Abstract
Selenocysteine incorporation in eukaryotes occurs cotranslationally at UGA codons via the interactions of RNA-protein complexes, one comprised of selenocysteyl (Sec)-tRNA[Ser]Sec and its specific elongation factor, EFsec, and another consisting of the SECIS element and SECIS binding protein, SBP2. Other factors implicated in this pathway include two selenophosphate synthetases, SPS1 and SPS2, ribosomal protein L30, and two factors identified as binding tRNA[Ser]Sec, termed soluble liver antigen/liver protein (SLA/LP) and SECp43. We report that SLA/LP and SPS1 interact in vitro and in vivo and that SECp43 cotransfection increases this interaction and redistributes all three proteins to a predominantly nuclear localization. We further show that SECp43 interacts with the selenocysteyl-tRNA[Ser]Sec-EFsec complex in vitro, and SECp43 coexpression promotes interaction between EFsec and SBP2 in vivo. Additionally, SECp43 increases selenocysteine incorporation and selenoprotein mRNA levels, the latter presumably due to circumvention of nonsense-mediated decay. Thus, SECp43 emerges as a key player in orchestrating the interactions and localization of the other factors involved in selenoprotein biosynthesis. Finally, our studies delineating the multiple, coordinated protein-nucleic acid interactions between SECp43 and the previously described selenoprotein cotranslational factors resulted in a model of selenocysteine biosynthesis and incorporation dependent upon both cytoplasmic and nuclear supramolecular complexes.
Significant strides have been made over the past 15 years in elucidating the mechanism and most of the players in eukaryotic selenoprotein biosynthesis. Key players in this process are the unique tRNA that decodes UGA as a selenocysteine codon (16), the specific secondary structures in the 3′ untranslated regions of selenoprotein mRNAs, termed SECIS elements, that are required for selenocysteine insertion (2), and protein factors that interact with the tRNA and SECIS element. Protein factors identified to date include an elongation factor specific for selenocysteyl (Sec)-tRNA[Ser]Sec, termed EFsec (10, 26), the SECIS binding protein, SBP2 (6), and most recently, a ribosomal protein, L30, that can also bind SECIS elements and may mediate the incorporation process at the ribosome (5). Two selenophosphate synthetases, SPS1 and SPS2, contribute to the selenoprotein synthesis pathway, in that they catalyze conversion of selenide and ATP to selenophosphate, the active selenium donor in selenocysteine biosynthesis (18). SPS2 is itself a selenoenzyme, thus serving a positive feedback role in selenoprotein synthesis. Recently, a kinase that phosphorylates Ser-tRNA[Ser]Sec has been identified in the genomes of organisms that encode other components of the selenoprotein synthesis machinery (4). However, its role in this process remains to be elucidated.
At least two activities crucial to selenocysteine incorporation have remained elusive, the factors(s) responsible for conversion of Ser-tRNA[Ser]Sec to Sec-tRNA[Ser]Sec, and the enzyme(s) catalyzing 2′-O-methylation of the 5-methoxycarbonylmethyl uridine at the wobble base position in Sec-tRNA[Ser]Sec. Two factors have emerged as candidates for these activities, both having been identified in association with tRNA[Ser]Sec. An autoimmune chronic hepatitis antigen was identified as a 48-kDa protein that coimmunoprecipitated with tRNA[Ser]Sec using autoimmune sera from chronic active hepatitis patients (11). This protein, also termed soluble liver antigen/liver protein (SLA/LP), was recently cloned, and its sequence is compatible with the architecture of the pyridoxal phosphate-dependent transferase superfamily (15). Intriguingly, bacterial selenocysteine synthase, the enzyme catalyzing conversion of Ser-tRNA[Ser]Sec to Sec-tRNA[Ser]Sec, is a pyridoxal phosphate-dependent enzyme. The second factor, SECp43, was identified in a degenerate PCR screen for RNA binding proteins (9). Immunoaffinity purification of SECp43 from HeLa cells revealed its association with tRNA[Ser]Sec, but direct binding to in vitro-transcribed tRNA[Ser]Sec could not be demonstrated. This suggested either that tRNA[Ser]Sec must be charged for binding to occur or that SECp43 requires other factors for association with tRNA[Ser]Sec. Protein overlay studies with SECp43 showed that it interacted with a 48-kDa protein in HeLa cell extracts, corresponding to the size of SLA/LP.
Herein, we have characterized the interactions between the factors described above and have investigated the role of SECp43 in selenoprotein synthesis. We report that in vitro, SLA/LP interacts with SPS1, and SECp43 interacts with Sec-tRNA[Ser]Sec and EFsec in a high-molecular-weight complex. In vivo, SECp43 coimmunoprecipitates with SPS1 and SLA/LP, and this association results in redistribution of SLA/LP to the nucleus. Thus, SECp43 appears to coordinate the activities of selenophosphate biosynthesis, seryl- to selenocysteyl-tRNA conversion, and tRNA methylation. SECp43 also coimmunoprecipitates with EFsec and enhances the association between EFsec and SBP2 in vivo, linking the factors involved in the early steps of selenocysteine biosynthesis and tRNA charging to the later steps resulting in the cotranslational incorporation of selenocysteine into selenoproteins. We show that SECp43 transfection increases selenocysteine incorporation and decreases UGA-mediated termination, implying a direct effect on the efficiency of selenoprotein synthesis. We further demonstrate that transfection of SECp43 increases the abundance of selenoprotein mRNAs in an intron-dependent manner, suggesting a role for this factor in circumventing nonsense-mediated decay of selenoprotein mRNAs. Finally, we examined the ribosome association of the various factors implicated in selenocysteine biosynthesis and incorporation, revealing the presence of complexes both in association with and separate from polysome fractions. The picture emerging from these findings is that of a dynamic pathway of complex assembly in the nuclear and cytoplasmic compartments to coordinate the multiple steps leading to translation of selenoproteins.
MATERIALS AND METHODS
Constructs.
SECp43 cDNA cloned into pBS-CMV (Stratagene, La Jolla, CA) in both the forward and reverse orientations was a generous gift of Paula Grabowski. SLA/LP cDNAs were obtained from Invitrogen (San Diego, CA) and from Xue-Ming Xu. SPS1 has been previously described (18). SBP2 in pcDNA3.1 was a generous gift of Paul Copeland and Donna Driscoll. Identification and characterization of the murine EFsec cDNA and subcloning into the pUHD10-3 vector for mammalian expression were described previously (26). All cDNAs were cloned into the Gateway vector system (Invitrogen, San Diego, CA) for bacterial expression with histidine and/or glutathione S-transferase (GST) tags, and for coupled in vitro transcription-translation (TnT T7 quick-coupled transcription/translation system; Promega, Madison, WI). Histidine-tagged Arabidopsis thaliana beta-glucuronidase (GUS) protein was used as a negative control for protein-protein interactions. GPX1 expression constructs, generous gifts of Roger Sunde, have been described previously (27).
Electrophoretic mobility shift and nitrocellulose filter binding assays.
