Abstract
In pituitary GH3B6 cells, signaling involving the protein kinase C (PKC) multigene family can self-organize into a spatiotemporally coordinated cascade of isoform activation. Indeed, thyrotropin-releasing hormone (TRH) receptor activation sequentially activated green fluorescent protein (GFP)-tagged or endogenous PKCβ1, PKCα, PKCɛ, and PKCδ, resulting in their accumulation at the entire plasma membrane (PKCβ and -δ) or selectively at the cell-cell contacts (PKCα and -ɛ). The duration of activation ranged from 20 s for PKCα to 20 min for PKCɛ. PKCα and -ɛ selective localization was lost in the presence of Gö6976, suggesting that accumulation at cell-cell contacts is dependent on the activity of a conventional PKC. Constitutively active, dominant-negative PKCs and small interfering RNAs showed that PKCα localization is controlled by PKCβ1 activity and is calcium independent, while PKCɛ localization is dependent on PKCα activity. PKCδ was independent of the cascade linking PKCβ1, -α, and -ɛ. Furthermore, PKCα, but not PKCɛ, is involved in the TRH-induced β-catenin relocation at cell-cell contacts, suggesting that PKCɛ is not the unique functional effector of the cascade. Thus, TRH receptor activation results in PKCβ1 activation, which in turn initiates a calcium-independent but PKCβ1 activity-dependent sequential translocation of PKCα and -ɛ. These results challenge the current understanding of PKC signaling and raise the question of a functional dependence between isoforms.
Space and time parameters are intimately linked to the biological function of proteins and are pivotal in the organization and functioning of living matter. Particularly important in the case of intracellular signaling networks, this spatiotemporal constraint determines the modalities by which a given protein interacts with its partners in order to exchange information. Efforts are currently being made to translate the experimental evidence of this plasticity into a visualization concept of dynamic signaling networks (4, 22). A degree of complexity is added when different isoforms of a protein coexist within the same cell and when they all potentially respond to the same stimulus, which is the case of the protein kinase C (PKC) family.
The PKC family is composed of at least 11 isoforms. Several isoforms usually coexist within a given cell type, and each isoform is thought to mediate distinct cellular functions leading to proliferation, differentiation, apoptosis, or secretion. PKC function requires its translocation to a membrane compartment. The current understanding of PKC translocation/activation has been largely based on the work of Oancea and Meyer (34), who presented conventional PKCs (cPKCs) and novel PKCs (nPKCs) as molecular machines responsible for decoding calcium and/or diacylglycerol (DAG)-mediated signals. Several observations suggest, however, that other, as yet unknown, parameters are involved in the temporal organization of PKC signaling. Indeed, despite similarities in sequence and cofactor regulation by DAG and Ca2+, the conventional PKCβI and PKCβII were shown to exhibit isoform-specific oscillation patterns following metabotropic glutamate receptor 1a (mGluR1) activation (3). This behavior was interpreted by the authors as a differential decoding of second messengers, although the key amino acids involved were located in the V5 region of PKCβII, which is not involved in binding to cofactors. Also, during mammalian egg activation, Eliyahu and Shalgi have shown the translocation of PKCα, PKCβ1, and PKCβ2 but noted that only PKCα translocation did not occur in the presence of ionomycin, a calcium ionophore known to elicit cPKC translocation in many systems (14). In a recent work, Braun et al. provided evidence that PKCα translocation was determined primarily by factors other than the localization of phorbol esters, unlike PKCδ translocation (5). Raghunath et al. (44) analyzed the translocation of PKCα, -β2, -δ, and -ɛ in neuroblastoma cells upon exposure to the muscarinic receptor agonist carbachol. These authors observed sustained translocations of PKCɛ, while PKCδ translocated on rare occasions. Similar translocation kinetics were observed for PKCα and -β2, and PKCα was translocated a few seconds after an individual C2 domain, the translocation of which coincided exactly with calcium variations (44). Concerning the work we performed with the GH3B6 cell line, several observations also question the general organization of PKC signaling. We have shown that upon stimulation by phorbol myristate acetate (PMA), PKCα (a cPKC) and PKCɛ (an nPKC) were selectively translocated to the cell-cell contact, whereas PKCδ (an nPKC) is recruited to the entire plasma membrane (43). Hence, PKC isoforms that belong to the same PKC subclass (PKCɛ and -δ), and are supposedly regulated similarly, are targeted differently when cells are stimulated by PMA (43). Another interesting finding was the lack of correlation between the rise in the intracellular concentration of Ca2+ ([Ca2+]i) and the subcellular localization of the PKCα isoform. Indeed, [Ca2+]i increased similarly in single cells and cells in contact upon thyrotropin-releasing hormone (TRH) stimulation, but PKCα did not translocate to the plasma membranes of isolated cells (52). In GH3B6 cells, the fact that PKCα relocated to the cytoplasm before the [Ca2+]i decreased to baseline levels (52) also indicates that cPKC activation is not always phase locked with variations in calcium concentration as suggested in many studies, such as that of Violin et al., who also showed the coincidence of PKC substrate phosphorylation with calcium oscillations (54). In addition, we have shown that the selective targeting of PKCα to cell-cell contacts is probably finely regulated at the molecular level, since the natural D294G point mutation (located in the V3 region of the enzyme and thus not involved in calcium or DAG binding), previously identified in human pituitary and thyroid tumors (2, 40, 41), abolished the selective accumulation of PKCα at the cell-cell contact (51). The same result was obtained when this mutation was introduced in the PKCɛ coding sequence (43). Together, these observations suggest the existence of a complex spatiotemporal organization of PKC signaling.
In order to understand the links between PKC signaling and the physiological roles of a given stimulus, one approach consists in first defining the functional relationships between the different PKC isoforms. The GH3B6 cell line is a suitable model system to study these links, since it has conserved essential characteristics of growth hormone- and prolactine-secreting cells. In particular, it is still physiologically regulated by hypothalamic hormones, such as TRH, and the cell-cell contact targeting of PKCα is similar to what is observed in pituitary tissue (43). The present study was aimed at clarifying the calcium dependence/independence of PKCα recruitment to the cell-cell contact and establishing the precise spatiotemporal dynamics of translocation of PKCα, -β1, -ɛ, and -δ (these isoforms are constitutively expressed in the GH3B6 cell line). Results show that PKCα translocation is indeed calcium independent in this cell line and that a coordinated cascade of isoform activation exists that links PKCα and -ɛ subcellular localization at cell-cell contacts to PKCβ1 activity. Furthermore, results on the TRH-induced β-catenin relocation at cell-cell contacts suggest that the PKC cascade does not have a unique functional effector.
MATERIALS AND METHODS
Materials.
Ionomycin was purchased from Sigma (Saint Quentin Fallavier, France). Restriction enzymes were from Biolabs New England (Beverly, Mass.). DNA polymerase (DyNAzyme) was from Finnzyme (Espoo, Finland). Synthetic oligonucleotides were purchased from Sigma-Genosys (Haverhill, United Kingdom). pEGFP-N1 plasmid was purchased from Clontech (Palo Alto, Calif.). The plasmids containing the full-length PKCβ1, PKCɛ, and PKCδ cDNAs were generously provided by P. J. Parker (Imperial Cancer Research Fund, London, United Kingdom). The pcDNA-PKCα-RFP (red fluorescent protein) plasmid was a gift of T. Ng (Randall Division of Cell and Molecular Biophysics, Guy's Campus, King's College London, London, United Kingdom). Plasmid containing dominant-negative (DN) human PKCα (hPKCα) (K368M) was kindly provided by B. Mari (INSERM U526, IFR50, Faculté de Médecine Pasteur, Nice, France). The QuikChange site-directed mutagenesis kit was purchased from Stratagene. ExGen 500 (linear ethylenimine polymer) was purchased from Euromedex (Souffelweyersheim, France). Fetal bovine serum was from Bio Media (Boussens, France). Gö6976 [12-(2-cyanoethyl)-6,7,12,13-tetrahydro-13-methyl-5-oxo-5H-indolo(2,3-a)pyrrolo(3,4-c)-carbazole] was from Calbiochem (Meudon, France). Thyrotropin-releasing hormone was purchased from Bachem Biochimie Sarl (Voisins-le-Bretonneux, France). Ham F-10 medium and horse serum were obtained from Eurobio (Les Ulis, France). Polyclonal anti-PKCδ was purchased from Sigma, monoclonal anti-PKCα and polyclonal anti-PKCɛ were from Euromedex, and polyclonal anti-β-catenin and monoclonal anti-PKCβ1 were from Santa Cruz Biotechnology (Santa Cruz, Calif.).
Construction of plasmids.
Construction of hPKCα-pEGFP-N1, PKCα-pcDNA-RFP, mPKCɛ-pEGFP-N1, and rPKCδ-pEGFP-N1 has been described previously (38, 43, 52). In the constitutively active (CA) PKCβ1 (CAPKCβ1-pEGFP-N1), alanine 25 (codon GCC) was replaced by glutamic acid (codon GAA). In the dominant-negative PKCβ1 (DNPKCβ1-pEGFP-N1), lysine 371 (codon AAG) was replaced by methionine (codon ATG). CA and DN PKCβ1 and DN PKCα were then subcloned into the pcDNA-RFP plasmid. PKCβ1 was also subcloned into the pcDNA-RFP and pEGFP-N1 plasmids. In the calcium-insensitive hPKCα and hPKCβ1, aspartic acids 187/246/248 (codons GAT/GAC/GAT) were replaced by asparagines (codons AAT/AAC/AAT). All constructs were checked by sequencing.
Site-directed mutagenesis.
