Abstract
The yeast S-phase cyclins Clb5 and Clb6 are closely related proteins that are synthesized late in G1. Although often grouped together with respect to function, Clb5 and Clb6 exhibit differences in their ability to promote S-phase progression. DNA replication is significantly slowed in clb5Δ mutants but not in clb6Δ mutants. We have examined the basis for the differential functions of Clb5 and Clb6 and determined that unlike Clb5, which is stable until mitosis, Clb6 is degraded rapidly at the G1/S border. N-terminal deletions of CLB6 were hyperstabilized, suggesting that the sequences responsible for directing the destruction of Clb6 reside in the N terminus. Clb6 lacks the destruction box motif responsible for the anaphase promoting complex-mediated destruction of Clb5 but contains putative Cdc4 degron motifs in the N terminus. Clb6 was hyperstabilized in cdc34-3 and cdc4-3 mutants at restrictive temperatures and when S/T-P phosphorylation sites in the N terminus were mutated to nonphosphorylatable residues. Efficient degradation of Clb6 requires the activities of both Cdc28 and Pho85. Finally, hyperstabilized Clb6 expressed from the CLB6 promoter rescued the slow S-phase defect exhibited by clb5Δ cells. Taken together, these findings suggest that the SCFCdc4 ubiquitin ligase complex regulates Clb6 turnover and that the functional differences exhibited by Clb5 and Clb6 arise from the distinct mechanisms controlling their stability.
Progression through the eukaryotic cell cycle is mediated by cyclin-dependent kinase (Cdk) complexes. In the budding yeast Saccharomyces cerevisiae, nine cyclins can associate with and activate the kinase Cdc28 (Cdk1). Based on sequence, function, and timing of expression, these cyclins can be grouped into three categories: the G1 cyclins Cln1, Cln2, and Cln3, the S-phase cyclins Clb5 and Clb6, and the mitotic cyclins Clb1, Clb2, Clb3, and Clb4. Cyclin binding is required to activate Cdc28, and cyclins are also thought to confer substrate specificity to the kinase complex. Thus, the expression of different cyclin subgroups at different times throughout the cell division cycle is thought to activate distinct events in each stage of the cycle.
Transition from G1 into S phase is triggered by the S-phase cyclins Clb5 and Clb6 (1, 8, 23, 35). Clb5 and Clb6 are approximately 40% identical at the amino acid sequence level and are transcribed in a pulse in G1 (8, 23, 35). Transcription is controlled by similar regulatory elements in the promoters of both CLB5 and CLB6 (28). Although CLB5 and CLB6 are transcribed in G1, Clb5/Cdc28 (and presumably Clb6/Cdc28) complexes are not activated until the G1/S border when the Clb-specific Cdk inhibitor, Sic1, is degraded by the action of the ubiquitin ligase complex, SCFCdc4 (Skp1/Cullin/F-box) (33, 34, 41, 42).
Clb5 and Clb6 appear to play redundant roles in the initiation of S phase. Either Clb5 or Clb6 can promote entry into S phase on schedule (1, 8, 35). However, Clb5 but not Clb6 is required for timely progression through S phase (8, 35). Further studies showed that clb5Δ cells could not efficiently trigger DNA replication from replication origins that fire late in S phase but that replication initiation from late-firing origins in clb6Δ cells was unperturbed (5).
Phenotypic differences between clb5 and clb6 mutants were also observed in the regulation of spindle pole body duplication (17) and DNA replication (S. B. Haase and S. I. Reed, unpublished data). Moreover, Clb6 but not Clb5 is required to regulate the nuclear localization of the transcription factor Swi6 (12).
The differential phenotypes observed for clb5Δ and clb6Δ mutants could be explained by differences in posttranscriptional regulation, protein function, or both regulation and function. Here, we report that the stability of Clb5 and Clb6 is differentially regulated. Clb5 has been shown to persist throughout S phase and is marked for destruction in mitosis by the action of the ubiquitin ligase complex APCCdc20(anaphase- promoting complex) (20, 21, 36). We present evidence that Clb6 is targeted for destruction at the G1/S border by the activity of a functionally distinct ubiquitin ligase complex, SCFCdc4. We also show that the differential regulation of Clb5 and Clb6 protein stability can account for the differences observed in the ability of these S-phase cyclins to promote timely progression through S phase.
MATERIALS AND METHODS
Plasmid and strain construction.
All strains used in this study are derivatives of BF264-15DU (MATa ade1 his2 leu2-3,112 trp1-1 ura3Δns) (31) except where noted. Relevant genotypes of strains are described in Table 1. CLB6 and CLB5 were tagged at their respective genomic loci by PCR-mediated homologous recombination (oligonucleotides are listed in Table 2) using pFA6KANMX2 (43) as a template. Haploid CLB6-9MYC-KAN cells (SBY464) were mated to haploid CLB5-9MYC-KAN cells (SBY466). Diploids were then sporulated to obtain haploid cells bearing both the CLB5-9MYC and the CLB6-9MYC alleles (SBY468).
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| SBY147 | MATabar1 clb6Δ::ADE1 | S. B. Haase |
| SBY317 | MATabar1 GAL-CLB6Δ135-HA3-LEU2 | This study |
| SBY334 | MATabar1 CLB6-HA-KAN | This study |
| SBY464 | MATabar1 CLB6-9MYC-KAN | This study |
| SBY466 | MATabar1 CLB5-9MYC-KAN | This study |
| SBY468 | MATabar1 CLB6-9MYC-KAN CLB5-9MYC-KAN | This study |
| SBY476 | MATabar1 GAL-CLB6-T39A-S6A-HA3-LEU2 | This study |
| SBY478 | MATabar1 GAL-CLB6Δ100-HA3-LEU2 | This study |
| SBY502 | MATabar1 GAL-CLB6-S6A-HA3-LEU2 | This study |
| SBY506 | MATabar1 GAL-CLB6-T39A-HA3-LEU2 | This study |
| SBY510 | MATabar1 GAL-CLB6-HA3-LEU2 | This study |
| SBY518 | MATabar1 GAL-CLB6Δ41-HA3-LEU2 | This study |
| SBY523 | MATabar1 GAL-CLB6-HA3-LEU2 apc2Δ::KAN apc2-4 | This study |
| SBY525 | MATabar1 GAL-CLB6-T39A-S6A-HA3-LEU2 apc2Δ::KAN apc2-4 | This study |
| SBY557 | MATabar1 CLB6-HA3-URA3 | This study |
| SBY573 | MATabar1 GAL-CLB6Δ3P-HA3-LEU2 | This study |
| SBY577 | MATabar1 GAL-CLB6-S147A-HA3-LEU2 | This study |
| SBY597 | MATabar1 CLB6Δ3P-HA3-URA3 | This study |
| SBY603 | MATaclb5Δ::ARG CLB6Δ3P-HA3-URA3 | This study |
| SBY605 | MATaclb5Δ::ARG CLB6-HA3-URA3 | This study |
| SBY648 | MATabar1 cdc4-3 sic1Δ::KAN GAL-CLB6Δ3P-HA3-LEU2 | This study |
| SBY653 | MATabar1 cdc34-3 sic1Δ::KAN GAL-CLB6-HA3-LEU2 | This study |
| SBY654a | MATabar1 cdc28Δ::cdc28-as1 GAL-CLB6-HA3-LEU2 | This study |
| SBY696 | MATabar1 GAL-CLB6-HA3-LEU2 pho85Δ::KAN | This study |
| SBY702 | MATabar1 GAL-CLB6-HA3-LEU2 sic1Δ::KAN | This study |
| SBY715 | MATabar1 cdc4-3 sic1Δ::KAN GAL-CLB6-HA3-LEU2 | This study |
| SBY722 | MATabar1 GAL-CLB6-HA3-LEU2 pdr5Δ::KAN | This study |
| SBY744 | MATabar1 cdc28Δ::cdc28-as1 | This study |
| SBY752 | MATabar1 GAL-CLB6-HA3-LEU2 pho85Δ::KAN cdc28Δ::cdc28-as1 | This study |
In W303 (ura3-1 trp1-1 leu2-3,112 his3-1 ade2-1 can1-100) background.
