Abstract
In higher eukaryotes, the large subunit of the general transcription factor TFIIA is encoded by the single TFIIAαβ gene and posttranslationally cleaved into α and β subunits. The molecular mechanisms and biological significance of this proteolytic process have remained obscure. Here, we show that TFIIA is a substrate of taspase 1 as reported for the trithorax group mixed-lineage leukemia protein. We demonstrate that recombinant taspase 1 cleaves TFIIA in vitro. Transfected taspase 1 enhances cleavage of TFIIA, and RNA interference knockdown of endogenous taspase 1 diminishes cleavage of TFIIA in vivo. In taspase 1−/− MEF cells, only uncleaved TFIIA is detected. In Xenopus laevis embryos, knockdown of TFIIA results in phenotype and expression defects. Both defects can be rescued by expression of an uncleavable TFIIA mutant. Our study shows that uncleaved TFIIA is transcriptionally active and that cleavage of TFIIA does not serve to render TFIIA competent for transcription. We propose that cleavage fine tunes the transcription regulation of a subset of genes during differentiation and development.
In eukaryotes, initiation of RNA polymerase II transcription requires the assembly of a preinitiation complex. Specific binding of TBP to promoters is a key step in the formation of PIC, which is followed by recruitment of general transcription factors and polymerase II. The basal transcription factor TFIIA interacts with TBP and stabilizes its binding to DNA (26, 28). TFIIA has also been shown to interact with several activators (11, 12, 18, 27) and is required for transcriptional activation of certain genes (10, 13, 14, 20, 21).
In higher eukaryotes, purified TFIIA is composed of three subunits, α, β, and γ. TFIIAαβ is encoded by a single gene and cleaved posttranslationally into α and β subunits. The γ subunit is conserved among different species, whereas sequence similarity in TFIIAαβ is limited mostly to the N-terminal region of the α subunit and the C terminus covering most of the β subunit (19). Recently, the cleavage recognition site (CRS) that is essential for TFIIA cleavage has been identified as QVDG (amino acids [aa] 272 to 275), and the N terminus of the β subunit was determined to be at D278, located 3 amino acids downstream of the CRS (Fig. 1B) (6). The CRS is remarkably similar in different evolutionarily distinct species and is embedded in an otherwise nonconserved and probably unstructured region (1, 4, 24). The germ cell-specific TFIIA-like factor ALF, a TFIIA variant that contains the CRS, was also shown to be cleaved (5, 6). TFIIA cleavage was first reported more than a decade ago (26), and it has been generally assumed that uncleaved TFIIA is the precursor and cleavage occurs to activate TFIIA for transcription. Both uncleaved αβ and the cleaved α and β subunits can be found in association with the TFIIAγ subunit in vivo (15, 16), and both forms interact with TBP on DNA and support transcription to similar extents in vitro and in reporter assays (6, 22). TFIIA is mainly found in the cleaved form (α plus β plus γ) in differentiated cells. In embryonal carcinoma P19 cells, a substantial amount of uncleaved TFIIA (αβ plus γ) is detected and stably interacts with TBP in the TAC complex to mediate transcription (15, 16). Therefore, uncleaved and cleaved forms of TFIIA may have distinct gene regulatory functions in differentiation. The observation that cleavage is the prerequisite for proteasome-mediated degradation of TFIIA (6) indicates that cleavage regulates TFIIA protein levels and may play a role in transcription. Elucidation of the biological function of TFIIA cleavage is hampered because the protease(s) that specifically cleaves TFIIA has not been identified.
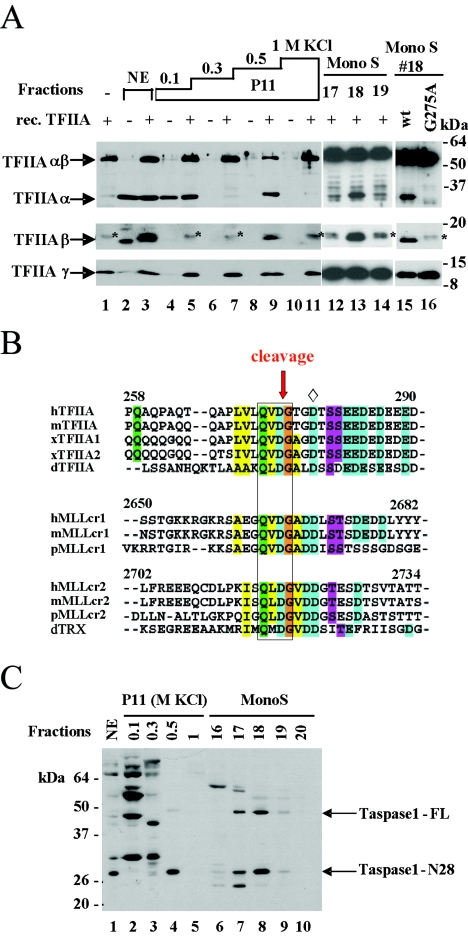
FIG. 1.
Taspase 1 activity in HeLa nuclear extracts. (A) A protease assay was performed to detect TFIIA cleavage activity in HeLa nuclear extracts. Highly purified recombinant TFIIA (rec. TFIIA) was used as the substrate as indicated, and Western blotting analysis was used to detect the cleavage products. The protease activity was further fractionated on a P11 column (lanes 4 to 11), followed by a Mono S column (lanes 12 to 14). The specificity of the protease activity was tested by incubation of purified wild-type (wt) TFIIA or the cleavage site G275A mutant with Mono S fraction 18 (lanes 15 and 16). Nonspecific bands are indicated by asterisks. (B) Alignment of the CRS and the cleavage site of TFIIA and the MLL protein from different organisms, i.e., humans (h), mice (m), Xenopus (x), pufferfish (p), and Drosophila (d). The conserved CRS is boxed. Cleavage of TFIIA and the MLL protein by taspase 1 is at D/G (red arrow). D278 (⋄) is the identified N-terminal end of the β subunit of TFIIA purified from mammalian cells. The acidic stretch (residues in blue and purple) is relatively conserved in TFIIA and the MLL protein. (C) Endogenous taspase 1, full-length taspase 1 (Taspase1-FL), and the autocleaved N-terminal part (Taspase1-N28) were detected by a taspase 1-specific antibody in all fractions containing the protease activity. NE, nuclear extracts.
