprolin (TTP), also known as Nup 475, TIS11, G0S24, or ZFP36, mediates TNF mRNA instability in macrophages by directly binding to its ARE. TTP-deficient mice have a systemic inflammatory syndrome with severe polyarticular arthritis and autoimmunity, as well as medullary and extramedullary myeloid hyperplasia (11). The syndrome is predominantly due to excess circulating TNF, resulting from the increased stability of the TNF mRNA and subsequent higher rates of secretion of the cytokine. The myeloid hyperplasia might be due in part to increased stability of granulocyte-macrophage colony-stimulating factor (GM-CSF). TTP controls its expression by binding to the 3′UTR of its own mRNA, which also contains AREs (6, 34). Following stimulation of macrophages by LPS, TTP is increasingly expressed in multiple, differentially phosphorylated isoforms. While increased expression of TTP is dependent on the activation of the p38 pathway (9, 27), TTP is a direct substrate for MK2 in vitro and in vivo, and the mouse protein is phosphorylated at two major sites: serine 52 (S52) and S178 (12). Furthermore, TTP can be directly phosphorylated by p38 (10, 35). TTP is recruited to stress granules (SGs) that are dynamic cytoplasmic foci at which stalled translation initiation complexes accumulate in cells subjected to environmental stress (19). Phosphorylation of TTP by MK2 at S52 and S178 excludes TTP from arsenite-induced SGs and promotes the assembly of TTP-14-3-3 complexes, leading to the inhibition of TTP-dependent degradation of ARE-containing transcripts (32).
The mechanism by which TTP regulates specific ARE-dependent mRNA degradation is not completely understood so far. In a cell-free system, TTP stimulates deadenylation of ARE-containing transcripts (22). Recently, in TTP and its homolog BRF-1, two domains which are necessary for its activity, an N-terminal domain and a C-terminal domain, have been identified (26). While the N-terminal domain was shown to recruit mRNA decay enzymes, the C-terminal domain obviously binds other trans-acting factors. Interestingly, TTP could also be involved in a microRNA16-mediated mechanism of ARE-dependent mRNA degradation and directly interact with components of the RISC complex but not with the microRNA itself (17).
Here we provide genetic evidence that TTP is the major target of MK2 involved in the posttranscriptional regulation of TNF. The removal of TTP from an MK2-deficient background completely reverses the low-TNF phenotype of MK2 knockout, TNF mRNA becomes stable, and high TNF levels are produced, leading to a phenotype similar to that of TTP deficiency alone. We demonstrate that MK2 is required for maintenance of normal TTP levels, and phosphorylation by MK2 leads to stabilization of TTP and also to inhibition of its mRNA destabilization activity.
MATERIALS AND METHODS
Cell lines, vectors, and materials.
WT and MK2−/− macrophage cell lines were produced as previously described (3). The expression of F4/80, Mac3, Gr-1,B7.1, B7.2, and FcR was tested by fluorescence-activated cell sorting. The MK2 deficiency in MK2−/− cells was confirmed by reverse transcription-PCR. TNF and IL-6 production were about 10% of WT in MK2−/−, similar to what was previously described for primary cells (20). The AU-rich element of the 3′UTR of TNF was cloned by the annealing of primers 5′CTA GAT ATT TAT ATT TGC ACT TAT TAT TTA TTA TTT ATT TAT TAT TTA TTT ATT TGC TTA TGA ATG TAT TTA TTT G3′ and 5′GAT CCA AAT AAA TAC ATT CAT AAG CAA ATA AAT AAA TAA TAA ATA AAT AAT AAA TAA TAA GTG CAA ATA TAA ATA T3′ and cloning into pBluescript KS+ BamHI and XbaI sites. Vectors pcDNA3.1-mycHisA, pcDNA3-TTP-mycHisA, pcDNA3-TTP-S52A-mycHisA, pcDNA3-TTP-S178A-mycHisA, and pcDNA3-TTP-AA-mycHisA were provided by G. Stoecklin (32). SAK21A anti-TTP antibody was previously described (27). 9E10 anti-myc antibody was obtained from supernatant of hybridoma cell culture. Recombinant murine macrophage colony-stimulating factor (M-CSF) was from R&D systems (Minneapolis, Minn.). SB203580 and SB202190 were from Calbiochem (La Jolla, Calif.).
Generation of mutant mice.
All mice used in this study were maintained under specific-pathogen-free conditions. MK2 and TTP knockout mice were generated as described previously (20, 33). TTP+/− mice were bred to MK2−/− mice to obtain MK2+/− TTP+/− mice; F2 littermates were genotyped by PCR and used for the experiments described below.
Mouse pathology.
During mouse autopsy, all tissues were collected and fixed in 10% neutral-buffered formalin. Samples of skin, heart, lung, spleen, liver, kidney, pancreas, peripheral lymph nodes, foreleg with joints, hind leg with joints, eyes, and conjunctival membranes were routinely embedded in paraffin and stained with hematoxylin and eosin. Each organ was examined for pathological changes by light microscopy using an Olympus BX41 microscope.
Bone marrow-derived macrophage (BMDM) preparation.
Bone marrow-derived macrophages were obtained from marrow plugs flushed from mice femurs with 2.5 ml of ice-cold complete medium. Typically, material from one mouse was cultured in two 10-cm plates in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS), 10 ng of M-CSF/ml, 100 μg of streptomycin/ml, and 100 U of penicillin/ml at 37°C. After 2 and 6 days, the cells were washed and grown in fresh medium for a total of 9 to 10 days. The cells appeared to be 100% macrophages according to morphology.
Affinity purification.