TABLE 2.
Oligoucleotides used in this study
| Purpose | Oligonucleotide | Sequence |
|---|---|---|
| Sequencing | CLB6 seq2 | CACTGCTTACCTGAAACATT |
| CLB6 seq2.5 | TTCGATAGAGATGGATGATCC | |
| CLB6 seq3 | CCCCCTAAATTTCATTAGGA | |
| CLB6revseq1 | GATAGGGAGATTGCGTGTCC | |
| CLB6 5′ flank | CTCTGATATTCTCTCCCTCC | |
| CLB6 3′ flank | TGATATTTAAGATGCAGGGGG | |
| Deletion | 5′ CLB6Δ135 | AGATCTATGTCCCTACCGACACATAACTATTTA |
| 5′ CLB6Δ41 | GGATCCAAAATGTCTACGAATGAAAAAAAAGTTCTATCC | |
| 5′ CLB6Δ100 | AGATCTATGCATCAATGGAAAAATTTGGAT | |
| 5′ SIC1_KAN | ATTTTGACCCTTGAAGCAGGGACTATTACACGAAAGCTTGCCTCGTCCCC | |
| 3′ SIC1_KAN | TATAATCGTTCCAGAAACTTTTTTTTTTCATTTCTGACACTGGATGGCGGC | |
| 5′ PDR5_KAN | TTCGTATCCGCTCGTTCGAAAGACTTTAGACAAAAGCTTGCCTCGTCCCC | |
| 3′ PDR5_KAN | TAAGTTTCTTTTCTTAACCAAATTCAAAATTCTAGACAGTGGATGGCGGC | |
| 5′ pho85_KAN | TAGCGCGGCAAACTGGGCAAACTTGAGCAATACCAGCTTGCCTCGTCCCC | |
| 3′ pho85_KAN | ATACATGGCTACGGTTTTTCGCTGACGGGCTGCGGACACTGGATGGCGGC | |
| Phosphorylation | 5′ CLB6S6A | AGATCTATGAATTGTATCCCTGCTCCAATTAGTGAAAGGAAA |
| site mutation | CLB6HA 3′ BglII | AGATCTTCAGCGGCCGCACTGAGCAGCGT |
| 5′ CLB6T39A | GAGAAAGTTCAATTAACTTGGCACCTCACTCTACGAATG | |
| 3′ CLB6T39A | CATTCGTAGAGTGAGGTGCCAAGTTAATTGAACTTTCTC | |
| 5′ CLB6S147A | CTATTTAATGGACACGCAAGCTCCCTATCATTTGAAAAGC | |
| 3′ CLB6S147A | GCTTTTCAAATGATAGGGACGTTGCGTGTCCATTAAATAG | |
| Integration | 5′ CLB6_replace | ATATTCTCTCCCTCCTTTTAAATTTTTAAAATGAATTGTATCCCTAGTCC |
| 5′ CLB6S6A_replace | ATATTCTCTCCCTCCTTTTAAATTTTTAAAATGAATTGTATCCCTGCTCC | |
| 3′ CLB6flank_3′ URA3 | CAGGGGGTTAGCTGGCTATAATTTTGATCTATGTTTTAGTTTTGCTGGCCG | |
| Tagging | CLB6MYCKAN5′ | ATGGTTTATTTCAAGGTTTTTGACTGGTGTAAACAAAAACGTGTCGACGGTGAACAAAAG |
| CLB6MYCKAN3′ | TAAGATGCAGGGGGTTAGCTGGCTATAATTTTGATCTATGTTGCTTGCCTCGTCCCCGCC | |
| CLB5MYCKAN5′ | TCCGAAATGCATAGCAACTTTCAAAATCTATTTAATCTTAAGGTCGACGGTGAACAAAAG | |
| CLB5MYCKAN3′ | ATGTAAAGAGTATGCGAATTCATGAGCATTACTAGTACTAATGCTTGCCTCGTCCCCGCC |
CLB6-HA3 was PCR amplified from the genome of SBY334, cloned into the pCRII vector (Invitrogen), and sequenced. CLB6-HA3 was removed from PCRII by digestion with BamHI and ligated into the integrating GAL1 expression vector YIpG2 (40). CLB6 N-terminal deletions were constructed by PCR amplification using pCRII-CLB6-HA3 as a template and oligonucleotides described in Table 2. PCR products were ligated into pCRII, sequenced, and then subcloned into YIpG2 as described above. Phosphorylation site mutations were generated by PCR-based site-directed mutagenesis of YIpG2-CLB6-HA3 using primers described in Table 2. PCR products were then ligated into pDRIVE (QIAGEN) and sequenced before being subcloned into YIpG2 on 5′ BamHI and 3′ BglII ends. All strains containing GAL-CLB6-HA3 and its derivatives were constructed by EcoRV digestion of the YIpG2-based plasmids and integration by homologous recombination at the LEU2 locus.
pCRII-CLB6-HA3-URA3 was constructed by cloning the URA3 open reading frame from pUCHURA3 into pCRII-CLB6-HA3 on HindIII ends. pUCHURA3 was constructed by PCR amplification of URA3 from yeast genomic DNA and ligation into the HindIII site of pUCH (18). PCR-mediated gene replacement was used to replace the CLB6 open reading frame with mutated CLB6 alleles. Oligonucleotides described in Table 2 were used to amplify CLB6-HA3-URA3 and its derivatives from pCRII-URA3 vectors. The resultant PCR products were integrated into yeast cells by homologous recombination. Mutations were verified by sequencing PCR fragments generated from genomic DNA. All gene deletions were made by PCR-mediated gene replacement using oligonucleotides listed in Table 2. The kanamycin resistance gene was amplified from pFA6KANMX2 with oligonucleotides complementary to sequence flanking the gene being replaced on their 3′ ends. PCR products were then integrated into yeast cells by homologous recombination, and transformants were selected by screening for kanamycin resistance.
Cell growth and arrest conditions.
Yeast cultures were grown in YEP medium (1% yeast extract, 2% Bacto peptone, 0.001% adenine, 0.001% uracil) supplemented with 2% sugar (dextrose, sucrose, galactose, or raffinose). Cells were grown at 30°C except as noted. Temperature-sensitive mutants were grown at 25°C and shifted to 34°C for arrest, except where otherwise noted. Mating pheromone synchrony experiments were performed by adding alpha factor (α-factor) (2 μg/ml α-factor for BAR1 strains and 40 to 60 ng/ml for bar1 strains) to the growth medium for 2 h. Synchronous populations were released from the arrest into medium without α-factor.
Protein stability assays.
GAL1-driven expression of all CLB6 alleles was induced by adding 10% galactose solution to mid-log-phase cultures in YEP-sucrose or YEP-raffinose to a final concentration of 2% galactose or by harvesting cells by centrifugation and resuspending them in YEP-galactose (except as noted). Galactose concentrations and pulse durations were adjusted as necessary to achieve protein expression levels similar to that of endogenous Clb6 at the G1/S border. In order to terminate transcription and translation, cells were harvested and resuspended in equal volumes of YEP-dextrose medium containing 1 mg/ml cycloheximide at t = 0 min. Proteins were extracted from whole-cell lysates, and results were analyzed by Western blotting as described previously (17).
Phosphatase assay.
Clb6-HA expression was induced in YEP-galactose medium for 1.5 h. Whole-cell lysates were prepared as described above and then treated with phosphatase. Phosphatase reaction mixtures were assembled as recommended by the vendor and either left on ice for 45 min or incubated at 30°C for 45 min. Where indicated, 400 U of lambda phosphatase was used per reaction, and 10 mM sodium orthovanadate and 50 mM sodium fluoride were used to inhibit phosphatase activity.
Flow cytometry.
Flow cytometric analysis of DNA content was performed as described previously (16). The percentage of cells in S phase was determined by first fitting normal curves to the 1C and 2C peaks and integrating under these curves to quantitate the percentage of cells in G1 and G2/M phases. The remaining cells were judged to be in S phase.