The recently identified cleavage site in TFIIAαβ, G277/D278 (6), did not match known consensus sequences of proteases. The CRS, QVDG, of TFIIA is, however, virtually identical to the cleavage sites of the MLL (mixed-lineage leukemia) protein, QVD/G (aa 2664 to 2667) and QLD/G (aa 2716 to 2719) (Fig. 1B) (8, 17). The MLL protein is a 500-kDa nuclear protein of the trithorax (Trx) group of proteins and is required for maintenance of proper HOX gene expression. Chromosomal translocation results in different MLL fusion proteins that are involved in various leukemias (3). The MLL protein is proteolytically cleaved at two adjacent cleavage sites by a single protease, taspase 1, an endopeptidase with an asparaginase 2 homology domain (7). Moreover, there is an acidic stretch downstream of the cleavage site in both the MLL protein and TFIIA. These similarities strongly indicate a molecular and/or functional link between TFIIA and MLL protein processing.
Here, we show that TFIIA is a genuine substrate of taspase 1. Taspase 1 cleaves TFIIA in vitro and in vivo. RNA interference (RNAi) knockdown of taspase 1 reduces cleavage of TFIIA, and TFIIA cleavage is undetectable in taspase 1−/− knockout mouse embryonal fibroblasts (MEFs). In Xenopus laevis, TFIIA is required in early development and gene expression, and an uncleavable mutant was able to rescue the phenotype in development and transcriptional defects in TFIIA-dependent genes.
MATERIALS AND METHODS
Plasmids and antibodies.
Mammalian expression vectors, myc-tagged TFIIAαβ (pSG5-myc-TFIIAαβ), its CRS mutants (alanine mutants from L271 to T279), hemagglutinin-tagged TFIIAγ (pSG5-HA-TFIIAγ), a green fluorescent protein (GFP) construct (pEGFP-N1) (6), and pcDNA-taspase 1 and its T234A mutant form (7) were described previously. TFIIAαβ and -γ genes were subcloned from their mammalian expression vectors into a single polycistronic vector, pST39 (23), between the SacI and KpnI sites and between the XbaI and BamHI sites, respectively, to generate pST-IIAγαβ for expression in Escherichia coli. The CRS mutants (alanine mutants from L271 to T279) in the pST vector were generated by excision of the MscI and NotI fragments containing the mutated sequences from the mammalian vectors and replacement of the wild-type sequence in pST-IIAγαβ. By comparative expressed sequence tag analysis, we identified a cDNA encoding a previously uncharacterized X. laevis TFIIAαβ gene (GenBank accession no. BE575947), referred to as αβ3, that is closely related to Xenopus TFIIAαβ1 and was previously described (5). Xenopus TFIIAαβ3 was PCR amplified and cloned into the BglII and EcoRV sites of pT7TS (29). A G269A mutation was introduced into the TFIIAαβ3 construct by site-directed mutagenesis and confirmed by sequencing. αβMO (morpholino)-resistant TFIIAαβ constructs were obtained by introducing seven silent mutations within the target sequence. Capped RNAs for injections were transcribed with an in vitro RNA synthesis kit (Ambion).
Protein C antibody was purchased from Roche Molecular Biochemicals. Monoclonal myc antibody; polyclonal GFP antibody; polyclonal TFIIAα-, TFIIAβ-, and TFIIA-γ-specific (16) antibodies; and taspase 1 (7) antibodies were previously described. To detect endogenous TFIIA, immunopurified polyclonal TFIIAα-, TFIIAβ-, and TFIIAγ-specific antibodies were used.
Cell culture, transient transfection, RNAi, protein extraction, and immunoprecipitation.
Maintenance and transfection of U2OS cells and extract preparation were performed as previously described (6). RNAi knockdown of taspase 1 was carried out with duplex RNAi oligonucleotides as described previously (7). After 48 h of RNAi treatment of U2OS cells, RNAi oligonucleotides were removed and cells were transfected with TFIIA plasmids. To detect the effect of taspase 1 RNAi knockdown on endogenous TFIIA, U2OS cells were treated with RNAi oligonucleotides for 3 and 4 days.
Protein expression and purification and Edman sequencing.
Polycistronic expression plasmid pST-IIAγαβ (and its CRS mutants) carrying both the TFIIAαβ (and its mutants) and TFIIAγ genes was transformed into BL21(DE3)plysS cells, and induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Overexpressed wild-type TFIIAαβ and TFIIAγ proteins were purified as a complex through Ni-nitrilotriacetic acid (NTA), Mono Q, and Mono S columns to nearly homogeneity. The purified TFIIA complex was functionally assayed in an electrophoretic mobility shift assay after each purification step. TFIIAγαβ CRS mutants expressed from pST constructs were semipurified with Ni-NTA resin and eluted with 250 mM imidazole before being subjected to in vitro protease assays. To purify the protease activity for TFIIA cleavage, HeLa nuclear extracts were prepared in high-salt buffer containing 20 mM HEPES (pH 7.8), 50 mM KCl, 300 mM NaCl, 0.1 mM EDTA, 1 mM dithiothreitol, 10% glycerol, 0.1 mM, phenylmethylsulfonyl fluoride, and 1× complete protease inhibitors (Roche Molecular Biochemicals) and fractionated on a P11 column, followed by step elutions with 100, 300, 500, and 1,000 mM KCl. The PC-C fraction (500 mM fraction) containing the protease activity was further fractionated on a Mono S column. For the Mono S column, a gradient of 10 to 1,000 mM KCl was applied and the activity eluted at 300 mM KCl. Protease activity was monitored with the in vitro protease assay. To perform Edman N-terminal sequencing analysis, recombinant TFIIA (rTFIIA) was digested with recombinant taspase 1, followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and staining with Coomassie. The band corresponding to the β subunit was excised and subjected to Edman N-terminal sequencing analysis as described before (8). Around 3 to 4 million taspase 1−/− MEF cells were extracted in 400 μl of low-salt buffer as described previously (7).
In vitro protease assay.
