Abstract
The precise machineries required for two aspects of eukaryotic DNA replication, Okazaki fragment processing (OFP) and telomere maintenance, are poorly understood. In this work, we present evidence that Saccharomyces cerevisiae Pif1 helicase plays a wider role in DNA replication than previously appreciated and that it likely functions in conjunction with Dna2 helicase/nuclease as a component of the OFP machinery. In addition, we show that Dna2, which is known to associate with telomeres in a cell-cycle-specific manner, may be a new component of the telomere replication apparatus. Specifically, we show that deletion of PIF1 suppresses the lethality of a DNA2-null mutant. The pif1Δ dna2Δ strain remains methylmethane sulfonate sensitive and temperature sensitive; however, these phenotypes can be suppressed by further deletion of a subunit of pol δ, POL32. Deletion of PIF1 also suppresses the cold-sensitive lethality and hydroxyurea sensitivity of the pol32Δ strain. Dna2 is thought to function by cleaving long flaps that arise during OFP due to excessive strand displacement by pol δ and/or by an as yet unidentified helicase. Thus, suppression of dna2Δ can be rationalized if deletion of POL32 and/or PIF1 results in a reduction in long flaps that require Dna2 for processing. We further show that deletion of DNA2 suppresses the long-telomere phenotype and the high rate of formation of gross chromosomal rearrangements in pif1Δ mutants, suggesting a role for Dna2 in telomere elongation in the absence of Pif1.
Yeast Pif1 is the founding member of the Pif1 subfamily of superfamily 1 DNA helicases (3). While other organisms, such as Caenorhabditis elegans and Homo sapiens, have only one identified Pif1 family member, in yeast, there is a second, closely related, protein, Rrm3p (3). In yeast, neither of these helicases is essential and mutants lacking both are viable and repair proficient. Both yeast proteins have 5′-to-3′ DNA helicase activity (21, 30, 31). The region of similarity between Pif1 and Rrm3 is limited to the seven helicase motifs, which exhibit 40% identity and 60% similarity (3). This may indicate that the two helicases have structurally similar DNA substrates. Nevertheless, the two helicases differ in their biological functions, and these differences are likely mediated not only by the helicase domain but also by the divergent N termini, which are not required for helicase activity (4). To date, it has been impossible to determine which helicase, Rrm3 or Pif1, is the functional homolog of the single ortholog in other eukaryotes.
One difference between Rrm3 and Pif1 is in their function at the rRNA gene. In rrm3 mutants, there is an increase in replisome pausing at the Fob1 protein-bound replication fork barrier (RFB) in the rRNA gene (22). The hypothesis is that Rrm3 is required to remove proteins that block the fork at that point, since Rrm3 is required for promoting fork movement at over 1,400 loci in the yeast genome, in addition to the RFB. In contrast to Rrm3, Pif1 seems to be required for pausing at the rRNA gene RFB, since pif1Δ mutants show a threefold-reduced number of forks accumulating at the RFB. Pif1 could itself be an inhibitor of fork movement, or it could be required for Fob1p to exert its blocking action (22, 50).
Pif1 and Rrm3 also appear to have different roles at telomeres. Two-dimensional gel analysis of telomere replication in rrm3Δ mutants suggests that Rrm3 is required for replication through Y′ elements and telomeric repeats, and similar experiments in rrm3Δ rap1 strains further suggest that Rrm3 specifically removes Rap1 protein, or proteins that associate with telomeres in the absence of Rap1, to allow progression of the telomere-proximal replication fork (21, 35). Pif1, on the other hand, is thought to be an inhibitor of telomerase, since pif1Δ mutants have long telomeres, 160 to 240 bp longer than normal telomeres, because overproduction of Pif1 shortens telomeres and since Pif1 decreases the processivity of telomerase in vitro (5). pif1Δ mutants also show a 200-fold increase in the de novo addition of telomeres at HO-endonuclease-induced double-strand breaks (DSBs) and a high level of gross chromosomal rearrangements (GCRs) in which there is excessive telomere addition to nontelomeric sequences at sites of intrachromosomal DNA damage, presumably primarily DSBs (36, 42, 50). By analogy with Rrm3, it has been proposed that Pif1 might remove a protein such as telomerase from the chromosome ends. pif1Δ mutants also have decreased silencing in the subtelomeric repeats.
A third possible difference between Pif1 and Rrm3 is that Pif1, but not Rrm3, is required for mitochondrial DNA recombination and genome maintenance (30, 31, 42). The pif1Δ mutant grows slowly on nonfermentable carbon sources and loses functional mitochondria rapidly (42). RRM3 does contain a mitochondrial import signal; however, it has not yet been shown to function in mitochondria (1, 3, 18, 23).
In this work, we report an additional difference between Pif1 and Rrm3. The rrm3Δ mutant is synthetically lethal with a mutation in the Dna2 helicase/nuclease, required for Okazaki fragment processing (OFP), telomere stability, and DNA repair (6-10, 14, 17, 47, 48). In contrast, we show here that pif1Δ suppresses both the DNA replication and repair defects of dna2 mutants and even the lethality of deletion of DNA2. The dna2Δ pif1Δ mutant retains some defects in replication and repair, but these are further suppressed by deletion of the POL32 subunit of polymerase δ (pol δ). Conversely, deletion of DNA2 suppresses the telomere phenotype of pif1Δ mutants. We describe and characterize here genetic interactions between PIF1, DNA2, and POL32 that present new insights into the functions of each gene in OFP, telomere function, and chromatin modification and into the evolutionarily conserved functions of Pif1 helicase.
MATERIALS AND METHODS
Strains.
The strains used in this study were derived either from BY4741, in which the yeast deletion collection is housed (Invitrogen, Carlsbad, Calif.), or from W303 RAD5+. Individual experiments compared only isogenic strains in either background. The full strain list is shown in Table 1. Standard genetic techniques were used for tetrad analysis. Gene disruptions were carried out by standard PCR-based methods and verified by PCR. Sequences of primers not shown below are available on request.
TABLE 1.
Strains used in this study
| Strain | Characteristics | Source or reference |
|---|---|---|
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Invitrogen |
| BY4742 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Invitrogen |
| W303 | MATatrp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 RAD5 | H. Klein |
| 4741dna2-1-6D | MATadna2-1 his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | This study |
| 4741dna2-2-11D | MATα dna2-2::LEU2 his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | This study |
| MB110 | MATadna2Δ::kanMX his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 trp1Δ (pSEY18GALDNA2) | |
| MB201 | BY4741 MATapif1Δ::HIS3 trp1Δ | This study |
| MB202 | BY4742 MATapif1Δ::HIS3 trp1Δ | This study |
| 10509 | BY4742 MATα pif1Δ::kanMX | This study |
| MB203.3 | BY4741 MATadna2Δ::kanMX pif1Δ::HIS3 trp1Δ | This study |
| MB203.5 | BY4741 MATadna2Δ::kanMX pif1Δ::HIS3 trp1Δ | This study |
| MB203.6 | BY4741 MATadna2Δ::kanMX pif1Δ::HIS3 trp1Δ | This study |
| MB204 | BY4742 MATα dna2Δ::kanMX pif1Δ::HIS3 trp1Δ | This study |
| MB205 | BY4741 MATapol32Δ::natR trp1Δ | This study |
| MB206 | BY4742 MATα pol32Δ::natR trp1Δ pif1Δ::HIS3 | This study |
| MB207 | BY4741 dna2Δ::kanMX pif1Δ::HIS3 pol32Δ::natR trp1Δ | This study |
| MB208 | BY4741 MATα dna2-2 pif1Δ::kanMX | This study |
| 579-6D | MATatrp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 | H. Klein |
| 580-10D | MATatrp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 | H. Klein |
| 3615 | MATaura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2 Bgl hom3-10 ade2-Δ1 ade8 hxt13::URA3 | 36 |
| 4400 | MATapif1Δ::kanMX ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2 Bgl hom3-10 ade2-Δ1 ade8 hxt13::URA3 | 36 |
| 4393 | MATapif1-m1 ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2 Bgl hom3-10 ade2-Δ1 ade8 hxt13::URA3 | 36 |
| 4344 | MATapif1-m2 ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2 Bgl hom3-10 ade2-Δ1 ade8 hxt13::URA3 | 36 |
| 4344dna2Δ | 4344 dna2Δ::HIS3 | This study |
| 3615-2-1 | 3615 dna2-1::natR | This study |
| 3615-2-2 | 3615 dna2-2::natR | This study |
| 3615-K1080A | 3615 dna2-K1080A::natR | This study |
| 5030 | MATapif1-m2 ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2 Bgl hom3-10 ade2-Δ1 ade8 hxt13::URA3 | 36, 49 |
| 5030dna2Δ | 5030 dna2Δ::HIS3 | |
| 509 | MATapif1Δ::kanMX his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Invitrogen |
| MB90-7A | MATadna2-1 trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 | This study |
| MB91 | MATadna2-1pif1Δ::HIS5 trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 | This study |
| U953-61A | MATamec1Δ::TRP1 sml1::HIS trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 | 49 |
| MB92-M | MATadna2-1 mec1Δ::TRP1 sml1::HIS3 trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 | 11 |
| SPY40 | MATatel1::URA3 trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 | 40 |
| MB92-31C | MATadna2-2 mec1Δ::TRP1 sml1Δ::HIS3 trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 | 11 |
| MB92-35A | MATα dna2-2 tel1Δ::URA3 sml1Δ::HIS3 trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 | 11 |
| U960-5C | MATatrp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 rad53::HIS3 sml1-1 | 49 |
| MB92-1C | MATadna2-1 tel1Δ::URA3 sml1Δ::HIS3 trp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 | 11 |
| MB504 | MATa/α PIF/pif1Δ::HIS3 DNA2/dna2Δ::kanMX CDC13/cdc13-1 | This study |
Oligonucleotides.
