Abstract
Generally, histone deacetylase (HDAC) inhibitor-induced p21Waf1/Cip1 expression is thought to be p53 independent. Here we found that an inhibitor of HDAC, depsipeptide (FR901228), but not trichostatin A (TSA), induces p21Waf1/Cip1 expression through both p53 and Sp1/Sp3 pathways in A549 cells (which retain wild-type p53). This is demonstrated by measuring relative luciferase activities of p21 promoter constructs with p53 or Sp1 binding site mutagenesis and was further confirmed by transfection of wild-type p53 into H1299 cells (p53 null). That p53 was acetylated after depsipeptide treatment was tested by sequential immunoprecipitation/Western immunoblot analysis with anti-acetylated lysines and anti-p53 antibodies. The acetylated p53 has a longer half-life due to a significant decrease in p53 ubiquitination. Further study using site-specific antiacetyllysine antibodies and transfection of mutated p53 vectors (K319/K320/K321R mutated and K373R/K382R mutations) into H1299 cells revealed that depsipeptide specifically induces p53 acetylation at K373/K382, but not at K320. As assayed by coimmunoprecipitation, the K373/K382 acetylation is accompanied by a recruitment of p300, but neither CREB-binding protein (CBP) nor p300/CBP-associated factor (PCAF), to the p53 C terminus. Furthermore, activity associated with the binding of the acetylated p53 at K373/K382 to the p21 promoter as well as p21Waf1/Cip1 expression is significantly increased after depsipeptide treatment, as tested by chromatin immunoprecipitations and Western blotting, respectively. In addition, p53 acetylation at K373/K382 is confirmed to be required for recruitment of p300 to the p21 promoter, and the depsipeptide-induced p53 acetylation at K373/K382 is unlikely to be dependent on p53 phosphorylation at Ser15, Ser20, and Ser392 sites. Our data suggest that p53 acetylation at K373/K382 plays an important role in depsipeptide-induced p21Waf1/Cip1 expression.
p53 is a short-lived protein and is sustained at low levels in normal physiological conditions (40, 45). In unstressed mammalian cells p53 is continually ubiquitinated by interacting with MDM2 (23, 57, 62), COP1 (15), Pirh2 (44), and ARF-BP1 (9). Subsequently p53 protein is degraded by the 26S proteasome (5, 30). However, p53 is maintained at a relative high level by posttranslational modifications in response to various stresses. The principal posttranslational modifications of p53 in response to DNA damage include phosphorylation and acetylation (22, 25, 31, 43, 67), through which p53 exerts its biochemical functions. Transcriptional coactivators p300/CREB-binding protein (CBP) and p300/CBP-associated factor (PCAF) were reported to acetylate p53 at K373/K382 and K320, respectively (25, 48, 49), and the lysine acetylation at these sites is linked to its ability to regulate cell cycle arrest and apoptosis (26, 34, 52). Furthermore, these two processes of p53 posttranslational modifications are interrelated (5, 29, 66). For example, in response to UV or irradiation, the N terminus of p53 firstly becomes phosphorylated at Ser33 and Ser37 and, in turn, phosphorylated p53 activates p300 and PCAF to induce p53 acetylation at K373/K382 and K320, respectively (43, 66). In addition, phosphorylation of p53 at the Ser20 or Thr18 site plays a critical role in stabilizing the p300-p53 complex (12, 53), and phosphorylation of p53 at Ser15 increases binding to CBP (43) and p300 (16). Recently, it was reported that p53 C-terminal phosphorylation induced by CHK1 and CHK2 also modulates C-terminal acetylation in responding to DNA damage (63). These data indicate that p53 modulation is a complex process, and the biological consequences of p53 activation induced by certain stimuli may be dependent on p53 posttranslational modifications at multiple sites.
There is controversy generated by reports regarding the functions of the acetylated p53 (3, 17, 25, 49, 56, 66). Whether the acetylated p53 increases its DNA binding as well as downstream transcriptional activity is the central question of this controversy. It has been hypothesized that p53 is latent in normal conditions and becomes active when cells are exposed to DNA damage or other genotoxic agents, during which p53 is phosphorylated and acetylated and, in turn, accumulates in the nucleus at its target genes (25, 31, 49, 66, 68). Stress-induced activation of p53 is due to a modification of its C terminus leading to the release of negative regulation of DNA binding exerted by the C terminal region (25, 49, 66). This model has been demonstrated in the assays with C terminus deletion (1, 61), lysine site mutations (18), and posttranslational modifications (25, 66, 70). In contrast, Espinosa and Emerson indicated that binding of p53 to its target site (such as the p21 promoter) does not require C-terminal modification by acetylation (17). Dornan et al. reported that site-specific acetylation of p53 was DNA dependent; deletion of the p53 proline repeat allows p53 to bind to p21, but p53 was unable to be acetylated, indicating that proline-directed acetylation of p53 is a post-DNA binding event (13, 14). Furthermore, phosphorylation of the C-terminal regulatory domain of p53 (such as at Ser392) by casein kinase II (CK2) promotes DNA binding and induces a site-specific DNA- and p300-dependent acetylation (7, 32). In addition, other reports showed that p53 binds to the p21 promoter in vitro and in vivo in the absence of DNA damage or extensive modifications of the C terminus (3, 39). However, the above hypotheses are based on experimental data from deletions of p53 or DNA damage, neither of which is a physiological condition. It is thus important to test whether p53 acetylation influences DNA binding by intact p53 and the transcriptional activity of p53's target genes in the absence of DNA damage.
Histone deacetylase (HDAC) inhibitors have been extensively studied in basic biological research to gain an understanding of basic chromatin structure and transcriptional control and have recently been introduced as potential clinical treatments for cancer (36, 54, 55, 71, 74). Generally, HDAC inhibitors induce accumulation of hyperacetylated nucleosome core histones and cause transcriptional activation of genes (36). In addition, HDAC inhibitors are reported to induce acetylation of nonhistone proteins (8, 25). However, although the commonly laboratory HDAC inhibitor trichostatin A (TSA) can acetylate p53, it primarily acetylates p53 under the conditions in which cells were irradiated with gamma rays (69) and UV (10, 34) or were irradiated in combination with addition of another HDAC inhibitor, nicotinamide (51). Therefore, a new and more powerful HDAC inhibitor is needed for the study of p53 acetylation. Depsipeptide is a novel and effective HDAC inhibitor (59), and its efficacy of suppressing histone deacetylases is 10 times that of TSA in human cancer cell lines (65, 73, 75). Depsipeptide therefore is a potential candidate for the study of p53 acetylation due to its characteristics of greater efficacy and duration of action.
In this study, human lung cancer cell lines A549 (with wild-type p53) and H1299 (null p53) were treated with depsipeptide to test changes of p53 acetylation. When assayed for relative luciferase activity using mutagenized p21 promoters and transfection of wild-type p53 into H1299 cells, depsipeptide-induced p21Waf1/Cip1 expression is shown to be partly through p53 acetylation. Furthermore, the depsipeptide-induced p53 acetylation is site specific: depsipeptide acetylates p53 at K373/K382 only, not at K320. Finally, depsipeptide-induced p53 acetylation is sufficient to increase its DNA binding as well as transcriptional activity at the p21 promoter.
MATERIALS AND METHODS
Cell culture and chemical treatments.
Human lung cancer cell lines A549 and H1299 were grown in RPMI 1640 supplemented with 10% fetal bovine serum (heat inactivated at 56°C for 45 min) and the appropriate amount of penicillin/streptomycin in a 37°C incubator with a humidified, 5% CO2 atmosphere. HDAC inhibitor depsipeptide at 0.05 μM to 0.2 μM or TSA at 0.25 μM to 2 μM was added into cells for 6 to 24 h, and then cells were washed with cold phosphate-buffered saline (PBS) twice. The HDAC inhibitor-treated cells were then incubated at 37°C for 0 to 24 h. Control cells were treated with dimethyl sulfoxide for similar time periods.
Western blotting.