RESULTS
Stability of Clb5 and Clb6 is differentially regulated through the cell cycle.
In order to understand why clb5Δ and clb6Δ mutants exhibit distinct phenotypes, we began by examining the expression of the two proteins in a synchronous population of cells as they traversed the cell cycle.
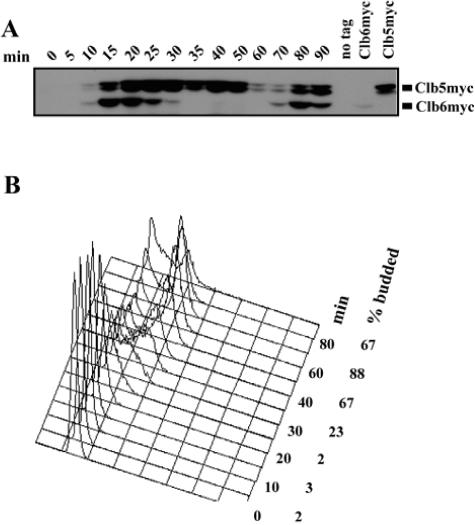
Both Clb5 and Clb6 were fused at the C terminus with a nine-myc tag at their respective genomic loci. CLB5-9MYC CLB6-9MYC cells were synchronized in G1 by treatment with the mating pheromone α-factor and then released into the cell cycle. Clb5 and Clb6 protein levels were then monitored at several time points by Western blotting (Fig. 1A). Cell cycle progression was assessed by monitoring budding index and DNA content (Fig. 1B).
FIG. 1.
Abundance of Clb5 and Clb6 during the cell cycle. (A) Clb5 and Clb6 protein levels over time in a synchronous population of cells released from α-factor arrest. (B) Budding index and flow cytometric analysis of DNA content for the same time points. Times indicated are minutes after release from α-factor.
As expected, Clb5 appeared in G1 cells upon release from α-factor arrest and persisted until 60 min after release, when cells entered mitosis (Fig. 1A and B). Clb6 also appeared in G1 but disappeared rapidly between 20 min and 30 min after release (Fig. 1A). Analysis of budding index and DNA content (Fig. 1B) indicates that Clb6 disappears near the G1/S transition. Because CLB5 and CLB6 are transcribed simultaneously in G1, these observations suggest that the turnover of Clb5 and Clb6 is regulated differently.
The N terminus of Clb6 confers instability on the protein.
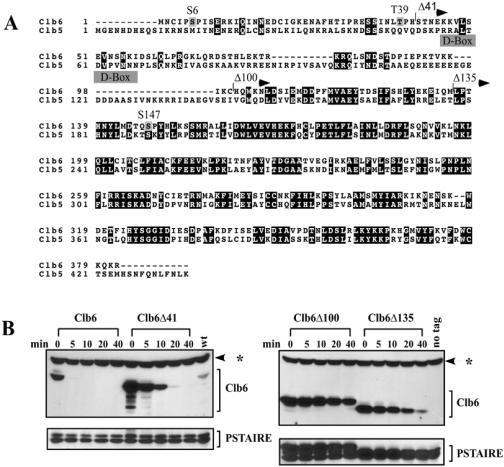
Clb5 and Clb6 are closely related at the protein sequence level. The proteins are approximately 40% identical throughout their lengths but show the highest degree of homology and identity in their C-terminal regions (Fig. 2A). The degron sequence motif recognized by the APCCdc20 (RRALTDVPVN) has been mapped to the N-terminal region of Clb5 (20, 21, 36). Although Clb6 and Clb5 share some homology in this region, a critical amino acid change in the degron sequence (RXXL to KXXL) (Fig. 2A) is likely to prevent recognition of Clb6 by the APC. Since degradation signals for Clb5 are located in the N terminus and Clb5 and Clb6 protein sequences are the most divergent in this region, we reasoned that degradation signals specific for Clb6 might also reside in its N-terminal region.
FIG. 2.
N-terminal deletions hyperstabilize Clb6. (A) ClustalW alignment of Clb5 and Clb6. Black boxes indicate identities. Gray boxes indicate putative phosphorylation sites (S/T-P) in Clb6. Clb5 D-box is shown. Arrows demarcate the start of Clb6 N-terminal deletions. (B) The relative stability of HA-tagged Clb6 N-terminal deletions. CLB6 transcription was induced from the GAL1 promoter by treating asynchronous cultures (in YEP-raffinose) with 0.1% galactose for 25 min. Cells were then harvested, and at t = 0 min, cells were resuspended in YEP-dextrose containing 1 mg/ml cycloheximide. Wild type (wt), peak levels of endogenous Clb6 (cells collected from synchronous population expressing CLB6-HA from its own promoter, 15 min after release from α-factor); no tag, cell lysates from cells expressing Clb6 without an HA epitope tag; PSTAIRE, loading control. The asterisk indicates an HA background band.
In order to investigate this hypothesis, we made three N-terminal deletions of various lengths and examined their stability by carrying out a promoter shutoff experiment (Fig. 2B). N-terminal deletions of CLB6 were fused at the C terminus to a triple hemagglutinin (HA) tag and then expressed from the inducible GAL1 promoter in asynchronous wild-type cells. Transcription was terminated by the addition of dextrose to the medium, and translation was blocked by the addition of cycloheximide. Cell populations were collected at several time points following shutoff and then examined for Clb6 protein levels (Fig. 2B).
Wild-type Clb6 appears to be very unstable in asynchronous cells, exhibiting a half-life of less than 5 min. The Clb6Δ41 deletion is significantly stabilized compared to wild-type Clb6, indicating that sequences important for degradation are contained in the first 41 amino acids. However, Clb6Δ100 and Clb6Δ135 appear to be more stable than Clb6Δ41, suggesting that additional degron sequences exist downstream. We have shown that expression of Clb6Δ100 is capable of promoting S phase in Δclb1,2,3,4,5,6 cells, indicating that Clb6Δ100 can still bind Cdc28 and activate DNA replication (data not shown) and suggesting that hyperstabilization is not the indirect result of a failure to form normal cyclin/Cdk complexes.
Clb6 is hyperstabilized in SCF mutants and is ubiquitinated.
Examination of the N terminus of Clb6 revealed sequences similar to SCFCdc4 phospho-degron motifs found in the Cdk inhibitor Sic1 (29), including three potential Cdk phosphorylation sites (no SP or TP sequences are found in Clb5). Moreover, like Sic1 (34), Clb6 is destroyed at the G1/S transition. Taken together, these observations suggest that SCF complexes may regulate Clb6 stability. Therefore, we tested the stability of Clb6 in cells bearing temperature-sensitive mutations in SCF components.
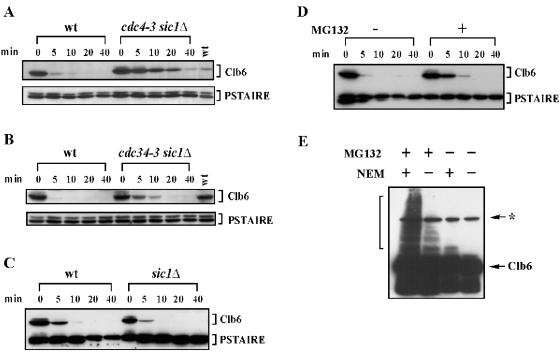
Using the GAL1 promoter shutoff protocol described in the legend to Fig. 2, we found that Clb6 is significantly stabilized in cdc34-3 sic1Δ and cdc4-3 sic1Δ cells at restrictive temperatures, compared to that in wild-type cells at the same temperature (Fig. 3A and B). These findings suggest that SCFCdc4 complexes regulate the turnover of Clb6.
FIG. 3.
