Abstract
Overexpression of transferrin receptor 1 (TFRC1), a major mediator of iron uptake in mammalian cells, is a common feature of human malignancies. Therapeutic strategies designed to interfere with tumor iron metabolism have targeted TFRC1. The c-Myc oncogenic transcription factor stimulates proliferation and growth by activating thousands of target genes. Here we demonstrate that TFRC1 is a critical downstream target of c-Myc. Using in vitro and in vivo models of B-cell lymphoma, we show that TFRC1 expression is activated by c-Myc. Chromatin immunoprecipitation experiments reveal that c-Myc directly binds a conserved region of TFRC1. In light of these findings, we sought to determine whether TFRC1 is required for c-Myc-mediated cellular proliferation and cell size control. TFRC1 inhibition decreases cellular proliferation and results in G1 arrest without affecting cell size. Consistent with these findings, expression profiling reveals that TFRC1 depletion alters expression of genes that regulate the cell cycle. Furthermore, enforced TFRC1 expression confers a growth advantage to cells and significantly enhances the rate of c-Myc-mediated tumor formation in vivo. These findings provide a molecular basis for increased TFRC1 expression in human tumors, illuminate the role of TFRC1 in the c-Myc target gene network, and support strategies that target TFRC1 for cancer therapy.
Dysregulated expression of c-Myc is a central event in the development of a diverse array of human malignancies (1, 10). c-MYC encodes a helix-loop-helix transcription factor that dimerizes with its partner Max, binds the consensus sequence 5′-CACGTG-3′, and regulates transcription of target genes. Through its myriad direct and indirect target genes, the c-Myc oncoprotein is linked to nearly all cellular processes (1, 12, 20, 40, 43, 48). An emerging view holds that c-Myc regulates 10 to 15% of genes in the human or Drosophila melanogaster genome, including noncoding microRNAs (18, 30, 38, 39). Despite the growing number of target genes being identified, the precise mechanism by which c-Myc orchestrates cell growth and division in normal and neoplastic cells is not fully understood.
It has become increasingly evident that c-Myc regulates the expression of several genes required for iron-dependent cellular processes, such as energy metabolism and mitochondrial homeostasis (29, 36, 56). The link between c-Myc function and iron metabolism was directly demonstrated through the identification of IRP2 and H-ferritin as coordinately regulated targets of c-Myc (58). Nramp1, which encodes a divalent cation transporter and removes iron from the cytosol, was recently shown to be repressed by c-Myc (5, 6). Taken together, these studies suggest that c-Myc functions to increase the intracellular iron pool.
The transferrin receptor (TFRC1/TFR/p90/CD71) is a key cell surface molecule that regulates uptake of iron-bound transferrin by receptor-mediated endocytosis (8). For more than 20 years, there has been a known correlation between the number of cell surface transferrin receptors and the rate of cell proliferation (9, 27, 28, 35, 45). Transferrin receptor expression is higher in cancer cells than in normal cells (19). Furthermore, TFRC1 is among a select group of genes that is overexpressed in a murine Myc-induced prostate cancer model as well as in primary human prostate cancers (15). Despite these findings, the mechanism by which transferrin receptor expression is increased in neoplastic cells remains poorly characterized.
Given that transferrin receptors are highly expressed in cancer cells and that TFRC1 has been reported as a c-Myc-responsive gene, we sought to determine if TFRC1 is directly regulated by the c-Myc oncoprotein. In the current study, we demonstrate that TFRC1 is a direct transcriptional target of c-Myc in a human B lymphocyte model system. TFRC1 expression also parallels inducible c-Myc expression in an in vivo murine model of lymphoma. RNA interference and iron chelation with the drug desferrioxamine (DFX) were used to evaluate the role of TFRC1 in c-Myc-driven cell cycle progression and cell size control. Here, we report that TFRC1-depleted lymphocytes undergo G1 arrest while maintaining normal cell size. Conversely, ectopic TFRC1 expression confers a significant growth advantage to cells grown in limiting serum conditions. We also demonstrate that enforced expression of TFRC1 significantly enhances the rate of c-Myc-mediated tumor formation in nude mice. Taken together, these results support a critical role for TFRC1 in c-Myc-mediated tumorigenesis.
MATERIALS AND METHODS
Cell culture.
P493-6 cells were the generous gift of D. Eick at the Institute for Clinical Molecular Biology und Tumor Genetics, GSF-Research Centre, Munich, Germany. Cells were cultured in RPMI 1640, 10% fetal calf serum, and penicillin-streptomycin (Pen-Strep). To repress c-MYC expression, cells were treated with RPMI 1640-10% fetal calf serum-Pen-Strep supplemented with 0.1 μg/ml tetracycline (Sigma) for 72 h, washed two to three times in 1× phosphate-buffered saline (PBS), and then restimulated with regular RPMI medium for various time points. For desferrioxamine treatment, P493-6 cells were placed in RPMI medium containing 50 or 100 μM desferrioxamine (Sigma) at the same time as removal of tetracycline and collected at various time points. TGR-1 (wild-type rat fibroblasts) and HO15.19 (c-MYC knockout rat fibroblasts) cells (gift of J. Sedivy, Brown University) and c-MYC null cells reconstituted with W135E (gift of L. Z. Penn, University of Toronto) were cultured in Dulbecco's modified Eagle's medium (Gibco/Life Tech)-10% fetal bovine serum-Pen-Strep. HO15-MYC cells were made by stable transfection of a MYC expression construct driven by a murine leukemia virus promoter. For growth rate experiments under limiting conditions, 2.5 × 104 cells were plated and subsequently starved in 0.1% serum for 48 h. Cell were then washed, grown in 1% serum, and counted in triplicate wells every 24 h for 6 to 7 days.
Wright staining.
Wright staining was performed according to the manufacturer's protocol using the HEMA 3 stain set (Fisher Scientific).
Western blot analysis.
For immunoblot analysis, cells were collected and lysed in incomplete Laemmli buffer at 95°C and protein was quantitated using a bicinchoninic acid kit (Pierce). Fifteen to twenty micrograms of protein was loaded on either a 8% or 10% polyacrylamide gel (for TFRC1 or c-MYC, respectively) and transferred to a nitrocellulose membrane. 9E10 c-Myc and TFRC1 mouse monoclonal antibodies were obtained from Zymed, Inc. (clone H68.4). α-Tubulin mouse monoclonal antibody was obtained from Oncogene Research Products.
In vivo analysis of TFRC1 in murine B-cell lymphomas.
Tumor-derived neoplastic lymphocytes were washed twice with PBS and resuspended in fluorescence-activated cell sorting (FACS) buffer (0.5% bovine serum albumin in PBS) at a concentration of 1 × 106 cells/ml. One-hundred-microliter aliquots were incubated on ice for 45 min with 2 μl phycoerythrin-labeled anti-mouse CD71 (clone RI7 217.1.4; CALTAG, Burlingame CA). Samples were washed with PBS and subjected to flow cytometry. Unstained cells were used as a reference.
Chromatin immunoprecipitation.
P493-6 cells untreated or treated with 0.1 μg/ml tetracycline for 72 h were used for chromatin immunoprecipitation (ChIP) assays. Cells were cross-linked with formaldehyde, and chromatin was immunoprecipitated as described previously (7). The rabbit polyclonal c-Myc (sc-764; Santa Cruz Biotechnology) and human hepatocyte growth factor (sc-7949; Santa Cruz) antibodies were used to immunoprecipitate chromatin fragments. Total input controls were collected from the “no antibody” control supernatant. Mock control samples lacked chromatin but were treated the same as other samples.
Retroviral production and infection.
The TRS1 vector was a gift from Jim Basilion (Massachusetts General Hospital, Harvard University). The insert was first cloned into the pSG5 vector using the EcoRV and SalI restriction sites and then cloned into the pMSCV retroviral vector using the BamHI and BglII restriction sites. For retroviral transduction, the pMSCV vector containing the TRS1 insert was transfected into Phoenix amphotropic cells (at 80 to 90% confluence) using the CaPO4 method. Viral supernatant was collected every 24 h for 3 days. For infection of c-MYC-null cells, approximately 2 × 105 cells were plated per 10-cm dish and infected with 8 ml viral supernatant and 8 μl/ml polybrene 24 h later. Two subsequent rounds of infection were performed, and cells were then selected with puromycin for 14 days. Cell lysates were then collected 7 days after removal from puromycin.
RNA interference.
Preannealled small interfering RNA (siRNA) duplexes directed against TFRC1 (and a scrambled control) were purchased from Dharmacon, Inc., and used according to the manufacturer's instructions (Option C). TFRC1 oligonucleotides used were GGAUGGUAACCUCAGAAAGdTdT (sense) and dTdTCCUACCAUUGGAGUCUUUC (antisense). Scrambled JTV1 control oligonucleotides were CACGCUCGGUCAAAAGGUUdTdT (sense) and dTdTGUGCGAGCCAGUUUUCCAA (antisense). RNA interference (RNAi) oligonucleotides were transfected by electroporation. siRNA duplexes were first added to a 4-mm-gap cuvette (BTX/Cambrige Pharmaceuticals) and then mixed with 3 × 106 P493-6 cells (in a total volume of 500 μl). Electroporation settings used for P493-6 cells were as follows: 240 V and 1500 μF. Cell viability was assessed immediately following electroporation by trypan blue exclusion (and determined to be approximately 30%). Mock-transfected cells and cells transfected with a scrambled siRNA duplex were used as controls. Cells were collected and assayed for gene function approximately 72 h after electroporation.
