Abstract
Processing from pre-mRNA introns is a widespread mechanism to generate human box C/D and H/ACA snoRNAs. Recent studies revealed that an optimal position relative to the 3′ splice site is important for efficient processing of most box C/D snoRNAs and that assembly of box C/D snoRNPs is stimulated by splicing factors likely bound to the branch point region. Here we have investigated the processing of another major class of human intron-encoded RNAs, the box H/ACA snoRNAs. Analysis of 80 H/ACA RNA genes revealed that human H/ACA RNAs possess no preferential localization close to the 3′ or 5′ splice site. In vivo processing experiments confirmed that H/ACA intronic snoRNAs are processed in a position-independent manner, indicating that there is no synergy between H/ACA RNA processing and splicing. We also showed that recognition of intronic H/ACA snoRNAs and assembly of pre-snoRNPs is an early event that occurs during transcription elongation parallel with pre-mRNA splice site selection. Finally, we found that efficient processing and correct nucleolar localization of the human U64 H/ACA snoRNA requires RNA polymerase II-mediated synthesis of the U64 precursor. This suggests that polymerase II-associated factors direct the efficient assembly and determine the correct subnuclear trafficking of human H/ACA snoRNPs.
Mammalian cells contain a large number of small non-protein-coding RNAs (ncRNAs), including small nuclear RNAs (snRNAs), nucleolar RNAs (snoRNAs), Cajal body-specific RNAs (scaRNAs), cytoplasmic RNAs (scRNAs), and short microRNAs (miRNAs) (1, 5, 6, 30, 44, 66). Biogenesis of small ncRNAs follows diverse pathways. Some ncRNAs are synthesized from independent transcription units (26), but the majority of mammalian ncRNA genes are located within introns of protein-coding genes, and they lack their own transcriptional regulatory elements (5, 20, 34, 54, 62). The intron-encoded RNAs are cotranscribed with and processed from the intron region of the host pre-mRNA. This genetic arrangement has an innate regulatory potential to coordinate the expression of the intron-encoded ncRNA and the protein product of the host gene. The intronic ncRNAs often show an apparent functional relationship with their host genes. The majority of mammalian snoRNAs required for rRNA processing are within genes that specify proteins involved in ribosome biogenesis or function (45, 54). Likewise, cotranscription with regulated protein-coding genes can ensure the tissue-specific and/or developmentally controlled expression of some intronic snoRNAs and miRNAs (2, 5).
Although many miRNAs seem to be processed from pre-mRNA introns, box C/D and H/ACA snoRNAs represent the most abundant groups of mammalian intron-borne ncRNAs. The majority of box C/D and H/ACA snoRNAs guide 2′-O-methylation and pseudouridylation of rRNAs in the nucleolus, respectively (reviewed in references 2, 15, 35, and 53). A minor group of intron-encoded box C/D 2′-O methylation and box H/ACA pseudouridylation guide RNAs, called scaRNAs, direct modification of spliceosomal snRNAs in the nucleoplasmic Cajal bodies (12, 32). While all box C/D RNAs are associated with fibrillarin (the 2′-O-methyltransferase), Nop56, Nop58, and 15.5-kDa proteins, the box H/ACA RNAs are complexed with dyskerin (the pseudouridine synthase), Gar1p, Nhp2p, and Nop10p to form functional modification guide snoRNPs or scaRNPs (15, 53). The RNP proteins specifically recognize the box C/D or box H/ACA structural core motifs which are composed of the conserved box sequences and associated stem structures.
Little is known about the processing of mammalian intron-encoded snoRNAs. The great majority of box C/D and H/ACA snoRNAs seem to be processed directly from the excised and debranched host introns by exonucleolytic trimmings (36, 37, 56). In a minor pathway, endonucleolytic cleavages of flanking intron sequences provide the entry sites for the processing exonucleases (9, 10, 42). The RNP proteins bound to the box C/D and H/ACA structural core motifs are believed to delineate the termini of mature snoRNAs by protecting them from exonucleolytic trimming (7, 10, 21, 61, 63).