Clb6p hyperstabilized in cdc4-3 and cdc34-3 mutant cells at restrictive temperatures and in the presence of a proteosome inhibitor. (A and B) The relative stability of HA-tagged Clb6 in (A) wild-type (wt) cells versus cdc4-3 sic1Δ mutants at 33°C and (B) wild-type cells versus cdc34-3 sic1Δ mutants at 35°C. (C) The relative stability of HA-tagged Clb6 in wild-type cells versus sic1Δ cells. The promoter shutoff experiments whose results are shown in panels A and B were performed as described in the legend to Fig. 2 except that cultures were shifted to restrictive temperatures for 30 min and then treated with galactose at restrictive temperatures for 30 min as follows: wt/CLB6-HA cells, 0.2% galactose; cdc4-3 sic1Δ/CLB6-HA cells, 0.8% galactose; and cdc34-3 sic1Δ/CLB6 cells, 1% galactose. In panel C, cells were harvested from YEP-sucrose and grown in YEP-galactose for 30 min. Times indicated are minutes after resuspension in YEP-dextrose containing cycloheximide. wt, peak levels of endogenous Clb6. (D) Stability of HA-tagged Clb6 in cells treated or untreated with MG132. Mid-log-phase cultures in YEP-sucrose were treated with 50 μM MG132 in dimethyl sulfoxide (+) or dimethyl sulfoxide alone (−) and then grown at 30°C for 1 h. Galactose (0.5%) was then added to induce Clb6 expression from the GAL1 promoter for 30 min. Times indicated are minutes after resuspension in YEP-dextrose containing cycloheximide. (E) Slow-mobility forms of Clb6 appear in the presence of the proteasome inhibitor MG132 and the deubiquitinase inhibitor NEM. Cells expressing an HA-tagged Clb6 from the GAL1 promoter were untreated (−) or treated (+) with 100 μM MG132 in YEP-sucrose for 45 min. Clb6-HA expression was then induced by the addition of galactose for 45 min. Total protein was then isolated in the presence (+) or absence (−) of NEM. (D and E) Strains had the gene encoding the ABC transporter Pdr5 deleted to decrease the rate at which MG132 was pumped out of cells. PSTAIRE, loading control. The asterisk indicates an HA background band.
We examined Clb6 stability in SCF mutants in which the SIC1 gene was deleted because cdc34-3 and cdc4-3 cells arrest at the restrictive temperature at the G1/S border due to hyperstabilization of Sic1 (27, 42). Thus, in the presence of the SIC1 gene, any observed stabilization of Clb6 in these mutants is not necessarily due to direct stabilization of Clb6 but could reflect an indirect stabilization of Clb6, resulting from an arrest in a specific cell cycle stage or from binding of Sic1 to Clb6/Cdc28 complexes. By deleting SIC1 from these strains, we eliminated the possibility that any observed stabilization in these SCF mutants is an indirect effect of Sic1 stabilization. We have also observed that the deletion of SIC1 (sic1Δ) in otherwise wild-type cells does not affect the stability of Clb6 (Fig. 3C), indicating that the hyperstabilization of Clb6 in cdc4-3 sic1Δ and cdc34-3 sic1Δ mutants is not an indirect effect of the loss of Sic1.
Clb6 does not appear to be fully stabilized in cdc34-3 sic1Δ mutants (compare Fig. 3B to Fig. 2B), which might suggest that other mechanisms are required for efficient Clb6 degradation. However, we have observed that cdc34-3 SIC1 cells can enter S phase (data not shown), suggesting that the cdc34-3 allele is partially functional even at 35°C. Furthermore, we have determined that Clb6 is significantly less stable at higher temperatures, even in wild-type cells (see Fig. S1 in the supplemental material). Thus, the general instability of Clb6 at high temperatures is likely to offset the stabilizing effects of the SCFts mutants. Consistent with these observations, the stability of Clb6 in cdc4-3 sic1Δ cells at 33°C (a partially restrictive temperature for the cdc4-3 allele) is comparable to that in the N-terminal deletions (compare Fig. 3A to Fig. 2B).
If Clb6 is a direct target of SCFcdc4, then it should be ubiquitinated and destroyed by the proteasome. Thus, inhibition of the proteasome should stabilize Clb6. Using the GAL1 promoter shutoff protocol described above, we found that HA-tagged Clb6 is stabilized in cells treated with the proteasome inhibitor MG132 (Fig. 3D). Ubiquitinated proteins are highly unstable and normally appear as a “ladder” of slow-mobility species on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. A prominent ladder of slower-mobility Clb6 species, indicative of ubiquitination, was observed on long exposures of Western blots in extracts from cells treated with MG132 (Fig. 3E). Moreover, the intensity of slower-mobility bands was also increased in extracts that were treated with the deubiquitinase inhibitor N-ethylmaleimide (NEM). Slower-mobility forms were not observed in untreated cells (Fig. 3E).
Clb6 is a phospho-protein, and mutation of putative phosphorylation sites leads to hyperstabilization.
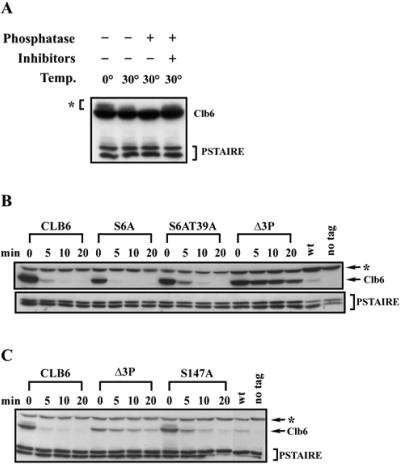
The SCF complex normally recognizes phosphorylated substrates (37, 44). Thus, we first determined whether Clb6 is a phospho-protein. Slower-mobility forms of Clb6 were observed under a number of conditions (see Fig. 2 and 3). In order to determine whether the slower-mobility forms of Clb6 represented phosphorylated species, we treated cell lysates with lambda phosphatase. Slow-mobility forms of Clb6 disappeared in lysates treated with phosphatase but remained in untreated lysates and in lysates containing phosphatase and phosphatase inhibitors (Fig. 4A).
FIG. 4.
Clb6 is a phospho-protein and is hyperstabilized when putative phosphorylation sites are mutated to nonphosphorylatable residues. (A) Multiple species of Clb6 are observed in whole-cell lysates. Lysates were untreated (−) or treated (+) with lambda phosphatase (phosphatase) in the presence (+) or absence (−) of sodium fluoride and of sodium orthovanadate (inhibitors). The asterisk indicates slow-mobility bands. (B and C) The relative stability of wild-type Clb6 and Clb6 proteins bearing phosphorylation site mutations. The experiment was carried out as described in the legend to Fig. 2 except that cultures were treated with 0.05% galactose for 20 min. Clb6 (wt) and the S6A, S6AT39A, and Δ3P mutants are shown (B). Clb6 (wt) and the Δ3P and S147A mutants are shown (C). Times indicated are minutes after resuspending cells in YEP-dextrose containing cycloheximide. wt, peak levels of endogenous Clb6; no tag, cell lysates from cells expressing Clb6 without an HA epitope tag; PSTAIRE, loading control. The asterisk in panels B and C indicates an HA background band.
SCF targets are often phosphorylated by proline-directed serine/threonine kinases. We mutated the serines or threonines of the three (S/T)P sites in Clb6 to the nonphosphorylatable residue alanine and then tested the stability of Clb6 alleles bearing individual or multiple phosphorylation site mutations using a GAL1 promoter shutoff assay (Fig. 4B and C). Mutation of either S6 to A (S6A) (Fig. 4B) or T39 to A (T39A) (data not shown) had no significant effect on the stability of Clb6. However, the allele bearing the S147A mutation was significantly stabilized (Fig. 4C). The S6A T39A double mutant was also significantly stabilized (Fig. 4B). Clb6 mutated at all three (S/T)P sites (Clb6Δ3P) was the most stable, exhibiting a half-life comparable to that of the N-terminal deletions (Fig. 4B and C). These results suggest that all three putative phosphorylation sites contribute to the instability of Clb6. SCF-regulated proteins, such as Sic1 in budding yeast (29) and cyclin E in human cells (39, 46), exhibit similar behaviors when putative phosphorylation sites are mutated. In both cases, mutation of a single phosphorylation site has some marginal stabilizing effect, but mutation of multiple sites is required for maximum stabilization.
Both Cdc28 and Pho85 contribute to the destruction of Clb6.