Transferrin uptake.
HO15 and TGR cells were first trypsinized and counted. Approximately 1 × 106 cells were washed two times in 1× PBS and then resuspended in 2 ml serum-free Dulbecco's modified Eagle's medium. Samples were then serum starved for 2 h with rotation at 37°C to clear transferrin (present in culture medium) from the transferrin receptors. Alexa633-labeled transferrin (Molecular Probes) was then added to each sample in a final concentration of 5 μg/ml, followed by incubation for 30 min, rotating at 37°C. Cells were then washed three times with 1× PBS, resuspended in chilled 1% paraformaldehyde, and analyzed using a Beckman Dickinson FACScalibur machine.
Nude mouse experiments.
Cells (5 × 106) in 100 μl of sterile Hanks balanced salt solution (Gibco) were injected subcutaneously into the right flank of male homozygous nude mice at 4 to 6 weeks of age. Tumor volume was measured using calipers every 3 to 5 days until the tumor mass reached 1,500 mm3. Tumor volume was calculated using the following formula: [length (mm) × width (mm)]2/2. Two independent experiments were performed (n = 4 to 5 mice per cell line for experiment 1 and n = 15 mice per cell line for experiment 2). All experiments were approved by the Johns Hopkins School of Medicine Animal Care and Use Committee.
Correlation of TFRC1 and c-MYC mRNA levels using the Atlas Gene Expression database.
TFRC1 and c-MYC mRNA levels were examined in 91 human tissues and cell lines using the Gene Expression Atlas database (http://expression.gnf.org/cgi-bin/index.cgi) (52). We first log transformed the expression values and then performed linear regression analysis (using SPSS version 11.0 for Windows) on log-transformed mRNA expression values with log TFRC1 expression levels as the dependent variable and log c-MYC expression values as the independent variable. An r2 value was determined which measured the extent to which expression of c-MYC and TFRC1 were correlated.
Flow cytometry.
For cell size analysis, P493 cells were collected, washed two to three times, and then resuspended in 1× PBS. For propidium iodide staining, approximately 1 × 106 cells were washed in 1× PBS, trypsinized for 10 min at room temperature (RT) (solution A), neutralized with trypsin inhibitor-RNase A for 10 min at RT (solution B), and then stained with propidium iodide/spermine tetrahydrochloride (solution C) for 10 min at RT (54). Cells were filtered and then analyzed using a Becton Dickinson FACScan or FACSCalibur flow cytometer.
For anti-human CD71 labeling of P493 lymphocytes, cells were washed in 1× PBS and then resuspended in staining buffer (1× PBS, 2% fetal bovine serum) to a concentration of 2 × 107 cells per ml. Twenty microliters fluorescein isothiocyanate (FITC)-labeled CD71 antibody (BD Biosciences) was added to 1 × 106 cells (in 50-μl aliquots) and incubated on ice in the dark for 30 min. After washing with cold 1× PBS, cells were resuspended in 500 μl 1× PBS and analyzed by flow cytometry.
mRNA quantitation (real-time PCR).
Total RNA was extracted from P493-6 cells using Trizol (Invitrogen) or the RNeasy kit (QIAGEN). Quantitative real-time PCR expression was performed using the ABI 7700 sequence detection system. TFRC1, p53, and p21 mRNA expression levels were determined using predeveloped mixtures of specific probe and primers (PE Applied Biosystems) and the TaqMan One-step RT-PCR Master Mix kit (PE Applied Biosystems). A predeveloped probe and primers specific to 18S rRNA levels were used for normalization. All PCRs were performed in triplicate.
Quantitation of ChIP fragments was performed using the SYBR Green core reagent kit (PE Applied Biosystems) according to the manufacturer's instructions. TFRC1-specific primers were designed using the OMIGA program or Primer Express software (listed in Table S1 in the supplemental material). PCR parameters were optimized using the Failsafe Real-time PCR PreMix selection kit (EpiCentre, Madison, WI). Known quantities of 10-fold dilutions of total input DNA were used to generate standard curves for each primer pair. Relative amounts of each ChIP sample (expressed as a percentage of total input) were determined in the linear range according to their CT value. For each primer set, melting curves were used to verify the correct PCR product.
For microarray validation, mRNA samples were first reverse transcribed and then subjected to SYBR Green quantitative real-time PCR. Primers were designed to cross exon-exon junctions and span less than 400 bp of the target mRNA. Primer sequences are shown in Table S6 in the supplemental material.
Expression profiling using DNA microarrays.
Seventy-two hours after TFRC1 siRNA transfection, P493 cells were stained with the CD71-FITC antibody as described above and then sorted into TFRC1-negative and -positive populations using a Becton Dickinson cell sorter. Approximately 40 independent TFRC1 siRNA transfections were pooled for sorting. Samples from two independent biological replicates were then processed at the JHMI microarray facility and used to probe an Affymetrix U133 Plus 2.0 array. Total RNA was isolated from cells using the RNeasy minikit (QIAGEN). Five micrograms of starting total RNA from control and experimental cell preparations (two independent replicates for each) was processed using single-round RNA amplification protocols, following Affymetrix specifications (Affymetrix GeneChip Expression Analysis Technical Manual). Briefly, 5 μg of total RNA was used to synthesize first-strand cDNA using oligonucleotide probes with 24 oligo(dT) plus T7 promoter as a primer (Proligo LLC) and the SuperScript Choice system (Invitrogen). Following double-stranded cDNA synthesis, the product was purified by phenol-chloroform extraction, and biotinylated antisense cRNA was generated through in vitro transcription using the BioArray RNA High Yield transcript labeling kit (ENZO Life Sciences Inc). Fifteen micrograms of the biotinylated labeled cRNA was fragmented at 94°C for 35 min (100 mM Tris-acetate, pH 8.2, 500 mM potassium acetate, 150 mM magnesium acetate), and 10 μg of total fragmented cRNA was hybridized to the Affymetrix Human Genome GeneChip array U133Plus 2.0 for 16 h at 45°C with constant rotation (60 rpm). An Affymetrix Fluidics Station 450 instrument was then used to wash and stain the chips, removing the nonhybridized target and incubating with a streptavidin-phycoerythrin conjugate to stain the biotinylated cRNA. The staining was then amplified using goat immunoglobulin G as a blocking reagent and biotinylated antistreptavidin antibody (goat), followed by a second staining step with a streptavidin-phycoerythrin conjugate. Fluorescence was detected using the Affymetrix GeneChip scanner (GS 3000), and image analysis of each GeneChip was done through the GeneChip operating system software from Affymetrix (GCOS1.1.1), using the standard default settings. For comparison between different chips, global scaling was used, scaling all probe sets to a user-defined target intensity of 150.
To ascertain the quality control of the total RNA from the samples, we used the Agilent Bioanalyzer, Lab on a Chip technology and confirmed that all the samples had optimal rRNA ratios and clean run patterns. Likewise, this technology is used to confirm the quality of the RNA in the form of cRNA and fragmented cRNA. To assess the quality control of the hybridization, GeneChip image, and comparison between chips, we confirmed the following parameters: resulting scaling factor values within comparable range, background within average, significant percentage of present calls, 3′/5′ ratios of glyceraldehyde-3-phosphate dehydrogenase as a representation of housekeeping genes, and presence of internal spike controls.
The initial analysis of the expression results was based on pairwise comparisons among the different experimental conditions represented by the samples. Any transcript that showed an arbitrary value of at least a twofold change in expression level between the experimental sample and the control sample was considered significantly differentially expressed. All analysis was perfomed using the Li and Wong method. A lower bound of 2.0 was used as the cutoff for a significant change in gene expression.
RESULTS
Analysis of transferrin receptor 1 expression in response to c-Myc in P493-6 cells.