So far, no factors involved in the processing of mammalian intronic snoRNAs have been identified. Recent studies revealed that assembly of most intronic box C/D snoRNPs is facilitated by the splicing machinery, and it occurs during a late step of splicing (27, 28). The 15.5-kDa protein, the first binding box C/D protein (60), binds to the box C/D core motif in a splicing-dependent manner at the C1 complex stage (27). Most likely, some splicing factor(s) recruits and deposits the 15.5-kDa protein on the intronic snoRNA that, as confirmed by mutational analysis, is located optimally about 70 nucleotides (nt) upstream of the 3′ splice site (3′ss) (28). Consistently, the majority of mammalian intronic box C/D snoRNAs are positioned about 60 to 90 nt upstream of the 3′ splice site of the host intron (28). In a few cases, suboptimal intronic location is compensated for by a stable external intronic stem which facilitates the correct folding of the snoRNA and thereby stimulates binding of the 15.5-kDa core protein (13, 27, 58).
Here we have investigated the relationship between processing of human intron-encoded box H/ACA RNAs and splicing of host pre-mRNAs. We provide evidence that recognition of box H/ACA intronic snoRNA sequences and selection of pre-mRNA splice sites are independent processes, although they both occur on the newly synthesized nascent pre-mRNA and both are dependent on RNA polymerase II (pol II) transcription.
MATERIALS AND METHODS
Plasmid construction.
All expression constructs were generated by PCR amplification, using standard reaction conditions and appropriate oligodeoxynucleotide primers (sequences are available upon request). In brief, to generate pCMV-RPS2, part of the human ribosomal protein S2 gene containing exons 3 and 4 was PCR amplified from human HeLa genomic DNA, and the resulting fragment was inserted into the HindIII and EcoRI sites of expression vector pCMV-globin (12). The pCMV-RPS2Δ10, pCMV-RPS2+21, and pCMV-RPS2+57 constructs were generated by the megaprimer amplification approach (14), using pCMV-RPS2 as a template and appropriately designed mutagenic oligonucleotides as PCR primers. To obtain pCMV-globin-a, pCMV-globin-b, and pCMV-globin-c expression vectors, ClaI and XhoI restriction sites were introduced into the second intron of the human β-globin gene 70 bp (globin-a) and 454 bp (globin-b) downstream of exon 2 or 70 bp (globin-c) upstream of exon 3 by using a two-step PCR approach essentially as described earlier (36). The HindIII-EcoRI fragments of the resulting mutant globin-a, globin-b, and globin-c genes, encompassing the first and second exons and the 5′ half of the third exon, were used to replace the HindIII-EcoRI fragment of pCMV-globin (12). The coding regions of human U64, U17, U19, and U92 box H/ACA RNAs with (U17f, U19f, and U64f) or without (U17, U19, U64, and U92) their natural flanking sequences were amplified from HeLa genomic DNA and inserted into the ClaI-XhoI cloning sites of pCMV-globin-a, pCMV-globin-b, and pCMV-globin-c. Site-directed mutagenesis of pCMV-globin-b-U92 to obtain U92m1, U92m2, U92m3, and U92m4 constructs was performed by the megaprimer amplification approach (14). Construction of p7SK expression vector carrying the human 7SK snRNA gene with its promoter (224 bp) and terminator (5 bp) regions has been reported previously (18). p7SK-U64, generated by PCR, carried the promoter (224 bp) and the first 6 bp of the coding region of the 7SK snRNA, the human U64 gene flanked by 28-bp upstream and 48-bp downstream sequences fused to the terminator of the 7SK gene. To obtain pW-U64, the U64 gene with its 28- and 48-bp flanking regions was inserted into the XbaI and XhoI sites of pW (24). The identity of all constructs was verified by sequence analysis. Transfection of simian COS7 was performed with FuGENE 6 transfection reagent (Roche) according to the instructions of the manufacturer. Mouse L929 cells were transfected by the DEAE-dextran method (52).
RNA analysis.
RNA from HeLa cells and transfected COS7 and L929 cells was isolated by the guanidine thiocyanate/phenol-chloroform extraction method 48 h after transfection (38). RNase A/T1 protection analysis was performed as described previously (23). To generate sequence-specific antisense RNA probes, the appropriate pCMV-globin expression plasmids were linearized with HindIII and used as templates for in vitro transcription by SP6 RNA polymerase in the presence of [α-32P]CTP (specific activity, 30 Ci/mmol) as a label. To obtain 7SK and 7SK-U64 RNA probes, the p7SK and p7SK-U64 plasmids were linearized with PstI and transcribed with T7 RNA polymerase. To generate an antisense probe for W-U64, the PstI-EcoRI fragment of pW-U64 encompassing the full-length ribosomal minigene was cloned into the same sites of pBluescript (KS), linearized with PstI, and transcribed by T3 RNA polymerase. Before utilization, all probes were purified on a 6% polyacrylamide sequencing gel.