The point mutated CA PKCβ1 (A25E) and DN PKCβ1 (K371M) were created by using the QuikChange site-directed mutagenesis kit according to the manufacturer's standard protocol. The pairs of synthetic oligonucleotides used to obtain the mutated PKCs were as follows: forward 5′-GCCCGCAAAGGCGAACTCCGGCAGAAGAACG-3′ and reverse 5′-CGTTCTTCTGCCGGAGTTCGCCTTTGCGGGC-3′ for CA PKCβ1 (A25E); forward 5′-GCTCTATGCTGTGATGATCCTGAAGAAGG-3′ and reverse 5′-CCTTCTTCAGGATCATCACAGCATAGAGC-3′ for DN PKCβ1 (K371M).
For D187/246/248N hPKCα- and hPKCβ1-pEGFP-N1, aspartic acids 187/246/248 (codons GAT/GAC/GAT) were mutated to asparagines (codons AAT/AAC/AAT). The pairs of synthetic oligonucleotides used to obtain the mutated PKCs were as follows: forward 5′-CTAATCCCTATGAATCCAAACGGGCTTTCAGATCC-3′ and reverse 5′-GGATCTGAAAGCCCGTTTGGATTCATAGGGATTAG-3′ for D187N hPKCα; forward 5′-CTGTAGAAATCTGGAACTGGAATCGAACAACAAGG-3′ and reverse 5′-CCTTGTTGTTCGATTCCAGTTCCAGATTTCTACAG-3′ for D246/248N hPKCα; forward 5′-CTTGTACCTATGAACCCCAATGGCCTGTCAGATCC-3′ and reverse 5′-GGATCTGACAGGCCATTGGGGTTCATAGGTACAAG-3′ for D187N hPKCβ1; and forward 5′-CAGTAGAGATTTGGAATTGGAATTTGACCAGCAGG-3′ and reverse 5′-CCTGCTGGTCAAATTCCAATTCCAAATCTCTACTG-3′ for D246/248N hPKCβ1.
Cell culture and transfection of fusion protein.
GH3B6 cells were cultured in Ham F-10 medium supplemented with 2.5% (vol/vol) fetal bovine serum and 15% (vol/vol) horse serum both heat inactivated at 56°C for 1 hour. Transient transfection of GH3B6 cells was performed with ExGen as described previously (43). Briefly, the cells were seeded at 300,000 cells per well in 24-well dishes (Falcon) 18 h before transfection. Immediately before transfection, fresh culture medium (1 ml) was added to the cells. ExGen 500 stock solution (1.25 μl) was diluted in 12.5 μl of 150 mM NaCl. Plasmid DNA (250 ng/well) was diluted in 12.5 μl of 150 mM NaCl. These solutions were then mixed. After 30 min, the transfection mixture was added to the cells. The 24-well dishes were then centrifuged for 5 min at 280 × g and maintained for 48 h at 37°C. To coexpress PKCs, equal quantities of DNA plasmids (0.250 μg) were cotransfected according to the manufacturer's instructions. In order to inhibit cPKC activity, GH3B6 cells were incubated with 100 nM Gö6976 for 15 min before and during TRH stimulation. Stimulation by TRH was achieved by applying TRH to single cells.
siRNA transfection.
Small interfering RNAs (siRNAs), targeting rat PKCα, PKCβ, and PKCɛ, were synthesized and purified by Eurogentec (Liège, Belgium). The sequences of each siRNA pair were as follows: for PKCα-siRNA, 5′-UCUUGCAAAGUGCAGUAUGtt-3′ and 5′-CAUACUGCACUUUGCAAGAtt-3′; for PKCβ-siRNA, 5′-GUUUAAGAUCCACACCUACtt-3′ and 5′-GUAGGUGUGGAUCUUAAACtt-3′; and for PKCɛ-siRNA, 5′-CUUGAAAACAACAUCCGGATT-3′ and 5′-UCCGGAUGUUGUUUUCAAGtt-3′. A scrambled siRNA was used as a control. For PKC expression analyses, GH3B6 cells were plated at 7 × 105 per well in six-well plates. After 24 h, siRNAs (final concentrations, 50, 100, and 200 nM) were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After 72 h, siRNA efficacy on PKCα, -β, and -ɛ expression was analyzed by Western blotting. For immunocytochemical experiments, GH3B6 cells were plated at 3 × 105 per well in 24-well plates. After 24 h, siRNAs targeting PKCβ, -α, and -ɛ were transfected as described above and 72 h later, translocation of endogenous PKCs upon TRH stimulation for 15 s, 60 s, or 5 min was observed. The TRH-induced β-catenin relocalization was analyzed by immunocytochemistry after transfection of siRNAs against PKCα and -ɛ and stimulation with TRH for 1 h. Efficiency of siRNA transfection was higher than 80% as assessed by the transfection of a control nonsilencing siRNA labeled with rhodamine (QIAGEN).
Immunocytochemistry.
GH3B6 cells were seeded on 12-mm round coverslips in 1 ml medium. They were washed three times with phosphate-buffered saline (PBS), pH 7.4, before being fixed for 10 min with 4% paraformaldehyde in PBS, pH 7.4 (vol/vol). The cells were then washed twice with PBS, permeabilized in 0.2% Triton X-100 for 5 min, washed in PBS, and incubated for 30 min in PBS supplemented with 1% bovine serum albumin. The coverslips were then incubated overnight at 4°C with the primary monoclonal anti-PKCα (diluted 1/250), polyclonal anti-PKCɛ (diluted 1/500), polyclonal anti-PKCδ (diluted 1/500), polyclonal anti-β-catenin (diluted 1/100), or monoclonal anti-PKCβ1 (diluted 1/100) antibodies. They were then washed three times for 10 min with PBS supplemented with 1% bovine serum albumin and further incubated for 1 h at room temperature with the secondary antibodies, which were either a Cy3-conjugated goat anti-mouse (Fab′)2 antibody or an Alexa 488-conjugated goat anti-rabbit antibody (diluted 1/1,000). The coverslips were then mounted in Mowiol and examined by conventional microscopy (Zeiss) by using a 63× objective.
Real-time fluorescence microscopy.
The localization of fusion proteins in living cells was examined by conventional or confocal fluorescence microscopy. Conventional microscopy was performed with an Axiophot 2.0 from Zeiss. Cells were viewed at room temperature with a 63× 0.9-numerical-aperture Achroplan water immersion objective lens (Zeiss). At the time of observation, the culture medium was replaced by a buffer composed of 140 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 10 mM HEPES, 6 mM glucose, pH 7.4. The camera was a CoolSNAP camera from Photometrics, a division of Roper Scientific. The acquisition software was MetaVue version 6.1 from Universal Imaging Corporation. The confocal laser-scanning microscope was equipped with an Ar/Kr laser (Odyssey XL with InterVision 1.4.1 software; Noran Instruments, Inc., Middleton, WI) as described by Guerineau et al. (18). Cells were viewed with a 63× 0.9-numerical-aperture Achroplan water immersion objective lens (Zeiss). At the time of observation, the culture medium was replaced by a prewarmed buffer, at 37°C, identical to the one used for the conventional microscopic observations. Images were acquired continuously with intervals between frames of 0.533 s. The ImageJ 1.32j software (National Institutes of Health, Bethesda, Md.) was used to quantify PKC translocation to plasma membrane. When movies were used for illustrations, five consecutive images were averaged in order to decrease pixellization.
See Fig. S1 and S2 in the supplemental material for the subcellular distribution of green fluorescent protein (GFP)-tagged PKCα, -β1, -ɛ, and -.δ upon PMA stimulation and the various subcellular distributions of PKCδ upon PMA stimulation, respectively. See Videos S1 to S4 in the supplemental material (they correspond to Fig. 3) for recordings of GFP-tagged isoforms before and after stimulation with TRH that starts at image 20. One image was taken every 0.533 s, with a Kallman average of 64 images. See Video S5 in the supplemental material for cells transfected with CA PKCβ1 fused to GFP. In this case, there is no TRH stimulation. Note that in this case, translocation is oscillatory.
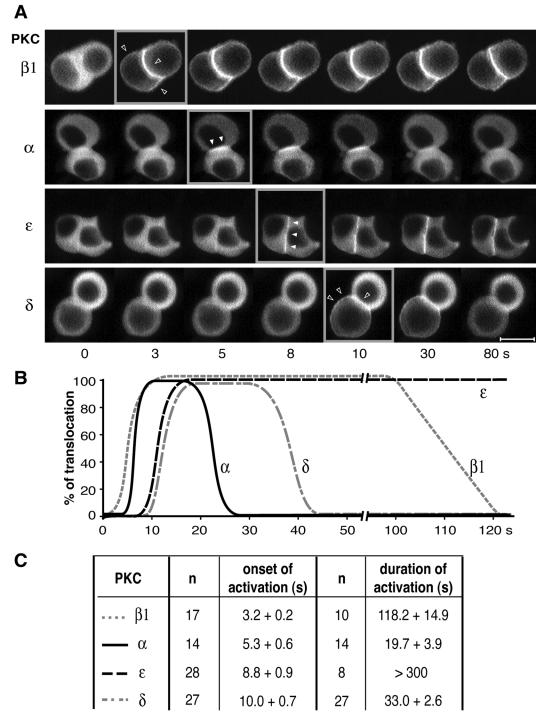
FIG. 3.
Analysis of the translocation kinetics of PKCα, -β1, -ɛ, and -δ. (A) Examples of real-time recordings of spatiotemporal activation of PKCβ1, -α, -ɛ, and -δ fused to GFP in GH3B6 cells stimulated with 100 nM TRH. Recordings were performed with a Noran confocal microscope. Images were acquired continuously, with intervals of 0.533 s between frames. Time zero is defined as the time of TRH application. Videos can be seen in the supplemental material. Bar, 5 μm. (B) Schematic representation of the data showing the different onsets and durations of PKCβ1, -α, -ɛ, and -δ translocations. (C) Statistical analysis of the data, with the number of cells analyzed. The P values were calculated with the Student t test.
RESULTS
Different spatial organizations of activated PKCα, -β1, -ɛ, and -δ.
