Abstract
The mammalian genome contains tens of thousands of CG and TG repeat sequences that have high potential to form the nonclassical left-handed double-helical Z-DNA structure. Previously we showed that activation of the colony-stimulating factor 1 (CSF1) gene by the chromatin remodeling enzyme, BRG1, results in formation of Z-DNA at the TG repeat sequence located within the promoter. In this report, we show that the TG repeats are assembled in a positioned nucleosome in the silent CSF1 promoter and that activation by BRG1 disrupts this nucleosome and results in Z-DNA formation. Active transcription is not required for the formation of Z-DNA but does result in an expanded region of Z-DNA. Formation of sequences by both BRG1 and the Z-DNA is required for effective chromatin remodeling of the CSF1 promoter. We propose the Z-DNA formation induced by BRG1 promotes a transition from a transient and partial remodeling to a more extensive disruption of the canonical nucleosomal structure. The data presented in this report establish that Z-DNA formation is an important mechanism in modulating chromatin structure, in similarity to the activities of ATP-dependent remodelers and posttranslational histone modifications.
DNA sequences with high potential for forming an alternate DNA structure, Z-DNA, are found frequently throughout the mammalian genome (6, 39). Despite extensive investigations over the past two decades, the biological function of Z-DNA structure has not been well established (34). Z-DNA-forming sequences, such as TG or GC repeats, are detected more frequently in the 5′ regulatory region of a gene than in other regions (36), suggesting that Z-DNA structure may play a role in the regulation of transcription. The formation of Z-DNA structure is well correlated with the transcriptional activity of the c-Myc gene (42). Studies of Saccharomyces cerevisiae with an artificial promoter suggest that Z-DNA structure in the promoter region can act as a cis element in gene regulation (33, 35). An analysis of human chromosome 22 indicates that Z-DNA-forming regions and the nuclear factor I (NFI) target sites are well correlated with the locations of known and predicted genes across the chromosome and accumulate around the transcription start sites (3). It was reported recently that the Z-DNA-binding activity of the E3L gene of the vaccinia virus product may regulate its pathogenicity, possibly by regulating transcription from cellular genes involved in fighting viral infection (17). We have previously shown that Z-DNA formation at the promoter of the colony-stimulating factor 1 (CSF1) gene is accompanied by transcriptional activation by a chromatin remodeling complex (25).
Mammalian DNA is organized in a highly ordered chromatin structure, with the nucleosome as its basic repeat unit. The local chromatin architecture affects the accessibility of regulatory sequences and, thus, the expression potential of a gene. Structural changes result from posttranslational histone modifications (1, 4, 20, 21, 37, 38, 44) and from the activity of ATP-dependent remodelers, prototyped by the SWI/SNF complex (9, 12, 14, 19, 27-29, 31, 32, 40). BRG1 is the essential ATPase subunit of the mammalian SWI/SNF-like BAF complexes (10, 15, 41). Mutations of BRG1 have been identified in numerous cancerous cell lines (43), and inactivation of BRG1 in mice is lethal (2). In cell lines devoid of BRG1, reexpression activates many genes, including the CSF1 gene (25, 45).
We have shown that the TG repeat sequence and a binding site for the NFI or CAAT-box transcription factor (CTF) in the CSF1 promoter are required for activation by BRG1 (25). After activation of the CSF1 gene by BRG1, a Z-DNA structure is detected in the promoter region (25), suggesting that Z-DNA formation may be involved in the activation process. However, there is no experimental evidence regarding the mechanism of Z-DNA function in the expression of the CSF1 gene. The extensive negative supercoiling generated by transcription from the promoter can stabilize Z-DNA conformation in cells (24, 42); thus, it is not clear whether Z-DNA formation is a result of transcriptional activation or whether it plays an active role in the chromatin remodeling and transcriptional activation. In this report, we provide critical evidence that formation of Z-DNA structure plays an active role in modulating inhibitory chromatin structure. We show that Z-DNA formation facilitates productive chromatin remodeling by the BRG1-containing complexes by promoting the transition from a transient and partial remodeling to a more complete disruption of the nucleosomal structure. We propose that formation of Z-DNA structure is an important mechanism in regulating chromatin structure.
MATERIALS AND METHODS
Cell culture and transfection.
SW-13 and MG63 cells were maintained as described previously (25). Transfections were performed with QIAGEN SuperFect reagent according to manufacturer instructions. The luciferase activity was assayed with the dual luciferase system from Promega.
Constructs.
pREP7-BRG1 and its control vector pREP7 were described previously (13). The CSF1 promoter constructs WT, noTG, and 18GC have been described previously (25). The mTA construct was generated by introducing point mutations in the two TATA boxes by use of a QuickChange kit from Stratagene.
ChIP.
SW-13 cells grown in 10-cm petri dishes were cotransfected with the CSF1 reporter constructs and pREP7-BRG1 or pREP7 by use of SuperFect reagent. The cells were selected with 50 μg/ml hygromycin B for 3 days before cross-linking with formaldehyde and harvesting for chromatin immunoprecipitation (ChIP) were performed as described previously (25). The antibody for BRG1 was described previously (25); the antibody for RNA polymerase II (Pol II) was from Santa Cruz Biotech.
Detection of Z-DNA structure by ZaaFOK digestion.
SW-13 cells were transfected and selected as for the ChIP experiments. MG63 cells were treated with 40 μg/ml of actinomycin D for 6 h. The cells were cross-linked with 1% formaldehyde for 5 min at room temperature, followed by washing once with 1 M glycine and once with 1× phosphate-buffered saline (PBS). Following treatment with 0.5% Triton X-100 in 1× PBS for 2 min, the cells were resuspended in 100 μl of 1 × NEB4 (20 mM Tris-acetate [pH 7.9], 10 mM magnesium acetate, 50 mM potassium acetate, 1 mM dithiothreitol) and digested with 5 ng/μl of ZaaFOK for 20 min at 37°C. The reaction was stopped by addition of 100 μl of 1 mg/ml proteinase K and 1% sodium dodecyl sulfate in 1× Tris-EDTA buffer (TE). After reverse cross-linking at 65°C for 6 h, the DNA was purified and analyzed by ligation-mediated PCR (LM-PCR) using CSF1 promoter primers as described previously (23, 25, 30).