The purified rTFIIA complex and semipurified TFIIA (and its mutant forms) were incubated at 37°C with 10 ng of recombinant taspase 1 for 1 h or with HeLa nuclear fractions for 12 h. Protease reaction buffer P contained 20 mM Tris (pH 8.0), 100 mM KCl, 0.2 mM EDTA, 2 mM dithiothreitol, and 10% glycerol. The reaction mixture was subsequently analyzed by Western blotting and probed for TFIIAαβ α, β, and γ subunits with the respective antibodies. One microliter of extract of taspase 1−/− MEF cells was incubated with 10 ng of recombinant taspase 1 in protease reaction buffer P, and cleavage was detected by Western blotting with immunopurified TFIIAα-, β-, and γ-specific antibodies.
Modified oligonucleotides for Xenopus injection.
Twenty nanograms of TFIIAαβ antisense MO-modified oligonucleotide (αβMO; 5′-GCGGCCTCGGCTAACGCAAACCCCG; Gene Tools) was generally injected per embryo. The same amount of standard control MO (cMO) was injected in parallel.
Western blotting for Xenopus extracts.
Xenopus embryo extracts were prepared as previously described (25). Usually, 2 egg equivalents was used for Western blotting.
TFIIA-TBP binding assay.
Band shift assays were performed as described previously (5), with 1 ng of recombinant TBP (rTBP), 0.5 to 1 embryo equivalent of Xenopus embryo extracts, and the adenovirus major late promoter TATA box radiolabeled probe.
RNA isolation and reverse transcription-quantitative PCR (RT-qPCR).
Xenopus embryo RNA was isolated by Trizol (Invitrogen) extraction and LiCl precipitation. RT was performed with 1 to 2 embryo equivalents of total mRNA with Superscript II (Amersham) and a combination of random hexanucleotides and pT21V. Levels of cDNA were quantified by qPCR. Designed primer sets for qPCR were as follows: for Hmg1, 5′-TCGAAAGGAAAGCTGCCAAG-3′ (forward) and 5′-GCAGGTTCTGGCTTTCCCTTA-3′ (reverse); for 1A11, 5′-CGAAGATGCTTTTCCTGCCA-3′ (forward) and 5′-CACAATCACAGGCTGCATGTG-3′ (reverse); for Hoxb4, 5′-CAAGAGATCCCGCACAGCTTA-3′ (forward) and 5′-CAAAGTGTGCGCAATTTCCA-3′ (reverse). Primer sets for Gsc, Gs17, Xbra, MyoDb, and Eflα are described at http://www.hhmi.ucla.edu/derobertis/index.html, and that for retinoblastoma (Rb) was described previously (9).
RESULTS
Specific protease activity for TFIIA cleavage in HeLa cell nuclear extracts.
To identify the protease for TFIIA, we set up an in vitro cleavage assay with purified rTFIIA composed of uncleaved αβ and γ subunits as the substrate to test for a cleavage activity for TFIIA in HeLa cell extracts. In crude nuclear extracts, cleavage could readily be monitored by the appearance of the His-tagged, exogenous β subunit (Fig. 1A, compare lanes 2 and 3); the cleaved, exogenous α subunit cannot be discriminated from endogenous TFIIA. Fractionation of nuclear extracts on a P11 column showed that the cleavage activity was eluted at 500 mM KCl (PC-C fraction) (Fig. 1A, lanes 8 and 9). The PC-C fraction was further fractionated on a Mono S column, and the cleavage activity was recovered at approximately 300 mM KCl in fractions 17, 18, and 19 (Fig. 1A, lanes 12 to 14). To assess whether the observed cleavage activity is specific and displays the same amino acid sequence requirements as in vivo, we tested a G275A cleavage site mutant of TFIIA (6) in our in vitro assay. The protease activity in Mono S fraction 18 cleaved wild-type TFIIA but not the G275A mutant (Fig. 1A, lanes 15 and 16), showing that the cleavage activity for TFIIA in HeLa nuclear extracts is specific.
Having identified the CRS for TFIIA (6), we noticed that the CRS in TFIIA, QVDG (aa 272 to 275), is identical or similar to the cleavage sites in the MLL protein, QVDG (aa 2664 to 2667) and QLDG (aa 2716 to 2719), which are both cleaved at D/G by taspase 1 (Fig. 1B). This similarity indicated that TFIIA and the MLL protein might be cleaved by the same protease. To test whether HeLa nuclear fractions enriched for TFIIA cleavage activity contain taspase 1, Western blotting analysis was performed with an anti-taspase 1 antibody (7). Figure 1C shows that autocleaved taspase 1 is present in fractions with cleavage activity for TFIIA, including the nuclear extracts (lane 1), the PC-C fraction (lane 4), and Mono S fractions 17 to 19 (lanes 7 to 9). Full-length taspase 1 could not be observed in whole-cell extracts (7) or nuclear extracts (lane 1) but was detectable in the PC-C fraction and was further enriched in Mono S fractions 17 to 19. These data strongly suggest that taspase 1 is the protease for TFIIA cleavage.
Cleavage of TFIIA by taspase 1 in vitro.
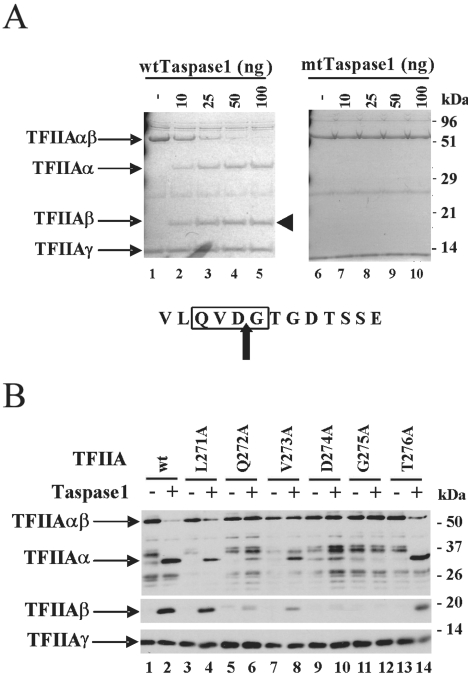
To directly assess whether TFIIA is a substrate of taspase 1, we first tested cleavage in vitro with recombinant TFIIA and taspase 1. Recombinant wild-type taspase 1 cleaved TFIIA efficiently (Fig. 2A, lanes 1 to 5), while the T234A active-site mutant of taspase 1 did not cleave wild-type TFIIA (Fig. 2A, lanes 6 to 10). Although the in vitro assays showed that taspase 1 cleaves TFIIA, the determined cleavage site of the MLL protein is different from that of TFIIA purified from cell extracts and analyzed by Edman degradation. Edman sequencing showed that cleavage in the MLL protein occurs at D/G within the conserved CRS, QVDG or QLDG (8), whereas in TFIIA, the most N-terminal amino acid of the β subunit was determined to be D278, 3 amino acids downstream of the CRS (Fig. 1B) (6). To resolve this ambiguity, the N terminus of the TFIIAβ generated in vitro by recombinant taspase 1 (Fig. 2A, the marked bands on the left side) was subjected to Edman degradation. The analysis yielded the amino acid sequence GTGDTSSE, showing that cleavage of TFIIA by taspase 1 occurred at D274/G275 (Fig. 2A). This cleavage site is within the conserved CRS that is essential for TFIIA cleavage, and it is consistent with the sites of MLL protein cleavage by taspase 1 (8).