75Se-Sec-tRNA[Ser]Sec was prepared by labeling HeLa cells with 75Se-sodium selenite. Purification of the labeled Sec-tRNA[Ser]Sec isoforms and of 3H-Ser-tRNA[Ser]Sec by reverse-phase high-performance liquid chromatography was as described previously (13). Bacterial expression and purification of EFsec have been described previously (26). Expressed, purified SBP2 and SECp43 were generous gifts of Paul Copeland and Paula Grabowski, respectively. 75Se-Sec-tRNA[Ser]Sec was incubated with purified recombinant EFsec in binding buffer containing 0.1 mM GTP for 10 min at 30°C, followed by addition of the indicated proteins and incubation for a further 10 min. Complexes were electrophoresed on a 5% acrylamide-Tris-borate-EDTA gel (Ready Gel; Bio-Rad) in Tris-borate-EDTA adjusted to pH 7.3 with glacial acetic acid, followed by autoradiography. Nitrocellulose filter binding assays were performed as described previously (26).
Bacterial expression, in vitro translations and pulldown assays.
Proteins were expressed in Escherichia coli BL21pLysS and purified via the respective tags on either glutathione-Sepharose beads (GST fusion proteins) or BD TALON (BD Biosciences-Clontech, Palo Alto, CA) metal affinity resin (histidine-tagged proteins). Purified bacterially expressed proteins were incubated with lysates of in vitro-translated [35S]methionine-labeled histidine-tagged or GST fusion proteins at room temperature. After one hour of incubation, the mixture was added to beads corresponding to the bait tag and allowed to incubate for an additional hour. The bead and protein mixture was eluted with extraction buffer (25 mM Tris, pH 7.5, 1 mM EDTA, 20 mM NaCl, 20% glycerol, 1× type II protease inhibitor cocktail [EMD Bioscience]) to disrupt the binding between beads and tagged protein. Samples were analyzed on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, followed by autoradiography.
Cell culture, transfections, and 75Se labeling.
Transient transfections in human embryonic kidney (HEK-293) cells were carried out using either calcium phosphate as described previously (17) or Mirus TransIT LT1 reagent (Mirus Bio, Madison, WI) according to the manufacturer's instructions. Cells were plated onto 60-mm culture dishes in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Cells were supplemented with 100 nM sodium selenite for 6 to 24 h prior to transfection reactions, unless otherwise specified. Cells were transfected with 5 to 7 μg of the expression plasmids and in the case of EFsec, cotransfected with 4 μg of the pUHD15 plasmid, which encodes a protein necessary for transcriptional activation of the pUHD10-3 promoter (12). To monitor transfection efficiencies, cells were cotransfected with 3 μg of an expression vector containing the human growth hormone cDNA under control of the herpes simplex virus thymidine kinase promoter. 75Se labeling was carried out by addition of sodium [75Se]selenite (1,000 μCi/μg or 3 to 6 μCi/ml) to the media, followed by incubation for 24 h.
Coimmunoprecipitations and Western analysis.
After harvesting the HEK-293 cells, lysates were prepared by one of two methods. For total protein lysates, cells were sonicated in a total lysis buffer containing 10 mM Tris-Cl, pH 7.6, 150 mM NaCl, 0.5% NP-40, 5% glycerol, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 0.2 U/ml aprotinin, and precleared with preimmune or normal serum plus Pansorbin (CalBiochem, San Diego, CA). For cytoplasmic and nuclear protein fractions, cells were first lysed in a cytosolic lysis buffer containing 50 mM HEPES, pH 7.4, 75 mM NaCl, 20 mM NaF, 10 mM iodoacetamide, 0.5% (wt/vol) Triton X-100, and 1 mM PMSF on ice for 30 min. The intact nuclei were pelleted by centrifugation (10,000 × g, 5 min, 4°C), thus, clearing the cytoplasmic fraction (a postnuclear lysate). The intact nuclear pellet was washed twice in cytosolic lysis buffer, and the nuclear pellet was extracted in a nuclear lysis buffer containing 50 mM HEPES, pH 7.4, 500 mM NaCl, 20 mM NaF, 10 mM iodoacetamide, 0.5% (wt/vol) Triton X-100, and 1 mM PMSF on ice for 30 min. The increased osmotic pressure from the elevated NaCl concentration in the nuclear lysis buffer ruptures the nuclear membranes and releases nuclear proteins. Nuclear lysates were cleared by centrifugation (10,000 × g, 5 min, 4°C) and diluted in cytosolic lysis buffer without any NaCl (but with equal concentrations of the other components) to achieve a final concentration of 75 mM NaCl for the immunoprecipitation reaction mixtures. For immunoprecipitation of total cellular lysates, supernatants were incubated with the indicated antibodies, followed by the addition of Pansorbin. For immunoprecipitation of cytosolic and nuclear fractions, lysates were incubated with glutathione-Sepharose beads (GST fusion proteins) or TALON metal affinity resin (histidine fusion proteins) or the indicated antibodies, followed by addition of protein A or protein G (depending on the isotype of the given antibody) Sepharose (Amersham Biosciences, Piscataway, NJ). Pellets were harvested, washed three times in lysis buffer, and analyzed by SDS-PAGE, followed by Western blotting with the indicated antibodies. SECp43 antiserum was a generous gift of Paula Grabowski. SBP2 antiserum was a generous gift of Paul Copeland and Donna Driscoll. FLAG and GST antibodies were from Sigma (St. Louis, MO). V5 antibody was from Invitrogen (Carlsbad, CA) or Serotec (Raleigh, NC). Pentahistidine antibody was from QIAGEN (Valencia, CA). Green fluorescent protein (GFP) antibody was from Molecular Probes (Eugene, OR). GRB2 antibody was obtained from BioSource International (Camarillo, CA). Histone H1 antibody was obtained from Upstate Cell Signaling Solutions and Serologicals Company (Charlottesville, VA). Glutathione-Sepharose resin was used for anti-GST coimmunoprecipitations according to the manufacturer's instructions (Amersham Biosciences). TALON metal affinity resin was used for antihistidine coimmunoprecipitations according to the manufacturer's instructions (Clontech, Palo Alto, CA). Expression levels were verified in each experiment either by acetone precipitation of cytosolic and nuclear lysates followed by Western analysis or by immunoprecipitation with antibodies corresponding to the epitope tags on the transfected factors, followed by western analysis with the same antibody. Western analysis with antibodies to cytoplasmic (GRB2) and nuclear (histone H1) markers were carried out to assess cross-contamination of the subcellular fractions.
Selenoprotein mRNA quantitation by macroarray.