The following oligonucleotides were used for gene disruption by standard genetic techniques (34): dna2NΔ (CAAGTGAGTACTCATTTTGTGCAAGCAAACACTGACAATTGAAGAGATCGTCAGGCGGATCCCCGGGTTAATTAA), dna2CΔ (TATTTTATGCTGTGATAGCTTTCCTGTTATGGAGAAGCTCTTCTTATTCCCCCTGGAATTCGAGCTCGTTTAAAC), dna2Nseq (CAATAAAGCAATTCCGTGCGGCAGA), pif1NΔ (ATTTTGATATATTATCCATTGAGCGATTAGCTTACTTGTATCAATCAATTTTACCGGATCCCCGGGTTAATTAA), pif1CΔ (GATTATTATAGCAGTTTGTATTCTATATAACTATGTGTATTAATATGTTACGAATTCGAGCTCGTTTAAAC), pTEF kan (CTCGCAGGTCTGCAGCGAGGAGCCG), pif1-95 (GGCCAGACATTGAAACTGG), and dna2-seq (CAATAATGCAATTCCGTGCGGCAGA).
Verification of deletions of PIF1 or DNA2.
We verified our earlier results and those of others that DNA2 is essential (6, 8, 17, 43). For the studies reported here, a heterozygous dna2Δ strain was obtained from Research Genetics as clone 22858, and dissection after sporulation shows that in the strain, DNA2 is essential. Complementation with a plasmid containing a functional DNA2 gene rescues the deletion, and transformation with constructs containing nonfunctional dna2 genes, dna2K1080E, dna2E675A, dna2E657A, and dna2E693A, does not rescue the deletion (10; unpublished data). Therefore, the dna2Δ pif1Δ mutant derived from a haploid segregant of clone 22858 is unlikely to contain any aberrant mutations, rearrangements, or unknown suppressors which are linked to DNA2. The DNA2 gene is also essential in Schizosaccharomyces pombe (26). All constructs containing deletions of these genes were checked in multiple ways. For physical analysis of strains used in Fig. 1B, colony PCR was used, permitting application to large numbers of transformants in a single experiment. The oligonucleotides used in PCR are available on request. The pif1Δ strain was confirmed using oligonucleotides pTEF kan and pif1-95. The resulting PCR product was 350 bp. The dna2Δ strains were confirmed using the oligonucleotides pTEF kan and dna2-seq. The resulting PCR product was 450 bp. All strains used in the subsequent experiments presented were confirmed in this way.
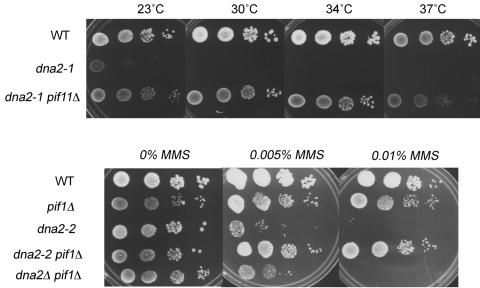
FIG. 1.
Suppression of dna2 mutant phenotypes by deletion of PIF1. (A) Serial dilutions of identical numbers of cells were carried out at the temperatures indicated. The following strains were used: W303, WT; MB90-7A, dna2-1; MB91, dna2-1 pif1Δ. (B) Serial dilutions of identical numbers of each strain were plated on medium containing the indicated amounts of MMS. Relevant genotypes are indicated. The following strains were used: BY4741, WT; 10509, pif1Δ; 4741dna2-2, dna2-2; MB208, dna2-2 pif1Δ; and MB203, dna2Δ pif1Δ (Table 1).
For verification of strains used in generating and in analyzing the genotype of strain MB504 (the double heterozygote used in the Results section to test lethality of dna2Δ and its suppression by deletion of PIF1), colony PCR was used; the primers are available on request.
Five strains were obtained from Richard Kolodner: 3615 (PIF1), 4393 (pifm1), 4344 (pifm2), 5030 (pif1m2), and 4400 (pif1Δ). The PCR product used to delete the DNA2 gene contained the HIS3 gene flanked by 55 bp of DNA homologous to DNA2 on the N- and C-terminal regions. The His+ transformants were picked and tested for temperature sensitivity, growth on glycerol, and methylmethane sulfonate (MMS) sensitivity. The temperature-sensitive (recessive, ts; dominant, TS) transformants were further tested by either colony PCR for the dna2Δ::HIS3 deletion or complementation using a dna2 strain. Strain 4400 (pif1Δ) had 26 transformants: 7 ts and 19 TS+. All ts transformants were dna2Δ. 4344 (pif1m2) had 31 transformants: 20 ts and 11 TS+. All ts transformants were dna2Δ. 5030 (pif1m2) had 32 transformants: 24 ts and 8 TS+. All ts transformants were dna2Δ. 4393 (pif1m1) had 32 transformants: 0 ts and 32 TS+. 3615 had 12 transformants: 0 ts and 12 TS+. None of these were dna2Δ. The PCR product is quite efficient in deleting the Dna2 gene in the pif1Δ or pif1m2 background, but not the PIF1+ or pif1m1 background. dna2Δ pif1-m2 transformants were also checked by complementation. The dna2Δ::G418 pif1-m2 strain was mated to a dna2-1 LEU2 strain, and G418R Leu+ diploids were selected. A dna2-1/dna2Δ PIF1/pif1-m2 strain fails to grow at 37°C, while a dna2-1/DNA2 PIF1/pif1-m2 diploid does grow at 37°C.
Additional phenotypic analysis of mutant constructs is described in the text where appropriate.
Determination of telomere length.
Chromosomal DNA was cleaved with XhoI, loaded onto a 1% agarose gel, and electrophoresed for 6 h at 60 V. The gel was blotted onto Gene Screen Plus and hybridized using a 300-bp double-stranded GT probe as described previously (14). Since telomere length is clonal, three independent isolates were analyzed for each strain. The hybridization “smear” at about 1.3 kb represents the terminal fragment of Y′ telomeres cut by XhoI and is indicative of telomere length in each strain.
RESULTS
Suppression of dna2-1 by pif1Δ.
We have reported that dna2-2 is synthetically lethal with rrm3Δ (47). RRM3 is highly homologous to PIF1, and they affect to some extent similar regions of the genome. To further understand the rrm3Δ dna2-2 synthetic lethality, we wished to examine dna2-2 pif1Δ double mutant strains. dna2-2 mutants (R1235Q) have a mutation in the helicase domain and are expected to have a substantial defect only in the helicase activity, although the dna2-2 protein has never been directly studied (17). When we constructed a dna2-2 pifΔ double mutant, instead of the expected synthetic lethality, we found that dna2-2 pif1Δ double mutants were viable. dna2-2 cells are viable but grow slowly, and when grown on plates most of the cells are budded, often with large buds, consistent with a previously established G2/M delay (17; unpublished observations). We noticed that dna2-2 pif1Δ had the same morphology as wild-type (WT) cells, small and unbudded (not shown), suggesting that pif1Δ might suppress the dna2-2 G2/M delay.
To further test the proposed suppression, we investigated the effect of deletion of PIF1 on a dna2 mutant with a more defined phenotype, the dna2-1 strain. dna2-1 is a temperature-sensitive allele (P504S) that maps in the nuclease domain of DNA2 and drastically reduces the nuclease activity of the Dna2 protein (10). Given that pif1Δ reduces pausing at the rRNA gene RFB, we expected pif1Δ mutants might show some suppression of dna2-1, since fob1Δ, which also decreases pausing at the RFB, suppresses the DNA damage sensitivity in dna2 mutants (47, 48). The PIF1 gene was deleted by PCR-mediated mutagenesis in a W303 dna2-1 strain, which was confirmed as described in Materials and Methods. As expected, deletion of PIF1 suppressed the growth defect of dna2-1 strains at 23°C and the lethality of dna2-1 strains at 34°C (Fig. 1A). Unexpectedly, the dna2-1 pif1Δ strain was even viable at 37°C, although the wild-type growth rate was not completely restored at 37°C (Fig. 1A). (The dna2-1 strain grows slowly even at 23°C, and plates at 23°C were photographed before they were fully grown so that the other strains would not be overgrown, accounting for the appearance of Fig. 1A. Further incubation verified that the dna2-1 cells were fully viable [data not shown]).