Protein expression was detected by Western blotting as previously described with minor modifications (73). Equal amounts of proteins (100 to 150 μg) were size fractionated by 9 to 15% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. The antibodies used are anti-p21Waf1/Cip1 (F-5; Santa Cruz), anti-p53 (DO-1 and Bp53-12; Santa Cruz), PCAF (C-16; Santa Cruz), p300 (H-272; Santa Cruz), CBP (A-22; Santa Cruz), HDAC1 (H-11; Santa Cruz), ubiqitin (P4D1; Santa Cruz), α-tubulin (Santa Cruz), and anti-acetyl-p53 (Lys373 and -382 and Lys320; Upstate). Other antibodies used in this study include anti-p53 (P240, Calbiochem) and anti-Ser15, anti-Ser20, and anti-Ser392 (Cell Signaling).
Site-directed mutagenesis.
A p53 mutant (K373R/K382R) construct was generated using a site-directed mutagenesis kit (QuikChange; Stratagene, La Jolla, CA). A wild-type p53 expression vector (pCIneo with full-length p53 cDNA) (73) was used as the mutagenesis template. Wild-type p53 was mutated at K373R/K382R sites, following the manufacturer's directions. Primers used for the mutagenesis are the following sequences: p53-373R-up, 5′-CAC CTG AAG TCC AAA AG(A)G GGT CAG TCT ACC TC-3′; p53-373R-down, 5′-GA GGT AGA CTG ACC CC(T)T TTT GGA CTT CAG GTG-3′; p53-382R-up, 5′-CTA CCT CCC GCC ATA AAA G(A)AC TCA TGT TCA AGA-3′; p53-382R-down, 5′-TCT TGA ACA TGA GTC(T) TTT TAT GGC GGG AGG TAG-3′. In these primers, underlined italic nucleotides indicate the replaced nucleotides and the nucleotides in parentheses indicate the original nucleotides.
Transient transfection and measurement of relative luciferase activity.
Vectors used for transfections in this study include the wild-type p53 vector (73), pWWP-Luc, pWWP-p53 mut1-Luc, pWWP-p53 mut2-Luc (60, 72), the Sp1-3 mutated p21 promoter (a gift from Christian Seiser), mutant lys320/373/381/382 p53, and mutant lys319/320/321 p53 (lysine codons at these sites are replaced with arginine codons) (gifts from Shelley Berger). The human wild-type p21 promoter luciferase fusion plasmid, pWWP-Luc, contains two p53 binding sites with 2.4 kb upstream of the translational start sites. pWWP-p53 mut1-Luc, which contains a mutated version of the first p53 binding site, GAACA (−2234 to −2230 relative to the translational start site), was replaced with GAAAC, and pWWP-p53mut2-Luc, which contains a mutated version of the second binding site, AGACT (−1344 to −1340 relative to the translational start site), was replaced with AGAAT (72). The Sp1-3-mutated p21 promoter contains two intact p53 binding sites and the Sp1-3 mutation site (42).
Extraction of nuclear proteins.
Nuclear protein was extracted as described previously with modifications (75). Briefly, 107 A549 cells were scraped into a 1.5-ml tube and centrifuged at room temperature for 5 min at 1,000 rpm. The cell pellet was washed with 1 to 2 ml of cold PBS and then centrifuged at 1,500 rpm for 20 to 30 s at 4°C. The resulting pellet was incubated in buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF], protease inhibitor cocktail) and then incubated on ice for 15 min. The cells were centrifuged at 1,500 rpm for 5 min at 4°C, and the resulting pellet was resuspended in buffer A and homogenized with a glass homogenizer (Kontes Glass Co., Vineland, N.J.). The cells were checked under a microscope with trypan blue, and the presence of >90% free nuclei was confirmed. After centrifugation at 1,000 rpm and 4°C, the supernatant was discarded, and the pellet was suspended in 1/2 volume of buffer B (20 mM HEPES, 0.2 mM EDTA, 1.5 mM MgCl2, 0.02 M KCl, 25% glycerol, 0.5 mM DTT, 0.5 mM PMSF, protease inhibitors). The suspension was then gently resuspended in 1/2 volume of buffer C (20 mM HEPES, 0.2 mM EDTA, 1.5 mM MgCl2, 1.2 M KCl, 25% glycerol, 0.5 mM DTT, 0.5 mM PMSF, protease inhibitors), incubated at 4°C for 30 min with rotation, and then centrifuged at 4°C at 14,000 rpm for 30 min. The nuclear protein was then dialyzed three times against dialysis buffer (20 mM HEPES, 0.2 mM EDTA, 0.1 M KCl, 20% glycerol, 0.5 mM DTT, 0.5 mM PMSF, protease inhibitors) for 2 h each time. Finally, the concentration of nuclear protein was determined and saved at −80°C for experiments.
Chromatin immunoprecipitation (ChIP) assay.
A549 cells were cross-linked with 1% formaldehyde for 10 min at 37°C and then washed with cold PBS. The cell pellet was resuspended in 0.3 ml of lysis buffer (1% SDS, 100 mM NaCl, 50 mM Tris-HCl, pH 8.1, 5 mM EDTA), followed by sonication to an average DNA length of 500 to 1,000 bp. Antibodies were added to each of the samples, which were then rotated at 4°C overnight. After interaction with protein A beads and incubation overnight at 65°C to reverse the cross-links, the DNA was dissolved in Tris-EDTA buffer and analyzed by PCR. The antibodies anti-p53 (Bp53-12), anti-acetylated p53 (K373/382 and K320), anti-p300, anti-CBP, and anti-PCAF were added separately into reaction solutions. Primers used for PCR were from p21Waf1/Cip1 promoter sequences: 5′-CTCACATCCTCCTTCTTCAG-3′ (sense) and 5′-CACACACAGAATCTGACTCCC-3′ (antisense).
Measurement of the half-life of endogenous p53.
A549 cells were treated with cycloheximide (CHX; 10 μg/ml) in the presence or absence of depsipeptide at 0.1 μM for different times. The treated cells were then harvested and extracted with radioimmunoprecipitation assay buffer (2.5 mM Tris, pH 7.4, 150 mM KCl, 5 mM EDTA, 1% NP-40, 0.5% Na deoxycholate, 0.1% SDS). The proteins were analyzed by Western blotting with anti-p53 (DO-1).
Coimmunoprecipitation (Co-IP).
Cells were harvested and then lysed in lysis buffer (1% NP-40, 150 mM NaCl, 50 mM Tris, 0.05% SDS, 1 mM PMSF, and a 1% cocktail of protease inhibitors) on ice for 20 min. After centrifugation at 4°C at 13,000 rpm for 10 min, antibodies were added to the supernatant on ice for 1 h. Agarose G was then added to the samples, and the samples were rolled at 4°C for 1 h. After the beads were washed three times with lysis buffer, the pellets were dissolved into 2× SDS loading buffer after centrifugation. The protein was analyzed by Western blotting with different antibodies.
Gamma ray irradiation.
A549 cells were irradiated with gamma rays at 1 Gy/min at different intervals. The irradiated cells were washed immediately after irradiation, fresh medium was added, and then cells were incubated at 37°C. Proteins were then extracted for further analysis.
RESULTS
Depsipeptide induces p21Waf1/Cip1 expression through p53 and Sp1.
