Abstract
Blood and vascular cells are generated during early embryogenesis from a common precursor, the hemangioblast. The stem cell leukemia gene (SCL/tal 1) encodes a basic helix-loop-helix transcription factor that is essential for the normal development of blood progenitors and blood vessels. We have previously characterized a panel of SCL enhancers including the +19 element, which directs expression to hematopoietic stem cells and endothelium. Here we demonstrate that SCL is expressed in bone primordia during embryonic development and in adult osteoblasts. Despite consistent expression in cells of the osteogenic lineage, SCL protein is not required for bone specification of embryonic stem cells. In transgenic mice, the SCL +19 core enhancer directed reporter gene expression to vascular smooth muscle and bone in addition to blood and endothelium. A 644-bp fragment containing the SCL +19 core enhancer was active in both blood and bone cell lines and was bound in vivo by a common array of Ets and GATA transcription factors. Taken together with the recent observation that a common progenitor can give rise to blood and bone cells, our results suggest that the SCL +19 enhancer targets a mesodermal progenitor capable of generating hematopoietic, vascular, and osteoblastic progeny.
Hematopoietic and vascular cells are generated during embryogenesis from a common progenitor, the hemangioblast (5, 23). These cells constitute a subpopulation of the primitive streak mesoderm and migrate into the yolk sac to establish the primitive hematopoietic program (48). Definitive hematopoiesis is established subsequently in the para-aortic splanchnopleure region by regional hemangioblasts or hemogenic endothelium, which supports the development of hematopoietic stem cells (HSCs) (7, 28, 34). The basic helix-loop-helix transcription factor encoded by the stem cell leukemia gene (SCL/tal 1) is expressed in blood progenitors and endothelium and is pivotal to the normal development of these tissues (reviewed in reference 30). Specification of hematopoietic stem cells from mesoderm during embryonic development requires SCL and mice lacking this gene product die at embryonic day 9.5 (E9.5) owing to a failure of hematopoiesis (42, 49).
Embryonic stem (ES) cell differentiation studies have identified the blast colony-forming cell (BL-CFC), a progenitor with hematopoietic, endothelial, and vascular smooth muscle potential (5, 28). Hemangioblast colonies isolated from the posterior primitive streak of E7.5 mouse embryos express SCL and have a similar cell lineage potential (23). Increased SCL expression in BL-CFCs has been shown to increase hematopoietic potential at the expense of smooth muscle differentiation (13). Conversely, loss of SCL expression favors smooth muscle differentiation (13). During embryonic development SCL is expressed in the early mesoderm and forced expression of SCL in zebra fish induces overproduction of hematopoietic and endothelial precursors from the lateral mesoderm at the expense of paraxial (somitic) mesoderm (15). Therefore SCL expression in mesodermal progenitors appears to favor hematopoiesis over other potential cell fates.
During embryogenesis, most of the head skeleton develops from mesenchymal cells derived from the cranial neural crest and the limb and axial skeleton from trunk mesoderm. There is considerable overlap in the origin of bones in the neck and shoulder region (33). The paraxial (somitic) mesoderm contributes to the axial skeleton, whereas the limb skeleton develops from the parietal (somitic) lateral plate mesoderm. Mesenchymal cells initially undergo condensation followed by differentiation within these condensations into osteoblasts (dermal or intramembranous bones) or chondrocytes (endochondral bones) (reviewed in reference 37). In endochondral bones, chondrocytes then proliferate and produce an extracellular matrix to form primordial cartilage. Proliferating chondrocytes in the central region of the cartilage undergo terminal differentiation to hypertrophic chondrocytes, which then exit the cell cycle. The hypertrophic cartilage is invaded by blood vessels along with osteoblasts, osteoclasts, and hematopoietic cells to form primary ossification centers.
Bone formation and maintenance through life are performed by a balance between matrix formation by osteoblasts and resorption by osteoclasts. Osteoblasts are also a component of the HSC niche (3, 59) and along with marrow stromal and endothelial cells (29) help maintain quiescence of HSCs, which is integral to their long-term repopulating capacity (reviewed in reference 53). Interestingly, adult hematopoietic cells and osteoblasts have also been reported to share a common marrow precursor (9, 36).
We have previously characterized a panel of SCL cis-regulatory elements (17, 18, 45, 52) and have demonstrated that the SCL +19 core enhancer directs reporter gene expression to adult and fetal HSCs, mast cells, megakaryocytes, and endothelium (20, 46, 51). Here we report that SCL is expressed in adult osteoblasts but not mature osteocytes and that the SCL +19 core enhancer was active not only in lineages derived from the hemangioblast but also in adult and developing skeleton and vascular smooth muscle. These data suggest that the SCL +19 stem cell enhancer targets a mesodermal progenitor capable of generating blood, endothelial, smooth muscle, and bone progeny. We also show that, consistent with its primary role in establishing the hematopoietic program, SCL is not required to establish the bone program in ES cell differentiation assays.
MATERIALS AND METHODS
Transgenic mice.
The generation and maintenance of SV/PLAP/+19 transgenic mice have been detailed elsewhere (20, 51). The transgenic reporter consisted of the following fragments: the simian virus 40 minimal promoter, the second rabbit β-globin intron, the human placental alkaline phosphatase (hPLAP) gene and simian virus 40 polyadenylation sequence and the SCL +19 core enhancer (a 644-bp fragment of mouse genomic DNA located 19 kb downstream of SCL promoter 1a). Nontransgenic littermates were analyzed as controls.
Histology.
Histochemical detection of PLAP in tissue sections was performed as previously described (51). For adult skeletal sections, bones were fixed in neutral buffered formalin for 24 h and decalcified for 48 h at 4°C in 0.5 M EDTA, pH 7.0, supplemented with 10% glycerol, 5 mM beta-mercaptoethanol, and 1 tablet of protease inhibitor cocktail (Roche). The decalcified bones were processed into paraffin wax and tissue sections were incubated in preheated phosphate-buffered saline at 75°C for 35 min to inactivate endogenous phosphatases, then stained with 5-bromo-4-chloro-3-indolylphosphate (BCIP)-nitroblue tetrazolium (NBT) (Roche, Basel) and counterstained with brazilin (Anachem, Bedfordshire) as previously described (51). For frozen sections, E16.5 embryos were bisected and fixed first in 2% paraformaldehyde for 4 h on ice and then in 30% sucrose-4% paraformaldehyde overnight at 4°C and embedded in Tissue-Tek OCT (Sakura, Torrance, CA). The tissue sections were heat treated, stained with BCIP-NBT, and counterstained with brazilin as described above.
For in situ RNA hybridization, SCL sense and antisense digoxigenin-labeled RNA probes were hybridized using the Ventana Discovery platform according to the manufacturer (Tucson, Arizona). A 6-hour, 60°C hybridization was performed, followed by three washes in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 75°C. Immunohistochemical staining for SCL was performed on mouse sections with a rabbit polyclonal anti-mouse SCL antibody (24) and on human bone marrow trephine sections with the anti-human SCL antibody 2TL242 (39) following protocols detailed elsewhere (8, 24). Immunohistochemical staining of hPLAP was performed with a rabbit anti-human PLAP primary antibody (Serotec, AHP 537) followed by a biotinylated sheep anti-rabbit secondary antibody (Serotec, 2AB02B), detected with horseradish peroxidase-extravidin (Sigma) and visualized with 3-amino-9-ethylcarbazole according to the manufacturer (Serotec, BUF019A).