Since dna2-2 cells are sensitive to MMS and bleomycin (14, 17, 19), we next tested the ability of pif1Δ to suppress the repair defect. The pif1Δ efficiently suppressed the MMS sensitivity of dna2-2 (Fig. 1B).
In sum, the phenotypes of the dna2-1 pif1Δ and dna2-2 pif1Δ show that deletion of PIF1 can suppress DNA replication and repair defects due to lesions in either the nuclease or the helicase domain of DNA2.
Deletion of PIF1 suppresses the lethality of a dna2Δ strain.
The suppression of temperature-sensitive growth of dna2-1 and of the DNA damage sensitivity of dna2-2 by deletion of PIF1 suggested that pif1Δ might suppress the inviability of a deletion of DNA2. PIF1 was disrupted in a dna2Δ strain containing a DNA2-complementing plasmid, pSEY18GALDna2 (7). Potential dna2Δ pif1Δ/pDNA2 double mutants were screened by testing for inability to grow on glycerol, since pif1Δ strains form petites at a high frequency. Deletion of PIF1 was then confirmed by PCR and by complementation as described in Materials and Methods. dna2Δ PIF1/pDNA2 and dna2Δ pif1Δ/pDNA2 strains were then streaked on 5-fluoroorotic acid (5-FOA) plates to evict the Dna2-complementing plasmid, which carries URA3. The dna2Δ pif1Δ strain grew on the 5-FOA plates, whereas dna2Δ PIF1 transformants did not. Serial dilutions of dna2Δ pif1Δ cells growing at 30°C are shown in Fig. 1B. Analysis of the dna2Δ strain in numerous crosses and tetrad dissections, as well as passage of the dna2Δ pif1Δ strain through genetic crosses described in the following experiments have never revealed an independently segregating suppressor. Therefore, deletion of PIF1 suppresses the inviability of a deletion of DNA2.
Since suppression of a deletion of DNA2 was unexpected, we used an alternative approach to confirm our results. A PIF1/pif1Δ heterozygote was transformed with a kanMX PCR product designed to disrupt the dna2 gene (the kanMX gene flanked by 55 bp N terminal to the ATG and 55 bp C terminal to the stop codon of the DNA2 gene) (34). DNA2/dna2Δ::kanMX heterozygotes were detected by PCR. The double heterozygote, ++/pif1Δ dna2Δ (ΜΒ504), was sporulated, and 36 tetrads were dissected. Thirty of the 94 viable spores recovered were pif1Δ dna2Δ, and none were dna2Δ. This is the expected number of spores if DNA2 is essential and if pif1Δ suppresses the lethality of dna2Δ.
To test the efficiency of suppression, the doubling time of the dna2Δ pif1Δ double mutant was compared to that of a wild-type yeast. The doubling time at 30°C was 2.5 h, compared to 2 h for pif1Δ, suggesting that suppression was not due to complete bypass of the need for Dna2 (not shown). We also analyzed the ability of the dna2Δ pif1Δ mutant to grow at 37°C. The double mutant failed to form colonies at 37°C (data not shown here, but see Fig. 4 below), and cells arrested with dumbbell morphology characteristic of an S phase or G2/M checkpoint (not shown). Therefore, deletion of PIF1 suppresses the essential function of DNA2 at 30°C but not at 37°C. We conclude that there is not complete bypass of Dna2 function, but suppression is nevertheless very strong.
FIG. 4.
Suppression of the temperature sensitivity of the dna2Δ pif1Δ strain by pol32Δ. The following strains were incubated on a YPD plate at 37°C for 3 days: MB202, pif1Δ; MB203.6, dna2Δ pif1Δ; MB206, pif1Δ pol32Δ; and MB207, dna2Δ pif1Δ pol32Δ.
Disruption of the nuclear, but not the mitochondrial function of PIF1 suppresses dna2Δ.
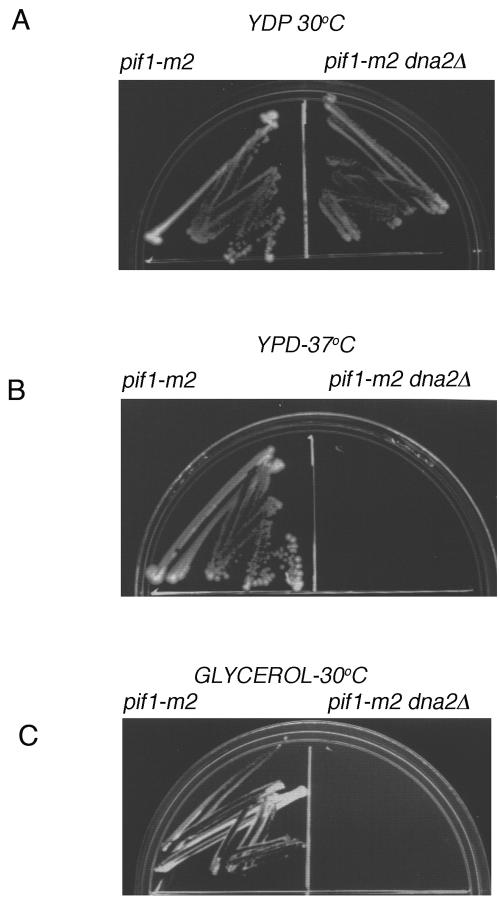
We wished to know whether it was disruption of the nuclear function and/or of the mitochondrial function of PIF1 that allowed cells to grow in the absence of DNA2. Two alleles of pif1, pif1-m1, and pif1-m2, separate the nuclear and mitochondrial function of PIF1 (50). pif1-m1 has a mutation in the first ATG, so the protein is missing the mitochondrial import function and the strain is petite. pif1-m1 appears to have its nuclear function intact, however, as evidenced by wild-type-length telomeres. pif1-m2 has a mutation in the second ATG. The pif1-m2 strain has its mitochondrial function intact but is deficient in its nuclear functions, leading to long telomeres. pif1-m2 mutants also show increases in the rate of gross chromosomal rearrangements (GCRs) involving de novo telomere addition (36). DNA2 was deleted from a set of isogenic strains containing mutations in pif1: 3615 (WT), 4400 (pif1Δ), 4393 (pif1-m1), 4344 (pif1-m2), and 5030 (pif1-m2) (36) by transforming each strain with a PCR product containing the HIS3 gene and 55 bp N terminal to the ATG and 55 bp C terminal to the stop codon of the DNA2 gene. The PCR product is expected to delete the DNA2 gene in strains that do not require Dna2 for viability. No viable dna2Δ pif1-m1 or dna2Δ PIF1 transformants were recovered. Thus, disruption of the mitochondrial function of Pif1 does not suppress dna2Δ lethality. Conversely, dna2Δ pif1Δ and dna2Δ pif1-m2 transformants, lacking the nuclear function of Pif1, were obtained and grew well, suggesting that it is the nuclear function of PIF1 that creates a requirement for DNA2 (Fig. 2A).
FIG. 2.
Suppression of dna2Δ lethality by inactivation of Pif1 nuclear function but not by inactivation of Pif1 mitochondrial function. The following strains were used: 4344, pif1-m2; and 4344dna2Δ, pif1-m2 dna2Δ. (A and B) Growth on glucose-containing medium. (C) Growth on nonfermentable glycerol medium.
In order to evaluate the fitness of the dna2Δ pif1-m2 mutant, its survival was tested at 37°C. As shown, the mutant was temperature sensitive, similar to the dna2Δ pif1Δ strain (Fig. 2B). The mutant was also replica plated to media containing 0.005% MMS, and the strain is also sensitive to MMS at 30°C (not shown). Thus, there is not total bypass of the requirement for Dna2 in the pif1-m2 mutant in DNA replication or in repair.
Both the dna2Δ pif1-m2 and the dna2Δ pif1Δ mutants grew more slowly than the pif1-m2 mutant at 30°C. In studying the slow growth, 50 to 100 of the dna2Δ pif-m2 transformants were replica plated to glycerol medium to determine if Dna2 deficiency led to production of petites. In fact, although the pif1-m2 mutant grows normally on glycerol, the dna2Δ pif1-m2 double mutants failed to grow on glycerol (Fig. 2C). This result may uncover a previously undiscovered role for Dna2 in mitochondrial maintenance, although further work is required to demonstrate that this is a direct effect. When a dna2Δ pif1-m2 petite mutant was crossed with DNA2 PIF1 grande, the diploids were all grande, suggesting that the dna2Δ pif1-m2 mutant carried either defective (but not suppressive) mitochondrial genomes or lacked mitochondrial DNA entirely.