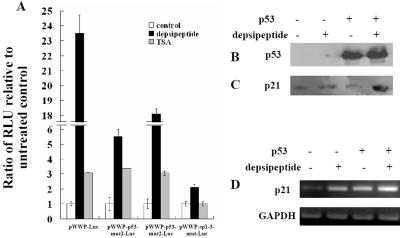
A line of evidence has confirmed that HDAC inhibitors induce p21Waf1/Cip1 expression through Sp1 (20, 60). In this study, depsipeptide induced significant p21Waf1/Cip1 expression in a dose-dependent manner in A549 cells and maximum p21Waf1/Cip1 expression was observed at 18 h after depsipeptide treatment, as shown in Fig. 1A and B. To determine the role of p53 in depsipeptide-induced p21Waf1/Cip1 expression, a wild-type human p21 promoter luciferase fusion plasmid and two p21 promoter plasmids with mutated p53 binding sites were transfected into A549 cells, followed by depsipeptide treatment. Figure 2A clearly shows that the depsipeptide-induced increase in the relative luciferase activity of the p21 promoter was much decreased when p53 binding sites of the p21 promoter are mutated, when compared to the wild-type p21 promoter. It is likely that the first p53 binding site (−2234 to −2230 relative to the translational start site) is more important than the second p53 binding site (−1344 to −1340 relative to the translational start site) in activating depsipeptide-induced p21Waf1/Cip1 expression (for example, compared to the wild-type p21 promoter, depsipeptide-induced relative luciferase activity was decreased 4.2-fold in the cells transfected with the plasmid having the first p53 binding site mutated whereas it was decreased only 1.2-fold in the cells transfected with the plasmid having the second p53 binding site mutated). In addition, a full-length p21 promoter luciferase fusion plasmid with a mutated Sp1-3 binding site but intact p53 binding sites was transfected into A549 cells to confirm the role of p53 in depsipeptide-induced p21Waf1/Cip1 expression. Depsipeptide still induces a 2.2-fold increase in the luciferase activity when the plasmid with the Sp1-3-mutated p21 promoter was transfected (Fig. 2A). This result indicates that the Sp1-3 binding site is critical for p21Waf1/Cip1 expression, and p53 also plays a role in depsipeptide-induced p21Waf1/Cip1 expression. Although TSA, a specific inhibitor of HDAC, has been reported to induce significant p21Waf1/Cip1 expression (21, 60), we found this effect to be p53 independent in this study. For example, TSA-induced relative luciferase activity of the p21 promoter was not changed when A549 cells were transfected with the plasmid with the wild-type p21 promoter or two p21 plasmids with mutated p53 binding sites (mut1-Luc and mut2-Luc) (Fig. 2A). (The relative luciferase activities are increased about 3.1-fold, 3.36-fold, and 3.05-fold when cells were transfected with the full-length p21 promoter, mut1-Luc, and mut2-Luc, respectively, compared to untreated cells after cells were treated with depsipeptide at 0.1 μM for 6 h.) Interestingly, the luciferase activity is not changed by TSA compared to untreated cells when cells were transfected with the plasmid having a p21 promoter with a mutated Sp1-3 binding site (Fig. 2A), indicating that TSA may induce p21Waf1/Cip1 expression only by the Sp1-3 site. Next, p53's role in depsipeptide-induced p21Waf1/Cip1 expression was further confirmed by using p53-transfected H1299 cells (Fig. 2B). Depsipeptide could induce a moderate increase in p21Waf1/Cip1 expression in H1299 cells; however, p21Waf1/Cip1 expression was obviously enhanced by depsipeptide when cells were transfected with wild-type p53 (Fig. 2C). Since p21Waf1/Cip1 can also be subject to posttranscriptional modification, a reverse transcription-PCR (RT-PCR) was performed. Results indicate that p21 mRNA is also significantly increased by depsipeptide treatment in a p53-dependent manner (Fig. 2D). These data suggest that depsipeptide may induce p21Waf1/Cip1 expression through both p53 and Sp1 pathways.
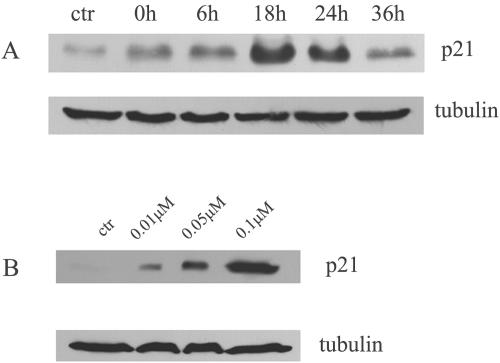
FIG. 1.
Depsipeptide induces p21Waf1/Cip1 expression with time- and dose-dependent manner. Representative Western blots indicate that A549 cells were treated with depsipeptide at 0.1 μM for 6 h and proteins were then extracted at different times after treatment (A), or cells treated at different concentrations of depsipeptide for 6 h then incubated at 37°C for 18 h (B). α-Tubulin as a loading control is shown under every panel. ctr, control.
FIG. 2.
Depsipeptide induces p21Waf1/Cip1 expression through both p53 and Sp1. A wild-type p21 promoter luciferase fusion plasmid (pWWP-Luc) and three plasmids with mutated p21 promoters (pWWP-p53 mut1-Luc, pWWP-p53 mut2-Luc, and pWWP-Sp1-3-mut-Luc), were transfected into A549 cells. At 24 h after transfection, depsipeptide (0.1 μM) or TSA (2 μM) was added onto the cells for 6 h. Eighteen hours after depsipeptide or TSA treatment, cells were harvested and relative luciferase activity was measured (A). The luciferase activity was normalized for the amount of protein in the cell lysate. All of the luciferase experiments were carried out at least three times in triplicate. Plasmids with wild-type p53 were transfected into H1299 cells, and depsipeptide (0.1 μM) was added into cells for 6 h. Eighteen hours after depsipeptide treatment, cells were harvested and protein was extracted for Western immunoblot analysis with anti-p53 (P240) (B) and anti-p21 (C). In addition, H1299 cells were treated as for panel B, and RNA was extracted for RT-PCR assay to detect changes in p21 mRNA (D). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) is a loading control for the RT-PCR.
Depsipeptide induces p53 acetylation and prolongs p53 half-life through posttranslational modifications.
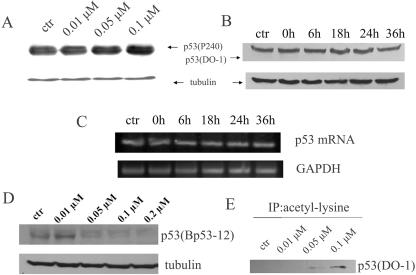
To investigate whether depsipeptide-induced p21Waf1/Cip1 expression results from an enhancement of p53 expression, Western blotting was performed to detect changes of p53 amount in the depsipeptide-treated A549 cells. Figure 3A and B show that depsipeptide does not induce an increase in p53 expression in either a dose- or time-dependent manner when analyzed with anti-full-length p53 (P240) and anti-N terminus of p53 (DO-1), respectively. To further determine whether p53 activity in gene expression is changed by depsipeptide treatment, RT-PCR was performed to see the readout of p53 mRNA in depsipeptide-treated cells. Figure 3C shows that p53 mRNA is not obviously changed upon depsipeptide treatment in A549 cells. However, a significant decrease in detectable p53 expression was observed by using anti-C terminus of p53 (Bp53-12) when A549 cells were treated with depsipeptide (Fig. 3D). These data imply that a modification in the C terminus of p53 induced by depsipeptide may interfere with the recognition of anti-p53 (Bp53-12) by the p53 molecule. Subsequently, an antiacetyllysine antibody was used for detecting total lysine acetylation of depsipeptide-treated A549 cells. By performing Co-IP with an antilysine antibody and then probing with an anti-p53 antibody (DO-1), a significant increase in p53 acetylation in a dose-dependent manner was confirmed in the depsipeptide-treated cells (Fig. 3E). Therefore, the apparent decrease in p53 levels seen with Bp53-12 (but not seen with DO1 or p240) is actually due to a change in the epitope (by lysine acetylation) recognized by Bp53-12, leading to decreased binding by the antibody.
FIG. 3.
Depsipeptide induces p53 acetylation. Representative Western blots show changes of p53 amount, in which A549 cells were treated with depsipeptide at different concentrations for 6 h (A) or at 0.1 μM for different incubation times after treatment (B). Proteins were extracted for Western blotting by using anti-p53 (P240 for panel A and DO-1 for panel B; these antibodies recognize full-length p53 and the N terminus of p53, respectively). A further RT-PCR assay was performed for testing changes in p53 mRNA at different intervals after treatment with depsipeptide (C). GAPDH is a loading control for the RT-PCR. (D) Cells were also treated with depsipeptide at different concentrations, and Western blotting was performed by using anti-p53 (Bp53-12, recognizing the unmodified C terminus). α-Tubulin, as a loading control, is shown under panels A, B, and D. (E) Cells were treated with depsipeptide at different concentrations, and protein was extracted for Co-IP by using antiacetyllysine antibody, followed by Western immunoblotting with anti-p53 (DO-1). ctr, control.