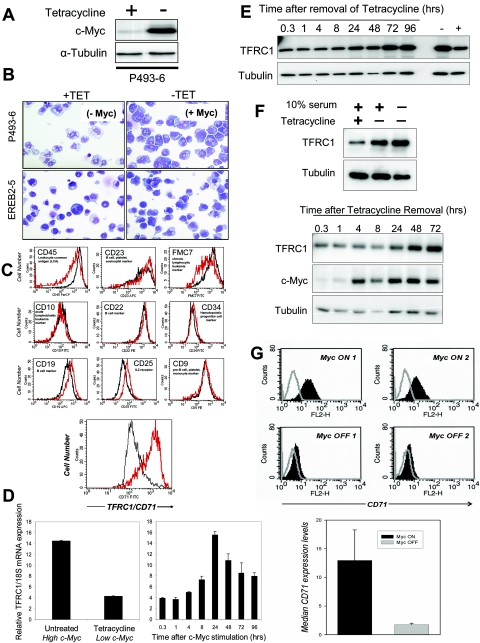
We used a previously described human B lymphocyte cell line, P493-6, that expresses a tetracycline-repressible c-MYC transgene to characterize c-Myc target genes. In the presence of tetracycline (Tet), P493-6 cells exhibit low levels of c-Myc protein, as evidenced by Western blotting (Fig. 1A) (42). P493-6 cells treated with tetracycline are small with pale blue cytoplasmic staining, whereas c-Myc induction leads to intense basophilic cytoplasmic staining and vacuoles that resemble Burkitt's lymphoma cells (Fig. 1B). In contrast, tetracycline-treated EREB2-5 cells, from which P493-6 cells were derived and which lack a Tet-repressible c-MYC allele, resemble untreated cells (Fig. 1B).
FIG.1.
TFRC1 Expression is responsive to c-Myc in B cells in vitro and in vivo. (A) Immunoblot analysis of c-Myc expression in P493-6. Cells were untreated or treated with 0.1 μg/ml tetracycline for 72 h. (B) Wright staining of P493-6 and EREB2-5 cells in the presence or absence of tetracycline. (C) FACS analysis of P493-6 cells using a panel of leukemia markers. Histograms represent expression of each cell surface marker in B cells with high c-Myc (red line; untreated) or low c-Myc (black line; Tet treated) expression levels. (D) Real-time PCR analysis of TFRC1 mRNA expression in untreated or Tet-treated P493-6 cells (left) and in P493-6 cells following induction of c-Myc by Tet removal (right). Bar graphs represent TFRC1 mRNA expression relative to 18S rRNA control. Error bars represent standard deviations derived from three independent measurements. (E) Immunoblot analysis of TFRC1 after Tet removal. α-Tubulin is used as a loading control. (F) Immunoblot analysis for TFRC1 and c-Myc expression in the absence of serum. P493-6 cells were treated with tetracycline for 72 h and then deprived of serum (0.1%) at the same time as tetracycline withdrawal to induce c-Myc expression. (G) Expression levels of transferrin receptor (CD71) on the surfaces of MycER-induced B-cell tumors. Top panel, flow cytometric analysis of CD71 expression. Cells were either left unstained (gray lines) or stained with the anti-CD71 antibody. Myc ON 1 and 2 refer to tumors from two different mice continuously treated with 4-OHT. Myc OFF 1 and 2 refer to tumors from two different mice initially treated with 4-OHT and then deprived of the hormone for 96 h. Bottom panel, quantitative analysis of CD71 expression. Median expression values plotted on the yaxis are differences between median fluorescence intensities of stained and unstained cells. Error bars represent standard deviations derived from two independent measurements in Myc ON and Myc OFF groups.
We sought to characterize c-Myc-induced phenotypes by examining alterations of cell surface markers typically used in clinical flow cytometric phenotyping of lymphomas. Because P493-6 cells serve as a model for Burkitt's lymphoma, we examined markers that have prognostic value to determine if any were indicative of c-Myc status. We analyzed the expression of 10 cell surface markers in tetracycline-treated (low c-Myc) or untreated (high c-Myc) cells. In contrast to other markers, TFRC1 (also known as CD71 or p90) was highly upregulated (sevenfold) in response to c-Myc (Fig. 1C).
Using the MYC cancer gene database (www.myccancergene.org), we found that TFRC1 is responsive to c-Myc in several previously reported microarray screens (11, 37, 47). Comprehensive analysis of gene expression using the Gene Expression Atlas database (www.expression.gnf.org) revealed that TFRC1 and c-MYC mRNA levels are expressed similarly in 91 human tissues and cell lines (52). We performed linear regression analysis (as described in Methods) on log-transformed mRNA expression values and found that c-MYC expression levels significantly correlated with TFRC1 expression values (r2 = 0.334; P value <0.00023). We also determined that c-Myc alters the expression of several well-characterized genes that regulate iron metabolism in three independent microarray analyses (see Table S1 in the supplemental material). These results are consistent with previous reports that link c-Myc to iron metabolism and provide further support that c-Myc regulates multiple components of the iron metabolome (5, 6, 58).
During the course of our study, one report using ChIP showed an area of binding for c-Myc in the TFRC1 gene in P493-6 cells (18). However, this study did not detect increased levels of TFRC1 mRNA at 8 h after c-Myc induction. In contrast, we discovered that TFRC1 mRNA levels are significantly increased 24 h after c-Myc induction in these cells (Fig. 1D). TFRC1 protein levels were also upregulated in response to c-Myc (Fig. 1E).
Because TFRC1 expression may reflect the proliferative status rather than the c-Myc status of P493-6 cells, we determined whether c-Myc could induce TFRC1 in serum-deprived cells. Western blotting confirms TFRC1 induction by c-Myc in the absence of cell proliferation (Fig. 1F). We also determined that c-Myc induction increased cell size in the absence of serum (data not shown).
Since TFRC1 expression is tightly coupled to c-Myc expression, we sought to determine whether TFRC1 is activated by c-Myc in vivo. We investigated whether c-Myc regulates TFRC1 expression levels in a murine B-cell lymphoma model, which is based on retroviral transduction of p53-null bone marrow cells with a Myc-encoding retrovirus (25, 59, 61). Using MycER retroviruses, which express a chimeric protein of c-Myc and the hormone binding domain of the estrogen receptor, neoplasms were generated in animals whose exponential growth was contingent upon continuous administration of 4-hydroxytamoxifen (4-OHT) (13, 60). Growing (Myc ON) and stagnant (Myc OFF) neoplasms from mice were used to prepare single-cell suspensions of neoplastic B-lymphocytes. Viable neoplastic cells were stained with a TFRC1 antibody, and expression levels were assessed from two different tumors (Fig. 1G). Following inactivation of c-Myc, we observed a sixfold downregulation of TFRC1. Thus, maintenance of transferrin receptor expression is dependent on the presence of active MycER in vivo.
Phylogenetic footprinting and chromatin immunoprecipitation validate TFRC1 as a direct MYC target gene.
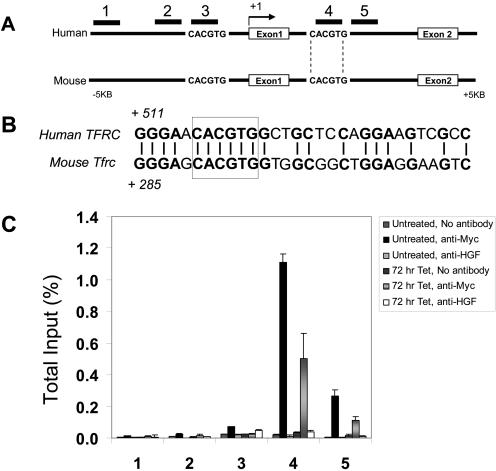
Although TFRC1 mRNA and protein levels are increased by c-Myc, whether TFRC1 is a direct or indirect c-Myc target gene has not been fully established. We and others use ChIP to determine where c-Myc binds within target genes (7, 18, 21, 62). We previously observed that c-Myc binds to phylogenetically conserved canonical E boxes, 5′-CACGTG-3′, in the promoter regions or intron 1 of its target genes (22, 63). A detailed analysis of the genomic locus of the human and murine transferrin receptor 1 gene was performed to identify conserved canonical E boxes. Sequences were downloaded using the UCSC and NCBI databases and analyzed using OMIGA software (Oxford Molecular Limited, Oxford, United Kingdom) (22). An alignment of the human and mouse TFRC1 loci, with two canonical E boxes found flanking exon 1, is shown in Fig. 2A. While both E boxes are conserved, only the intron 1 E box is contained within a 30-bp window of 65% nucleotide identity, a criterion we have previously used to infer functional conservation (Fig. 2B) (22).
FIG. 2.
Chromatin immunoprecipitation validates TFRC1 as a direct c-Myc target gene. (A) Sequence alignment of the human and mouse transferrin receptor genomic locus (from 5 kb upstream of the transcriptional start site through intron 1) reveals two canonical E boxes. The E box flanked by vertical lines falls within a window of 65% nucleotide identity between human and mouse. PCR amplicons that were analyzed for c-Myc binding are indicated by numbers. (B) Alignment of the human and mouse sequences of the intron 1 E box (boxed). (C) Quantification of c-Myc binding by real-time PCR analysis (expressed as a percentage of total input DNA). ChIP was performed on untreated P493-6 cells or cells treated with Tet for 72 h using anti-Myc antibody or anti-hepatocyte growth factor as a control antibody. Error bars represent standard deviations derived from three independent measurements. A representative experiment is shown.