Affinity purification was performed as previously described (4) with the following modifications. Linearized vector was used as template for in vitro transcription using the T7-MEGAshortscript kit (Ambion, Huntingdon, United Kingdom) according to instructions of the manufacturer, except that CTP was mixed 2:1 with biotin-14-CTP (Gibco/Life Technologies, Inchinnan, Scotland). Cells were grown to 70 to 80% confluence on 10-cm plates, washed with cold phosphate-buffered saline (PBS), and lysed by resuspension in 200 μl low-salt lysis buffer (20 mM HEPES, pH 7.5, 50 mM KCl, 2 mM MgCl2, 0.5 mM dithiothreitol [DTT], 10% protease inhibitor cocktail complete mini EDTA-free [Roche, Mannheim, Germany], phosphatase inhibitor cocktails I and II at 10% each [Sigma], and 0.2% Nonidet P-40), and incubated on ice for 10 min. The lysates were then centrifuged at 800 × g. Affinity purification was carried out in batch at 4°C. Streptavidin agarose beads (30 μl/assay; Gibco/Life Technologies, Inchinnan, Scotland) were equilibrated by three washes with binding buffer (20 mM HEPES, pH 7.5, 150 mM KCl). The RNA pellet from transcription was resuspended in binding buffer, and 10 to 20% of the transcription reaction in 20 μl was added to 30-μl beads. KCl (1 M) was added to extracts to reach a salt concentration of 160 mM. After addition of RNase inhibitor (0.4 U/μl), heparin, and tRNA (final concentrations of 5 mg/ml and 0.2 mg/ml, respectively), 60 μl of material was incubated with RNA-loaded beads for 90 min at 4°C. After extensive washing with buffer (20 mM HEPES, pH 7.5, 150 mM KCl, 2 mM MgCl2, 0.5 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride, 5% glycerol, and 0.1% Nonidet P-40), 2× sodium dodecyl sulfate (SDS) loading buffer was directly added to beads and incubated for 5 min at 95°C. After a short centrifugation, the supernatant containing the loading buffer and proteins was directly loaded on SDS-polyacrylamide gel electrophoresis (PAGE) gels.
Alkaline phosphatase treatment.
Macrophage lysates were prepared in low-salt lysis buffer as described for affinity purification. Calf intestinal alkaline phosphatase (CIP) (NEB Biolabs) was used for dephosphorylation. Twenty-six microliters of lysate in 1× NEB buffer 3 was incubated with 10 U CIP at 37°C for 15 min. Control lysates were incubated under the same conditions but without CIP.
Western blot.
For Western blot analysis, the protein concentration was measured and 4× Laemmli's sodium dodecyl sulfate (SDS) sample buffer was added (40% glycerol, 4% SDS, 4% β-mercaptoethanol, 0.4 M Tris-HCl, pH 6.7, and 2 mg/ml bromphenol blue). Samples were boiled, briefly centrifuged, and resolved by SDS-polyacrylamide gel electrophoresis (10% acrylamide), and proteins were transferred from the gels to Hybond ECL membranes (Amersham Pharmacia Biotech). Blots were incubated for 2 h in PBS-1% Tween 20 (PBST) containing 5% powdered skim milk. After three washes with PBST, membranes were incubated for 1 h with the primary antibody and for 1 h with horseradish peroxidase-conjugated secondary (2,000-fold diluted). Bound antibodies were detected with an ECL detection kit (Santa Cruz Biotechnology).
TTP protein stability experiments in macrophages.
Cells were induced with 1 μg/ml LPS for 2 h. Subsequently, SB203580 (10 μM final concentration), cycloheximide (7 μM final concentration), or SB203580 and cycloheximide together were added. Because SB203580 stock was dissolved in dimethyl sulfoxide (DMSO), in control samples lacking SB203580 equal volumes of DMSO were added. After different times, lysates were obtained by direct resuspension in 2× SDS loading buffer. Quantification of protein was done according to reference 18, and equal or, where indicated, different amounts of protein were applied to Western blots using the SAK21A anti-TTP antibody.
Green fluorescent protein (GFP)-TTP stability.
Cells were seeded at a 1:2 ratio and grown overnight in DMEM plus 10% FCS. The next day the cells were harvested by gentle scraping. A total of 2 × 106 cells/point were nucleofected with 5 μg DNA containing 500 ng pEGFPc1-TTP using a nucleofector with solution T and program T-20 (Amaxa, Cologne, Germany). Replicate transfections were combined before seeding into 6-well plates to control for differences in transfection efficiency. The cells were cultured in RPMI plus 20% FCS for 2 h and then either left untreated or stimulated with LPS (10 ng/ml) for 1.75 h. Cycloheximide (5 μg/ml) was added to the LPS-stimulated cells for 15 min, and then the unstimulated and 1× LPS-stimulated points were harvested and cytoplasmic extracts were prepared (27). The remaining cells were treated with either 0.1% DMSO or 1 μM SB202190 for 2 h and then harvested as described above. One-hundred micrograms of cytoplasmic extract was applied to Western blots using the SAK21A anti-TTP antibody.
Detection of mRNA and determination of RNA stability.
Macrophages were treated with 1 μg/ml LPS (Escherichia coli; Sigma L-6529) for the time points indicated before addition of actinomycin D (2 μg/ml) for the indicated time. Total cellular RNA was obtained from macrophage cultures using a QIAGEN mini RNA kit, typically from 80% confluent 6-cm tissue culture plates. RNA concentration was determined by spectrophotometrical measurement at 260 nm. Appropriate amounts of RNA were separated on 1.2% agarose-formaldehyde gels and blotted onto nitrocellulose membrane. The filters were probed with 33P-labeled mouse cDNA using RAPID-HYBbuffer (Amersham Biosciences).
TNF measurement.
BMDM were scratched from 10-cm plates and counted, and 3 × 104 cells were transferred to a 48-well plate and incubated in 0.5 ml medium for 2 h before addition of 1 μg/ml LPS for 4 h. TNF was measured in macrophage supernatant by enzyme-linked immunosorbent assay (Quantikine M mouse TNF immunoassay kit; R&D Systems).
Transfection of HEK293 cells.
HEK293 cells were cultured in DMEM containing 10% FCS. These cells were maintained in 5% CO2 at 37°C. Cells at about 60% confluence were transfected by the Lipofectamine method (Life Technologies, Inc.). After incubation for 12 h, the transfected cells were washed with DMEM and incubated in fresh growth medium for 24 h.
RESULTS
TTP is the main downstream element of MK2 in the posttranscriptional regulation of TNF.