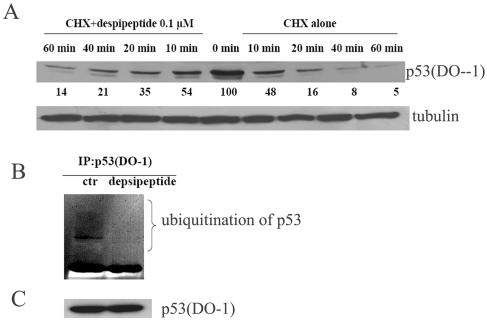
One of the consequences of p53 acetylation is a decrease in p53 degradation (33, 64). To evaluate the mechanism for this, A549 cells were treated with CHX, an inhibitor of protein synthesis, at 10 μM for up to 2 h alone or with depsipeptide at 0.1 μM. Western immunoblots shown in Fig. 4A indicate that depsipeptide significantly prolongs the half-life of p53. This extension of p53's half-life comes from a decrease in the ubiquitination of p53. As shown in Fig. 4B, proteins of cells with or without depsipeptide treatment were immunoprecipitated with anti-p53 (DO-1) and then incubated with antiubiquitin antibody for performing Western immunoblotting. p53-conjugated ubiquitin in the depsipeptide-treated cells is greatly decreased compared to that in the cells without depsipeptide treatment (Fig. 4B), although total p53 expression was not changed after depsipeptide treatment by using anti-p53 (DO-1) (Fig. 4C). These data demonstrate that the acetylated p53 induced by depsipeptide has dramatically decreased ubiquitination and, therefore, a much more protracted half-life in the treated cells. The lag time in sustained p53-induced expression of its downstream targets, such as p21, may be dependent on received stimuli. For example, p21Waf1/Cip1 expression is increased at 6 h after irradiation-induced p53 overexpression and at 72 h after hydroxyurea-induced p53 expression (24). The depsipeptide-induced p21Waf1/Cip1expression is maximally increased at 18 h after p53 acetylation in this study (Fig. 1 and Fig. 3), which may come from the p53 stabilization.
FIG.4.
Depsipeptide prolongs p53 half-life. (A) Representative Western blots for A549 cells treated with CHX (10 μg/ml) alone or with depsipeptide at 0.1 μM for different times as indicated. Nuclear proteins were extracted for Western blotting with anti-p53 (DO-1). α-Tubulin as a loading control is shown in the lower panel. Protein bands were scanned with a phosphorimager, and relative band intensities were normalized for each α-tubulin band. The band intensity at 0 minutes after depsipeptide treatment was set as 100%. The numerical value of each band intensity represents the percentage of band intensity with respect to that at zero time. (B) A549 cells were treated with or without depsipeptide at 0.1 μM for 6 h with proteasome inhibitors N-acetyl-L-leucyl-L-leucyl-L-norleucine and MG132, both at 25 μM. The treated cells were then lysed and immunoprecipitated (IP) with anti-p53 (DO-1), immunoprecipitated proteins were size fractionated by SDS-polyacrylamide gel electrophoresis, and then Western immunoblotting was performed with antiubiquitin. ctr, control. (C) Western blotting with p53 (DO-1) as a control, indicating the same amount of p53 in both lanes.
Depsipeptide specifically acetylates p53 at K373/K382, but not at K320.
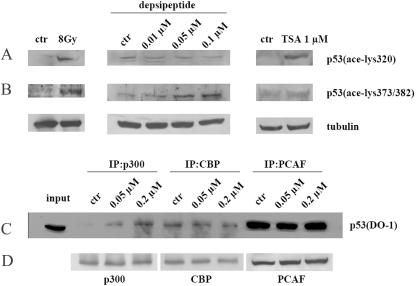
It is well demonstrated that ionizing irradiation activates p53 by phosphorylating p53 at specific sites and acetylating p53 at lysine 320 and lysine 373/382 through PCAF and p300, respectively (48, 49). To detect whether depsipeptide induces p53 acetylation at the same sites as irradiation does, antibodies specific for p53 acetylation at K320 and K373/K382 were used in this study. Gamma ray irradiation (8 Gy) induces acetylation of p53 at K320 and at K373/K382 (Fig. 5A and B). Interestingly, depsipeptide could induce acetylation of p53 at K373/K382 but not at K320 (Fig. 5A and B). With TSA, a significant increase in p53 acetylation at K320 was observed; however, TSA could not induce an increase in p53 acetylation at K373/K382 in A549 cells (Fig. 5B). To further investigate the reason for depsipeptide-induced lysine acetylation at these specific residues, a Co-IP assay was performed to test the interaction of coactivators (PCAF and p300/CBP, with activity of histone acetylases) and p53 in the treated cells. Figure 5C shows that depsipeptide induces a recruitment of p300 to p53 (DO-1); however, depsipeptide could not enhance interactions of CBP and PCAF with p53. These data suggest that depsipeptide may specifically enhance recruitment of p300 to p53 and thus induce p53 acetylation at K373/K382.
FIG. 5.
Depsipeptide specifically enhances recruitment of p300 to the C terminus of p53 and acetylates p53 at K373/K382. Representative Western blots indicate changes of p53 acetylation at K320 (A) and K373/K382 (B), in which A549 cells were exposed to gamma rays at 8 Gy (left panel), depsipeptide (0.01 to 0.1 μM, for 6 h) (middle panel), and TSA (1 μM for 12 h) (right panel). α-Tubulin as a loading control is shown under every panel. ctr, control. (C) A549 cells also were treated with different concentrations of depsipeptide for 6 h, and protein was extracted for performing Co-IP with anti-CBP, -p300, and -PCAF, followed by Western immunoblotting with anti-p53 (DO-1). (D) Western blots produced by reprobing with the same anti-p300, anti-CBP, and anti-PCAF as those used for determining IP efficiency.
p53 acetylation at K373/K382 is required for depsipeptide-induced p21Waf1/Cip1 expression.
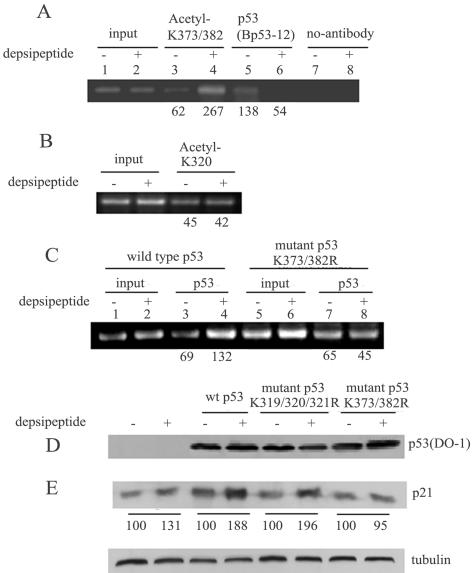
To investigate the role of p53 acetylation at K373/K382 in the activation of the p21 promoter, a ChIP assay was performed. Figure 6A shows that depsipeptide (0.1 μM for 6 h) significantly increases the binding of acetylated p53 at K373/K382 to the p21 promoter (4.3-fold increase) in A549 cells. In contrast, a significant decrease in the binding of the C terminus of p53 to the p21 promoter was observed in the ChIP assay with anti-Bp53-12 (anti-C terminus) (2.5-fold decrease), indicating that acetylated p53 preferentially binds to the p21 promoter under these conditions. Consistent with the results above, depsipeptide could not increase the binding of acetylated p53 at K320 to the p21 promoter (Fig. 6B). To further demonstrate that depsipeptide-induced p21 expression is through p53 acetylation at K373/K382, a plasmid encoding p53 with K373R/K382R mutations was generated by mutagenesis PCR. The wild-type p53 plasmid and the mutated p53 plasmid (K373R/K382R) were transfected into H1299 cells, and then a ChIP assay was performed after depsipeptide treatment. Figure 6C shows that depsipeptide could not induce an increased binding to the p21 promoter when the p53 plasmid encoding the K373R/K382R mutations was transfected into cells. These results clearly indicate that p53 K373/K382 sites are specific sites for depsipeptide-induced p21Waf1/Cip1 expression.