We localized c-Myc binding to TFRC1 using scanning ChIP in P493-6 cells (Fig. 2C) (62). Using real-time PCR quantitation, we obtained evidence for in vivo association of c-Myc with the genomic region containing the conserved intron 1 CACGTG sequence 516 nucleotides downstream of the transcriptional start site (Fig. 2C, amplicon 4). A decrease in signal intensity was observed upon scanning away from the canonical E box to amplicon 5 (approximately 106 bp away). Because amplicon 3, which contained the canonical E box −2703 to −2698 bp upstream of the TFRC1 transcriptional start site, was extraordinarily amplified from total input DNA, we examined the sequence of this region and found that it lies within a repetitive short interspersed nucleotide element. We then scanned approximately 115 bp upstream to a region just outside the short interspersed nucleotide element and observed that amplicon 2 also demonstrated background levels of binding to c-Myc. (Primer sequences used in our ChIP experiments are shown in Table S2 in the supplemental material). These data suggest that c-Myc binds exclusively to the conserved canonical E-box sequence in intron 1 of the TFRC1 genomic region and provide strong evidence that TFRC1 is a direct c-Myc target gene.
Transferrin receptor 1 is required for cellular proliferation and cell cycle progression in P493-6 cells.
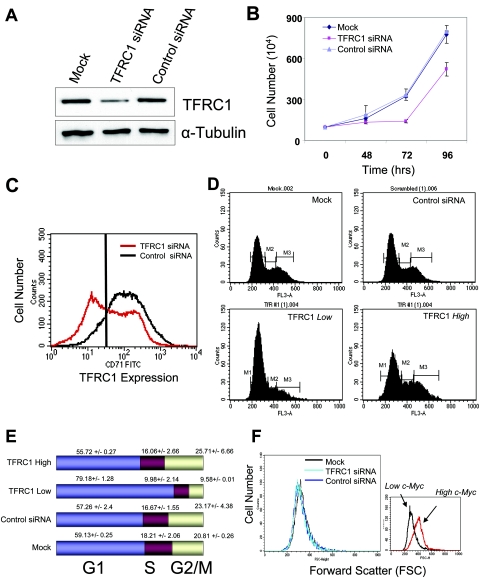
Since TFRC1 expression is elevated in lymphomas and is regulated by c-Myc, we sought to determine whether TFRC1 is required for cell cycle proliferation and cell size control. With the P493-6 system in place to study these c-Myc-mediated phenotypes, we utilized RNAi to inhibit TFRC1 expression in these cells (14). siRNA duplexes were designed to inhibit expression of TFRC1 and introduced into P493-6 cells by electroporation. After 72 h, TFRC1 protein levels were diminished by approximately 60% (Fig. 3A). Inhibition of TFRC1 also resulted in decreased rates of cell proliferation (Fig. 3B).
FIG. 3.
RNA interference of TFRC1 expression abrogates cell proliferation and cell cycle progression. (A) Immunoblot analysis of TFRC1 levels 72 h after transfection with siRNA oligonucleotides in unsorted cell populations. α-Tubulin is used as a loading control. (B) Growth rates are diminished after suppression of TFRC1. Cell proliferation was measured in mock-electroporated cells or in cells electroporated with TFRC1 or control siRNAs. Error bars represent standard deviations from two independent measurements. At least three independently performed experiments yielded similar results. (C) FACS analysis of anti-TFRC1-FITC labeled cells 72 h after transfection with siRNA oligonucleotides. (D) Cell cycle profiles after knockdown of TFRC1. Cells were double labeled with 7-amino-actinomycin and an anti-TFRC1-FITC antibody. (E) Bar graphs representing mean values and standard deviations of each cell cycle phase derived from three independent experiments. (F) Cell size remains unaffected after RNAi-mediated suppression of TFRC1. Forward light scattering (FSC) histograms obtained by FACS analysis are shown.
Staining of P493-6 cells with a FITC-conjugated anti-TFRC1 antibody following electroporation revealed two cell populations, one with low TFRC1 expression and one with normal levels of TFRC1 expression (Fig. 3C), reflecting the transfection efficiency of this cell line. By double labeling cells with an anti-TFRC1 antibody and the DNA binding dye 7-amino-actinomycin, we sought to compare cell cycle profiles of the cell population with low TFRC1 expression to control cell populations (Fig. 3D). Inhibition of TFRC1 expression resulted in a significant G1 accumulation (Fig. 3E) (79.18% ± 0.27% compared to 55.72% ± 2.4% of cells in G1 for TFRC1-negative and TFRC1-positive populations, respectively). In contrast, cell size did not change with TFRC1 gene silencing (Fig. 3F). Taken together, these data demonstrate that TFRC1 is necessary for progression through the G1-S phase transition and cell proliferation.
To further characterize the cell cycle arrest phenotype following silencing of TFRC1, we examined mRNAs that are involved in cell cycle regulation. In order to select for a homogenous population of cells, we sorted for TFRC1-low-expressing cells. Real-time PCR quantitation detected a twofold increase in p21 mRNA levels in TFRC1-low-expressing cells compared to controls, whereas p53 mRNA levels remained the same (see Fig. S2 in the supplemental material). Immunoblot analysis further revealed increased levels of the p53 and p21 proteins, whereas p27 levels remained constant (see Fig. S2 in the supplemental material). We also performed TFRC1 knockdown in the human erythroleukemia K562 cells. Despite the absence of functional p53 in these cells, TFRC1 siRNA-treated cells maintain a G1 phase arrest (our unpublished observations).
As an additional control, we also silenced the c-Myc target gene JTV1. Diminished JTV1 expression, however, did not affect P493-6 cell proliferation or cell size (data not shown). These data provide evidence that TFRC1 expression is necessary for c-Myc-mediated cell proliferation but not cell size increase.
Iron chelation inhibits cell proliferation and cell cycle progression but not cell size increase in P493-6 cells.
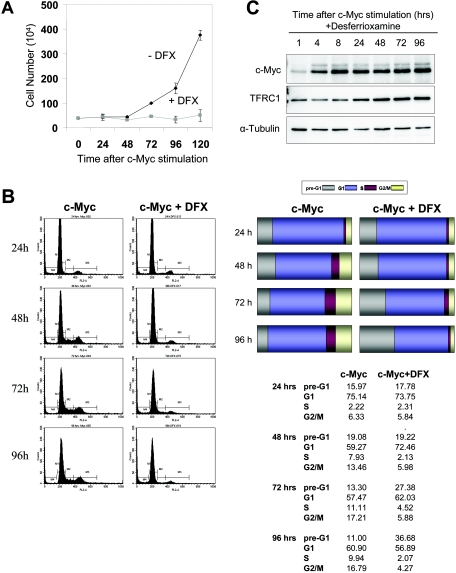
In order to determine whether the cell cycle arrest we observed following TFRC1 knockdown was secondary to depletion of intracellular iron pools, we directly diminished intracellular iron by chemical chelation. Desferrioxamine (DFX) is a potent iron chelator that has been shown to inhibit the growth of aggressive tumors, such as neuroblastoma and leukemia (3, 4, 46) and is currently used to treat iron-overload disorders (45). By 120 h after tetracycline removal, cells stimulated by c-Myc in the presence of DFX failed to proliferate, in contrast to control cells stimulated by c-Myc alone (Fig. 4A). Cell cycle analysis revealed that Tet-treated cells are arrested in G1, whereas induction of c-Myc allows progression of cells through the G1-S transition. In contrast, DFX-treated cells accumulated in G1 phase at early time points (Fig. 4B). With longer exposure to DFX (72 to 96 h), the lack of cell cycle progression in the c-Myc-plus-DFX cells was associated with an increase in cells undergoing apoptosis. These data demonstrate that short-term iron deprivation mimics TFRC1 knockdown and inhibits cellular proliferation in P493-6 cells. Previous studies have shown that DFX treatment of neuroblastoma cells inhibits N-Myc expression (16). We thus sought to determine whether DFX treatment resulted in nonspecific inhibition of c-Myc or TFRC1 expression. Western blot analysis confirmed that c-Myc and TFRC1 are expressed normally in the presence of DFX (Fig. 4C).
FIG. 4.
Iron depletion inhibits c-Myc-mediated cell proliferation and cell cycle progression. (A) Growth rates of P493-6 cells grown in the presence or absence of 100 μM DFX after tetracycline withdrawal. (B) Cell cycle profiles obtained by propidium iodide labeling of cells stimulated by c-Myc in the presence or absence of DFX. c-Myc stimulation results in cell cycle progression. In contrast, DFX-treated cells accumulate in G1 at early time points and subsequently undergo apoptosis at later time points. Bar graphs represent the percentage of cells in each phase of the cell cycle. Results of a representative experiment are shown. One independent experiment yielded similar results. (C) Immunoblot analysis of c-Myc and TFRC1 after DFX treatment. α-Tubulin is used as a loading control. Error bars for all panels represent standard deviations derived from at least two independent experiments.
Since iron is involved in multiple metabolic pathways and attaining a threshold cell size in G1 is required for cell proliferation, it is plausible that iron deprivation could inhibit c-Myc-mediated cell size increase. Flow cytometric analysis revealed that DFX treatment had no significant effect on cell size (data not shown). Thus, these experiments underscore the requirement of iron for cellular proliferation and clearly demonstrate that disruption of iron metabolism has a profound effect on cell cycle progression without affecting cell size. Moreover, these data highlight the importance of TFRC1 in maintaining sufficient intracellular iron levels for proliferation.