FIG. 2.
Cleavage of TFIIA by taspase 1 in vitro. (A) Coomassie staining was performed to detect cleavage of the recombinant TFIIA by recombinant taspase 1. Wild-type (wt) TFIIA was incubated with different amounts of wild-type taspase 1 (lanes1 to 5) or mutant (mt) taspase 1 (T234A) (lanes 6 to 10) as indicated. The β subunit of TFIIA (arrowhead) was cut out of the gel and subjected to Edman analysis. Edman analysis showed that G275 is the N-terminal end of the β subunit. (B) Western blot analysis was performed to test the cleavage of mutant TFIIAs covering the CRS. These TFIIA mutants were expressed in complex with the γ subunit in E. coli, and one-step Ni-NTA purification was applied to obtain semipurified proteins.
Having shown that TFIIA is cleaved by taspase 1 in vitro and the cleavage site is identical to that of the MLL protein, we tested whether cleavage of TFIIA by taspase 1 has the same amino acid requirement as cleavage of TFIIA in vivo, as shown previously (6). A panel of mutants covering the CRS which was tested previously in vivo were expressed in E. coli and purified with Ni-NTA resin, and subsequently, the Ni-NTA eluates were analyzed in our in vitro assay with recombinant taspase 1. In this assay, wild-type TFIIA (Fig. 2B, lanes 1 and 2) and mutant forms with changes flanking the CRS, L271A (lanes 3 and 4) and T276A (lanes 13 and 14), were readily cleaved by taspase 1. Cleavage of the CRS mutant forms was either completely blocked (D274A and G275A) or occurred weakly (Q272A and V273A) (Fig. 2B, lanes 9 to 12 and 5 to 8, respectively), which matches the cleavage profile observed for the endogenous protease (Fig. 3B) (6). In conclusion, our in vitro data show that cleavage by taspase 1 requires the CRS and that taspase 1 cleaves TFIIA at D274/G275.
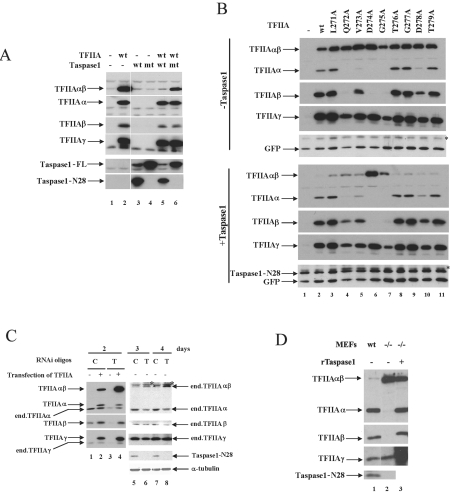
FIG. 3.
Cleavage of TFIIA by taspase 1 in vivo. (A) Wild-type (wt) TFIIA was transfected either alone or together with either wild-type or mutant (mt) taspase 1 (T234A) in U2OS cells, and cleavage was analyzed by Western blotting. This experiment was performed more than 10 time, and the ratio of uncleaved to cleaved TFIIA was consistent. (B) TFIIA mutants covering the CRS were tested either alone or together with taspase 1 for their cleavage in U2OS. GFP was cotransfected as the internal control. Nonspecific bands detected by taspase 1 antibody are indicated by asterisks. (C) Endogenous (end.) taspase 1 was knocked down by RNAi duplex oligonucleotides (oligos). Control oligonucleotides (C) and taspase 1 oligonucleotides (T) were used in this experiment. To test the effect on transiently transfected TFIIA, oligonucleotides were transfected for 48 h and removed, followed by transfection of TFIIA constructs. To test the effect on endogenous TFIIA, U2OS cells were treated with oligonucleotides for 3 and 4 days as indicated. Nonspecific bands detected by TFIIAα-specific antibody are marked by asterisks. (D) TFIIA cleavage was tested in taspase 1−/− MEF cells. Extracts from wild-type (lane1) and taspase 1−/− MEF cells incubated without (lane 2) and with recombinant taspase 1 (rTaspase1) (lane 3) were subjected to Western blot analysis. The nonspecific signal masking the TFIIAγ subunit in lane 3 is from cross-reaction of the TFIIAγ antibody with the recombinant taspase 1 preparation.
Cleavage of TFIIA by taspase 1 in vivo.
To corroborate and extend our in vitro observations, we tested whether TFIIA is cleaved by taspase 1 in vivo in transient-transfection assays. In U2OS cells, expression of wild-type taspase 1, followed by Western blot analysis with taspase 1 antibody against the N-terminal region of taspase 1 (7), revealed two polypeptides of approximately 50 kDa and 28 kDa (Fig. 3A, lanes 3 and 5) corresponding to full-length taspase 1 (taspase 1-FL) and the autocleaved N terminus (taspase 1-N28) (7). Coexpression of TFIIA and taspase 1 led to complete cleavage of TFIIAαβ (lanes 5), while expression of T234A mutant taspase 1, which cannot undergo autocleavage, did not change the ratio of uncleaved and cleaved TFIIA (compare lanes 6 and 2). These data show that TFIIA is cleaved specifically by taspase 1 in vivo. Interestingly, we did not observe a clear increase in the TFIIAα and TFIIAβ subunits upon complete cleavage of TFIIAαβ (compare lanes 5 and 6), suggesting that in vivo the levels of cleaved TFIIA are measured and maintained in cells. To assess whether the CRS is essential for taspase 1 cleavage in vivo, we again utilized the alanine scanning mutants covering the CRS. Without overexpression of taspase 1, mutations in the CRS either completely abolished cleavage of TFIIA (Q272A, D274A, G275A) (Fig. 3B, top, lanes 4, 6, and 7) or yielded only small amounts of the cleaved products (V273A) (lane 5), as observed previously (6). Coexpression of taspase 1 with wild-type TFIIA and mutants with amino acid changes outside the CRS resulted in significant reduction of the uncleaved αβ subunits (Fig. 3B, bottom, lanes 2 and 3 and 8 to 11). Q272A and V273A mutant forms showed elevated cleavage in the presence of overexpressed taspase 1, but the cleaved products remained at low levels (lanes 4 and 5). Importantly, mutant forms with changes at the cleavage site, D274A and G275A, cannot be cleaved, even upon overexpression of taspase 1 (lanes 6 and 7), demonstrating that D274 and G275 are absolutely essential for cleavage by taspase 1, which is consistent with the requirement of TFIIA cleavage by the endogenous protease (6).