FIG. 6.
Depsipeptide-induced p53 acetylation increases DNA binding to the p21 promoter as well as transcriptional activities. (A) ChIP assay with antibodies against p53 acetylated at K373/K382 (lanes 3 and 4) or anti-Bp53-12 (lanes 5 and 6) for a specific sequence of the p21 promoter in A549 cells treated with or without depsipeptide at 0.1 μM for 6 h. Bands without antibody show negative controls (lanes 7 and 8). Lanes 1 and 2 show an input signal. (B) ChIP assay with antibodies against p53 acetylated at K320 for a specific sequence of the p21 promoter in A549 cells treated with or without depsipeptide at 0.1 μM for 6 h as indicated. (C) H1299 cells were transfected with plasmids containing wild-type p53 (lanes 1 to 4) or mutant p53 K373R/K382R (lanes 5 to 8) and then treated with depsipeptide at 0.1 μM for 6 h. A ChIP assay was then performed with anti-p53 (DO-1) for the p21 promoter. The input PCR signals were set at 100%, and the numerical values of the ChIP signal represent the percentages of input. (D and E) Representative Western blots for plasmids with p53 mutated at K319/K320/K321R or K373R/K382R. Plasmids were transfected into H1299 cells, and then depsipeptide at 0.1 μM was added into the cells for 6 h for testing the changes of p53 (D) or p21Waf1/Cip1 (E), respectively. α-Tubulin as a loading control is shown under panel E. The numerical values of band intensity from Western blots represent the percentages of the untreated control.
Subsequently, for further functional testing of whether depsipeptide-induced p21 activation is due to p53 acetylation at K373/K382, two p53 plasmids encoding mutations at specific sites (p53 mutations at K319/K320/K321R and p53 mutations at K373R/K382R) were transfected into H1299 cells to detect depsipeptide-induced p21Waf1/Cip1 expression. Depsipeptide-induced p21Waf1/Cip1 expression is dependent upon lysines at K373/K382 (Fig. 6E). For example, depsipeptide could induce a significant increase in p21Waf1/Cip1 expression in the transfected cells with wild-type p53 and p53 mutations at K319/K320/K321R (Fig. 6E), but not in the cells with p53 mutations at K373R/K382R (Fig. 6E) although the p53 levels are not obviously changed by depsipeptide in the both types of plasmid-transfected cells (Fig. 6D). These data imply that p53 acetylation-induced p21Waf1/Cip1 expression may be dependent on specific acetylation of p53 lysine sites, such as K373/K382.
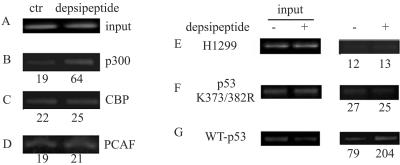
Enhancement of p300 recruitment to the p21 promoter after depsipeptide treatment is dependent on p53 acetylation.
To further determine whether depsipeptide-induced p21Waf1/Cip1 expression may be partly due to a direct recruitment of coactivators to the p21 promoter, a ChIP assay with anti-p300, -CBP, and -PCAF was performed. Figure 7A to D show that depsipeptide could induce a recruitment of p300, but not CBP and PCAF, to the p21 promoter (increased 3.3-fold) in A549 cells. This recruitment of p300 to the p21 promoter is likely dependent on p53 acetylation. For example, the binding of p300 to the p21 promoter is 2.6-fold increased in the depsipeptide-treated H1299 cells transfected with wild-type p53 (Fig. 7G), but not in untransfected cells or in the cells transfected with p53 mutated at K373R/K382R (Fig. 7E and F). This demonstrates that p53 acetylation at K373/K382 is required for p300 recruitment to the p21 promoter in depsipeptide-treated cells.
FIG. 7.
p53 acetylated at K373/382 is required for recruitment of p300 to the p21 promoter. A549 cells were treated with depsipeptide at 0.1 μM for 6 h and then harvested for ChIP assay with a special sequence of the p21 promoter by using anti-p300 (B), -CBP (C), and -PCAF (D), respectively. (A) Inputs. (E to G) A ChIP assay with the p21 promoter was also performed by using anti-p300 in H1299 cells (E) or in H1299 cells transfected with p53 mutated at K373R/K382R (F) or wild-type p53 (G), with or without depsipeptide treatment. Inputs are shown at the left. The numerical values of ChIP signal represent the percentages of inputs.
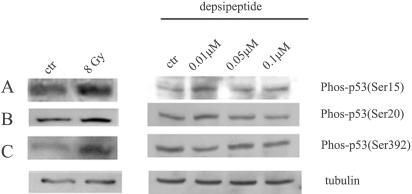
Depsipeptide-induced p53 acetylation is not dependent on p53 phosphorylation at Ser15, Ser20, and Ser392 sites.
It is well known that in certain cases a stress-induced increase in p53 activity comes from a phosphorylation-acetylation cascade (12, 16, 43, 63, 66). There thus is a possibility that depsipeptide activates kinases and thereafter induces p53 acetylation in a postphosphorylation pattern. Phosphorylation of p53 at Ser15 (reflecting ATM activity), Ser20/Thr18 (reflecting CHK2 activity), and Ser392 (reflecting CK2 activity) was reported to be closely related to p53 acetylation. Herein, anti-phos-Ser15, anti-phos-Ser20, and anti-phos-Ser392 were selected to test whether phosphorylation of these sites is changed by depsipeptide. A549 cells were irradiated with gamma rays (at 8 Gy) as positive phosphorylation controls. Cells were treated with depsipeptide at different doses (0 to 0.1 μM) for 6 h and then harvested immediately for Western blots. Ionizing irradiation could induce a significant induction of phosphorylated p53 at all sites tested (Fig. 8A to C). However, p53 phosphorylation did not occur when cells were treated with depsipeptide at different doses, as indicated by Western blotting using anti-phos-p53-Ser15, -Ser20, and -Ser392. These data indicate that depsipeptide-induced p53 acetylation at K373/K382 is not dependent upon prior phosphorylation at these key kinase sites.
FIG. 8.
Depsipeptide does not increase p53 phosphorylation at Ser15, Ser20, and Ser392 sites. A549 cells were treated with depsipeptide (0 to 0.1 μM) for 6 h, and then protein was extracted for Western blots by using anti-Ser15-p53 (A), anti-Ser20-p53 (B), and anti-Ser392-p53 (C). A549 cells were irradiated with gamma rays at 8 Gy in the left panel of each figure as p53 phosphorylation controls. α-Tubulin as a loading control is shown under every panel.
DISCUSSION
The presented data provide evidence that the HDAC inhibitor depsipeptide induces p21Waf1/Cip1 expression through both Sp1/Sp3 and p53 pathways. Posttranslational modifications of p53 play an important role in depsipeptide-induced p21Waf1/Cip1 expression, in which depsipeptide specifically acetylates p53 at K373/K382, which is required for p53-induced p21Waf1/Cip1 expression.
Although most reports indicate that HDAC inhibitors induce p21Waf1/Cip1 expression mainly by activating the Sp1/Sp3 pathway independent of p53 (21, 60), recent reports clearly show that multiple factors, such as ATM (37) and c-myc (46), are involved in the HDAC inhibitor-induced p21Waf1/Cip1 expression in several human cancer cell lines. A direct role for p53 in HDAC-associated p21Waf1/Cip1 expression has also been reported (42). In response to DNA damage, the p53 protein binds directly to the C terminus of Sp1, a domain that was known as a site for interaction with HDAC1 (42). HDAC1 competes with p53 to bind to the Sp1 domain, indicating that HDAC inhibitors may play a role in p53-associated p21Waf1/Cip1 expression. Our data are consistent with those of others mentioned above, such that depsipeptide may have mechanisms other than Sp1 alone to induce p21Waf1/Cip1 expression. For example, p53 is also a key regulator for depsipeptide-induced p21Waf1/Cip1 expression (Fig. 2A). This p53 dependence was further supported by evidence that depsipeptide-induced p21Waf1/Cip1 expression is much enhanced, compared to untransfected cells, only when wild-type p53 is transfected into H1299 cells (Fig. 2B and C). We also tested whether the interactions between Sp1/Sp3 and p53 or HDAC1 are enhanced after depsipeptide treatment in A549 cells by Co-IP. However, the interactions of Sp1/Sp3-p53 or Sp1-HDAC1 are not changed after depsipeptide treatment (data not shown). This implies that there may be an alternative mechanism for p53's role in depsipeptide-nduced p21Waf1/Cip1 expression.
