Abstract
Faithful chromosome segregation depends on the opposing activities of the budding yeast Glc7/PP1 protein phosphatase and Ipl1/Aurora protein kinase. We explored the relationship between Glc7 and Ipl1 and found that the phosphorylation of the Ipl1 substrate, Dam1, was altered by decreased Glc7 activity, whereas Ipl1 levels, localization, and kinase activity were not. These data strongly suggest that Glc7 ensures accurate chromosome segregation by dephosphorylating Ipl1 targets rather than regulating the Ipl1 kinase. To identify potential Glc7 and Ipl1 substrates, we isolated ipl1-321 dosage suppressors. Seven genes (SDS22, BUD14, GIP3, GIP4, SOL1, SOL2, and PEX31) encode newly identified ipl1 dosage suppressors, and all 10 suppressors encode proteins that physically interact with Glc7. The overexpression of the Gip3 and Gip4 suppressors altered Glc7 localization, indicating they are previously unidentified Glc7 regulatory subunits. In addition, the overexpression of Gip3 and Gip4 from the galactose promoter restored Dam1 phosphorylation in ipl1-321 mutant cells and caused wild-type cells to arrest in metaphase with unsegregated chromosomes, suggesting that Gip3 and Gip4 overexpression impairs Glc7's mitotic functions. We therefore propose that the overexpression of Glc7 regulatory subunits can titrate Glc7 away from relevant Ipl1 targets and thereby suppress ipl1-321 cells by restoring the balance of phosphatase/kinase activity.
The accurate partitioning of the genome during mitosis requires the precise regulation of the connection between chromosomes and the mitotic spindle. This fundamental interaction is mediated by the kinetochore, a specialized protein complex that assembles on centromeric DNA and facilitates the capture of dynamic spindle microtubules that arise from opposite poles (for reviews, see references 5, 13, and 17). Bipolar attachments promote accurate chromosome segregation by ensuring that the spindle forces on the replicated chromosomes (sister chromatids) are directed toward opposite sides of the cell. Once all chromosomes make proper bipolar attachments, the cell transitions to anaphase where sister chromatids are pulled to opposite poles. Failure to achieve bipolar attachments results in chromosome missegregation, and this aneuploid state predisposes multicellular organisms to the development of a variety of diseases. To prevent the premature segregation of improperly attached chromosomes, the spindle checkpoint monitors kinetochore-microtubule interactions and delays the metaphase to anaphase transition until bipolar attachments are achieved (for a review, see reference 42).
An important regulator of both kinetochore attachment and the spindle checkpoint is the conserved Ipl1/Aurora B protein kinase, a component of the chromosomal passenger complex that localizes to kinetochores, spindles, and the spindle midzone and midbody (for reviews, see references 25 and 69). Ipl1/Aurora B facilitates proper attachments by destabilizing inappropriate kinetochore-microtubule interactions, such as monopolar attachments in which kinetochores bind microtubules emanating from the same pole (4, 12, 39, 54, 63). Despite the presence of improper attachments that should activate the spindle checkpoint, cells with impaired Ipl1/Aurora B function proceed through the cell cycle (3, 10, 19, 26, 40). Ipl1 is thought to promote proper chromosome segregation, in part, by phosphorylating components of the Dam1/DASH/DDD complex, an essential regulator of kinetochore-microtubule interactions and microtubule function (15, 16, 34, 35, 43, 44, 48, 59, 73).
Ipl1 activity is opposed by Glc7, the sole essential protein phosphatase 1 (PP1) catalytic subunit in budding yeast (21, 22, 32, 58, 76). Glc7 regulates numerous cellular processes including mitosis, meiosis, glycogen and sugar metabolism, transcription, translation, and mRNA processing (for a review, see reference 11). The regulation of these processes is guided by Glc7 interactions with specific regulatory subunits that target the phosphatase to appropriate substrates. Many glc7 alleles cause cells to arrest in mitosis (1, 6, 29, 46), suggesting that Glc7 substrates must be dephosphorylated to allow cell cycle progression. Furthermore, impairing Glc7 function suppresses the ipl1 temperature-sensitive growth defect and restores the phosphorylation of the Ipl1 targets Ndc10 and histone H3, indicating that Glc7 antagonizes Ipl1-mediated phosphorylation (21, 22, 32, 58). In addition, genetic interactions between glc7 mutants and mutants that alter the phosphorylation status of the Ipl1 substrate Dam1 also support this idea (15, 76). Consistent with this, some glc7 mutants activate the spindle checkpoint and exhibit reduced kinetochore binding to microtubules in vitro (7, 58). Despite these observations, the precise relationship between the kinase and phosphatase is not well understood, and Glc7 regulation of Ipl1 function has not been examined.
Here, we further explore the relationship between Ipl1 and Glc7. We found that Glc7 does not appear to directly modulate Ipl1 and likely opposes the essential functions of Ipl1 by dephosphorylating common substrates. We identified proteins that physically interact with Glc7 as dosage suppressors of an ipl1 mutant and found that two of these proteins, Gip3 and Gip4, are previously unidentified Glc7 regulatory subunits. Consistent with this, phosphorylation of the essential Ipl1 substrate, Dam1, is restored in ipl1 mutant cells when Glc7 is relocalized out of the nucleus by Gip3 and Gip4 overexpression. We propose that Glc7 regulatory subunits restore the kinase/phosphatase balance in ipl1 mutants by titrating Glc7 away from essential mitotic substrates.
MATERIALS AND METHODS
Microbial techniques and yeast strain construction.
Media and microbial techniques were essentially as previously described (57). Nocodazole was used at 10 μg/ml. Galactose was added to a final concentration of 4%. Yeast strains are listed in Table 1 and were constructed by standard genetic techniques. The glc7-10 (58) and glc7-12 (46) (gifts from Michael Stark, University of Dundee, Dundee, United Kingdom) and ipl1-321 (4) alleles were crossed to make strains for this study. Strains containing TUB1-CFP::URA3 were obtained by integrating plasmid pSB375 (a gift from Kerry Bloom, University of North Carolina, Chapel Hill, NC) digested with StuI at the URA3 locus. GLC7-GFP3 strains were made by integration of plasmid pSB881 digested with EcoRI at the GLC7 locus. Insertion of the pGAL promoter and the HA3, myc13, and FLAG3 epitope tags and construction of gip3 and gip4 deletion strains were made using a PCR-based integration system (45) and were confirmed by PCR. Specific primer sequences are available upon request. All fusion proteins are fully functional.
TABLE 1.
Yeast strains used in this studya
| Strain | Genotype |
|---|---|
| SBY3 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ |
| SBY214 | MATaura3-1 leu2,3-112 his3-11::pCUP1-GFP12-lacI12::HIS3 trp1-1::256lacO::TRP1 lys2Δ ade2-1 can1-100 bar1Δ |
| SBY322 | MATaura3-1 leu2,3-112 his3-11::pCUP1-GFP12-lacI12::HIS3 trp1-1::256lacO::TRP1 lys2Δ ade2-1 can1-100 bar1Δ ipl1-321 |
| SBY625 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ GLC7-HA3::HIS3 |
| SBY818 | MATaura3-1 leu2,3-112 his3-11::pCUP1-GFP12-lacI12::HIS3 trp1-1::256lacO::TRP1 lys2Δ ade2-1 can1-100 bar1Δ PDS1-myc18::LEU2 |
| SBY1063 | MATaura3-1 leu2,3-112::pGAL-ipl1(R343A)::LEU2 his3-11::pCUP1-GFP12-lacI12::HIS3 trp1-1::256lacO::TRP1 lys2Δ ade2-1 can1-100 bar1Δ ipl1-321 |
| SBY1258 | MATaura3-1 leu2,3-112 his3-11 trp1-1::glc7-10::TRP1 ade2-1 can1-100 glc7::LEU2 ipl1ΔKAN::ipl1T260A::LEU2 |
| SBY1264 | MATaura3-1 leu2,3-112 his3-11 trp1-1::GLC7::TRP1 ade2-1 can1-100 glc7::LEU2 ipl1ΔKAN::ipl1T260A::LEU2 |
| SBY1306 | MATaura3-1 leu2,3-112 his3-11 trp1-1::glc7-10::TRP1 ade2-1 can1-100 glc7::LEU2 bar1Δ |
| SBY1994 | MATaura3-1 leu2,3-112 his3-11 trp1-1::glc7-10::TRP1 ade2-1 can1-100 bar1Δ glc7::LEU2 ipl1-321 |
| SBY2055 | MATaura3-1 leu2,3-112 his3-11 trp1-1 lys2Δ ade2-1 can1-100 bar1Δ DAM1-myc9::TRP1 |
| SBY2833 | MATaura3-1::TUB1-CFP::URA3 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ IPL1-GFP3::HIS3 |