The role of endogenous taspase 1 in TFIIA cleavage was further tested by an RNAi approach. In an experiment with transfected TFIIA, treatment of U2OS cells with RNAi oligonucleotides for 2 days led to a clear accumulation of the uncleaved TFIIAαβ subunit and a small decrease in the cleaved products (Fig. 3C, compare lanes 2 and 4). To investigate the effect on endogenous TFIIA, U2OS cells were treated with RNAi oligonucleotides for 3 and 4 days, respectively. This treatment gave rise to a clear decrease in the endogenous taspase 1 level and, concomitantly, accumulation of the uncleaved form of endogenous TFIIAαβ and a slight decrease in the cleaved α and β subunits (Fig. 3C, compare lanes 5 and 6 and lanes 7 and 8). To gain insights into the role of taspase 1 in TFIIA cleavage, we used MEF cells established from taspase 1−/− mice (J. J. Hsieh, unpublished data). In extracts of these MEFs, only uncleaved TFIIAαβ could be detected (Fig. 3D, compare lanes 1 and 2) and cleavage could be recovered in vitro by adding recombinant taspase 1 to taspase 1−/− MEF cell extracts (lane 3). In summary, our in vivo results show unambiguously that TFIIA is a genuine substrate for taspase 1.
Uncleaved TFIIA is transcriptionally active during early stages of Xenopus development.
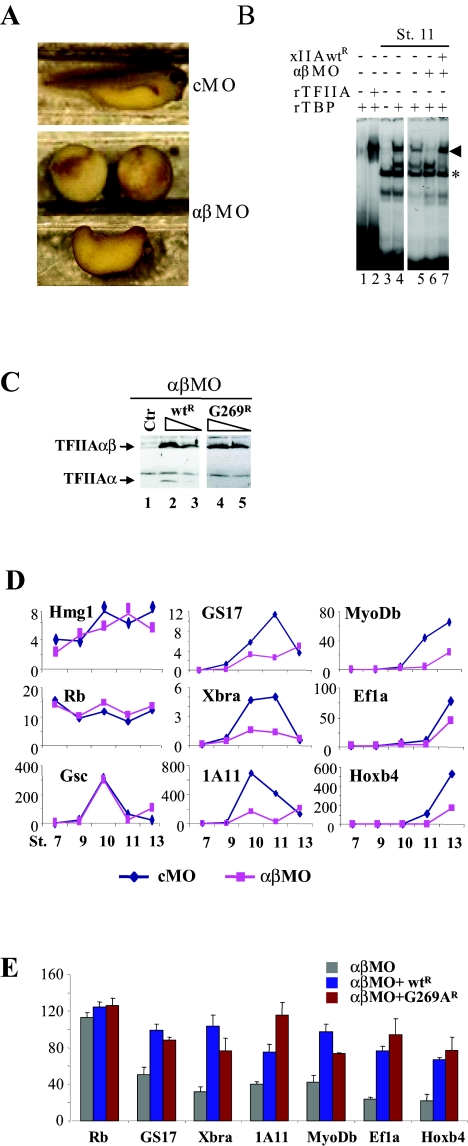
It has long been assumed that the uncleaved form is a nonfunctional precursor and that cleavage renders TFIIA functional for transcription because cleaved TFIIA is the major form present in most cells. The fact that taspase 1−/− MEF cells could be established and maintained in culture indicates that uncleaved TFIIA is most probably functional and sufficient for bulk transcription. To provide further evidence, we turned to X. laevis as a model organism where knockdown and rescue experiments can be performed more easily. As Xenopus TFIIAαβ mRNA is maternally contributed and TFIIA is detected after maturation (5), we used a morpholino antisense oligonucleotide directed against the three TFIIAαβ isoforms (αβMO) identified in X. laevis to knock down the endogenous TFIIA. Injection of αβMO, but not cMO, into one-cell-stage embryos gave rise to a variety of phenotypes, ranging from complete developmental arrest during late gastrulation to severe axial defects resulting in shortened and twisted tadpoles (Fig. 4A and data not shown). We took advantage of the TATA box binding property of the TBP-TFIIA complex (TA complex) to assess the efficiency of TFIIA knockdown. Extracts from stage 11 embryos supplemented with rTBP yielded a TA complex migrating at the same position as a TA complex from rTFIIA and rTBP (Fig. 4B, compare lane 4 and lane 2). Extracts from embryos injected with the αβMO antisense oligonucleotide did not yield a TA band shift, showing that endogenous TFIIA is efficiently knocked down (Fig. 4B, lane 6). To rescue TFIIA expression, we utilized an antisense-resistant TFIIAαβ synthetic mRNA (hereafter xIIAwtR) in which silent mutations blocked knockdown by the antisense oligonucleotide. Coinjection of xIIAwtR mRNA together with αβMO restored the TA complex (Fig. 4B, lane 7), showing that αβMO specifically knocked down endogenous xTFIIA but not morpholino-resistant xIIAwtr. To investigate whether uncleaved TFIIA is functional, silent mutations were also introduced into an uncleavable mutant form of xTFIIA (G269A; corresponding to human G275A) mRNA (G269AR), and the xIIAwtR and G269AR mRNAs were tested in two different amounts (150 ng and 300 ng) in rescue experiments. In two independent experiments, phenotypes of TFIIA knockdown by αβMO were largely rescued by injection of xIIAwtr mRNA, 66.1% and 78.8% at amounts of 150 ng and 300 ng, respectively (Table 1, Total), indicating that the observed phenotypic defects are specific for TFIIA knockdown. Importantly, injection of G269AR mRNAs resulted in a similar rescue of the αβΜΟ phenotype, 66.1% and 69.5% at amounts of 150 ng and 300 ng, respectively (Table 1, Total), showing that G269A is able to replace endogenous TFIIA in embryonal development. Note that wild-type TFIIA and the G269A mutant were expressed at similar levels (Fig. 4C). Therefore, the uncleavable G269A mutant form of TFIIA is functional during the early stages of Xenopus development.