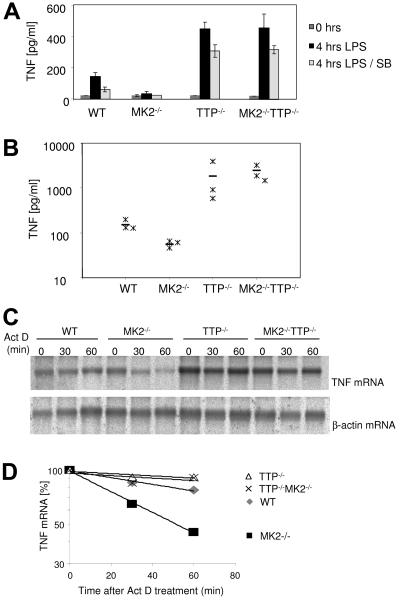
TTP+/− mice were crossed with MK2−/− mice and backcrossed with the F1 generation, leading to MK2−/− TTP−/− double knockout animals. Bone marrow-derived macrophages were isolated from these animals and stimulated with 1 μg/ml LPS, and the TNF content of culture supernatants was analyzed by enzyme-linked immunosorbent assay (ELISA) 4 h after stimulation. Remarkably, MK2 TTP double knockout macrophages show strongly increased amounts of TNF almost identical to that of TTP-deficient mice (Fig. 1A). We also analyzed TNF production in vivo after LPS challenge of mice with the different genotypes. It can be seen that TNF levels in both TTP-deficient and TTP- and MK2-deficient mice is comparably increased compared to the wild type, while the TNF level is reduced in MK2-deficient animals (Fig. 1B). Hence, TTP deficiency completely abolishes the deleterious effect of MK2 deficiency on TNF production, indicating that TTP is genetically downstream of MK2.
FIG. 1.
MK2 TTP double knockout macrophages produce TNF protein and mRNA levels comparable to those of TTP-deficient cells. (A) TNF ELISA of macrophage culture supernatants. BMDM from three mice were pooled, grown for 10 days, harvested, and counted, and equal numbers of cells were transferred to a 48-well plate. Cells were induced with LPS or left untreated as control (C) for 4 h. TNF levels in the supernatants were measured by ELISA. Where indicated, 10 μM SB203580 (SB) was added 1 h prior to LPS stimulation. For each genotype, three independent measurements were made, and averages and standard deviations are shown. The experiment is representative for four similar experiments. (B) Three mice of each genotype were injected intraperitoneally with LPS (5 mg per kg of body weight) diluted in PBS. The TNF level in serum was measured before and 90 min after injection. Before injection, serum TNF levels were below the detection limit of ELISA. Levels after 90 min for each mouse (asterisk) and mean values for each genotype (−) are indicated. (C) Bone marrow from three WT, three MK2−/−, three TTP−/−, and three MK2−/− TTP−/− mice were pooled, and macrophages were grown in 10-cm plates for 10 days. After induction for 2 h with 1 μg/ml LPS, 2 μg/ml actinomycin D (Act D) was added for 30 and 60 min. Total RNA was prepared, and 10 μg RNA was used for Northern blot analysis with a TNF probe. The same blot was reused for the β-actin control. This experiment is representative of two independently performed experiments. (D) Quantification of the Northern blot shown in panel C. The intensity of the TNF mRNA bands was quantified using TINA software (Raytest, Staubenhardt, Germany) and normalized to actin mRNA. The calculated half-lives for TNF mRNA are the following: WT, 130 min; MK2−/− TTP−/−, 307 min; TTP−/−, 350 min; and MK2−/−, 51 min.
TNF mRNA levels and stability were analyzed by stimulating macrophages for 2 h with LPS and arresting transcription by actinomycin D. The level of TNF mRNA is comparable in TTP−/− and MK2−/− TTP−/− macrophages and is higher than that in WT or MK2−/− cells (Fig. 1C). In MK2-deficient cells, the stability of TNF mRNA is lower that in the WT control, while in the TTP-deficient background the lack of MK2 does not alter TNF mRNA stability significantly (Fig. 1D). This indicates that the action of MK2 is necessary for stabilization of TNF mRNA upon LPS treatment and that the differences in TNF mRNA stability depend on the presence of TTP.
Pathology of MK2 TTP double knockout mice.
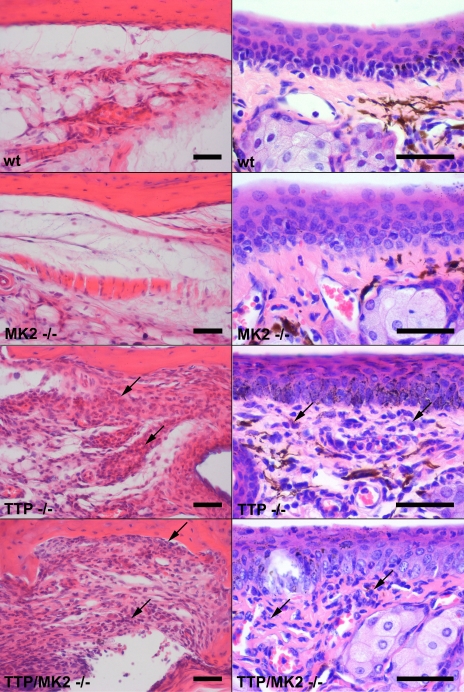
We were interested to learn whether the similarity in the increase in TNF production in TTP- and TTP- and MK2-deficient mice results in a comparable phenotype. First, like TTP knockout mice, TTP MK2 double knockout mice had reduced growth, associated with cachexia and the “Kangaroo” hunched posture which is characteristic for many inflammatory syndromes in mice. Like TTP knockout mice, TTP MK2 double knockout mice showed reddened and swollen paw joints. Light microscopic examinations revealed that both TTP knockout and TTP MK2 double knockout mice had a moderate to severe inflammatory infiltration with neutrophils and plasma cells in numerous tissues, including the periarticular soft tissue and the conjunctival membranes, while WT and MK2-deficient mice did not show such infiltrations (Fig. 2). Our observations indicated that TTP MK2 double knockout mice show a phenotype comparable to that of TTP knockout mice described previously (33).
FIG. 2.
MK2 TTP double knockout mice share the pathology of TTP knockout mice. Both the TTP knockout mice and the TTP MK2 double knockout mice had moderate to severe infiltrations with neutrophils and plasma cells (arrows) in numerous tissues, including the periarticular soft tissue (left panels) and the conjunctival membranes (right panels). WT and MK2 knockout mice showed no infiltrations. Bar, 100 μm.
TTP expression is down-regulated in MK2-deficient cells.