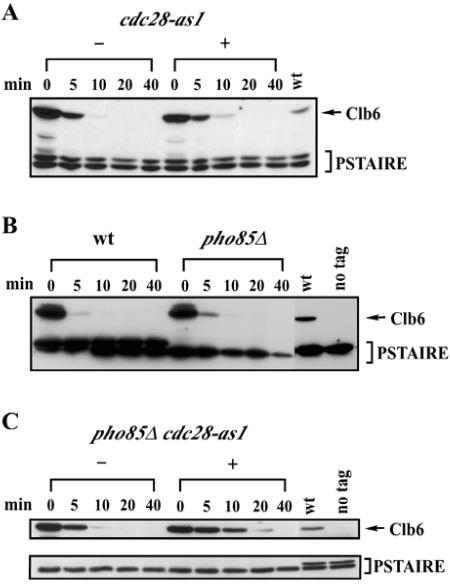
Many SCF targets in both yeast and mammalian cells are phosphorylated by cyclin/Cdk complexes (19, 24, 37, 44, 46) and are hyperstabilized when cyclin/Cdk activities are inhibited (46). Therefore, we asked whether inhibition of cyclin/Cdk activity would stabilize Clb6. Using the GAL1 promoter shutoff protocol described above, we examined Clb6 stability in cells bearing a cdc28as-1 allele that can be inhibited by the ATP analogue 1-NM-PP1 (Fig. 5A) (2).
FIG. 5.
Both Cdc28 and Pho85 regulate the stability of Clb6. (A) Stability of Clb6-HA in cells expressing the analogue-sensitive allele cdc28-as1. Log-phase asynchronous cultures in YEP-sucrose were harvested, and Clb6-HA expression was induced in YEP-galactose medium for 45 min. Cultures were then either untreated (−) or treated (+) with 5 μM 1-NM-PP1 for 30 min. Times indicated are minutes after terminating expression by resuspending cells in YEP-dextrose containing cycloheximide. (B) Stability of Clb6-HA in cells with the PHO85 gene deleted. Cultures were treated as described in the legend to Fig. 2. (C) Stability of Clb6-HA in cdc28-as1 pho85Δ cells. cdc28-as1 pho85Δ cultures were either untreated (−) or treated (+) with 5 μM 1-NM-PP1 as described for panel A. wt, wild type; PSTAIRE, loading control.
We found that Clb6 was partially stabilized in cdc28as-1 cells, even in the absence of 1-NM-PP1 (compare Fig. 5A to Fig. 5B). The cdc28as-1 allele is known to be hypomorphic, and the reduction in Cdc28 kinase activity may have contributed to the increased stability of Clb6 even in the absence of analogue. However, Clb6 was only marginally more stable in cdc28as-1 cells treated with 1-NM-PP1 than that in untreated cells (Fig. 5A). We confirmed that this dose of 1-NM-PP1 rapidly arrested the cell cycle (data not shown) as previously reported (2). Moreover, Clb6Δ3P was significantly more stable than wild-type Clb6 in cdc28as-1 cells treated with 1-NM-PP1 (data not shown), confirming that these putative phosphorylation sites are important for the degradation of Clb6. Taken together, these results suggest that an additional kinase is required for efficient degradation of Clb6.
Efficient degradation of the SCFcdc4 target, Sic1, requires the combined action of Cdc28 and Pho85 kinases (30). We therefore examined the stability of Clb6 in cells with the PHO85 gene deleted, both alone (Fig. 5B) and in combination with the cdc28as-1 allele. Again, Clb6 was only partially stabilized in pho85Δ cells (Fig. 5B) and in pho85Δ cdc28as-1 double mutants in the absence of 1-NM-PP1 (Fig. 5C). However, Clb6 was significantly hyperstabilized in pho85Δ cdc28as-1 double mutants in the presence of 1-NM-PP1 (Fig. 5C). These results suggest that Pho85 and Cdc28 have redundant functions in the regulation of Clb6 stability.
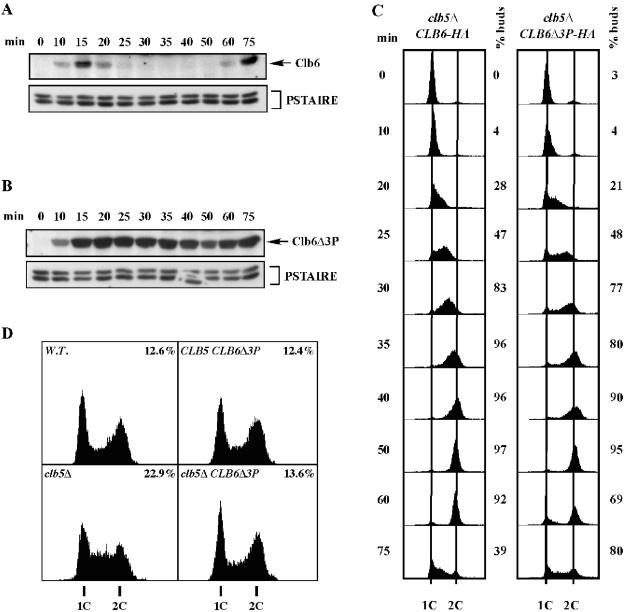
Clb6Δ3P expressed from the endogenous CLB6 promoter is not destroyed at the G1/S border and suppresses the S-phase defect of clb5Δ cells.
Having established that mutation of putative phosphorylation sites stabilizes Clb6, we examined whether Clb6Δ3P was destroyed at the G1/S border. We replaced the endogenous wild-type copy of CLB6 with an HA-tagged CLB6Δ3P allele in a strain with the CLB5 gene deleted (clb5Δ). clb5Δ CLB6Δ3P cells were arrested in G1 with α-factor. The synchronized cell population was then released into the cell cycle and sampled at various time points after release for the analysis of both Clb6 protein levels and DNA content (Fig. 6B and C). Results were compared to those of clb5Δ cells bearing a wild-type allele of CLB6 tagged with HA (Fig. 6A and C). In the clb5Δ CLB6-HA control strain, Clb6 protein appeared in late G1, disappeared at the G1/S border, and reappeared in G1 of the next cell cycle (Fig. 6A). Clb6Δ3P also appeared in late G1, but Clb6Δ3P levels persisted beyond the G1/S border, remaining high throughout S phase (Fig. 6B). Thus, mutation of the phosphorylation sites bypassed the controls that normally trigger the destruction of Clb6 at the G1/S border.
FIG. 6.
Clb6Δ3P expressed from the endogenous CLB6 promoter is hyperstable and suppresses the S-phase defect of clb5Δ cells. Wild-type Clb6 at its genomic locus was replaced by Clb6-HA or Clb6Δ3P-HA in clb5Δ cells. Clb6-HA (A) and Clb6Δ3P-HA (B) protein levels over time in a synchronous population of cells released from α-factor arrest. Times indicated are minutes after release from alpha factor. (C) Flow cytometric analysis of DNA content and budding index for the experiments whose results are shown in panels A and B. (D) Flow cytometric analysis of asynchronous, log-phase cultures. The percentage of cells in S phase is shown for each histogram. PSTAIRE, loading control.
Although Clb6Δ3P persisted throughout the time course, protein levels dropped somewhat around the time of mitosis (Fig. 6B). From this experiment, we could not determine whether the decrease in Clb6Δ3P levels was cell cycle regulated (e.g., an APC-mediated event). Therefore, we performed a similar G1 arrest/release experiment but released clb5Δ CLB6Δ3P cells into medium containing nocodazole in order to arrest cells before anaphase and prevent any APC-mediated destruction of Clb6Δ3P. We found that Clb6Δ3P levels declined in a continuous manner in nocodazole-arrested cells (see Fig. S2 in the supplemental material), suggesting that the decrease in Clb6Δ3P levels observed in cycling cells (Fig. 6B) is not due to a regulated mitotic destruction mechanism.