| SBY3672 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ IPL11-FLAG3::KAN |
| SBY3675 | MATaura3-1 leu2,3-112 his3-11 trp1-1::glc7-12::TRP1 ade2-1 can1-100 bar1Δ glc7::LEU2 |
| SBY4114 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ gip3ΔKAN |
| SBY4175 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ gip4ΔKAN |
| SBY4179 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ pGAL-HA3-GIP3::HIS3 |
| SBY4209 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ GLC7-HA3::HIS3 GIP4-myc13::KAN |
| SBY4292 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 lys2Δ can1-100 bar1Δ pGAL-HA3-GIP4::HIS3 PDS1-myc18::LEU2 |
| SBY4541 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ ipl1-321-FLAG3::KAN |
| SBY4764 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ DAM1-myc9::TRP1 ipl1-321 |
| SBY4801 | MATaura3-1 leu2,3-112 his3-11 trp1-1::glc7-10::TRP1 ade2-1 can1-100 bar1Δ glc7::LEU2 ipl1-321 DAM1-myc9::TRP1 |
| SBY4822 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ GLC7-HA3::HIS3 IPL1-myc13::KAN |
| SBY4826 | MATaura3-1 leu2,3-112 his3-11 trp1-1::glc7-10::TRP1 ade2-1 can1-100 bar1Δ glc7::LEU2 DAM1-myc9::TRP1 |
| SBY4892 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ GLC7-3GFP::HIS3 NIC96-CFP::KAN |
| SBY4874 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ GLC7-HA3::HIS3 SOL1-myc13::KAN |
| SBY4920 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ GLC7-HA3::HIS3 PEX31-myc13::KAN |
| SBY4995 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ GLC7-3GFP::HIS3 NIC96-CFP::KAN pGAL-HA3-GIP3::HIS3 |
| SBY4999 | MATaura3-1::TUB1-CFP::URA3 leu2,3-112 his3-11 trp1-1::glc7-10::TRP1 ade2-1 can1-100 bar1Δ glc7::LEU2 IPL1-3GFP::HIS3 |
| SBY5004 | MATaura3-1 leu2,3-112 his3-11 trp1-1::glc7-12::TRP1 ade2-1 can1-100 bar1Δ glc7::LEU2 ipl1-321 |
| SBY5070 | MATa/α ura3-1/ura3-1 leu2,3-112/leu2,3-112 his3-11::pCUP1-GFP12-lacI12::HIS3/his3-11,15 trp1-1::GLC7::TRP1/ trp1-1::256lacO::TRP1 ade2-1/ade2-1 lys2Δ/LSY2 can1-100/can1-100 bar1Δ/bar1Δ ipl1-321/IPL1 GLC7/glc7::LEU2 |
| SBY5072 | MATa/α ura3-1/ura3-1 leu2,3-112/leu2,3-112 his3-11::pCUP1-GFP12-lacI12::HIS3/his3-11,15 trp1-1::GLC7::TRP1/ trp1-1::256lacO::TRP1 ade2-1/ade2-1 lys2Δ/LSY2 can1-100/can1-100 bar1Δ/bar1Δ ipl1-321/ipl1-321 GLC7/glc7::LEU2 |
| SBY5074 | MATa/α ura3-1/ura3-1 leu2,3-112/leu2,3-112 his3-11::pCUP1-GFP12-lacI12::HIS3/his3-11,15 trp1-1::glc7-12::TRP1 trp1-1::256lacO::TRP1 ade2-1/ade2-1 lys2Δ/LSY2 can1-100/can1-100 bar1Δ/bar1Δ ipl1-321/ipl1-321 GLC7/glc7::LEU2 |
| SBY5127 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ DAM1-myc9::TRP1 ipl1-321 pGAL-HA3-GIP3::HIS3 |
| SBY5128 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ DAM1-myc9::TRP1 ipl1-321 pGAL-HA3-GIP4::HIS3 |
| SBY5032 | MATaura3-1 leu2,3-112 his3-11 trp1-1::glc7-12::TRP1 lys2Δ ade2-1 can1-100 bar1Δ glc7::LEU2 DAM1-myc9::TRP1 ipl1-321 |
| SBY5034 | MATaura3-1 leu2,3-112 his3-11 trp1-1::glc7-12::TRP1 lys2Δ ade2-1 can1-100 bar1Δ glc7::LEU2 DAM1-myc9::TRP1 |
| SBY5274 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ gip3ΔKAN DAM1-myc9::TRP1 |
| SBY5275 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ DAM1-myc9::TRP1 |
| SBY5276 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ gip4ΔKAN DAM1-myc9::TRP |
| SBY5277 | MATaura3-1 leu2,3-112 his3-11 trp1-1::glc7-10::TRP1 ade2-1 can1-100 bar1Δ glc7::LEU2 ipl1-321-3FLAG::KAN |
| SBY5278 | MATaura3-1 leu2,3-112 his3-11 trp1-1::glc7-10::TRP1 ade2-1 can1-100 bar1Δ glc7::LEU2 IPL1-3FLAG::KAN |
| SBY5279 | MATaura3-1 leu2,3-112 his3-11::pCUP1-GFP12-lacI12::HIS3 trp1-1::256lacO::TRP1 ade2-1 can1-100 bar1Δ pGAL-HA3-GIP3::HIS3 |
| SBY5284 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ GLC7-3GFP::HIS3 gip3ΔKAN |
| SBY5285 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 bar1Δ GLC7-3GFP::HIS3 |
| SBY5287 | MATaura3-1 leu2,3-112 his3-11::pCUP1-GFP12-lacI12::HIS3 trp1-1::256lacO::TRP1 ade2-1 can1-100 gip3ΔKAN bar1Δ |
| SBY5288 | MATaura3-1 leu2,3-112 his3-11::pCUP1-GFP12-lacI12::HIS3 trp1-1::256lacO::TRP1 ade2-1 can1-100 gip4ΔKAN bar1Δ |
| SBY5294 | MATaura3-1 leu2,3-112 his3-11::pCUP1-GFP12-lacI12::HIS3 trp1-1::256lacO::TRP1 ade2-1 can1-100 bar1Δ pGAL-HA3-GIP4::HIS3 |
All strains are isogenic with the W303 background and were generated for this study.
Plasmid construction.
The GLC7-GFP3 integrating plasmid was made by PCR amplification of the C-terminal 400 bp of GLC7 from pKC1048 (a gift from John Cannon, University of Missouri, Columbia, MO) using primers SB1047 and SB1048 that have ClaI and BamHI restriction sites engineered, respectively. The resulting PCR product was digested with ClaI and BamHI and ligated into the ClaI and BamHI sites of PB1585 (a gift from David Pellman, Harvard Medical School, Boston, MA) to create pSB881. Glutathione S-transferase (GST)-Dam1 was constructed by PCR amplification of Dam1 using primers SB283 and SB284 that have BamHI sites engineered. The PCR product was digested with BamHI and ligated into the BamHI site of pGEX-2T (Pharmacia) to create pSB449.
ipl1-321 dosage suppressor screen.
The ipl1-321 strain (SBY1063) was transformed with a 2μm URA3-marked genomic yeast library, plated on selective medium at a permissive temperature (23°C) for 3 days, and then replica printed to the restrictive temperature (35.5°C) for 1 day. Of the 48 temperature-resistant colonies identified, 29 showed temperature resistance that was plasmid dependent and were subjected to plasmid rescue and retransformation. The 26 remaining positive colonies were grouped based on restriction mapping, and representatives from each group were sequenced with primers SB359 and SB360 (sequences available upon request). A total of 12 genomic regions containing the following genes were identified: IPL1 (four times), GLC8 (two times), SCD5 (two times), SDS22 (one time), BUD14 (two times), FUN21/GIP4 (two times), PEX31 (one times), SOL1 (two times), SOL2 (one time), YPL137C/GIP3 (seven times), YOR342C (one time), and glc7Δ186-312. glc7Δ186-312, GLC8, and SCD5 were previously identified as ipl1-1 dosage suppressors, so their genomic regions were not further dissected. FUN21/GIP4 and YPL137C/GIP3 were confirmed to encode the dosage suppressors by generation of a series of plasmid deletions that were retested for temperature resistance in SBY1063. To determine which genes encoded the remaining dosage suppressors, we obtained strains from the GST-ORF collection (a gift from Stan Fields, University of Washington, Seattle, WA) for each of the open reading frames in the above genomic regions. We isolated the GST-ORF plasmids, retransformed them into SBY1063, and screened them for temperature resistance. By this method, we identified SDS22, BUD14, SOL1, and PEX31 as dosage suppressors. Although SOL2 is 78% identical to SOL1, we have not eliminated the possibility that another gene in the genomic region is the dosage suppressor. We have not determined which gene in the YOR342C genomic region suppresses ipl1-321.
Microscopy.
Live microscopy was performed as described previously (9). More than 100 cells were analyzed for all reported experiments.
Protein and immunological techniques.
Protein extracts were made and immunoblotted as described previously (47). 9E10 antibodies that recognize the myc tag and 12CA5 antibodies that recognize the hemagglutinin (HA) tag were obtained from Covance and used at a 1:10,000 dilution. GST-Dam1 was purified as previously described (36). To analyze Dam1 phosphorylation, 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels with decreased bisacrylamide were used.
For immunoprecipitations, 50-ml cultures of mid-log-phase cells were collected, and lysates were prepared as previously described (9). A total of 450 μl of supernatant was incubated with 5 μl protein G-coated Dynabeads (Dynal Biotech, Inc.) and 2 μl of M2 anti-flag antibody (Sigma) or 5 μl of A-14 anti-myc antibody (Santa Cruz Biotechnology) for 2 h at 4°C. The beads were washed five times with 500 μl lysis buffer, and the immunoprecipitates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted as above.
Kinase assays were performed as previously described (9), except that 5 μg histone H3 (Roche) or GST-Dam1 was used as a substrate.
RESULTS
Glc7 does not regulate Ipl1 levels or localization.