For comparison of selenoprotein mRNA levels in HepG2, HEK-293 and HeLa cells in the absence and presence of transfected SECp43, two 60-nucleotide-long oligonucleotide probes (37 to 60% GC) were designed for each selenoprotein gene on the custom selenoprotein macroarrays, with positioning as close as possible to the 3′ end. Sequences were analyzed for absence of secondary structure and cross-hybridization elsewhere in the genome. Probes were spotted onto GeneScreen Plus nylon membranes by using a V&P Scientific (San Diego, CA) 1,536-pin replicator and immobilized by alkali treatment. Total RNA (2 μg) was labeled via oligo(T)-directed first-strand cDNA synthesis using 400 units murine leukemia virus reverse transcriptase (Invitrogen) and [α-33P]dCTP (40 μCi). cDNA was purified using QiaQuick PCR columns (QIAGEN), heat denatured, and hybridized in triplicate to arrays in MicroHyb buffer (Research Genetics, Huntsville, AL) overnight at 60°C. Arrays were washed with 2× SSC, 0.5% SDS at 50°C, followed by 1 × to 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.5% SDS at 65°C, and then exposed to phosphor storage screens and signals quantified using a Molecular Dynamics PhosphorImager (Sunnyvale, CA). Signal readings were taken for each spot, and background readings were taken at empty spots. Raw data were further automatically processed using Microsoft Excel. Spot readings that failed to exceed the average background value by more than 3 standard deviations were excluded from the analysis. The remaining readings were scaled by the average signal in selected steadily expressed genes and then averaged among triplicate measurements.
Selenoprotein mRNA quantitation by reverse transcriptase PCR (RT-PCR).
For comparison of mouse GPX1 mRNA levels in HEK-293 cells in the absence and presence of transfected SECp43, two sets of primers which could distinguish the mRNA products of the transfected mouse GPX1 constructs (GPX plus intron or GPX without intron) from endogenous human GPX1 were designed. Reverse transcription was carried out using a Superscript III first-strand synthesis system (Invitrogen) followed by PCR using a Platinum SYBR green qPCR SuperMix uracil DNA glycosylase (Invitrogen) in a Light Cycler II (Roche). The primers sets used in the quantitative PCR were either a 5′ primer corresponding to a unique sequence in the FLAG epitope tag at the N terminus of the GPX1 coding region in combination with a 3′ primer unique to mouse GPX1 or a 5′ primer derived from a unique sequence in the mouse GPX1 coding region, in combination with a distinct 3′ primer unique to mouse GPX1. Hypoxanthine phosphoribosyltransferase primers were used as an internal reference for cDNA synthesis reactions. Data were analyzed using Light Cycler software, version 4 (Roche), and were presented as picograms of starting material, calculated based on an external standard curve using an equivalent amplicon.
Sucrose gradient fractionation.
Sucrose gradient separation of polysomes was carried out on 15 to 50% (wt/vol) gradients as previously described (20), followed by fractionation and monitoring for absorbance at 254 nm by using an ISCO syringe pump with UV-6 detector (Brandel). Fractions were divided in two parts: half were extracted with TRIzol reagent (Invitrogen) to isolate total RNA and the other half was acetone precipitated to isolate proteins. Western blot analysis was performed on the protein fractions as before, except that samples were normalized by loading equal volumes from each fraction instead of normalization by adding equivalent cell numbers (as in all of the other analyses).
RESULTS
Interactions between SECp43 and the Sec-tRNA[Ser]Sec-EFsec complex.
SECp43 was initially identified in association with Sec-tRNA[Ser]Sec in vivo, but binding could not be demonstrated in vitro (9), suggesting the requirement of other factors for the interaction. To investigate possible candidates mediating this interaction, we performed electrophoretic mobility shift assays using 75Se-Sec-tRNA[Ser]Sec and purified recombinant SECp43, alone or in combination with the selenoprotein translation factors, EFsec and SBP2. SECp43 did not directly bind Sec-tRNA[Ser]Sec in this assay (not shown), in agreement with previously published findings (9). EFsec binds Sec-tRNA[Ser]Sec in vitro, as shown by its shift in mobility in the presence of EFsec compared to the absence of EFsec (Fig. 1A, lane 2 versus lane 1), and this is in agreement with previous filter binding assays (26). We anticipated that the addition of SBP2 might result in “supershifting” of the Sec-tRNA[Ser]Sec-EFsec complex, based on our previous demonstrations that EFsec and SBP2 coimmunoprecipitate from transfected-cell lysates (26), and that Sec-tRNA[Ser]Sec greatly enhances EFsec-SBP2 interaction (29). Surprisingly, the addition of SBP2 in vitro resulted in release of Sec-tRNA[Ser]Sec from EFsec (Fig. 1A, lane 3). Preincubation with the nonhydrolyzable analog, guanosine 5′-O-(3-thiotriphosphate) did not prevent release of Sec-tRNA[Ser]Sec (Fig. 1A, lane 4), implying that release was not due to GTP hydrolysis and resultant conformational changes in EFsec. This suggests that other factors present in vivo, but not in the in vitro reaction, may be necessary for formation or stabilization of the Sec-tRNA[Ser]Sec-EFsec-SBP2 complex. Association of SECp43 with tRNA[Ser]Sec in vivo but not in vitro pointed to this factor as a possible candidate for modulating the interactions of the other factors. Strikingly, inclusion of SECp43 in the Sec-tRNA[Ser]Sec-EFsec-SBP2 reaction resulted in shifting of the Sec-tRNA[Ser]Sec-EFsec complex to a high-molecular-weight aggregate that did not enter the gel (Fig. 1A, lanes 5 and 6). In addition, release of Sec-tRNA[Ser]Sec by SBP2 was significantly decreased. Formation of a high-molecular-weight aggregate containing Sec-tRNA[Ser]Sec was also seen in the absence of SBP2 (Fig. 1A, lane 7), indicating that SBP2 was not required for this association to occur. Thus, SECp43 can both interact with the Sec-tRNA[Ser]Sec-EFsec complex in the absence of SBP2, and block the SBP2-dependent release of Sec-tRNA[Ser]Sec from EFsec. Finally, the addition of HEK-293 cell lysate instead of SECp43 produced a similar shift to a high-molecular-weight aggregate (Fig. 1A, lane 8). This suggests that SECp43 or other factors that associate with Sec-tRNA[Ser]Sec-EFsec are abundant in HEK-293 cells.
FIG. 1.

Complex formation between EFsec, Sec-tRNA[Ser]Sec, and SECp43 in vitro. (A) 75Se-Sec-tRNA[Ser]Sec was incubated with purified recombinant EFsec, followed by the addition of the indicated proteins. Complexes were electrophoresed under nondenaturing conditions, followed by autoradiography. GTP-γ-S, guanosine 5′-O-(3-thiotriphosphate). (B) Nitrocellulose filter binding assays were performed with 75Se-Sec-tRNA[Ser]Sec or 3H-Ser-tRNA[Ser]Sec and the indicated bacterially expressed purified proteins.
To further investigate interactions between SECp43, Sec-tRNA[Ser]Sec, and EFsec, nitrocellulose filter binding assays were carried out using the two protein factors with individual isoforms of Sec-tRNA[Ser]Sec differing by the presence or absence of a 2′-O-methylation of the 5-methoxycarbonylmethyl uridine at the wobble base (mcm5Um or mcm5U, respectively). The precursor to Sec-tRNA[Ser]Sec biosynthesis, Ser-tRNA[Ser]Sec, was also included in binding studies as a negative control. SECp43 alone exhibited only nonspecific binding to all three of the amino acyl-tRNAs (Fig. 1B) comparable to levels of binding seen with bovine serum albumin. Thus, the charging of tRNA[Ser]Sec with either serine or selenocysteine is not sufficient for SECp43 binding to occur. EFsec alone bound both the mCm5U and mCm5Um forms of Sec-tRNA[Ser]Sec, but not Ser-tRNA[Ser]Sec (Fig. 1B), in agreement with our previously published findings (26). Upon theaddition of SECp43, the amount of Sec-tRNA[Ser]Sec associated with protein increased (Fig. 1B), further supporting the ability of SECp43, EFsec, and Sec-tRNA[Ser]Sec to form a complex.