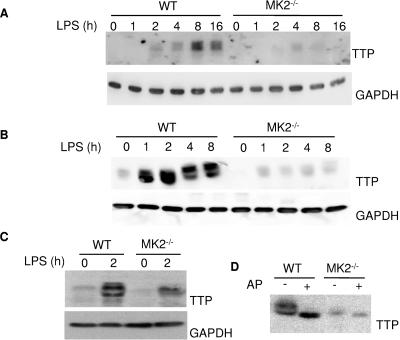
After having established the genetic relationship between TTP and MK2, we were interested in the molecular mechanism responsible for this relationship. Inhibition of p38 by SB208530 leads to a significant reduction of the LPS-induced TTP protein expression in RAW264.7 macrophages (27, 34). Since it was not clear whether this effect is MK2 dependent, we analyzed TTP levels in bone marrow-derived primary macrophages, in immortalized macrophage cell lines, and in spleen from MK2-deficient mice after LPS stimulation by Western blotting. It can be seen that protein levels of TTP are reduced in all MK2-deficient samples (Fig. 3A to C). In contrast, in WT cells, TTP was induced by LPS stimulation and expressed in multiple differentially phosphorylated forms that show different mobilities in SDS-PAGE. Treatment with alkaline phosphatase leads to the collapse of the phosphorylated bands to one, probably nonphosphorylated band (Fig. 3D). In MK2-deficient cells, alkaline phosphatase treatment leads only to a weak shift of the already weaker phosphorylated TTP population.
FIG. 3.
TTP protein expression is reduced in MK2-deficient cells. (A) TTP levels in total lysates from WT and MK2-deficient bone marrow-derived primary macrophages that were induced with 1 μg/ml LPS for the times indicated were analyzed by Western blot. (B) Total lysates from WT and MK2-deficient immortalized macrophage cell lines induced with LPS and analyzed by Western blot. (C) TTP Western blot analysis of spleen lysates from mice that were injected with 100 μg LPS for 2 h. (D) Alkaline phosphatase causes a collapse of TTP from lysate of LPS induced immortalized WT macrophages to a single band, indicating that the slower motility is due to phosphorylation.
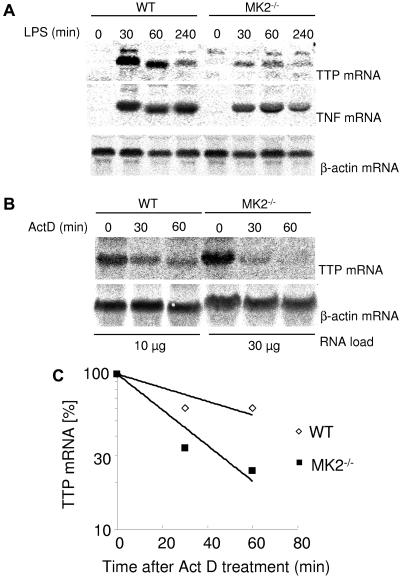
Northern blot analysis revealed a significant reduction of TTP mRNA after LPS induction in immortalized MK2−/− macrophages compared to WT macrophages, whereas TNF and β-actin mRNA levels were comparable between MK2−/− and WT mice under the same conditions (Fig. 4). RNA stability experiments indicate that TTP mRNA is less stable in MK2−/− cells, suggesting an MK2-dependent stabilizing effect on TTP mRNA (Fig. 4C). Taken together, we suggest that MK2 is an important downstream kinase of p38 responsible for the regulation of TTP at the posttranscriptional level.
FIG. 4.
TTP mRNA is unstable in MK2-deficient macrophages. (A) WT and MK2−/− immortalized macrophages were induced with 1 μg/ml LPS for the times indicated and used for total RNA preparation and Northern blotting (10 μg total RNA per lane). (B) WT and MK2−/− immortalized macrophages were induced with 1 μg/ml LPS for 45 min before addition of 2 μg/ml actinomycin D (ActD) for the times indicated. Ten micrograms of total RNA was loaded per lane for WT and 30 μg total RNA per lane for MK2−/−. After development with the TTP probe, Northern blots were stripped and rehybridized with a TNF probe and/or β-actin probe as control. (C) Quantification of the Northern blot shown in panel B. The intensity of the TTP mRNA bands was quantified and normalized to actin mRNA. A half-life for TTP mRNA of 34 min in MK2-deficient macrophages and 63 min in WT cells could be calculated.
The p38/MK2 pathway can stabilize TTP by phosphorylation.
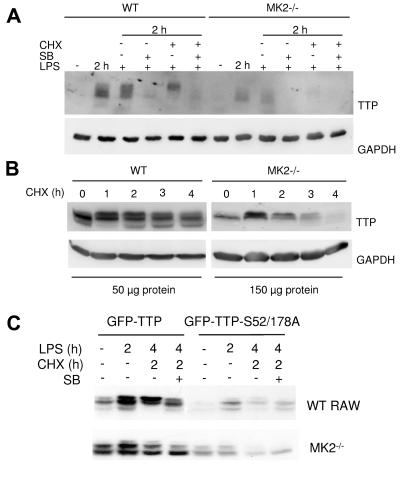
The p38/MK2 signaling pathway regulates the levels of mRNA, protein, and phosphorylation of TTP (10, 12, 27, 35). We decided to analyze whether this pathway also regulates the stability of TTP. Bone marrow-derived primary macrophages were grown to 80% confluence and induced with LPS for 2 h, leading to expression and phosphorylation of TTP (Fig. 5A, lane 2 h LPS). Subsequently, cycloheximide and/or SB203580 was added to inhibit protein translation and/or p38 MAPK. Interestingly, addition of SB203580 reduced the TTP protein level compared to that of the control. This indicates that the p38 pathway has a stabilizing function on TTP protein. In addition, the TTP band that accumulates in the cycloheximide control migrates slower compared to the TTP band in the presence of SB203580, indicating higher stability of the phosphorylated forms of TTP. To assay the role of MK2 for this stabilization, the TTP level was analyzed at different times after blocking protein translation in immortalized MK2-deficient macrophages and in control WT macrophages (Fig. 5B). It can be seen that TTP is less stable in MK2−/− macrophages than in WT macrophages. The estimated half-life of TTP in WT cells is about 9 h, while in MK2−/− cells it is about 2.5 h.
FIG. 5.
Phosphorylation by MK2 stabilizes TTP. (A) Bone marrow-derived primary macrophages were induced for 2 h with 1 μg/ml LPS. At this time point, either SB203580 (SB), cycloheximide (CHX), or SB and cycloheximide were added for 2 h. The addition of SB destabilizes TTP protein; in the absence of SB and the presence of cycloheximide, a higher phosphorylation band accumulates. (B) Immortalized WT and MK2−/− macrophages were induced for 2 h with LPS. Subsequently, cycloheximide was added to stop translation. Cells were lysed after an additional 1, 2, 3, and 4 h of incubation. Fifty micrograms of total lysate from WT cells and 150 μg of total lysate from MK2−/− cells was loaded. (C) Stability of transfected GFP-TTP and GFP-TTP-S52/178A mutant. RAW cells and MK2−/− immortalized macrophages were transfected with GFP-TTP and GFP-TTP-S52/178A. Four hours after transfection, LPS was added for 2 h and, subsequently, SB202190 (SB) and cycloheximide (CHX) were added as indicated for a further 2 h.