Regardless of changes in total p53 amount after depsipeptide treatment, acetylated p53 was significantly increased in this study (Fig. 3E). Intriguingly, the pattern of depsipeptide-induced p53 acetylation is different from that of gamma ray-induced p53 acetylation. Firstly, depsipeptide induces p53 acetylation in a relative physiological condition without detectable DNA damage. Secondly, depsipeptide-induced p53 acetylation is site specific; for example, depsipeptide induces p53 acetylation at K373/K382 only, whereas irradiation induces p53 acetylation at K320 and K373/K382 sites (Fig. 5A and B). In support of these results, the histone acetyltransferase p300 (for acetylating p53 at K373/382), but not PCAF (for acetylating p53 at K320), was recruited to the p53 C terminus after depsipeptide treatment, which is also different from radiation-induced changes. However, there is a similarity between p53 posttranslational modifications induced by depsipeptide and other DNA damage agents (34, 38), namely, significant extension of p53's half-life (Fig. 4). As is well known, several specific proteins such as MDM2 possess an E3-like ubiquitin ligase activity (30), which rapidly promotes degradation of p53 so as to retain p53 at a low level in unstressed cells (19, 27, 41). Upon irradiation or UV, posttranslational modifications of p53 result in conformational changes of p53. Specifically, p53 acetylation sites K373/K382 were reported to be the same as the sites for MDM2 binding (6, 33, 47). Consequently, MDM2 cannot bind to p53 for degradation, and thus p53 is kept at a high level (2, 34). These data provide a clear picture that acetylation of p53 actually influences the function of ubiquitin-associated p53 degradation. Consistent with this hypothesis, depsipeptide-induced p53 acetylation significantly prolongs the half-life of p53 (Fig. 4A) by decreasing p53 ubiquitination (Fig. 4B).
Another important consequence of acetylation of p53 is an enhancement of activity for binding to its target genes, which induces an increase in the transcriptional activities of its downstream targets after DNA damage (25, 49, 50, 58, 66). There are at least two models explaining why the acetylation of p53 induces enhanced transcriptional activation towards its target genes. The allosteric model states that the C terminus of p53 is a negative regulator and may lock the DNA binding domain in a latent conformation (32, 35). If the interaction between the C terminus and the core binding domain is disrupted by posttranslational modification (such as phosphorylation and acetylation), the DNA binding domain will become active, thus inducing enhanced transcriptional activity (25, 32, 35). Consistent with this model, several reports confirmed that the acetylation of p53 significantly increases its sequence-specific DNA binding activity in vivo, probably due to the acetylation-induced p53 conformational changes (11, 25, 29, 49, 66). However, a recent report argues against this hypothesis by showing that a C terminally deleted p53 is unable to bind and transactivate target genes in vivo, indicating that the C terminus of p53 is a positive regulator of DNA binding and transactivation (56). We don't know how to explain the discrepancy between these data; however, both sets of experiments have confirmed that the C terminus of p53 is required for efficient binding and transcriptional activation of its target promoters in vivo. In our study, abundant acetylated p53 at K373/K382 but not at K320 is bound to the p21 promoter after depsipeptide treatment (Fig. 6A and B), indicating that the binding of acetylated p53 to the p21 promoter plays an important role in activating p21Waf1/Cip1 expression. Another model proposed by Barlev et al. (3) focuses on recruitment of coactivators to the p21 promoter but not binding activity. In this model, p53 acetylation promotes recruitment of coactivators to their target promoters and induces histone acetylation around the target promoters, thereby activating transcription of target genes (3) which is consistent with a later study (28). For example, upon irradiation, levels of binding of CBP and TRRAP (transcriptional activator) to the p21 promoter were increased two- and fivefold, respectively, and this increased binding is due to p53 acetylation in U2OS cells (3). Subsequently, acetylated histones H3 and H4 are tightly bound to the p21 promoter and thus induce increased p21Waf1/Cip1 expression. Our data here show that depsipeptide could recruit p300 to the p21 promoter, but only when p53 is acetylated at K373/K382 sites (Fig. 7B, F, and G). In addition p53 acetylation at K373/K382 enhanced p21 promoter binding though mutated p53 and p53 acetylated at K320 clearly do not. The differences between Barlev's data and ours may come from a difference of stimuli for inducing p53 acetylation. However, our data suggest that depsipeptide-induced p53 acetylation is an important factor for regulating p21Waf1/Cip1 expression and furthermore support the existence of multiple mechanisms involved in p53-regulated p21Waf1/Cip1 expression, including enhancement of the binding of acetylated p53 to the p21Waf1/Cip1 promoter and recruitment of p300 to the p21Waf1/Cip1 promoter.
Finally, although both depsipeptide and TSA are well known as HDAC inhibitors, their abilities to induce p53 acetylation are different. TSA-induced p53 acetylation is less frequently reported in the literature. It appears that TSA could increase p53 acetylation together with other stress stimuli (10, 25, 50). In contrast, in this study depsipeptide alone at low doses could significantly induce p53 acetylation (Fig. 3E), through which p21Waf1/Cip1 expression is increased. The basis of the differences between the consequences of these two HDAC inhibitors in the p53/p21Waf1/Cip1 pathway may come from the evidence that depsipeptide but not TSA could specifically acetylate p53 at K373/K382 and recruit p300 to the p53 C terminus (Fig. 5). Similarly, there is another example to explain the difference between TSA and other HDAC inhibitors in inducing p53 acetylation at K382: a yeast homologue of Sir2, SIRT1, was reported to be a deacetylase of p53, and the SIRT1-induced p53 deacetylation could be released by the HDAC inhibitor nicotinamide but not by TSA (51, 69). Whether depsipeptide acetylates p53 by inhibiting the activity of SIRT1 is an interesting subject for testing in the future. This also points out the possibility and, in fact, probability that different HDAC inhibitors may have distinct activities, perhaps related to preferential inhibition of specific HDAC classes or HDAC enzymes (4). In addition, although it is likely that depsipeptide has no ability to activate the well-known kinase-induced phosphorylation of p53 at Ser15, Ser20, and Ser392, which reflects ATM, CHK2, and CK2 activity, respectively, assayed in this study (Fig. 8), depsipeptide but not TSA may function on an undefined kinase or phosphorylation sites by inducing cellular stress that is not through an HDAC-dependent mechanism. This possibility will also be further studied in the future.
Depsipeptide induces p53 acetylation at K373/K382 sites, which seems to be a key factor for activating p21Waf1/Cip1 expression. Our results may provide a useful clue for explaining the different roles of different HDAC inhibitors in treatment of cancers and may help set an appropriate therapeutic strategy for cancer treatment.
Acknowledgments
We greatly appreciate Shelley Berger, Christian Seiser, and T. Sakai for kindly providing us vectors used in this study.
This work was supported by grants 2005CB522403 from the Ministry of Science and Technology of China and 30425017 and 30171613 from National Natural and Scientific Foundation of China.