Nucleosome mapping.
SW-13 cells grown in one 10-cm petri dish were cross-linked with 1% formaldehyde for 5 min at room temperature. Following two washes with 1× PBS, the cells were incubated in 5 ml of 1%Triton X-100 in 1× PBS for 2 min. The cells were harvested into two Eppendorf tubes and were resuspended in 100 μl of 1× TE in each tube followed by digestion with 0.05 and 0.01 U of micrococcal nuclease (Sigma) for 5 min at 37°C in 1 mM CaCl2 buffer. The reaction was stopped by addition of 100 μl of 1 mg/ml proteinase K—5 mM EGTA—1% sodium dodecyl sulfate in 1× TE. After reverse cross-linking at 65°C for 6 h, 10 μg of purified DNA was resolved on a 2% agarose gel in 1× TBE buffer followed by Southern blotting using a 32P-labeled CSF1 promoter probe (KpnI/NheI fragment).
For mapping nucleosome boundaries, dinucleosome-sized DNA was isolated from an agarose gel and subjected to LM-PCR analysis. A first round of PCR to amplify regions of interest was performed followed by primer extension with a labeled primer. Locations of the labeled primers used in primer extension are denoted in the figures. The primer sequence for forward PCR primer −361F was 5′-ACGATCATAGAGCGCCAGCACTGA-3′, for forward labeling primer −318F was 5′-AAGGAAAGGGTCGGTCCGCAGA, for reverse PCR primer −40R was 5′-GGCATGTGGTTTATGGGAAAT, and for reverse labeling primer −58R was 5′-AATCACCCTGGCCAGGCGCCA (see Fig. 2B). The primer sequence for PCR primer A (−203R) was 5′-CTTCCTAGTCACCCTCTGTCTT, for labeling primer A (−225R) was 5′-CTGCGCTGCACTTCCAAGCCT, for PCR primer B (−39R) was 5′-GGCATGTGGTTTATGGGAA, for labeling primer B (−58R) was 5′-AATCACCCTGGCCAGGCGCCA, for PCR primer C (+127R) was 5′-GCTCGCTCGCTGGCTGCTGC, for labeling primer C (+107R) was 5′-GGAGTGGGCGCCCCGGCCGA, for PCR primer D (−223F) was 5′-GAAGACAGAGGGTGACTAGGAAGA, for labeling primer D (−161F) was 5′-GAGGCGGGGGAAGGCGGCTGA, for PCR primer E (−349F) was 5′-GAGCGCCAGCACTGAATCAGCCTGG, and for labeling primer E (−329F) was 5′-GAGAGCGCGGAAGGAAAGGGT (see Fig. 3A).
FIG. 2.
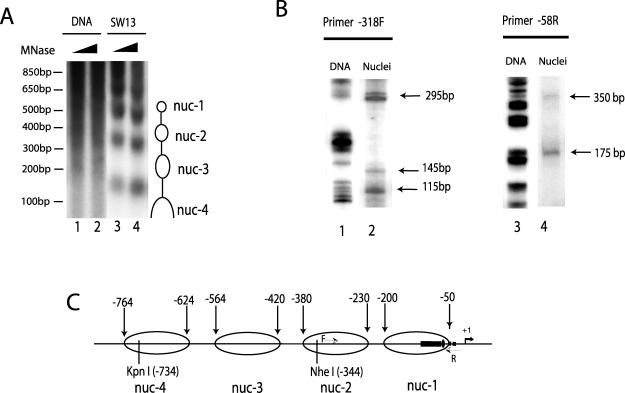
The Z-DNA-forming sequence in the CSF1 promoter is contained within a positioned nucleosome. (A) The CSF1 promoter region is organized into several positioned nucleosomes. SW-13 nuclei or purified genomic DNA was subjected to limited digestion with micrococcal nuclease (MNase) followed by complete digestion with KpnI enzyme, which recognizes a site at −734 relative to the transcription start site as +1. The products were analyzed by Southern blotting using the KpnI/NheI fragment as a probe. The nucleosomal ladder protected from the MNase digestion is indicated by open ovals on the right side of the panel. The DNA size markers are indicated on the left. (B) Mapping the nucleosomal boundaries by LM-PCR. A universal linker was ligated to the dinucleosome-sized DNA isolated from the MNase-digested nuclei and analyzed by PCR using a linker primer and CSF1-specific primers. The PCR products were detected by primer extension with 32P-labeled nested primer F or R in the promoter region (as indicated in panel C by thin arrows) and analyzed by denaturing acrylamide gel. The boundaries of the two positioned nucleosomes, named nuc-1 and nuc-2, are indicated by arrows. (C) The CSF1 promoter structure. The filled rectangle indicates the TG repeat sequence (−126 to −75); the filled oval indicates the NFI/CTF binding site; the filled squares indicate the two potential TATA boxes; the thick arrow and +1 indicate the transcription start site. The KpnI and NheI sites are indicated by vertical lines. The four nucleosomes and their boundaries are indicated by the open ovals.
FIG. 3.