To exclude transcriptional effects of the p38 inhibition and to understand the role of the identified phosphorylation sites for MK2, S52, and S178, we also analyzed regulation and stability of a cytomegalovirus promoter-driven GFP-TTP fusion protein and the appropriate mutant GFP-TTP-S52,178A in RAW 264.7 cells and in MK2-deficient macrophages (Fig. 5C). First, the GFP-TTP-S52,178A mutant is weakly detectable and significantly less stable than WT GFP-TTP, whether expressed in RAW 264.7 or MK2−/− macrophages, indicating that these phosphorylation sites are essential for stabilization. Second, LPS induction leads to an up-regulation of GFP-TTP in RAW cells but not in MK2-deficient cells, suggesting that phosphorylation by MK2 in RAW cells stabilizes TTP. Third, SB203580 treatment in the presence of cycloheximide leads to a down-regulation of GFP-TTP in RAW 264.7- but not MK2-deficient cells (Fig. 5C, compare GFP-TTP lanes for LPS at 4 h, CHX at 2 h, and SB− and SB+), indicating that part of the p38-dependent TTP stabilization is due to MK2. Taken together, these findings support the notion that phosphorylation by MK2 at S52 and/or S178 contributes to an LPS-induced stabilization of TTP.
Phosphorylation of TTP decreases its ability to bind the TNF mRNA 3′UTR.
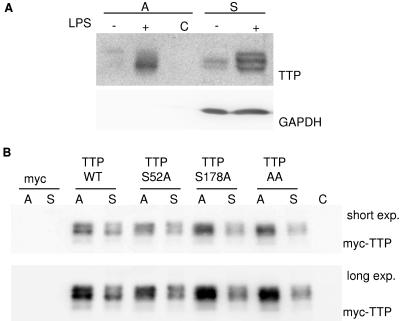
MK2 is able to phosphorylate TTP both in vivo and in vitro, and major sites of phosphorylation were identified as serine residues S52 and S178 (12, 27). After LPS induction, TTP is detected in multiple differentially phosphorylated forms that migrate with different mobilities in SDS-PAGE (see, for example, Fig. 3). To test whether some of the phosphorylated forms of TTP display higher levels of binding to TNF-ARE, we performed affinity purification experiments using in vitro-transcribed biotinylated ARE of TNF mRNA coupled to streptavidin beads and protein lysates from LPS-induced WT macrophages. In the supernatant (Fig. 6A, S), differently phosphorylated isoforms of TTP can be detected, especially after LPS treatment (lane S+). Affinity purification (Fig. 6A, lane A+) demonstrates preferential binding of a band with faster mobility and, thus, lower phosphorylation to the 3′UTR of TNF, while TTP does not bind to streptavidin agarose itself (lane C). To analyze whether phosphorylation of TTP by MK2 at the serine residues identified is responsible for the apparent inhibition of binding, we performed affinity purification experiments using HEK293 cells transfected with myc-tagged phosphorylation site mutants TTP-S52A, TTP-S178A, and TTP-S52,178A. Before lysis, the transfected cells were incubated for 1 h with 100 μM arsenite to stimulate p38. Protein lysates were used for affinity purification on TNF-ARE as described above. Comparison of the intensity of affinity purified TTP bands (A) with the bands from the supernatant (S) gives an indication for the strength of binding (Fig. 6B). Interestingly, TTP mutants containing the S178A substitution (TTP-S178A and TT-AA) have a slightly stronger binding to TNF-ARE than WT or TTP-S52A (Fig. 6B, lane TTP S52A, A), suggesting a reduction in ARE binding of TTP by phosphorylation, especially at S178. This effect is rather subtle, and apparent differences in binding of the differently phosphorylated isoforms between those shown in Fig. 6A and B exist, which might arise from the fact that in Fig. 6A endogenous protein from LPS-treated macrophages is analyzed, while in Fig. 6B myc-tagged forms of TTP overexpressed in HEK293 cells stimulated with arsenite are used.
FIG. 6.
Phosphorylation reduces the affinity of TTP to TNF-ARE. Shown are affinity purification experiments using biotin-labeled in vitro-transcribed TNF-ARE coupled to streptavidin beads. (A) Protein lysates from immortalized WT macrophages that were induced with 1 μg/ml LPS were used for affinity purification (A, affinity purified; S, supernatant). The least phosphorylated TTP band with high mobility shows the strongest binding. C−, negative control using beads that are not coupled to RNA and lysates of LPS-induced macrophages. (B) myc-tagged WT TTP and mutants with serine-to-alanine substitutions at serine 52 (TTP-S52A) and/or at serine 178 (TTP-S178A) and TTP-AA were overexpressed in HEK293 cells. One hour prior to lysis, 100 μM arsenite was added to activate the p38 pathway. Lysates were used for affinity purification as described for panel A. C−, negative control using beads that are not coupled to RNA and lysates from TTP transfected cells. myc stands for the empty vector control. This experiment is representative of two independently performed experiments.
Inhibition of p38 reduces TTP levels in MK2−/− macrophages and TNF levels in TTP−/− MK2−/− macrophages.
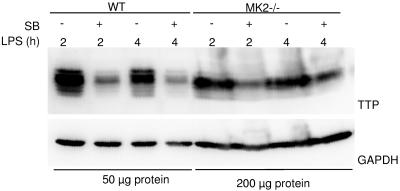
We were interested to determine whether MK2 is the only downstream element necessary and sufficient for the p38-dependent regulation of TTP expression. TTP expression levels were analyzed in MK2−/− macrophages after SB203580 treatment and LPS induction. p38 inhibition reduced TTP levels in WT macrophages and, to a somewhat lesser extent, also in MK2−/− macrophages to basal levels, indicating the presence of another p38 downstream element in the regulation of TTP expression or a direct p38 effect on TTP levels (Fig. 7).
FIG. 7.