REFERENCES
- 1.Anderson, M. E., B. Woelker, M. Reed, P. Wang, and P. Tegtmeyer. 1997. Reciprocal interference between the sequence-specific core and nonspecific C-terminal DNA binding domains of p53: implications for regulation. Mol. Cell. Biol. 17:6255-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appella, E., and C. W. Anderson. 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268:2764-2772. [DOI] [PubMed] [Google Scholar]
- 3.Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8:1243-1254. [DOI] [PubMed] [Google Scholar]
- 4.Blagosklonny, M. V., R. Robey, D. L. Sackett, L. Du, F. Traganos, Z. Darzynkiewicz, T. Fojo, and S. E. Bates. 2002. Histone deacetylase inhibitors all induce p21 but differentially cause tubulin acetylation, mitotic arrest, and cytotoxicity. Mol. Cancer Ther. 1:937-941. [PubMed] [Google Scholar]
- 5.Bode, A. M., and Z. Dong. 2004. Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer 4:793-805. [DOI] [PubMed] [Google Scholar]
- 6.Brooks, C. L., and W. Gu. 2003. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr. Opin. Cell Biol. 15:164-171. [DOI] [PubMed] [Google Scholar]
- 7.Bruins, W., E. Zwart, L. D. Attardi, T. Iwakuma, E. M. Hoogervorst, R. B. Beems, B. Miranda, C. T. van Oostrom, J. van den Berg, G. J. van den Aardweg, G. Lozano, H. van Steeg, T. Jacks, and A. de Vries. 2004. Increased sensitivity to UV radiation in mice with a p53 point mutation at Ser389. Mol. Cell. Biol. 24:8884-8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, H. M., M. Krstic-Demonacos, L. Smith, C. Demonacos, and N. B. La Thangue. 2001. Acetylation control of the retinoblastoma tumour-suppressor protein. Nat. Cell Biol. 3:667-674. [DOI] [PubMed] [Google Scholar]
- 9.Chen, D., N. Kon, M. Li, W. Zhang, J. Qin, and W. Gu. 2005. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell 121:1071-1083. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, H. L., R. Mostoslavsky, S. Saito, J. P. Manis, Y. Gu, P. Patel, R. Bronson, E. Appella, F. W. Alt, and K. F. Chua. 2003. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. USA 100:10794-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dornan, D., M. Ecker, M. Wallace, H. Shimizu, E. Ramsay, T. R. Hupp, and K. L. Ball. 2004. Interferon regulatory factor 1 binding to p300 stimulates DNA-dependent acetylation of p53. Mol. Cell. Biol. 24:10083-10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dornan, D., and T. R. Hupp. 2001. Inhibition of p53-dependent transcription by BOX-I phospho-peptide mimetics that bind to p300. EMBO Rep. 2:139-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dornan, D., H. Shimizu, L. Burch, A. J. Smith, and T. R. Hupp. 2003. The proline repeat domain of p53 binds directly to the transcriptional coactivator p300 and allosterically controls DNA-dependent acetylation of p53. Mol. Cell. Biol. 23:8846-8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dornan, D., H. Shimizu, N. D. Perkins, and T. R. Hupp. 2003. DNA-dependent acetylation of p53 by the transcription coactivator p300. J. Biol. Chem. 278:13431-13441. [DOI] [PubMed] [Google Scholar]
- 15.Dornan, D., I. Wertz, H. Shimizu, D. Arnott, G. D. Frantz, P. Dowd, K. O'Rourke, H. Koeppen, and V. M. Dixit. 2004. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429:86-92. [DOI] [PubMed] [Google Scholar]
- 16.Dumaz, N., and D. W. Meek. 1999. Serine 15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J. 18:7002-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Espinosa, J. M., and B. M. Emerson. 2001. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell 8:57-69. [DOI] [PubMed] [Google Scholar]
- 18.Feng, L., T. Lin, H. Uranishi, W. Gu, and Y. Xu. 2005. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol. Cell. Biol. 25:5389-5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freedman, D. A., L. Wu, and A. J. Levine. 1999. Functions of the MDM2 oncoprotein. Cell. Mol. Life Sci. 55:96-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gartel, A. L., and S. Radhakrishnan. 2005. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 65:3980-3985. [DOI] [PubMed] [Google Scholar]
- 21.Gartel, A. L., and A. L. Tyner. 1999. Transcriptional regulation of the p21(WAF1/CIP1) gene. Exp. Cell Res. 246:280-289. [DOI] [PubMed] [Google Scholar]
- 22.Giaccia, A. J., and M. Kastan. 1998. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 12:2973-2983. [DOI] [PubMed] [Google Scholar]
- 23.Gottifredi, V., and C. Prives. 2001. Molecular biology. Getting p53 out of the nucleus. Science 292:1851-1852. [DOI] [PubMed] [Google Scholar]
- 24.Gottifredi, V., S. Shieh, Y. Taya, and C. Prives. 2001. p53 accumulates but is functionally impaired when DNA synthesis is blocked. Proc. Natl. Acad. Sci. USA 98:1036-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu, W., and R. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 26.Guo, A., P. Salomoni, J. Luo, A. Shih, S. Zhong, W. Gu, and P. P. Pandolfi. 2000. The function of PML in p53-dependent apoptosis. Nat. Cell Biol. 2:730-736. [DOI] [PubMed] [Google Scholar]
- 27.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 28.Ho, J. S., W. Ma, D. Y. Mao, and S. Benchimol. 2005. p53-dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol. Cell. Biol. 25:7423-7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hofmann, T. G., A. Moller, H. Sirma, H. Zentgraf, Y. Taya, W. Droge, H. Will, and M. L. Schmitz. 2002. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 4:1-10. [DOI] [PubMed] [Google Scholar]
- 30.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25-27. [DOI] [PubMed] [Google Scholar]
- 31.Hupp, T. R., and D. P. Lane. 1994. Allosteric activation of latent p53 tetramers. Curr. Biol. 4:865-875. [DOI] [PubMed] [Google Scholar]
- 32.Hupp, T. R., A. Sparks, and D. P. Lane. 1995. Small peptides activate the latent sequence-specific DNA binding function of p53. Cell 83:237-245. [DOI] [PubMed] [Google Scholar]
- 33.Ito, A., Y. Kawaguchi, C. H. Lai, J. J. Kovacs, Y. Higashimoto, E. Appella, and T. P. Yao. 2002. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 21:6236-6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito, A., C. H. Lai, X. Zhao, S. Saito, M. H. Hamilton, E. Appella, and T. P. Yao. 2001. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 20:1331-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jayaraman, J., and C. Prives. 1995. Activation of p53 sequence-specific DNA binding by short single strands of DNA requires the p53 C-terminus. Cell 81:1021-1029. [DOI] [PubMed] [Google Scholar]
- 36.Johnstone, R. W. 2002. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat. Rev. Drug Discov. 1:287-299. [DOI] [PubMed] [Google Scholar]
- 37.Ju, R., and M. Muller. 2003. Histone deacetylase inhibitors activate p21WAF1 expression via ATM. Cancer Res. 63:2891-2897. [PubMed] [Google Scholar]
- 38.Juan, L. J., W. J. Shia, M. H. Chen, W. M. Yang, E. Seto, Y. S. Lin, and C. W. Wu. 2000. Histone deacetylases specifically down-regulate p53-dependent gene activation. J. Biol. Chem. 275:20436-20443. [DOI] [PubMed] [Google Scholar]
- 39.Kaeser, M. D., and R. D. Iggo. 2002. Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc. Natl. Acad. Sci. USA 99:95-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ko, L. J., and C. Prives. 1996. p53: puzzle and paradigm. Genes Dev. 10:1054-1072. [DOI] [PubMed] [Google Scholar]