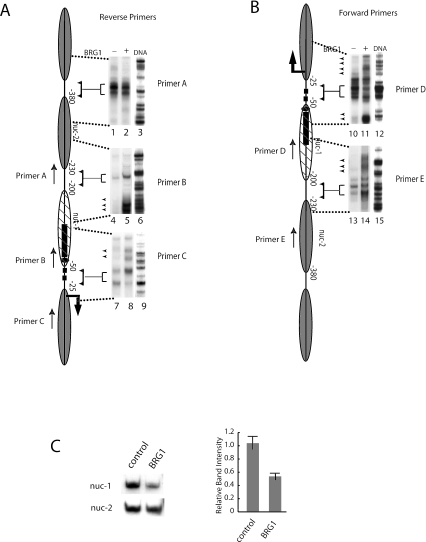
Activation of the CSF1 gene by BRG1 disrupted the −1 nucleosome and induced Z-DNA formation. (A and B) Nuclei were isolated from SW-13 cells transfected with a BRG1 or a control expression vector and were subjected to MNase digestion followed by LM-PCR analysis using nested primers specific for the CSF1 promoter. Total genomic DNA was used as a control. The location and directionality of the primers are indicated by arrows next to a schematic depicting the nucleosomal organization of the CSF1 promoter. LM-PCR products were resolved on an acrylamide gel. Brackets are used to indicate linker regions corresponding to the promoter schematic. The arrowheads indicate the cleavage sites induced by BRG1. (C) Nuclei were isolated from SW-13 cells transfected with a BRG1 or control expression vector and were digested with MNase mostly to mononucleosomes. The mononucleosome-sized DNA isolated from agarose gel was analyzed by PCR in the presence of [α-32P]dATP with primers specific to the nuc-2 nucleosome or the nuc-1 nucleosome. After 18 cycles of amplification, the products were resolved on polyacrylamide gel and quantified by phosphorimager analysis. The graph was derived by normalizing the band intensity of the nuc-1 nucleosome with that of the nuc-2 nucleosome. The error bars indicate the standard deviations of data from three separate experiments.
Restriction enzyme accessibility assay.
The SW-13 cells were transfected with pREP4-CSF1-luc reporter constructs and pREP7-BRG1 or pREP7 and with SuperFect reagent. The transfected cells were selected with hygromycin B. The restriction enzyme accessibility assay was performed as described previously (22). The primers used for the analysis of the CSF1 promoter were PCR primer 176R (5′-TCATAGCCTTATGCAGTTGCTCTCCAG-3′) and labeling primer +152R (5′-CAGCGGTTCCATCTTCCAGCGGATAGA-3′).
RESULTS
BRG1-mediated activation of the CSF1 promoter results in Z-DNA formation.
We have shown previously that the chromatin remodeling BAF or hSWI/SNF complexes are required for activation of the CSF1 gene (25). We used reverse transcription-PCR analysis to measure CSF1 gene expression in a Brg1-deficient cell line, SW-13 (Fig. 1A) (25). Transient expression of BRG1 resulted in increased CSF1 expression, while expression of an ATPase-deficient BRG1 mutant, mutBRG1, had no effect on CSF1 expression (Fig. 1A).
FIG. 1.
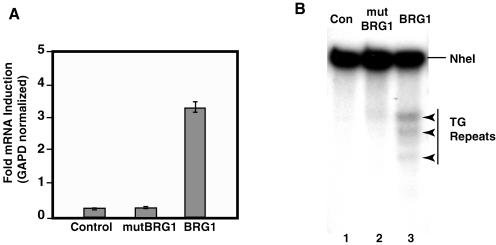
BRG1 activates expression of CSF1 and induces Z-DNA formation in the promoter. (A) SW-13 cells were transfected with a control, a BRG1 ATPase mutant (mutBRG1), or a BRG1 expression vector. Total RNA was isolated 24 h posttransfection, reverse transcribed, and analyzed by real-time PCR. CSF1 expression was normalized to GAPD. Data were plotted from three independent experiments; error bars represent standard deviations. (B) SW-13 cells transfected as described for panel A were cross-linked with formaldehyde followed by treatment with ZaaFOK protein. The cleavage sites were detected by LM-PCR using CSF1 promoter-specific primers. The TG repeat region in the promoter is indicated on the right side of the panel. The BRG1-induced cleavage sites are indicated by arrowheads on the right. Con, control.
The CSF1 promoter contains a long stretch of TG repeats, which have high potential for forming Z-DNA (8). The Z-DNA conformation, which is energetically less favorable than B-DNA, is stabilized in negatively supercoiled plasmid DNA (8). The negative supercoils generated by an actively transcribing RNA polymerase allow for the Z-DNA conformation. In addition, chromatin remodeling generates negative superhelical torsion (7, 24) and, thus, may also stabilize Z-DNA. Upon transcriptional activation of the CSF1 gene by BRG1, the TG repeat sequence within the promoter may be converted to Z-DNA conformation. We have previously shown that the CSF1 promoter does adopt a Z-DNA conformation in cells constitutively expressing BRG1 in vivo but not in cells lacking the protein. To detect Z-DNA structure, we cross-linked the cells with formaldehyde, which may prevent the conversion of Z-DNA to a more stable B-DNA conformation when the cells are disrupted. Following permeabilization, the cells were treated with a fusion protein that contains a Z-DNA binding domain and the nuclease domain from the restriction endonuclease FokI. This protein, ZaaFOK, specifically binds Z-DNA via two copies of the Z-DNA-binding domain (Zα) of the editing enzyme ADAR1 (16, 25). The ZaaFOK nuclease domain then makes cleavages within or around the Z-DNA region to which it is bound. The generation of cleavage sites in regions of interest can be detected by LM-PCR. We assayed the induction of Z-DNA structure at the CSF1 promoter in vivo by first formaldehyde cross-linking SW-13 cells transfected with either an expression vector for BRG1 for mutBRG1 or a control expression vector. The cells were then permeabilized and treated with ZaaFOK. The LM-PCR products with CSF1 promoter-specific primers are shown in Fig. 1B. Several additional bands were generated when BRG1 was expressed compared to the results seen with cells expressing mutBRG1 or no BRG1. These new cleavage sites are located in or near the region of the CSF1 promoter containing the TG repeat sequence. These data indicate that a B-DNA-to-Z-DNA transition was induced by BRG1 in the endogenous CSF1 promoter region, and this transition required the ATP-dependent chromatin remodeling activity. Importantly, the Z-DNA-forming TG repeat sequence is required for the BRG1-mediated activation (25).
The Z-DNA-forming TG repeat sequences are assembled in a positioned nucleosome.