Inhibition of p38 down-regulates the level of TTP protein in MK2-deficient macrophages. Western blot for TTP and GAPDH as a control of total protein lysates from macrophages that were induced with 1 μg/ml LPS for 2 or 4 h. SB203580 (SB) was added 1 h prior to addition of LPS to a final concentration of 10 μM. Addition of SB203580 leads to a reduction of TTP protein levels in WT cells compared to the LPS-stimulated control and, to a lesser extent, also in MK2−/− cells compared to the LPS-stimulated MK2-deficient control.
It has been described that TTP-deficient cells have a decreased sensitivity to p38 inhibitors (10). We were interested to know if the MK2/TTP pathway is sufficient and necessary to exert the effect of p38 on TNF production. We analyzed TNF production of MK2 TTP double knockout macrophages in the presence or absence of the p38 inhibitor. Interestingly, TNF expression in TTP−/− and TTP MK2 double knockout macrophages is significantly inhibited by SB203580 (Fig. 1A). These results support the above notion that p38 is directly involved or has a downstream element other than the MK2-TTP module that also contributes to the regulation of TNF protein expression.
DISCUSSION
Genetic evidence that TTP is downstream of MK2 in posttranscriptional regulation.
Upon LPS stimulation, MK2-deficient macrophages produce levels of TNF mRNA similar to those of WT macrophages but express only about 10% of the WT level of TNF protein. In contrast, TTP-deficient cells produce elevated levels of TNF mRNA and protein. Here, we demonstrated that the removal of TTP from an MK2-deficient genotype completely reversed the MK2-deficient phenotype and that the resulting MK2- and TTP-double-deficient macrophages produce levels of TNF mRNA and protein similar to those of the TTP knockout. This, together with previous reports that TTP can be phosphorylated by MK2 (12, 27), provides strong evidence that TTP is downstream of MK2 in the regulation of both stability and translation of TNF mRNA.
MK2 stabilizes and inactivates TTP by phosphorylation.
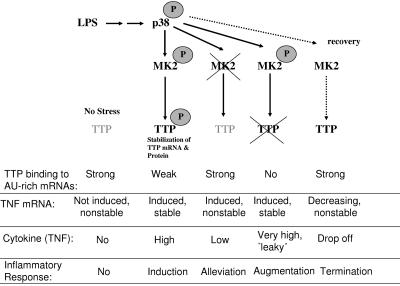
The results obtained suggest the following mechanism for the posttranscriptional regulation of TNF biosynthesis by the p38 MAPK signaling pathway (Fig. 8). Upon LPS, p38 is stimulated, and it phosphorylates and activates MK2. Activity of MK2 is essential for stabilization of TTP mRNA and protein as well as for stabilization of TNF mRNA. In addition to being essential for TTP expression, MK2 could also inhibit TTP's destructive activity against TNF mRNA by the same phosphorylations that stabilize the protein. This effect could be based on reduced binding of TTP to the ARE of TNF mRNA and/or on its increased binding to 14-3-3 proteins (10, 32). As a result, TNF mRNA is efficiently expressed and translated. In a second phase of the response to LPS (designated “recovery” in Fig. 8), where p38 and MK2 activity is back to basal levels, TTP could then be dephosphorylated and activated, and it can down-regulate TNF mRNA by destabilization in parallel to its own degradation. One may speculate that binding of TTP to ARE or components of the degradation machineries is a prerequisite for its destabilization and that mRNA destabilizing activity and degradation of TTP itself is coupled at the exosome or RISC/micro RNP complex (17). Alternatively, a codegradation of TTP and ARE mRNAs in a proteasome-dependent manner seems possible (23, 24). Apparently, in MK2-deficient cells, codegradation of TTP together with TNF mRNA becomes the default pathway, resulting in low levels of both components. However, in WT cells the above scenario requires the presence of a phosphatase(s) that possibly activates TTP. The stabilization of TTP by the p38 pathway and its accumulation in an inactive form, which can be activated by dephosphorylation when the p38 pathway is turned off, is of plausible physiological relevance for limiting the inflammatory response. Interestingly, preliminary results indicate a significant positive effect of phosphatase inhibitors on TTP expression and stability (E. Hitti and M. Gaestel, unpublished data).
FIG. 8.
Model of posttranscriptional regulation by the p38/MK2/TTP pathway in response to LPS in the different genetic situations. TTP is expressed in small amounts in nonstimulated cells (no stress), is not phosphorylated, and is able to bind and destabilize rare TNF mRNA occasionally transcribed and, hence, elevates the threshold of inflammation. In the case of stimulation by LPS, the rate of TNF and TTP transcription increases, and MK2 gets activated, phosphorylates TTP, and reduces its binding to ARE, making ARE-containing mRNA stable. Phosphorylated TTP protein is stable, and its accumulation can be important in the recovery step, when MK2 is no longer active and TTP is rapidly dephosphorylated. Dephosphorylated TTP binds to and destabilizes TNF mRNA, resulting in decreasing expression of TNF. Apparently, in MK2-deficient cells, codegradation of nonphosphorylated TTP bound to TNF mRNA becomes the default pathway, resulting in low levels of both components, and causes alleviation of inflammatory response. In contrast, TTP-deficient mice which are compromised in degradation of ARE-containing mRNAs produce increased levels of TNF upon LPS stimulation and, even in the absence of stimulation, develop TNF excess syndrome. Since TTP is downstream of MK2, TNF excess syndrome is not modulated by MK2 deficiency in the absence of TTP.
The nonphosphorylated form of TTP is the active form.
The idea that the nonphosphorylated form of TTP is active in posttranscriptional repression of TNF expression is supported by two independent observations. First, the high levels of TNF production in the TTP MK2 double knockout compared to reduced TNF production in MK2 knockout mice indicates that the remaining TTP in MK2 knockout mice, which is not phosphorylated by MK2, actively contributes to repression of TNF production. In contrast, the high level of phosphorylated TTP in the WT has a reduced ability to inhibit TNF expression. Second, affinity purification suggests that TTP mutants that cannot be phosphorylated at serine 178 have a slightly increased ability to bind ARE compared to WT TTP. This finding is in accordance with the previously described stronger ability of TTP to bind ARE after alkaline phosphatase treatment (10). Phosphorylation of TTP at serine 52 and serine 178 also increases its binding to 14-3-3 protein and prevents its entry into stress granules, where TNF mRNA is stored (27). Hence, the reduced binding of phospho-TTP in the affinity purification can also be due to competitive binding to 14-3-3 proteins in the lysate.