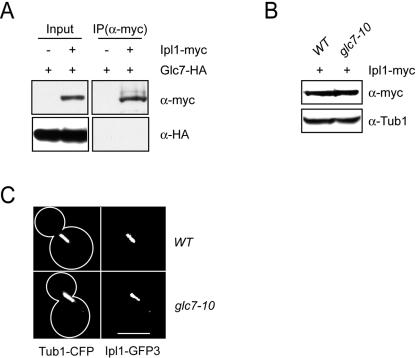
Though the phosphorylation status of several proteins is modulated by the opposing activities of Glc7 and Ipl1 (15, 32, 35, 58), the precise relationship between the yeast phosphatase and kinase has not been studied. Because glc7 mutants suppress ipl1 mutants (21, 22, 32), one possibility is that Glc7 directly inhibits Ipl1 activity. We therefore tested whether Glc7 physically interacts with Ipl1. Using strains coexpressing endogenous COOH-terminal fusions of Glc7-HA3 and Ipl1-myc13, we were unable to detect the physical association of Glc7 with Ipl1 (Fig. 1A). In addition, Glc7 did not coprecipitate with the Ipl1 activator, Sli15 (data not shown). These data indicate that Ipl1 and Glc7 do not form a detectable complex in vivo under these conditions.
FIG. 1.
Glc7 does not regulate Ipl1 levels or localization. (A) Glc7 and Ipl1 do not form a detectable complex. Extracts from cells expressing Glc7-HA3 alone (SBY625) or in combination with Ipl1-myc13 (SBY4822), were immunoprecipitated with anti-myc antibody. Extracts (Input) and immunoprecipitates (IP) were analyzed by anti-HA and anti-myc immunoblotting. (B) Glc7 does not alter Ipl1 protein levels. Wild-type (SBY4767) and glc7-10 mutant cells (SBY4766) expressing Ipl1-myc13 were arrested in mitosis with nocodazole and shifted to 37°C for 2 h, and the extracts were analyzed by anti-myc and anti-Tub1 immunoblotting as a loading control. (C) Glc7 does not regulate Ipl1's metaphase localization. Wild-type (SBY2833) and glc7-10 cells (SBY4999) expressing Ipl1-GFP3 and Tub1-CFP were grown at 37°C for 2 h. Scale bar, 5 μm.
We next examined whether Glc7 alters Ipl1 protein levels in strains carrying the temperature-sensitive glc7-10 allele. If Glc7 were a negative regulator of Ipl1 levels, Glc7 mutant cells would express more Ipl1 protein. The glc7-10 mutant suppresses ipl1-321 at high temperatures (see Fig. 3B) and is defective in Glc7's known mitotic functions (1, 58). Wild-type and glc7-10 cells expressing Ipl1-myc13 were arrested in mitosis with nocodazole to eliminate cell cycle variation, shifted to the restrictive temperature (37°C) for 2 h, and monitored for Ipl1 protein levels by α-myc immunoblotting (Fig. 1B). Wild-type and glc7-10 cells expressed equal amounts of Ipl1, indicating that Glc7 does not regulate Ipl1 levels. We obtained similar results using cells asynchronously shifted to the restrictive temperature (data not shown).
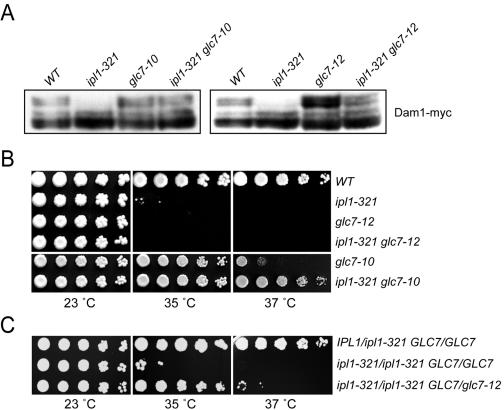
FIG. 3.
Ipl1 and Glc7 activities must be precisely balanced. (A) The balance of Ipl1 kinase and Glc7 phosphatase controls Dam1 phosphorylation. Wild-type (SBY2055), ipl1-321 (SBY4764), glc7-10 (SBY4826), ipl1-321 glc7-10 (SBY4801), glc7-12 (SBY5034), and ipl1-321 glc7-12 (SBY5032) cells expressing Dam1-myc9 were grown at 35°C for 3 h. Extracts were analyzed by anti-myc immunoblotting for changes in Dam1 gel mobility and showed that the upper Dam1 phosphoforms missing in ipl1-321 cells were restored in ipl1-321 glc7-10 and ipl1-321 glc7-12 mutant cells. (B) ipl1-321 temperature sensitivity is suppressed by glc7-10 but not glc7-12 in haploid cells. Fivefold serial dilutions of wild-type (SBY214), ipl1-321 (SBY322), glc7-12 (SBY3675), glc7-12 ipl1-321 (SBY5004), glc7-10 (SBY1306), and ipl1-321 glc7-10 (SBY1994) cells were incubated for 3 days at 23°C and 2 days at 35 and 37°C. (C) The ipl1-321 temperature sensitivity is suppressed by glc7-12 in the presence of wild-type GLC7. Fivefold serial dilutions of ipl1-321/IPl1 GLC7/GLC7 (SBY5070), ipl1-321/ipl1-321 GLC7/GLC7 (SBY5072), and ipl1-321/ipl1-321 glc7-12/GLC7 (SBY5074) cells were incubated for 3 days at 23°C and 2 days at 35 and 37°C.
Since the metaphase kinetochore localization of Ipl1 is thought to reflect its role in chromosome segregation, we analyzed Glc7 effects on this localization. We visualized triple green fluorescent protein (GFP) epitope-tagged Ipl1 (Ipl1-GFP3) in wild-type and glc7-10 cells coexpressing cyan fluorescent protein (CFP)-tagged tubulin to mark spindles (Tub1-CFP) after they were shifted to the restrictive temperature for 2 h (37°C) (Fig. 1C). In wild-type cells, Ipl1-GFP3 localized to kinetochores and microtubules on short (1.5- to 3.0-μm) metaphase spindles, as previously described (9). Ipl1-GFP3 localization was similar in that observed with glc7-10 cells, indicating that Glc7 does not regulate Ipl1's metaphase localization.
Glc7 does not regulate Ipl1 kinase activity.
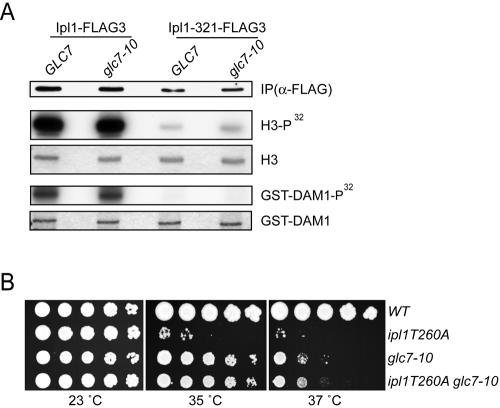
We next tested whether Glc7 negatively regulates Ipl1 kinase activity as previously described (9). We analyzed the activity of both the wild-type Ipl1 protein and the temperature-sensitive Ipl1-321 protein, which has reduced catalytic activity (4). Wild-type and glc7-10 cells expressing Ipl1-FLAG3 or Ipl1-321-FLAG3 were arrested in mitosis with nocodazole and shifted to the restrictive temperature (37°C) for 2 h. Ipl1 and Ipl1-321 were immunoprecipitated from cell lysates and used in kinase assays in vitro with the substrates histone H3 and Dam1 (Fig. 2A). There were equivalent amounts of wild-type Ipl1 kinase activity against H3 and Dam1 in both wild-type and glc7-10 mutant cells, indicating that Glc7 does not regulate bulk Ipl1 activity. Cells asynchronously shifted to the restrictive temperature also contained equal amounts of Ipl1 kinase activity (data not shown). Although the kinase activity of Ipl1-321 was much lower than that of wild-type Ipl1, it was also similar in wild-type and glc7-10 cells. Because ipl1-321 glc7-10 cells are viable at the nonpermissive temperature but ipl1-321 cells are nonviable, it is highly unlikely that the glc7-10 suppression of ipl1-321 is due to direct regulation of Ipl1 kinase activity.
FIG. 2.
Glc7 does not regulate Ipl1 kinase activity. (A) Ipl1 kinase activity is normal in glc7 mutant cells. Wild-type and glc7-10 mutant cells expressing Ipl1-FLAG3 (SBY3672 and SBY5278) or Ipl1-321-FLAG3 (SBY4541 and SBY5277) were arrested in mitosis with nocodazole and shifted to 37°C for 2 h. Extracts were immunoprecipitated (IP) with α-FLAG antibody, and the IPs were analyzed by α-FLAG immunoblotting and used in kinase assays in vitro with the substrates histone H3 or GST-Dam1. (B) glc7-10 suppresses ipl1T260A. Fivefold serial dilutions of wild-type (SBY2055), ipl1T260A (SBY1264), glc7-10 (SBY1306), and ipl1T260 glc7-10 (SBY1258) cells were incubated for 3 days at the temperatures indicated.
As a second test of Ipl1 regulation by Glc7, we analyzed genetic interactions between the glc7-10 allele and an ipl1 allele that cannot be phosphorylated on the activating residue. The Aurora kinases are activated by phosphorylation of a threonine residue in the activation loop that corresponds to threonine 260 (T260) in Ipl1. This residue was phosphorylated in vivo, and a mutation of T260 to alanine in Ipl1 (ipl1-T260A) resulted in a temperature-sensitive phenotype (Fig. 2B) (15, 22). If Glc7 dephosphorylated T260, a reduction in Glc7 activity would not alter the temperature sensitivity of the ipl1-T260A mutant because it cannot be phosphorylated. In contrast, a reduction in Glc7 activity would be predicted to suppress the temperature sensitivity of the ipl1-T260A mutant if Glc7 acts on Ipl1 targets. We found that glc7-10 did suppress the temperature-sensitive phenotype of the ipl1-T260A allele (Fig. 2B), indicating that it is unlikely that Glc7 opposes Ipl1 activity by dephosphorylating the kinase directly. Taken together, these data show that negative regulation of Ipl1 by Glc7 is unlikely to explain the relationship between the kinase and phosphatase.