Failure of Rad53 checkpoint kinase activation in the dna2-1 pif1Δ double mutant.
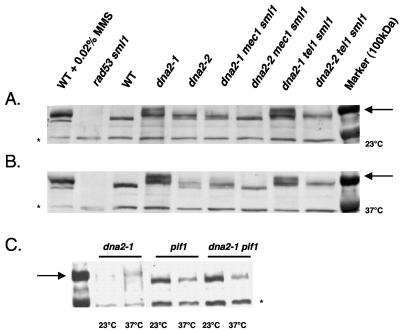
To investigate the extent to which pif1Δ removed the requirement for Dna2 in the cell, we indirectly measured in vivo DNA damage in dna2 mutants in the presence and absence of Pif1 by determining the level of Rad53 phosphorylation. Rad53 is a protein kinase that is phosphorylated and activated in response to DNA damage and replication stress. In both dna2-2 and in dna2-1 mutants at permissive and nonpermissive temperatures, Rad53 is indeed phosphorylated (Fig. 3A and B), consistent with dna2 mutants suffering endogenous DNA damage. The phosphorylation, and implied DNA damage, seen at 23°C in the dna2-1 mutant was not surprising since dna2-1 mutants are sick even at this temperature. We have previously shown that the upstream master checkpoint kinase Mec1 is not essential for viability of dna2-1 cells at the permissive temperature, and that dna2-2 mec1Δ sml1-1 mutants grow more rapidly than dna2-2 mutants (11). (Note that the sml1-1 strain is included in the strains because it is required for mec1Δ viability.) We were therefore interested in whether MEC1 was required for Rad53 phosphorylation in the dna2-1 and dna2-2 mutants. As shown in Fig. 3A and B, full Rad53 phosphorylation in the mutants is dependent on checkpoint induction by Mec1, since Rad53-P is drastically reduced in a dna2-1 mec1Δ sml1-1 strain and absent in a dna2-2 mec1Δ sml1-1 strain.
FIG. 3.
Reduced DNA damage in dna2-1 pif1Δ. The strains used are isogenic derivatives of strain W303—MB90-7A, MB91, U953-61, MB92-M, SPY40, MB92-31C, MB92-35A, and U960-5C (Table 1)—with one exception, the pif1Δ strain MB509. The relevant genotypes are indicated in the figure. Strains were grown to approximately 1 × 107 per ml. Extracts were prepared and Western blots performed using antibody against Rad53 (gift of John Diffley, Clare Hall, England) as described previously (16). Wild-type W303 was also treated with 0.02% MMS where indicated to show phosphorylated Rad53. (A) Rad53 is phosphorylated in dna2-1 and dna2-2 strains at 23°C in a MEC1-dependent and TEL1-independent fashion. The 100-kDa marker runs with fully phosphorylated Rad53 and is indicated by the arrow in the figure. The asterisk denotes a nonspecific, cross-reacting species. (B) Rad53 is phosphorylated in dna2-1 and dna2-2 strains at 37°C in a MEC1-dependent and TEL1-independent manner. (C) pif1Δ suppresses Rad53 phosphorylation in the dna2-1 strain.
We have also previously shown that dna2-2 mutants grow poorly in the absence of TEL1, a kinase related to MEC1 (11). It is therefore interesting that Rad53 phosphorylation in dna2-1 and dna2-2 mutants is relatively independent of TEL1, compared to dependence on MEC1 (Fig. 3A and B).
To see if there is less damage in a dna2-1 pif1Δ mutant, we measured Rad53 phosphorylation in the double mutant. As shown in Fig. 3C, the dna2-1 mutant allele did not lead to induction of Rad53 phosphorylation, at either 23°C or 37°C, when PIF1 was deleted. (The weaker, but clear signals in the dna2-1 controls are due to loading less protein than in adjacent lanes, as indicated by the nonspecific protein indicated by the asterisk. Equivalent controls are also shown in Fig. 3A and B.) Since deletion of PIF1 suppresses Rad53 phosphorylation in dna2-1 mutants, we tentatively propose that there is less replication fork failure in the dna2-1 pif1Δ mutant than in the dna2-1 mutant or that the checkpoint signal is somehow masked in the double mutant.
Suppression of the residual temperature sensitivity and MMS sensitivity in the dna2Δ pif1Δ mutant by pol32Δ.
If Pif1 helicase creates 5′ flaps via its 5′-to-3′ helicase activity, then deletion of Pif1 should lead to reduced occurrence of 5′ flaps in the cell, which might, in part, account for the reduced requirement for the 5′ flap helicase/nuclease activity of Dna2 in a pif1Δ mutant. POL32 is a subunit of pol δ that leads to decreased 5′ flap strand displacement by pol δ when mutated (25). Since deletion of POL32 also suppresses dna2-1 temperature sensitivity (11), we asked whether deleting POL32 could suppress the residual temperature-sensitive growth of the dna2Δ pif1Δ double mutant. Suppression was efficient, since the triple mutant now grew at 37°C (Fig. 4).
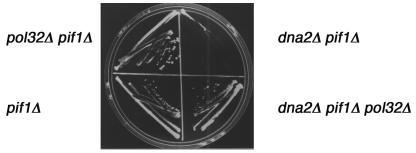
Genetic interaction between PIF1 and POL32.
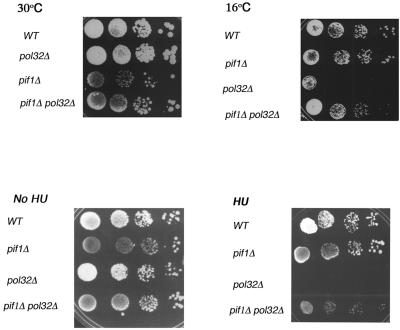
In the course of these studies, we also obtained a pifΔ pol32Δ strain. Although the slow growth of the pif1Δ strain has always been attributed exclusively to the generation of petites, we noticed that the slow growth of the pif1Δ strain was suppressed by pol32Δ (Fig. 4; note colony sizes, for instance). While the pif1Δ strain grows more slowly than the pol32Δ strain, pif1Δ pol32Δ double mutants appear to have an intermediate growth rate between the pol32Δ strain, which grows like the wild type, and pif1Δ strains (Fig. 5, top, left). The putative suppression of pif1Δ strain slow growth by pol32Δ suggested that the pif1Δ mutation affects a nuclear function involving pol δ in addition to its mitochondrial defect. To further investigate the possible functional interaction between Pif1 and pol δ and to determine if it was related to the replication function of POL32, we tested if deletion of PIF1 affects the cold-sensitive lethality and/or the hydroxyurea (HU; an inhibitor of DNA replication) sensitivity of pol32Δ strains. As shown in Fig. 5, the pol32Δ strain grows normally at 30°C. At 16°C, the pol32Δ strain is inviable, but viability is restored by deletion of PIF1 (Fig. 5, top right). In addition, the pol32Δ strain is sensitive to inhibition of DNA replication by 38 mM HU, while the pif1Δ pol32Δ strain is resistant (Fig. 5, bottom). The suppression of the cold sensitivity and HU sensitivity of the pol32Δ strain by deletion of PIF1 provides further evidence that Pif1 interacts with pol δ, most likely during chromosomal DNA replication.
FIG. 5.
Suppression of the cold sensitivity and HU sensitivity of the pol32Δ strain by pif1Δ. The following BY4741 isogenic strains were used: BY4741, WT; MB205, pol32Δ; MB201, pif1Δ; and MB206, pol32Δ pif1Δ. YPD plates were spotted with these strains and incubated at 30°C for 2 days (top, left panel) or at 16°C for 7 days (top right panel) as indicated. YPD plates were spotted with the same strains in the absence (lower left) or presence (lower right) of 38 mM HU and incubated at 23°C.
Does Pif1 act through Dna2 to inhibit telomerase?