MK2 and TTP are not the only factors in regulation of LPS-induced TNF production.
Inhibition of p38 in MK2-deficient cells further reduces the level of TTP. This observation demonstrates that although MK2 plays a significant role in regulation of TTP expression and stabilization, other downstream targets of p38 that influence TTP expression and/or stability could exist. Since p38 has been demonstrated to phosphorylate TTP directly at several sites (8, 10, 35), direct modification of TTP by p38 is probably one additional mechanism involved. It is also possible that p38 plays a role in the regulation of TTP transcription. Interestingly, it is known that MK2 stabilizes p38 MAPK and that the p38 level is reduced in MK2-deficient cells (21). Hence, direct contributions of p38 can be at least partially inhibited in the MK2-deficient cells.
Interestingly, TNF production can partially be inhibited by SB203580 also in TTP- and MK2-deficient cells. Since it is unlikely that SB203580 inhibits transcriptional activation of the TNF gene (5), a role of further downstream targets of p38 on posttranscriptional regulation of TNF expression seems possible, while the involvement of direct targets of MK2, such as hnRNP-A0 (29) or PABP-1 (4), can be excluded. Interestingly, a role of MAPK-interacting kinases, another group of kinases downstream of p38, in posttranscriptional regulation of TNF via hnRNP A1 has been recently described (7).
Regulation of other ARE-containing transcripts.
TTP regulates it own expression by binding to AU-rich elements of its own mRNA (6, 34). It is therefore probable that the posttranscriptional regulation of TTP itself is similar to the regulation of TNF. We observed that the half-life of TTP mRNA is short in LPS-stimulated WT macrophages (about 60 min) and further decreased in MK2-deficient cells (about 30 min). An explanation could be that directly after translation, nonphosphorylated TTP binds to the 3′UTR of its own mRNA due to spatial vicinity, a situation that does not occur with other AU-rich transcripts. This could also explain the potency of the ARE of TTP which belongs to group IV AREs containing only two AUUUA pentamers. We have observed increased levels and stability of other transcripts that contain AREs in both TTP−/− and MK2 TTP double knockout cells after stress induction (data not shown). Hence, it is unlikely that TTP has different functions on different transcripts. TTP itself does not have nuclease domains and probably acts as a scaffolding protein (26). The differences in stability between ARE-containing transcripts, as in the case of IL-6 and TNF, are probably due to other ARE-binding factors, such as HuR, AUF1, AUF2, FBP1, FBP2 (KSRP), TIA-1, and TIAR (14, 28). Together with some of these proteins, TTP not only destabilizes ARE-containing transcripts but could also inhibit translation. Because of the relatively high level of transcription of TNF mRNA in LPS-stimulated cells, translational control of TNF mRNA is the best known example so far.
Interestingly, there are examples where no effects of the p38 pathway were detected on ARE-containing transcripts (16, 20). It is also possible that the p38 pathway acts differently on the posttranscriptional level in different cell types depending on the expression of ARE binding proteins (9, 16, 25). Differences in the same cell type on different ARE-containing transcripts could be due to differences in kinetics of mRNA transcription and activation of the p38 pathway. In the case of TNF, upon LPS treatment both TNF mRNA transcription and p38 activity in WT and MK2−/− macrophages peaks at approximately the same time between 30 to 60 min after induction (data not shown), which results in major p38 effects. However, when the induction of transcription is slow and the activation of the p38 pathway is not maintained, its effect on other ARE-containing transcripts would be only marginal.
Acknowledgments
We thank Svetlana Gromova, Berlin, Germany, for technical advice, and Georg Stoecklin (Boston, Mass.) for sharing TTP constructs.
This work was supported by European Community grant RTN-HPRN-CT-2002-00255 and by Deutsche Forschungsgemeinschaft grant SFB 566-TP B12.
REFERENCES
- 1.Bakheet, T., M. Frevel, B. R. Williams, W. Greer, and K. S. Khabar. 2001. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 29:246-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakheet, T., B. R. Williams, and K. S. Khabar. 2003. ARED 2.0: an update of AU-rich element mRNA database. Nucleic Acids Res. 31:421-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasi, E., D. Radzioch, L. Merletti, and L. Varesio. 1989. Generation of macrophage cell line from fresh bone marrow cells with a myc/raf recombinant retrovirus. Cancer Biochem. Biophys. 10:303-317. [PubMed] [Google Scholar]
- 4.Bollig, F., R. Winzen, M. Gaestel, S. Kostka, K. Resch, and H. Holtmann. 2003. Affinity purification of ARE-binding proteins identifies polyA-binding protein 1 as a potential substrate in MK2-induced mRNA stabilization. Biochem. Biophys. Res. Commun. 301:665-670. [DOI] [PubMed] [Google Scholar]
- 5.Brook, M., G. Sully, A. R. Clark, and J. Saklatvala. 2000. Regulation of tumour necrosis factor alpha mRNA stability by the mitogen-activated protein kinase p38 signalling cascade. FEBS Lett. 483:57-61. [DOI] [PubMed] [Google Scholar]
- 6.Brooks, S. A., J. E. Connolly, and W. F. C. Rigby. 2004. The role of mRNA turnover in the regulation of tristetraprolin expression: evidence for an extracellular signal-regulated kinase-specific, AU-rich element-dependent, autoregulatory pathway. J. Immunol. 172:7263-7271. [DOI] [PubMed] [Google Scholar]
- 7.Buxade, M., J. L. Parra, S. Rousseau, N. Shapiro, R. Marquez, N. Morrice, J. Bain, E. Espel, and C. G. Proud. 2005. The Mnks are novel components in the control of TNF alpha biosynthesis and phosphorylate and regulate hnRNP A1. Immunity 23:177-189. [DOI] [PubMed] [Google Scholar]