For endothelial and vascular smooth muscle detection, anti-CD31 (BD Biosciences) and anti-smooth muscle α-actin (Sigma) primary antibodies were used with appropriate biotinylated secondary antibodies followed by extravidin-horseradish peroxidase (Sigma) conjugation and visualized with diaminobenzidine. Immunohistochemical staining of collagen types I and II to detect osteoblasts and chondrocytes, respectively, was performed with rabbit anticollagen primary antibodies (Chemicon) and visualized using a biotinylated goat anti-rabbit secondary antibody followed by peroxidase-conjugated avidin with NovaRED as a substrate (9). The sections were counterstained with Harris hematoxylin (Sigma). As controls, specimens were also incubated with isotype rabbit immunoglobulin G (Vector Laboratories) as the primary antibody and visualized as described above.
Cell culture and in vitro osteogenic differentiation.
The human bone cell lines MG63 and SAOS2 were maintained in Dulbecco's modified Eagle's medium (GIBCO), the murine osteocytic cell line MLO-Y4 in alpha minimal essential medium (αMEM; GIBCO), and the murine early myeloid progenitor line 416B in RPMI medium, each supplemented with 10% fetal bovine serum and 1:100 penicillin-streptomycin. The murine preosteoblastic cell line MC3T3-E1 (subclone 4) was maintained in αMEM without ascorbic acid, with ribonucleosides, deoxyribonucleosides, 2 mM l-glutamine, 1 mM sodium pyruvate supplemented with 10% fetal bovine serum, and 1:100 penicillin-streptomycin. SCL−/− and wild-type J1 murine ES cells were maintained in Dulbecco's modified Eagle's medium (GIBCO) with standard supplements (ES medium) including 100 U/ml leukemia inhibitory factor to inhibit differentiation. For osteogenic differentiation of ES cells, embryoid bodies (EBs) were generated using the hanging-drop technique, treated with 100 nM all-trans retinoic acid for 3 days, and cultured in ES medium (without leukemia inhibitory factor) supplemented with 50 μg/ml ascorbic acid, 10 mM β-glycerophosphate, and 10 ng/ml BMP-2 (38). MC3T3-E1 cells were differentiated with ascorbic acid and β-glycerophosphate at the same concentrations. Alizarin red staining and assessment of cellular alkaline phosphatase were performed as described elsewhere (2).
Stable transfection and luciferase reporter assay.
We electroporated 10 × 106 MC3T3 and 416B cells growing in log phase with 1 μg of linearized pGK Neo and 10 μg of either linearized SV/luc or SV/luc/+19. Resistant clones were selected at 24 h with neomycin (500 μg/ml for MC3T3 and 750 μg/ml for 416B) and luciferase activity was assayed at about day 10 as previously described (19).
Reverse transcription-PCR (RT-PCR).
RNA was extracted using the RNeasy kit (QIAGEN) and reverse transcribed following a standard protocol, using 2 μg of RNA per sample, random hexamer primers (Amersham), and Moloney murine leukemia virus reverse transcriptase (Invitrogen). The SCL (5′-TATGAGATGGAGATTTCTGATG-3′; 5′-GCTCCTCTGTGTAACTGTCC-3′), hypoxanthine-guanine phosphoribosyltransferase (HPRT). (5′-CCAGCAAGCTTGCAACCTTAACCA-3′; 5′-GTAATGATCAGTCAACGGGGGAC-3′), Ets1 (5′-CTACGGTATCGAGCATGCTCAGTG-3′; 5′-AAGGTGTCTGTCTGGAGAGGGTCC-3′), Ets2 (5′-ACTCTCACCTCAACGCGGTTCCTC-3′; 5′-GGAAGTCCTGGCTGATGGAACAGT-3′), Fli-1 (5′-CCAGAACATGGATGGCAAGGA-3′; 5′-CCCGGAATCTGATAAGGATCTGGC-3′), Elf-1 (5′-ACAGTGCCACTCACAACGGT-3′; 5′-CGCTCCATTGCAAATGGACTG-3′), Erg (5′-CAGTATATCCCGAAGCTACGC-3′; 5′-TTTGGACTGAGGGGTGAGGTGGCT-3′), and GATA2 (5′-CGGAATTCGACACACCACCCGATACCCACCTAT-3′; 5′-CGGAATTCGCCTACGCCATGGCAGTCACCATGCT-3′) transcripts were amplified by PCR as previously described.
SCL, Lyl1, CBFA1, and osteocalcin expression relative to β-actin was quantified using SYBR green real-time PCR (Applied Biosystems 7700) with the following primer sets: SCL (5′-CATGTTCACCAACAACAACCG-3′; 5′GGTGTGAGGACCATCAGAAATCTC-3′), Lyl1 (5′-AGATGAGGAAACGCCCTGTA-3′; 5′AGCCACTGCAAGTAGCCTGT-3′), CBFA1 (5′-GGACGAGGCAAGAGTTTCAC-3′; 5′-GGACCGTCCACTGTCACTTT-3′), osteocalcin (5′-CCATCTTTCTGCTCACTCTGC-3′; 5′-TGGACATGAAGGCTTTGTCA-3′), and β-actin (5′-TCCTGGCCTCACTGTCCAC-3′; 5′-GTCCGCCTAGAAGCACTTGC-3′). Details of transcript amplification and quantification are listed at http://hscl.cimr.cam.ac.uk/supplementary_pimanda05.html.
Quantitative analysis.
Chromatin immunoprecipitation assays were performed as detailed elsewhere (57). Briefly, 416B and MC3T3-E1 cells were treated with 0.4% formaldehyde and the cross-linked chromatin was retrieved by nuclear isolation and lysis. The chromatin was sonicated to yield an average fragment size of ∼500 bp, precleared with rabbit serum, and immunoprecipitated with antibodies to Ets1 (sc-350x), Ets2 (sc-351x), Fli-1 (sc-356x), Elf-1 (sc-631x), Erg (sc-354x) and GATA2 (sc-9008x) from Santa Cruz Biotechnology to recover the DNA-bound transcription factors and an anti-acetylated histone 3 antibody (06-599) from Upstate (Lake Placid, N.Y.) to recover acetylated histones.
Enrichment was measured by real-time PCR using SYBR green (Stratagene) as previously described (57). The levels of enrichment were normalized to that obtained with a control rabbit antibody and were calculated as the fold increase over that measured at the promoter region of α-fetoprotein, a gene not expressed in blood or bone, which was used as a control. The following target regions were amplified (forward and reverse primer sets, designed using Beacon Designer 4.0 software [Premier Biosoft International]): α-fetoprotein promoter region (5′-TGTTTGCTCACTGAAGGTTACTAG-3′; 5′-AGTGCTGGAAGTGGGATGTTTC-3′) and SCL +19 region (5′-CCATACTCTTGCCAAGGCTACC-3′; 5′-AGCAGTCCTACATGGGCCTAAA-3′).
RESULTS
SCL is expressed in the developing and adult skeleton.
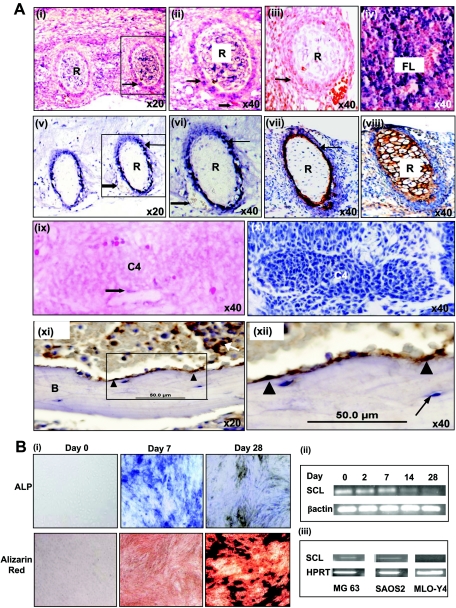
To examine SCL expression in the developing skeleton, we performed in situ RNA hybridization and immunohistochemistry on E14.5 mouse embryos (Fig. 1A). At E14.5, SCL RNA was expressed in cells within the perichondrium (Fig. 1A, compare panels i and ii with panel iii) and, as previously described, in the fetal liver (Fig. 1A, iv) and brain (data not shown). SCL protein was also detected in cells within the perichondrium (Fig. 1A, v and vi, arrow). SCL RNA and protein were also detected in endothelial cells (block arrows in Fig. 1A, ii, v, and vi). The expression of SCL protein corresponds with the specification of osteoblasts in the perichondrium (Fig. 1A, compare arrows in panels vi and vii) and not chondrocytes in the centrum (Fig. 1A, compare panels vi and viii). At E11.5, SCL RNA is expressed in endothelium (block arrow in Fig. 1A, ix) but not in bone primordia (C4) and corresponds with the absence of collagen 1-positive osteoblasts (no brown-stained cells in Fig. 1A, x).