SECp43 interactions with EFsec and SBP2 in vivo.
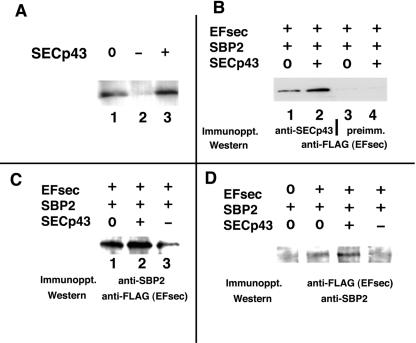
To begin to investigate the function of SECp43 in vivo, we examined the levels of endogenous SECp43 in HEK-293 cells, the enhancement upon SECp43 plasmid transfection, and the inhibition of expression upon antisense plasmid transfection. Significant levels of expression were seen in the absence of transfection (Fig. 2A, lane 1), and the enhancement upon transfection was minimal (Fig. 2A, lane 3), suggesting abundant endogenous SECp43. Transfection of antisense plasmid reduced expression to undetectable levels (Fig. 2A, lane 2). To assess interactions between SECp43 and the selenocysteine incorporation factors in vivo, SECp43 was cotransfected with FLAG-tagged EFsec and native SBP2, and coimmunoprecipitation assays were performed. Transfected-cell homogenates were immunoprecipitated with antibodies against the FLAG tag, SBP2 or SECp43, and immunoprecipitates analyzed by Western blotting. Immunoprecipitation with anti-SECp43 antisera, followed by Western analysis for EFsec, showed that the two proteins coimmunoprecipitated. Coprecipitation was detected with endogenous levels of SECp43 (Fig. 2B, lane 1) but was enhanced upon SECp43 cotransfection (Fig. 2B, lane 2), indicating that these two factors associate in vivo. Immunoprecipitation with anti-SBP2 antisera followed by Western analysis for EFsec showed increased EFsec coprecipitation upon SECp43 cotransfection (Fig. 2C, lane 2 versus lane 1). Expression of antisense SECp43 plasmid decreased coprecipitation of EFsec and SBP2 (Fig. 2C, lane 3 versus lanes 1 and 2, respectively). The converse experiment, in which anti-FLAG (EFsec) antisera were used in the immunoprecipitation, confirmed the enhanced coprecipitation of EFsec and SBP2 upon SECp43 cotransfection (Fig. 2D, lane 3 versus lane 2). Expression of antisense SECp43 again decreased coprecipitation of the other two factors (Fig. 2D, lane 4). Taken together, the in vivo effect of SECp43 on enhancing coprecipitation of EFsec and SBP2 and also the inhibitory effect on dissociation of the Sec-tRNA[Ser]Sec-EFsec-SBP2 complex in vitro strongly implicate a role for SECp43 in formation or stabilization of the Sec-tRNA[Ser]Sec-EFsec-SBP2 complex.
FIG. 2.
SECp43 coimmunoprecipitates with EFsec and enhances interaction between EFsec and SBP2 in vivo. (A) HEK-293 cells were either untransfected (0) or transfected with either SECp43 antisense plasmid (−) or SECp43 sense expression plasmid (+). Expression levels were assessed by Western blotting with anti-SECp43 antisera. (B) HEK-293 cells were transfected with cDNAs encoding the indicated factors. Immunoprecipitation (Immunoppt.) was carried out with anti-SECp43 antisera, followed by Western blotting for EFsec by using anti-FLAG antibody. preimm., preimmune serum. (C) Immunoprecipitation (Immunoppt.) was carried out with anti-SBP2 antisera, followed by Western blotting for EFsec by using anti-FLAG antibody. (D) Immunoprecipitation (Immunoppt.) was carried out for EFsec by using anti-FLAG antisera, followed by Western blotting using anti-SBP2 antisera.
In vitro interactions among selenoprotein biosynthesis factors.
To characterize additional potential interactions among SECp43 and the other factors implicated in selenoprotein biosynthesis, we carried out pulldown assays, with each factor (SPS1, SLA/LP, SECp43, EFsec, and SBP2) expressed as a histidine-tagged or GST fusion protein in bacteria, to be used as bait, and conversely, with each produced in vitro in reticulocyte lysates and labeled with [35S]methionine, as test interaction partners. Pulldown experiments were performed using glutathione-Sepharose or TALON metal affinity resin. SPS1 with either a histidine or GST tag was able to efficiently pull down SLA/LP, whereas the negative-control target, GUS, was not pulled down (Fig. 3, left and center panels). Reciprocal pulldown experiments using histidine-tagged SLA/LP as bait protein efficiently pulled down in vitro-translated SPS1-GST fusion (Fig. 3, right panel). Thus, SPS1 and SLA/LP interact independent of any other factors. No other interactions between the factors were detected in vitro, in either pairwise or multifactor combinations. Yeast two-hybrid studies with SECp43 were not informative, as the protein exhibited a high background level of transactivation on its own. Thus, interactions with unknown partners could not be assessed in this system.
FIG. 3.

Coimmunoprecipitation analysis of factors involved in selenocysteine incorporation. Bacterially expressed histidine- or GST-tagged SLA/LP or SPS1 was incubated with the indicated [35S]methionine-labeled in vitro-translated proteins. Coimmunoprecipitations were performed with pentahistidine antibody and TALON metal affinity resin, and pulldowns were performed with glutathione-agarose, followed by SDS-PAGE and autoradiography. Aliquots (10%) of input in vitro translation reactions were loaded in the left lane of each panel (input; lanes 1, 4, and 7), followed by negative-control pulldowns (GUS; lanes 2, 5, and 8) and pulldowns using SLA/LP or SPS1 as bait (SLA/LP or SPS1; lanes 3, 6, and 9).
Subcellular localization and in vivo interactions among SPS1, SLA/LP, and SECp43.
Examination of the sequences of SPS1, SLA/LP, and SECp43 revealed one putative nuclear localization sequence each in SLA/LP and SECp43 and three putative nuclear export sequences in SLA/LP. No putative localization sequences are predicted in SPS1. The SLA/LP and SECp43 nuclear localization sequences are conserved among vertebrate species but not in insects or nematodes. To investigate the interactions of SLA/LP and SPS1 in vivo and to examine the role of SECp43 in this interaction, we carried out subcellular fractionation and coprecipitation studies following transient expression of the proteins in HEK-293 cells. When the factors were expressed individually in the presence of tRNA[Ser]Sec, SLA/LP was detected only in the cytoplasmic fraction, whereas SPS1 was found in both the cytoplasmic and nuclear fractions (Fig. 4A). Upon the coexpression of multiple factors, a different picture emerged. Coexpression of SLA/LP and SPS1 resulted in a slight redistribution of SLA/LP into the nuclear fraction (Fig. 4B through D, upper panels, lanes 5 versus lanes 4). In the presence of cotransfected SECp43, nuclear localization of SLA/LP was greatly increased (Fig. 4B through D, lanes 6), and SLA/LP, SPS1 and SECp43 were found to coprecipitate in both nuclear and cytosolic fractions (Fig. 4B through D, lanes 3 and 6), indicating the formation of complexes. This was observed whether the initial precipitation was directed at SLA/LP or SPS1.