To investigate the interplay between Z-DNA and chromatin remodeling in activation of the CSF1 gene, we first mapped the nucleosomal structure of the promoter. SW-13 nuclei or purified genomic DNA was briefly treated with micrococcal nuclease (MNase) followed by complete digestion with KpnI, which recognizes a site at −734 relative to the transcription start site (+1) in the CSF1 promoter. Southern blot analysis using a probe from KpnI to the NheI sites detected a ladder of nucleosome-sized DNA from nuclei but not from purified DNA samples (Fig. 2A; compare lanes 1 and 2 with lanes 3 and 4), indicating that the promoter region is organized into several positioned nucleosomes. The nucleosome boundaries for nuc-4 and nuc-3 were estimated to be at −764 to −624 and at −564 to −420, respectively. We mapped the nucleosome boundaries of nuc-1 and nuc-2 by LM-PCR analysis of MNase-digested SW-13 nuclei (Fig. 2B). Using a primer specific to the CSF1 promoter at −318, LM-PCR yielded three bands (Fig. 2B, lane 2). The approximate size of each product is indicated in the figure. To determine the location of the protected regions, we subtracted the size of the universal linker (25 bp) to yield 90-bp, 120-bp, and 270-bp products. The protected region of approximately 150 bp corresponds to a nucleosome (nuc-1) positioned between about −200 and −50 relative to the transcription start site. A reverse primer starting at −58 revealed another protected region corresponding to a second nucleosome (nuc-2) (Fig. 2B, lane 4). We can assign a general positioning estimate for the nuc-2 boundaries at −230 and −380. Although at this resolution we cannot precisely map the nucleosome boundaries, the data strongly indicate the presence of four nucleosomal regions within the indicated ranges. Interestingly, the long stretch of Z-DNA-forming TG repeat sequence (−125 to −76) lies within nuc-1. The results are summarized in Fig. 2C.
The TG-containing nucleosome was disrupted upon activation of the CSF1 gene by BRG1.
The BRG1-containing BAF complexes use energy derived from ATP hydrolysis to disrupt the extensive DNA histone interactions in a nucleosome. This disruption allows for greater accessibility of nucleosomal DNA to transcription factors. Because the DNA sequence associated with nuc-1 in the CSF promoter is required for BRG1-mediated activation of the gene, a chromatin reorganization event likely occurs in this region. We therefore examined the fate of the two promoter proximal nucleosomes after expression of BRG1. We transfected BRG1-deficient SW-13 cells with a BRG1 or control expression vector. Nuclei were isolated and subjected to MNase digestion to yield mostly dinucleosome sized material. The dinucleosome bands were purified from an agarose gel and analyzed by LM-PCR. As a control for potential bias in MNase cleavage, DNA was used. Based on the coordinates generated from the MNase mapping data, CSF1 promoter-specific primers were designed to analyze boundary regions of nuc-1 and nuc-2. The linker regions around and between these two nucleosomes were accessible to MNase both in the absence and presence of BRG1 (Fig. 3A and B; linker regions are indicated by brackets which designate the region being mapped). Activation of the CSF1 gene by BRG1 did not significantly change the digestion pattern of nuc-2, as demonstrated by the LM-PCR products generated from a primer within nuc-2 and from a primer at the 3′ edge of nuc-1 (Fig. 3A, lanes 1 and 2 [Primer A] and lanes 4 and 5 [Primer B]). However, the expression of BRG1 induced several new bands in the otherwise protected region of nuc-1 that were detected by two forward primers (Fig. 3B, lanes 10 and 11 [Primer D] and lanes 13 and 14 [Primer E]) as well as by the reverse primers B and C (Fig. 3A, lanes 4 and 5 and lanes 7 and 8). Interestingly, there were more digestions in the linker regions both upstream and downstream of nuc-1 in the presence of BRG1, which is consistent with an overall more accessible chromatin structure accompanying the remodeling of nuc-1 by BRG1. These results provide strong evidence for nucleosome disruption by BRG1 at nuc-1 but not at the more distal nuc-2.
To confirm the disruption of nuc-1 by BRG1, we employed an assay based on the increased accessibility of a disrupted nucleosome to nucleases such as MNase. The accessibility to MNase was determined by measuring the relative abundance of a specific DNA sequence using quantitative PCR after digestion by the enzyme. A decrease in the total amount of DNA would be expected when the nucleosome is disrupted at the site being analyzed. We isolated nuclei from SW-13 cells transfected with either a BRG1 or control expression vector and digested with MNase to mostly mononucleosome-sized DNA, which was then isolated from an agarose gel. The amount of nuc-2 and nuc-1 DNA contained within the isolated mononucleosomal DNA was measured by quantitative PCR using specific primers. As shown in Fig. 3C (left panel), expression of BRG1 did not allow for increased accessibility of the DNA contained within nuc-2 to MNase. In contrast, a significant decrease in the amount of nuc-1 DNA contained within the mononucleosome fraction was observed in the presence of BRG1. The quantified data are presented graphically in the lower panel of Fig. 3C. Normalization of the nuc-1 DNA band to the nuc-2 DNA band indicates that there is a roughly twofold decrease of the nuc-1 DNA level (Fig. 3C, right panel). The decrease of the nuc-1 DNA level in the mononucleosome fraction could be caused by a partial disruption or complete ejection of nuc-1 upon remodeling by BRG1. Though we cannot distinguish between the two mechanisms, the data provide further evidence that the nuc-2 nucleosome in the CSF1 promoter is stable upon activation by BRG1 whereas the nuc-1 nucleosome is disrupted.
BRG1 associated with transcriptionally inactive CSF1 promoters in vivo.