As mentioned above, cells with the CLB5 gene deleted normally progress slowly through S phase compared to wild-type or clb6Δ cells. Because Clb6Δ3P persisted throughout S phase, we were able to determine whether the inability of Clb6 to promote normal S-phase kinetics on its own was related to its destruction at the G1/S transition. S-phase kinetics of clb5Δ cells bearing wild-type CLB6 or the CLB6Δ3P allele in the synchrony/release experiments shown in Fig. 6A and B were analyzed by flow cytometry (Fig. 6C).
A synchronous population of clb5Δ cells bearing wild-type CLB6 moved slowly through S phase (Fig. 6C). Cells entered S phase on schedule (coincident with bud emergence), approximately 20 min after release from the α-factor arrest. Cells then progressed through the middle of S phase slowly, with DNA contents intermediate between 1C and 2C at the 25-min and 30-min time points. The majority of the population reached 2C content of DNA between 35 min and 40 min.
clb5Δ CLB6Δ3P cells entered S phase with similar kinetics, indicating that the phosphorylation site mutation is competent for initiating S phase on schedule. However, the clb5Δ CLB6Δ3P mutants completed S phase significantly faster than clb5Δ CLB6 cells. Though clb5Δ CLB6Δ3P cells appeared to release from the G1 arrest somewhat less synchronously than the clb5Δ CLB6 cells, the bulk of the population had nearly completed S phase by 30 min. Furthermore, a significant fraction of clb5Δ CLB6Δ3P cells completed mitosis and reached G1 (1C DNA content and unbudded) again at 60 min, while very few clb5Δ CLB6 cells reappeared in G1 until 75 min. These results suggest that the inability of Clb6 to promote normal S-phase kinetics in the absence of Clb5 is due to its destruction at the G1/S border.
When Clb6Δ3P was expressed from the CLB6 promoter in wild-type cells, we observed no increase in the speed of S phase (data not shown). Thus, the rescue of the slow S-phase defect observed in clb5Δ mutants is likely to be a suppression of the defect in the clb5Δ mutant rather than a general increase in S-phase velocity.
We also examined S-phase kinetics in asynchronous cell populations by flow cytometric analysis of DNA content (Fig. 6D). In all strains shown, Clb6 and Clb6 mutants are epitope tagged with HA. As shown previously (8, 23, 35), a significant increase in the population of S-phase cells is observed in clb5Δ cells compared to that in wild-type cells, indicative of a slow S phase. The cell cycle profile of an asynchronous population of clb5Δ CLB6Δ3P cells (13.6% S-phase cells) looks similar to that of the wild-type cells (12.6% S-phase cells) and has noticeably fewer cells in S phase than clb5Δ CLB6 cells (22.9% S-phase cells). These results are consistent with the finding in synchronous populations (Fig. 6C) that Clb6Δ3P can suppress the slow S-phase defect of clb5Δ cells. Furthermore, asynchronous wild-type cells expressing Clb6Δ3P exhibit a cell cycle distribution (12.4% S-phase) similar to that of cells expressing wild-type CLB6 (Fig. 6D), indicating that Clb6Δ3P does not accelerate cells through S phase in wild-type cells.
The decrease in the S-phase fraction observed for asynchronous clb5Δ CLB6Δ3P cells compared to that for clb5Δ CLB6 cells (Fig. 6D) does not necessarily reflect a more rapid progression through S phase. A similar cell cycle profile could result from extending the time required to traverse both G1 and G2/M phases. However, no evidence for slow progression through G1 or S phase was observed in synchronized clb5Δ CLB6Δ3P cell populations (Fig. 6C). In fact, clb5Δ CLB6Δ3P cells entered S phase at the same time and returned to G1 and S phase on the second cycle more rapidly than did clb5Δ CLB6 cells (Fig. 6C), indicating no delay in progression through G1 or G2/M.
DISCUSSION
The stability of Clb5 and Clb6 is regulated by distinct mechanisms.
Although Clb5 and Clb6 are often grouped together with respect to function, previous findings suggest that the S-phase cyclins are in fact functionally distinct (1, 5, 17, 35). We set out to determine whether the phenotypic differences observed for clb5Δ and clb6Δ mutants are related to differential posttranscriptional regulation of the two proteins. Our findings indicate that unlike Clb5, which persists until anaphase, Clb6 is destroyed at the G1/S border via N-terminal motifs by the SCFCdc4 complex. Three putative Cdk phosphorylation sites in Clb6 contribute to its instability, and both Cdc28 and the related kinase Pho85 promote the degradation of Clb6. The observation that Clb6 is not quite as stable in pho85Δ cdc28as-1 double mutants (see Fig. 5C) as in the triple phosphorylation site mutant (see Fig. 5C and Fig. 4B and C) indicates that an additional kinase(s) may play a minor redundant role in the phosphorylation of Clb6.
The differential regulation of Clb5 and Clb6 stability appears to have arisen from the acquisition of SCF degron sequences in Clb6, as well as a mutation at a critical residue in the destruction box (D-box) (RXXL to KXXL). However, in Drosophila melanogaster, the Pim protein apparently has a functional D-box with a KXXL sequence (25), suggesting that the D-box in Clb6 could still be functional. Furthermore, the stability of the S-phase cyclin in Schizosaccharomyces pombe, Cig2, appears to be controlled collaboratively by the SCF and APC ligase complexes (47). We found that Clb6 was not measurably stabilized in cells treated with nocodazole or in cells bearing a temperature-sensitive allele of APC2 (see Fig. S2 in the supplemental material). Taken together, these findings suggest that the APC plays a very minor role, if any, in regulating the stability of Clb6.
Differential regulation of S-phase cyclins and the progression of S phase.
Entry into S phase is triggered by the destruction of Clb-specific Cdk inhibitor Sic1 at the G1/S border (29, 33, 41, 42). Our findings indicate that Clb6 is also destroyed very close to the G1/S border. Because Clb6 can promote timely S-phase entry in the absence of Clb5, active Clb6/Cdc28 complexes must be present at G1/S, but such complexes are likely to exist for only a brief period.
Although Clb6-associated kinase is present for only a very short period at the G1/S border, the conservation of Clb6 among the Saccharomyces species (see below) suggests an important function. Clb6 has been shown to be involved in the regulation of Swi6 localization (12), although this regulation does not appear to be critical for cell cycle progression. It has also been reported that clb6Δ mutants actually grow and divide faster than wild-type cells (1), which would suggest that cells bearing a wild-type allele of CLB6 should be at a selective disadvantage. For now, the conserved function of Clb6 remains a mystery.
Although either Clb5 or Clb6 can trigger the timely entry into S phase, Clb6 cannot promote normal progression through S phase without Clb5. The basis for the S-phase defect in clb5Δ cells is not yet fully understood. Nonetheless, a defect in late origin firing in clb5Δ cells has been reported (5) and could account for the significant slowing of DNA replication in clb5Δ mutants. The fact that Clb6 is normally destroyed at the G1/S border suggests that this defect is simply due to the lack of Clb6 availability late in S phase. Consistent with this hypothesis, Clb/Cdc28-dependent Cdc45 loading at late-firing origins occurs late in S phase (48, 49). Moreover, overexpression of Clb6 both promotes normal S-phase kinetics and restores the firing of late origins in clb5Δ mutants (13), suggesting that slow progression of cells through S phase in these cells is in fact linked to the defect in late origin firing.
Like Clb5 and Clb6, the related mitotic cyclins Clb1 and Clb2 also appear to have evolved divergent functions. Clb2 but not Clb1 is required for efficient mitotic progression in vegetative cells (10, 31), and Clb1 is essential for meiotic progression while Clb2 is dispensable (14). The basis for the functional diversity in this case appears to be differential transcription during mitosis and meiosis (14). It is interesting that both Clb1/Clb2 and Clb5/Clb6 diverged with respect to regulation of protein expression.
Gene deletion studies indicate a significant functional redundancy between the members of the B-cyclin family. In an extreme example, the essential functions of all six B-type cyclins in a sextuple deletion strain can be rescued by expressing Clb1 constitutively (15), arguing that differential expression patterns are the major distinguishing feature of B-cyclin functions. However, biochemical studies indicate that cyclin/Cdk complexes can phosphorylate a broad spectrum of Cdk targets but that distinct complexes vary in their intrinsic activities and substrate affinities (26). Thus, the specialization of cyclins for diverse cell cycle functions may reflect a suite of evolutionary changes that affect cyclin expression as well as localization, activity, and substrate affinity.