TFRC1 depletion alters cell cycle and apoptosis-promoting genes.
To globally assess alterations in expression of cell cycle regulatory genes, we examined expression profiles in TFRC1 siRNA-treated B cells. Our oligonucleotide microarray studies of TFRC1-low cells revealed that the levels of 849 out of approximately 10,000 expressed transcripts were altered by decreased TFRC1 expression. The 202 downregulated transcripts included cell division cycle genes, cyclins, and other factors required for DNA replication. The 647 upregulated transcripts include p53-responsive and apoptosis-promoting genes. A summary of the various gene categories affected by TFRC1 inhibition is shown in Table 1.
TABLE 1.
Transcripts responsive to TFRC1 knockdowna
| Class | Gene product (gene name) | Fold change
|
|
|---|---|---|---|
| Microarray value | Real-time PCR validation | ||
| Cell cycle | Cell division cycle 20 (CDC20) | −3.29 | −3.17 |
| Cyclin B1 (CCNB1) | −3.24 | −2.53 | |
| Cyclin A2 (CCNA2) | −2.79 | −3.62 | |
| Cell division cycle (CDC2) | −2.86 | ||
| Growth arrest and DNA damage-inducible 45 alpha (GADD45A) | +2.66 | +2.90 | |
| Cyclin-dependent kinase inhibitor 1A (CDKN1A) | +2.56 | +1.51 | |
| Inhibitor of DNA binding 2 (ID2) | +2.24 | ||
| Apoptosis | BCL2 binding component 3 (BBC3/PUMA) | +3.05 | +2.18 |
| Annexin A4 (ANXA4) | +3.91 | +3.89 | |
| Annexin A1 (ANXA1) | +2.12 | ||
| Tumor necrosis factor receptor superfamily, member 6 (TNFRSF6) | +2.25 | ||
| p53 regulated | Cyclin-dependent kinase inhibitor 1A (CDKN1A) | +2.56 | +1.51 |
| p53 target zinc finger protein (WIG1) | +2.42 | +2.27 | |
| BCL2 binding component 3 (BBC3/PUMA) | +3.05 | +2.18 | |
| TP53-activated protein 1 (TP53AP1) | +2.43 | +2.13 | |
| Tumor protein p53-inducible nuclear protein 1 (TP53INP) | +3.72 | +4.68 | |
| DNA replication | DNA2 DNA replication helicase 2-like (DNA2L) | −2.49 | |
| Topoisomerase (DNA) II alpha (TOP2A) | −2.32 | −3.72 | |
| Polymerase (DNA directed), epsilon 2 (POLE2) | −2.29 | −2.79 | |
| Chromosome organization | BUB1 budding uninhibited by benzimidazoles 1 homolog (BUB1) | −3.05 | −3.16 |
| Centromere protein A (CENPA) | −2.51 | ||
| Centromere protein F (CENPF) | −2.45 | ||
Fold change represents expression difference between scrambled control siRNA-treated cells and TFRC1 (low)-depleted cells.
To analyze our microarray data set, we utilized Expression Analysis Systematic Explorer (EASE), a gene ontology software application (24). Each biologic theme is determined from a list of genes and assigned an EASE score, a measure of statistical significance that is similar to a one-tailed Fisher exact probability test. Remarkably, each of the top 15 gene categories downregulated by inhibition of TFRC1 is associated with the regulation of the cell cycle (see Table S3 in the supplemental material). Using an EASE score cutoff of 1.84E-07, the annotated list of down-regulated genes that fall within a cell cycle regulation category is shown in Table S4 in the supplemental material. Analysis of upregulated genes using EASE further revealed that many fall into apoptotic and programmed cell death categories (see Table S5 in the supplemental material). This is consistent with the observed increase in apoptotic cells after prolonged iron deprivation in P493-6 cells (Fig. 4B).
To validate our microarray results, we performed quantitative real-time PCR analysis to measure expression of diverse genes that are p53 targets or are involved in cell cycle control or apoptosis (see Table 1, last column; also see Fig. S3 in the supplemental material). In summary, 13 out of 15 tested genes from our microarray experiments showed concordant expression changes by real-time PCR. (Primer sequences used in our microarray confirmation are shown in Table S6 in the supplemental material).
To further delineate the role of TFRC1 within the c-Myc target gene network, we sought to compare our microarray data set described above with that of all c-Myc-responsive genes in P493-6 cells. To this end, we performed expression profiling with P493-6 cells under conditions of high versus low c-Myc expression (using untreated versus Tet-treated cells). Out of approximately 10,000 expressed transcripts, 2,679 genes were c-Myc responsive in P493-6 cells (29). Of these c-Myc-responsive genes, 175 were also altered by TFRC1 knockdown. Upon further analysis of these 175 genes, 59 c-Myc-induced genes are downregulated by TFRC1 inhibition (see Table S7B in the supplemental material). Consistent with our previous observation that TFRC1 depletion resulted in a G1 arrest but did not affect cell size, many of the genes that responded to both c-Myc and TFRC1 levels are involved in cell cycle regulation. In fact, EASE analysis reveals that the “mitotic cell cycle” is the most statistically significant gene category (with a score of 3.92E-14) from this list. The remaining c-Myc-responsive genes, including those involved in metabolism and ribosomal biogenesis, were predominantly unaffected by TFRC1 knockdown. Examples of these genes are represented in Table S8 in the supplemental material.
Ectopic TFRC1 expression confers a growth advantage in limiting serum conditions.
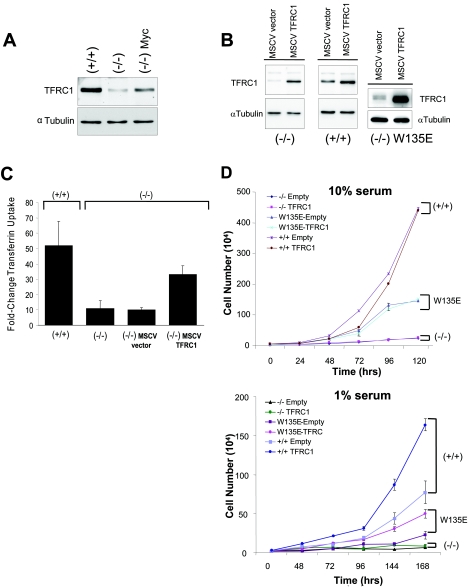
Having demonstrated that TFRC1 is necessary for cellular proliferation, we next sought to determine whether TFRC1 overexpression is sufficient to provide a growth advantage under conditions with limiting growth factors (including transferrin) that may reflect the physiologic milieu of cancers (49, 57). We reasoned that cells grown in limiting 1% serum conditions may mimic the physiologic context of tumor growth, where cells must compete for limiting growth factors and nutrients. We chose to study wild-type Rat1 TGR fibroblasts, c-Myc null Rat1 cells, and null cells that have been reconstituted with a hypomorphic mutant MYC W135E allele (23, 33, 41). We first determined the expression of TFRC1 in c-MYC null cells (HO15.19), compared to the wild type (TGR), and null cells reconstituted with murine leukemia virus-driven wild-type c-MYC (HO15.19-MYC). We found that TFRC1 expression is greatly diminished in null fibroblasts, demonstrating that c-Myc is necessary for full physiologic expression of TFRC1 (Fig. 5A).
FIG. 5.
Ectopic TFRC1 expression in Rat1 fibroblasts confers growth advantage in limiting (1%) serum conditions. (A) Immunoblot analysis of endogenous TFRC1 expression in c-MYC−/− (HO15.19), c-MYC+/+ (TGR), and c-MYC−/− cells reconstituted with human c-Myc (−/− Myc) Rat1 fibroblasts. α-Tubulin is used as a loading control. (B) Immunoblot analysis demonstrating TFRC1 overexpression in c-MYC null (HO15.19), wild-type (TGR), and W135E mutant cells infected with a human TFRC1 retrovirus. α-Tubulin is used as a loading control. (C) Transferrin uptake in +/+ (TGR) and −/− (HO15.19) cells and in −/− cells infected with an empty vector or a TFRC1 retrovirus. Quantitation was determined by FACS analysis after incubating cells with fluorescence-labeled transferrin for 30 min. Fold changes shown refer to median Tf uptake levels compared to those for unlabeled cells. The bar graph represents mean values and standard deviations derived from four independent experiments. (D) Growth rates under normal (10%) and limiting (1%) serum conditions. Error bars represent standard deviations derived from three independent measurements. The graph is representative of two independent experiments.
To assess whether ectopic expression of TFRC1 affected the growth of wild-type or knockout cells, we introduced a human TFRC1-expressing vector or empty vector alone into both cell lines by retroviral transduction. Western blot analysis confirms retroviral expression of TFRC1 (Fig. 5B). Anti-TFRC1 staining by flow cytometry verified functional TFRC1 expression at the cell surface (data not shown).