FIG. 4.
Subcellular localization and interactions among SLA/LP, SPS1, and SECp43. (A) Subcellular fractionation and localization of SLA/LP and SPS1. HEK-293 cells were transfected with tRNA[Ser]Sec alone, or tRNA[Ser]Sec and GST-tagged SLA/LP or GST-tagged SPS1 (as indicated). The cells were lysed and fractionated. Cytosolic and nuclear lysates were acetone precipitated (precip.) (total protein; lanes 1, 2, 3, 6, 7, and 8) or immunoprecipitated using glutathione-Sepharose (Seph.) (lanes 4, 5, 9, and 10). Western blot analysis was performed using anti-GST. (B) Coimmunoprecipitation of SLA with SPS1. HEK-293 cells were transfected with combinations of the following, as indicated: tRNA[Ser]Sec (no tag), GST-tagged SPS1, His-tagged SLA/LP, and SECp43 (no tag). The cells were lysed and fractionated. Cytosolic and nuclear lysates were immunoprecipitated (Immunoppt.) by using glutathione-Sepharose. Western blot analysis was performed using anti-His (top) and anti-GST (bottom). (C) Coimmunoprecipitation of SPS1 with SLA. HEK-293 cells were transfected with combinations of the following, as indicated: tRNA[Ser]Sec (no tag), His-tagged SLA/LP, V5-tagged SPS1, and SECp43 (no tag). The cells were lysed and fractionated. Cytosolic and nuclear lysates were immunoprecipitated (Immunoppt.) using antipentahistidine. Western blot analysis was performed using anti-V5 (top) and anti-His (bottom). (D) Coimmunoprecipitation of SLA with SPS1. HEK-293 cells were transfected with combinations of the following, as indicated: tRNA[Ser]Sec, V5-tagged SPS1, His-tagged SLA/LP, and SECp43 (no tag). The cells were lysed and fractionated. Cytosolic and nuclear lysates were immunoprecipitated (Immunoppt.) using anti-V5. Western blot analysis was performed using anti-His (top) and anti-V5 (bottom).
Effects of SECp43 on selenoprotein synthesis and selenoprotein mRNA levels.
We and others have previously shown that several of the factors involved in selenocysteine incorporation, including tRNA[Ser]Sec, SPS1, and SBP2, are limiting for selenocysteine incorporation in mammalian cell lines and enhance incorporation upon their overexpression (3, 6, 17, 18). Given the effects of SECp43 expression on the interactions of the other factors discussed above, we examined the effects of SECp43 expression on selenoprotein synthesis. Zebra fish selenoprotein Pa was expressed in the absence or presence of SECp43 cotransfection. SECp43 coexpression, however, significantly increased the levels of zebra fish selenoprotein Pa production (Fig. 5A). We previously showed that expression of SBP2 results in an increase in premature termination products from zebra fish selenoprotein Pa. We speculated that this may be due to an increased incorporation rate at early UGA codons, resulting in inability to outcompete termination at downstream UGAs. We also showed that this effect was reversed upon expression of EFsec (26). Strikingly, expression of SECp43 resulted in a similar reversal of termination of zebra fish selenoprotein Pa (Fig. 5B), further supporting its role in UGA decoding.
FIG. 5.
Effects of SECp43 expression on selenoprotein synthesis and selenoprotein mRNA levels. (A) HEK-293 cells were transfected with zebra fish selenoprotein Pa expression plasmid (Zf selP) in the absence (0) or presence (+) of SECp43 plasmid. Triplicate samples for each condition were labeled with 75Se and the media analyzed by SDS-PAGE and autoradiography. (B) Transfections were carried out as described for panel A, in the presence of SBP2 plasmid, followed by 75Se labeling, SDS-PAGE, and autoradiography. 75Se Zf selP, zebra fish selenoprotein Pa expression plasmid; UGA term, UGA-mediated termination. (C) Transfections of SECp43 plasmid were carried out and mRNA isolated two days after transfection. mRNAs for SECp43 and the indicated selenoproteins were quantitated by hybridization to oligonucleotides spotted on nylon membrane arrays as described in Materials and Methods. TR1, thioredoxin reductase 1; Sep 15, a 15-kDa selenoprotein; D1, deiodinose 1; *, P ≤ 0.05; **, P ≤ 0.01. (D) Cotransfections of HEK-293 cells with Sec-tRNA[Ser]Sec and SBP2-GFP with or without SECp43 plasmid and with one of two GPX expression plasmids. The mRNA levels of the transfected mouse GPX1 constructs with (GPX + intron) or without (GPX − intron) a genomic intron downstream of the UGA codon were quantitated by real-time quantitative PCR analysis using primers specific for the FLAG tag and GPX1 (arrowheads) or unique GPX1 (arrows) sequences. Hypoxanthine phosphoribosyltransferase (HPRT) mRNA levels were quantitated as an internal control for the cDNA synthesis reactions. **, P ≤ 0.01; no symbol, P ≥ 0.05.
As increased efficiency of selenoprotein synthesis also has implications for the ability of selenoprotein mRNAs to elude nonsense-mediated decay, we examined the responses of selenoprotein mRNAs to SECp43 overexpression in three cell lines, HepG2, HeLa, and HEK-293; transfection of SECp43 resulted in ∼1.5-, ∼8-, and 18-fold increases in the levels of SECp43, respectively, in these three lines (Fig. 5C). In each case, selenoprotein mRNA levels correlated with the increases in SECp43 mRNA levels, i.e., minimal changes in HepG2 cells, modest changes in HeLa, and more significant changes in HEK-293 cells. Intriguingly, the endogenous levels of SECp43 mRNA were lowest in HEK-293 and highest in HepG2. Notably, the mRNA for thioredoxin reductase 1 exhibited the smallest increases, and these are not likely to be due to a nonsense-decay-related effect, as the UGA selenocysteine codon and the termination codon are not separated by an intron in the pre-mRNA. Stabilization of the thioredoxin reductase 1 mRNA may be due to increased recruitment of factors to the 3′ untranslated region, potentially masking the AU-rich elements in this region.
