Abstract
Telomerase reverse transcriptase (TERT) and telomerase RNA (TER) assemble as part of a holoenzyme that synthesizes telomeric repeats at chromosome ends. Genetic approaches have identified proteins that are required for in vivo association of TERT and TER, including the Tetrahymena telomerase holoenzyme protein p65. Here, we use quantitative assays to define the mechanisms underlying p65 function in holoenzyme biogenesis. We demonstrate that four modules of p65 contribute affinity for TER, including a C-terminal domain that recognizes the conserved dinucleotide bulge of central stem IV. This C-terminal domain is necessary and sufficient for p65's function in enhancing the recruitment of TERT to TER. Finally, we show that p65 and TERT assemble on TER with hierarchical rather than cooperative binding. These findings elucidate an extensive network of p65-TER recognition specificity and define a novel p65 RNA binding domain that initiates telomerase holoenyzme biogenesis.
The telomerase ribonucleoprotein (RNP) restores the telomeric simple sequence repeats that are lost as a consequence of cellular proliferation. A telomerase reverse transcriptase, TERT, and a functional telomerase RNA, TER, can be combined in cell lysate to form catalytically active RNP (4, 6, 14). The reconstitution of the minimal enzyme in vitro is limited by inefficient RNP assembly, which must bypass the complex in vivo process of telomerase holoenzyme biogenesis. The physiological pathway for telomerase RNP assembly in diverse species appears to initiate with TER binding by proteins that promote TER accumulation (3, 5). Telomerase-associated proteins that have been shown to be required for TER stability in vivo include H/ACA motif binding proteins in vertebrates, Sm proteins in yeasts, and a La motif protein in ciliates (9, 13, 15). These proteins must identify nonabundant TER primary transcripts and package them into biologically stable forms while leaving the template and other TER regions accessible for association with TERT and telomeres.
Tetrahymena thermophila p65 is one of five protein subunits of an endogenously assembled telomerase holoenzyme (15). The genetic depletion of p65 dramatically and specifically inhibits the accumulation of both TER and TERT, revealing a crucial role for p65 in the biogenesis of active telomerase RNP. Purified recombinant p65 binds TER directly and promotes the formation of a p65-TER-TERT ternary complex (12). These biochemical activities of p65 likely account for its biological functions in protecting TER and TERT from degradation. We previously investigated TER sequence requirements for p65 interaction in vitro (12). Variant TERs with sequence substitutions in a base-paired region of stem IV were crippled in their competition with wild-type (WT) TER for p65 binding, suggesting that p65 could use recognition of stem IV to discriminate TER from other RNAs.
A primary sequence analysis of p65 suggests two likely RNA binding domains (RBDs), a La motif and a putative RNA recognition motif (RRM), which are flanked by N- and C-terminal extensions (15). Adjacent La and RRM motifs in eukaryotic La proteins have been shown to constitute the high-affinity binding site for the poly(U) tract that is present at the 3′ end of RNA polymerase III transcripts (16). Although p65 La and RRM-like motifs diverge considerably from consensus, at least the La motif is conserved in p43, the Euplotes ortholog of p65 (1). In curious contrast with La, however, both Euplotes p43 and Tetrahymena p65 show only a minor dependence on the ciliate TER 3′ poly(U) tract for binding (2, 12).
To develop a better understanding of p65-TER interaction, we expressed and purified recombinant full-length and domain-truncated variants of p65. Using electrophoretic mobility shift assays (EMSAs), we defined contributions from several regions of the protein to its TER binding and TERT RNP assembly activities. Our findings reveal a modular organization of p65 with domains providing specificity for TER stem I, stem IV, the central stem IV dinucleotide bulge and the 3′ poly(U) tract. A novel C-terminal domain of p65 recognizes the dinucleotide bulge of central stem IV and mediates p65 function in TERT RNP assembly. These studies elucidate molecular mechanisms that are underlying telomerase holoenzyme protein function in the cellular biogenesis of RNP.
MATERIALS AND METHODS
RNA and protein expression and purification.
Wild-type TER and all TER variants were synthesized by using T7 RNA polymerase, gel purified, and examined by denaturing gel electrophoresis to verify RNA quality (10). Sequence substitutions were to the complement of the wild type unless indicated otherwise. The linker added in TER stem I/IV has the sequence UUAAUUCAUU. The tetraloop substitution of positions 121 to 146 used the sequence UUCG (12). Synthetic genes were used to express Tetrahymena p65 (15) and TERT (4). Protein expression in rabbit reticulocyte lysate was done by standard methods (7). Escherichia coli expression of p65 was performed using pET28 that was engineered to include an N-terminal His6 tag followed by a tobacco etch virus protease cleavage site. Bacterial expression and purification of p65 and TERT polypeptides were carried out by chromatography on nickel agarose as previously described (12). Only minor variations in TER binding affinity were observed with independent preparations of each protein. The amino acids of p65 that were included in the truncated polypeptides are as follows (N, N-terminal domain; L, La motif; R, putative RRM; C, C-terminal domain): 1 to 239 for NL, 1 to 340 for NLR, 240 to 542 for RC, and 302 to 542 for C.