Glc7 and Ipl1 activity must be precisely balanced.
Another possibility is that Ipl1 and Glc7 regulate a common set of substrates, as proposed (21, 22). We therefore determined whether the balance of kinase and phosphatase regulates the phosphorylation of an essential Ipl1 substrate, Dam1 (15). To do this we monitored ipl1 and glc7 mutants, as well as ipl1 glc7 double mutants, for Dam1 gel mobility as previously described (35, 44). In addition to the glc7-10 allele, we also analyzed glc7-12, another mitotic defective allele (46). Wild-type, ipl1-321, glc7-10, ipl1-321 glc7-10, glc7-12, and ipl1-321 glc7-12 cells expressing an endogenous COOH-terminal fusion of Dam1-myc9 were asynchronously shifted to the restrictive temperature (35°C) for 3 h. Dam1 displayed a series of slower-migrating phosphoforms in wild-type cells that were abolished in ipl1-321 mutant cells as previously reported (Fig. 3A) (35, 44). Importantly, the Dam1 phosphoforms were more similar to the wild type in both the glc7-10 ipl1-321 and glc7-12 ipl1-321 double mutant cells than in the ipl1-321 cells. This restoration of phosphorylation indicates that the ipl1-321 allele retains some enzymatic activity at higher temperatures and is consistent with Dam1 phosphorylation being regulated by a balance of Ipl1 kinase and Glc7 phosphatase activity in vivo.
Although the Dam1 phosphoforms appeared to be restored in both glc7-10 ipl1-321 and glc7-12 ipl1-321 mutant cells at 35°C, the glc7-10 allele suppressed the temperature sensitivity of ipl1-321 at 35°C, while glc7-12 did not (Fig. 3B). The Dam1 phosphoforms were more intense in glc7-12 cells than in glc7-10 cells (Fig. 3A), indicating that glc7-12 likely retained less residual phosphatase activity at the restrictive temperature. If this were true, a possible explanation for the inability of glc7-12 to suppress ipl1-321 is that there is not enough residual phosphatase activity to oppose the remaining Ipl1-321 kinase activity. To test this hypothesis, we analyzed growth in diploids where the balance of Ipl1 and Glc7 could be altered by changing allele copy numbers. Similar to ipl1-321 haploid cells, ipl1-321/ipl1-321 homozygous diploid cells were temperature sensitive at 35°C and 37°C (Fig. 3C). However, unlike the haploid cells, the ipl1-321/ipl1-321 homozygous mutants were suppressed by a single copy of the glc7-12 allele. Because glc7-12 suppressed ipl1-321 in the presence of a wild-type copy of GLC7, it strongly supported our hypothesis that glc7-12 retains too little phosphatase activity at the restrictive temperature to balance the remaining ipl1-321 kinase activity. Although the Dam1 phosphoforms appeared to be restored to wild-type levels in glc7-12 ipl1-321 cells, it may be that the Dam1 gel mobility assay was not sensitive enough to distinguish small differences in the phosphorylation state. Taken together, these data provide further evidence that the Ipl1 kinase and Glc7 phosphatase activities must be precisely balanced.
Ipl1-321 high-copy-number suppressor screen.
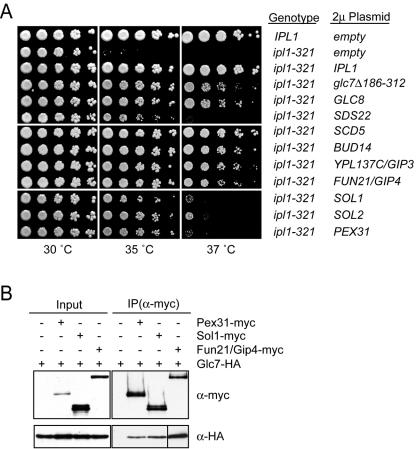
To identify potential Ipl1 and Glc7 substrates or Ipl1 regulators, we carried out a dosage suppressor screen of the ipl1-321 temperature-sensitive growth defect at 35.5°C (4). We found 10 genes (Table 2) that suppressed ipl1-321 when present on a high-copy-number 2μm plasmid (Fig. 4A). Consistent with previous dosage suppressor screens using the temperature-sensitive ipl1-1 allele, we identified the dominant negative PP1 allele glc7Δ186-312, as well as the GLC8 and SCD5 genes (22, 66, 72). In addition to these known suppressors, we identified seven novel ipl1-321 dosage suppressors: SDS22, BUD14, YPL137C, FUN21, SOL1, SOL2, and PEX31. At the restrictive temperature of 35°C, all of the dosage suppressors restored ipl1-321 growth to near-wild-type levels, while at 37°C there were various levels of suppression (Fig. 4A).
TABLE 2.
ipl1-321 dosage suppressors
| Gene name | Description | Localizationa | Glc7 interactiona | Reference(s) |
|---|---|---|---|---|
| glc7Δ 186-312 | Dominant negative GLC7 allele | NA | NA | 21, 72 |
| GLC8 | GLC7 regulatory subunit | Cytoplasm, nucleus | 2H, AP | 30, 33, 38, 55, 70 |
| SCD5 | Multicopy suppressor of clathrin deficiency | Cell cortex | 2H, AP, Co-IP | 14, 28, 30, 50, 68, 70 |
| SDS22 | GLC7 regulatory subunit | Nucleus, cytoplasm | 2H, AP, Co-IP | 23, 27, 31, 33, 38, 53, 55, 75 |
| BUD14 | Involved in bud site selection | Bud site and neck | 2H, Co-IP | 18, 33, 37, 38, 41, 51, 68 |
| YPL137c/GIP3 | Uncharacterized open reading frame | Cytoplasm, endoplasmic reticulum | AP | 33, 71 |
| FUN21/GIP4 | Uncharacterized open reading frame | Cytoplasm | Co-IPb | 33 |
| SOL1 | Regulator of tRNA function | Cytoplasm, nucleus | Co-IPb | 33, 38 |
| SOL2 | Regulator of tRNA function | Cytoplasm | Unknown | 33 |
| PEX31 | Peroxisomal integral membrane protein | Peroxisomes, cytoplasm | Co-IPb | 38 |
NA, not applicable; 2H, two hybrid; AP, affinity precipitation; Co-IP, coimmunoprecipitation.
This work.
FIG. 4.
ipl1-321 dosage suppressors encode Glc7-interacting proteins. (A) Glc7 regulators are high-copy-number ipl1-321 suppressors. Fivefold serial dilutions of wild-type cells (SBY214) with an empty 2μm plasmid, ipl1-321 cells (SBY1063) with an empty 2μm plasmid, or ipl1-321 cells with 2μm plasmids containing the indicated dosage suppressors were incubated for 2 days at 30, 35, and 37°C. (B) Glc7 physically associates with Pex31, Sol1, and Fun21/Gip4. Extracts from cells expressing Glc7-HA3 alone (SBY625) or in combination with Pex31-myc13 (SBY4920), Sol1-myc13 (SBY4874), and Fun21/Gip4-myc13 (SBY4209) were immunoprecipitated with α-myc antibody. Extracts (Input) and immunoprecipitates (IP) were analyzed by anti-HA and anti-myc immunoblotting. The anti-HA immunoblot of the Fun21/Gip4 IP was exposed for 1/10 of the time used for the Pex31 and Sol1 IPs.
Fun21, Sol1, and Pex31 physically interact with Glc7.
It was originally proposed that reduced Glc7 activity suppresses Ipl1 mutations by restoring the balance of kinase/phosphatase activity (22). The ipl1-321 suppressors Glc8, Scd5, Sds22, Bud14, and Ypl137C physically interact with Glc7 (14, 23, 27, 30, 31, 37, 41, 53, 55, 65, 68, 70, 71, 75). We therefore tested whether the remaining dosage suppressors (Pex31, Sol1, and Fun21) also interacted with Glc7. We generated strains coexpressing endogenous COOH-terminal fusions of Glc7-HA3 with Pex31-myc13, Sol1-myc13, and Fun21-myc13 and found that all three proteins immunoprecipitated Glc7 (Fig. 4B). Therefore, like the known ipl1 dosage suppressors, Pex31, Sol1, and Fun21 physically interacted with Glc7. Since the Ypl137C and Fun21 proteins physically interacted with Glc7, we named the genes that encode them GIP3 and GIP4, respectively, for Glc7-interacting protein (65).
Gip3 and Gip4 do not regulate chromosome segregation.