Why are Clb5 and Clb6 differentially regulated?
Our observations suggest that Clb5 and Clb6 are functionally similar. Why then is it important for Clb6 to be destroyed at the G1/S border while Clb5 persists until mitosis?
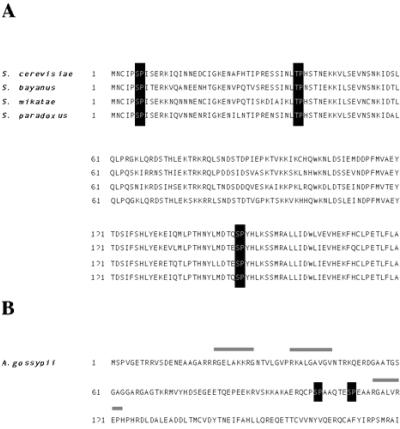
The destruction of Clb6 at the G1/S transition may be conserved through the evolution of Saccharomyces species. Protein sequence alignments with Clb6 orthologs from other Saccharomyces species (S. bayanus, S. mikatae, and S. paradoxus) indicate that all three putative Cdk phosphorylation sites and the sequences surrounding them are highly conserved (Fig. 7A) (3, 22). Clb5 orthologs are highly conserved in the region of the D-box degron sequences that mediates destruction by the APC. These observations suggest that the distinct regulatory processes for Clb5 and Clb6 are evolutionarily conserved.
FIG. 7.
Alignments of the N-terminal sequences of Clb6 for Saccharomyces species and the N-terminal sequences of A. gossypii. (A) The N-terminal amino acid sequence of Clb6 is shown for S. cerevisiae, S. bayanus, S. mikatae, and S. paradoxus. Sequences were downloaded from the Saccharomyces Genome Database (http://db.yeastgenome.org/fungi/YGR109C.html). Black boxes highlight putative S/TP phosphorylation sites. (B) The N-terminal amino acid sequence for AAR100C, the Clb5/Clb6 homolog from A. gossypii. The sequence was downloaded from the Ashbya Genome Database (http://agd.unibas.ch/Ashbya_gossypii/geneview?gene=AAR100C). Black boxes highlight putative S/TP phosphorylation sites, and gray bars indicate D-box-like sequences.
Many duplicated genes in the S. cerevisiae genome, including the cyclin gene pairs CLN1/CLN2, CLB1/CLB2, and CLB5/CLB6, are thought to have arisen from an ancient whole-genome doubling event (45). Ashbya gossypii, a filamentous fungus, diverged from the budding yeasts before the genome duplication event and thus is likely to be a good model for the structure and function of ancestral yeast proteins before the genome duplication and evolutionary divergence of gene pairs (4). Interestingly, the Clb5/Clb6 homolog in Ashbya contains D-box-like sequences in the N terminus, as well as two putative (S/T-P) phosphorylation sites (Fig. 7B) (4). However, these phosphorylation sites are not embedded in consensus SCFCdc4 degron sequences, suggesting that the ancestral S-phase cyclin may be degraded at mitosis and not at the G1/S border.
It has been reported that Clb6 inhibits progression through G2 (1). Thus, destruction of Clb6 at the G1/S border may be required for timely progression through G2. However, we have shown that expression of the CLB6Δ3P mutant from the endogenous CLB6 promoter in clb5Δ cells (Fig. 6C and D) or wild-type cells (data not shown) causes no measurable change in cell division time or defects in cell cycle distribution (Fig. 6D) as measured by flow cytometry. It is likely that Clb6 levels in the constitutive overexpression experiment reported by Basco et al. are significantly higher than the levels of Clb6Δ3P expressed from the CLB6 promoter (which are similar to wild-type levels) and that the significant G2 delay they observed is a result of elevated Clb6 levels rather than the persistence of Clb6 late in the cell cycle. Furthermore, Basco et al. favored a model in which Clb6 exerts its inhibitory effect on G2, at least in part, via regulation of CDC6 transcription (1). Because CDC6 is transcribed in G1, our finding that Clb6 is destroyed at the G1/S border is not necessarily inconsistent with the premise that Clb6 influences progression through G2 by affecting CDC6 transcription. However, our findings do indicate that destruction of Clb6 at the G1/S border is not likely to be necessary for maintaining normal cell cycle kinetics through G2/M.
Clues to the importance of destroying Clb6 at the G1/S border may be found in studies of the mammalian S-phase cyclin cyclin E. In many respects, budding yeast Clb6 and mammalian cyclin E are strikingly similar. Like Clb6, cyclin E is first expressed in G1 and then destroyed soon after cells traverse the G1/S transition (7). Cyclin E contains two consensus Cdk sites, and its stability is also regulated by the SCFhCdc4 complex (39).
Improper regulation of cyclin E expression and turnover appears to be associated with tumorigenesis and genome instability. Mutations in hCdc4 in a breast cancer cell line have been correlated with high expression levels of cyclin E (39). In fact, cyclin E levels are elevated in many tumor types (6, 32), and deregulated cyclin E expression has been shown to promote chromosome instability (38). However, the mechanism for the induction of chromosome instability by deregulated expression of cyclin E has not yet been fully elucidated.
Reminiscent of Clb5, mammalian cyclin A is expressed at the G1/S border (9) and persists until mitosis when it is degraded by the APC (11). Thus, there appear to be parallel regulatory mechanisms controlling the persistence of S-phase cyclins (Clb5 ≅ cyclin A and Clb6 ≅ cyclin E) in both yeast and mammalian cells. The biological significance of the differential regulatory mechanisms for cyclin E and cyclin A has yet to be determined.