Binding and activity assays.
Radiolabeled TER was synthesized by T7 RNA polymerase by using 32P-UTP and gel purified. EMSAs were performed as described previously (10), including control experiments to confirm that the radiolabeled probe was substoichiometric and that binding reactions reached equilibrium before gel electrophoresis. Ten-microliter binding reaction mixtures contained 20 mM Tris-HCl (pH 8.0), 1 mM MgCl2, 10% glycerol, 150 mM KCl, 5 mM dithiothreitol, 5 μg bovine serum albumin, 0.25 μ l RNasin, 500 ng E. coli tRNA as nonspecific competitor, bromophenol blue, and xylene cyanol. Competitor RNA was added before the addition of the ∼0.1-nM-or-less final concentration of radiolabeled TER. Affinities were calculated with ImageQuant software, following data acquisition by a phosphorimager (Amersham). The stated affinity is the protein concentration at which 50% of the probe is bound.
For RNP assembly in rabbit reticulocyte lysate, protein expression reaction mixtures were combined with purified TER in an approximately fivefold excess of TERT and incubated at 30°C for 15 min. For immunopurification, the tagged p65 polypeptide was recovered using immunoglobulin G (IgG) agarose (Sigma) at 4°C for 30 min in buffer containing 20 mM Tris (pH 8.0), 1 mM MgCl2, 10% glycerol, 5 mM dithiothreitol, and 10 μg/ml each of E. coli tRNA (Sigma) and bovine serum albumin. Primer extension activity assays were carried out for 30 min at room temperature using 0.6 μM 32P-dGTP, 5 μM unlabeled dGTP, 200 μM TTP, and 1 μM of the DNA primer (TG)8T2G3 in reaction buffer with 50 mM Tris-acetate (pH 8.0), 10 mM spermidine, 5 mM beta-mercaptoethanol, and 2 mM MgCl2. Products were precipitated and resolved by denaturing gel electrophoresis.
RESULTS
Multiple p65 domains contribute to TER binding.
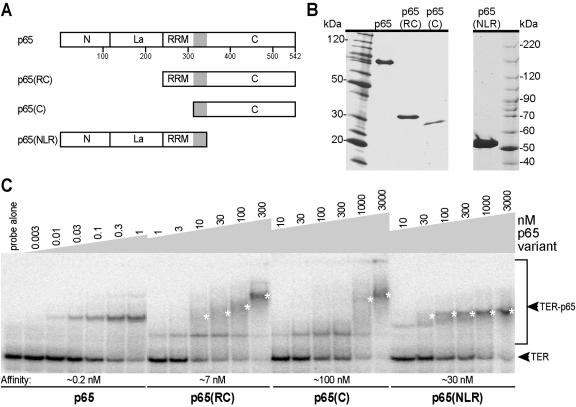
The adjacent La and RRM-like motifs of p65 are reminiscent of eukaryotic La proteins, which use tandem La and RRM domains to recognize the 3′ poly(U) tract of RNA polymerase III primary transcripts and other small RNAs (16). However, the La and RRM-like motifs of p65 have insertions and deviations from consensus and bacterially expressed p65 requires a duplex region of TER for binding (12). To resolve whether the La and RRM-like regions of p65 establish its binding specificity for TER, we assayed purified, bacterially expressed p65 truncation variants for RNA binding activity. We tested a panel of polypeptides containing one or more of the p65 regions (N, L, R, and C) delineated by primary sequence. Because the C-terminal boundary of the putative RRM was ambiguous (Fig. 1A), p65(NLR) and p65(C) were designed to share a region of overlap. All of the p65 polypeptides were expressed with an N-terminal His6 tag, which allowed for their purification to near homogeneity (Fig. 1B). The removal of the tag by specific protease cleavage had no impact on RNA binding activity (data not shown).
FIG. 1.
Identification of p65 domains with TER binding activity. (A) Schematic of full-length p65 and p65 truncation variants with TER binding activity. Amino acid numbering is indicated for full-length p65. The shaded region indicates that the C-terminal boundary of the putative RRM was ambiguous. (B) SDS-PAGE and Coomassie blue staining of purified polypeptides. Molecular mass markers are indicated in kilodaltons. (C) EMSA analysis of p65 and p65 truncation variants using radiolabeled wild-type TER. Results are shown over different ranges of protein concentration due to large differences in binding affinity. Asterisks indicate the heterogeneous supershift of p65-TER that results from protein aggregation.