To prepare cellular extract, about 2 × 107 mouse L929 cells were collected in NET2 buffer containing 50 mM Tris-HCl (pH 7.4), 250 mM NaCl, and 0.05% Nonidet NP-40 and sonicated three times for 30 s each with 30-s intervals at a setting of 300 W using a Branson B15 sonifier. The sonicate was centrifuged for 10 min at 10,000 × g and precleared by incubation with protein A-agarose (Sigma) for 1 h. The precleared supernatant was used for immunoprecipitation with anti-human NAP57 (kindly provided by U. T. Meier, Albert Einstein College of Medicine, Bronx, N.Y.), anti-human GAR1 (17), and antifibrillarin (72B9) (55) antibodies bound to protein A-agarose. Before mixing with nuclear extracts, the agarose beads were blocked by incubation for 90 min in NET2 supplemented with 0.2 mg/ml salmon sperm DNA and 0.5 mg/ml bovine serum albumin. After immunoprecipitation, beads were washed five times in 1 ml of NET2, and RNA was recovered by proteinase K treatment and phenol-chloroform extraction followed by ethanol precipitation. For Northern blot analysis, RNAs were separated on a 6% denaturing polyacrylamide gel, electroblotted onto a Hybond-N nylon membrane (Amersham Biosciences), and probed with terminally labeled U17-specific (5′-TTCCTGCATGGTTTGTCTCC-3′) and U75-specific (5′-GTCCACTACTCTCATACCATCATGGGCTT-3′) deoxyoligonucleotide probes. Reverse transcription-PCR (RT-PCR) was performed as reported previously (57). To detect wild-type RPS2 (5′-CAACAAATGCCTGCGAAAAG-3′) and mutant RPS2-3′ss (5′-CAACAAATGCGGGCGAAAAG-3′) pre-mRNAs, we utilized sequence-specific 3′ primers in combination with a common 5′ primer (5′-CAGGTTCAAGGTACCCGGCT-3′). The amplified DNA products were analyzed on 2% agarose gels.
Florescent in situ hybridization and image acquisition and processing.
In situ hybridization of COS7 and mouse L929 cells with a fluorescent oligonucleotide probe (TT*ACGAAAGTCACACGGGT*GAAGCCAAGTGCAACT*T; asterisks indicate aminoallyl-T residues) specific for the human U64 snoRNAs was performed as described earlier (reference 12 and http://singerlab.aecom.yu.edu). The oligonucleotide probe was labeled with FluoroLink Cy3 monofunctional reactive dye (Amersham). Fibrillarin was detected by monoclonal antifibrillarin antibody (72B9; 1/200 dilution) (50), followed by incubation with anti-mouse antibodies conjugated to fluorescein (1/100 dilution; Sigma). Nuclear DNA was stained by 0.1 μg/ml DAPI (4′,6′-diamidino-2-phenylindole).
RESULTS
Human intronic box H/ACA RNAs possess no preferential localization close to the 3′ splice site.
The majority of mammalian intron-encoded box C/D snoRNA genes are positioned about 60 to 90 bp from the 3′ splice site of the host intron (28). Recent identification of a large number of human box H/ACA snoRNAs and scaRNAs enabled us to perform a comprehensive analysis of the genomic localization pattern of human box H/ACA intronic RNA genes (33). The distances between box H/ACA RNA genes and the 5′ and 3′ splice sites of their host introns were defined by alignment of genomic sequences with cDNA sequences of known cognate mRNAs or reported expressed sequence tags.