We next sought to assess whether the increases in selenoprotein mRNA levels observed in Fig. 5C could be due to circumvention of the nonsense-mediated-decay pathway, as a result of increased UGA decoding efficiency when SECp43 is overexpressed. Targeting of an mRNA to the nonsense-mediated-decay pathway occurs when a nonsense codon is found upstream of an intron in the pre-mRNA. Under conditions of limited selenium, the UGA selenocysteine codon is interpreted as a nonsense codon (21). In selenoprotein genes where the selenocysteine codon occurs upstream of an intron, e.g., glutathione peroxidase 1 (GPX1), selenium deprivation has been shown to trigger nonsense-mediated decay of the selenoprotein mRNA (21). Constructs encoding GPX1, either containing or lacking the intron downstream of the UGA codon, were transfected in the presence or absence of SECp43 cotransfection. All of the treatment groups were also cotransfected with Sec-tRNA[Ser]Sec and SBP2 expression plasmids to ensure that these factors were not limiting; however, these cells were not supplemented with sodium selenite. GPX1 mRNA levels were quantitated by real-time RT-PCR using two sets of primer pairs specific to the exogenous mouse GPX1 constructs but not the endogenous human GPX1 gene product (Fig. 5D, upper panels). SECp43 cotransfection produced ∼6.4- and 11.7-fold increases in GPX1 mRNA derived from the intron-containing construct (Fig. 5D), whereas no change was detected in the levels of GPX1 mRNA derived from the intronless construct (Fig. 5D), strongly supporting a nonsense-mediated-decay effect on the mRNA that is partially overcome by SECp43. Further, GPX1 mRNA levels derived from the intron-containing construct in the absence of SECp43 were lower than from the intronless construct, suggesting that the nonsense-mediated-decay pathway decreased mRNA levels derived from the former, while the latter is insensitive to this pathway.
Defining the supramolecular complexes required for selenocysteine incorporation.
To assess the interactions of the major factors involved in selenoprotein biosynthesis and the effects of these interactions on subcellular localization of the complexes, we carried out additional single-factor or multifactor transfections, subcellular fractionation and coimmunoprecipitions. HEK-293 cells were transfected with single-epitope-tagged factors: SPS1-GST, SLA/LP-pentahistidine, SECp43-GST, EFsec-FLAG, and SBP2-GFP. Cytosolic and nuclear lysates were prepared, immunoprecipitated with antibody corresponding to the epitope tag on the transfected factor, and electrophoresed under nondenaturing conditions, followed by Western blot analysis. SPS1-GST, SECp43-GST, and SBP2-GFP proteins were detected both in large (>250-kDa) supramolecular complexes and as monomer-sized proteins in cytosolic samples (Fig. 6A, left panel, and data not shown) but not in the corresponding nuclear samples. In contrast, SLA/LP-His (Fig. 6A, right panel) and EFsec-FLAG (data not shown) were detected as free monomer-sized factors in cytosolic samples. SLA/LP was not detected in the nuclear samples (Fig. 6A, right panel). EFsec was detected as a 150-kDa, EFsec-containing complex in nuclear samples, corresponding to the size of an EFsec-SBP2 heterodimer (data not shown). Larger supramolecular complexes (>250 kDa) were not detected in nuclear fractions analyzed under nondenaturing conditions. However, the osmolarity increase used to extract nuclear proteins might disrupt these complexes. Conversely, if nuclear supramolecular complexes were larger than those detected in the cytosol, these very large supramolecular complexes would be unlikely to enter the 4 to 20% gradient gels used in these experiments.
FIG. 6.
Supramolecular complexes mediate selenocysteine incorporation. (A) HEK-293 cells were transfected with vector alone (V), SPS1-GST (left blot) or SLA/LP-His (right blot), cytoplasmic (C) and nuclear (N) lysates prepared as described in Materials and Methods. Lysates were immunoprecipitated with antibody corresponding to the epitope tag, followed by nondenaturing gel electrophoresis and Western blot analysis using the same antibody. (B) HEK-293 cells were transfected with the Sec-tRNA[Ser]Sec gene alone (−lanes) or with Sec-tRNA[Ser]Sec, SPS1-V5, SLA/LP-His, SECp43-GST, EFsec-FLAG, and SBP2-GFP (+lanes). The cells were lysed and fractionated, and aliquots of the cytosolic and nuclear lysates were immunoprecipitated (Immunoppt.) using the antibody to each epitope in a separate reaction. Western blot analysis was performed using each antibody in succession. In the lower panels, Western analysis was carried out with antibodies to cytoplasmic (GRB2) and nuclear (histone H1) markers to verify the lack of cross-contamination of the subcellular fractions. (C) Sucrose gradient fractionation was used to separate mRNP, ribosome subunit, and monosome and polysome fractions, followed by Western analysis to localize selenoprotein synthesis factors and RT-PCR to localize 18S rRNA as a marker for 40S ribosomal subunits. Black dots indicate the boundaries of collected fractions on the absorbance tracing, with the numbers in between indicating the fraction numbers.
Another approach to identify the supramolecular interaction partners responsible for selenocysteine biosynthesis and incorporation involved multiconstruct cotransfections of HEK-293 cells. HEK-293 cells were transfected with either a Sec-tRNA[Ser]Sec-encoding plasmid alone or a Sec-tRNA[Ser]Sec-encoding plasmid and all five of the tagged factors (see the list above, but SPS1 was tagged with V5 instead of GST, to ensure that each gene was uniquely tagged). Cytosolic and nuclear lysates were prepared and equal portions of each immunoprecipitated with one of the five tagged-protein detection systems (e.g., glutathione-Sepharose beads for GST-tagged proteins or TALON metal affinity resin for histidine-tagged proteins), followed by electrophoresis under denaturing conditions and Western blot analysis. Western blots using the antibody corresponding to the pulldown antibody verified the “bait” in these coimmunoprecipitations (Fig. 6B). Western analysis with antibodies to cytoplasmic (GRB2) and nuclear (His1) markers verified that there was no detectable cross-contamination of the subcellular fractions (Fig. 6B, lower panels). Those factors implicated in Sec-tRNA[Ser]Sec biosynthesis, SPS1, SLA/LP, and SECp43, are associated in both the cytosol and the nucleus, but the factors implicated in selenocysteine incorporation, EFsec and SBP2, were not present in these coimmunoprecipitates. Conversely, complexes consisting of SPS1, SECp43, EFsec, and SBP2 were also found in both cytosol and nucleus, but SLA/LP was not detected in these complexes. Thus, the association of SLA/LP with SECp43 and SPS1 and the association of EFsec and SBP2 with SECp43 and SPS1 appear to be mutually exclusive (see Fig. 7 below). Interestingly, SBP2 and SLA/LP were immunoprecipitated from the cytosolic, but not the nuclear, lysates. However, they were detected in nuclear lysates when other factors were used as the bait in coimmunoprecipitations. This pattern may reflect differences in antibody affinity for the epitope-tags in the bound conformation of a supramolecular complex. Alternatively, these proteins may be less abundant in nuclear lysates than the bait proteins used; therefore, the coimmunoprecipitation may serve as an enrichment process.
FIG. 7.
Selenocysteine biosynthesis and incorporation. A model that incorporates previous and new experimental evidence regarding selenium incorporation and selenoprotein biosynthesis is shown. Aminoacylation of Ser-tRNA[Ser]Sec and its conversion to Sec-tRNA[Ser]Sec, via the actions of SPS1 (green) and the putative Sec-tRNA[Ser]Sec synthase (aqua) are depicted along the top. Recruitment of SECp43 (purple) to the tRNA is depicted at the top right. Shuttling of the complex of Sec-tRNA[Ser]Sec and enzymes into the nucleus and association with EFsec (blue), SBP2 (red), and the SECIS element are depicted along the right side, and cytoplasmic export and translation are shown along the bottom. Finally, release of selenium from selenoproteins for recycling is shown on the left. Me, the methyl group added at the wobble base of Sec-tRNA[Ser]Sec.