We first determined the affinities of variously truncated p65 polypeptides for full-length TER. The mobility shift of substoichiometric radiolabeled TER was monitored over a range of protein concentrations (Fig. 1C). Particularly high protein concentrations induced nonspecific protein aggregation that was evident in EMSA as a heterogeneous supershift of the p65-TER complex (Fig. 1C). Full-length p65 bound TER with an affinity of ∼0.2 nM under the conditions used here (Fig. 1C, outer left). The truncation of the C-terminal domain to create p65(NLR) reduced TER binding affinity by 150-fold to ∼30 nM (Fig. 1C, outer right). Truncations of the N-terminal domain reduced binding affinity by up to fivefold (data not shown). Countering our initial expectation, however, combined truncation of the N-terminal and C-terminal domains to create a polypeptide with only the p65 La and putative RRM domains completely abolished TER binding (data not shown). These findings indicate that the two previously predicted RNA binding domains of p65 are not sufficient to support protein-RNA interaction.
We next determined whether any two-domain p65 polypeptide could retain TER binding activity. We found that the p65(RC) polypeptide formed a TER mobility shift complex with an affinity of ∼7 nM (Fig. 1C, middle left), which was reduced only 35-fold from the binding affinity of full-length p65. To our surprise, further truncation of the putative RRM to create p65(C) did not destroy TER binding activity. Despite lacking any known RNA binding motifs, the p65 C-terminal domain bound TER with an affinity of ∼100 nM (Fig. 1C, middle right). No other domain of p65 that was expressed in isolation formed a discrete mobility shift complex with TER (data not shown). Curiously, however, the deletion of this same C-terminal domain from the full-length protein did not prevent p65(NLR) interaction with TER (Fig. 1C, right). Together, these results indicate that multiple domains of p65 interact with TER and can bind TER in an at least partially independent fashion.
Distinct TER binding specificities of p65 domains.
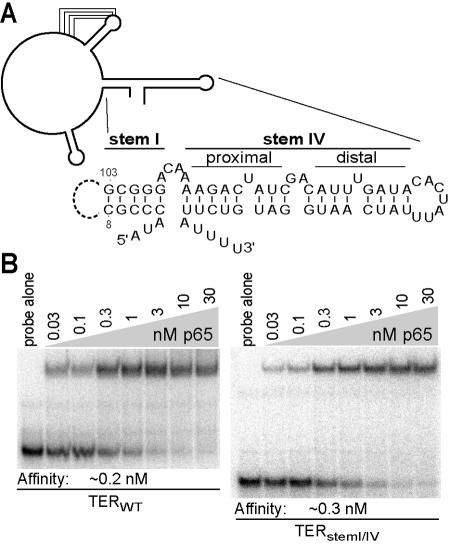
If p65 RNA binding domains make distributed interactions with TER, we would expect multiple regions of TER to participate in p65 binding. The sequence and structure of template-proximal stem IV were previously shown to impact p65 binding (12), but we found that stem IV alone was not sufficient for p65 interaction (data not shown). Previous mobility shift competition experiments also suggested some compromise of p65-TER interaction from substitution or deletion changes in stem I, the central stem IV dinucleotide bulge (GA121-122), or the 3′ poly(U) tract (12). To determine whether the combination of all of these regions was sufficient for high-affinity p65 binding, we replaced the template and surrounding sequences with a 10-nucleotide linker to create TER stem I/IV (Fig. 2A). Unlabeled TER stem I/IV competed effectively with radiolabeled wild-type TER for binding to p65 (data not shown). We next compared p65 binding to the radiolabeled wild-type and stem I/IV TERs. EMSAs using radiolabeled wild-type or stem I/IV TER revealed an almost indistinguishable affinity of p65 interaction (Fig. 2B). Changing the linker sequence in TER stem I/IV did not affect p65 binding (data not shown). We conclude that the major determinants of p65 binding are contained within the TER stem I/IV region.
FIG. 2.
A region of TER sufficient for p65 binding. (A) Schematic of T. thermophila TER and sequence of the p65 binding region. The dashed line indicates the linker in TER stem I/IV, which replaces wild-type TER positions 9 to 102, including stem II, the template, and the stem III pseudoknot. (B) EMSA analysis of p65 using radiolabeled WT and stem I/IV TERs. The binding assays were performed in parallel and electrophoresed on the same gel.