FIG. 4.
An uncleavable G269A mutant of xTFIIA is transcriptionally active in early Xenopus development. (A) TFIIA is required for early development. Embryos at the one-cell stage were injected with 20 ng of cMO or αβMO, and pictures were taken at stage 37 (tadpole). (B) Knockdown of endogenous xTFIIA was assessed by a TBP-TFIIA band shift assay. Embryos were injected with αβMO alone or together with αβMO-resistant xIIAwtR mRNA and collected at stage (St.) 11. A 32P-labeled TATA box probe was incubated in the presence of rTBP (except for lane 3) and either rTFIIA or extracts from stage 11 embryos and analyzed as described in Materials and Methods. The TA complex is represented by the symbol ◂, and nonspecific bands are marked by asterisks. (C) Embryos were injected with αβMO alone or together with xIIAwtR or G269AR mRNA and analyzed at stage 11. αβMO-resistant xIIAwtR (lanes 2 and 3) and G269AR (lanes 4 and 5) were expressed at similar levels. Ctr, control. (D) TFIIA is required for gene expression during early stages of Xenopus development. Expression levels of several genes were analyzed at the indicated stages (St.) by RT-qPCR from extracts of embryos injected with either cMO or αβMO. (E) An uncleavable G269A mutant form is able to rescue the expression of TFIIA-dependent genes. Embryos were injected with αβMO alone or together with xIIAwtR or G269AR mRNA and analyzed at stage 11 by RT-qPCR. The expression values of αβMO plus xIIAwtR versus αβMO or αβMO plus G269AR versus αβMO are significantly different (P < 0.05), except for Rb (P > 0.05). The difference in expression values between αβMO plus xIIAwtR and αβMO plus G269AR was not significant (P > 0.05).
TABLE 1.
Rescue of TFIIA function by the G269A mutant in X. laevis embryosa
| Parameter | cMO | αβMO | αβMO + xIIAwtR (150 ng) | αβMO + xIIAwtR (300 ng) | αβMO + G269AR (150 ng) | αβMO + G269AR (300 ng) |
|---|---|---|---|---|---|---|
| No. of injected embryos in expt 1 | 11 | 32 | 17 | 20 | 29 | 26 |
| % Abnormalb | 0 | 100 | 29.4 | 25.0 | 24.1 | 34.6 |
| % Normal | 100 | 0 | 70.6 | 75.0 | 75.9 | 65.4 |
| No. of injected embryos in expt 2 | 31 | 48 | 45 | 32 | 30 | 33 |
| % Abnormalb | 0 | 100 | 35.6 | 18.6 | 43.3 | 27.3 |
| % Normal | 100 | 0 | 64.4 | 81.3 | 56.7 | 72.7 |
| Total no. of injected embryos | 42 | 80 | 62 | 52 | 59 | 59 |
| % Abnormalb | 0 | 100 | 33.9 | 21.2 | 33.9 | 30.5 |
| % Normal | 100 | 0 | 66.1 | 78.8 | 66.1 | 69.5 |
Phenotypes were scored at stage 37.
The abnormal phenotypes observed in MO-injected embryos ranged from arrested gastrulation to severe axial defects.
To study the transcriptional role of TFIIA, we set out to identify TFIIA-dependent genes. We screened a low-density X. laevis cDNA microarray and identified several candidate genes that were consistently down-regulated upon αβMO injection (unpublished data). To verify that these genes are TFIIA dependent, we analyzed the expression of these genes at different stages of development after the onset of embryonic transcription at the mid-blastula transition (stage 8.5) by RT-qPCR (Fig. 4D). Several maternally contributed mRNAs, such as those encoding the high-mobility group 1 (Hmg1) and Rb proteins, were not affected by TFIIA knockdown. In contrast, a number of genes transcribed de novo during early embryogenesis (GS17, Xbra, 1A11, MyoDb, Ef1α, and Hoxb4) were down-regulated at different stages of development. Induction of the homeobox transcription factor Goosecoid (Gsc), however, was not affected upon TFIIA knockdown, which excludes the possibility that the observed effects on gene expression resulted from a general developmental delay. Therefore, the defects in gene-specific expression caused by TFIIA knockdown showed that TFIIA is essential for gene expression in early Xenopus development. Rescue of gene expression of the TFIIA-dependent genes was tested by injection of xIIAwtr or uncleavable G269AR mRNA together with αβMO and subsequent RT-qPCR analysis of these genes (GS17, Xbra, 1A11, MyoDb, Ef1α, and Hoxb4) from extracts of embryos at stage 11. As shown in Fig. 4E, wild-type and G269A mutant TFIIA could rescue expression defects in these genes caused by αβMO to significant levels (P < 0.05 in both cases), while the levels of rescue by xIIAwt and G269A were not significantly different (P > 0.05), showing that the uncleavable G269A mutant is fully functional in transcription during early stages of Xenopus development.
DISCUSSION
In this study, we have provided several lines of evidence that taspase 1 is the protease for TFIIA. First, taspase 1 cleaves TFIIA efficiently in vitro and in vivo, whereas the TFIIA cleavage site mutants D274A and G275A cannot be cleaved by taspase 1. Second, knockdown of endogenous taspase 1 by RNAi reduces cleavage of overexpressed, as well as endogenous, TFIIA, and most conclusively, uncleaved TFIIA is the only form detected in taspase 1−/− MEFs. The fact that taspase 1−/− MEF cells could be established and maintained in culture indicates that the uncleaved TFIIA is transcriptionally active rather than a nonfunctional precursor. This conclusion was corroborated and extended by MO knockdown experiments with X. laevis. An uncleavable G269A mutant of TFIIA (corresponding to human G275A) was able to rescue phenotypic and transcriptional defects caused by TFIIA knockdown, showing that uncleaved TFIIA is sufficient for bulk transcription.