Previously, we employed a transient reporter assay to determine the role of the TG repeat sequence on transcription from the CSF1 promoter (25). Using this assay, we showed that the CSF1 promoter cloned in the episomal pREP4 vector adopts Z-DNA structure when transiently cotransfected with BRG1 into SW-13 cells. The TG repeat sequence was required for activation of the promoter by BRG1 (25) (shown in Fig. 4A [noTG]). Replacement of the TG repeat sequence with a GC repeat fully supported the activation of the promoter by BRG1 (25) (shown in Fig. 4A [18GC]). In this study, we have used the approach to assay the effect of active transcription on Z-DNA formation. We generated a construct that was transcriptionally inactive but still contained the TG repeats required for Z-DNA formation by mutating the two potential TATA boxes within the CSF1 promoter (Fig. 4A [mTA]). We first asked whether the BRG1-mediated transcription from the noTG or from the mTA promoter constructs was inhibited due to decreased binding of BRG1. We used ChIP assays to test this possibility. SW-13 cells were cotransfected with the BRG1 expression vector or a control and with one of the CSF1 promoter reporter constructs. After cross-linking with formaldehyde was performed, cells were sonicated to generate mostly mono- and dinucleosome-sized DNA fragments and an anti-BRG1 antibody was used for immunoprecipitation. The relative amount of CSF1 promoter associated with BRG1 was measured by PCR. As shown in Fig. 4B, the anti-BRG1 antibody pulled down the CSF1 promoter sequence from all three constructs in a BRG1-dependent manner (lanes 2, 4, and 6), indicating that BRG1 was associated with all three promoters in vivo. However, when ChIP was performed on the same cells by use of an anti-RNA polymerase II antibody, only the wild-type promoter was associated with RNA polymerase (Fig. 4B, lane 2 [α-Pol II]). Furthermore, the endogenous CSF1 promoter was also associated with BRG1 and Pol II when activated in SW-13 cells by expressing BRG1 (data not shown). The loss of transcriptional activity at the mutant promoters is not due to decreased association of BRG1. There is, however, a decreased association with Pol II, even at the “noTG” construct which has an intact TATA box. These data indicate that the Z-DNA-forming potential is necessary for BRG1 activity, which is required for activation of the CSF1 gene.
FIG. 4.
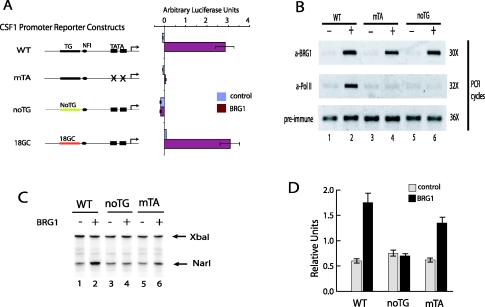
Chromatin remodeling by BRG1 requires the Z-DNA-forming sequence in the CSF1 promoter. (A) The two potential TATA boxes or the TG repeat sequences of the CSF1 promoter in pREP4-luc reporter vector were mutated as indicated. The constructs were cotransfected with a BRG1 or a control expression vector into SW-13 cells for 48 to 72 h. The luciferase activity was measured with the dual luciferase system from Promega. The error bars indicate the standard deviations of data from three experiments. WT, wild-type CSF1 promoter; mTA, the two TATA boxes are mutated; noTG, the TG repeat is replaced with a random sequence; 18GC, the TG repeat is replaced with GC repeat sequence. (B) Mutation of the TATA boxes and TG repeat did not inhibit the BRG1 binding in vivo. The promoter constructs described for panel A were cotransfected with BRG1 or control vector into SW-13 cells for 3 days. Chromatin lysates were prepared by sonication from formaldehyde-cross-linked cells. Chromatin immunoprecipitation was performed using preimmune serum or antibodies against BRG1 and RNA polymerase II. The immunoprecipitated DNA was analyzed by PCR using primers recognizing the CSF1 promoter. The transfected cells used for chromatin preparation are indicated on the top of the panel. The antibodies used for ChIP are indicated on the left. The numbers of PCR cycles for each ChIP are indicated on the right. (C) Restriction enzyme accessibility assay showing that mutation of the TG repeat sequence inhibited chromatin remodeling by BRG1. Nuclei were isolated from SW-13 cells transfected as described for panel B and were digested briefly with NarI enzyme followed by complete digestion with XbaI. The cleavages sites were detected by LM-PCR. (D) The data described for panel C were quantified by phosphorimager analysis. The NarI band was normalized to the XbaI control band. The error bars indicate the standard deviations of data from three independent experiments.
The Z-DNA-forming region was required for productive chromatin remodeling by BRG1.
BRG1-mediated activation of the CSF1 gene and association of BRG1 with the CSF1 promoter (Fig. 4B) strongly suggests that chromatin remodeling occurs at the promoter. We performed restriction enzyme accessibility assays on the different CSF1 promoter constructs to determine the effect of the sequence on the chromatin remodeling activity of BRG1. We cotransfected the reporter constructs shown in Fig. 4A with either BRG1 or control vector into SW-13 cells for 3 days. The nuclei isolated from the cells were subjected to digestion with NarI enzyme, whose recognition site (−71) is within nuc-1 immediately downstream of the TG repeat sequence. The purified DNA was completely digested at an XbaI site in the vector and was analyzed by LM-PCR (Fig. 4C). The data are presented graphically in Fig. 4D. BRG1 caused significant nucleosome remodeling at the wild-type and TATA-mutated promoters. BRG1 expression resulted in an increase of about threefold in NarI accessibility in the wild-type CSF1 promoter (Fig. 4C, lanes 1 and 2; Fig. 4D, WT). At the TATA-mutated promoter, a twofold increase in NarI accessibility was observed in response to BRG1 expression (Fig. 4C, lanes 5 and 6; Fig. 4D, mTA). In striking contrast, no increase in NarI accessibility was detected in the noTG promoter (Fig. 4C and D). These results demonstrate that even though BRG1 was associated with the noTG promoter construct, there was no observed nucleosome remodeling. The remodeling was only observed on the promoter constructs containing the TG repeat sequence, even with the promoter that was transcriptionally inactive. Thus, the Z-DNA-forming sequence was critical for the BRG1-mediated chromatin remodeling and subsequent transcriptional activation of the CSF1 promoter.
Chromatin remodeling- but not transcription-initiated Z-DNA formation.