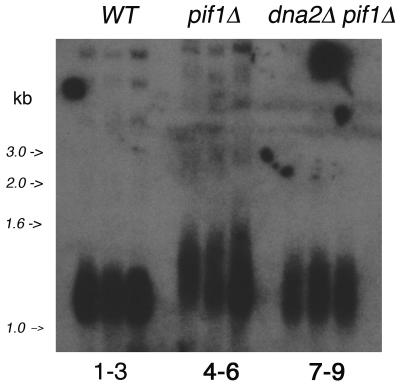
While pif1Δ suppresses the requirement for Dna2, we also addressed the question of whether deletion of DNA2 suppressed the requirement for Pif1. pif1 mutants have long telomeres, overproduction of Pif1 results in short telomeres, and in vitro Pif1 reduces the processivity of telomerase, suggesting Pif1 acts as an inhibitor of telomerase (5, 50). Dna2 affects telomere metabolism in several ways. Dna2 is localized to telomeres in the G1 and G2 phases of the cell cycle but relocalizes during S phase to internal sites in the chromosomes, including but not limited to the rRNA gene (14). In addition, Dna2 is mobilized from telomeres upon treatment of cells with genotoxic agents such as bleomycin (14). Suggesting that telomeric localization implies a telomeric function for Dna2, we have shown that dna2-2 est1Δ mutants, lacking telomerase activity, senesce much more rapidly than est1Δ mutants, and dna2-2 est1Δ mutants show altered pathways of telomerase-independent survival (14). dna2 mutants have normal or slightly long telomeres, but overproduction of DNA2 leads to increased occurrence of single-stranded regions at telomeres (17, 39). Figure 6 compares telomere length in wild-type, pif1Δ, and dna2Δ pif1Δ strains. The pif1Δ telomere is longer than the wild-type telomere, as previously demonstrated (50). Interestingly, the pif1Δ dna2Δ telomere is dramatically shorter than the pif1Δ telomere and similar in size to the wild-type telomere. This suggests that the long telomere phenotype of pif1Δ is dependent on Dna2.
FIG. 6.
Deletion of DNA2 reduces telomere length in pif1Δ strains. Lanes 1, 2, and 3, WT strains 47421, BY4741, and BY4742, respectively. Lanes 4, 5, and 6, three colonies of pif1Δ strain 10509. Lanes 7, 8, and 9, dna2Δ pif1Δ strains MB203.3, MB203.5, and MB203.6, respectively. MB203.3, MB203.5, and MB203.6 are three independent pif1Δ transformants of MB110.
Given that dna2-2 est1Δ and dna2-2 est2Δ strains show increased rates of senescence compared to either single mutant, and given the reduction in telomere length in the pif1Δ dna2Δ mutant, we hypothesized that Dna2 might somehow aid telomerase in its function. If so, then we would expect dna2Δ to reduce the frequency of GCRs in a pif1Δ strain. We therefore investigated the effect of deletion of DNA2 on the increased frequency of GCRs observed in pif1-m2 mutants. The assay measures the rate of deletion of a region of chromosome V that is nonessential and that contains CAN1 and URA3, by scoring the simultaneous appearance of canavanine-resistant (CanR) 5-FOAR cells. The pif1-m2 mutant shows a 137-fold increase over wild-type strains in appearance of GCRs characterized primarily by the addition of new telomeres to internal chromosomal sequences (Table 2) (36). The rate of GCR formation was reduced to a 70-fold increase over the wild type in the dna2Δ pif1-m2 strain (Table 2). This result is also consistent with an interaction between Pif1 and Dna2 in telomere biogenesis.
TABLE 2.
Frequency of GCRs
| Relevant genotypea | GCR rate (CanR 5-FOAR)b |
|---|---|
| Wild type | 3.5 × 10−10 (1) |
| pif1-m2 | 4.8 × 10−8 (137) |
| pif1-m2 dna2Δ | 2.5 × 10−8 (71) |
| dna2-1 | >3.5 × 10−10 (1) |
| dna2-2 | 7.0 × 10−9 (20) |
| dna2-K1080A | 1.8 × 10−9 (5) |
All strains were constructed for this study by standard genetic techniques and are isogenic with the wild-type strain 3615 MATa ura3-52 leu2Δ1 trp1Δ63 his3 Δ200 lys2 Bgl hom3-10 ade2-Δ1 ade8 hxt13::URA3:pif1-m2, strain 4344; pif1-m2 dna2Δ, strain 4344dna2Δ; dna2-1, strain 3615-2-1; dna2-2, strain 3615-2-2; and dna2-K1080A, strain 3615-K1080A. The dna2-K1080A mutant is defective in helicase motif I.
GCR rates were determined by fluctuation analysis of at least two experiments consisting of 5 or 11 cultures, and average values are reported. Numbers in parentheses indicate fold increase in GCRs compared to wild type.
Mutations in genes that function to suppress GCRs usually lead to a greater than 100-fold increase in GCR frequency compared to the wild type (13, 36). Interestingly, dna2-1 (a nuclease mutation), dna2-2, or dna2 K1080A (two different helicase mutations) caused only a 2- to 20-fold increase in GCRs compared to the wild type.
Interaction of DNA2 and PIF1 with other DNA2-interacting genes.
We have recently completed a synthetic lethality screen with dna2-1 and dna2-2 and found that DNA2 interacts with 56 genes affecting at least seven different genome maintenance pathways (11). We next tested whether the pif1Δ suppressed the synthetic effects previously observed between dna2-1 and 27 of the 44 nonessential genes identified in the screen (Table 3). The genes chosen are representatives of each of the pathways identified. Deletion of PIF1 failed to suppress the lethality of dna2 with many genes involved in OFP, with other helicases, and with genes governing chromatin dynamics. The lethality of the triple mutants rules out a potentially trivial explanation of the synthetic lethality of these genes with dna2-1, namely, that transcription of dna2-1 was reduced by the mutations. The lethality of the triple mutants also is consistent with other results presented here that indicate that pif1Δ does not fully bypass the need for Dna2.
TABLE 3.
Growth of various triple mutants in the absence of Dna2 and Pif1a
| Effect | dna2Δ pif1Δ genes |
|---|---|
| Lethal | hst3, rtf1, sae2, mre11, yen1, exo1, rnh35, rad27, rrm3, ctf4, sgs1, bre1 |
| Sick | est2,b rpd3, rtt103, sap30, lge1, swd3, swd1, vid21 |
| No effect | cdc73, caf20, hir3, htz1, paf1, rad18, rad51 |
All mutations are synthetic lethal with dna2-1 and/or dna2-2. All mutations are deletions in the BY4741 background and were obtained by crossing MB203.3 with strains carrying the respective single mutations obtained from Invitrogen. Tetrad analysis was performed by standard techniques. At least 10 tetrads were dissected for each cross. Synthetic lethality was scored as the absence of viable triple mutant spores. Synthetic sickness was also scored and represents fewer than the expected number of triple mutants and slower growth than either the single mutant or the dna1Δ pif1Δ strain.
Enhanced senescence.
Usefulness of dna2Δ pif1Δ: synthetic lethality of rtf1Δ but not of paf1Δ with dna2Δ pif1Δ.
Recently, we reported that dna2-1 was synthetically lethal with members of the Rad6 epistasis group specifically involved in histone modification. dna2-1 was synthetically lethal with rtf1, a gene encoding a member of the PAF1 transcription factor/histone modification complex (11). dna2Δ pif1Δ rtf1Δ mutations are also synthetically lethal (Table 3). To try to distinguish whether the effect was indirect and due to reduced transcription of another replication gene, we tested whether dna2Δ pif1Δ was synthetic lethal with another member of the pathway, paf1Δ. We had previously found that dna2-1 paf1Δ mutant was viable, but were concerned that the viability might result from residual Dna2 protein activity in the dna2-1 strain. Indeed, dna2Δ pif1Δ paf1Δ triple mutants were viable. The difference between the effects of deletion PAF1 and RTF1 shows that synthetic sickness with one member of a pathway does not imply synthetic sickness with additional members, likely due to the participation of individual components in multiple, nonoverlapping pathways. The difference between the effect of paf1 and rtf1 correlates, not with an effect on transcription, but with their quantitative effect on methylation. paf1 mutants reduce dimethylation of histone H3 by 75%, while dimethylation is nearly completely eliminated in the rtf1 mutant. Therefore, the synthetic lethality of rtf1 with dna2 is more likely due to a histone methylation defect than to a transcription defect. This interpretation is also supported by recent evidence that Rtf1 has an additional role in methylation in a process other than transcription elongation (38). Because dna2Δ pif1Δ is synthetically lethal with several other histone modification gene deletions (Table 2) (11), we propose that the synthetic lethality of dna2Δ pif1Δ rtf1Δ is due to failure of dimethylation of histone H3 K4.