Supplementary Material
Acknowledgments
We thank Danny Lew, Peter Kaiser, and Dave Stuart for yeast strains and plasmids, Kevan Shokat for a generous supply of 1-NM-PP1, and David Orlando and Todd Wasson for computational assistance. We also thank Duncan Clarke, Peter Kaiser, Danny Lew, and members of the Haase lab for helpful discussions and Danny Lew and Mark Chee for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Basco, R. D., M. D. Segal, and S. I. Reed. 1995. Negative regulation of G1 and G2 by S-phase cyclins of Saccharomyces cerevisiae. Mol. Cell. Biol. 15:5030-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop, A. C., J. A. Ubersax, D. T. Petsch, D. P. Matheos, N. S. Gray, J. Blethrow, E. Shimizu, J. Z. Tsien, P. G. Schultz, M. D. Rose, J. L. Wood, D. O. Morgan, and K. M. Shokat. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407:395-401. [DOI] [PubMed] [Google Scholar]
- 3.Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton, J. Majors, R. Waterston, B. A. Cohen, and M. Johnston. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301:71-76. [DOI] [PubMed] [Google Scholar]
- 4.Dietrich, F. S., S. Voegeli, S. Brachat, A. Lerch, K. Gates, S. Steiner, C. Mohr, R. Pohlmann, P. Luedi, S. Choi, R. A. Wing, A. Flavier, T. D. Gaffney, and P. Philippsen. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304:304-307. [DOI] [PubMed] [Google Scholar]
- 5.Donaldson, A. D., M. K. Raghuraman, K. L. Friedman, F. R. Cross, B. J. Brewer, and W. L. Fangman. 1998. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol. Cell 2:173-182. [DOI] [PubMed] [Google Scholar]
- 6.Donnellan, R., and R. Chetty. 1999. Cyclin E in human cancer. FASEB J. 13:773-780. [DOI] [PubMed] [Google Scholar]
- 7.Ekholm, S. V., and S. I. Reed. 2000. Regulation of G(1) cyclin-dependent kinases in the mammalian cell cycle. Curr. Opin. Cell Biol. 12:676-684. [DOI] [PubMed] [Google Scholar]
- 8.Epstein, C. B., and F. R. Cross. 1992. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 6:1695-1706. [DOI] [PubMed] [Google Scholar]
- 9.Erlandsson, F., C. Linnman, S. Ekholm, E. Bengtsson, and A. Zetterberg. 2000. A detailed analysis of cyclin A accumulation at the G(1)/S border in normal and transformed cells. Exp. Cell Res. 259:86-95. [DOI] [PubMed] [Google Scholar]
- 10.Fitch, I., C. Dahmann, U. Surana, A. Amon, K. Nasmyth, L. Goetsch, B. Byers, and B. Futcher. 1992. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell 3:805-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geley, S., E. Kramer, C. Gieffers, J. Gannon, J. M. Peters, and T. Hunt. 2001. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol. 153:137-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geymonat, M., A. Spanos, G. P. Wells, S. J. Smerdon, and S. G. Sedgwick. 2004. Clb6/Cdc28 and Cdc14 regulate phosphorylation status and cellular localization of Swi6. Mol. Cell. Biol. 24:2277-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson, D. G., J. G. Aparicio, F. Hu, and O. M. Aparicio. 2004. Diminished S-phase cyclin-dependent kinase function elicits vital Rad53-dependent checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:10208-10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grandin, N., and S. I. Reed. 1993. Differential function and expression of Saccharomyces cerevisiae B-type cyclins in mitosis and meiosis. Mol. Cell. Biol. 13:2113-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haase, S. B., and S. I. Reed. 1999. Evidence that a free-running oscillator drives G1 events in the budding yeast cell cycle. Nature 401:394-397. [DOI] [PubMed] [Google Scholar]
- 16.Haase, S. B., and S. I. Reed. 2002. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle 1:132-136. [PubMed] [Google Scholar]
- 17.Haase, S. B., M. Winey, and S. I. Reed. 2001. Multi-step control of spindle pole body duplication by cyclin-dependent kinase. Nat. Cell Biol. 3:38-42. [DOI] [PubMed] [Google Scholar]
- 18.Heinzel, S. S., P. J. Krysan, M. P. Calos, and R. B. DuBridge. 1988. Use of simian virus 40 replication to amplify Epstein-Barr virus shuttle vectors in human cells. J. Virol. 62:3738-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henchoz, S., Y. Chi, B. Catarin, I. Herskowitz, R. J. Deshaies, and M. Peter. 1997. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 11:3046-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irniger, S., and K. Nasmyth. 1997. The anaphase-promoting complex is required in G1 arrested yeast cells to inhibit B-type cyclin accumulation and to prevent uncontrolled entry into S-phase. J. Cell Sci. 110:1523-1531. [DOI] [PubMed] [Google Scholar]
- 21.Jacobson, M. D., S. Gray, M. Yuste-Rojas, and F. R. Cross. 2000. Testing cyclin specificity in the exit from mitosis. Mol. Cell. Biol. 20:4483-4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kellis, M., N. Patterson, M. Endrizzi, B. Birren, and E. S. Lander. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423:241-254. [DOI] [PubMed] [Google Scholar]
- 23.Kuhne, C., and P. Linder. 1993. A new pair of B-type cyclins from Saccharomyces cerevisiae that function early in the cell cycle. EMBO J. 12:3437-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanker, S., M. H. Valdivieso, and C. Wittenberg. 1996. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science 271:1597-1601. [DOI] [PubMed] [Google Scholar]
- 25.Leismann, O., A. Herzig, S. Heidmann, and C. F. Lehner. 2000. Degradation of Drosophila PIM regulates sister chromatid separation during mitosis. Genes Dev. 14:2192-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loog, M., and D. O. Morgan. 2005. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature 434:104-108. [DOI] [PubMed] [Google Scholar]
- 27.Mathias, N., S. L. Johnson, M. Winey, A. E. Adams, L. Goetsch, J. R. Pringle, B. Byers, and M. G. Goebl. 1996. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S-phase transition and identifies a conserved family of proteins. Mol. Cell. Biol. 16:6634-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moll, T., E. Schwob, C. Koch, A. Moore, H. Auer, and K. Nasmyth. 1993. Transcription factors important for starting the cell cycle in yeast. Philos. Trans. R. Soc. Lond. B Biol. Sci. 340:351-360. [DOI] [PubMed] [Google Scholar]
- 29.Nash, P., X. Tang, S. Orlicky, Q. Chen, F. B. Gertler, M. D. Mendenhall, F. Sicheri, T. Pawson, and M. Tyers. 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414:514-521. [DOI] [PubMed] [Google Scholar]
- 30.Nishizawa, M., M. Kawasumi, M. Fujino, and A. Toh-e. 1998. Phosphorylation of Sic1, a cyclin-dependent kinase (Cdk) inhibitor, by Cdk including Pho85 kinase is required for its prompt degradation. Mol. Biol. Cell 9:2393-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richardson, H., D. J. Lew, M. Henze, K. Sugimoto, and S. I. Reed. 1992. Cyclin-B homologs in Saccharomyces cerevisiae function in S phase and in G2. Genes Dev. 6:2021-2034. [DOI] [PubMed] [Google Scholar]
- 32.Sandhu, C., and J. Slingerland. 2000. Deregulation of the cell cycle in cancer. Cancer Detect. Prev. 24:107-118. [PubMed] [Google Scholar]
- 33.Schneider, B. L., Q. H. Yang, and A. B. Futcher. 1996. Linkage of replication to start by the Cdk inhibitor Sic1. Science 272:560-562. [DOI] [PubMed] [Google Scholar]
- 34.Schwob, E., T. Bohm, M. D. Mendenhall, and K. Nasmyth. 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79:233-244. [DOI] [PubMed] [Google Scholar]
- 35.Schwob, E., and K. Nasmyth. 1993. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 7:1160-1175. [DOI] [PubMed] [Google Scholar]
- 36.Shirayama, M., A. Toth, M. Galova, and K. Nasmyth. 1999. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature 402:203-207. [DOI] [PubMed] [Google Scholar]
- 37.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209-219. [DOI] [PubMed] [Google Scholar]
- 38.Spruck, C. H., K. A. Won, and S. I. Reed. 1999. Deregulated cyclin E induces chromosome instability. Nature 401:297-300. [DOI] [PubMed] [Google Scholar]
- 39.Strohmaier, H., C. H. Spruck, P. Kaiser, K. A. Won, O. Sangfelt, and S. I. Reed. 2001. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature 413:316-322. [DOI] [PubMed] [Google Scholar]
- 40.Stueland, C. S., D. J. Lew, and S. I. Reed. 1993. Full activation of p34CDC28 histone H1 kinase activity is unable to promote entry into mitosis in checkpoint-arrested cells of the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 13:3744-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verma, R., R. S. Annan, M. J. Huddleston, S. A. Carr, G. Reynard, and R. J. Deshaies. 1997. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278:455-460. [DOI] [PubMed] [Google Scholar]
- 42.Verma, R., R. M. Feldman, and R. J. Deshaies. 1997. SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities. Mol. Biol. Cell 8:1427-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 44.Willems, A. R., T. Goh, L. Taylor, I. Chernushevich, A. Shevchenko, and M. Tyers. 1999. SCF ubiquitin protein ligases and phosphorylation-dependent proteolysis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354:1533-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolfe, K. H., and D. C. Shields. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387:708-713. [DOI] [PubMed] [Google Scholar]
- 46.Won, K. A., and S. I. Reed. 1996. Activation of cyclin E/CDK2 is coupled to site-specific autophosphorylation and ubiquitin-dependent degradation of cyclin E. EMBO J. 15:4182-4193. [PMC free article] [PubMed] [Google Scholar]
- 47.Yamano, H., K. Kominami, C. Harrison, K. Kitamura, S. Katayama, S. Dhut, T. Hunt, and T. Toda. 2004. Requirement of the SCFPop1/Pop2 ubiquitin ligase for degradation of the fission yeast S phase cyclin Cig2. J. Biol. Chem. 279:18974-18980. [DOI] [PubMed] [Google Scholar]
- 48.Zou, L., and B. Stillman. 2000. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol. Cell. Biol. 20:3086-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zou, L., and B. Stillman. 1998. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science 280:593-596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.