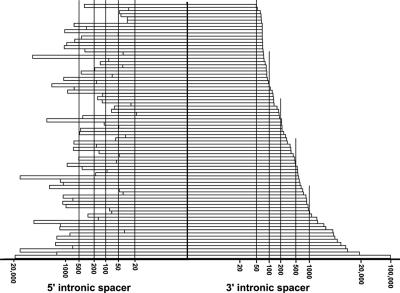
A plot of length distribution of the upstream and downstream intronic spacer sequences separating 80 human intronic box H/ACA RNA genes from the neighboring 5′ and 3′ splice sites is shown in Fig. 1. Apparently, the lengths of human pre-mRNA introns hosting box H/ACA RNAs show a great variation between 218 and about 120,000 nucleotides, indicating that box H/ACA RNAs can be efficiently processed from both short and long introns. More importantly, the majority of intronic box H/ACA RNA genes are separated from the adjacent 3′ splice site by more than a hundred or, sometimes, even by several thousand base pairs of intronic sequences. The upstream intronic spacer sequences of human box H/ACA RNA genes also show a broad length distribution, ranging from 19 to almost 20,000 bp. Hence, we conclude that human intronic box H/ACA RNA genes, in contrast to box C/D snoRNA genes, possess no preferential localization close to the 3′ or 5′ splice site of the host intron.
FIG. 1.
Distribution of the length of intronic spacer sequences separating 80 human intron-encoded box H/ACA RNAs from adjacent 5′ and 3′ splice sites. The numbers of upstream and downstream intronic flanking nucleotides are indicated on logarithmic scales.
Alteration of the downstream intronic spacer length has no effect on U64 snoRNA processing.
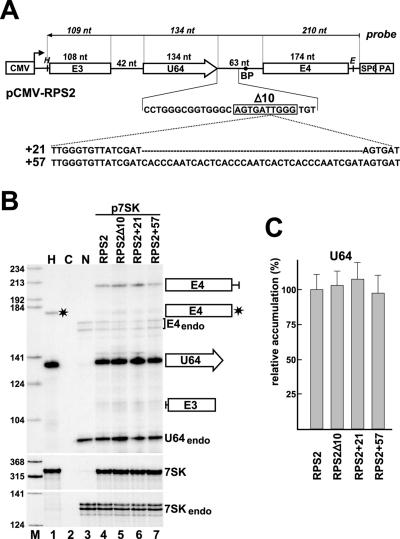
An optimal position of human intronic box C/D snoRNAs relative to the branch point is crucial for efficient snoRNA processing, because the splicing machinery stimulates the recruitment of box C/D core proteins (27, 28). The finding that human intronic box H/ACA RNA genes are randomly positioned within their host introns suggests that there is no synergy between H/ACA RNA processing and host pre-mRNA splicing. To test this assumption, we first investigated the in vivo processing of the human U64 intronic H/ACA snoRNA (Fig. 2). The U64 gene has an optimal intronic position defined for efficient processing of box C/D intronic snoRNAs; it is located 63 bp upstream of the 3′ splice site of the third intron of ribosomal protein S2 (RPS2) gene (21). A fragment of the RPS2 gene encompassing the host intron of U64 together with flanking exons E3 and E4 was placed under the control of the cytomegalovirus (CMV) promoter (Fig. 2A). To assay the potential importance of the intronic position of U64, the downstream spacer region separating U64 from the branch point was either shortened by deletion of 10 bp or elongated by insertion of 21- and 57-bp DNA fragments.
FIG. 2.
In vivo processing of human U64 box H/ACA snoRNA from the third intron of ribosomal protein S2 pre-mRNA. (A) Schematic structure of expression constructs. A fragment of the human ribosomal protein S2 gene encompassing exons 3 and 4 (E3 and E4) and the third intron carrying the U64 gene (open arrow) was fused to the cytomegalovirus promoter (CMV). The polyadenylation region (PA) and the promoter of the SP6 RNA polymerase are indicated. Relevant restriction sites are shown (H, HindIII; E, EcoRI). The antisense RNA probe used for RNase A/T1 mapping and the expected sizes of protected fragments (in italics) are shown. The downstream intronic flanking sequences of U64 as well as sequences deleted from RPS2-Δ10 (boxed) or inserted into the RPS2+21 and RPS2+57 genes are indicated. (B) RNase A/T1 protection. RNAs obtained from mouse L929 transfected with the indicated expression constructs were mapped with a sequence-specific antisense RNA probe, and the protected fragments were separated on a 6% sequencing gel. Control mappings with HeLa (H) and nontransfected L929 cellular RNA (N) and with Escherichia coli tRNA (C) are shown. Positions and structures of protected fragments are indicated on the right. Asterisks indicate a probe fragment protected by the fourth exon of the human HeLa RPS2 mRNA. E4endo, U64endo, and 7SKendo fragments represent endogenous mouse RPS2 exon 4, U64 snoRNA, and 7SK snRNA sequences that partially protect human antisense RNA probes. Lane M, molecular size markers (HaeIII- and TaqI-digested pBR322). (C) Relative accumulation of U64 snoRNA. The intensities of protected probe fragments were quantified by PhosphorImager. The relative levels of U64 were normalized to the transiently expressed 7SK snRNA.