FIG. 1.
(A) SCL is expressed in bone. (i) Section through ribs of an E14.5 embryo hybridized with an SCL antisense probe. SCL RNA is expressed in the perichondrium (arrow) of ribs (R). (ii) Magnified view of boxed area in panel i showing RNA expression in the perichondrium (arrow) and endothelium (block arrow) of blood vessels. (iii) Tissue section corresponding to panel i, probed with a sense SCL probe as a negative control. (iv) Section through fetal liver (FL) showing SCL expression in blood progenitors. (v and vi) Immunohistochemical studies of SCL expression in E14.5 mouse embryos with SCL-positive cells stained blue-black. (v) Ribs in cross section showing expression of SCL in cells in the perichondrium (arrow) and endothelial cells (block arrow). (vi) Magnified view of boxed area in panel v. (vii and viii) Immunohistochemical staining of osteoblasts and chondrocytes in rib sections of E14.5 mouse embryos. (vii) Osteoblasts (detected using a collagen 1 antibody and stained brown) are seen predominantly in the perichondrium (arrow). (viii) Chondrocytes (detected using a collagen 2 antibody and stained brown) fill the cartilaginous template. (ix) Section through the centrum of the C4 vertebra in an E11.5 mouse embryo hybridized with an SCL antisense probe. SCL RNA is expressed in endothelium (block arrow) but not in the vertebral template. (x) An E11.5 section corresponding to panel ix, stained for osteoblasts using a collagen 1 antibody. Collagen 1-expressing osteoblasts are also not present at this stage. (xi and xii) Immunohistochemical studies of SCL expression in adult human bone marrow with SCL-positive cells stained brown. (xii) SCL was expressed by osteoblasts (arrowhead) lining the endosteal surface of bone (B) and a subset of hematopoietic cells (arrow). (xiii) Magnified view of the boxed area in panel xi showing SCL expression in osteoblasts (arrowhead) but not in osteocytes (arrow). (B) SCL expression falls with bone differentiation. (i) Osteogenic differentiation of the preosteoblast cell line MC3T3-E1. Cells cultured in bone differentiation medium were stained at days 0, 7, and 28 for alkaline phosphatase activity (ALP, upper panel) and alizarin red staining (lower panel). Alkaline phosphatase activity (intense blue) was prominent at day 7 and decreased with the acquisition of a mature osteocytic phenotype at day 28. There was a corresponding increase in the degree of matrix mineralization as assessed by alizarin red staining (intense red) from days 0 to 28. (ii) SCL expression in MC3T3 cells was assessed by RT-PCR at various time points and decreased with bone differentiation. (iii) SCL expression in the osteoblastic cell lines MG63 and SAOS2 and the osteocytic cell line MLO-Y4 was assessed by RT-PCR. SCL was expressed in the osteoblastic cell lines but was absent in MLO-Y4 cells, which have the phenotype of terminally differentiated osteocytes.
To examine SCL expression in adult bone, we performed immunohistochemistry on human bone marrow trephines (Fig. 1A, xi to xii). SCL protein was expressed in osteoblasts lining the endosteal surface of bone (Fig. 1A, xi and xii, arrowhead) but not in mature osteocytes (Fig. 1A, xii, arrow), suggesting that SCL protein levels fall during osteocyte differentiation.
SCL expression falls with osteogenic differentiation of MC3T3-E1 cells.
The murine preosteoblast cell line MC3T3-E1 differentiates along the osteoblast lineage, expressing markers in a manner that mimics primary cultures of osteoblasts (55). As shown in Fig. 1B, i, differentiation of MC3T3-E1 cells results in staining for alkaline phosphatase (a marker of osteoblasts) by day 7, followed by staining with alizarin red, a marker of matrix mineralization, by day 28. MC3T3-E1 cells express SCL RNA in the undifferentiated state and the levels fall with bone differentiation (Fig. 1B, ii). Consistent with these results SCL transcripts were also detected in the immature osteoblastic cell lines MG63 and SAOS2 but were absent in MLO-Y4 cells, which have a mature phenotype (26) (Fig. 1B, iii).
SCL is not required for the osteogenic differentiation of embryonic stem cells in vitro.
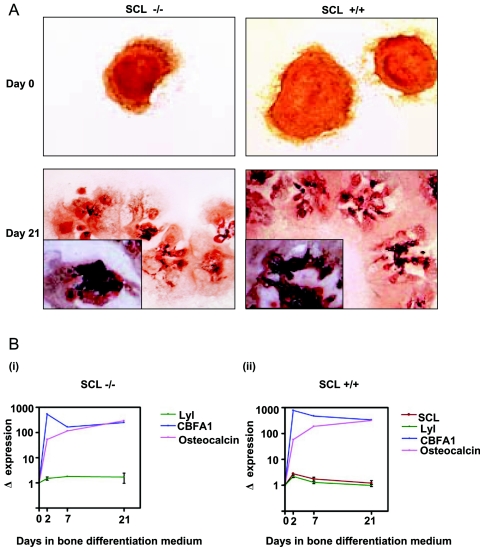
SCL−/− mice die in utero at E9.5 (42, 49) around the time when precartilaginous mesenchymal condensations first appear (58). To examine the role of SCL in bone differentiation, embryoid bodies formed from SCL+/+ and SCL−/− ES cells were pretreated with all-trans-retinoic acid and cultured in bone differentiation medium for 3 weeks. There was no difference in the mineralization of SCL−/− and SCL+/+ EBs assessed by alizarin red staining (Fig. 2A, compare panel on left with panel on right). There was also no difference in the gene expression profiles of osteocalcin, a matrix-associated protein produced by osteoblasts (10), or the transcription factor CBFA1, an early determinant of bone specification (Fig. 2B, compare panels i and ii).
FIG. 2.
(A) SCL−/− embryonic stem cells cultured in bone differentiation medium produce a mineralized matrix. EBs generated from SCL−/− ES cells were stained with alizarin red before and after culture in bone differentiation medium. The degree of matrix mineralization (black stain in insets) at day 21 was similar in EBs generated from either SCL−/− ES cells or SCL+/+ ES cells. (B) Osteogenic markers are up-regulated during differentiation of SCL−/− and SCL+/+ embryonic stem cells in RT-PCR analysis of ES cells undergoing bone differentiation. (i) SCL−/− EBs. (ii) SCL+/+ EBs.
We considered the possibility that Lyl1, an SCL paralogue, might compensate for the absence of SCL; however, the levels of Lyl1 expression were similar in SCL−/− and SCL+/+ EBs (Fig. 2B). These data suggest that SCL is not required to establish the bone program under the osteoinductive conditions used in this in vitro differentiation assay (38). There is a small relative increase in SCL expression in SCL+/+ EBs (Fig. 2B, ii) following the addition of bone differentiation medium at day 0 and a progressive reduction thereafter. The fall in SCL expression during bone differentiation of ES cells is consistent with our observation in MC3T3-E1 cells (Fig. 1B, ii) and the absence of SCL in mature osteocytes (Fig. 1A, xii).