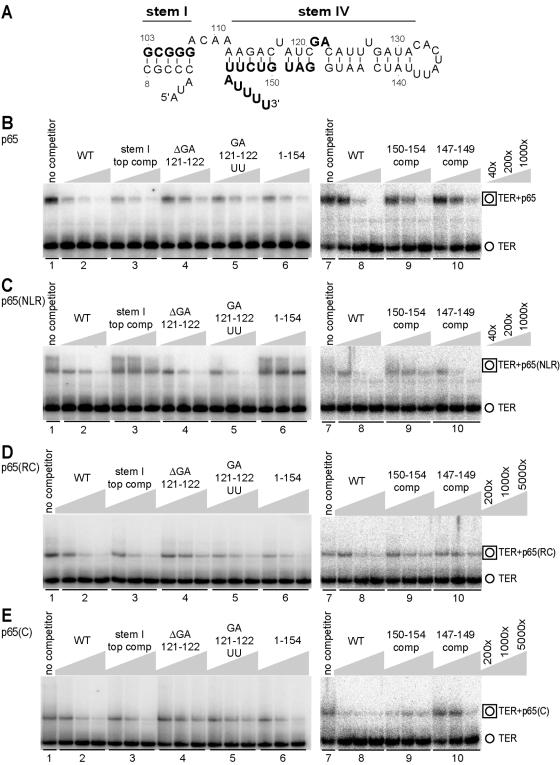
We next investigated whether p65 domains make distinguishable contributions to TER recognition specificity. We compared the specificity of TER recognition among p65 polypeptides that are capable of forming discrete mobility shift complexes as follows: full-length p65, p65(NLR), p65(RC), and p65(C), which are assayed in Fig. 3 B through E, respectively. We competed the interactions of these proteins with radiolabeled wild-type TER by the addition of fivefold increasing steps of wild-type TER and TER variants that unpair stem I or template-proximal stem IV, delete or substitute the central stem IV dinucleotide bulge, or remove the 3′ poly(U) tract (Fig. 3A). Competition for TER binding to full-length p65 was optimal with only wild-type TER (Fig. 3B, lane sets 1 and 2). However, each variant retained some ability to compete with wild-type TER, which is consistent with a distributed set of p65-TER interactions. Among the TER variants, unpairing stem IV or altering the central stem IV dinucleotide bulge imposed the most severe loss of competition (Fig. 3B, compare lane sets 9 and 10 versus lane set 8 and lane sets 4 and 5 versus lane set 2). Unpairing stem I or removing the 3′ poly(U) tract had less impact (Fig. 3B, compare lane sets 3 and 6 versus lane set 2).
FIG. 3.
Distinct TER binding specificities of p65 domains. (A) TER variants in the stem I/IV region. TER sequence in the stem I/IV region is shown with residues in bold type indicating the positions of sequence substitution or deletion described in the text. (B through E) EMSA competition with WT and variant TERs. Unlabeled TERs were added at the excess concentrations over radiolabeled wild-type TER indicated by the keys on the right. For the binding assays shown in lane sets 1 through 6 or lane sets 7 through 10, the p65, p65(NLR), p65(RC), and p65(C) competitions were performed in parallel with additional controls not shown. We note that, for lane set 9 of panel E, the first lane has less of the mobility shift complex than was detected in a duplicate set of reaction mixtures electrophoresed on the same gel. EMSAs in panels 1 through 6 used 0.2 nM p65, 40 nM p65(NLR), 5 nM p65(RC), and 50 nM p65(C). EMSAs in panels 7 through 10 used 0.4 nM p65, 50 nM p65(NLR), 5 nM p65(RC), and 50 nM p65(C). ΔGA 121-122, deletion of GA 121 and 122.
In assays with p65(NLR), equally effective competition was observed by using wild-type TER and TER variants with deletion or substitution of the central stem IV bulge (Fig. 3C, compare lane sets 4 and 5 versus lane set 2). In contrast, the loss of competition imposed by unpairing stem I, the immediately adjacent region of stem IV, or the 3′ poly(U) tract was severe (Fig. 3C, compare lane sets 3 and 6 versus lane set 2 and lane set 9 versus lane set 8). This specificity is notably opposite that of the full-length protein (Fig. 3B). In comparison, p65(RC) and p65(C) both showed a competition specificity similar to that of full-length p65 and complementary to that of p65(NLR). For EMSAs with p65(RC) and p65(C), the most severe loss of competition was imposed by unpairing stem IV or altering the central stem IV bulge. For p65(RC), stem IV unpairing appeared most deleterious along with deletion of the central stem IV bulge (Fig. 3D, compare lane sets 9 and 10 versus lane set 8 and lane set 4 versus lane set 2). For p65(C), the deletion or substitution of the central stem IV bulge severely compromised competition (Fig. 3E, compare lane sets 4 and 5 versus lane set 2). Notably, unpairing stem I or removing the 3′ poly(U) tract had no impact on TER competition for binding to p65(RC) or p65(C): these variants were as effective competitors as was the wild type (Fig. 3D and E, compare lane sets 3 and 6 versus lane set 2). Together, these results support a distributed mode of p65-TER interaction in which the N-terminal and La motif domains of p65 recognize stem I and the 3′ poly(U) tract, and the p65 C-terminal domain recognizes the central stem IV bulge. The precise role of the RRM-like region remains speculative. We hypothesize that it could have a direct interaction with stem IV, based on competition specificity and the ability of this region to improve TER binding affinity when fused to either p65(NL) or p65(C).