DISCUSSION
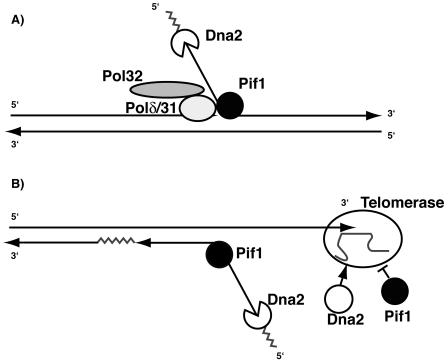
We have shown that deletion of PIF1 or inactivation of its nuclear function suppresses the temperature-sensitive phenotype of dna2-1 mutants, the MMS sensitivity of dna2-2 mutants, and even the inviability of dna2Δ mutants. Our main conclusions are summarized in Fig. 7, namely that Pif1 is involved in OFP and that Dna2 is involved, along with Pif1, in regulating telomere length. We will discuss three primary insights underlying these conclusions. First, the suppression of the lethality of the dna2Δ mutation by pif1Δ shows that the nuclear function of Pif1 creates the essential requirement for Dna2, a helicase/nuclease involved in Okazaki fragment processing. In molecular terms, this first suggests that the Pif1 helicase generates structures that are lethal to the cell if Dna2 is not present. Similarly, the absence of Dna2 may lead to structures that are lethal if processed by Pif1. Second, the genetic interaction between PIF1 and DNA2 implies that the nuclear form of Pif1 may play a more general role in chromosomal replication than previously thought. The related observation that pif1Δ also suppresses the cold sensitivity and the HU sensitivity of pol32Δ, a subunit of pol δ, a major DNA polymerase at the replication fork, supports this interpretation. Third, the suppression of the major pif1Δ phenotype, long telomeres, by dna2Δ shows that the long telomeres in pifΔ mutants require Dna2 for synthesis.
FIG. 7.
Interpretation of genetic interactions between PIF1 and DNA2. (A) Pif1 may help to make a flap during RNA removal by Dna2 during OFP. (B) Dna2 may be required for optimum elongation of telomeres by telomerase, whereas Pif1 has been shown to inhibit telomerase processivity (5). Pif1 may have a second role at telomeres in which it aids Dna2 in removal of RNA from the Okazaki fragments at the telomere. The last Okazaki fragment might have a special requirement for Pif1 helicase as there is no pol δ/PCNA present to strand displace and to recruit FEN1.
PIF1 creates a requirement for DNA2.
Dna2 is thought to participate in primer removal from Okazaki fragments, but only in cases where pol δ strand displaces more than 30 bases of the downstream Okazaki fragment before ligation (2, 24, 28, 29). Dna2 is a 5′-to-3′ helicase and a nuclease that requires a single-stranded DNA end for activity (29). Dna2 appears to load onto a single-stranded terminus and to translocate by a threading mechanism (27). Thus, Dna2 has evolved to be compatible with processing displaced flaps but not to cleave single-stranded regions endonucleolytically. In addition to the biochemical properties of Dna2, genetic interactions with other genes that function on 5′ flaps, such as those coding for FEN1, Exo1, and Yen1, and with genes involved in removing RNA, such as that coding for RNase H2, strengthen the notion that Dna2 is required for OFP (8, 11). Thus, although Okazaki fragments have not formally been demonstrated to accumulate in dna2 mutants, there is overwhelming biochemical and genetic evidence that the essential function of Dna2 lies in some aspect of OFP. Taking that as our assumption, our new results suggest that one substrate of Pif1 is an Okazaki fragment. There are several ways in which Pif1 might function: either through a mechanism intrinsic to Okazaki fragment formation and maturation or, more indirectly, in repairing faulty synthesis or processing. In OFP, reconstitution of reaction mixtures containing pol δ, replication protein A (RPA), Dna2, FEN1, DNA ligase, and model substrates representing intermediates in OFP, suggests that Dna2 is required for OFP only when there is excessive (greater than 30 nucleotides) strand displacement of a 5′ flap on the previously synthesized Okazaki fragment by the pol δ extending the nascent fragment. The role of Dna2 helicase/nuclease is to participate in removal of the flap. This situation probably does not arise at every Okazaki fragment, however, since pol δ and FEN1 normally act in concerted fashion producing only transient flaps shorter than 4 nucleotides in vitro (18). Furthermore, Dna2 is not essential in vitro even to remove long flaps in the presence of FEN1, but is only stimulatory to FEN1, which appears to be the major nuclease. Our studies suggest that in this strand displacement pathway for OFP, there may be an additional participant, Pif1 helicase. Pif1 might assist pol δ in strand displacement, and a pif1 mutation may reduce requirement for Dna2 by reducing levels of strand displacement.
We have previously shown that dna2 mutants grow better in the absence of MEC1 but have increased growth defects in the absence of TEL1 (11). This implies that there is sufficient damage to induce the Mec1-dependent checkpoint in dna2 cells, but that the checkpoint is not necessary for viability. Instead, the checkpoint slows growth, perhaps to allow repair. We verify that result here with physical evidence that Rad53 becomes phosphorylated in dna2-1 and dna2-2 mutants and that Rad53 phosphorylation is dependent on Mec1 kinase but not on Tel1 kinase. However, in the dna2-1 mec1Δ sml1-1 strain (Fig. 3A), there is still some phosphorylation, unlike in the dna2-2 mec1Δ sml1-1 strain. The two different mutations could generate different types of DNA damage. In any case, deletion of PIF1 resulted in drastic reduction of the Rad53 phosphorylation, which we propose would be due to a reduction in extended flaps below the threshold required to activate the checkpoint, also suggesting a tight connection between Pif1 and Dna2 function in DNA replication.
It remains possible that Pif1 is only required for repair, although we think it less likely. In this scenario, Pif1 would create a DNA structure at sites where DNA replication forks on the lagging strand are blocked by endogenous DNA damage, and this structure is lethal if not resolved. Dna2 helicase/nuclease would then be required for removal of this structure.
Genetic interactions between PIF1, DNA2, and POL32 suggest a general (but not universal) role for PIF1 in replication fork progression.
Several of our recent results support the model just presented in which Dna2 acts on flaps produced by both pol δ and Pif1. First, the temperature sensitivity of dna2-1 mutants and the DNA damage sensitivity of dna2-2 mutants, but not the lethality of dna2Δ, are suppressed by the deletion of POL32 (11). The POL32 gene encodes a subunit of pol δ that is required for optimum processivity, and its deletion can be expected to reduce strand displacement in vivo as it has been demonstrated to do in vitro (12, 25). This would explain the reduced requirement for DNA2. Second, the dna2-1 mutation is synthetically lethal with pol3-01 (11), a pol δ mutation expected to increase strand displacement (18). This would explain the synthetic lethality. Third, the observation presented in this work of the suppression of residual defects in the dna2Δ pif1Δ strain by deletion of POL32 is consistent with a model in which deletion of Pif1 reduces but does not abolish strand displacement, while further deletion of POL32 eliminates strand displacement and shifts the course of Okazaki fragment processing to a pathway that is Dna2 independent, such as an RNase H/FEN1-alone pathway (33). As would be anticipated in such a model, dna2-1 and dna2Δ pif1Δ are synthetically lethal with genes encoding RNase H2 subunits (rnh35Δ and rnh202Δ) (11) (Table 2). This putative Dna2-independent pathway might be inhibited by flap production by Pif1, since we have shown here that deletion of PIF1 also suppresses the cold-sensitive lethality of pol32Δ, which in itself might favor the alternative Dna2-independent pathway. Finally, suppression of the slow growth of pif1Δ by deletion of a purely nuclear function, POL32, suggests that PIF1 is required for optimal nuclear replication and not just for mitochondrial DNA stability, as previously proposed. In a more general sense, the nature of the genetic interactions between PIF1, POL32, and DNA2 suggest that a delicate balance of these three activities is required for maintaining chromosomal stability in a range consistent with viability. Since such a role in nuclear replication for PIF1 has been overlooked to date, further biochemical reconstitution experiments incorporating Pif1 helicase and pol δ lacking Pol32 will be required to fully understand this pathway.
Although it is widely held that Pif1 functions in chromosomal DNA replication primarily at telomeres, the genetic interactions we report, if indeed they do imply nontelomeric activities, are not the first evidence that PIF1 does not function solely at telomeres in S. cerevisiae. Ivessa et al. showed clearly that Pif1 is required for replication fork pausing at the Fob1-dependent replication fork barrier in the rRNA gene (22). Dna2, in contrast, is required to prevent pausing, and in its absence, there is not only increased pausing but also an increased frequency of DSBs at the RFB (47, 48). Thus, Dna2 and Pif1 may have opposing functions at the RFB, just as they appear to have at telomeres (Fig. 6).
DNA2 in chromosomal, mitochondrial, and telomere replication.
Although deletion of PIF1 suppresses the lethality of a dna2Δ mutation and the DNA damage/replication stress-induced Rad53 phosphorylation seen in dna2 mutants, mutants retain defects in DNA replication and repair since they are temperature sensitive and sensitive to MMS. We have begun to explore the residual DNA damage in the dna2Δ pif1Δ double mutant. Analysis of cell cycle progression and DNA replication in synchronized cells after ectopic expression of PIF1 in a dna2Δ pif1Δ background revealed that cells arrested within the first cell cycle as dumbbells, a G2/M arrest (data not shown). Further work should allow a more sensitive evaluation of the precise steps in DNA replication that require Dna2 than were possible with conditional alleles, which are by definition leaky.