Using labeled transferrin (Tf), we found a direct correlation between TFRC1 expression and the functional uptake of Tf in c-MYC null and wild-type cells (Fig. 5C). Quantitation of labeled-transferrin uptake revealed that the magnitude of Tf uptake is 4.7-fold greater in wild-type cells than in c-MYC knockout cells. Additionally, we observed that transferrin uptake increased by 3.3-fold in c-MYC knockout cells expressing ectopic TFRC1 (Fig. 5C).
It was previously reported that reconstitution of c-MYC null cells with a hypomorphic mutant MYC W135E allele resulted in an intermediate growth rate between those of wild-type cells and c-MYC null cells (41). Intriguingly, TFRC1 expression was diminished in W135E cells compared to that in wild-type rat fibroblasts (data not shown). W135E cells expressing a TFRC1 or control retrovirus were generated as demonstrated by Western blotting (Fig. 5B, right panel). Despite the ability to increase levels of transferrin uptake, TFRC1 overexpression was insufficient to rescue the slow growth rates of these cells in normal (10%) serum culture conditions (Fig. 5D, top panel). However, ectopic TFRC1 expression significantly enhanced the growth rates not only of W135E c-MYC mutant cells but also of wild-type cells when grown in limiting (1%) serum conditions (Fig. 5D, bottom panel). These data suggest that upregulation of TFRC1 by c-Myc may be advantageous to cells growing in limiting nutrients, the typical environment of tumor cells.
TFRC1 promotes tumorigenicity in Rat1a-Myc cells.
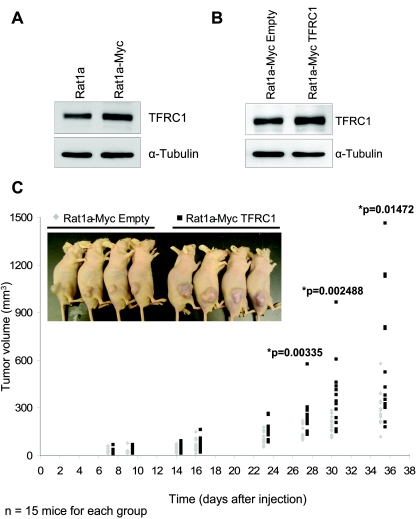
Last, we tested directly whether TFRC1 could enhance the rate of tumorigenesis in a well-characterized model system, Rat1a fibroblasts overexpressing c-Myc. Rat1a cells are immortalized, nontransformed fibroblasts that do not form tumors when injected subcutaneously into nude mice. Ectopic expression of select oncogenes, including c-Myc, results in cellular transformation as indicated by their ability to form tumors in nude mice and promote the anchorage-independent growth of colonies in soft agar (50, 51). We observed that TFRC1 is expressed at approximately twofold-higher levels in Rat1a-Myc cells than in Rat1a cells (Fig. 6A).
FIG. 6.
TFRC1 enhances tumorigenesis in Rat1a-Myc cells. (A) Western blot analysis of TFRC1 in Rat1a and Rat1a-Myc cells. α-Tubulin is used as a loading control. (B) Western blot analysis demonstrating TFRC1 overexpression in Rat1a-Myc cells. α-Tubulin is used as a loading control. (C) Analysis of tumor volume in nude mice. A two-tailed Student t test (type 3, unequal variance) was used to determine whether the difference between groups was statistically significant. A representative experiment is shown (n = 15 mice for each group). One independent tumorigenesis experiment yielded similar results.
In order to determine whether TFRC1 cooperates with c-Myc in promoting tumorigenesis, we generated a stable cell line with enforced TFRC1 expression in Rat1a-Myc cells. Western blot analysis confirms retroviral expression of TFRC1 in these cells, which already express high levels of endogenous TFRC1 (Fig. 6B). We examined the growth rates of these cells under normal (10%) and limiting (1%) serum culture conditions. Consistent with our previous results using W135E mutant and TGR wild-type rat fibroblasts, enforced TFRC1 expression enhanced the growth rates of Rat1a-Myc cells under limiting conditions (see Fig. S4A in the supplemental material). We then subcutaneously injected Rat1a-Myc (Empty) and Rat1a-Myc (TFRC1) cells into 4-week-old nude mice and assessed tumor volume over time. As demonstrated in Fig. 6C, Rat1a-Myc cells with ectopic TFRC1 expression exhibited a statistically significant increase in the rate of tumorigenesis compared to Rat1a-Myc (Empty) cells (P values = 0.00335, 0.00248, and 0.01472 at days 27, 30, and 35, respectively). These results provide compelling evidence that TFRC1 enhances tumorigenesis in this system.
DISCUSSION
TFRC1 is a critical gene in the c-Myc target gene network.
c-Myc is one of the most frequently dysregulated proteins in human malignancies, underscoring the importance of achieving a more complete understanding of the mechanisms through which this oncoprotein drives tumorigenesis. The fact that human or Drosophila c-Myc regulates between 10 and 15% of genes presents a formidable challenge. Thus, it is critical to identify downstream targets and pathways that are essential for each of its biologic functions in both normal and neoplastic cells.
The results reported above lead to three main conclusions relevant to the role of TFRC1 in the c-Myc target gene network and B-cell lymphoma. First, we have demonstrated that TFRC1 is a direct transcriptional target of the c-MYC proto-oncogene. This likely explains why TFRC1 is commonly overexpressed in tumors. Second, TFRC1 is necessary for B-lymphocyte proliferation based on experiments using RNA interference to inhibit its expression. Enforced TFRC1 expression also confers a distinct growth advantage to cells grown under limiting serum conditions, suggesting that upregulation of this pathway by c-Myc promotes cell proliferation under conditions that are analogous to the environment of a tumor cell. Third, we present compelling evidence that TFRC1 promotes tumorigenesis in Rat1a fibroblasts overexpressing c-Myc.
Our knockdown experiments and microarray studies revealed that many cell cycle regulatory genes and downstream p53 target genes are affected by reduction of TFRC1 expression. Since iron depletion is known to affect many cellular processes, the underlying mechanism for these alterations in gene expression and the observed cell cycle arrest is likely complex and dependent on multiple pathways. One possible mechanism contributing to p53 activation is inhibition of ribonucleotide reductase (RR), an iron-dependent enzyme required for DNA replication. Treatment with the RR inhibitor hydroxyurea has been shown to cause a p53-dependent G0/G1 arrest (31). Similarly, knockdown of TFRC1 may render RR inactive by depriving cells of adequate intracellular iron, thus depleting deoxyribonucleotide pools and activating p53. Consistent with this model, we observed increased levels of p53 protein but not mRNA, as is known to occur following inhibition of DNA synthesis.
Based on our experiments with K562 lymphocytes which lack wild-type p53, cell cycle arrest persists in TFRC1 siRNA-treated cells. These studies reveal that although induced if present (as in P493-6 cells), wild-type p53 is not required for the G1 phase arrest in response to TFRC1 inhibition. These data support the idea that iron depletion has pleiotropic effects on the cell cycle regulatory apparatus. Nevertheless, our findings firmly establish that TFRC1 is a critical downstream target of c-Myc that is necessary for cellular proliferation and promotes tumorigenesis. Moreover, analysis of gene expression changes following TFRC1 depletion in the context of all genes that are responsive to c-Myc has allowed us to pinpoint the role of TFRC1 within the c-Myc target gene network as a whole. This type of analysis should also prove useful for identifying specific pathways affected by other c-Myc target genes.
Our microarray data also revealed that in addition to TFRC1, c-Myc controls several well-characterized genes that either regulate iron metabolism or require iron for their normal function (see Table S1 in the supplemental material). These include divalent metal transporter 1 (DMT1 or Nramp2), which transports iron out of endosomes into the cytoplasm, and succinate dehydrogenase B, which catalyzes the oxidation of succinate to fumarate during the citric acid cycle. Frataxin, which encodes a mitochondrial membrane protein with putative roles in iron-sulfur cluster synthesis and mitochondrial iron metabolism, was also upregulated by c-Myc. These results provide further support that c-Myc regulates multiple components of the iron metabolome. Future studies are necessary to determine the contribution of these additional factors to c-Myc-mediated proliferation and transformation.
TFRC1 as a clinical marker and therapeutic target for human cancer.
For many years, clinical studies have unknowingly taken advantage of altered TFRC1 expression for the detection of tumors. 67Gallium uptake, now known to be through TFRC1, has been used clinically to image lymphomas in patients (17, 55). Since c-Myc is known to play an important role in lymphomagenesis and both c-Myc and TFRC1 have been linked to the development of aggressive diffuse large B-cell lymphoma (2, 32), it is not surprising that TFRC1 has served as a useful clinical marker.
Moreover, several lines of evidence demonstrate that TFRC1 expression levels are indicative of c-Myc expression. TFRC1 is among a select group of genes that are overexpressed in a murine c-Myc-induced prostate cancer model and in primary human prostate cancers (15). Numerous expression profiling studies have documented increased TFRC1 expression in cancers of the blood, lung, colon, and skin (44). A recent study using a correlation-based model to characterize the gene interaction network of MycER-stimulated cells identified TFRC1 as one such c-Myc-activated gene (43a). Our studies, however, not only are correlative but also reveal a direct link between c-MYC expression and TFRC1 expression. Our experiments also demonstrate for the first time that TFRC1 can enhance in vivo tumor growth by cooperating with c-Myc in Rat1a fibroblasts.