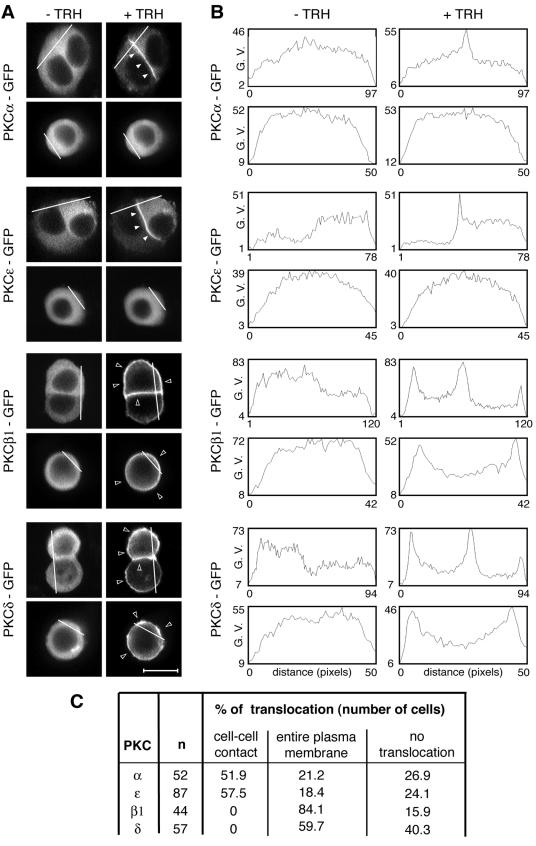
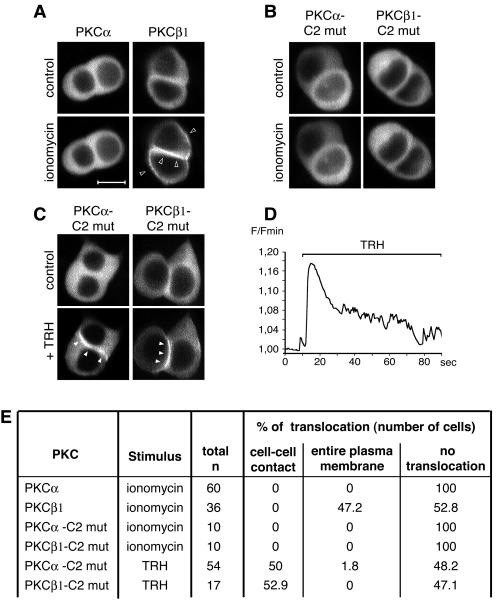
We have previously described the spatial organization of TRH-activated PKCα and those of PMA-stimulated PKCα, -ɛ, and -δ in GH3B6 cells (43, 52). Figure 1A illustrates the spatial organization of PKCα, -ɛ, -β1, and -δ in contacting and isolated cells upon TRH stimulation. While all isoforms were located in the cytoplasm in unstimulated cells, GFP-tagged PKCα and -ɛ accumulated selectively at the cell-cell contact in 52 and 57%, respectively, of the cells analyzed, whereas GFP-tagged PKCδ never did, accumulating instead at the entire plasma membrane in 60% of cases (Fig. 1A and C). In the case of PKCβ1, the native endogenous protein (see Fig. 8) as well as the GFP-tagged fusion protein accumulated without selectivity at the entire plasma membrane in 84% of cells. Furthermore, in TRH-stimulated isolated cells, PKCα and -ɛ remained in the cytoplasm, whereas PKCβ1 and -δ accumulated at the plasma membrane (Fig. 1A). Analysis of pixel values along a line that crosses the cell without going through the nucleus illustrates these various spatial patterns and confirms that the pixel values increased only at the cell-cell contacts for PKCα and -ɛ, which was not the case for PKCβ1 and δ (Fig. 1A, right panels). The same analyses performed with isolated cells show an increase in the pixel values at the plasma membrane only in the case of activated PKCβ1 and -δ, demonstrating that isoforms that selectively accumulated at cell-cell contacts do not translocate in isolated cells, in contrast to isoforms that accumulated without selectivity at the plasma membrane, which also translocated in isolated cells. Therefore, although belonging to different PKC subclasses, PKCα and -ɛ on the one hand and PKCβ1 and -δ on the other display identical subcellular distributions after activation. In many cases where a PKC isoform accumulated at the entire plasma membrane, the cell-cell contact appeared more fluorescent than the remainder of the plasma membrane (see Fig. 3A, PKCβ1, and 4A, C, and D, PKCα, + TRH + Gö-6976), probably due to the apposition of the plasma membranes of the two contacting cells.
FIG. 1.
Analysis of PKCα, -β1, -ɛ, and -δ targeting in the GH3B6 cell line. (A) Micrographs depicting what we define as a targeting to the entire plasma membrane or to cell-cell contacts. PKCα, -β1, -ɛ, and -δ were subcloned in frame with GFP at the C-terminal end of their sequence in the pEGFP-N1 vector (Clontech). PKCβ1 and -δ accumulated at the entire plasma membrane in the presence (+) of 100 nM TRH, whereas PKCα and -ɛ accumulated selectively at the cell-cell contact. In isolated cells, PKCβ1 and -δ behaved as in cells in contact, whereas PKCα and -ɛ remained in the cytoplasm. Observations were performed with a confocal microscope. In Fig. 1 to 10, the empty arrowheads show translocation at the entire plasma membrane, whereas the filled arrowheads show selective translocation to cell-cell contacts. Bar, 5 μm. (B) Analysis with Image J 1.32j software (National Institutes of Health, Bethesda, Md.) was used to quantify PKC translocation and demonstrated that the pixel values increased only at the cell-cell contacts for PKCα and -ɛ, which was not the case for PKCβ1 and -δ. G. V., gray values. (C) Summary of the results as the percentages of cells where TRH-induced translocation was observed either selectively at cell-cell contacts or at the entire plasma membrane. The percentage of cells where no translocation was observed is also indicated.
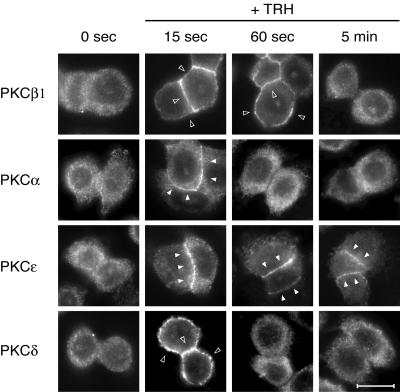
FIG. 8.
Endogenous PKCs are also sequentially activated by TRH. GH3B6 cells were stimulated with TRH for the indicated times and then immediately fixed with paraformaldehyde. Immunocytochemistry performed with selective PKC antibodies showed that endogenous PKCs behave like the GFP-tagged ones. After 15 s of TRH stimulation, all PKCs were translocated. After 60 s, only PKCβ1 and -ɛ were still activated, whereas at 5 min of stimulation, only PKCɛ was activated. Bar, 5 μm.
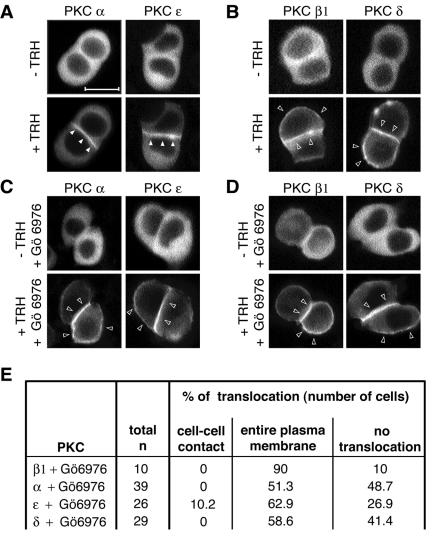
FIG. 4.
Cell-cell contact targeting of PKCα and -ɛ is dependent on PKC activity, unlike the PKCβ1 and -δ targeting to the entire plasma membrane. (A to D) GH3B6 cells were preincubated (A and B) or not (C and D) with the cPKC activity inhibitor Gö6976 and then stimulated (+) with 100 nM TRH. Real-time recordings were performed as described in the legend to Fig. 2. (C) When preincubated with 100 nM Gö6976 for 15 min, PKCα and -ɛ behaved like PKCβ1 and -δ: they accumulated at the entire plasma membrane. Gö6976 had no effect on PKCβ1 and -δ targeting (D). Bar, 5 μm. (E) Summary of the number of cells analyzed.
Upon PMA stimulation, the distribution of activated enzymes was very similar to that in the presence of TRH (see Fig. S1 in the supplemental material) (43), except that, in some cells, PKCδ also accumulated in a perinuclear region as well as at the nuclear membrane as reported for other cellular systems (5) (see Fig. S2 in the supplemental material). It is noteworthy that such nuclear and perinuclear localizations have never been observed upon TRH stimulation.
Existence of a calcium-independent translocation mechanism for a cPKC.
In order to analyze the calcium dependence of PKCα translocation to cell-cell contacts, we first studied the effects of a calcium ionophore on the subcellular localization of PKCα, while PKCβ1 was used for comparison. We found that whereas PKCβ1 was translocated and accumulated at the entire plasma membrane in the presence of 2 μM ionomycin, PKCα remained in the cytoplasm (Fig. 2A). A similar lack of PKCα translocation was described before in eggs incubated in the presence of 2 μM ionomycin (14). The differences of sensitivity of the two isoforms towards ionomycin cannot be explained by different calcium binding affinities, since it has been shown that the 50% inhibitory concentrations of PKCα and -β1 for calcium are very similar (23). This calcium independence of PKCα in GH3B6 cells was further demonstrated by using a PKCα-GFP fusion protein bearing the D187/246/248N mutations, known to abolish calcium binding to the C2 domain (10). This calcium-insensitive PKCα-GFP was still able to accumulate selectively at the cell-cell contact in the presence of TRH (Fig. 2C), providing the first convincing evidence that a cPKC can be translocated by a different mechanism than the one currently accepted today, which is that an increase in the [Ca2+]i is the leading event in cPKC translocation. For a control, we introduced the same three point mutations in the C2 domain of PKCβ1. This mutant did not translocate in the presence of ionomycin, as expected (Fig. 2B). Surprisingly, we found that in the presence of TRH, the calcium-insensitive PKCβ1 does not accumulate at the entire plasma membrane, but like native PKCα (Fig. 2C), accumulates selectively at the cell-cell contact. Conversely, we had shown before that D294G PKCα, like native PKCβ1, accumulates at the entire plasma membrane instead of being selectively translocated to the cell-cell contact (51). Thus, affecting either amino acid 294 in PKCα or abolishing calcium binding in PKCβ1 switches the mode of translocation of PKCα to that of PKCβ1 and vice versa. Figure 2D confirms that TRH induced the expected variation in intracellular calcium under these experimental conditions. The unambiguous nature of these results is emphasized when they are expressed as percentages of cells where PKC translocation is observed or PKC translocation is not observed (Fig. 2E).