SCL +19 stem cell enhancer directs reporter gene expression to the adult and developing skeleton.
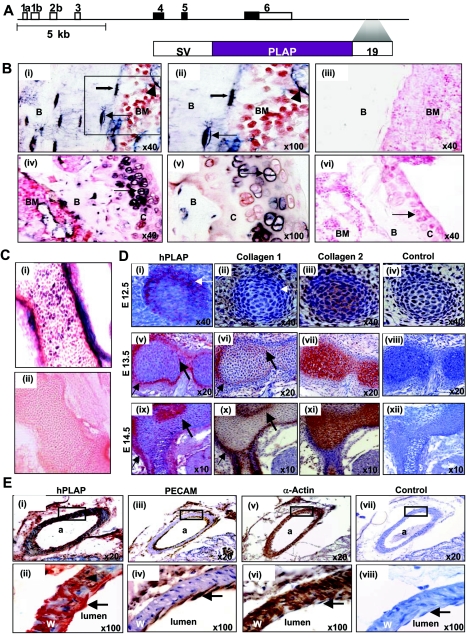
A 5.5-kb fragment containing the SCL +19 enhancer directs expression to hemangioblasts and to their hematopoietic and endothelial progeny during development (45, 46). We have also shown that a 644-bp SCL +19 core enhancer directs expression to adult and fetal HSCs, mast cells, megakaryocytes, and endothelium (51). Given recent reports that blood and bone can be regenerated from a common precursor in adult bone marrow (9, 36), we investigated whether the SCL +19 core enhancer also directs the reporter gene for human placental alkaline phosphatase (see Fig. 3A) expression to bone by surveying the skeletons of developing and adult mice.
FIG. 3.
(A) Schematic diagram of the mouse SCL locus and the SV/PLAP/+19 transgenic reporter. SV, simian virus 40 minimal promoter; hPLAP, human placental alkaline phosphatase; 19, a 644-bp fragment of the mouse SCL locus, +19 kb from promoter 1a. (B) The SCL +19 core enhancer directs expression to the adult skeleton. Histological analysis of PLAP expression in mice carrying the SV/PLAP/+19 transgene (line 1772). (i) Longitudinal section of a femur from an adult transgenic mouse showing expression of PLAP in osteocytes (arrow), osteoblasts (solid arrow), and endothelium (arrowhead). (ii) Magnified view of the boxed area in panel i. (iii) There was no PLAP staining in the femur of wild-type littermate controls. (iv) Longitudinal section of the distal femur showing PLAP expression in chondrocytes (arrow) in the articular cartilage of adult transgenic mice. (v) Magnified view of articular chondrocytes from panel iv. (vi) There was no staining in articular chondrocytes (arrow) of littermate controls. B, bone; BM, bone marrow; C, cartilage. (C) The SCL +19 core enhancer directs expression to the developing skeleton. (i and ii) Cryosections of E16.5 embryos. (i) A longitudinal section of vertebrae from a transgenic embryo showing PLAP-positive cells (blue-purple) in the perichondrium and within ossifying cartilage. (ii) Vertebrae of E16.5 wild-type embryos do not stain for PLAP. (D) The SCL +19 core enhancer targets osteoblasts but not chondrocytes in the developing skeleton. (i to iv) Immunohistochemistry on serial sections of E12.5 embryos. (i) Section of a cervical vertebral body showing expression of the PLAP transgene (detected by immunohistochemistry and stained red) in cells within the perichondrium (arrow). (ii) Osteoblasts (detected using a collagen 1 antibody and stained brown) are not seen in the perichondrium (arrow) at this stage. (iii) Chondrocytes (detected using a collagen 2 antibody and stained brown) fill the vertebral anlage. (iv) The isotype control shows no staining. (v to viii) Immunohistochemistry on serial sections of E13.5 embryos. (v) Section through a cervical vertebral body showing PLAP-positive cells (red) in the perichondrium (arrow) and within the bone anlage (solid arrow). (vi) Osteoblasts (stained brown) are present in the perichondrium and correspond to the distribution of PLAP-positive cells (arrow and solid arrow) in panel v. (vii) Chondrocytes (stained brown) fill the cartilaginous core of the vertebral body. (viii) The isotype control shows no staining. (ix to xii) Immunohistochemistry on serial sections of E14.5 embryos, with sections corresponding to those of E13.5 embryos in panels v to viii. (E) The SCL +19 core enhancer also directs expression to the endothelium and vascular smooth muscle. (i) Section of a kidney from an adult transgenic mouse showing expression of the PLAP transgene (detected by immunohistochemistry and stained red) in the endothelium and smooth muscle wall of a branch of the renal artery (a). (ii) Magnified view of the boxed area in panel i showing PLAP expression in endothelial cells (arrow) and smooth muscle cells (arrowhead). W, wall. (iii) An adjacent section stained with an anti-CD31 (PECAM) antibody showing staining (brown) of the endothelium but not smooth muscle. (iv) Magnified view of the boxed area in panel iii showing endothelial staining (arrow). (v) An adjacent section stained with an anti-smooth muscle actin antibody showing staining (brown) of the smooth muscle wall. (vi) Magnified view of the boxed area in panel v showing staining of the wall but no staining in the endothelium (arrow). (vii) An adjacent section used as a negative (isotype) control. (viii) Magnified view of the boxed area in panel vii showing no staining in either the vessel wall or the endothelial lining.
In adult mice the +19 core enhancer directed PLAP expression to osteocytes and to osteoblasts lining the bony trabeculae of long bones (Fig. 3B, compare panels i and ii with panel iii). PLAP expression was also observed in the cranial vault, facial bones, and axial skeleton (data not shown) and in a fraction of chondrocytes within the articular cartilage (Fig. 3B, compare panels iv and v with panel vi). During development, PLAP was expressed in cells within the cartilaginous template of future bone and in the perichondrium (Fig. 3C, compare panels i and ii). This pattern of staining was seen in two independent transgenic lines, 1772 (data shown) and 1791 (data not shown).
During mouse embryonic development, cells within mesenchymal condensations begin to differentiate into chondrocytes around E11 and the first osteoblasts appear in the perichondrium around E13 (25, 58). In limb bud mesenchyme, Sox 9, a transcription factor that activates Col2a1, a chondrocyte-specific marker gene, can be detected at E10 (1), whereas Runx2 and Osterix, transcription factors that activate Col1a1, an osteoblast-specific marker gene, appear later, at E11 and E13, respectively (1). During development, PLAP-positive cells appear in the perichondrium of cervical vertebrae at ∼E12.5 (Fig. 3D, compare panels i and iv). At later time points PLAP-positive cells appear deeper within the perichondrium and within the cartilaginous core (Fig. 3D, compare panels v and viii and compare panels ix and xii). The appearance of PLAP-positive cells in the perichondrium precedes by approximately a day the appearance of collagen 1-positive osteoblasts in the perichondrium (Fig. 3D, compare panels i and ii). The distribution of PLAP-positive cells (arrows in Fig. 3D, v and ix) matches the distribution of collagen 1-positive osteoblasts (stained brown in Fig. 3D, vi and x), which appear to migrate from the perichondrium into the cartilaginous core (stained brown in Fig. 3D, vii and xi) during endochondral ossification.
SCL +19 stem cell enhancer also directs reporter gene expression to vascular smooth muscle.