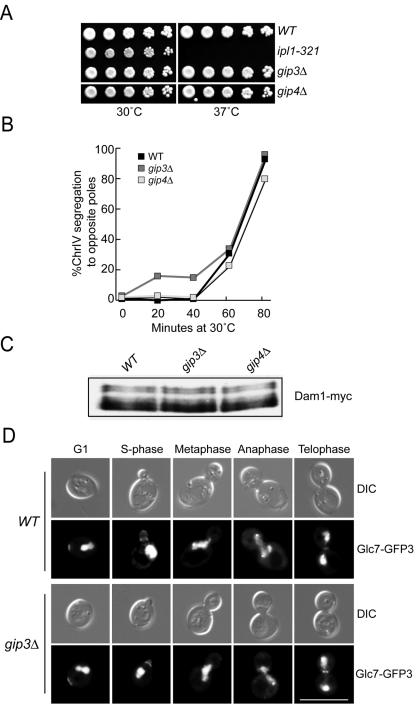
We further characterized the functions of Gip3 and Gip4 to determine how they suppress ipl1-321 temperature-sensitive cells when expressed from a 2μm plasmid. First, we tested whether the Gip3 and Gip4 proteins regulate chromosome segregation in a manner similar to Ipl1. The GIP3 and GIP4 genes were deleted, and the corresponding strains were viable as previously reported (24, 60) and did not exhibit growth defects at higher temperatures (Fig. 5A). To analyze chromosome segregation, wild-type, gip3Δ, and gip4Δ cells that contained fluorescently marked chromosome IV (ChrIV) were arrested in G1 and released into the cell cycle. All three strains began budding at 40 min after release and remained synchronous throughout the time course (data not shown). Similar to wild-type cells, the gip3Δ and gip4Δ mutant cells segregated ChrIV to opposite poles, indicating that Gip3 and Gip4 do not have apparent roles in chromosome segregation (Fig. 5B).
FIG. 5.
Gip3 and Gip4 do not regulate chromosome segregation. (A) gip3Δ and gip4Δ strains grow normally at all temperatures. Serial dilutions of wild-type (SBY3), ipl1-321 (SBY322), gip3Δ (SBY4114), and gip4Δ (SBY4175) cells were incubated for 2 days at 30 and 37°C. (B) Gip3 and Gip4 are not required for chromosome segregation. Wild-type (SBY818), gip3Δ (SBY5287), and gip4Δ (SBY5288) cells were arrested in G1 and released into the cell cycle. Fluorescently tagged ChrIV was monitored over the time course and segregated to opposite poles in all strains by 80 min after release. (C) Dam1 phosphorylation is normal in gip3Δ and gip4Δ strains. Wild-type (SBY5275), gip3Δ (SBY5274), and gip4Δ (SBY5276) strains containing Dam1-myc9 were grown at 23°C. Extracts were analyzed by α-myc immunoblotting for changes in Dam1 gel mobility and show that the Dam1 phosphoforms are not altered in the absence of Gip3 or Gip4. (D) Glc7 localization is normal in the absence of Gip3 and Gip4. Wild-type (SBY5285) and gip3Δ (SBY5284) cells containing Glc7-GFP3 were analyzed throughout the cell cycle for Glc7 localization. Scale bar, 10 μm.
We next determined whether Gip3 and Gip4 affect the phosphorylation status of the Dam1 protein that is regulated by Ipl1 and Glc7. Extracts prepared from asynchronously growing wild-type, gip3Δ and gip4Δ cells containing Dam1-myc9 were analyzed for Dam1 phosphorylation (Fig. 5C). There was no change in Dam1 phosphorylation in either mutant strain, indicating that Gip3 and Gip4 do not regulate Dam1. Taken together, these data suggest that unlike Ipl1 and Glc7, Gip3 and Gip4 do not have functions related to chromosome segregation.
Because Gip3 and Gip4 physically interact with Glc7, we considered the possibility that they were previously unidentified Glc7 regulatory subunits that control Glc7 localization. Consistent with this hypothesis, both proteins contain the R/K-V/I-X-F motif that targeting subunits use to bind to protein phosphatase 1 (20). We therefore analyzed the localization of a fully functional endogenous COOH-terminal fusion of Glc7 to triple green fluorescent protein (Glc7-GFP3) in wild-type, gip3Δ, and gip4Δ strains throughout the cell cycle (Fig. 5D and data not shown). In wild-type cells, Glc7-GFP3 localized to the nucleus throughout the cell cycle, as previously reported (8, 77). In addition, Glc7 localized to the presumptive bud site during G1 phase and then the bud neck and bud cortex during S phase through telophase (8). As previously noted, anaphase and some telophase cells contained two dots of Glc7 at opposite ends of the nucleus (8). This localization was reported to be spindle pole body (SPB) staining because it colocalized with the Nuf2 protein that was originally thought to be an SPB component (52). However, it was subsequently shown that Nuf2 is a kinetochore protein (74), indicating that Glc7 localizes to kinetochores instead of SPBs during anaphase. Because there were no differences in Glc7 localization at any of these cellular sites in the absence of Gip3 (Fig. 5D) and Gip4 (data not shown), these proteins cannot be the sole regulators of Glc7 localization to any of these locations.
Gip3 and Gip4 overexpression is lethal and prevents chromosome segregation.
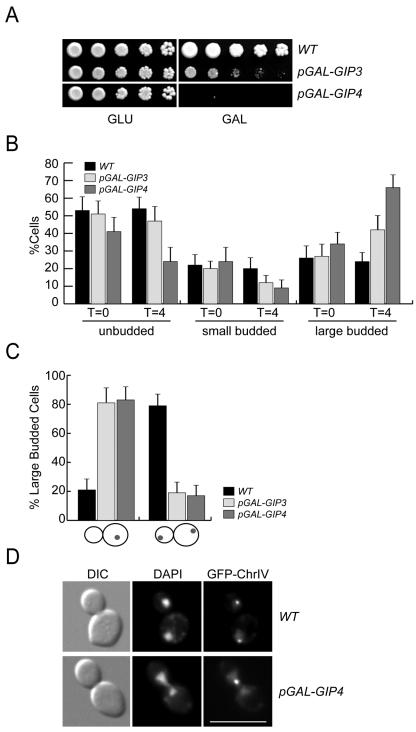
Although we did not detect growth defects when Gip3 and Gip4 were deleted, it was previously reported that Gip3 overexpression is lethal (60). We therefore analyzed the phenotypes of cells expressing Gip3 and Gip4 from the highly inducible galactose promoter. Although wild-type cells grow on both glucose and galactose media, cells expressing pGAL-GIP4 cannot grow on galactose medium, and cells expressing pGAL-GIP3 are severely compromised for growth as previously reported (Fig. 6A) (60).
FIG. 6.
The overexpression of Gip3 and Gip4 inhibits cell growth and prevents chromosome segregation. (A) Gip3 and Gip4 overexpression from the galactose promoter inhibits cell growth. Serial dilutions of wild-type (SBY3), pGAL-GIP3 (SBY4179) and pGAL-GIP4 (SBY4292) cells were plated onto glucose (GLU) and galactose (GAL) media and incubated for 2 days at 30°C. (B) Gip3 and Gip4 overexpression increases the population of large budded cells. Wild-type (SBY818), pGAL-GIP3 (SBY5279), and pGAL-GIP4 (SBY5294) cells were induced with galactose for 4 h, and the percentage of cells at each stage of the cell cycle was quantified. Error bars indicate the 95% confidence interval. (C) Gip3 and Gip4 overexpression prevents chromosome segregation. Cells containing fluorescently tagged ChrIV were grown as described in the legend to panel B. Large budded cells were scored for either unsegregated ChrIV (left) or segregation of ChrIV to opposite poles (right). (D) Examples of wild-type cells (SBY818) where ChrIV segregates to opposite poles and cells overexpressing Gip4 (SBY5294) where ChrIV does not segregate. Scale bar, 10 μm.
To better understand this growth inhibition, we assessed cell cycle progression when Gip3 and Gip4 were overexpressed. Wild-type, pGAL-GIP3, and pGAL-GIP4 cells were grown in galactose for 4 h and analyzed for cell cycle morphology (Fig. 6B). Prior to induction, the distribution of cells throughout the cell cycle was similar in all three strains. However, after 4 h in galactose medium, there was an increase in large budded cells in the pGAL-GIP3 and pGAL-GIP4 strains. The increase was greater in pGAL-GIP4 strains, consistent with the stronger growth defect observed when this gene was overexpressed. We next analyzed chromosome segregation in the large budded cells after 4 h of galactose induction (Fig. 6C). We monitored both the overall DNA by staining with DAPI (4′,6′-diamidino-2-phenylindole), as well as a single chromosome by using fluorescently tagged ChrIV. In the majority of wild-type cells (79%), the DNA and ChrIV segregated to opposite poles (Fig. 6C and D). However, in large budded cells overexpressing Gip3 and Gip4, neither the total DNA nor ChrIV segregated. Strikingly, these data are consistent with the phenotypes of glc7 mutant cells that arrest in metaphase (1, 2, 6, 29, 46).
Overexpression of Gip3 and Gip4 titrates Glc7 from the nucleus.
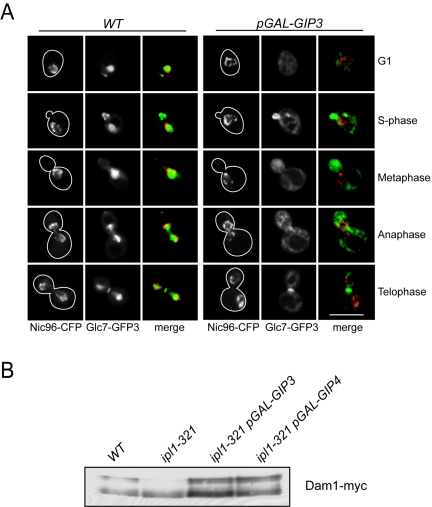
Because the overexpression of Gip3 and Gip4 led to a phenotype that resembled a decrease in the mitotic functions of Glc7, we considered the possibility that they decreased the nuclear pool of Glc7. We first analyzed Glc7 levels when Gip3 and Gip4 were overexpressed and found that they were not altered (data not shown). We therefore analyzed Glc7 localization when they were overexpressed. Glc7-GFP3 was localized in wild-type cells and cells overexpressing galactose-inducible Gip3 and Gip4, which are reported to be in the cytoplasm (Fig. 7A; Table 2) (33 and data not shown). Nuclei were visualized by coexpressing the nuclear pore component Nic96 fused to cyan fluorescent protein (Nic96-CFP). In cells overexpressing Gip3 and Gip4, Glc7-GFP3 nuclear localization disappeared, and the phosphatase was predominantly cytoplasmic at all cell cycle stages. However, localization to the bud neck and bud cortex was unaltered. Because Gip3 and Gip4 altered Glc7 localization when overexpressed, they are likely to be previously unidentified regulatory subunits.