The DNA2 gene can be deleted in the pif1-m2 strain, which has a mutated nuclear and functional mitochondrial Pif1, but cannot be deleted in a pif1-m1 strain, which has a functional nuclear and mutated mitochondrial Pif1. We noticed, however, that all of the dna2Δ pif1-m2 strains are petite. Since pif1-m2 is not deficient in mitochondrial DNA functions, Dna2 might be required for mitochondrial DNA stability. The mechanism of yeast mitochondrial DNA replication is not well enough understood, however, to speculate on whether the effect is due to a direct role for Dna2 in mitochondrial DNA metabolism or if it is indirect.
Pif1 has been proposed to inhibit telomerase, since pif1Δ mutants have long telomeres and show increased frequency of de novo telomere synthesis at internal DSBs (50). DSBs give rise to a high frequency of GCRs in pif1-m2 strains (36). Although Pif1 inhibits telomerase in vitro (5), the mechanism of telomerase inhibition in vivo is not fully understood. The fact that we show that deletion of DNA2 in a pif1Δ strain restores nearly normal telomere length suggests that Dna2 may need to be functional in order to observe long telomeres in the absence of Pif1. Dna2 may function at telomeres to stimulate telomerase-dependent telomere lengthening (Fig. 4 and 7). This interpretation is consistent with our previous work (see the introduction and reference 14), but the putative activation role of Dna2 on telomerase synthesis is more directly unmasked by deletion of PIF1, as shown here. The reduction in GCR rate from a 137-fold increase over wild type in the pif1-m2 strain to a 70-fold increase in the pif1-m2 dna2Δ double mutants also is consistent with this interpretation.
The dna2Δ pif1Δ telomere phenotype is similar to that of rrm3Δ pif1Δ. The long telomeres of the pif1Δ mutant and the frequency of telomere addition to DSBs in pif1Δ cells require RRM3 (20). Also, the GCR rate of pif1Δ is reduced by deletion of RRM3 (20). The dna2Δ rrm3Δ pif1Δ triple mutant is inviable (Table 2), obviating an experiment to test if telomeres are even shorter or the GCR rate is further reduced than in either double mutant. Outside of rrm3Δ and mutations that reduce telomerase activity (e.g., est2Δ and stn1-13), very few mutations have been found that reduce the GCR rate in pif1-m2 strains (36, 37). These comprise spindle checkpoint genes (mad2, mad3, and bub3), a mitotic exit network gene (bub2), and sister chromatid cohesion genes (ctf8, ctf18, and dcc1) (37). These genes may be involved in allowing cells with pre-GCR lesions to survive and generate GCRs. dna2Δ is not known to affect the spindle checkpoint. Thus, Dna2 suppression of GCRs in the pif1-m2 strain is more similar to suppression by rrm3Δ, stn1-13, and est2Δ. A further connection between Dna2 and telomerase-dependent telomere elongation is provided by the fact that dna2Δ pif1Δ est2Δ cells form small colonies and senesce more rapidly than est2Δ mutants (Table 2).
Diede and Gottschling (15) have shown that telomerase does not synthesize telomeres de novo in the absence of Okazaki fragment synthesis. The effects of Dna2 and/or Pif1 on telomere synthesis may be a consequence of a role in OFP (Fig. 7). If Pif1 inhibits telomerase indirectly as well as directly, it might do so by functioning on the lagging strand during telomere maturation. Pif1 may displace the strand on the Okazaki fragment at the end of the telomere, exposing G-rich DNA, which may potentially form a G-quartet structure that inhibits telomerase. These G-quartets might not hybridize to the telomerase RNA, for instance. The absence of Pif1 may minimize formation of such structures. If Pif1 functions as an inhibitor of telomerase independently of Dna2, then a telomerase lacking Pif1 inhibition still requires Dna2 for optimal function, since telomeres are shortened in the dna2Δ pif1Δ mutant. If a major role of Dna2 is at telomeres, this might explain some puzzling observations in C. elegans. All fungal versions of DNA2 are essential. C. elegans dna2 mutants show a more complex phenotype. They appear deficient in DNA replication but show 90% embryonic viability in F1. However, they are embryonic lethal in F2 (32). Late-generation lethality is a phenotype of telomere deficiency in other organisms.
In S. pombe, pfh1+ is an essential helicase whose helicase domain is closely related to both the nonessential S. cerevisiae Pif1 and Rrm3 proteins, but which diverges from both in the N terminus. It has been difficult to make a one-to-one comparison with respect to function between pfh1+ and either RRM3 or PIF1 to date. Our demonstration that S. cerevisiae PIF1 shows genetic interaction with DNA2 and POL32 may shed light on the conserved functions of PIF1, since S. pombe pfh1+, dna2+, and cdc27+ (the ortholog of POL32) also interact genetically. A mutant allele of pfh1+, pfh1-R20, affecting the ATPase domain, suppresses the temperature-sensitive growth of the dna2-C2 mutant, which has a mutation affecting the Dna2 helicase domain of the S. pombe dna2+ strain (41, 45). Whether the pfh1 mutant suppresses nuclease-deficient S. pombe dna2 strains, as pif1Δ suppresses the dna2Δ and dna2-1 (a lesion in the nuclease) mutants of S. cerevisiae, has not been reported. Nor has it been shown that deletion of pfh1 bypasses the essential function of DNA2, as no nuclease-deficient dna2 alleles have been described in S. pombe. Therefore, a direct interaction between the two is less well established in S. pombe. S. pombe pfh1+ and cdc27+ also interact. In contrast to the suppression of S. cerevisiae pol32Δ by pif1Δ that we observe, however, in S. pombe, cdc27 conditional mutants are synthetically lethal with pfh1-R20 and pfh1-R23 mutations (44). The pfh1 mutations may affect a function performed in S. cerevisiae by RRM3 rather than by PIF1, accounting for the difference between the two organisms. Another difference between the two organisms is that an additional essential gene, cdc24+, which has not been identified in S. cerevisiae, appears to interact with these genes in S. pombe. Despite the differences in alleles tested to date in the two organisms, it is clear that the essential pfh1+ gene in S. pombe shows similar genetic interactions to PIF1, and therefore it appears that it is S. cerevisiae PIF1, and not RRM3, as previously believed, that conserves the putative essential role of pfh1+ in interactions with DNA2 and with pol δ in OFP.
Acknowledgments
The work done by K. Myung's laboratory was supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health. The work carried out in J. L. Campbell's laboratory was supported by GM25508, a grant from the Research Management Group, and a predoctoral fellowship from Fundacao para a Ciencia e Technologia (SFRH/BD/9612/2002), Portugal, to C.C.R.