- 8.Cao, H., L. J. Deterding, J. D. Venable, E. A. Kennington, J. R. Yates III, K. B. Tomer, and P. J. Blackshear. 2006. Identification of the anti-inflammatory protein tristetraprolin as a hyperphosphorylated protein by mass spectrometry and site-directed mutagenesis. Biochem. J. 394:285-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao, H., J. S. Tuttle, and P. J. Blackshear. 2004. Immunological characterization of tristetraprolin as a low abundance, inducible, stable cytosolic protein. J. Biol. Chem. 279:21489-21499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carballo, E., H. Cao, W. S. Lai, E. A. Kennington, D. Campbell, and P. J. Blackshear. 2001. Decreased sensitivity of tristetraprolin-deficient cells to p38 inhibitors suggests the involvement of tristetraprolin in the p38 signaling pathway. J. Biol. Chem. 276:42580-42587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrick, D., W. Lai, and P. Blackshear. 2004. The tandem CCCH zinc finger protein tristetraprolin and its relevance to cytokine mRNA turnover and arthritis. Arthritis Res. Ther. 6:248-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chrestensen, C. A., M. J. Schroeder, J. Shabanowitz, D. F. Hunt, J. W. Pelo, M. T. Worthington, and T. W. Sturgill. 2004. MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14-3-3 binding. J. Biol. Chem. 279:10176-10184. [DOI] [PubMed] [Google Scholar]
- 13.Clark, A. R., J. L. Dean, and J. Saklatvala. 2003. Post-transcriptional regulation of gene expression by mitogen-activated protein kinase p38. FEBS Lett. 546:37-44. [DOI] [PubMed] [Google Scholar]
- 14.Dean, J. L., G. Sully, A. R. Clark, and J. Saklatvala. 2004. The involvement of AU-rich element-binding proteins in p38 mitogen-activated protein kinase pathway-mediated mRNA stabilisation. Cell Signal. 16:1113-1121. [DOI] [PubMed] [Google Scholar]
- 15.Fechir, M., K. Linker, A. Pautz, T. Hubrich, U. Forstermann, F. Rodriguez-Pascual, and H. Kleinert. 2005. Tristetraprolin regulates the expression of the human inducible nitric-oxide synthase gene. Mol. Pharmacol. 67:2148-2161. [DOI] [PubMed] [Google Scholar]
- 16.Frevel, M. A., T. Bakheet, A. M. Silva, J. G. Hissong, K. S. Khabar, and B. R. Williams. 2003. p38 mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol. Cell. Biol. 23:425-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jing, Q., S. Huang, S. Guth, T. Zarubin, A. Motoyama, J. Chen, F. Di Padova, S. C. Lin, H. Gram, and J. Han. 2005. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 120:623-634. [DOI] [PubMed] [Google Scholar]
- 18.Karlsson, J. O., K. Ostwald, C. Kabjorn, and M. Andersson. 1994. A method for protein assay in Laemmli buffer. Anal. Biochem. 219:144-146. [DOI] [PubMed] [Google Scholar]
- 19.Kedersha, N., and P. Anderson. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963-969. [DOI] [PubMed] [Google Scholar]
- 20.Kotlyarov, A., A. Neininger, C. Schubert, R. Eckert, C. Birchmeier, H. Volk, and M. Gaestel. 1999. MAPKAP kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat. Cell Biol. 1:94-97. [DOI] [PubMed] [Google Scholar]
- 21.Kotlyarov, A., Y. Yannoni, S. Fritz, K. Laass, J.-B. Telliez, D. Pitman, L.-L. Lin, and M. Gaestel. 2002. Distinct cellular functions of MK2. Mol. Cell. Biol. 22:4827-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai, W. S., E. A. Kennington, and P. J. Blackshear. 2003. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol. Cell. Biol. 23:3798-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laroia, G., R. Cuesta, G. Brewer, and R. J. Schneider. 1999. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284:499-502. [DOI] [PubMed] [Google Scholar]
- 24.Laroia, G., B. Sarkar, and R. J. Schneider. 2002. Ubiquitin-dependent mechanism regulates rapid turnover of AU-rich cytokine mRNAs. Proc. Natl. Acad. Sci. USA 99:1842-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu, J.-Y., and R. J. Schneider. 2004. Tissue distribution of AU-rich mRNA-binding proteins involved in regulation of mRNA decay. J. Biol. Chem. 279:12974-12979. [DOI] [PubMed] [Google Scholar]
- 26.Lykke-Andersen, J., and E. Wagner. 2005. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 19:351-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahtani, K. R., M. Brook, J. L. E. Dean, G. Sully, J. Saklatvala, and A. R. Clark. 2001. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol. Cell. Biol. 21:6461-6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neininger, A., D. Kontoyiannis, A. Kotlyarov, R. Winzen, R. Eckert, H.-D. Volk, H. Holtmann, G. Kollias, and M. Gaestel. 2002. MK2 targets AU-rich elements and regulates biosynthesis of tumor necrosis factor and interleukin-6 independently at different post-transcriptional levels. J. Biol. Chem. 277:3065-3068. [DOI] [PubMed] [Google Scholar]
- 29.Rousseau, S., N. Morrice, M. Peggie, D. G. Campbell, M. Gaestel, and P. Cohen. 2002. Inhibition of SAPK2a/p38 prevents hnRNP A0 phosphorylation by MAPKAP-K2 and its interaction with cytokine mRNAs. EMBO J. 21:6505-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saklatvala, J. 2004. The p38 MAP kinase pathway as a therapeutic target in inflammatory disease. Curr. Opin. Pharmacol. 4:372-377. [DOI] [PubMed] [Google Scholar]
- 31.Saklatvala, J., J. Dean, and A. Clark. 2003. Control of the expression of inflammatory response genes. Biochem. Soc. Symp. 70:95-106. [DOI] [PubMed] [Google Scholar]
- 32.Stoecklin, G., T. Stubbs, N. Kedersha, S. Wax, W. F. Rigby, T. K. Blackwell, and P. Anderson. 2004. MK2-induced tristetraprolin:14-13-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23:1313-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor, G., E. Carballo, D. Lee, W. Lai, M. Thompson, D. Patel, D. Schenkman, G. Gilkeson, H. Broxmeyer, B. Haynes, and P. Blackshear. 1996. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4:445-454. [DOI] [PubMed] [Google Scholar]
- 34.Tchen, C. R., M. Brook, J. Saklatvala, and A. R. Clark. 2004. The stability of tristetraprolin mRNA is regulated by mitogen-activated protein kinase p38 and by tristetraprolin itself. J. Biol. Chem. 279:32393-32400. [DOI] [PubMed] [Google Scholar]
- 35.Zhu, W., M. A. Brauchle, F. Di Padova, H. Gram, L. New, K. Ono, J. S. Downey, and J. Han. 2001. Gene suppression by tristetraprolin and release by the p38 pathway. Am. J. Physiol. Lung Cell Mol. Physiol. 281:L499-L508. [DOI] [PubMed] [Google Scholar]