FIG. 7.
Gip3 and Gip4 overexpression redistributes Glc7 from the nucleus and alters Dam1 phosphorylation. (A) Overexpression of GIP3 from the galactose promoter reduces nuclear Glc7. Wild-type (SBY4892) and pGAL-GIP3 (SBY4995) cells expressing Glc7-GFP3 and Nic96-CFP were grown in galactose for 4 h at 30°C. A representative cell from each stage of the cell cycle is shown, as well as a merge of the CFP and GFP channels. Scale bar, 5 μm. (B) GIP3 and GIP4 overexpression restores Dam1 phosphorylation in ipl1-321 cells. Wild-type (SBY2055), ipl1-321 (SBY4764), pGAL-GIP3 ipl1-321 (SBY5127) and pGAL-GIP4 ipl1-321 (SBY5128) cells containing Dam1-myc9 were grown in galactose at 23°C for 30 min and shifted to 35°C in the presence of galactose for 3 h. Extracts were analyzed by anti-myc immunoblotting for changes in Dam1 gel mobility and show that the upper Dam1 phosphoforms missing in ipl1-321 cells are restored when Gip3 and Gip4 are overexpressed.
The overexpression of Gip3 and Gip4 restores Dam1 phosphorylation in ipl1 mutants.
The observation that Gip3 and Gip4 overexpression reduces the nuclear pool of Glc7 suggests that ipl1-321 dosage suppression may occur by spatially separating the phosphatase from its relevant nuclear targets opposing Ipl1 function. Although all of the ipl1-321 dosage suppressors physically interacted with Glc7, they were involved in disparate cellular processes and exhibited different localization patterns (Table 2). We therefore hypothesized that the suppressors may inhibit Glc7's mitotic functions opposing Ipl1 kinase activity by titrating Glc7 away from the relevant Ipl1 substrates. To test whether Gip3 and Gip4 overexpression alters the phosphorylation of important Ipl1 substrates, we analyzed Dam1 phosphorylation. Wild-type, ipl1-321, and ipl1-321 cells expressing galactose-inducible Gip3 and Gip4 were grown under inducing conditions for 30 min, shifted to 35°C for 3 h, and analyzed for Dam1-myc9 phosphorylation. Strikingly, the Dam1 mobility shift that was abolished in ipl1-321 mutants was restored when either Gip3 or Gip4 was overexpressed (Fig. 7B). Therefore, the overexpression of these proteins likely suppressed ipl1-321 by titrating Glc7 away from important Ipl1 targets, such as Dam1, thus restoring the balance of kinase and phosphatase activity.
DISCUSSION
The opposing activities of the Ipl1/Aurora protein kinase and Glc7/PP1 protein phosphatase are required for accurate chromosome segregation. Decreased Glc7 activity affected the phosphorylation of the Ipl1 substrate Dam1 but did not alter Ipl1 levels, localization, or bulk kinase activity, supporting the proposal that Glc7 opposes Ipl1 function by regulating the phosphorylation of common targets. We show here that at least two ipl1 dosage suppressors encode Glc7 interacting proteins that, when overexpressed, likely restore the kinase/phosphatase balance by reducing Glc7 access to relevant substrates.
Ipl1 and Glc7 regulate a common set of substrates.
Three simple models have been proposed that could account for the functional interaction between Glc7 and Ipl1. (i) Glc7 negatively regulates Ipl1. (ii) Ipl1 negatively regulates Glc7. (iii) Ipl1 and Glc7 modulate the phosphorylation status of a common set of substrates (21, 22, 32, 58). Attempts to distinguish between these models have not yet been carried out with budding yeast. Here, we show that Ipl1 activity was not affected by decreased Glc7 activity. Because our kinase assay can only measure bulk Ipl1 activity, the possibility remains that a subset of Ipl1 is directly regulated by Glc7. However, an Ipl1 activation loop mutant that cannot be phosphorylated was still suppressed by a reduction in Glc7 activity, making it highly unlikely that Glc7 regulates Ipl1 activity through dephosphorylation of this residue. To date, the only other site where phosphorylation has been detected on Ipl1 in vivo is S76 (15). Although the S76 site is predicted to be a CDK phosphorylation site and is phosphorylated by Cdc28 in vitro (67), mutation of S76 to alanine does not result in any growth defects when integrated into the genome (data not shown). Therefore, even if S76 was regulated by Glc7, it is unlikely to explain the nature of the essential interaction between Ipl1 and Glc7. We cannot exclude the possibility that phosphorylation on other unidentified Ipl1 sites is regulated by Glc7. However, our data contrast with results from cultured vertebrate cells and Xenopus chromatin, where incubation with PP1 inhibitors resulted in elevated Aurora B kinase activity (49, 62). We also did not observe a change in the levels or localization of Ipl1 in mitotic cells with reduced Glc7 function, as has been described for Aurora B in meiotic Caenorhabditis elegans cells treated with PP1 RNA interference (56). It is possible that these results reveal true differences in Ipl1 and Aurora B regulation among organisms, although they may also represent a lack of inhibitor specificity or the limitations of our Ipl1 kinase and localization assays. In addition, we were unable to detect a physical interaction between Glc7 and Ipl1 when expressed at endogenous levels. However, Aurora B interacted with each of the three PP1 isoforms (α, δ, and γ) when they were co-overexpressed in cultured cells (62). It is not clear if the potential association of Glc7 with Ipl1 escaped our detection due to a weak, transient, or cell cycle stage-specific interaction, whether the overexpression studies promoted an interaction that is not present under normal conditions, or whether the interactions between Ipl1 and Glc7 were also organism specific. Consistent with our results indicating that Glc7 likely does not directly regulate Ipl1, the kinetochore-associated PP1γ isoform localizes to a domain distinct from Aurora B in cultured cells (64). We have not eliminated the possibility that Ipl1 negatively regulates Glc7 or its mitotic regulatory subunit(s), though Ipl1 does not phosphorylate Glc7 in vitro (data not shown) and Glc7 is not phosphorylated in vivo in budding yeast (61).
It was previously shown that Ipl1/Aurora B and Glc7/PP1 regulate the phosphorylation of the histone H3 and Ndc10 proteins (32, 58). Although these studies did not differentiate between the models described above, these results and genetic studies (15, 76) are consistent with a role for the kinase and the phosphatase working in parallel to control the phosphorylation level of a common set of substrates. Similarly, we found that impairing Glc7 function restores the phosphorylation of the Dam1 protein in ipl1 mutant cells, consistent with previously reported genetic interactions (15). Taken together, these results suggest that in the budding yeast, Ipl1 and Glc7 act on common targets to promote proper chromosome segregation.
A genetic screen for ipl1 dosage suppressors identifies Glc7 regulatory subunits.
Protein phosphatase 1 catalytic subunits, such as Glc7, control numerous cellular processes through their interaction with specialized regulatory subunits that target the phosphatase to appropriate substrates (for a review, see reference 11). We show here that the ipl1 temperature-sensitive growth defect is suppressed by the increased dosage of genes encoding Glc7-interacting proteins (Table 2). These genes include previously described ipl1 dosage suppressors GLC8 and SCD5, as well as SDS22, BUD14, and GIP3, newly identified dosage suppressors that encode known Glc7-interacting proteins (14, 23, 27, 30, 31, 37, 41, 53, 55, 65, 68, 70, 71, 75). In addition, we identified GIP4, SOL1, SOL2, and PEX31 as ipl1 dosage suppressors and showed that these genes also encode proteins that physically interact with Glc7. Given the exquisite sensitivity of ipl1 mutant cell growth to the dosage of genes encoding Glc7 interacting proteins, the careful evaluation of changes in the levels of Glc7 interactors should be considered for any ipl1 suppressor.
Because the Gip3 and Gip4 proteins physically interact with Glc7 and cause its relocalization when overexpressed, we propose that they are previously unidentified Glc7 regulatory subunits. Although we did not detect changes in Glc7 localization when Gip3 and Gip4 were deleted, this may have been due to redundant functions with other Glc7 regulatory subunits. Though gip3Δ strains are viable, a gip3Δ strain is inviable when combined with a deletion of the open reading frame YOR227W (60), suggesting that these two genes act in parallel pathways to regulate a common, essential function. The protein product of the YOR227W gene has been affinity purified with Glc7 (30), consistent with the possibility that it is also a previously unidentified regulatory subunit that could have an overlapping function with Gip3. Because we were not able to detect any defects in chromosome segregation or Dam1 phosphorylation in the absence of Gip3 and Gip4, it is unlikely that these proteins participate in the essential functions of Ipl1. Future characterization of the functions of these genes should therefore reveal additional cellular roles for the Glc7 phosphatase.
Overexpression of Glc7 regulatory subunits can restore the kinase/phosphatase balance by relocalizing Glc7.