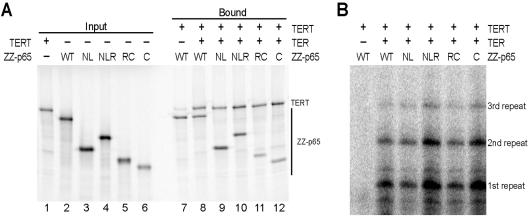
The physiological specificity of p65-TER interaction must leave the template and other important motifs available for association with TERT and for participation in the catalytic cycle. For a control for this crucial aspect of TER interaction specificity, we assembled p65 truncation variants with full-length TER and TERT in rabbit reticulocyte lysate, immunopurified the ternary complexes, and assayed their catalytic activity. We expressed full-length p65 (WT), p65(NL), p65(NLR), p65(RC), and p65(C) in N-terminal fusion with the tandem protein A tag (ZZ-tag) that was used previously (12). These epitope-tagged p65 proteins and full-length TERT were expressed in separate synthesis reaction mixtures containing radiolabeled methionine (Fig. 4A, lanes 1 through 6), combined with or without TER, and then immunopurified using IgG agarose under low-ionic-strength conditions to preserve low-affinity p65-TER interactions (see Materials and Methods). Aliquots of each bound sample were analyzed in parallel by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and primer extension activity assay.
FIG. 4.
Assembly and activity of TER and TERT with p65 and p65 truncation variants. Active ternary complexes formed by p65 polypeptides, TER and TERT. TERT and ZZ-tagged p65 polypeptides were expressed in reticulocyte lysate. Aliquots of TERT expression reaction mixture were combined with either full-length p65 (WT) or the p65 truncation variant indicated and then supplemented with buffer (−TER lanes) or wild-type TER (+TER lanes). Ternary complexes bound to IgG agarose were split for analysis of 35S-labeled proteins by SDS-PAGE (A) or analysis of catalytic activity by primer extension and denaturing gel electrophoresis (B). In panel A, SDS-PAGE was performed on both the input expression reaction mixtures (input) and purified samples (bound). In panel B, products representing sequential rounds of complete repeat synthesis to the template 5′ end are indicated.
All of the truncated p65 polypeptides that were examined could copurify TERT in the presence of TER (Fig. 4A, lanes 8 to 12), while only a nonspecific background level of TERT was recovered with p65 in the absence of TER (lane 7; also data not shown). All of these ternary complexes were catalytically active (Fig. 4B), with no obvious difference in product synthesis when normalized to the recovery of TERT. We conclude that the truncated p65 polypeptides retain some sequence specificity for TER, allowing each to assemble with TER in a manner that is permissive of functional TER-TERT interaction. We note, however, that the catalytic activity reconstituted by Tetrahymena TERT and TER in reticulocyte lysate does not recapitulate the high repeat addition processivity of the endogenously assembled holoenzyme (4). Therefore, p65 influence on catalytic activity in the holoenzyme context remains to be investigated after the development of a holoenzyme reconstitution system.
p65 stimulates TER association with a single domain of TERT.
In addition to binding TER, p65 promotes TERT assembly into RNP. Previous experiments have not provided evidence for a direct protein-protein interaction between p65 and TERT (12). Instead, perhaps p65-TER interaction reduces TER structural heterogeneity, favoring a TER conformation that is optimal for TERT binding. Previous assays of the p65-TER-TERT ternary complex assembly used either full-length TERT expressed in rabbit reticulocyte lysate or the N-terminal half of TERT, TERT(1-516), expressed in E. coli (12). TERT(1-516) harbors two domains that can each independently associate with TER: a high-affinity RNA binding domain and an extreme N-terminal domain with much lower affinity for TER (10). It was therefore plausible that p65-TER interaction promotes TERT RNP assembly by promoting synergy in the binding of the two TERT RNA interaction domains.