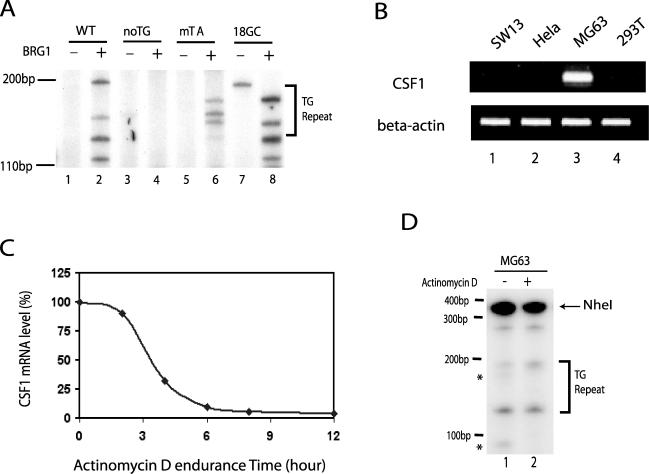
To understand how the Z-DNA-forming sequence facilitates chromatin remodeling by BRG1, we used ZaaFOK digestion to examine the induction of Z-DNA structure in the different promoter constructs in the absence and presence of BRG1. As shown in Fig. 5A, the expression of BRG1 resulted in several new ZaaFOK cleavage sites within or near the TG repeat sequence in the wild-type promoter (compare lanes 1 and 2), indicating the formation of Z-DNA structure. Similar results were observed for the promoter containing GC repeats in place of the TG repeats (compare lanes 7 and 8). However, no significant Z-DNA structure was detected in the promoter without TG repeats even in the presence of BRG1 (lanes 3 and 4). Interestingly, in the presence of BRG1, several bands were detected within the TG repeat region in the TATA-mutated promoter, indicating that Z-DNA structure was induced even in the absence of transcription. These results strongly support the idea that the two factors required for Z-DNA formation at the CSF1 promoter are BRG-1-mediated chromatin remodeling and DNA sequence which has the potential to form Z-DNA.
FIG. 5.
Formation of the Z-DNA structure does not require active transcription from the CSF1 promoter. (A) The different promoter constructs described for panel A of Fig. 4 were cotransfected with a BRG1 or control expression vector into SW-13 cells for 3 days. Following cross-linking with formaldehyde, the cells were permeabilized and treated with ZaaFOK. The cleavage sites were detected by LM-PCR. The TG repeat region is indicated on the right. (B) Reverse transcription-PCR analysis of total RNA isolated from HeLa, SW-13, MG63, and 293T cells by use of primers specific to CSF1. (C) MG63 cells were treated with 40 μg/ml of actinomycin D for up to 12 h. RNA was purified at 0, 2, 4, 6, 8, and 12 h posttreatment and analyzed by real-time PCR using CSF1-specific primers. The value at the 0 h time point is set as 100% CSF1 mRNA, and the data are plotted as percent CSF1 mRNA against time of actinomycin D treatment. (D) MG63 cells were treated with 40 μg/ml of actinomycin D for 6 h. Following cross-linking with formaldehyde, the cells were permeabilized and treated with ZaaFOK. The DNA was purified and digested completely with NheI, which recognizes a site upstream of the TG repeat sequence in the CSF1 promoter. The cleavage sites were detected by LM-PCR using CSF1 promoter-specific primers downstream of the TG repeat sequence. The TG repeat region is indicated on the right.
It is worth noting that the BRG1-induced ZaaFOK cleavage bands were detected beyond the TG repeat region in both the wild-type promoter and in the 18GC promoter, while the cleavage occurred only within the TG repeat region in the TATA-mutated promoter (Fig. 5A; compare lanes 2 and 8 with lane 6). These results indicate that the Z-DNA formation is more extensive in the transcriptionally competent promoters, which is consistent with the notion that the negative supercoiling from transcription may extend the Z-DNA structure, induced by chromatin remodeling by BRG1, to a broader region.
To confirm that the induction of Z-DNA structure at the CSF1 promoter does not require active transcription, we used actinomycin D to inhibit transcription in a cell line (MG63) that constitutively expresses CSF1 (Fig. 5B) (25). We treated MG63 cells with 40 μg/ml of actinomycin D for 6 h, which reduced the amount of CSF1 mRNA to less than 10% of the results seen with untreated cells (Fig. 4C). The cells were then fixed, permeabilized, and subjected to ZaaFOK treatment. LM-PCR of the resulting cleaved DNA revealed that the Z-DNA structure persisted even in the presence of actinomycin D (Fig. 4D; compare lanes 1 and 2), providing further evidence that the formation of Z-DNA structure in the CSF1 promoter does not require transcriptional activity. However, the presence of actinomycin D did inhibit two bands (as indicated by the asterisks), suggesting that, as seen with the promoter reporter constructs, the extent of Z-DNA structure was more restricted in the absence of transcription.
DISCUSSION
In this report, we show that the Z-DNA-forming sequence in the inactive CSF1 promoter is organized in a positioned nucleosome and that this nucleosome was disrupted upon activation of the CSF1 gene by BRG1. A Z-DNA structure was formed accompanying the BRG1-mediated nucleosome disruption. Both the Z-DNA-forming sequence and the BRG1 nucleosome remodeling event were required for the B- to Z-DNA transition. Both were also required for transcription from the CSF1 promoter, which raised the following question: does BRG1-mediated nucleosome remodeling induce Z-DNA formation or does active transcription resulting from the BRG1 remodeling event induce Z-DNA formation? We have shown that BRG1 did induce Z-DNA formation on a transcriptionally incompetent promoter, though the region that formed Z-DNA was more extensive on a transcriptionally active promoter. Finally, the BRG1-mediated activation of CSF1 required Z-DNA-forming sequence. Thus, we conclude that Z-DNA structure plays a role in the BAF-induced chromatin remodeling required for activation of the CSF1 gene.
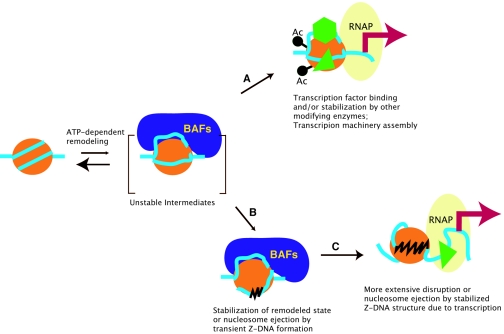
Z-DNA may aid in chromatin remodeling by stabilizing transient nucleosome intermediates.