REFERENCES
- 1.Ayyagari, R., X. V. Gomes, D. A. Gordenin, and P. M. J. Burgers. 2003. Okazaki fragment maturation in yeast. I. Distribution of functions between Fen1 and Dna2. J. Biol. Chem. 278:1618-1625. [DOI] [PubMed] [Google Scholar]
- 2.Bae, S. H., K.-H. Bae, J. A. Kim, and Y. S. Seo. 2001. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412:456-461. [DOI] [PubMed] [Google Scholar]
- 3.Bessler, J. B., J. Z. Torre, and V. A. Zakian. 2001. The Pif1p subfamily of helicases: region-specific DNA helicases? Trends Cell Biol. 11:60. [DOI] [PubMed] [Google Scholar]
- 4.Bessler, J. B., and V. A. Zakian. 2004. The amino terminus of the Saccharomyces cerevisiae DNA helicase Rrm3p modulates protein function altering replication and checkpoint activity. Genetics 168:1205-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boule, J.-B., L. R. Vega, and V. A. Zakian. 2005. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature 438:57-61. (First published 3 November 2005; doi: 10.1038/nature04091.) [DOI] [PubMed] [Google Scholar]
- 6.Budd, M. E., and J. L. Campbell. 1995. A new yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc. Natl. Acad. Sci. USA 92:7642-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budd, M. E., and J. L. Campbell. 2000. The pattern of sensitivity of yeast dna2 mutants to DNA damaging agents suggests a role in DSB and postreplication repair pathways. Mutat. Res. 459:173-186. [DOI] [PubMed] [Google Scholar]
- 8.Budd, M. E., and J. L. Campbell. 1997. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 17:2136-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Budd, M. E., W.-C. Choe, and J. L. Campbell. 1995. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J. Biol. Chem. 270:26766-26769. [DOI] [PubMed] [Google Scholar]
- 10.Budd, M. E., W.-C. Choe, and J. L. Campbell. 2000. The nuclease activity of the yeast Dna2 protein, which is related to the RecB-like nucleases, is essential in vivo. J. Biol. Chem. 275:16518-16529. [DOI] [PubMed] [Google Scholar]
- 11.Budd, M. E., A. Tong, X. Peng, P. Polaczek, C. Boone, and J. L. Campbell. 2 December 2005. A network of multi-tasking proteins at the DNA replication fork preserves genome stability. PLoS Genet. doi: 10.1371/journal.pgen.0010061. [DOI] [PMC free article] [PubMed]
- 12.Burgers, P. M. J., and K. J. Gerik. 1998. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 273:19756-19762. [DOI] [PubMed] [Google Scholar]
- 13.Chen, C., and R. D. Kolodner. 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23:81-85. [DOI] [PubMed] [Google Scholar]
- 14.Choe, W., M. Budd, O. Imamura, L. Hoopes, and J. L. Campbell. 2002. Dynamic localization of an Okazaki fragment processing protein suggests a novel role in telomere replication. Mol. Cell. Biol. 22:4202-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diede, S. G., and D. E. Gottschling. 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerase α and δ. Cell 99:723-733. [DOI] [PubMed] [Google Scholar]
- 16.Elsasser, S. E., Y. Chi, P. Yang, and J. L. Campbell. 1999. Phosphorylation controls timing of Cdc6p destruction: a biochemical analysis. Mol. Biol. Cell 10:3263-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Formosa, T., and T. Nitiss. 1999. Dna2 mutants reveal interactions with DNA polymerase alpha and Ctf4, a Pol α accessory factor, and show that full DNA2 helicase activity is not essential for growth. Genetics 151:1459-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garg, P., C. M. Stith, N. Sabouri, E. Johansson, and P. M. Burgers. 2004. Idling by DNA polymerase δ maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 18:2764-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imamura, O., and J. L. Campbell. 2003. The human Bloom syndrome gene suppresses the DNA replication and repair defects of yeast dna2 mutants. Proc. Natl. Acad. Sci. USA 100:8193-8198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivessa, A. S., B. A. Lenzmeier, J. B. Bessler, L. K. Goudsouzian, S. L. Schnakenberg, and V. A. Zakian. 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell 12:1525-1536. [DOI] [PubMed] [Google Scholar]
- 21.Ivessa, A. S., J.-Q. Zhou, V. P. Schulz, E. K. Monson, and V. A. Zakian. 2002. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 16:1383-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivessa, A. S., J.-Q. Zhou, and V. Zakian. 2000. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in the ribsomal DNA. Cell 100:479-489. [DOI] [PubMed] [Google Scholar]
- 23.Jin, Y. H., R. Ayyagari, M. A. Resnick, D. A. Gordenin, and P. M. J. Burgers. 2003. Okazaki fragment maturation in yeast. II. Cooperation between the polymerase and 3′ to 5′ exonuclease activities of Pol δ in the creation of a ligatable nick. J. Biol. Chem. 278:1626-1633. [DOI] [PubMed] [Google Scholar]
- 24.Jin, Y. H., R. Obert, P. M. J. Burgers, T. A. Kunkel, M. A. Resnick, and D. A. Gordenin. 2001. The 3′-5′ exonuclease of DNA polymerase δ can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc. Natl. Acad. Sci. USA 98:5122-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansson, E., P. Garg, and P. M. J. Burgers. 2004. The Pol32 subunit of DNA polymerase δ contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J. Biol. Chem. 279:1907-1915. [DOI] [PubMed] [Google Scholar]
- 26.Kang, J.-Y., E. Choi, S.-H. Bae, K.-H. Lee, B.-S. Gim, H.-D. Kim, C. Park, S. A. MacNeill, and Y.-S. Seo. 2000. Genetic analyses of Schizosaccharomyces pombe dna2+ reveal that Dna2 plays an essential role in Okazaki fragment metabolism. Genetics 155:1055-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kao, H.-I., J. L. Campbell, and R. A. Bambara. 2004. Dna2p helicase/nuclease is a tracking protein, like FEN1, for flap cleavage during Okazaki fragment maturation. J. Biol. Chem. 279:50840-50849. [DOI] [PubMed] [Google Scholar]
- 28.Kao, H. I., L. A. Henricksen, Y. Liu, and R. A. Bambara. 2002. Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J. Biol. Chem. 277:14379-14389. [DOI] [PubMed] [Google Scholar]
- 29.Kao, H. I., J. Veeraraghavan, P. Polaczek, J. L. Campbell, and R. A. Bambara. 2004. On the roles of Saccharomyces cerevisiae Dna2p and FEN1 in Okazaki fragment processing. J. Biol. Chem. 279:15014-15024. [DOI] [PubMed] [Google Scholar]
- 30.Lahaye, A., S. Leterme, and F. Foury. 1993. PIF1 DNA helicase from Saccharomyces cerevisiae. Biochemical characterization of the enzyme. J. Biol. Chem. 268:26155-26161. [PubMed] [Google Scholar]
- 31.Lahaye, A., H. Stahl, D. Thine-Sempoux, and F. Foury. 1991. PIF1: a DNA helicase in yeast mitochondria. EMBO J. 10:997-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, K. H., M. H. Lee, T. H. Lee, J. W. Han, Y. J. Park, J. Ahnn, Y. S. Seo, and H. S. Koo. 2003. Dna2 requirement for normal reproduction of Caenorhabditis elegans is temperature-dependent. Mol. Cell 15:81-86. [PubMed] [Google Scholar]
- 33.Liu, Y., H.-I. Kao, and R. A. Bambara. 2004. Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 73:589-615. [DOI] [PubMed] [Google Scholar]
- 34.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 35.Makovets, S., I. Herskowitz, and E. H. Blackburn. 2004. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol. Cell. Biol. 24:4019-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myung, K., C. Chen, and R. D. Kolodner. 2001. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411:1073. [DOI] [PubMed] [Google Scholar]
- 37.Myung, K., S. Smith, and R. D. Kolodner. 2004. Mitotic checkpoint function in the formation of gross chromosomal rearrangements in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101:15980-15985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng, H. H., S. Dole, and K. Struhl. 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278:33625-33628. [DOI] [PubMed] [Google Scholar]
- 39.Parenteau, J., and R. J. Wellinger. 1999. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell. Biol. 19:4143-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ritchie, K. B., J. C. Mallory, and T. D. Petes. 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6065-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryu, G.-H., H. Tanaka, D.-H. Kim, J.-H. Kim, S.-H. Bae, Y.-N. Kwon, J. S. Rhee, S. A. MacNeill, and Y.-S. Seo. 2004. Genetic and biochemical analyses of Pfh1 DNA helicase function in fission yeast. Nucleic Acids Res. 32:4205-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schulz, V. P., and V. A. Zakian. 1994. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76:145-155. [DOI] [PubMed] [Google Scholar]
- 43.Singer, M. S., A. Kahana, A. J. Wolf, L. L. Meisinger, S. E. Peterson, C. Goggin, M. Nahowald, and D. E. Gottschling. 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150:613-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka, H., G.-H. Ryu, Y.-S. Seo, and S. A. MacNeill. 2004. Genetics of lagging strand DNA synthesis and maturation in fission yeast: suppression analysis links the Dna2-Cdc24 complex to DNA polymerase δ. Nucleic Acids Res. 32:6367-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka, H., G.-H. Ryu, Y.-S. Seo, K. Tanaka, H. Okayama, S. A. MacNeill, and Y. Yuasa. 2002. The fission yeast pfh1+ gene encodes an essential 5′ to 3′ DNA helicase required for the completion of S-phase. Nucleic Acids Res. 30:4728-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomita, K., T. Kibe, H.-Y. Kang, Y.-S. Seo, M. Uritani, T. Ushimaru, and M. Ueno. 2004. Fission yeast Dna2 is required for generation of the telomeric single-strand overhang. Mol. Cell. Biol. 24:9557-9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weitao, T., M. Budd, and J. L. Campbell. 2003. Evidence that yeast SGS1, DNA2, SRS2, and FOB1 interact to maintain rDNA stability. Mutat. Res. 532:157-172. [DOI] [PubMed] [Google Scholar]
- 48.Weitao, T., M. Budd, L. L. Mays Hoopes, and J. L. Campbell. 2003. Dna2 helicase/nuclease causes replicative fork stalling and double-strand breaks in the ribosomal DNA of Saccharomyces cerevisiae. J. Biol. Chem. 278:22513-22522. [DOI] [PubMed] [Google Scholar]
- 49.Zhao, X., E. G. D. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2:329-340. [DOI] [PubMed] [Google Scholar]
- 50.Zhou, J.-Q., E. K. Monson, S.-C. Teng, V. P. Schulz, and V. A. Zakian. 2000. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science 289:771-774. [DOI] [PubMed] [Google Scholar]