- 41.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 42.Lagger, G., A. Doetzlhofer, B. Schuettengruber, E. Haidweger, E. Simboeck, J. Tischler, S. Chiocca, G. Suske, H. Rotheneder, E. Wintersberger, and C. Seiser. 2003. The tumor suppressor p53 and histone deacetylase 1 are antagonistic regulators of the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 gene. Mol. Cell. Biol. 23:2669-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lambert, P. F., F. Kashanchi, M. F. Radonovich, R. Shiekhattar, and J. N. Brady. 1998. Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 273:33048-33053. [DOI] [PubMed] [Google Scholar]
- 44.Leng, R. P., Y. Lin, W. Ma, H. Wu, B. Lemmers, S. Chung, J. M. Parant, G. Lozano, R. Hakem, and S. Benchimol. 2003. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 112:779-791. [DOI] [PubMed] [Google Scholar]
- 45.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 46.Li, H., and X. Wu. 2004. Histone deacetylase inhibitor, Trichostatin A, activates p21WAF1/CIP1 expression through downregulation of c-myc and release of the repression of c-myc from the promoter in human cervical cancer cells. Biochem. Biophys. Res. Commun. 324:860-867. [DOI] [PubMed] [Google Scholar]
- 47.Li, M., J. Luo, C. L. Brooks, and W. Gu. 2002. Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 277:50607-50611. [DOI] [PubMed] [Google Scholar]
- 48.Lill, N. L., S. Grossman, D. Ginsberg, J. DeCaprio, and D. M. Livingston. 1997. Binding and modulation of p53 by p300/CBP coactivators. Nature 387:823-827. [DOI] [PubMed] [Google Scholar]
- 49.Liu, L., D. Scolnick, R. C. Trievel, H. B. Zhang, R. Marmorstein, T. D. Halazonetis, and S. L. Berger. 1999. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19:1202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo, J., M. Li, Y. Tang, M. Laszkowska, R. G. Roeder, and W. Gu. 2004. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc. Natl. Acad. Sci. USA 101:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo, J., A. Y. Nikolaev, S. Imai, D. Chen, F. Su, A. Shiloh, L. Guarente, and W. Gu. 2001. Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107:137-148. [DOI] [PubMed] [Google Scholar]
- 52.Luo, J., F. Su, D. Chen, A. Shiloh, and W. Gu. 2000. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 408:377-381. [DOI] [PubMed] [Google Scholar]
- 53.MacPherson, D., J. Kim, T. Kim, B. K. Rhee, C. T. Van Oostrom, R. A. DiTullio, M. Venere, T. D. Halazonetis, R. Bronson, A. De Vries, M. Fleming, and T. Jacks. 2004. Defective apoptosis and B-cell lymphomas in mice with p53 point mutation at Ser 23. EMBO J. 23:3689-3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marks, P. A., V. M. Richon, R. Breslow, and R. A. Rifkind. 2001. Histone deacetylase inhibitors as new cancer drugs. Curr. Opin. Oncol. 13:477-483. [DOI] [PubMed] [Google Scholar]
- 55.Marks, P. A., V. M. Richon, and R. A. Rifkind. 2000. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J. Natl. Cancer Inst. 92:1210-1216. [DOI] [PubMed] [Google Scholar]
- 56.McKinney, K., M. Mattia, V. Gottifredi, and C. Prives. 2004. p53 linear diffusion along DNA requires its C terminus. Mol. Cell 16:413-424. [DOI] [PubMed] [Google Scholar]
- 57.Momand, J., G. P. Zambetti, D. C. Olson, D. George, and A. J. Levine. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69:1237-1245. [DOI] [PubMed] [Google Scholar]
- 58.Mujtaba, S., Y. He, L. Zeng, S. Yan, O. Plotnikova, Sachchidanand, R. Sanchez, N. J. Zeleznik-Le, Z. Ronai, and M. M. Zhou. 2004. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol. Cell 13:251-263. [DOI] [PubMed] [Google Scholar]
- 59.Nakajima, H., Y. B. Kim, H. Terano, M. Yoshida, and S. Horinouchi. 1998. FR901228, a potent antitumor antibiotic, is a novel histone deacetylase inhibitor. Exp. Cell Res. 241:126-133. [DOI] [PubMed] [Google Scholar]
- 60.Nakano, K., T. Mizi, Y. Sowa, T. Orita, T. Yoshino, Y. Okuyama, T. Fujita, N. Ohtani-Fujita, Y. Matsukawa, T. Tokino, H. Yamagishi, T. Oka, H. Nomura, and T. Sakai, T. 1997. Butyrate activates the WAF1/Cip1 gene promoter through Sp1 sites in a p53-negative human colon cancer cell line. J. Biol. Chem. 272:22199-22206. [DOI] [PubMed] [Google Scholar]
- 61.Nie, Y., H. H. Li, C. M. Bula, and X. Liu. 2000. Stimulation of p53 DNA binding by c-Abl requires the p53 C terminus and tetramerization. Mol. Cell. Biol. 20:741-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oliner, J. D., K. W. Kinzler, P. S. Meltzer, D. L. George, and B. Vogelstein. 1992. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358:80-83. [DOI] [PubMed] [Google Scholar]
- 63.Ou, Y. H., P. H. Chung, T. P. Sun, and S. Y. Shieh. 2005. p53 C-terminal phosphorylation by CHK1 and CHK2 participates in the regulation of DNA-damage-induced C-terminal acetylation. Mol. Biol. Cell 16:1684-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prives, C., and P. A. Hall. 1999. The p53 pathway. J. Pathol. 187:112-126. [DOI] [PubMed] [Google Scholar]
- 65.Rajgolikar, G., K. K. Chan, and H. C. Wang. 1998. Effects of a novel antitumor depsipeptide, FR901228, on human breast cancer cells. Breast Cancer Res. Treat. 51:29-38. [DOI] [PubMed] [Google Scholar]
- 66.Sakaguchi, K., J. E. Herrera, S. Saito, T. Miki, M. Bustin, A. Vassilev, C. W. Anderson, and E. Appella. 1998. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12:2831-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shieh, S. Y., J. Ahn, K. Tamai, Y. Taya, and C. Prives. 2000. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 14:289-300. [PMC free article] [PubMed] [Google Scholar]
- 68.Takenaka, I., F. F. Morin, B. R. Seizinger, and N. Kley. 1995. Regulation of the sequence-specific DNA binding function of p53 by protein kinase C and protein phosphatases. J. Biol. Chem. 270:5405-5411. [DOI] [PubMed] [Google Scholar]
- 69.Vaziri, H., S. K. Dessain, E. Ng Eaton, S. I. Imai, R. A. Frye, T. K. Pandita, L. Guarente, and R. A. Weinberg. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149-159. [DOI] [PubMed] [Google Scholar]
- 70.Wang, Y., and C. Prives. 1995. Increased and altered DNA binding of human p53 by S and G2/M but not G1 cyclin-dependent kinases. Nature 376:88-91. [DOI] [PubMed] [Google Scholar]
- 71.Yu, X., Z. S. Guo, M. G. Marcu, L. Neckers, D. M. Nguyen, G. A. Chen, and D. S. Schrump. 2002. Modulation of p53, ErbB1, ErbB2, and Raf-1 expression in lung cancer cells by depsipeptide FR901228. J. Natl. Cancer Inst. 94:504-513. [DOI] [PubMed] [Google Scholar]
- 72.Zhu, W. G., T. Hileman, Y. Ke, P. Wang, S. Lu, W. Duan, Z. Dai, T. Tong, M. A. Villalona-Calero, C. Plass, and G. A. Otterson. 2004. 5-Aza-2′-deoxycytidine activates the p53/p21Waf1/Cip1 pathway to inhibit cell proliferation. J. Biol. Chem. 279:15161-15166. [DOI] [PubMed] [Google Scholar]
- 73.Zhu, W. G., R. R. Lakshmanan, M. D. Beal, and G. A. Otterson. 2001. DNA methyltransferase inhibition enhances apoptosis induced by histone deacetylase inhibitors. Cancer Res. 61:1327-1333. [PubMed] [Google Scholar]
- 74.Zhu, W. G., and G. A. Otterson. 2003. The interaction of histone deacetylase inhibitors and DNA methyltransferase inhibitors in the treatment of human cancer cells. Curr. Med. Chem. Anti-Cancer Agents 3:187-199. [DOI] [PubMed] [Google Scholar]
- 75.Zhu, W. G., K. Srinivasan, Z. Dai, W. Duan, L. J. Druhan, H. Ding, L. Yee, M. A. Villalona-Calero, C. Plass, and G. A. Otterson. 2003. Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in the p21Cip1 promoter. Mol. Cell Biol. 23:4056-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]