FIG. 2.
Existence of a calcium-independent targeting of PKCα. (A) PKCα and -β1 behaved differently when GH3B6 cells were incubated with 2 μM ionomycin. PKCβ1 is activated and accumulated at the entire plasma membrane similar to what is seen in the presence of 100 nM TRH. PKCα was not activated and remained in the cytoplasm. (B) PKCα and -β bearing the triple point mutations D187/246/248N do not translocate in the presence of ionomycin. mut, mutant. (C) D187/246/248N PKCα, unable to bind calcium, was still targeted to the cell-cell contacts upon stimulation, indicating that its translocation can be calcium independent. The same mutations in the PKCβ1 sequence show that PKCβ1 behaved like PKCα when unable to bind calcium: it accumulated at cell-cell contacts. Bar, 5 μm. (D) An example of intracellular calcium modification upon TRH stimulation. Image acquisition was performed on a confocal microscope, with 0.533-s intervals between frames. Calcium monitoring was described previously (48). F/Fmin, fluorescence/minimum fluorescence. (E) Summary of the results as described in the legend to Fig. 1C.
Existence of a cascade of isoform activation.
Our second goal was to uncover a possible hierarchy in the kinetics of PKC isoform activation. Figure 3 shows a statistically relevant analysis of PKC isoform targeting kinetics. It demonstrates that upon TRH activation, PKCβ1 is the first isoform to be translocated to the plasma membrane (starting 3.2 ± 0.2 s [n = 17] after TRH application), followed 2 seconds later by the selective translocation of PKCα to cell-cell contacts (starting 5.3 ± 0.6 s [n = 14]; statistically different from PKCβ1 onset of activation, P < 0.05). PKCɛ was translocated another 3.5 s later, immediately followed by PKCδ. The duration of activation was also found to vary significantly between the different isoforms, independently of the subclass to which these isoforms belong (Fig. 3B and C): 118 s for PKCβ1, 20 s for PKCα, 15 to 20 min for PKCɛ, and 33 s for PKCδ (see the videos in the supplemental material). Strikingly, the time during which PKCβ1 remained activated was identical to that of the D294G PKCα mutant (51). The fact that PKCα mutant activation is phase locked with [Ca2+]i variations (51) suggests that the PKCα mutant has indeed switched from a Ca2+-independent to a Ca2+-dependent translocation as hypothesized above. Conversely, the D187/246/248N PKCβ1 mutant, which accumulated at cell-cell contacts (see above), displayed kinetic characteristics that were different from those of PKCβ1 but close to those of PKCα (onset of activation, 5.8 ± 0.6 s; duration of activation, 46 ± 10 s [n = 3]), indicating that spatial and temporal dimensions are indeed undissociable clues for understanding PKC signaling.
The selective accumulation of PKCα and -ɛ at cell-cell contacts is dependent on PKC activity.
Since there is a statistically significant difference between the onsets of activation of PKCβ1 and -α, we asked whether PKCβ1, which is the first to be activated, could play the role of “leader” in this cascade of sequential translocation/activation. We started by looking at the effect of Gö6976, an inhibitor of conventional PKCs at the dose used in our experiments (100 nM) (28), even though it can also inhibit other kinases but at much higher doses (>1 μM) (42). As shown in Fig. 4A to D, when cells were incubated in the presence of Gö6976 and TRH, PKCα and -ɛ were translocated to the entire plasma membrane instead of accumulating at the cell-cell contact. This suggested that selective translocation of PKCα and -ɛ to cell-cell contacts was dependent on the activity of at least one conventional PKC isoform. PKCβ1 was a more obvious candidate than PKCα, since we had previously shown that deleting the catalytic domain of PKCα does not abolish its selective accumulation at cell-cell contacts (52). Although PKD is also inhibited by Gö6976 (17), it is probably not the isoform involved here, as it is not sensitive to calcium or DAG variations.
Coordination of the cascade of PKC isoform activation. (i) PKCα translocation depends on PKCβ1 activity.
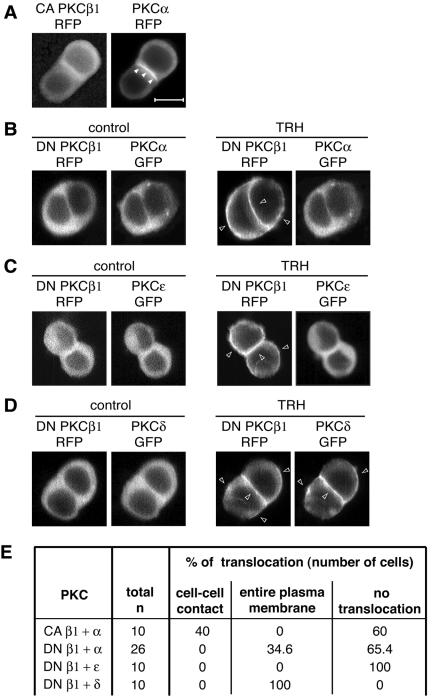
If PKCβ1 were the leader in the sequential PKC isoform activation cascade and if it conditions PKCα translocation, then bypassing activation of the TRH receptor by using a constitutively active PKCβ1 (resulting in no increases in Ca2+ and DAG concentrations within the cell) should elicit a spontaneous accumulation of PKCα at the cell-cell contact. Figure 5A shows that indeed, in the absence of TRH, PKCα accumulated at the cell-cell contact of cells transfected with CA PKCβ1-RFP. This does not mean, however, that CA PKCβ1-RFP is sufficient to fully activate PKCα at the cell-cell contact, only that PKCβ1 activity is sufficient to elicit PKCα translocation. By contrast, in cells transfected with a dominant-negative PKCβ1-RFP plasmid, PKCα-GFP remained in the cytoplasm, even in the presence of TRH (Fig. 5B). Figure 5B shows also that DN PKCβ1-RFP (as well as DN PKCβ1-GFP transfected alone [data not shown]) localizes at the entire plasma membrane upon TRH stimulation as did the native enzyme. These observations demonstrated that PKCβ1 plays an essential role in the initiation and regulation of PKCα translocation to the cell-cell contact.
FIG. 5.
PKCα and -ɛ translocation depends on PKCβ1 activity. CA PKCβ1-RFP was first cotransfected with PKCα-GFP in GH3B6 cells, and observations were performed 48 h later. (A) PKCα-GFP still accumulated at cell-cell contacts in the presence of CA PKCβ1, which bypasses receptor activation and therefore does not induce any increase in intracellular Ca2+ and DAG concentrations. Observations were performed with a conventional microscope. A video in the supplemental material shows that CA PKCβ1-GFP oscillates spontaneously, without TRH, between the cytoplasm and plasma membrane. In the presence of DN PKCβ1-RFP (B) and in the presence of 100 nM TRH, PKCα-GFP remained in the cytoplasm of most cells or accumulated at the entire plasma membrane in 9/26 cells, as did PKCɛ (C). PKCδ translocation was not affected by the cotransfection of DN PKCβ1-RFP (D). All the data presented in panels A to D were obtained 48 h after the cotransfection by real-time recordings. (E) Summary of the total number of cells analyzed in each situation and the subcellular localizations of the different GFP fusion proteins. As previously shown by others for DN PKCγ and -δ (49), we found that in the presence of 100 nM TRH, DN PKCβ1 accumulated at the same site as the native enzyme, namely, at the entire plasma membrane (B, C, and D), while the CA PKCβ1 keeps oscillating between the cytoplasm and plasma membrane (see the video in the supplemental material). In each experiment, it was checked (n = 5 cells for each PKC) that PKCα, PKCɛ, and PKCδ-GFP transfected alone were localized exclusively at cell-cell contacts.
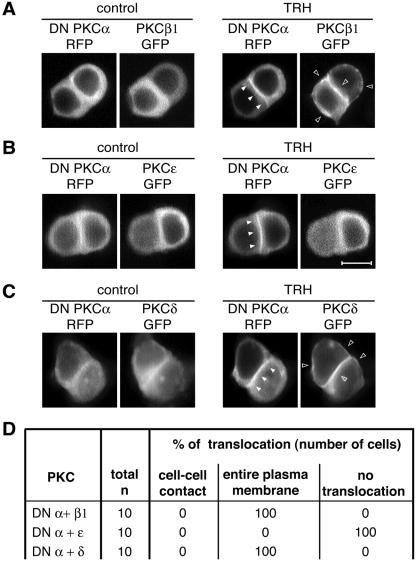
(ii) PKCɛ, but not PKCδ, translocation depends on PKCβ1 and PKCα activities.