Together with recent reports that a common progenitor in adult bone marrow can give rise to both blood and bone (9, 36), our results suggest that the SCL +19 stem cell enhancer is activated in a mesodermal progenitor capable of giving rise to blood, endothelium and bone. Flk1, one of the receptors for vascular endothelial growth factor, is a marker for lateral plate mesoderm and the earliest differentiation marker for endothelial and blood cells. Flk1+ cells have been shown by single-cell in vitro culture and chicken/mouse in vivo chimeric assays to contain common progenitors for endothelial and mural cells. Furthermore, in ES cell differentiation assays, BL-CFCs are single cells with trilineage (blood, endothelium, and smooth muscle) potential (5, 28). We hypothesized therefore that an element which targets a mesodermal progenitor with potential to give rise to blood, endothelium, and bone might also target vascular smooth muscle cells.
As shown in a series of kidney sections in Fig. 3E, the +19 enhancer directs PLAP expression (red precipitate in panels i and ii) to both the endothelium (arrow in panel ii) and smooth muscle of a branch of the renal artery (arrowhead in panel ii). Adjacent sections were stained with anti-CD31 (platelet endothelial cell adhesion molecule) as a positive control for the endothelium (brown precipitate in Fig. 3E, iii, and arrow in panel iv) and anti-smooth muscle α-actin for mural cells (brown precipitate in Fig. 3E, v and vi). The isotype controls in Fig. 3E, vii and viii, were negative.
SCL +19 stem cell enhancer is active in MC3T3-E1 cells.
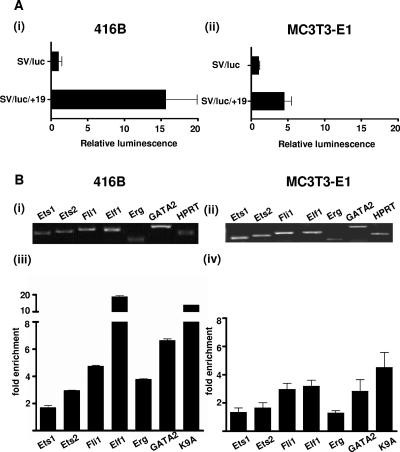
To assess the in vitro transcriptional activity of the SCL +19 core enhancer, we subcloned the 644-bp fragment into a luciferase reporter plasmid (SV/luc/+19) and tested the construct in stable transfection assays using the MC3T3-E1 preosteoblastic cell line (Fig. 4A, ii). The construct with the +19 enhancer had approximately fivefold higher activity than a control vector containing the simian virus minimal promoter (SV/luc). The 416B early myeloid progenitor cell line expresses approximately 100-fold more SCL by quantitative RT-PCR than MC3T3-E1 cells (results not shown). The SV/luc/+19 construct had approximately 15-fold higher activity than SV/luc in 416B cells (Fig. 4A, i) which corresponds to ∼3-fold higher activity than in MC3T3-E1 cells (Fig. 4A, compare panels i and ii).
FIG. 4.
(A) SCL +19 core enhancer is active in both blood and bone cell lines. (i) Stable transfection assay in 416B cells, an SCL-expressing cell line derived from a multipotent hematopoietic progenitor. (ii) Stable transfection assay in MC3T3-E1 cells, a preosteoblastic cell line. The luciferase activities are given as increase over the activity of the basic (SV/luc) vector. Each bar is the mean of the relative luciferase activity from at least two experiments performed in triplicate ± standard deviation. (B) Fli1, Elf1, and GATA2 bind the SCL +19 core enhancer in both blood and bone cell lines. The expression profiles of selected Ets and GATA factors were assessed by RT-PCR in (i) 416B and (ii) MC3T3-E1 cell lines. Chromatin immunoprecipitation assays were performed with anti-Ets1, -Ets2, -Fli1, -Elf1, -GATA2, -acetylated histone H3 (K9A), and control immunoglobulin G antibodies in the (iii) 416B and (iv) MC3T3-E1 cell lines. The level of enrichment with each antibody was normalized to the levels obtained with control immunoglobulin G and plotted as the increase over the level of enrichment at a control region (the promoter region of the α-fetoprotein gene). Histone acetylation status (K9A) was used as a surrogate marker of chromatin accessibility. The SCL +19 region is accessible in both cell lines and is bound by Fli1, Elf1, Erg, and GATA2 in 416B cells and Fli1, Elf1, and GATA2 in MC3T3-E1 cells. Histone acetylation and transcription factor binding at the SCL +19 region are lower in MC3T3-E1 cells than in 416B cells, in line with the lower expression of endogenous SCL in the former.
Fli1, Elf1, and GATA2 occupy the SCL +19 enhancer in MC3T3-E1 cells in vivo.
We have previously shown that the SCL +19 enhancer in 416B cells is bound in vivo by Fli1, Elf1, Ets2, Erg, and GATA2 (20, 40). Interestingly, 416B hematopoietic cells and MC3T3-E1 osteogenic cells share a common expression profile of selected Ets and GATA transcription factors (Fig. 4B, i and ii). To determine whether the expression of SCL and the in vitro activity of the +19 enhancer in MC3T3-E1 osteogenic cells reflect a transcriptional process that is common with 416B hematopoietic cells, we studied the endogenous SCL +19 region in MC3T3-E1 cells.
Histone acetylation can increase transcription factor access to chromatin (32). To determine whether the in vitro activity of the +19 enhancer in MC3T3-E1 cells correlates with histone modifications thought to signify active chromatin at the endogenous +19 region, we quantified the degree of histone acetylation at this region by a quantitative chromatin immunoprecipitation assay (see result for K9A in Fig. 4B, iv). The results demonstrated that histones in the +19 region were acetylated ∼5-fold higher than the promoter region of α-fetoprotein, a gene that is not expressed in blood or bone. In line with our previous observations with 416B cells, the histones in the +19 region were also highly acetylated (see result for K9A in Fig. 4B, iii).
To determine whether the transcription factors recruited to the +19 core enhancer in MC3T3-E1 cells were similar to those recruited to this region in 416B cells, we performed quantitative chromatin immunoprecipitation assays with antibodies to Ets1, Ets2, Fli1, Elf1, Erg, and GATA2 (Fig. 4B, compare panels iii and iv). Immunoprecipitated chromatin samples were analyzed by real-time PCR. The levels of enrichment were normalized to that obtained with a control antibody and are represented as the fold increase over that measured at a control region (the promoter region of the α-fetoprotein gene). As shown in Fig. 4B, iii and iv, there was specific 3- to 20-fold enrichment of Ets2, Fli1, Elf1, Erg, and GATA2 at the +19 enhancer in 416B cells and ∼3-fold enrichment of Fli1, Elf1, and GATA2 in MC3T3-E1 cells. Therefore, these cells utilize common transcription factors and cis regulatory elements to drive SCL transcription. The lower enrichments in MC3T3-E1 cells appear to correlate with lower expression of transcription factors in this cell line (Fig. 4B).
DISCUSSION
SCL is expressed in blood progenitors and endothelium and is essential for the normal development of these tissues. We report that SCL is also expressed in bone primordia during embryonic development and in adult osteoblasts. However, in contrast to its role in hematopoiesis, SCL is not required for bone specification of ES cells. We also show that the SCL +19 core enhancer, which directs expression to blood and vascular cells, is also active in osteoblasts, osteocytes, and articular chondrocytes and is bound in vivo by the Fli-1, Elf1, and GATA2 transcription factors in both osteoblastic and hematopoietic cell lines.
Our results demonstrate that SCL RNA and protein are expressed in developing and adult bone but that SCL is not required for bone differentiation of ES cells in vitro. However, during embryogenesis, bone develops from both mesoderm- and neural crest-derived mesenchyme and retinoic acid treatment of ES cells to induce bone differentiation in vitro is reported to promote mesenchyme development via the neural crest route while suppressing mesodermal differentiation (27). It is important to emphasize therefore that our data do not exclude a role for SCL in bone development in vivo. In this regard it is salient to note that, although SCL−/− ES cells can generate endothelial cells in vitro (43), vascular remodeling in SCL−/− embryos is abnormal (56).