Nucleosomal structure is energetically stabilized by extensive DNA-histone interactions (26), which can be disrupted by chromatin remodeling enzymes by use of ATP-derived energy (9, 12, 14, 19, 27-29, 31, 32, 40). Extensive studies of the mechanisms of chromatin remodeling suggest that the BAF complexes modify nucleosomal structure by transiently converting a stable nucleosome to several unstable intermediates, as illustrated in Fig. 6 (reviewed in references 31 and 19). If the unstable intermediates revert to the original, more stable state, the remodeling is unproductive. This may be the most frequent case in living cells, as the remodeling enzymes scan through the genome for specific target sites or partner proteins. However, the transient nucleosome disruption may provide an opportunity for transcription factors and/or other chromatin-modifying enzymes to access their target sites (18). The binding of transcription factors or other enzymes may capture the less stable intermediates and stabilize the remodeled state to some degree, as illustrated by step A in Fig. 6.
FIG. 6.
A model for stabilization of BRG1-induced unstable remodeling intermediates by Z-DNA formation. See text for details.
In addition to the mechanisms highlighted above, we speculate on the basis of the data presented in this report that the generation of Z-form DNA may stabilize the transient nucleosomal intermediates by either capturing a remodeling intermediate or ejecting the nucleosome (step B in Fig. 6). Disruption of the nucleosomal structure by ATP-utilizing enzymes generates superhelical torsion (5, 7, 11). TG or CG repeat sequences release negative superhelical torsion by converting to a Z-DNA conformation. At the CSF1 promoter, for example, remodeling of the nuc-1 nucleosome by BRG-1 generates torsional strain that is absorbed by the formation of Z-DNA through the TG repeats. The generation of Z-DNA, in turn, could result in a stable remodeled nucleosome. Indeed, mutating the sequence necessary for Z-DNA formation at the CSF1 promoter results in unproductive BRG1 activity.
Does transcription play a role in the formation of Z-DNA structure in the CSF1 promoter?
The negative superhelical torsion generated behind an actively transcribing RNA polymerase has been associated with Z-DNA formation (24, 42). The results presented in Fig. 5 are consistent with a transcription-independent induction of Z-DNA structure at the CSF1 promoter. However, the ZaaFOK cleavage patterns differ between the wild-type (active) and TATA-mutated (inactive) CSF1 promoters (Fig. 5A). The cleaved bands are only detected within the TG repeat region in the TATA-mutated promoter, while the cleavage appears more extensive at the wild-type promoter since bands are detected outside of the TG repeat region (Fig. 5A). These results suggest that the Z-DNA structure induced by the chromatin remodeling activity of BRG1 in the TATA-mutated promoter is less stable or spans a smaller region than in the wild-type promoter (step B in Fig. 6). More-extensive Z-DNA as seen in the wild-type promoter may result from more negative superhelical torsion generated by transcription (step C in Fig. 6). The zigzag structure may lead to a more extensive disruption of the nucleosomal structure or ejection of the nucleosome (step C in Fig. 6).
These data are consistent with a model suggesting that induction of a localized Z-DNA structure by chromatin remodeling is followed by formation of a more extensive Z-DNA structure promoted by active transcription. The more extensive Z-DNA can inhibit reformation of the canonical nucleosome, maintaining an open chromatin structure at the promoter. Therefore, the Z-DNA structure induced by chromatin remodeling and by transcription functions at different stages of promoter activation. Chromatin remodeling-induced Z-DNA formation functions early to promote the transition from a transient and partial remodeling to a more complete disruption of the nucleosomal structure, while the transcription-induced Z-DNA formation functions late to stabilize and maintain an open chromatin structure by inhibiting reformation of the nucleosome.
Acknowledgments
We thank Warren Leonard for critical reading of the manuscript. We thank the members of the W. Leonard and K. Zhao laboratories for stimulating discussions.
This work was supported by intramural grants to the National Heart, Lung, and Blood Institute, National Institutes of Health.
REFERENCES
- 1.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 2.Bultman, S., T. Gebuhr, D. Yee, C. La Mantia, J. Nicholson, A. Gilliam, F. Randazzo, D. Metzger, P. Chambon, G. Crabtree, and T. Magnuson. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6:1287-1295. [DOI] [PubMed] [Google Scholar]
- 3.Champ, P. C., S. Maurice, J. M. Vargason, T. Camp, and P. S. Ho. 2004. Distributions of Z-DNA and nuclear factor I in human chromosome 22: a model for coupled transcriptional regulation. Nucleic Acids Res. 32:6501-6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felsenfeld, G., and M. Groudine. 2003. Controlling the double helix. Nature 421:448-453. [DOI] [PubMed] [Google Scholar]
- 5.Gavin, I., P. J. Horn, and C. L. Peterson. 2001. SWI/SNF chromatin remodeling requires changes in DNA topology. Mol. Cell 7:97-104. [DOI] [PubMed] [Google Scholar]
- 6.Hamada, H., and T. Kakunaga. 1982. Potential Z-DNA forming sequences are highly dispersed in the human genome. Nature 298:396-398. [DOI] [PubMed] [Google Scholar]
- 7.Havas, K., A. Flaus, M. Phelan, R. Kingston, P. A. Wade, D. M. Lilley, and T. Owen-Hughes. 2000. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell 103:1133-1142. [DOI] [PubMed] [Google Scholar]
- 8.Herbert, A., and A. Rich. 1999. Left-handed Z-DNA: structure and function. Genetica 106:37-47. [DOI] [PubMed] [Google Scholar]
- 9.Horn, P. J., and C. L. Peterson. 2002. Molecular biology. Chromatin higher order folding—wrapping up transcription. Science 297:1824-1827. [DOI] [PubMed] [Google Scholar]
- 10.Imbalzano, A. N., H. Kwon, M. R. Green, and R. E. Kingston. 1994. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature 370:481-485. [DOI] [PubMed] [Google Scholar]