We then went on to investigate the roles of PKCβ1 and -α in the control of PKCɛ localization. Similar to DN PKCβ1, which localized at the same subcellular sites as the native enzyme, DN PKCα-RFP (or DN PKCα-GFP alone [data not shown]) translocated, as did native PKCα, to the cell-cell contact upon TRH stimulation (Fig. 6A). In cells transfected with either DN PKCβ1-RFP or DN PKCα-RFP (Fig. 5C and 6B), the TRH-induced PKCɛ targeting to the cell-cell contact was abolished and PKCɛ remained in the cytoplasm. The fact that the DN PKCα was sufficient to induce this effect indicates that PKCα is essential to the control of PKCɛ localization at the cell-cell contact. However, in contrast to PKCɛ, PKCδ was not found to be a part of the cascade controlled by PKCβ1, since neither the expression of DN PKCβ1-RFP nor that of DN PKCα-RFP prevented the TRH-induced PKCδ-GFP translocation (Fig. 5D and 6C).
FIG. 6.
PKCɛ translocation depends on PKCα activities. When cotransfected with DN PKCα-RFP, PKCɛ-GFP remained in the cytoplasm of all the TRH-stimulated cells analyzed (B). Under the same conditions, PKCβ1 and -δ translocation were not affected (A and C). (D) Summary of the total number of cells analyzed in each situation and the subcellular localizations of the different GFP fusion proteins. All the data presented in panels A, B, and C were obtained 48 h after the cotransfection by real-time recordings. In each experiment, it was checked (n = 5 cells for each PKC) that PKCβ1, PKCɛ, and PKCδ-GFP, transfected alone, were localized at cell-cell contacts for PKCɛ and at the entire membrane for PKCβ1 and -δ. Bar, 5 μm.
If PKCα were indeed activated following activation of PKCβ1, abolishing its activity should not affect PKCβ1 translocation. This was analyzed by cotransfecting DN PKCα-RFP with PKCβ1-GFP. Figure 6A shows that indeed PKCβ1-GFP translocation was not affected by the presence of the DN PKCα.
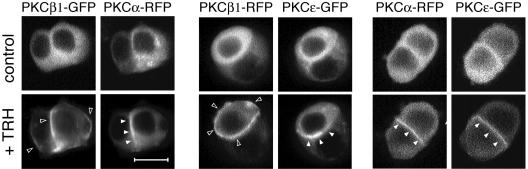
Finally, cotransfection experiments demonstrated that PKC isoforms implicated in the cascade do accumulate at their expected subcellular localizations in the same cell. Figure 7 shows that indeed, upon TRH stimulation, both PKCα and -ɛ accumulate at the cell-cell contact within the same cells and that PKCβ1 accumulates at the entire plasma membrane in cells where PKCα and -ɛ are present at the cell-cell contact.
FIG. 7.
The different subcellular compartment localizations of PKCα, -β1, and -ɛ occur within the same cells. Cotransfection experiments of PKCβ1 and PKCα subcloned in the pcDNA-RFP vector with either PKCβ- or PKCɛ-GFP indicated that PKCβ1 accumulated at the entire plasma membrane in cells where PKCα and -ɛ were translocated to cell-cell contacts. Cotransfection with PKCα-RFP and PKCɛ-GFP indicated that these two isoforms accumulated at the same subcellular location, the cell-cell contacts, within the same cells. Recordings were performed with a Noran confocal laser-scanning microscope equipped with an Ar/Kr laser (Odyssey XL with InterVision 1.4.1 software; Noran Instruments, Inc., Middleton, WI). Images were acquired with intervals of 17 s between the two frames. Bar, 5 μm.
Endogenous PKCs are also organized as a coordinated cascade.
In order to check that the organization of PKC signaling that we describe for the GFP-tagged enzymes reflects the physiological behavior of PKC isoforms in these cells and is not an artifact of the GFP tag or of the transient transfection, we checked that it also existed for the endogenous proteins. We performed immunocytochemistry experiments following stimulation of GH3B6 cells with TRH for 15 s, 60 s, or 5 min, these time points being chosen on the basis of the results of the time course experiments shown in Fig. 3. Figure 8 shows that indeed, after 15 s of stimulation, all endogenous PKCs are translocated either at the entire plasma membrane (PKCβ1 and -δ) or selectively at cell-cell contacts (PKCα and -ɛ). After 60 s of stimulation, only PKCβ1 and -ɛ remain translocated, and after 5 min, only PKCɛ is detected at cell-cell contacts. Thus, what we have shown for the GFP-tagged enzymes in transient-transfection experiments does reflect the behavior of the endogenous proteins.
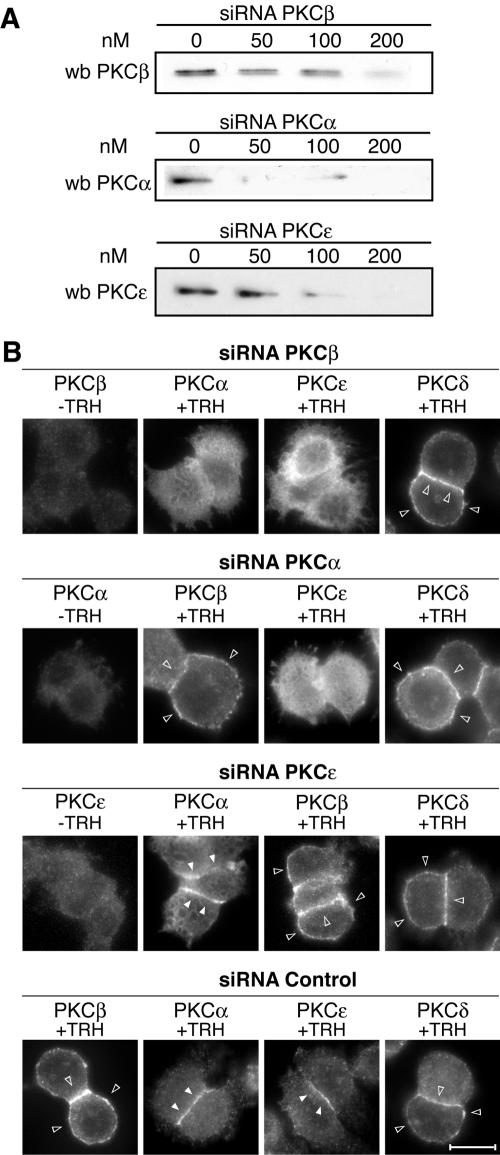
Then, RNA interference was used in order to knock down selectively the expression of PKCβ1, -α, and -ɛ, and the consequences were systematically analyzed on the translocation of endogenous PKCs. Figure 9A shows that all three siRNAs used were efficient in knocking down enzyme expression. Maximum efficacy was achieved at 200 nM, reaching 70%, 95%, and 97% inhibition for PKCβ1, -α, and -ɛ siRNAs, respectively. Figure 9B shows that knocking down PKCβ1 expression abolished the translocation of PKCα and -ɛ, but not that of PKCδ, after stimulation with TRH. In contrast, PKCα depletion affected PKCɛ translocation, but not translocation of PKCβ1 and -δ, and finally, knocking down PKCɛ expression did not have any effect on PKCβ1, PKCα, or PKCδ translocation. A scrambled siRNA was used as a control and did not affect translocation of any of the PKCs.
FIG. 9.
The sequential endogenous PKC activation is coordinated. By using siRNAs targeted to PKCβ1, -α, or -ɛ, we show that the sequential activation of endogenous PKCs is coordinated in the same way as it is for the GFP-tagged enzymes. (A) The efficacy of the siRNAs is dose dependent, and it is maximal for 200 nM. At this concentration, the siRNA targeted to PKCβ1 abolished translocation of PKCα and -ɛ but not that of PKCδ (B). The siRNA targeted to PKCα abolished translocation only of PKCɛ, and the siRNA targeted to PKCɛ did not affect the translocation of PKCβ1, -α, or -δ (B). wb, Western blot. Bar, 5 μm.
Thus, the data presented here argue in favor of a novel organization of endogenous PKC signaling into a coordinated cascade of activation in the pituitary GH3B6 cell line.
PKCα, but not -ɛ, is responsible for the TRH-induced β-catenin relocation.
In order to clarify the functional significance of such a coordinated cascade of PKC activation, we tried to establish whether PKC isoforms accumulating in the same location after stimulation would induce the same biological effect, implying that only the isoform situated in the most downstream position would act as the effector of the cascade. In order to answer this question, we analyzed the involvement of PKCα and -ɛ on the TRH-induced β-catenin relocation at cell-cell contacts previously reported for this cell line (51).
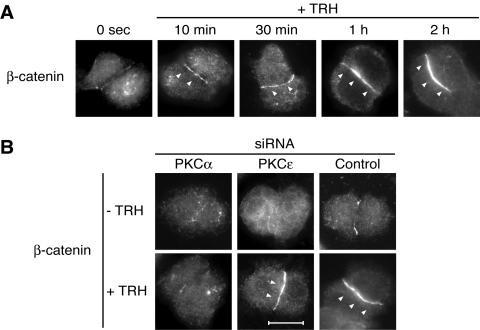
Figure 10A shows the time course of β-catenin relocation at cell-cell contacts upon TRH stimulation. Reorganization of β-catenin distribution was detected from 10 min after stimulation. After 30 min of stimulation, a more intense staining started to be homogenously distributed along the cell-cell contact, and after 1 h of stimulation, relocation of β-catenin was maximal, as shown by the very intense homogenous staining along cell-cell contacts. The involvement of PKCα and -ɛ in translocation was therefore studied after 1 h of TRH stimulation.
FIG. 10.
PKCα, but not PKCɛ, is responsible for the TRH-induced relocalization of β-catenin at cell-cell contacts. In order to clarify whether PKCα and -ɛ had distinct functions at the cell-cell contact, we analyzed the previously described TRH-induced β-catenin relocation to cell-cell contact (48) in the presence or absence of siRNAs targeted either to PKCα or to PKCɛ. (A) Relocation of β-catenin started as soon as 10 min after TRH stimulation, and the effect was maximal after 1 h of stimulation. The TRH-induced redistribution of β-catenin was abolished when cells were transfected with the siRNA targeted to PKCα, which was not the case in cells transfected with the siRNA targeted to PKCɛ (B). Bar, 5 μm.