Assessing the role of SCL in bone development in vivo is not straightforward because SCL null mice die at E9.5 (42, 49), before bone development is established. Conditional SCL knockout mice survive in excess of 6 months after loss of SCL expression with only mild anemia and thrombocytopenia (21). To our knowledge, bone development in these mice has not been assessed, and any potential abnormality would likely be missed unless the SCL alleles were deleted during prenatal or early postnatal growth, when bone turnover is high. Nonetheless, our results raise the possibility that low-level SCL expression in bone progenitors may reflect a requirement for SCL in precursors of bone progenitors.
HSCs have been reported to function as osteoblast precursors (36) and are known to express SCL. Similarly, HSCs and osteoblasts, both of which express SCL, are reported to originate from a common marrow precursor after bone marrow transplantation (9). High-level SCL expression in BL-CFCs favors hematopoietic and endothelial differentiation over smooth muscle differentiation, whereas low-level SCL promoted smooth muscle differentiation (13). It is possible that a fall in SCL expression is needed for osteoblastic differentiation in a manner analogous to vascular smooth muscle differentiation.
Bone development during embryogenesis is complex. Lineage labeling studies in chick (31) and more recently in mice (33) show that the cranial vault (except the occiput) and parts of the neck and shoulder are formed by mesenchymal stem cells (MSCs) of neural crest origin, whereas the limb and axial skeleton are formed by MSCs of mesodermal origin (33, 50). Our data demonstrate that the SCL +19 core enhancer targets osteoprogenitors in bones, which arise from both mesoderm- and neural crest-derived MSCs.
The activity of the enhancer in bone could be due to activity in MSCs, which subsequently contribute to bone formation. This however is unlikely for the following reasons. First, activity in MSCs would have resulted in most if not all cells in bone primordia and other tissues derived from MSCs, i.e., fat, cartilage, and skeletal muscle, expressing the transgene. This clearly is not the case, with only a subpopulation of cells in bone primordia expressing the transgene and the absence of activity in the cartilage anlagen prior to the onset of ossification at E13.5. Second, MSCs cultured from bone marrow, which have bone differentiating potential, do not express SCL (unpublished observations). It is however possible that the transgene targets subpopulations of MSCs and that SCL is expressed during osteoblastic differentiation of a common osteochondroprogenitor precursor from which most osteoblasts arise during embryogenesis (1). In our survey of E11.5 to E14.5 mouse embryos, PLAP-positive cells were first observed in the perichondrium at ∼E12, which precedes the appearance of osteoblasts at ∼E13. The movement of PLAP-positive cells from the perichondrium into the centrum at later time points mirrors the invasion of osteoblasts from the perichondrium during endochondral ossification.
Alternatively, osteoprogenitors with SCL +19 enhancer activity could be derived from circulating HSCs or a common progenitor. In this scenario we envisage HSCs or a common progenitor invading bone primordia during cartilage vascularization and differentiating into osteoblasts (with concomitant loss of hematopoietic potential) in response to local cues. In favor of this model, HSCs which are established in the aorta-gonad-mesonephros at ∼E10.5 (reviewed in reference 44) and amplified in the fetal liver are present in the circulation around the time bone is ossified at ∼E13 (25); HSCs or a common marrow precursor in adult bone marrow have been shown to contribute to blood and bone formation in vivo (9, 36). The SCL +19 enhancer targets HSCs (46), which as a circulating cell population would not be restricted by embryonic boundaries, explaining the presence of PLAP-positive cells in bones derived from the neural crest as well as mesoderm.
The other cell population in intimate contact with the endosteal bone surface is osteoclasts, which, although now widely regarded to be derived from hematopoietic stem cells (14), were long considered to be skeletal in origin (reviewed in reference 22). It is important to emphasize however that a contribution to osteoblasts in the perichondrium from a circulating pool of HSCs or a common progenitor during embryonic bone development is likely to be only an adjunct to bone development from MSCs in the perichondrium (6, 11).
The expression of the SV/PLAP/+19 transgene in mature osteocytes contrasts with a fall in endogenous SCL in MC3T3-E1 cells during bone differentiation and its absence in the phenotypically mature MLO-Y4 cells in vitro and in osteocytes in vivo. There are several potential explanations for this apparent discrepancy. PLAP expression in histological sections is not quantitative, PLAP and SCL may differ in the stability of their transcripts or proteins, or the transgene may lack cis elements required for inhibition of endogenous SCL expression in mature osteocytes. The expression of the SV/PLAP/+19 transgene in a subpopulation of chondrocytes in the articular cartilage of adult mice is interesting in view of the lack of expression in cartilage anlagen during development. It is salient to note that articular chondrocytes, which arise during joint development, are a biologically distinct population of chondrocytes (reviewed in reference 25).
The Ets transcription factors Fli-1 and Elf-1, which are enriched on the SCL +19 enhancer in blood and bone cell lines, are also expressed in bone primordia (reviewed in reference 41). Although specific roles for Fli-1 and Elf-1 in osteogenesis have not been described, Ets1 binds the osteopontin promoter and acts synergistically with the critical osteogenic transcription factor Cbfa1 in regulating the expression of osteopontin, an extracellular matrix protein secreted by osteoblasts (47). Furthermore, the promoter regions of osteocalcin, bone sialoprotein, osteonectin, and a number of other genes expressed in osteoblasts have Ets transcription factor binding sites (41), although their functional significance is unknown. GATA2, which was also enriched on the +19 enhancer in MC3T3-E1 cells, does not as yet have a known role in osteogenesis, although it is an important regulator of SCL in hematopoiesis (20) and down-regulation of its expression is required for preadipocyte-adipocyte transition (54).
The SCL +19 enhancer is proving to be a powerful tool for manipulating hematopoietic progenitors and stem cells (4, 12, 16, 35, 46), but the design and interpretation of such transgenic studies require a detailed understanding of enhancer activity in vivo. Our data suggest that the SCL +19 core enhancer targets a mesodermal precursor with the potential to differentiate into blood, endothelium, vascular smooth muscle, articular cartilage, and bone.
Acknowledgments
We thank Catherine Porcher and Stuart Orkin for the SCL−/− and wild-type J1 ES cells. The MLO-Y4 cells and the 2TL242 anti-hSCL antibody were gifts from Linda Bonewald and Karen Pulford, respectively. We are grateful for the assistance given us by Liz Delaney and Aileen Smith with ES cell culture, Sandie Piltz and Paula Braker with animal husbandry, Sharyn Bord with histology, Dongrong Chen with in situ hybridization, and Ian Donaldson for the web link.