The accessibility of TFRC1 as a membrane receptor has made it an attractive target for cancer therapy. Past therapeutic studies with anti-TFRC1 antibodies have met with limited clinical success, perhaps due to the lack of appropriate phenotyping for TFRC1. More recently a monoclonal TFRC1 antibody, A24, was shown to induce apoptosis in T lymphocytes from patients with CD71-positive T-cell leukemias (26, 34, 53). In fact, these studies are consistent with our microarray data where we identified apoptotic genes upregulated in response to TFRC1 inhibition in P493-6 B lymphocytes. Thus, our studies provide experimental support for anticancer therapies that target TFRC1. Future efforts to delineate the molecular mechanisms underlying the cell cycle arrest which occurs following TFRC1 depletion may allow the development of other therapies that complement the interruption of iron uptake in the treatment of cancers.
In conclusion, we have demonstrated that the transferrin receptor is a direct c-Myc target gene that is necessary for c-Myc-mediated cell cycle progression but not cell size increase. By conferring a significant growth advantage on cells growing in limiting nutrients, activation of TFRC1 contributes to the ability of c-Myc to reprogram the metabolic state of the cell such that growth in the severe environment of a tumor is promoted. Further definition of the c-Myc transcription factor-target gene network through the identification of other principal tumor-promoting targets will advance our understanding and perhaps control of the transformation process.
Supplementary Material
Acknowledgments
We thank D. Eick, J. Sedivy, L. Penn, and J. Basilion for valuable cell lines and reagents, D. Warren and Y. Yang for assistance with electroporation, L. Blosser and A. Tam for flow sorting, D. Arking for statistical analysis, and F. Martinez Murillo and C. Jie for microarray hybridization and analysis. We also thank J. Mendell, L. Gardner, M. McDevitt, R. Osthus, and L. Lee for critical reading of the manuscript.
This work was supported by NCI grants CA57341 (C.V.D.), CA097932 (A.T.-T.), and CA102709 (A.T.-T.) and NIH-NHLBI grant T32HL007525 (K.A.O.). C.V.D. is the Johns Hopkins Family Professor in Oncology Research. J. Kim is a Howard Hughes Predoctoral Fellow.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adhikary, S., and M. Eilers. 2005. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell. Biol. 6:635-645. [DOI] [PubMed] [Google Scholar]
- 2.Alizadeh, A. A., M. B. Eisen, R. E. Davis, C. Ma, I. S. Lossos, A. Rosenwald, J. C. Boldrick, H. Sabet, T. Tran, X. Yu, J. I. Powell, L. Yang, G. E. Marti, T. Moore, J. Hudson, Jr., L. Lu, D. B. Lewis, R. Tibshirani, G. Sherlock, W. C. Chan, T. C. Greiner, D. D. Weisenburger, J. O. Armitage, R. Warnke, R. Levy, W. Wilson, M. R. Grever, J. C. Byrd, D. Botstein, P. O. Brown, and L. M. Staudt. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403:503-511. [DOI] [PubMed] [Google Scholar]
- 3.Becton, D. L., and P. Bryles. 1988. Deferoxamine inhibition of human neuroblastoma viability and proliferation. Cancer Res. 48:7189-7192. [PubMed] [Google Scholar]
- 4.Blatt, J., and S. Stitely. 1987. Antineuroblastoma activity of desferoxamine in human cell lines. Cancer Res. 47:1749-1750. [PubMed] [Google Scholar]
- 5.Bowen, H., T. E. Biggs, S. T. Baker, E. Phillips, V. H. Perry, D. A. Mann, and C. H. Barton. 2002. c-Myc represses the murine Nramp1 promoter. Biochem. Soc. Trans. 30:774-777. [DOI] [PubMed] [Google Scholar]
- 6.Bowen, H., A. Lapham, E. Phillips, I. Yeung, M. Alter-Koltunoff, B. Z. Levi, V. H. Perry, D. A. Mann, and C. H. Barton. 2003. Characterization of the murine Nramp1 promoter: requirements for transactivation by Miz-1. J. Biol. Chem. 278:36017-36026. [DOI] [PubMed] [Google Scholar]
- 7.Boyd, K. E., J. Wells, J. Gutman, S. M. Bartley, and P. J. Farnham. 1998. c-Myc target gene specificity is determined by a post-DNAbinding mechanism. Proc. Natl. Acad. Sci. USA 95:13887-13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, Y., O. Zak, P. Aisen, S. C. Harrison, and T. Walz. 2004. Structure of the human transferrin receptor-transferrin complex. Cell 116:565-576. [DOI] [PubMed] [Google Scholar]
- 9.Chitambar, C. R., E. J. Massey, and P. A. Seligman. 1983. Regulation of transferrin receptor expression on human leukemic cells during proliferation and induction of differentiation. Effects of gallium and dimethylsulfoxide. J. Clin. Investig. 72:1314-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole, M. D., and S. B. McMahon. 1999. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene 18:2916-2924. [DOI] [PubMed] [Google Scholar]
- 11.Coller, H. A., C. Grandori, P. Tamayo, T. Colbert, E. S. Lander, R. N. Eisenman, and T. R. Golub. 2000. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. USA 97:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang, C. V. 1999. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 19:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eilers, M., D. Picard, K. R. Yamamoto, and J. M. Bishop. 1989. Chimaeras of myc oncoprotein and steroid receptors cause hormone-dependent transformation of cells. Nature 340:66-68. [DOI] [PubMed] [Google Scholar]
- 14.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 15.Ellwood-Yen, K., T. G. Graeber, J. Wongvipat, M. L. Iruela-Arispe, J. Zhang, R. Matusik, G. V. Thomas, and C. L. Sawyers. 2003. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell 4:223-238. [DOI] [PubMed] [Google Scholar]
- 16.Fan, L., J. Iyer, S. Zhu, K. K. Frick, R. K. Wada, A. E. Eskenazi, P. E. Berg, N. Ikegaki, R. H. Kennett, and C. N. Frantz. 2001. Inhibition of N-myc expression and induction of apoptosis by iron chelation in human neuroblastoma cells. Cancer Res. 61:1073-1079. [PubMed] [Google Scholar]
- 17.Feremans, W., W. Bujan, P. Neve, J. P. Delville, and L. Schandene. 1991. CD71 phenotype and the value of gallium imaging in lymphomas. Am. J. Hematol. 36:215-216. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez, P. C., S. R. Frank, L. Wang, M. Schroeder, S. Liu, J. Greene, A. Cocito, and B. Amati. 2003. Genomic targets of the human c-Myc protein. Genes Dev. 17:1115-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gatter, K. C., G. Brown, I. S. Trowbridge, R. E. Woolston, and D. Y. Mason. 1983. Transferrin receptors in human tissues: their distribution and possible clinical relevance. J. Clin. Pathol. 36:539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grandori, C., and R. N. Eisenman. 1997. Myc target genes. Trends Biochem. Sci. 22:177-181. [DOI] [PubMed] [Google Scholar]
- 21.Grandori, C., J. Mac, F. Siebelt, D. E. Ayer, and R. N. Eisenman. 1996. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 15:4344-4357. [PMC free article] [PubMed] [Google Scholar]
- 22.Haggerty, T. J., K. I. Zeller, R. C. Osthus, D. R. Wonsey, and C. V. Dang. 2003. A strategy for identifying transcription factor binding sites reveals two classes of genomic c-Myc target sites. Proc. Natl. Acad. Sci. USA 100:5313-5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanson, K. D., M. Shichiri, M. R. Follansbee, and J. M. Sedivy. 1994. Effects of c-myc expression on cell cycle progression. Mol. Cell. Biol. 14:5748-5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hosack, D. A., G. Dennis, Jr., B. T. Sherman, H. C. Lane, and R. A. Lempicki. 2003. Identifying biological themes within lists of genes with EASE. Genome Biol. 4:R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iritani, B. M., and R. N. Eisenman. 1999. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc. Natl. Acad. Sci. USA 96:13180-13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kemp, J. D., K. M. Smith, J. M. Mayer, F. Gomez, J. A. Thorson, and P. W. Naumann. 1992. Effects of anti-transferrin receptor antibodies on the growth of neoplastic cells. Pathobiology 60:27-32. [DOI] [PubMed] [Google Scholar]
- 27.Kuhn, L. C. 1989. The transferrin receptor: a key function in iron metabolism. Schweiz. Med. Wochenschr. 119:1319-1326. [PubMed] [Google Scholar]
- 28.Larrick, J. W., and P. Cresswell. 1979. Modulation of cell surface iron transferrin receptors by cellular density and state of activation. J. Supramol. Struct. 11:579-586. [DOI] [PubMed] [Google Scholar]