Figure 10B shows the incidence of siRNA-mediated depletion of PKCα or PKCɛ on β-catenin relocation at cell-cell contacts. While the siRNA targeted against PKCα prevented the β-catenin relocation at cell-cell contact, the siRNA targeted to PKCɛ had no effect, showing that both isoforms are clearly capable of exerting independent biological functions despite their participation in a common activation cascade.
DISCUSSION
There are many examples of signaling cascades involving series of kinases, the canonical example of which are the mitogen-activated protein kinase cascades (7, 35). In these cascades, phosphorylation of one kinase by the other is mandatory for the activation of the former and differences in terms of kinetics and localization have been reported; these differences depend on whether the cascade is activated via β-arrestin or via G proteins (1). The PKC family of isoforms has never been considered up to now as potentially organized into a cascade, although the sequential activation of several isoforms has been observed upon physiological stimulation of living cells (3, 19, 25, 26, 31, 44, 46, 47, 49, 50, 53, 55). The results of the present study demonstrate the existence of such an organization and unravels a new regulatory mechanism, a coordinated cascade of isoform activation controlling localization at a specific subcellular location.
Today, translocation of a PKC (a cPKC or nPKC) is considered essential for its activation and is understood to be regulated by calcium and/or DAG. The two modules C1 and C2 are responsible for translocation events. Recognition of calcium by the C2 module, in the case of a cPKC, occurs in the cytoplasm, and an increase in calcium concentration seems sufficient to induce translocation. Activation of a cPKC is thus considered “phase-locked with calcium oscillations” (54). Concerning DAG dependence, PKCs are supposed to act as sensors for a concentration gradient, which is, for example, higher at the membrane after activation of a G-protein-coupled receptor. However, we have found some discordant data in this general scheme of calcium- and DAG-dependent PKC activation. The change in subcellular location of the mutated D294G PKCα enzyme was accompanied by phase locking with calcium, which was not the case for the wild-type enzyme. From that observation, we could already suspect that a calcium-independent translocation of PKCα was possible, in addition to the well-documented calcium-dependent one. The following results presented in the present study demonstrate that such a calcium-independent mechanism occurs. (i) PKCα does not translocate in the presence of ionomycin. (ii) PKCα D187/246/268N, which is calcium insensitive, still translocates selectively to cell-cell contacts. Furthermore, PKCβ1 D187/246/268N also translocates to the cell-cell contacts. The latter observation did puzzle us and suggests that a cPKC might be able to switch from a calcium-dependent to a calcium-independent mechanism and vice versa. Consistent with this, we observed a switch in the PKCα translocation site from cell-cell contacts to the entire plasma membrane in the presence of the cPKC inhibitor Gö6976. Thus, an alternative control of PKC translocation regulation exists, independent of calcium and DAG, and associated with the cell-cell contact targeting. In favor of the hypothesis that two regulatory mechanisms can control PKC translocation is the fact that within a given cell population, not all cells behave identically. Indeed, upon stimulation with TRH, PKCα and PKCɛ selectively accumulate at cell-cell contacts in 55% of the cells, but in a substantial fraction of the cells (20%), PKCα and PKCɛ are translocated to the entire plasma membrane like native PKCβ1 (Fig. 1C).
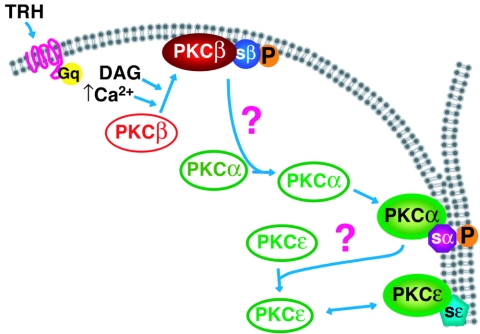
Several reports have documented the fact that activation of a membrane receptor, such as a G-protein-coupled receptor, may activate several PKC isoforms (3, 12, 44, 46, 50, 53). In these studies, the fact that all PKC isoforms involved translocated to the same subcellular location, the plasma membrane, did not suggest different, isoform-specific mechanisms of translocation. In the present work, we hypothesized that decoding the temporal pattern of activation, in addition to the spatial one, could be important to understand the organization of PKC signaling. Several reasons lead us to suggest that the cascade of activation could be coordinated. First, the fact that PKCα localization at cell-cell contacts was dependent on cPKC activity (the present study) but did not require its own kinase activity (52) and the fact that PKCβ1 was the first to be activated suggest that PKCα activation could be under PKCβ1 control. The results obtained in this work for both endogenous and GFP-tagged PKCs show that it is indeed the case and demonstrate that similarly, PKCɛ is under the control of PKCα. Thus, there is a close association between the control of localization and that of activation. However, the mechanism by which PKCβ1 controls the location of PKCα and the mechanism by which PKCα controls the location of PKCɛ are not known at present. Since an activated PKC isoform is not in the same subcellular compartment as an inactive PKC, the links between PKCβ1 and PKCα and between PKCα and PKCɛ are probably indirect. Also, since it is the activity of PKCβ1 and -α that controls cell-cell contact targeting of PKCα and -ɛ, respectively, then a substrate of PKCβ1 may serve as the link between PKCβ1 and PKCα, and a substrate for PKCα may serve as the link between PKCα and PKCɛ. These substrates would translocate from the membrane compartment to the cytoplasm upon phosphorylation. An example of such a substrate, able to translocate from the membrane to the cytoplasm upon phosphorylation by a PKC, was provided by the characterization of MARCKS (named MARCKS for myristoylated alanine-rich C-kinase substrate) (13, 31). These considerations led us to propose the hypothetical model presented in Fig. 11: upon stimulation of the Gq-coupled TRH receptor, intracellular Ca2+ and DAG concentrations increase and PKCβ1 is activated and accumulate at the plasma membrane, according to the currently accepted paradigm of PKC translocation/activation. Upon phosphorylation, a PKCβ1 substrate would then relocalize to the cytoplasm. This unknown substrate would, alone or in association with other partners, play the role of cargo to elicit the selective translocation of PKCα to the cell-cell contact. PKCα would in turn phosphorylate a substrate, itself mediating directly or indirectly PKCɛ translocation to the cell-cell contact. PKCδ does not belong to this cascade, since its translocation to the plasma membrane is not dependent on PKC activity. This hypothesis takes into account the current “dogma,” since PKCβ1 is calcium and DAG regulated and is the isoform that initiates the cascade. However, it also introduces the concept that without this coordinated cascade of activation, and without the existence of a calcium-independent but PKC activity-dependent control of PKC translocation, PKCα and -ɛ would not be targeted to their site of action. In a recent work, Denis and Cyert highlighted the complex regulation of the localization of the unique PKC isozyme present in Saccharomyces cerevisiae, Pkc1p (11). In particular, it was shown that functional Pkc1p activity is essential for its proper targeting, an observation which is in line with our results demonstrating that the cell-cell contact selective targeting of PKCα and -ɛ is dependent on PKC activity.
FIG. 11.
Schematic representation of the coordinated cascade of PKCβ1, -α, and -ɛ activation. Upon stimulation of the Gq-coupled TRH receptor, intracellular Ca2+ and DAG concentrations increase and PKCβ1 would be activated according to the currently accepted paradigm of PKC translocation/activation. It accumulates at the plasma membrane where it would phosphorylate a substrate that would relocalize to the cytoplasm, as already shown for example in the case of MARCKS (31). The unknown PKCβ1 substrate would elicit directly or indirectly the selective translocation of PKCα to the cell-cell contact. PKCα would in turn phosphorylate a substrate that would mediate directly or indirectly PKCɛ translocation to the cell-cell contact. PKCδ would not enter in this scheme, as its translocation to the plasma membrane is not dependent on PKC activity. sβ, PKCβ1 substrate; sα, PKCα substrate; sɛ, PKCɛ substrate.
Decoding the sequential and coordinated activation of PKCs has evidenced that PKCs display different activation profiles, in regards to the time of onset and the duration of activation, which are both unrelated to the subclass the isoform belongs to or to the subcellular targeting site. The various durations of activation could be interpreted in terms of different interaction modes at the membrane, either with a protein or with membrane phospholipids, and/or to the different phosphorylation status of each PKC isoform. Indeed, PKCs are known to interact with many partners (39), some of which are adaptor proteins able to anchor specific isoforms to selective subcellular locations and thus to maintain each PKC isoform next to a subset of proteins and away from the other substrates (29, 30, 48). A recent example of this has been provided by Park et al. who show that RACK1 (named RACK1 for receptor for activated C kinase I) anchors PKCβ on melanosomes where it phosphorylates tyrosinase (36). In PKCɛ-induced heart failure, PKCβ2-RACK1 interactions are enhanced (37). AKAPs (A-kinase anchoring proteins) are also important interacting proteins for PKC signaling (6), able for instance to associate with cadherin adhesion molecules (16). Other proteins also play the role of anchor for selective PKCs in selective subcellular sites, such as pericentrin for PKCβ2 in the centrosome (8). In GH3B6 cells, we know that RACK1 does not account for PKCα or PKCɛ cell-cell contact location, since RACK1 immunoreactivity can be evidenced at the plasma membrane but is excluded from the cell-cell contact (51). A search for the PKCα and -ɛ partners expressed in the GH3B6 cell line is currently under way. The various onsets and durations of activation could also be due to the phosphorylation state of each isoform. Concerning the onset of activation, if the cPKC isoform phosphorylation status does not seem to be involved, this might not be the case for the nPKCs isoform status. Under the usual experimental conditions, i.e., in the presence of serum, cPKC isoforms already bear the three priming phosphorylations necessary for their activation. This is in agreement with our previous observations showing that the regulatory domain of PKCα, which does not contain any of the three amino acids involved in the priming phosphorylations, is still able to accumulate at cell-cell contacts at a rate similar to that of PKCα (52). In the case of the nPKCs, these phosphorylations are completed together with the activation process and they can involve cross regulation between the nPKCs themselves (45). Interestingly, it has been shown that integrin engagement can induce PKCɛ phosphorylation (21). It is also known that the phosphorylation of the activation loop of nPKCs is involved in the duration of activation, since the concerned isoform remains activated for a longer time when phosphorylation is prevented by targeted mutation (15, 49).