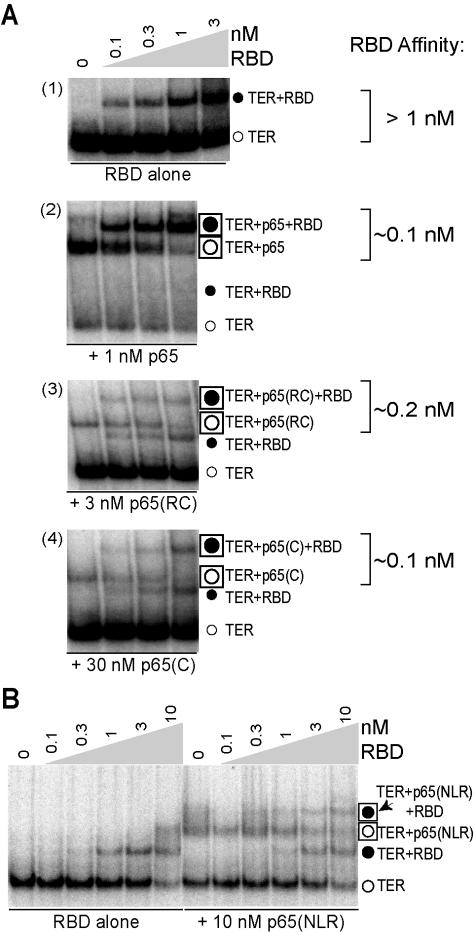
To investigate this possibility, we compared the influence of p65 on assembly of TER with TERT(1-516) or the isolated TERT RBD. If p65 arranges TER to promote cooperative binding of TERT domains, p65 should enhance TERT(1-516) interaction with TER but not RBD interaction with TER. We found that TERT(1-516) bound TER with an affinity of ∼6 nM (Fig. 5A, left) and showed an improved affinity for the p65-TER complex of ∼0.4 nM (Fig. 5A, right), an enhancement similar to previous findings (12). The TERT RBD bound TER with an affinity of ∼4 nM (Fig. 5B, left). Interestingly, and in contrast with the expectation described above, RBD assembly with TER was also enhanced in the presence of p65 (Fig. 5B, right). RBD bound to p65-TER with an affinity of ∼0.4 nM, a 10-fold improvement over its binding to TER alone (quantification of data from Fig. 5B is shown in Fig. 5C). Furthermore, p65-enhanced RBD assembly had the same TER sequence requirements that were shown previously for p65-enhanced assembly of TERT(1-516) (12). For example, replacing the template-distal region of stem IV with a tetraloop did not prevent p65-TER or TERT-TER interaction but did eliminate p65 enhancement of TERT assembly (Fig. 5D). Therefore, p65 can promote TERT-TER interaction through an impact on the TERT RBD alone by a mechanism not involving coordination of separable TERT domain interactions with TER.
FIG. 5.
A p65-dependent enhancement of TERT RBD assembly. (A, B, and D) Radiolabeled wild-type TER or TER with a tetraloop replacing distal stem IV (tetraloop 121-146) was combined with either buffer (left) or a final concentration of 4 nM p65 (right) and then the indicated concentrations of TERT(1-516) or TERT RBD. The binding assays with TERT RBD were performed in parallel and electrophoresed on the same gel. Symbols in panels B and D are defined in panel A. (C) Quantification of the EMSA in panel B is shown. We note that, above nanomolar p65 concentrations, we sometimes detected two closely migrating but discrete p65-TER mobility shifts, which are evident as a doublet in panel B. The major, slower-mobility p65-TER complex has preferential binding to TERT: it shifts into ternary complex at lower concentrations of RBD. The maximal p65-mediated enhancement of TERT RNP assembly will be underestimated in the presence of the faster-mobility p65-TER complex.
An RNP assembly mechanism directed by p65(C).
We next investigated which domains of p65 promote TERT assembly into RNP. We compared TERT RBD binding to that of TER alone or TER assembled with p65 or a p65 truncation variant (Fig. 6A and B). We used limiting concentrations of the p65 truncation variants to guard against nonspecific protein aggregation (Fig. 1C, high protein concentration lanes). We quantified the influence of the p65 polypeptides on RBD binding affinity by calculating the ratios of TER to TER-RBD and p65-TER to p65-TER-RBD as a function of increasing RBD concentration. To our surprise, both p65(RC) and p65(C) enhanced RBD binding affinity for TER, with an impact similar to that of full-length p65 (Fig. 6A). In contrast, p65(NLR) did not appear to substantially alter RBD binding affinity (Fig. 6B): the supershift of free TER to TER-RBD and p65(NLR)-TER to p65(NLR)-TER-RBD occurred in parallel, even within the same sample. Thus, the C-terminal RNA binding domain of p65 is necessary and sufficient to enhance TERT-TER interaction.
FIG. 6.
Ternary complex RNP assembly with p65 and p65 truncation variants. (A and B) Radiolabeled wild-type TER was combined with either buffer (RBD alone sections), p65, or a p65 truncation variant, and then various concentrations of TERT RBD. All binding assays in panels A and B were performed in parallel and electrophoresed on the same gel; note that EMSAs in panels A and B used different preparations of TERT RBD.