- 11.Imbalzano, A. N., G. R. Schnitzler, and R. E. Kingston. 1996. Nucleosome disruption by human SWI/SNF is maintained in the absence of continued ATP hydrolysis. J. Biol. Chem. 271:20726-20733. [DOI] [PubMed] [Google Scholar]
- 12.Kadam, S., and B. M. Emerson. 2002. Mechanisms of chromatin assembly and transcription. Curr. Opin. Cell Biol. 14:262-268. [DOI] [PubMed] [Google Scholar]
- 13.Kang, H., K. Cui, and K. Zhao. 2004. BRG1 controls the activity of the retinoblastoma protein via regulation of p21CIP1/WAF1/SDI. Mol. Cell. Biol. 24:1188-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katsani, K. R., T. Mahmoudi, and C. P. Verrijzer. 2003. Selective gene regulation by SWI/SNF-related chromatin remodeling factors. Curr. Top. Microbiol. Immunol. 274:113-141. [DOI] [PubMed] [Google Scholar]
- 15.Khavari, P. A., C. L. Peterson, J. W. Tamkun, D. B. Mendel, and G. R. Crabtree. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170-174. [DOI] [PubMed] [Google Scholar]
- 16.Kim, Y. G., K. Lowenhaupt, T. Schwartz, and A. Rich. 1999. The interaction between Z-DNA and the Zab domain of double-stranded RNA adenosine deaminase characterized using fusion nucleases. J. Biol. Chem. 274:19081-19086. [DOI] [PubMed] [Google Scholar]
- 17.Kim, Y. G., M. Muralinath, T. Brandt, M. Pearcy, K. Hauns, K. Lowenhaupt, B. L. Jacobs, and A. Rich. 2003. A role for Z-DNA binding in vaccinia virus pathogenesis. Proc. Natl. Acad. Sci. USA 100:6974-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 19.Kornberg, R. D., and Y. Lorch. 1999. Chromatin-modifying and -remodeling complexes. Curr. Opin. Genet. Dev. 9:148-151. [DOI] [PubMed] [Google Scholar]
- 20.Kouzarides, T. 2002. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12:198-209. [DOI] [PubMed] [Google Scholar]
- 21.Kurdistani, S. K., and M. Grunstein. 2003. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 4:276-284. [DOI] [PubMed] [Google Scholar]
- 22.Liu, H., H. Kang, R. Liu, X. Chen, and K. Zhao. 2002. Maximal induction of a subset of interferon target genes requires the chromatin-remodeling activity of the BAF complex. Mol. Cell. Biol. 22:6471-6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, H., and K. Zhao. 2004. Assay of Z-DNA induction by chromatin remodeling factors. Methods Enzymol. 377:412-420. [DOI] [PubMed] [Google Scholar]
- 24.Liu, L. F., and J. C. Wang. 1987. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 84:7024-7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, R., H. Liu, X. Chen, M. Kirby, P. O. Brown, and K. Zhao. 2001. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell 106:309-318. [DOI] [PubMed] [Google Scholar]
- 26.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 27.Lusser, A., and J. T. Kadonaga. 2003. Chromatin remodeling by ATP-dependent molecular machines. Bioessays 25:1192-1200. [DOI] [PubMed] [Google Scholar]
- 28.Martens, J. A., and F. Winston. 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13:136-142. [DOI] [PubMed] [Google Scholar]
- 29.Muchardt, C., and M. Yaniv. 1999. ATP-dependent chromatin remodelling: SWI/SNF and Co. are on the job. J. Mol. Biol. 293:187-198. [DOI] [PubMed] [Google Scholar]
- 30.Mueller, P. R., and B. Wold. 1989. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science 246:780-786. [DOI] [PubMed] [Google Scholar]
- 31.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 32.Neely, K. E., and J. L. Workman. 2002. The complexity of chromatin remodeling and its links to cancer. Biochim. Biophys. Acta 1603:19-29. [DOI] [PubMed] [Google Scholar]
- 33.Oh, D. B., Y. G. Kim, and A. Rich. 2002. Z-DNA-binding proteins can act as potent effectors of gene expression in vivo. Proc. Natl. Acad. Sci. USA 99:16666-16671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rich, A., and S. Zhang. 2003. Timeline: Z-DNA: the long road to biological function. Nat. Rev. Genet. 4:566-572. [DOI] [PubMed] [Google Scholar]
- 35.Rothenburg, S., F. Koch-Nolte, A. Rich, and F. Haag. 2001. A polymorphic dinucleotide repeat in the rat nucleolin gene forms Z-DNA and inhibits promoter activity. Proc. Natl. Acad. Sci. USA 98:8985-8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroth, G. P., P. J. Chou, and P. S. Ho. 1992. Mapping Z-DNA in the human genome. Computer-aided mapping reveals a nonrandom distribution of potential Z-DNA-forming sequences in human genes. J. Biol. Chem. 267:11846-11855. [PubMed] [Google Scholar]
- 37.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 38.Turner, B. M. 2000. Histone acetylation and an epigenetic code. Bioessays 22:836-845. [DOI] [PubMed] [Google Scholar]
- 39.Wang, A. H., G. J. Quigley, F. J. Kolpak, J. L. Crawford, J. H. van Boom, G. van der Marel, and A. Rich. 1979. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 282:680-686. [DOI] [PubMed] [Google Scholar]
- 40.Wang, W. 2003. The SWI/SNF family of ATP-dependent chromatin remodelers: similar mechanisms for diverse functions. Curr. Top. Microbiol. Immunol. 274:143-169. [DOI] [PubMed] [Google Scholar]
- 41.Wang, W., J. Cote, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370-5382. [PMC free article] [PubMed] [Google Scholar]
- 42.Wittig, B., S. Wolfl, T. Dorbic, W. Vahrson, and A. Rich. 1992. Transcription of human c-myc in permeabilized nuclei is associated with formation of Z-DNA in three discrete regions of the gene. EMBO J. 11:4653-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong, A. K., F. Shanahan, Y. Chen, L. Lian, P. Ha, K. Hendricks, S. Ghaffari, D. Iliev, B. Penn, A. M. Woodland, R. Smith, G. Salada, A. Carillo, K. Laity, J. Gupte, B. Swedlund, S. V. Tavtigian, D. H. Teng, and E. Lees. 2000. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 60:6171-6177. [PubMed] [Google Scholar]
- 44.Zhang, Y., and D. Reinberg. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343-2360. [DOI] [PubMed] [Google Scholar]
- 45.Zhao, K., W. Wang, O. J. Rando, Y. Xue, K. Swiderek, A. Kuo, and G. R. Crabtree. 1998. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95:625-636. [DOI] [PubMed] [Google Scholar]