This work was supported by the Leukemia Research Fund and the Wellcome Trust. J.E.P. is a C. J. Martin/R. G. Menzies Fellow of the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Akiyama, H., T. Kamitani, X. Yang, R. Kandyil, L. C. Bridgewater, M. Fellous, Y. Mori-Akiyama, and B. de Crombrugghe. 2005. The transcription factor Sox9 is degraded by the ubiquitin-proteasome system and stabilized by a mutation in a ubiquitin-target site. Matrix Biol. 23:499-505. [DOI] [PubMed] [Google Scholar]
- 2.Bodine, P. V., M. Trailsmith, and B. S. Komm. 1996. Development and characterization of a conditionally transformed adult human osteoblastic cell line. J. Bone Miner. Res. 11:806-819. [DOI] [PubMed] [Google Scholar]
- 3.Calvi, L. M., G. B. Adams, K. W. Weibrecht, J. M. Weber, D. P. Olson, M. C. Knight, R. P. Martin, E. Schipani, P. Divieti, F. R. Bringhurst, L. A. Milner, H. M. Kronenberg, and D. T. Scadden. 2003. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425:841-846. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C. Z., M. Li, D. de Graaf, S. Monti, B. Gottgens, M. J. Sanchez, E. S. Lander, T. R. Golub, A. R. Green, and H. F. Lodish. 2002. Identification of endoglin as a functional marker that defines long-term repopulating hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 99:15468-15473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, K., M. Kennedy, A. Kazarov, J. C. Papadimitriou, and G. Keller. 1998. A common precursor for hematopoietic and endothelial cells. Development 125:725-732. [DOI] [PubMed] [Google Scholar]
- 6.Colnot, C., C. Lu, D. Hu, and J. A. Helms. 2004. Distinguishing the contributions of the perichondrium, cartilage, and vascular endothelium to skeletal development. Dev. Biol. 269:55-69. [DOI] [PubMed] [Google Scholar]
- 7.Cumano, A., J. C. Ferraz, M. Klaine, J. P. Di Santo, and I. Godin. 2001. Intraembryonic, but not yolk sac hematopoietic precursors, isolated before circulation, provide long-term multilineage reconstitution. Immunity 15:477-485. [DOI] [PubMed] [Google Scholar]
- 8.Dekel, B., E. Hochman, M. J. Sanchez, N. Maharshak, N. Amariglio, A. R. Green, and S. Izraeli. 2004. Kidney, blood, and endothelium: developmental expression of stem cell leukemia during nephrogenesis. Kidney Int. 65:1162-1169. [DOI] [PubMed] [Google Scholar]
- 9.Dominici, M., C. Pritchard, J. E. Garlits, T. J. Hofmann, D. A. Persons, and E. M. Horwitz. 2004. Hematopoietic cells and osteoblasts are derived from a common marrow progenitor after bone marrow transplantation. Proc. Natl. Acad. Sci. USA 101:11761-11766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ducy, P., R. Zhang, V. Geoffroy, A. L. Ridall, and G. Karsenty. 1997. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89:747-754. [DOI] [PubMed] [Google Scholar]
- 11.Eames, B. F., L. de la Fuente, and J. A. Helms. 2003. Molecular ontogeny of the skeleton. Birth Defects Res. C 69:93-101. [DOI] [PubMed] [Google Scholar]
- 12.Eguchi, M., M. Eguchi-Ishimae, A. Green, T. Enver, and M. Greaves. 2005. Directing oncogenic fusion genes into stem cells via an SCL enhancer. Proc. Natl. Acad. Sci. USA 102:1133-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ema, M., P. Faloon, W. J. Zhang, M. Hirashima, T. Reid, W. L. Stanford, S. Orkin, K. Choi, and J. Rossant. 2003. Combinatorial effects of Flk1 and Tal1 on vascular and hematopoietic development in the mouse. Genes Dev. 17:380-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferron, M., and J. Vacher. 2005. Targeted expression of Cre recombinase in macrophages and osteoclasts in transgenic mice. Genesis 41:138-145. [DOI] [PubMed] [Google Scholar]
- 15.Gering, M., A. R. Rodaway, B. Gottgens, R. K. Patient, and A. R. Green. 1998. The SCL gene specifies haemangioblast development from early mesoderm. EMBO J. 17:4029-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gothert, J. R., S. E. Gustin, M. A. Hall, A. R. Green, B. Gottgens, D. J. Izon, and C. G. Begley. 2005. In vivo fate-tracing studies using the Scl stem cell enhancer: embryonic hematopoietic stem cells significantly contribute to adult hematopoiesis. Blood 105:2724-2732. [DOI] [PubMed] [Google Scholar]
- 17.Gottgens, B., L. M. Barton, J. G. Gilbert, A. J. Bench, M. J. Sanchez, S. Bahn, S. Mistry, D. Grafham, A. McMurray, M. Vaudin, E. Amaya, D. R. Bentley, A. R. Green, and A. M. Sinclair. 2000. Analysis of vertebrate SCL loci identifies conserved enhancers. Nat. Biotechnol. 18:181-186. [DOI] [PubMed] [Google Scholar]
- 18.Gottgens, B., C. Broccardo, M. J. Sanchez, S. Deveaux, G. Murphy, J. R. Gothert, E. Kotsopoulou, S. Kinston, L. Delaney, S. Piltz, L. M. Barton, K. Knezevic, W. N. Erber, C. G. Begley, J. Frampton, and A. R. Green. 2004. The SCL +18/19 stem cell enhancer is not required for hematopoiesis: identification of a 5′ bifunctional hematopoietic-endothelial enhancer bound by Fli-1 and Elf-1. Mol. Cell. Biol. 24:1870-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottgens, B., F. McLaughlin, E. O. Bockamp, J. L. Fordham, C. G. Begley, K. Kosmopoulos, A. G. Elefanty, and A. R. Green. 1997. Transcription of the SCL gene in erythroid and CD34 positive primitive myeloid cells is controlled by a complex network of lineage-restricted chromatin-dependent and chromatin-independent regulatory elements. Oncogene 15:2419-2428. [DOI] [PubMed] [Google Scholar]
- 20.Gottgens, B., A. Nastos, S. Kinston, S. Piltz, E. C. Delabesse, M. Stanley, M. J. Sanchez, A. Ciau-Uitz, R. Patient, and A. R. Green. 2002. Establishing the transcriptional programme for blood: the SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 21:3039-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall, M. A., N. J. Slater, C. G. Begley, J. M. Salmon, L. J. Van Stekelenburg, M. P. McCormack, S. M. Jane, and D. J. Curtis. 2005. Functional but abnormal adult erythropoiesis in the absence of the stem cell leukemia gene. Mol. Cell. Biol. 25:6355-6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanaoka, H. 1979. The origin of the osteoclast. Clin. Orthop. Relat. Res. 145:252-263. [PubMed] [Google Scholar]
- 23.Huber, T. L., V. Kouskoff, H. J. Fehling, J. Palis, and G. Keller. 2004. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature 432:625-630. [DOI] [PubMed] [Google Scholar]
- 24.Kallianpur, A. R., J. E. Jordan, and S. J. Brandt. 1994. The SCL/TAL-1 gene is expressed in progenitors of both the hematopoietic and vascular systems during embryogenesis. Blood 83:1200-1208. [PubMed] [Google Scholar]
- 25.Karsenty, G., and E. F. Wagner. 2002. Reaching a genetic and molecular understanding of skeletal development. Dev. Cell 2:389-406. [DOI] [PubMed] [Google Scholar]
- 26.Kato, Y., J. J. Windle, B. A. Koop, G. R. Mundy, and L. F. Bonewald. 1997. Establishment of an osteocyte-like cell line, MLO-Y4. J. Bone Miner. Res. 12:2014-2023. [DOI] [PubMed] [Google Scholar]
- 27.Kawaguchi, J., P. J. Mee, and A. G. Smith. 2005. Osteogenic and chondrogenic differentiation of embryonic stem cells in response to specific growth factors. Bone 36:758-769. [DOI] [PubMed] [Google Scholar]
- 28.Kennedy, M., M. Firpo, K. Choi, C. Wall, S. Robertson, N. Kabrun, and G. Keller. 1997. A common precursor for primitive erythropoiesis and definitive haematopoiesis. Nature 386:488-493. [DOI] [PubMed] [Google Scholar]
- 29.Kiel, M. J., O. H. Yilmaz, T. Iwashita, C. Terhorst, and S. J. Morrison. 2005. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121:1109-1121. [DOI] [PubMed] [Google Scholar]
- 30.Lecuyer, E., and T. Hoang. 2004. SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp. Hematol. 32:11-24. [DOI] [PubMed] [Google Scholar]
- 31.Le Douarin, N., and C. Kalcheim. 1999. The neural crest, 2nd ed. Cambridge University Press, Cambridge, United Kingdom.