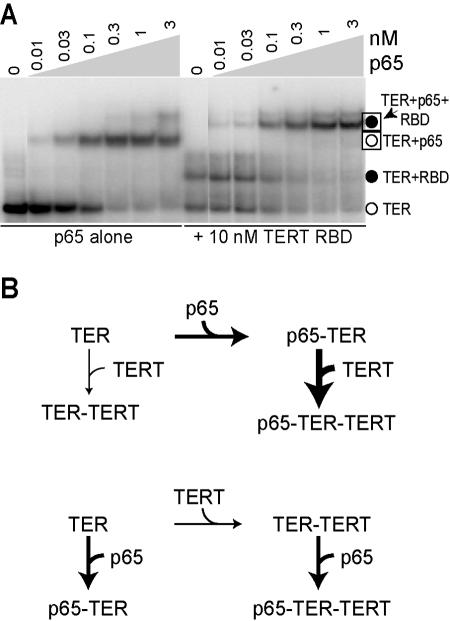
In vivo accumulation of p65 does not require its binding to TER or TERT, but the physiological stabilities of TER and TERT are dependent on their partners in ternary complex formation (15) (K. Witkin, Q. Tieu, and K. Collins, unpublished data). To determine whether p65 impacts TERT RNP assembly in a strictly hierarchical manner or, instead, whether TERT-TER interaction has a reciprocal impact on p65 RNP assembly, we assayed p65 binding affinity for TER by using either TER alone (Fig. 7A, left) or the RBD-TER complex (Fig. 7A, right). The affinity of p65 for TER was unaffected by the presence of TERT: p65 bound to TER alone or to the RBD-TER complex with similar affinity (∼0.1 nM in this experiment). The same result was obtained using TERT(1-516) instead of the isolated RBD (data not shown).
FIG. 7.
A hierarchical RNP assembly pathway. (A) Radiolabeled wild-type TER was combined with either buffer (p65 alone panel) or the TERT RBD, and then various concentrations of p65. (B) Schematic of pathways for ternary complex RNP assembly. The top pathway depicts the preferential assembly of TERT on p65-TER versus TER, illustrating the data shown in Fig. 5B. The bottom pathway depicts equivalent affinity of p65 for TER alone or TER-TERT, illustrating the data shown in panel A.
Together, the results above define a hierarchical assembly pathway for the initial ternary complex of Tetrahymena telomerase holoenzyme. We have shown that p65 enhances TERT assembly into RNP (Fig. 5B and 7B, top), but TERT does not enhance p65 assembly into RNP as would be expected with cooperative binding (Fig. 7A and B, bottom). At least in vitro, p65 affinity for TER is greater than that of TERT(1-516) or TERT RBD. When concentrations of both proteins are limiting, as would be expected in vivo for these nonabundant proteins, ternary complex assembly will follow a hierarchical pathway of TER-p65 interaction followed by the recruitment of TERT. Within the p65-TER complex, p65 C-terminal domain association with the central stem IV dinucleotide bulge induces the improvement in TERT binding affinity and thus directs the cellular biogenesis pathway of Tetrahymena telomerase holoenzyme.
DISCUSSION
Modular p65 domain organization and function.
The p65 domain organization suggested by primary sequence resembles the conserved architecture of a La protein. The biological functions of p65 and La are also similar in that both confer stability and direct RNP assembly of primary transcripts of RNA polymerase III. Truncated La proteins containing only the La motif and adjacent RRM retain a near wild-type RNA binding activity (11). Here we show that this is not true of p65. The binding affinity and competition specificity assays above demonstrate that p65 domains make unique contributions to the full-length protein interaction with TER. We propose a model for domain associations in which the p65 N-terminal domain contacts stem I, the La motif contacts the 3′ poly(U) tract, the putative RRM contacts duplex stem IV, and the C-terminal domain contacts central stem IV, including the dinucleotide bulge. This multimodule complexity of p65-TER interactions implies a substantial contact surface between p65 and TER. Such an extensive network of p65-TER interactions could have evolved to promote TER protection from degradation in vivo, consistent with the increase in TER protection from chemical modification in the holoenzyme RNP versus free RNA (17).
Truncation analysis pointed to an unanticipated significance of the p65 C-terminal domain. Of the individual p65 domains and the double-domain combinations, only p65(C) and p65(RC) bound TER with enough affinity and specificity to generate a single mobility shift complex under our EMSA conditions. Binding competition assays revealed that the p65 C-terminal domain provides recognition specificity for the central stem IV bulge. This bulge and its flanking base pairs are universally conserved in all TERs of Tetrahymena species, while stem I and other stem IV sequences are partially divergent (8). Finally, ternary complex assembly assays showed that p65(C) improves the assembly of TERT with TER. These properties of the C-terminal domain suggest that it contributes to p65 biological function by helping to protect TER from degradation and by initiating holoenzyme RNP assembly.
Telomerase holoenzyme assembly.
The studies above add strong support for hierarchical protein-RNA interactions in the endogenous pathway of telomerase enzyme biogenesis. Here we have exploited in vitro assays to investigate the initial steps of Tetrahymena telomerase holoenzyme assembly. We show that the influence of p65 on TERT RNP assembly can be recapitulated by using the p65 C-terminal domain alone, the major TERT RNA binding domain alone, and TER. The p65-enhanced assembly of TERT has a dependence on the TER sequence that extends beyond the requirements for p65-TER and TERT-TER interactions per se. We suggest that p65 binding and local TER conformational change favor a more global change in TER tertiary structure, which in turn favors TERT binding. The p65-induced TER conformation(s) could also influence the highly processive catalytic activity of the Tetrahymena telomerase holoenzyme, which is not reconstituted in rabbit reticulocyte lysate. Because TERT binding affinity for TER is influenced by p65 but not vice versa, we suggest that the pathway of RNP assembly in vitro affects ternary complex structure.