Finally, sucrose gradient fractionation was used to separate mRNP, ribosome subunit, and monosome and polysome fractions, followed by Western analysis to localize selenoprotein synthesis factors. Ribosome-containing fractions were verified by both the absorbance readings at 254 nm and RT-PCR using 18S primers on total RNA preparations from the fractions. In this study, SPS1, SLA/LP, and SECp43 were localized primarily to mRNP (Fig. 6C, lanes 1 and 2), with a small amount of SECp43 detected in the 40S subunit fraction (lane 3) and small amounts of SLA/LP trailing into the monosome and light polysome fractions (lanes 3 to 7). EFsec and SBP2 were detected in the mRNP and monosome and polysome fractions (Fig. 6C, lanes 1, 2, and 4 through 9). This indicates that complexes mediating selenocysteine biosynthesis and incorporation exist both in association with and free of ribosomes.
DISCUSSION
We have attempted to merge what is known about selenocysteine incorporation in eukaryotes with our experimental data to generate the working model shown in Fig. 7. Ser-tRNA[Ser]Sec serves as the precursor for Sec-tRNA[Ser]Sec, likely via a phosphoseryl-tRNA[Ser]Sec intermediate. Recently, a kinase that phosphorylates Ser-tRNA[Ser]Sec has been identified in the genomes of organisms that encode other components of the selenoprotein synthesis machinery (4). Conversion of Ser-tRNA[Ser]Sec to Sec-tRNA[Ser]Sec through a phosphoseryl-tRNA[Ser]Sec intermediate is postulated to occur via a putative selenocysteine synthase, perhaps in complex with either of the SPSs (Fig. 7, top) and their product, selenophosphate. SLA/LP is a likely candidate for the selenocysteine synthase (see below). SLA/LP and SECp43 associate with the charged tRNA (Fig. 7, top right), and the complex undergoes nuclear import (Fig. 4B through D and 6B), as do EFsec and SBP2 (Fig. 6B and 7, top right). Inside the nucleus, complexes consisting of SPS1, SECp43, and SLA/LP or SPS1, SECp43, EFsec, and SBP2 are detected. This suggests that EFsec may displace SLA/LP in association with Sec-tRNA[Ser]Sec (Fig. 7, lower right), prior to associating with SBP2 bound to the SECIS elements of selenoprotein mRNAs. The assembled complex on the mRNA then exits the nucleus, with translation ensuing concurrent with or following export. The many factors responsible for selenocysteine incorporation are likely to be recycled, either free or as complexes. In addition, free selenium may be released from selenoproteins by selenocysteine lyase, thus completing the selenium cycle.
The finding that SECp43 was associated with tRNA[Ser]Sec in vivo but that binding to either uncharged or charged tRNA[Ser]Sec could not be demonstrated in vitro (8; our results herein) suggested that association of SECp43 with Sec-tRNA[Ser]Sec may only occur in complexes with other macromolecules. Here we show that SECp43 cotransfection promotes formation of a complex consisting of SPS1, SLA/LP, and SECp43, and further, that these factors are redistributed to a nuclear localization. Further, SECp43 associates with the Sec-tRNA[Ser]Sec-EFsec complex in vitro, and with EFsec in vivo. We previously showed that EFsec-SBP2 complex formation requires the presence of Sec-tRNA[Ser]Sec (29), and we now demonstrate that SECp43 enhances association between EFsec and SBP2 in vivo and inhibits SBP2-dependent release of Sec-tRNA[Ser]Sec from EFsec in vitro.
Based on these findings and recent published studies, plausible functions for SLA/LP and SECp43 are roles in the selenocysteine synthase reaction and the 2′-O-methylation of the wobble base in Sec-tRNA[Ser]Sec, respectively. Evidence for the former includes the following. First, comparative sequence analysis identified SLA/LP as a member of the pyridoxal phosphate-dependent transferase superfamily (15) to which bacterial selenocysteine synthases belong. Second, recent studies identified a phosphoseryl-tRNA intermediate in the pathway for Cys-tRNA biosynthesis in archaea (24), analogous to the proposed Sec-tRNA[Ser]Sec pathway, implying similar functions for SLA/LP and the archaeal enzyme. Third, the fact that EFsec appears to displace SLA/LP in complexes containing SECp43, SPS1, and tRNA[Ser]Sec suggests a function for SLA/LP upstream of EFsec binding, e.g., Ser-tRNA[Ser]Sec to Sec-tRNA[Ser]Sec conversion.
Evidence for SECp43 functioning in the 2′-O-methylation reaction is as follows. SECp43 mRNA levels increased approximately fivefold in HEK-293 cells in response to selenium supplementation (unpublished data), and methylation of Sec-tRNA[Ser]Sec has previously been shown to increase in response to selenium (8). Methylation is known to increase selenocysteine incorporation (14), consistent with our findings herein, suggesting an adaptation to increase efficiency of incorporation when selenium is abundant. While methylation is not required for EFsec binding to Sec-tRNA[Ser]Sec (26), it may promote the association of EFsec with SBP2, a role that has not previously been examined. Finally, silencing of SECp43 expression using siRNA technology results in a decrease in this methylation (28), providing strong evidence for a role for SECp43 in this modification. Further investigation of the detailed functions of SLA/LP and SECp43 are required before their functions can be definitively assigned in selenocysteine biosynthesis and incorporation.
Previous studies have shown that other tRNAs undergo aminoacylation in the nucleus prior to nucleocytoplasmic export (19). Nuclear localization of the enzymes necessary for the generation of mature methylated Sec-tRNA[Ser]Sec, in combination with our recent finding that EFsec and SBP2 also undergo nucleocytoplasmic shuttling (7), would set the stage for assembly of selenocysteine incorporation complexes prior to nucleocytoplasmic export. Thus, selenoprotein mRNAs would be primed for translation when the first ribosomes associate on the cytoplasmic side of the nuclear pore, concomitant with export, thus facilitating circumvention of nonsense-mediated decay.
What are the additional implications for the formation of complexes consisting of the selenocysteine biosynthesis and selenocysteine incorporation factors? Several studies have provided evidence for the existence of supramolecular translation complexes, consisting of ribosomes, mRNA, elongation factors, aminoacyl tRNAs, and aminoacyl tRNA synthetases (1, 22, 23, 25). Similar supramolecular complexes for selenoprotein mRNAs, including the factors specific to Sec-tRNA[Ser]Sec charging and modification and to cotranslational selenocysteine incorporation, could contribute significantly to the efficiency of selenoprotein synthesis, which would be particularly important for the incorporation of multiple selenocysteines into selenoprotein P. Further investigation of these interactions will certainly yield new insights into this intriguing translation mechanism.
Acknowledgments
We thank Ilko Stoytchev for technical assistance and Rosa Tujebajeva for expert assistance with the sucrose gradient polysome fractionation. We would also like to thank Helen Turner at the Queen's Center for Biomedical Research in Honolulu, HI, for her generous donation of cell lines and other materials, for excellent technical advice, and for providing a safe haven for our research while our building was closed due to extensive flood damage in October of 2004.
This work was supported by Public Health Service grants DK52963 and DK47320 to M.J.B. from the NIH.