- 29.Li, F., Y. Wang, K. I. Zeller, J. J. Potter, D. R. Wonsey, K. A. O'Donnell, J. W. Kim, J. T. Yustein, L. A. Lee, and C. V. Dang. 2005. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell. Biol. 25:6225-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, Z., S. Van Calcar, C. Qu, W. K. Cavenee, M. Q. Zhang, and B. Ren. 2003. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc. Natl. Acad. Sci. USA 100:8164-8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linke, S. P., K. C. Clarkin, A. Di Leonardo, A. Tsou, and G. M. Wahl. 1996. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 10:934-947. [DOI] [PubMed] [Google Scholar]
- 32.Lossos, I. S., A. A. Alizadeh, M. Diehn, R. Warnke, Y. Thorstenson, P. J. Oefner, P. O. Brown, D. Botstein, and R. Levy. 2002. Transformation of follicular lymphoma to diffuse large-cell lymphoma: alternative patterns with increased or decreased expression of c-myc and its regulated genes. Proc. Natl. Acad. Sci. USA 99:8886-8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mateyak, M. K., A. J. Obaya, S. Adachi, and J. M. Sedivy. 1997. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 8:1039-1048. [PubMed] [Google Scholar]
- 34.Moura, I. C., Y. Lepelletier, B. Arnulf, P. England, C. Baude, C. Beaumont, A. Bazarbachi, M. Benhamou, R. C. Monteiro, and O. Hermine. 2003. A neutralizing monoclonal antibody (mAb A24) directed against the transferrin receptor induces apoptosis of tumor T lymphocytes from ATL patients. Blood 103:1838-1845. [DOI] [PubMed] [Google Scholar]
- 35.Neckers, L. M., and J. B. Trepel. 1986. Transferrin receptor expression and the control of cell growth. Cancer Investig. 4:461-470. [DOI] [PubMed] [Google Scholar]
- 36.Nikiforov, M. A., S. Chandriani, B. O'Connell, O. Petrenko, I. Kotenko, A. Beavis, J. M. Sedivy, and M. D. Cole. 2002. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol. Cell. Biol. 22:5793-5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Connell, B. C., A. F. Cheung, C. P. Simkevich, W. Tam, X. Ren, M. K. Mateyak, and J. M. Sedivy. 2003. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J. Biol. Chem. 278:12563-12573. [DOI] [PubMed] [Google Scholar]
- 38.O'Donnell, K. A., E. A. Wentzel, K. I. Zeller, C. V. Dang, and J. T. Mendell. 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435:839-843. [DOI] [PubMed] [Google Scholar]
- 39.Orian, A., B. van Steensel, J. Delrow, H. J. Bussemaker, L. Li, T. Sawado, E. Williams, L. W. Loo, S. M. Cowley, C. Yost, S. Pierce, B. A. Edgar, S. M. Parkhurst, and R. N. Eisenman. 2003. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 17:1101-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oster, S. K., C. S. Ho, E. L. Soucie, and L. Z. Penn. 2002. The myc oncogene: MarvelouslY Complex. Adv. Cancer Res. 84:81-154. [DOI] [PubMed] [Google Scholar]
- 41.Oster, S. K., D. Y. Mao, J. Kennedy, and L. Z. Penn. 2003. Functional analysis of the N-terminal domain of the Myc oncoprotein. Oncogene 22:1998-2010. [DOI] [PubMed] [Google Scholar]
- 42.Pajic, A., D. Spitkovsky, B. Christoph, B. Kempkes, M. Schuhmacher, M. S. Staege, M. Brielmeier, J. Ellwart, F. Kohlhuber, G. W. Bornkamm, A. Polack, and D. Eick. 2000. Cell cycle activation by c-myc in a burkitt lymphoma model cell line. Int. J. Cancer 87:787-793. [DOI] [PubMed] [Google Scholar]
- 43.Pelengaris, S., M. Khan, and G. Evan. 2002. c-MYC: more than just a matter of life and death. Nat Rev. Cancer 2:764-776. [DOI] [PubMed] [Google Scholar]
- 43a.Remondini, D., B. O’Connell, B. Intrator, J. M. Sedivy, N. Neretti, G. C. Castellani, and L. N. Cooper. 2005. Targeting c-Myc activated genes with a correlation method: detection of global changes in large gene expression network dynamics. Proc. Natl. Acad. Sci. USA 102:6902-6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhodes, D. R., J. Yu, K. Shanker, N. Deshpande, R. Varambally, D. Ghosh, T. Barrette, A. Pandey, and A. M. Chinnaiyan. 2004. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richardson, D. R. 2002. Iron chelators as therapeutic agents for the treatment of cancer. Crit. Rev. Oncol. Hematol. 42:267-281. [DOI] [PubMed] [Google Scholar]
- 46.Richardson, D. R. 2002. Therapeutic potential of iron chelators in cancer therapy. Adv. Exp. Med. Biol. 509:231-249. [DOI] [PubMed] [Google Scholar]
- 47.Schuhmacher, M., F. Kohlhuber, M. Holzel, C. Kaiser, H. Burtscher, M. Jarsch, G. W. Bornkamm, G. Laux, A. Polack, U. H. Weidle, and D. Eick. 2001. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 29:397-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Secombe, J., S. B. Pierce, and R. N. Eisenman. 2004. Myc: a weapon of mass destruction. Cell 117:153-156. [DOI] [PubMed] [Google Scholar]
- 49.Sherr, C. J., and R. A. DePinho. 2000. Cellular senescence: mitotic clock or culture shock? Cell 102:407-410. [DOI] [PubMed] [Google Scholar]
- 50.Small, M. B., N. Hay, M. Schwab, and J. M. Bishop. 1987. Neoplastic transformation by the human gene N-myc. Mol. Cell. Biol. 7:1638-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stone, J., T. de Lange, G. Ramsay, E. Jakobovits, J. M. Bishop, H. Varmus, and W. Lee. 1987. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol. Cell. Biol. 7:1697-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su, A. I., M. P. Cooke, K. A. Ching, Y. Hakak, J. R. Walker, T. Wiltshire, A. P. Orth, R. G. Vega, L. M. Sapinoso, A. Moqrich, A. Patapoutian, G. M. Hampton, P. G. Schultz, and J. B. Hogenesch. 2002. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. USA 99:4465-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taetle, R., J. M. Honeysett, and I. Trowbridge. 1983. Effects of anti-transferrin receptor antibodies on growth of normal and malignant myeloid cells. Int. J. Cancer 32:343-349. [DOI] [PubMed] [Google Scholar]
- 54.Vindelov, L. L., I. J. Christensen, and N. I. Nissen. 1983. A detergent-trypsin method for the preparation of nuclei for flow cytometric DNA analysis. Cytometry 3:323-327. [DOI] [PubMed] [Google Scholar]
- 55.Weiner, R. E. 1996. The mechanism of 67Ga localization in malignant disease. Nucl. Med. Biol. 23:745-751. [DOI] [PubMed] [Google Scholar]
- 56.Wonsey, D. R., K. I. Zeller, and C. V. Dang. 2002. The c-Myc target gene PRDX3 is required for mitochondrial homeostasis and neoplastic transformation. Proc. Natl. Acad. Sci. USA 99:6649-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woo, R. A., and R. Y. Poon. 2004. Activated oncogenes promote and cooperate with chromosomal instability for neoplastic transformation. Genes Dev. 18:1317-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, K. J., A. Polack, and R. Dalla-Favera. 1999. Coordinated regulation of iron-controlling genes, H-ferritin and IRP2, by c-MYC. Science 283:676-679. [DOI] [PubMed] [Google Scholar]
- 59.Yu, D., D. Allman, M. H. Goldschmidt, M. L. Atchison, J. G. Monroe, and A. Thomas-Tikhonenko. 2003. Oscillation between B-lymphoid and myeloid lineages in Myc-induced hematopoietic tumors following spontaneous silencing/reactivation of the EBF/Pax5 pathway. Blood 101:1950-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu, D., M. Dews, A. Park, J. W. Tobias, and A. Thomas-Tikhonenko. 2005. Inactivation of Myc in murine two-hit B lymphomas causes dormancy with elevated levels of interleukin 10 receptor and CD20: implications for adjuvant therapies. Cancer Res. 65:5454-5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu, D., and A. Thomas-Tikhonenko. 2002. A non-transgenic mouse model for B-cell lymphoma: in vivo infection of p53-null bone marrow progenitors by a Myc retrovirus is sufficient for tumorigenesis. Oncogene 21:1922-1927. [DOI] [PubMed] [Google Scholar]
- 62.Zeller, K. I., T. J. Haggerty, J. F. Barrett, Q. Guo, D. R. Wonsey, and C. V. Dang. 2001. Characterization of nucleophosmin (B23) as a Myc target by scanning chromatin immunoprecipitation. J. Biol. Chem. 276:48285-48291. [DOI] [PubMed] [Google Scholar]
- 63.Zeller, K. I., A. G. Jegga, B. J. Aronow, K. A. O'Donnell, and C. V. Dang. 2003. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 4:R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.