We showed that taspase 1 cleaves TFIIA at D274/G275 within the highly conserved CRS and that the N terminus G275 generated by taspase 1 is different from the N terminus of the β subunit identified by us purified from mammalian extracts (6). Our new finding that G275 rather than D278 is the primary N-terminal residue of the β subunit of TFIIA is supported by the observation that mutations of D274 and G275 prevented cleavage completely, even upon overexpression of taspase 1, whereas mutation of D278 diminished but did not abolish cleavage (Fig. 3B) (6). Furthermore, TFIIA cleavage could not be detected in taspase 1−/− MEF cells, which unequivocally demonstrates that taspase 1 is the primary protease for TFIIA. Studies on the germ cell-specific paralogue of TFIIAαβ, TFIIA-like factor (ALF), showed that the C terminus of the α subunit of endogenous mouse ALF is D341 (2), indicating that the cleavage site of ALF in vivo is at D341/G342 (corresponding to D274/G275 in human TFIIA). Mass spectrometric analysis to identify the C terminus of the human TFIIAα subunit is complicated due to the lack of arginine and lysine residues in the region around the cleavage site (unpublished data). N-terminal residue D278 in the TFIIAβ subunit reported previously (6) is probably generated by a secondary protease. This could be either an endo- or an exopeptidase activity that removes three more amino acids and yields D278 as the N terminus. The secondary cleavage generates a destabilizing N terminus for the destruction pathway and might be part of an intricate regulatory circuitry to fine tune the level of TFIIA (6). Support for tight regulation of the levels of TFIIA was obtained from our transient-transfection experiments, in which a clear increase in the levels of the cleaved α and β subunits could not be observed upon complete cleavage of TFIIAαβ. Moreover, we only observed a slight decrease in the levels of the cleaved subunits in the RNAi experiments while a clear increase in uncleaved TFIIAαβ was detected (Fig. 3). These in vivo observations suggest that the level of cleaved TFIIA is measured and maintained in cells. One possibility is that excessive amounts of the cleaved subunits are degraded through the proteasome-dependent pathway. However, upon proteasome inhibitor treatment, we could not observe an increase in the level of the cleaved α and β subunits, even when the uncleaved αβ form was completely processed by overexpressed taspase 1 (data not shown), which suggests that, apart from the proteasome-dependent pathway, there may be other mechanisms involved in maintaining the cleaved-protein levels.
TFIIA is the second substrate for taspase 1 identified so far. The CRS of TFIIA is evolutionarily conserved between different species (Fig. 2B), with the exception of the large subunit of yeast TFIIA, TOA1, which does not contain a CRS and is not cleaved (19). In addition to the CRS, a downstream acidic stretch is also conserved in TFIIA in different species, as well as in Trx group proteins (Fig. 2B). Apart from the CRS and the acidic stretch, there is little homology in surrounding regions in different TFIIA proteins and no overall homology between TFIIA and the MLL protein. These findings suggest that the CRS, together with the acidic stretch, is necessary and probably sufficient for cleavage by taspase 1. The acidic stretch may play a role in cleavage recognition or facilitate docking or positioning of the active site of taspase 1 on the CRS. Searching for the CRS sequence QV/LDG in the SwissProt database revealed about 150 proteins that contain the QV/LDG sequence, and about 1/10 of these proteins contain acidic stretches (data not shown). It will be of interest to test whether they are also substrates of taspase 1.
It has remained elusive for a long time whether the uncleaved, the cleaved, or both forms of TFIIA are transcriptionally competent. Since cleaved TFIIA is the major form detected in most cell lines, it has been assumed that the cleaved form is the active form in transcription. We have previously shown that uncleaved TFIIA interacts with TBP to form a distinct TAC complex in embryonal carcinoma P19 cells (15, 16), suggesting that uncleaved TFIIA is transcriptionally active. Taspase 1−/− knockout mice in which only the uncleaved TFIIA form is present (Fig. 3D) survived until birth and showed minor overall defects (Hsieh, unpublished), indicating that uncleaved TFIIA is transcriptionally competent and that cleavage of TFIIA does not serve to render TFIIA competent for transcription. Taking advantage of an uncleavable mutant form of TFIIA and the identification of the TFIIA protease, we provide evidence that uncleaved TFIIA is functional during Xenopus development. TFIIAαβ knockdown in Xenopus resulted in reduced expression of a number of genes induced during embryogenesis, such as those for GS17 and Xbra, whereas the expression pattern of Gsc, which is also regulated during embryogenesis, was not altered (Fig. 4D). Importantly, an uncleavable TFIIA mutant (G269A in Xenopus) was able to rescue phenotypic and transcriptional defects in TFIIA knockdown embryos, showing that the uncleaved form of TFIIA is functional in early embryogenesis. Our study shows that cleaved TFIIA is dispensable for bulk transcription and reinforces our hypothesis that the biological role of TFIIA cleavage is to regulate the levels of TFIIA by degradation through the proteasome-dependent pathway (6) and cleavage of TFIIA might be important for the expression of a subset of genes. Resolving these important issues will require the generation of conditional knock-in mice carrying an uncleavable mutant of TFIIA.
Acknowledgments
Salvatore Spicuglia was supported by a Marie Curie Fellowship of the European Community program Training and Mobility of Researchers under contract HPMF-CT-2002-01646. This work was supported by grants 812.08.006 NWO (ALW) and 700.53.311 WWO-CW-TOP in The Netherlands.
We thank Xavier Le Guezennec and Michiel Vermeulen for providing reagents.