Endogenous telomerase RNP biogenesis pathways in ciliates, yeasts, and vertebrates use different strategies for RNA processing and different sets of holoenzyme proteins for TER binding. The differences in identities of interacting factors may be masking a fundamentally conserved role for architectural TER binding proteins in the early steps of holoenzyme RNP assembly. Divergence in TER sequence and TER binding proteins may have met the demand for new strategies of telomerase regulation, possibly linked to evolutionary changes in nuclear organization. It is curious, however, that a relatively small ciliate telomerase holoenzyme uses a telomerase-specific protein to direct RNP biogenesis, while larger vertebrate and yeast telomerase holoenzymes employ a set of proteins shared with more abundant RNPs. Perhaps telomerase-specific RNP biogenesis factors for the larger holoenzymes remain to be discovered.
Acknowledgments
We thank Collins laboratory members and other colleagues, particularly Phillip Zamore, for experimental discussion and comments on the manuscript.
Funding was provided by a predoctoral fellowship from the National Science Foundation (C.M.O.) and by National Institutes of Health grant GM54198 (K.C.).
REFERENCES
- 1.Aigner, S., J. Lingner, K. J. Goodrich, C. A. Grosshans, A. Shevchenko, M. Mann, and T. R. Cech. 2000. Euplotes telomerase contains an La motif protein produced by apparent translational frameshifting. EMBO J. 19:6230-6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aigner, S., J. Postberg, H. J. Lipps, and T. R. Cech. 2003. The Euplotes La motif protein p43 has properties of a telomerase-specific subunit. Biochemistry 42:5736-5747. [DOI] [PubMed] [Google Scholar]
- 3.Chen, J. L., and C. W. Greider. 2004. Telomerase RNA structure and function: implications for dyskeratosis congenita. Trends Biochem. Sci. 29:183-192. [DOI] [PubMed] [Google Scholar]
- 4.Collins, K., and L. Gandhi. 1998. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc. Natl. Acad. Sci. USA 95:8485-8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins, K., and J. R. Mitchell. 2002. Telomerase in the human organism. Oncogene 21:564-579. [DOI] [PubMed] [Google Scholar]
- 6.Kelleher, C., M. T. Teixeira, K. Forstemann, and J. Lingner. 2002. Telomerase: biochemical considerations for enzyme and substrate. Trends Biochem. Sci. 27:572-579. [DOI] [PubMed] [Google Scholar]
- 7.Lai, C. K., M. C. Miller, and K. Collins. 2003. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol. Cell 11:1673-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCormick-Graham, M., and D. P. Romero. 1995. Ciliate telomerase RNA structural features. Nucleic Acids Res. 23:1091-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell, J. R., J. Cheng, and K. Collins. 1999. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol. 19:567-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Connor, C. M., C. K. Lai, and K. Collins. 2005. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J. Biol. Chem. 280:17533-17539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohndorf, U. M., C. Steegborn, R. Knijff, and P. Sondermann. 2001. Contributions of the individual domains in human La protein to its RNA 3′-end binding activity. J. Biol. Chem. 276:27188-27196. [DOI] [PubMed] [Google Scholar]
- 12.Prathapam, R., K. L. Witkin, C. M. O'Connor, and K. Collins. 2005. A telomerase holoenzyme protein enhances telomerase RNA assembly with telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 12:252-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seto, A. G., A. J. Zaug, S. G. Sobel, S. L. Wolin, and T. R. Cech. 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401:177-180. [DOI] [PubMed] [Google Scholar]
- 14.Weinrich, S. L., R. Pruzan, L. Ma, M. Ouellette, V. M. Tesmer, S. E. Holt, A. G. Bodnar, S. Lichsteiner, N. W. Kim, J. B. Trager, R. D. Taylor, R. Carlos, W. H. Andrews, W. E. Wright, J. W. Shay, C. B. Harley, and G. B. Morin. 1997. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 17:498-502. [DOI] [PubMed] [Google Scholar]
- 15.Witkin, K. L., and K. Collins. 2004. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev. 18:1107-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolin, S. L., and T. Cedervall. 2002. The La protein. Annu. Rev. Biochem. 71:375-403. [DOI] [PubMed] [Google Scholar]
- 17.Zaug, A. J., and T. R. Cech. 1995. Analysis of the structure of Tetrahymena nuclear RNAs in vivo: telomerase RNA, the self-splicing rRNA intron, and U2 snRNA. RNA 1:363-374. [PMC free article] [PubMed] [Google Scholar]