It is likely that the mechanism of ipl1 mutant dosage suppression involves the redistribution of Glc7 away from the targets relevant to Ipl1's essential functions. Consistent with this idea, cells overexpressing GIP3 and GIP4 have reduced Glc7 in the nucleus, and most of the other dosage suppressors encode cytoplasmic or membrane-bound proteins that would be predicted to mislocalize Glc7 away from nuclear Ipl1 targets when overexpressed (Table 2). In contrast, increased levels of a mitotic Glc7 regulatory subunit important for directing the phosphatase to Ipl1 substrates should exacerbate the ipl1 temperature sensitivity by further skewing the kinase/phosphatase balance toward a lack of phosphorylation. It is therefore unlikely that any of the dosage suppressors encode this Glc7 mitotic regulator. Although previous work suggested that Sds22 was the Glc7 mitotic regulator (53), we isolated SDS22 as an ipl1 dosage suppressor. Because sds22 mutants also suppress the ipl1 temperature sensitivity and result in Glc7 mislocalization, the proposal that Sds22 acts as a Glc7 chaperone is more consistent with our observations (53). The hypothesis that the Glc7 regulatory subunits titrate Glc7 away from essential Ipl1 targets is supported by two observations. First, the phosphorylation of an essential Ipl1 substrate, Dam1, was restored in ipl1-321 cells when Gip3 and Gip4 were overexpressed. Second, the overexpression of Gip3 and Gip4 from the galactose promoter caused lethality and prevented chromosome segregation in wild-type cells, phenotypes consistent with a reduction in the mitotic functions of Glc7 (1, 2, 6, 29, 46). Because these genes were not lethal when expressed on 2μm plasmids, it is likely that the levels of expression from the 2μm plasmid were lower than from the strong galactose promoter. We propose that the other dosage suppressors act in a manner similar to Gip3 and Gip4 overexpression and reduce the effective mitotic functions of Glc7. Although Sds22 is a nuclear protein, it could titrate Glc7 away from essential Ipl1 targets that presumably localize to kinetochores or kinetochore microtubules.
Our data indicate that yeast cells must carefully balance the levels of the numerous Glc7 regulatory subunits. In addition, our results emphasize the importance of maintaining the balance between the kinase and phosphatase to ensure accurate chromosome segregation. In the future, it will be critical to isolate the Glc7 mitotic regulators to elucidate the mechanisms that control Glc7 activity toward Ipl1 substrates.
Acknowledgments
We thank Trisha Davis, Bungo Akiyoshi, Kim Collins, and Suzanne Furuyama for critical reading of the manuscript and discussions. We thank Kerry Bloom, John Cannon, Stan Fields, David Pellman, and Michael Stark for generously providing strains and plasmids.
B.A.P. was supported by a Paul Allen Foundation fellowship, C.V.K. was supported by a Department of Defense Breast Cancer predoctoral fellowship, and S.B. was supported by Sidney Kimmel and Beckman Young Investigator awards. This work was supported by a National Institutes of Health grant (R01-GM64386). S.B. is a Scholar of the Leukemia and Lymphoma Society.
REFERENCES
- 1.Andrews, P. D., and M. J. Stark. 2000. Type 1 protein phosphatase is required for maintenance of cell wall integrity, morphogenesis and cell cycle progression in Saccharomyces cerevisiae. J. Cell Sci. 113:507-520. [DOI] [PubMed] [Google Scholar]
- 2.Baker, S. H., D. L. Frederick, A. Bloecher, and K. Tatchell. 1997. Alanine-scanning mutagenesis of protein phosphatase type 1 in the yeast Saccharomyces cerevisiae. Genetics 145:615-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biggins, S., and A. W. Murray. 2001. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 15:3118-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biggins, S., F. F. Severin, N. Bhalla, I. Sassoon, A. A. Hyman, and A. W. Murray. 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13:532-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biggins, S., and C. E. Walczak. 2003. Captivating capture: how microtubules attach to kinetochores. Curr. Biol. 13:R449-R460. [DOI] [PubMed] [Google Scholar]
- 6.Black, S., P. D. Andrews, A. A. Sneddon, and M. J. Stark. 1995. A regulated MET3-GLC7 gene fusion provides evidence of a mitotic role for Saccharomyces cerevisiae protein phosphatase 1. Yeast 11:747-759. [DOI] [PubMed] [Google Scholar]
- 7.Bloecher, A., and K. Tatchell. 1999. Defects in Saccharomyces cerevisiae protein phosphatase type I activate the spindle/kinetochore checkpoint. Genes Dev. 13:517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloecher, A., and K. Tatchell. 2000. Dynamic localization of protein phosphatase type 1 in the mitotic cell cycle of Saccharomyces cerevisiae. J. Cell Biol. 149:125-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buvelot, S., S. Y. Tatsutani, D. Vermaak, and S. Biggins. 2003. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 160:329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carvalho, A., M. Carmena, C. Sambade, W. C. Earnshaw, and S. P. Wheatley. 2003. Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J. Cell Sci. 116:2987-2998. [DOI] [PubMed] [Google Scholar]
- 11.Ceulemans, H., and M. Bollen. 2004. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol. Rev. 84:1-39. [DOI] [PubMed] [Google Scholar]
- 12.Chan, C. S., and D. Botstein. 1993. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics 135:677-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan, G. K., S. T. Liu, and T. J. Yen. 2005. Kinetochore structure and function. Trends Cell Biol. 15:589-598. [DOI] [PubMed] [Google Scholar]
- 14.Chang, J. S., K. Henry, B. L. Wolf, M. Geli, and S. K. Lemmon. 2002. Protein phosphatase-1 binding to Scd5p is important for regulation of actin organization and endocytosis in yeast. J. Biol. Chem. 277:48002-48008. [DOI] [PubMed] [Google Scholar]
- 15.Cheeseman, I. M., S. Anderson, M. Jwa, E. M. Green, J. Kang, J. R. Yates, C. S. Chan, D. G. Drubin, and G. Barnes. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111:163-172. [DOI] [PubMed] [Google Scholar]
- 16.Cheeseman, I. M., C. Brew, M. Wolyniak, A. Desai, S. Anderson, N. Muster, J. R. Yates, T. C. Huffaker, D. G. Drubin, and G. Barnes. 2001. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J. Cell Biol. 155:1137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cleveland, D. W., Y. Mao, and K. F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112:407-421. [DOI] [PubMed] [Google Scholar]
- 18.Cullen, P. J., and G. F. Sprague, Jr. 2002. The Glc7p-interacting protein Bud14p attenuates polarized growth, pheromone response, and filamentous growth in Saccharomyces cerevisiae. Eukaryot. Cell 1:884-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ditchfield, C., V. L. Johnson, A. Tighe, R. Ellston, C. Haworth, T. Johnson, A. Mortlock, N. Keen, and S. S. Taylor. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egloff, M. P., D. F. Johnson, G. Moorhead, P. T. Cohen, P. Cohen, and D. Barford. 1997. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 16:1876-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Francisco, L., and C. S. Chan. 1994. Regulation of yeast chromosome segregation by Ipl1 protein kinase and type 1 protein phosphatase. Cell. Mol. Biol. Res. 40:207-213. [PubMed] [Google Scholar]
- 22.Francisco, L., W. Wang, and C. S. Chan. 1994. Type 1 protein phosphatase acts in opposition to Ipl1 protein kinase in regulating yeast chromosome segregation. Mol. Cell. Biol. 14:4731-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 24.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 25.Giet, R., C. Petretti, and C. Prigent. 2005. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 15:241-250. [DOI] [PubMed] [Google Scholar]
- 26.Hauf, S., R. W. Cole, S. LaTerra, C. Zimmer, G. Schnapp, R. Walter, A. Heckel, J. van Meel, C. L. Rieder, and J.-M. Peters. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161:281-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hazbun, T. R., L. Malmstrom, S. Anderson, B. J. Graczyk, B. Fox, M. Riffle, B. A. Sundin, J. D. Aranda, W. H. McDonald, C. H. Chiu, B. E. Snydsman, P. Bradley, E. G. Muller, S. Fields, D. Baker, J. R. Yates III, and T. N. Davis. 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell 12:1353-1365. [DOI] [PubMed] [Google Scholar]
- 28.Henry, K. R., K. D'Hondt, J. Chang, T. Newpher, K. Huang, R. T. Hudson, H. Riezman, and S. K. Lemmon. 2002. Scd5p and clathrin function are important for cortical actin organization, endocytosis, and localization of Sla2p in yeast. Mol. Biol. Cell 13:2607-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hisamoto, N., K. Sugimoto, and K. Matsumoto. 1994. The Glc7 type 1 protein phosphatase of Saccharomyces cerevisiae is required for cell cycle progression in G2/M. Mol. Cell. Biol. 14:3158-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 31.Hong, G., R. J. Trumbly, E. M. Reimann, and K. K. Schlender. 2000. Sds22p is a subunit of a stable isolatable form of protein phosphatase 1 (Glc7p) from Saccharomyces cerevisiae. Arch. Biochem. Biophys. 376:288-298. [DOI] [PubMed] [Google Scholar]
- 32.Hsu, J. Y., Z. W. Sun, X. Li, M. Reuben, K. Tatchell, D. K. Bishop, J. M. Grushcow, C. J. Brame, J. A. Caldwell, D. F. Hunt, R. Lin, M. M. Smith, and C. D. Allis. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102:279-291. [DOI] [PubMed] [Google Scholar]