The coordinated cascade of PKC isoform activation revealed in the present study could be a hallmark of differentiated secreting pituitary cells. Although its function is yet unknown, targeting of selective PKC isoforms at cell-cell contacts has been shown to be functionally important in other cellular systems. Indeed, in a coculture of tumoral epithelial cells and healthy fibroblasts, PKCα and -ɛ strictly accumulate at the heterologous cell-cell contact. In this case, the establishment of these cell-cell contacts and the activation of PKCα and -ɛ are essential for the production of matrix metalloproteinase stromelysin-3 by the fibroblasts (27). In the macrophage-like cell line RAW 264.7, PKCα is targeted selectively at the site of phagocytosis induced by contact with beads covered with immunoglobulin G (24). PKCθ has also been shown to selectively localize at the immunologic synapse, which is the heterologous contact between the T cell and the antigen-presenting cell (32, 33). This selective PKCθ localization is dependent upon CD28 expression and is involved in T-cell activation (20). In the GH3B6 cell line, TRH is known to regulate hormone secretion and proliferation, but it does not have any well-established functions at cell-cell contacts. PKCα and -ɛ at cell-cell contacts may not serve the hormonal secretion, since it has been shown in primary cultured isolated lactotrophs that TRH elicits secretion (9). Indeed, in these isolated cells, PKCα and -ɛ activation is not expected, since we show (Fig. 1) that, in the presence of TRH, PKCα and -ɛ are not translocated, and thus not activated, in isolated pituitary GH3B6 cells. Rather, results from our group suggest that the selective targeting of PKCα and -ɛ to the cell-cell contact may regulate adherent junctions, since TRH was found to induce the relocalization of β-catenin at the cell-cell contact (51). In the present study, we show that PKCα is involved in the relocalization of β-catenin at cell-cell contacts, whereas PKCɛ is not. This suggests that although they are mechanistically associated into a cascade of activation and translocated to the same location after stimulation, each of these PKC isoforms might exert one or several specific functions. Furthermore, these results demonstrate that PKCɛ clearly does not behave as an “end-point” kinase and is not the unique functional effector of this activation cascade.
However, the fact that PKC signaling is organized into a coordinated cascade implies that knocking down PKCβ1 would affect the biological effects directly related to this isoform but also those connected to the isoforms located further downstream in the cascade, namely, PKCα and -ɛ. This also renders more complex the interpretation of the various phenotypes obtained in PKC isoform-specific knockout mice by questioning to which extent one can attribute a particular phenotype to a particular isoform.
In conclusion, signaling by the PKC family constitutes a good example of a space/time/isoform-determined process. It is capable of self-organization in order to achieve an appropriate spatiotemporal control of localization and activation of selective isoforms.
Supplementary Material
Acknowledgments
We thank Philippe Jay and Catherine Legraverend (IGF, Montpellier, France) and Daniel Fisher (IGMM, Montpellier, France) for their helpful criticisms.
This work was supported by the Ministère de la Recherche et de la Technologie, by the Association pour la Recherche contre le Cancer (ARC) (grant no. 5695). Barthélémy Diouf was supported by a grant from the ARC. Marion Peter was a recipient of a Long-Term Fellowship from the International Human Frontier Science Program Organization.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ahn, S., S. K. Shenoy, H. Wei, and R. J. Lefkowitz. 2004. Differential kinetic and spatial patterns of beta-arrestin and G protein-mediated ERK activation by the angiotensin II receptor. J. Biol. Chem. 279:35518-35525. [DOI] [PubMed] [Google Scholar]
- 2.Alvaro, V., L. Levy, C. Dubray, A. Roche, F. Peillon, B. Querat, and D. Joubert. 1993. Invasive human pituitary tumors express a point-mutated alpha-protein kinase-C. J. Clin. Endocrinol. Metab. 77:1125-1129. [DOI] [PubMed] [Google Scholar]
- 3.Babwah, A. V., L. B. Dale, and S. S. Ferguson. 2003. Protein kinase C isoform-specific differences in the spatial-temporal regulation and decoding of metabotropic glutamate receptor 1a-stimulated second messenger responses. J. Biol. Chem. 278:5419-5426. [DOI] [PubMed] [Google Scholar]
- 4.Brancho, D., J. J. Ventura, A. Jaeschke, B. Doran, R. A. Flavell, and R. J. Davis. 2005. Role of MLK3 in the regulation of mitogen-activated protein kinase signaling cascades. Mol. Cell. Biol. 25:3670-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, D. C., Y. Cao, S. Wang, S. H. Garfield, G. M. Hur, and P. M. Blumberg. 2005. Role of phorbol ester localization in determining protein kinase C or RasGRP3 translocation: real-time analysis using fluorescent ligands and proteins. Mol. Cancer Ther. 4:141-150. [PubMed] [Google Scholar]
- 6.Carnegie, G. K., F. D. Smith, G. McConnachie, L. K. Langeberg, and J. D. Scott. 2004. AKAP-Lbc nucleates a protein kinase D activation scaffold. Mol. Cell 15:889-899. [DOI] [PubMed] [Google Scholar]
- 7.Chang, L., and M. Karin. 2001. Mammalian MAP kinase signalling cascades. Nature 410:37-40. [DOI] [PubMed] [Google Scholar]
- 8.Chen, D., A. Purohit, E. Halilovic, S. J. Doxsey, and A. C. Newton. 2004. Centrosomal anchoring of protein kinase C betaII by pericentrin controls microtubule organization, spindle function, and cytokinesis. J. Biol. Chem. 279:4829-4839. [DOI] [PubMed] [Google Scholar]
- 9.Cochilla, A. J., J. K. Angleson, and W. J. Betz. 2000. Differential regulation of granule-to-granule and granule-to-plasma membrane fusion during secretion from rat pituitary lactotrophs. J. Cell Biol. 150:839-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corbalan-Garcia, S., J. A. Rodriguez-Alfaro, and J. C. Gomez-Fernandez. 1999. Determination of the calcium-binding sites of the C2 domain of protein kinase Calpha that are critical for its translocation to the plasma membrane. Biochem. J. 337:513-521. [PMC free article] [PubMed] [Google Scholar]
- 11.Denis, V., and M. S. Cyert. 2005. Molecular analysis reveals localization of Saccharomyces cerevisiae protein kinase C to sites of polarized growth and Pkc1p targeting to the nucleus and mitotic spindle. Eukaryot. Cell 4:36-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Disatnik, M. H., S. C. Boutet, C. H. Lee, D. Mochly-Rosen, and T. A. Rando. 2002. Sequential activation of individual PKC isozymes in integrin-mediated muscle cell spreading: a role for MARCKS in an integrin signaling pathway. J. Cell Sci. 115:2151-2163. [DOI] [PubMed] [Google Scholar]
- 13.Disatnik, M. H., S. C. Boutet, W. Pacio, A. Y. Chan, L. B. Ross, C. H. Lee, and T. A. Rando. 2004. The bi-directional translocation of MARCKS between membrane and cytosol regulates integrin-mediated muscle cell spreading. J. Cell Sci. 117:4469-4479. [DOI] [PubMed] [Google Scholar]
- 14.Eliyahu, E., and R. Shalgi. 2002. A role for protein kinase C during rat egg activation. Biol. Reprod. 67:189-195. [DOI] [PubMed] [Google Scholar]
- 15.Feng, X., and Y. A. Hannun. 1998. An essential role for autophosphorylation in the dissociation of activated protein kinase C from the plasma membrane. J. Biol. Chem. 273:26870-26874. [DOI] [PubMed] [Google Scholar]
- 16.Gorski, J. A., L. L. Gomez, J. D. Scott, and M. L. Dell'acqua. 2005. Association of an A-kinase-anchoring protein signaling scaffold with cadherin adhesion molecules in neurons and epithelial cells. Mol. Biol Cell. 16:3574-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gschwendt, M., S. Dieterich, J. Rennecke, W. Kittstein, H. J. Mueller, and F. J. Johannes. 1996. Inhibition of protein kinase C mu by various inhibitors. Differentiation from protein kinase c isoenzymes. FEBS Lett. 392:77-80. [DOI] [PubMed] [Google Scholar]
- 18.Guerineau, N. C., X. Bonnefont, L. Stoeckel, and P. Mollard. 1998. Synchronized spontaneous Ca2+ transients in acute anterior pituitary slices. J. Biol. Chem. 273:10389-10395. [DOI] [PubMed] [Google Scholar]
- 19.Halet, G. 2004. PKC signaling at fertilization in mammalian eggs. Biochim. Biophys. Acta 1742:185-189. [DOI] [PubMed] [Google Scholar]
- 20.Huang, J., P. F. Lo, T. Zal, N. R. Gascoigne, B. A. Smith, S. D. Levin, and H. M. Grey. 2002. CD28 plays a critical role in the segregation of PKC theta within the immunologic synapse. Proc. Natl. Acad. Sci. USA 99:9369-9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivaska, J., L. Bosca, and P. J. Parker. 2003. PKCepsilon is a permissive link in integrin-dependent IFN-gamma signalling that facilitates JAK phosphorylation of STAT1. Nat. Cell Biol. 5:363-369. [DOI] [PubMed] [Google Scholar]