The resulting pCMV-RPS2, pCMV-RPS2-Δ10, pCMV-RPS2+21, and pCMV-RPS2+57 expression constructs, together with the p7SK transfection control plasmid carrying the human 7SK snRNA gene, were transfected into mouse L929 cells. Excision of U64 snoRNA, splicing of the “wild-type” and mutant RPS2 pre-mRNAs, and expression of 7SK snRNA were monitored by RNase A/T1 mapping performed with sequence-specific antisense RNA probes (Fig. 2B). The U64 snoRNA was efficiently processed from all RPS2 transcripts (lanes 4, 5, 6, and 7), yielding an RNA identical in size to the authentic human U64 snoRNA (lane 1). The relative intensities of the U64-specific protected fragments obtained by PhosphorImager quantification and normalized to the 7SK external control are shown in Fig. 2C. No significant differences were observed in the accumulation levels of U64 when processed from the “wild-type” RPS2 (considered 100%) or from the mutant RPS2-Δ10, RPS2+21, and RPS2+57 primary transcripts. These results support the idea that efficient processing of human intron-encoded box H/ACA snoRNAs, in contrast to that of box C/D snoRNAs, is independent from the position of the snoRNA relative to the 3′ splice site.
Position-independent processing of U64, U19, and U17 H/ACA snoRNAs from the globin pre-mRNA.
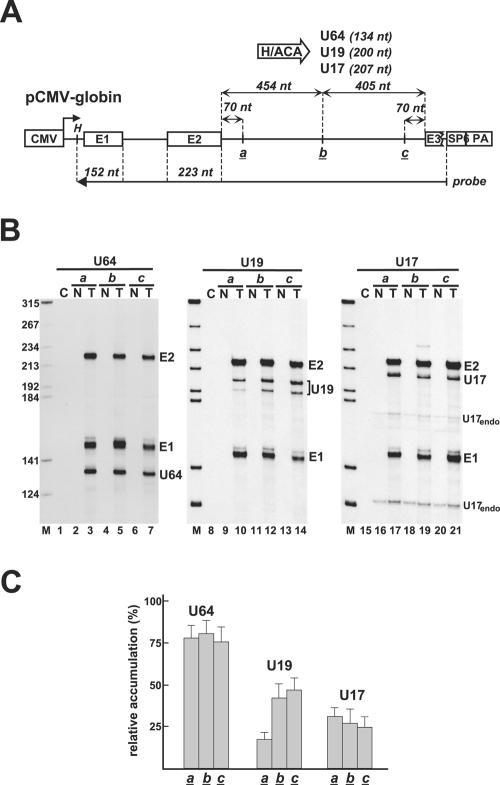
To further substantiate the notion that the position of intronic H/ACA snoRNAs has no effect on snoRNA expression, we investigated the in vivo processing of human U64, U17, and U19 box H/ACA snoRNAs imbedded in three different positions of the second intron of human β-globin pre-mRNA (Fig. 3A). The coding genes of U64, U17, and U19 devoid of flanking sequences were inserted either 70 bp downstream of the 5′ splice site (site a), 70 bp upstream of the 3′ splice site (site c), or in the middle of the second intron (site b) of the human β-globin gene that had been placed under the control of the CMV promoter. Each expression construct was transfected into COS7 cells, and processing of the intronic snoRNAs and splicing of the host globin pre-mRNAs were monitored by RNase A/T1 mapping (Fig. 3B). All globin transcripts carrying U64, U19, and U17 sequences were correctly and efficiently spliced, indicating that insertion of H/ACA intronic snoRNAs has no effect on pre-mRNA splicing. Likewise, the U64 and U17 snoRNAs located at the a, b, or c site of the second intron of the transiently expressed globin pre-mRNA were processed with similar efficiency (lanes 3, 5, 7, 17, 19, and 21). This conclusion was corroborated by PhosphorImager quantification of the protected probe fragments representing the excised snoRNAs and the spliced globin introns (Fig. 3C).