- 33.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 34.Janke, C., J. Ortiz, T. U. Tanaka, J. Lechner, and E. Schiebel. 2002. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21:181-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang, J., I. M. Cheeseman, G. Kallstrom, S. Velmurugan, G. Barnes, and C. S. Chan. 2001. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J. Cell Biol. 155:763-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kellogg, D. R., A. Kikuchi, T. Fujii-Nakata, C. W. Turck, and A. W. Murray. 1995. Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J. Cell Biol. 130:661-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knaus, M., E. Cameroni, I. Pedruzzi, K. Tatchell, C. De Virgilio, and M. Peter. 2005. The Bud14p-Glc7p complex functions as a cortical regulator of dynein in budding yeast. EMBO J. 24:3000-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar, A., S. Agarwal, J. A. Heyman, S. Matson, M. Heidtman, S. Piccirillo, L. Umansky, A. Drawid, R. Jansen, Y. Liu, K. H. Cheung, P. Miller, M. Gerstein, G. S. Roeder, and M. Snyder. 2002. Subcellular localization of the yeast proteome. Genes Dev. 16:707-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lampson, M. A., K. Renduchitala, A. Khodjakov, and T. M. Kapoor. 2004. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 6:232-237. [DOI] [PubMed] [Google Scholar]
- 40.Lens, S. M., R. M. Wolthuis, R. Klompmaker, J. Kauw, R. Agami, T. Brummelkamp, G. Kops, and R. H. Medema. 2003. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 22:2934-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lenssen, E., N. James, I. Pedruzzi, F. Dubouloz, E. Cameroni, R. Bisig, L. Maillet, M. Werner, J. Roosen, K. Petrovic, J. Winderickx, M. A. Collart, and C. De Virgilio. 2005. The Ccr4-Not complex independently controls both Msn2-dependent transcriptional activation—via a newly identified Glc7/Bud14 type I protein phosphatase module—and TFIID promoter distribution. Mol. Cell. Biol. 25:488-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lew, D. J., and D. J. Burke. 2003. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 37:251-282. [DOI] [PubMed] [Google Scholar]
- 43.Li, J. M., Y. Li, and S. J. Elledge. 2005. Genetic analysis of the kinetochore DASH complex reveals an antagonistic relationship with the Ras/protein kinase A pathway and a novel subunit required for Ask1 association. Mol. Cell. Biol. 25:767-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li, Y., J. Bachant, A. A. Alcasabas, Y. Wang, J. Qin, and S. J. Elledge. 2002. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 16:183-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 46.MacKelvie, S. H., P. D. Andrews, and M. J. Stark. 1995. The Saccharomyces cerevisiae gene SDS22 encodes a potential regulator of the mitotic function of yeast type 1 protein phosphatase. Mol. Cell. Biol. 15:3777-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minshull, J., A. Straight, A. Rudner, A. Dernburg, A. Belmont, and A. W. Murray. 1996. Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr. Biol. 6:1609-1620. [DOI] [PubMed] [Google Scholar]
- 48.Miranda, J. J., P. De Wulf, P. K. Sorger, and S. C. Harrison. 2005. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 12:138-143. [DOI] [PubMed] [Google Scholar]
- 49.Murnion, M. E., R. R. Adams, D. M. Callister, C. D. Allis, W. C. Earnshaw, and J. R. Swedlow. 2001. Chromatin-associated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J. Biol. Chem. 276:26656-26665. [DOI] [PubMed] [Google Scholar]
- 50.Nelson, K. K., M. Holmer, and S. K. Lemmon. 1996. SCD5, a suppressor of clathrin deficiency, encodes a novel protein with a late secretory function in yeast. Mol. Biol. Cell 7:245-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ni, L., and M. Snyder. 2001. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell 12:2147-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osborne, M. A., G. Schlenstedt, T. Jinks, and P. A. Silver. 1994. Nuf2, a spindle pole body-associated protein required for nuclear division in yeast. J. Cell Biol. 125:853-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peggie, M. W., S. H. MacKelvie, A. Bloecher, E. V. Knatko, K. Tatchell, and M. J. Stark. 2002. Essential functions of Sds22p in chromosome stability and nuclear localization of PP1. J. Cell Sci. 115:195-206. [DOI] [PubMed] [Google Scholar]
- 54.Pinsky, B. A., S. Y. Tatsutani, K. A. Collins, and S. Biggins. 2003. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev. Cell 5:735-745. [DOI] [PubMed] [Google Scholar]
- 55.Ramaswamy, N. T., L. Li, M. Khalil, and J. F. Cannon. 1998. Regulation of yeast glycogen metabolism and sporulation by Glc7p protein phosphatase. Genetics 149:57-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rogers, E., J. D. Bishop, J. A. Waddle, J. M. Schumacher, and R. Lin. 2002. The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J. Cell Biol. 157:219-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rose, M. D., F. Winston, and P. Heiter. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 58.Sassoon, I., F. F. Severin, P. D. Andrews, M. R. Taba, K. B. Kaplan, A. J. Ashford, M. J. Stark, P. K. Sorger, and A. A. Hyman. 1999. Regulation of Saccharomyces cerevisiae kinetochores by the type 1 phosphatase Glc7p. Genes Dev. 13:545-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shang, C., T. R. Hazbun, I. M. Cheeseman, J. Aranda, S. Fields, D. G. Drubin, and G. Barnes. 2003. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol. Biol. Cell 14:3342-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stevenson, L. F., B. K. Kennedy, and E. Harlow. 2001. A large-scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes. Proc. Natl. Acad. Sci. USA 98:3946-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stuart, J. S., D. L. Frederick, C. M. Varner, and K. Tatchell. 1994. The mutant type 1 protein phosphatase encoded by glc7-1 from Saccharomyces cerevisiae fails to interact productively with the GAC1-encoded regulatory subunit. Mol. Cell. Biol. 14:896-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sugiyama, K., K. Sugiura, T. Hara, K. Sugimoto, H. Shima, K. Honda, K. Furukawa, S. Yamashita, and T. Urano. 2002. Aurora-B associated protein phosphatases as negative regulators of kinase activation. Oncogene 21:3103-3111. [DOI] [PubMed] [Google Scholar]
- 63.Tanaka, T. U., N. Rachidi, C. Janke, G. Pereira, M. Galova, E. Schiebel, M. J. Stark, and K. Nasmyth. 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108:317-329. [DOI] [PubMed] [Google Scholar]
- 64.Trinkle-Mulcahy, L., P. D. Andrews, S. Wickramasinghe, J. Sleeman, A. Prescott, Y. W. Lam, C. Lyon, J. R. Swedlow, and A. I. Lamond. 2003. Time-lapse imaging reveals dynamic relocalization of PP1γ throughout the mammalian cell cycle. Mol. Biol. Cell 14:107-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tu, J., W. Song, and M. Carlson. 1996. Protein phosphatase type 1 interacts with proteins required for meiosis and other cellular processes in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4199-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tung, H. Y., W. Wang, and C. S. Chan. 1995. Regulation of chromosome segregation by Glc8p, a structural homolog of mammalian inhibitor 2 that functions as both an activator and an inhibitor of yeast protein phosphatase 1. Mol. Cell. Biol. 15:6064-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ubersax, J. A., E. L. Woodbury, P. N. Quang, M. Paraz, J. D. Blethrow, K. Shah, K. M. Shokat, and D. O. Morgan. 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425:859-864. [DOI] [PubMed] [Google Scholar]
- 68.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 69.Vagnarelli, P., and W. C. Earnshaw. 2004. Chromosomal passengers: the four-dimensional regulation of mitotic events. Chromosoma 113:211-222. [DOI] [PubMed] [Google Scholar]
- 70.Venturi, G. M., A. Bloecher, T. Williams-Hart, and K. Tatchell. 2000. Genetic interactions between GLC7, PPZ1 and PPZ2 in Saccharomyces cerevisiae. Genetics 155:69-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Walsh, E. P., D. J. Lamont, K. A. Beattie, and M. J. Stark. 2002. Novel interactions of Saccharomyces cerevisiae type 1 protein phosphatase identified by single-step affinity purification and mass spectrometry. Biochemistry 41:2409-2420. [DOI] [PubMed] [Google Scholar]
- 72.Wek, R. C., J. F. Cannon, T. E. Dever, and A. G. Hinnebusch. 1992. Truncated protein phosphatase GLC7 restores translational activation of GCN4 expression in yeast mutants defective for the eIF-2α kinase GCN2. Mol. Cell. Biol. 12:5700-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Westermann, S., A. Avila-Sakar, H. W. Wang, H. Niederstrasser, J. Wong, D. G. Drubin, E. Nogales, and G. Barnes. 2005. Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol. Cell 17:277-290. [DOI] [PubMed] [Google Scholar]
- 74.Wigge, P. A., and J. V. Kilmartin. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152:349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu, X., and K. Tatchell. 2001. Mutations in yeast protein phosphatase type 1 that affect targeting subunit binding. Biochemistry 40:7410-7420. [DOI] [PubMed] [Google Scholar]
- 76.Zhang, K., W. Lin, J. A. Latham, G. M. Riefler, J. M. Schumacher, C. Chan, K. Tatchell, D. H. Hawke, R. Kobayashi, and S. Y. Dent. 2005. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell 122:723-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang, S., S. Guha, and F. C. Volkert. 1995. The Saccharomyces SHP1 gene, which encodes a regulator of phosphoprotein phosphatase 1 with differential effects on glycogen metabolism, meiotic differentiation, and mitotic cell cycle progression. Mol. Cell. Biol. 15:2037-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]