- 32.Lee, D. Y., J. J. Hayes, D. Pruss, and A. P. Wolffe. 1993. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 72:73-84. [DOI] [PubMed] [Google Scholar]
- 33.Matsuoka, T., P. E. Ahlberg, N. Kessaris, P. Iannarelli, U. Dennehy, W. D. Richardson, A. P. McMahon, and G. Koentges. 2005. Neural crest origins of the neck and shoulder. Nature 436:347-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medvinsky, A., and E. Dzierzak. 1996. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell 86:897-906. [DOI] [PubMed] [Google Scholar]
- 35.Murphy, G. J., B. Gottgens, A. Vegiopoulos, M. J. Sanchez, A. D. Leavitt, S. P. Watson, A. R. Green, and J. Frampton. 2003. Manipulation of mouse hematopoietic progenitors by specific retroviral infection. J. Biol. Chem. 278:43556-43563. [DOI] [PubMed] [Google Scholar]
- 36.Olmsted-Davis, E. A., Z. Gugala, F. Camargo, F. H. Gannon, K. Jackson, K. A. Kienstra, H. D. Shine, R. W. Lindsey, K. K. Hirschi, M. A. Goodell, M. K. Brenner, and A. R. Davis. 2003. Primitive adult hematopoietic stem cells can function as osteoblast precursors. Proc. Natl. Acad. Sci. USA 100:15877-15882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olsen, B. R., A. M. Reginato, and W. Wang. 2000. Bone development. Annu. Rev. Cell Dev. Biol. 16:191-220. [DOI] [PubMed] [Google Scholar]
- 38.Phillips, B. W., N. Belmonte, C. Vernochet, G. Ailhaud, and C. Dani. 2001. Compactin enhances osteogenesis in murine embryonic stem cells. Biochem. Biophys. Res. Commun. 284:478-484. [DOI] [PubMed] [Google Scholar]
- 39.Pulford, K., N. Lecointe, K. Leroy-Viard, M. Jones, D. Mathieu-Mahul, and D. Y. Mason. 1995. Expression of TAL-1 proteins in human tissues. Blood 85:675-684. [PubMed] [Google Scholar]
- 40.Rainis, L., T. Toki, J. E. Pimanda, E. Rosenthal, K. Machol, S. Strehl, B. Gottgens, E. Ito, and S. Izraeli. 2005. The proto-oncogene ERG in megakaryoblastic leukemias. Cancer Res. 65:7596-7602. [DOI] [PubMed] [Google Scholar]
- 41.Raouf, A., and A. Seth. 2000. Ets transcription factors and targets in osteogenesis. Oncogene 19:6455-6463. [DOI] [PubMed] [Google Scholar]
- 42.Robb, L., I. Lyons, R. Li, L. Hartley, F. Kontgen, R. P. Harvey, D. Metcalf, and C. G. Begley. 1995. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc. Natl. Acad. Sci. USA 92:7075-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robertson, S. M., M. Kennedy, J. M. Shannon, and G. Keller. 2000. A transitional stage in the commitment of mesoderm to hematopoiesis requiring the transcription factor SCL/tal-1. Development 127:2447-2459. [DOI] [PubMed] [Google Scholar]
- 44.Robin, C., and E. Dzierzak. 2005. Hematopoietic stem cell enrichment from the AGM region of the mouse embryo. Methods Mol. Med. 105:257-272. [DOI] [PubMed] [Google Scholar]
- 45.Sanchez, M., B. Gottgens, A. M. Sinclair, M. Stanley, C. G. Begley, S. Hunter, and A. R. Green. 1999. An SCL 3′ enhancer targets developing endothelium together with embryonic and adult haematopoietic progenitors. Development 126:3891-3904. [DOI] [PubMed] [Google Scholar]
- 46.Sanchez, M. J., E. O. Bockamp, J. Miller, L. Gambardella, and A. R. Green. 2001. Selective rescue of early haematopoietic progenitors in Scl(-/-) mice by expressing Scl under the control of a stem cell enhancer. Development 128:4815-4827. [DOI] [PubMed] [Google Scholar]
- 47.Sato, M., E. Morii, T. Komori, H. Kawahata, M. Sugimoto, K. Terai, H. Shimizu, T. Yasui, H. Ogihara, N. Yasui, T. Ochi, Y. Kitamura, Y. Ito, and S. Nomura. 1998. Transcriptional regulation of osteopontin gene in vivo by PEBP2alphaA/CBFA1 and ETS1 in the skeletal tissues. Oncogene 17:1517-1525. [DOI] [PubMed] [Google Scholar]
- 48.Shalaby, F., J. Ho, W. L. Stanford, K. D. Fischer, A. C. Schuh, L. Schwartz, A. Bernstein, and J. Rossant. 1997. A requirement for Flk1 in primitive and definitive hematopoiesis and vasculogenesis. Cell 89:981-990. [DOI] [PubMed] [Google Scholar]
- 49.Shivdasani, R. A., E. L. Mayer, and S. H. Orkin. 1995. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature 373:432-434. [DOI] [PubMed] [Google Scholar]
- 50.Shubin, N., C. Tabin, and S. Carroll. 1997. Fossils, genes and the evolution of animal limbs. Nature 388:639-648. [DOI] [PubMed] [Google Scholar]
- 51.Silberstein, L., M. J. Sanchez, M. Socolovsky, Y. Liu, G. Hoffman, S. Kinston, S. Piltz, M. Bowen, L. Gambardella, A. R. Green, and B. Gottgens. 2005. Transgenic analysis of the stem cell leukemia +19 stem cell enhancer in adult and embryonic hematopoietic and endothelial cells. Stem Cells. 23:1378-1388. [DOI] [PubMed] [Google Scholar]
- 52.Sinclair, A. M., B. Gottgens, L. M. Barton, M. L. Stanley, L. Pardanaud, M. Klaine, M. Gering, S. Bahn, M. Sanchez, A. J. Bench, J. L. Fordham, E. Bockamp, and A. R. Green. 1999. Distinct 5′ SCL enhancers direct transcription to developing brain, spinal cord, and endothelium: neural expression is mediated by GATA factor binding sites. Dev. Biol. 209:128-142. [DOI] [PubMed] [Google Scholar]
- 53.Suda, T., F. Arai, and A. Hirao. 2005. Hematopoietic stem cells and their niche. Trends Immunol. 26:426-433. [DOI] [PubMed] [Google Scholar]
- 54.Tong, Q., G. Dalgin, H. Xu, C. N. Ting, J. M. Leiden, and G. S. Hotamisligil. 2000. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science 290:134-138. [DOI] [PubMed] [Google Scholar]
- 55.Towler, D. A., and R. St. Arnaud. 2002. Use of cultured osteoblastic cells to identify and characterize transcriptional regulatory complexes, p. 1503-1527. In J. P. Bilezikian and G. A. Rodan (ed.), Principles of bone biology, 2nd ed. Academic Press, London, England.
- 56.Visvader, J. E., Y. Fujiwara, and S. H. Orkin. 1998. Unsuspected role for the T-cell leukemia protein SCL/tal-1 in vascular development. Genes Dev. 12:473-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, J., F. Iwata, J. A. Grass, C. S. Osborne, L. Elnitski, P. Fraser, O. Ohneda, M. Yamamoto, and E. H. Bresnick. 2005. Molecular determinants of NOTCH4 transcription in vascular endothelium. Mol. Cell. Biol. 25:1458-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang, X., and G. Karsenty. 2002. Transcription factors in bone: developmental and pathological aspects. Trends Mol. Med. 8:340-345. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, J., C. Niu, L. Ye, H. Huang, X. He, W. G. Tong, J. Ross, J. Haug, T. Johnson, J. Q. Feng, S. Harris, L. M. Wiedemann, Y. Mishina, and L. Li. 2003. Identification of the haematopoietic stem cell niche and control of the niche size. Nature 425:836-841. [DOI] [PubMed] [Google Scholar]