- 22.Jenster, G. 2004. A visualisation concept of dynamic signalling networks. Mol. Cell. Endocrinol. 218:1-6. [DOI] [PubMed] [Google Scholar]
- 23.Kohout, S. C., S. Corbalan-Garcia, A. Torrecillas, J. C. Gomez-Fernandez, and J. J. Falke. 2002. C2 domains of protein kinase C isoforms alpha, beta, and gamma: activation parameters and calcium stoichiometries of the membrane-bound state. Biochemistry 41:11411-11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsen, E. C., J. A. DiGennaro, N. Saito, S. Mehta, D. J. Loegering, J. E. Mazurkiewicz, and M. R. Lennartz. 2000. Differential requirement for classic and novel PKC isoforms in respiratory burst and phagocytosis in RAW 264.7 cells. J. Immunol. 165:2809-2817. [DOI] [PubMed] [Google Scholar]
- 25.Lenz, J. C., H. P. Reusch, N. Albrecht, G. Schultz, and M. Schaefer. 2002. Ca2+-controlled competitive diacylglycerol binding of protein kinase C isoenzymes in living cells. J. Cell Biol. 159:291-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Looby, E., A. Long, D. Kelleher, and Y. Volkov. 2005. Bile acid deoxycholate induces differential subcellular localisation of the PKC isoenzymes beta 1, epsilon and delta in colonic epithelial cells in a sodium butyrate insensitive manner. Int. J. Cancer 114:887-895. [DOI] [PubMed] [Google Scholar]
- 27.Louis, K., N. Guerineau, O. Fromigue, V. Defamie, A. Collazos, P. Anglard, M. A. Shipp, P. Auberger, D. Joubert, and B. Mari. 2005. Tumor cell-mediated induction of the stromal factor stromelysin-3 requires heterotypic cell contact-dependent activation of specific protein kinase C isoforms. J. Biol. Chem. 280:1272-1283. [DOI] [PubMed] [Google Scholar]
- 28.Martiny-Baron, G., M. G. Kazanietz, H. Mischak, P. M. Blumberg, G. Kochs, H. Hug, D. Marme, and C. Schachtele. 1993. Selective inhibition of protein kinase C isozymes by the indolocarbazole Go 6976. J. Biol. Chem. 268:9194-9197. [PubMed] [Google Scholar]
- 29.Mochly-Rosen, D. 1995. Localization of protein kinases by anchoring proteins: a theme in signal transduction. Science 268:247-251. [DOI] [PubMed] [Google Scholar]
- 30.Mochly-Rosen, D., B. L. Smith, C. H. Chen, M. H. Disatnik, and D. Ron. 1995. Interaction of protein kinase C with RACK1, a receptor for activated C-kinase: a role in beta protein kinase C mediated signal transduction. Biochem. Soc. Trans. 23:596-600. [DOI] [PubMed] [Google Scholar]
- 31.Mogami, H., H. Zhang, Y. Suzuki, T. Urano, N. Saito, I. Kojima, and O. H. Petersen. 2003. Decoding of short-lived Ca2+ influx signals into long term substrate phosphorylation through activation of two distinct classes of protein kinase C. J. Biol. Chem. 278:9896-9904. [DOI] [PubMed] [Google Scholar]
- 32.Monks, C. R., B. A. Freiberg, H. Kupfer, N. Sciaky, and A. Kupfer. 1998. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature 395:82-86. [DOI] [PubMed] [Google Scholar]
- 33.Monks, C. R., H. Kupfer, I. Tamir, A. Barlow, and A. Kupfer. 1997. Selective modulation of protein kinase C-theta during T-cell activation. Nature 385:83-86. [DOI] [PubMed] [Google Scholar]
- 34.Oancea, E., and T. Meyer. 1998. Protein kinase C as a molecular machine for decoding calcium and diacylglycerol signals. Cell 95:307-318. [DOI] [PubMed] [Google Scholar]
- 35.Olson, J. M., and A. R. Hallahan. 2004. p38 MAP kinase: a convergence point in cancer therapy. Trends Mol. Med. 10:125-129. [DOI] [PubMed] [Google Scholar]
- 36.Park, H. Y., H. Wu, C. E. Killoran, and B. A. Gilchrest. 2004. The receptor for activated C-kinase-I (RACK-I) anchors activated PKC-beta on melanosomes. J. Cell Sci. 117:3659-3668. [DOI] [PubMed] [Google Scholar]
- 37.Pass, J. M., J. Gao, W. K. Jones, W. B. Wead, X. Wu, J. Zhang, C. P. Baines, R. Bolli, Y. T. Zheng, I. G. Joshua, and P. Ping. 2001. Enhanced PKC beta II translocation and PKC beta II-RACK1 interactions in PKC epsilon-induced heart failure: a role for RACK1. Am. J. Physiol. Heart Circ. Physiol. 281:H2500-H2510. [DOI] [PubMed] [Google Scholar]
- 38.Peter, M., S. M. Ameer-Beg, M. K. Hughes, M. D. Keppler, S. Prag, M. Marsh, B. Vojnovic, and T. Ng. 2005. Multiphoton-FLIM quantification of the EGFP-mRFP1 FRET pair for localization of membrane receptor-kinase interactions. Biophys. J. 88:1224-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poole, A. W., G. Pula, I. Hers, D. Crosby, and M. L. Jones. 2004. PKC-interacting proteins: from function to pharmacology. Trends Pharmacol. Sci. 25:528-535. [DOI] [PubMed] [Google Scholar]
- 40.Prevostel, C., V. Alvaro, F. de Boisvilliers, A. Martin, C. Jaffiol, and D. Joubert. 1995. The natural protein kinase C alpha mutant is present in human thyroid neoplasms. Oncogene 11:669-674. [PubMed] [Google Scholar]
- 41.Prevostel, C., V. Alvaro, A. Vallentin, A. Martin, S. Jaken, and D. Joubert. 1998. Selective loss of substrate recognition induced by the tumour-associated D294G point mutation in protein kinase Calpha. Biochem. J. 334:393-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qatsha, K. A., C. Rudolph, D. Marme, C. Schachtele, and W. S. May. 1993. Go 6976, a selective inhibitor of protein kinase C, is a potent antagonist of human immunodeficiency virus 1 induction from latent/low-level-producing reservoir cells in vitro. Proc. Natl. Acad. Sci. USA 90:4674-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quittau-Prevostel, C., N. Delaunay, A. Collazos, A. Vallentin, and D. Joubert. 2004. Targeting of PKCalpha and epsilon in the pituitary: a highly regulated mechanism involving a GD(E)E motif of the V3 region. J. Cell Sci. 117:63-72. [DOI] [PubMed] [Google Scholar]
- 44.Raghunath, A., M. Ling, and C. Larsson. 2003. The catalytic domain limits the translocation of protein kinase C alpha in response to increases in Ca2+ and diacylglycerol. Biochem. J. 370:901-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rybin, V. O., A. Sabri, J. Short, J. C. Braz, J. D. Molkentin, and S. F. Steinberg. 2003. Cross-regulation of novel protein kinase C (PKC) isoform function in cardiomyocytes. Role of PKC epsilon in activation loop phosphorylations and PKC delta in hydrophobic motif phosphorylations. J. Biol. Chem. 278:14555-14564. [DOI] [PubMed] [Google Scholar]
- 46.Schaefer, M., N. Albrecht, T. Hofmann, T. Gudermann, and G. Schultz. 2001. Diffusion-limited translocation mechanism of protein kinase C isotypes. FASEB J. 15:1634-1636. [DOI] [PubMed] [Google Scholar]
- 47.Schaefer, M., H. Mischak, S. Schnell, A. Griese, R. Iakubov, G. Riepenhausen, and C. Schofl. 2004. Mechanisms of arginine-vasopressin-induced Ca2+ oscillations in beta-cells (HIT-T15): a role for oscillating protein kinase C. Endocrinology 145:4635-4644. [DOI] [PubMed] [Google Scholar]
- 48.Schechtman, D., and D. Mochly-Rosen. 2001. Adaptor proteins in protein kinase C-mediated signal transduction. Oncogene 20:6339-6347. [DOI] [PubMed] [Google Scholar]
- 49.Seki, T., H. Matsubayashi, T. Amano, Y. Shirai, N. Saito, and N. Sakai. 2005. Phosphorylation of PKC activation loop plays an important role in receptor-mediated translocation of PKC. Genes Cells 10:225-239. [DOI] [PubMed] [Google Scholar]
- 50.Uchino, M., N. Sakai, K. Kashiwagi, Y. Shirai, Y. Shinohara, K. Hirose, M. Iino, T. Yamamura, and N. Saito. 2004. Isoform-specific phosphorylation of metabotropic glutamate receptor 5 by protein kinase C (PKC) blocks Ca2+ oscillation and oscillatory translocation of Ca2+-dependent PKC. J. Biol. Chem. 279:2254-2261. [DOI] [PubMed] [Google Scholar]
- 51.Vallentin, A., T. C. Lo, and D. Joubert. 2001. A single point mutation in the V3 region affects protein kinase Cα targeting and accumulation at cell-cell contacts. Mol. Cell. Biol. 21:3351-3363.11313461 [Google Scholar]
- 52.Vallentin, A., C. Prevostel, T. Fauquier, X. Bonnefont, and D. Joubert. 2000. Membrane targeting and cytoplasmic sequestration in the spatiotemporal localization of human protein kinase C alpha. J. Biol. Chem. 275:6014-6021. [DOI] [PubMed] [Google Scholar]
- 53.van Baal, J., J. de Widt, N. Divecha, and W. J. van Blitterswijk. 2005. Translocation of diacylglycerol kinase theta from cytosol to plasma membrane in response to activation of G protein-coupled receptors and protein kinase C. J. Biol. Chem. 280:9870-9878. [DOI] [PubMed] [Google Scholar]
- 54.Violin, J. D., J. Zhang, R. Y. Tsien, and A. C. Newton. 2003. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J. Cell Biol. 161:899-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, Q. J., G. Lu, W. A. Schlapkohl, A. Goerke, C. Larsson, H. Mischak, P. M. Blumberg, and J. F. Mushinski. 2004. The V5 domain of protein kinase C plays a critical role in determining the isoform-specific localization, translocation, and biological function of protein kinase C-delta and -epsilon. Mol. Cancer Res. 2:129-140. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.