REFERENCES
- 1.Bleichenbacher, M., S. Tan, and T. J. Richmond. 2003. Novel interactions between the components of human and yeast TFIIA/TBP/DNA complexes. J. Mol. Biol. 332:783-793. [DOI] [PubMed] [Google Scholar]
- 2.Catena, R., M. Argentini, I. Martianov, C. Parello, S. Brancorsini, M. Parvinen, P. Sassone-Corsi, and I. Davidson. 2005. Proteolytic cleavage of ALF into alpha- and beta-subunits that form homologous and heterologous complexes with somatic TFIIA and TRF2 in male germ cells. FEBS Lett. 579:3401-3410. [DOI] [PubMed] [Google Scholar]
- 3.Daser, A., and T. H. Rabbitts. 2004. Extending the repertoire of the mixed-lineage leukemia gene MLL in leukemogenesis. Genes Dev. 18:965-974. [DOI] [PubMed] [Google Scholar]
- 4.Geiger, J. H., S. Hahn, S. Lee, and P. B. Sigler. 1996. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science 272:830-836. [DOI] [PubMed] [Google Scholar]
- 5.Han, S., W. Xie, S. R. Hammes, and J. DeJong. 2003. Expression of the germ cell-specific transcription factor ALF in Xenopus oocytes compensates for translational inactivation of the somatic factor TFIIA. J. Biol. Chem. 278:45586-45593. [DOI] [PubMed] [Google Scholar]
- 6.Hoiby, T., D. J. Mitsiou, H. Zhou, H. Erdjument-Bromage, P. Tempst, and H. G. Stunnenberg. 2004. Cleavage and proteasome-mediated degradation of the basal transcription factor TFIIA. EMBO J. 23:3083-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsieh, J. J., E. H. Cheng, and S. J. Korsmeyer. 2003. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell 115:293-303. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh, J. J., P. Ernst, H. Erdjument-Bromage, P. Tempst, and S. J. Korsmeyer. 2003. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol. Cell. Biol. 23:186-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jallow, Z., U. G. Jacobi, D. L. Weeks, I. B. Dawid, and G. J. Veenstra. 2004. Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proc. Natl. Acad. Sci. USA 101:13525-13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi, N., T. G. Boyer, and A. J. Berk. 1995. A class of activation domains interacts directly with TFIIA and stimulates TFIIA-TFIID-promoter complex assembly. Mol. Cell. Biol. 15:6465-6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi, N., P. J. Horn, S. M. Sullivan, S. J. Triezenberg, T. G. Boyer, and A. J. Berk. 1998. DA-complex assembly activity required for VP16C transcriptional activation. Mol. Cell. Biol. 18:4023-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraemer, S. M., R. T. Ranallo, R. C. Ogg, and L. A. Stargell. 2001. TFIIA interacts with TFIID via association with TATA-binding protein and TAF40. Mol. Cell. Biol. 21:1737-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lieberman, P. 1994. Identification of functional targets of the Zta transcriptional activator by formation of stable preinitiation complex intermediates. Mol. Cell. Biol. 14:8365-8375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lieberman, P. M., J. Ozer, and D. B. Gursel. 1997. Requirement for transcription factor IIA (TFIIA)-TFIID recruitment by an activator depends on promoter structure and template competition. Mol. Cell. Biol. 17:6624-6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitsiou, D. J., and H. G. Stunnenberg. 2003. p300 is involved in formation of the TBP-TFIIA-containing basal transcription complex, TAC. EMBO J. 22:4501-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitsiou, D. J., and H. G. Stunnenberg. 2000. TAC, a TBP-sans-TAFs complex containing the unprocessed TFIIAαβ precursor and the TFIIAγ subunit. Mol. Cell 6:527-537. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura, T., T. Mori, S. Tada, W. Krajewski, T. Rozovskaia, R. Wassell, G. Dubois, A. Mazo, C. M. Croce, and E. Canaani. 2002. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell 10:1119-1128. [DOI] [PubMed] [Google Scholar]
- 18.Ozer, J., P. A. Moore, A. H. Bolden, A. Lee, C. A. Rosen, and P. M. Lieberman. 1994. Molecular cloning of the small (gamma) subunit of human TFIIA reveals functions critical for activated transcription. Genes Dev. 8:2324-2335. [DOI] [PubMed] [Google Scholar]
- 19.Ranish, J. A., N. Yudkovsky, and S. Hahn. 1999. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 13:49-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stargell, L. A., Z. Moqtaderi, D. R. Dorris, R. C. Ogg, and K. Struhl. 2000. TFIIA has activator-dependent and core promoter functions in vivo. J. Biol. Chem. 275:12374-12380. [DOI] [PubMed] [Google Scholar]
- 21.Stargell, L. A., and K. Struhl. 1995. The TBP-TFIIA interaction in the response to acidic activators in vivo. Science 269:75-78. [DOI] [PubMed] [Google Scholar]
- 22.Sun, X., D. Ma, M. Sheldon, K. Yeung, and D. Reinberg. 1994. Reconstitution of human TFIIA activity from recombinant polypeptides: a role in TFIID-mediated transcription. Genes Dev. 8:2336-2348. [DOI] [PubMed] [Google Scholar]
- 23.Tan, S. 2001. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr. Purif. 21:224-234. [DOI] [PubMed] [Google Scholar]
- 24.Tan, S., Y. Hunziker, D. F. Sargent, and T. J. Richmond. 1996. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature 381:127-151. [DOI] [PubMed] [Google Scholar]
- 25.Veenstra, G. J., O. H. Destree, and A. P. Wolffe. 1999. Translation of maternal TATA-binding protein mRNA potentiates basal but not activated transcription in Xenopus embryos at the midblastula transition. Mol. Cell. Biol. 19:7972-7982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weideman, C. A., R. C. Netter, L. R. Benjamin, J. J. McAllister, L. A. Schmiedekamp, R. A. Coleman, and B. F. Pugh. 1997. Dynamic interplay of TFIIA, TBP and TATA DNA. J. Mol. Biol. 271:61-75. [DOI] [PubMed] [Google Scholar]
- 27.Yokomori, K., A. Admon, J. A. Goodrich, J. L. Chen, and R. Tjian. 1993. Drosophila TFIIA-L is processed into two subunits that are associated with the TBP/TAF complex. Genes Dev. 7:2235-2245. [DOI] [PubMed] [Google Scholar]
- 28.Yokomori, K., M. P. Zeidler, J. L. Chen, C. P. Verrijzer, M. Mlodzik, and R. Tjian. 1994. Drosophila TFIIA directs cooperative DNA binding with TBP and mediates transcriptional activation. Genes Dev. 8:2313-2323. [DOI] [PubMed] [Google Scholar]
- 29.Zorn, A. M., and P. A. Krieg. 1997. The KH domain protein encoded by quaking functions as a dimer and is essential for notochord development in Xenopus embryos. Genes Dev. 11:2176-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]