REFERENCES
- 1.Barbarese, E., D. E. Koppel, M. P. Deutscher, C. L. Smith, K. Ainger, F. Morgan, and J. H. Carson. 1995. Protein translation components are colocalized in granules in oligodendrocytes. J. Cell Sci. 108:2781-2790. [DOI] [PubMed] [Google Scholar]
- 2.Berry, M. J., L. Banu, Y. Y. Chen, S. J. Mandel, J. D. Kieffer, J. W. Harney, and P. R. Larsen. 1991. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature 353:273-276. [DOI] [PubMed] [Google Scholar]
- 3.Berry, M. J., J. W. Harney, T. Ohama, and D. L. Hatfield. 1994. Selenocysteine insertion or termination: factors affecting UGA codon fate and complementary anticodon:codon mutations. Nucleic Acids Res. 22:3753-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson, B. A., X. M. Xu, G. V. Kryukov, M. Rao, M. J. Berry, V. N. Gladyshev, and D. L. Hatfield. 2004. Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc. Natl. Acad. Sci. USA 101:12848-12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chavatte, L., B. A. Brown, and D. M. Driscoll. 2005. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat. Struct. Mol. Biol. 12:408-416. [DOI] [PubMed] [Google Scholar]
- 6.Copeland, P. R., J. E. Fletcher, B. A. Carlson, D. L. Hatfield, and D. M. Driscoll. 2000. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 19:306-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Jesus, L. A., P. R. Hoffmann, T. Michaud, E. P. Forry, A. Small-Howard, R. J. Stillwell, N. Morozova, J. W. Harney, and M. J. Berry. 2006. Nuclear assembly of UGA decoding complexes on selenoprotein mRNAs: a mechanism for eluding nonsense mediated decay? Mol. Cell. Biol. 26:1795-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diamond, A. M., I. S. Choi, P. F. Crain, T. Hashizume, S. C. Pomerantz, R. Cruz, C. J. Steer, K. E. Hill, R. F. Burk, J. A. McCloskey, and D. L. Hatfield. 1993. Dietary selenium affects methylation of the wobble nucleoside in the anticodon of selenocysteine tRNA([Ser]Sec). J. Biol. Chem. 268:14215-14223. [PubMed] [Google Scholar]
- 9.Ding, F., and P. J. Grabowski. 1999. Identification of a protein component of a mammalian tRNA(Sec) complex implicated in the decoding of UGA as selenocysteine. RNA 5:1561-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fagegaltier, D., N. Hubert, K. Yamada, T. Mizutani, P. Carbon, and A. Krol. 2000. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 19:4796-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gelpi, C., E. J. Sontheimer, and J. L. Rodriguez-Sanchez. 1992. Autoantibodies against a serine tRNA-protein complex implicated in cotranslational selenocysteine insertion. Proc. Natl. Acad. Sci. USA 89:9739-9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatfield, D., B. J. Lee, L. Hampton, and A. M. Diamond. 1991. Selenium induces changes in the selenocysteine tRNA[Ser]Sec population in mammalian cells. Nucleic Acids Res. 19:939-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatfield, D. L., and V. N. Gladyshev. 2002. How selenium has altered our understanding of the genetic code. Mol. Cell. Biol. 22:3565-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kernebeck, T., A. W. Lohse, and J. Grotzinger. 2001. A bioinformatical approach suggests the function of the autoimmune hepatitis target antigen soluble liver antigen/liver pancreas. Hepatology 34:230-233. [DOI] [PubMed] [Google Scholar]
- 16.Lee, B. J., P. J. Worland, J. N. Davis, T. C. Stadtman, and D. L. Hatfield. 1989. Identification of a selenocysteyl-tRNA(Ser) in mammalian cells that recognizes the nonsense codon, UGA. J. Biol. Chem. 264:9724-9727. [PubMed] [Google Scholar]
- 17.Low, S. C., E. Grundner-Culemann, J. W. Harney, and M. J. Berry. 2000. SECIS-SBP2 interactions dictate selenocysteine incorporation efficiency and selenoprotein hierarchy. EMBO J. 19:6882-6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Low, S. C., J. W. Harney, and M. J. Berry. 1995. Cloning and functional characterization of human selenophosphate synthetase, an essential component of selenoprotein synthesis. J. Biol. Chem. 270:21659-21664. [DOI] [PubMed] [Google Scholar]
- 19.Lund, E., and J. E. Dahlberg. 1998. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 282:2082-2085. [DOI] [PubMed] [Google Scholar]
- 20.Martin, G. W., III, and M. J. Berry. 2001. Selenocysteine codons decrease polysome association on endogenous selenoprotein mRNAs. Genes Cells 6:121-129. [DOI] [PubMed] [Google Scholar]
- 21.Moriarty, P. M., C. C. Reddy, and L. E. Maquat. 1998. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol. Cell. Biol. 18:2932-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Negrutskii, B. S., V. F. Shalak, P. Kerjan, A. V. El'skaya, and M. Mirande. 1999. Functional interaction of mammalian valyl-tRNA synthetase with elongation factor EF-1a in the complex with EF-1H. J. Biol. Chem. 274:4545-4550. [DOI] [PubMed] [Google Scholar]
- 23.Sampath, P., B. Mazumder, V. Seshadri, C. A. Gerber, L. Chavatte, M. Kinter, S. M. Ting, J. D. Dignam, S. Kim, D. M. Driscoll, and P. L. Fox. 2004. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell 119:195-208. [DOI] [PubMed] [Google Scholar]
- 24.Sauerwald, A., W. Zhu, T. A. Major, H. Roy, S. Palioura, D. Jahn, W. B. Whitman, J. R. Yates III, M. Ibba, and D. Soll. 2005. RNA-dependent cysteine biosynthesis in archaea. Science 307:1969-1972. [DOI] [PubMed] [Google Scholar]
- 25.Stapulionis, R., and M. P. Deutscher. 1995. A channeled tRNA cycle during mammalian protein synthesis. Proc. Natl. Acad. Sci. USA 92:7158-7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tujebajeva, R. M., P. R. Copeland, X. M. Xu, B. A. Carlson, J. W. Harney, D. M. Driscoll, D. L. Hatfield, and M. J. Berry. 2000. Decoding apparatus for eukaryotic selenocysteine incorporation. EMBO Rep. 2:158-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss, S. L., and R. A. Sunde. 1998. Cis-acting elements are required for selenium regulation of glutathione peroxidase-1 mRNA levels. RNA 4:816-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu, X. M., H. Mix, B. A. Carlson, P. J. Grabowski, V. N. Gladyshev, M. J. Berry, and D. L. Hatfield. 2005. Evidence for direct roles of two additional factors, SECp43 and SLA, in the selenoprotein synthesis machinery. J. Biol. Chem. 280:41568-41575. [DOI] [PubMed] [Google Scholar]
- 29.Zavacki, A. M., J. B. Mansell, M. Chung, B. Klimovitsky, J. W. Harney, and M. J. Berry. 2003. Coupled tRNASec dependent assembly of the selenocysteine decoding apparatus. Mol. Cell 11:773-781. [DOI] [PubMed] [Google Scholar]