FIG. 3.
Processing of U64, U19, and U17 box H/ACA snoRNAs from the second intron of the human β-globin pre-mRNA. (A) Schematic structure of expression constructs. Exons (E1, E2, and E3) of the globin gene and the insertion sites of U64, U19, and U17 snoRNA genes (a, b, and c) are indicated. (B) RNase A/T1 mapping. RNAs from COS7 cells either nontransfected (N) or transfected (T) with the indicated expression vector were mapped with sequence-specific RNA probes. Protected probe fragments representing the spliced exons (E1 and E2) of the transiently expressed human globin mRNA and the excised U64, U19, and U17 snoRNAs are indicated. Fragments U17endo represent endogenous mouse U17 snoRNA partially protecting the human U17 snoRNA probe. (C) Accumulation of U64, U19, and U17 snoRNAs. The relative levels of U64, U19, and U17 were normalized to the levels of exons 1 and 2. For other details, see the legend to Fig. 2.
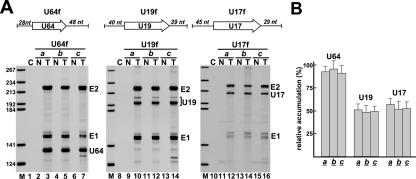
The U19 snoRNA, however, showed a reduced accumulation when its gene had been placed close to the 5′ splice site (site a) of the globin host intron (Fig. 3B, lane 10, and C). Since the globin mRNA was faithfully and efficiently expressed, we concluded that processing of the U19 snoRNA, rather than synthesis of the globin-a-U19 pre-mRNA, was compromised. We speculated that the intronic sequences flanking the a site of the globin-a pre-mRNA may accidentally interfere with correct folding of U19 and, thereby, may inhibit binding of box H/ACA RNP proteins. Previous works demonstrated that adjacent intronic sequences can facilitate the processing of box C/D intronic snoRNAs (13, 27, 58). To test this possibility, the U19, U64, and U17 snoRNA genes, together with short natural flanking sequences (U64f, U19f, and U17f), were inserted into the a, b, and c sites of pCMV-globin (Fig. 4A). Each expression construct was transfected into COS7 cells, and processing of the transiently expressed pre-mRNAs was analyzed by RNase A/T1 mapping. The presence of natural intronic flanking sequences fully restored the processing of U19 from the a site of the globin-a-U19f transcript (lane 10). Moreover, compared to the expression levels of U64, U19, and U17 snoRNAs devoid of natural flanking nucleotides (Fig. 3C), inclusion of cognate intronic flanking sequences significantly increased the efficacy of U19, U64, and U17 processing from the a, b, and c sites of transiently expressed globin pre-mRNAs (Fig. 4B). Based on the above results, we conclude that the nucleotide composition of the immediate intronic flanking regions can largely facilitate the processing of human H/ACA snoRNAs. However, in contrast to box C/D snoRNAs, efficient processing of H/ACA snoRNAs does not require a preferential intronic localization relative to the 5′ and 3′ splice sites of the host intron.
FIG. 4.
Processing of U64, U19, and U17 box H/ACA snoRNAs flanked with natural intronic sequences. (A) RNase A/T1 mapping. The U64, U19, and U17 snoRNAs genes together with their natural flanking sequences (U64f, U19f, and U17f) were inserted into the a, b, and c sites of pCMV-globin. RNAs obtained from COS7 cells nontransfected (N) or transfected (T) with the indicated expression construct were mapped with cRNA probes. (B) Relative accumulation of U64, U19, and U17 snoRNAs. For other details, see the legend to Fig. 3.
Recognition of U92 H/ACA intronic RNA and selection of the host pre-mRNA splice sites occur concomitantly.
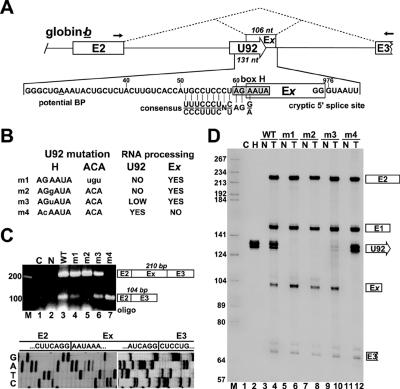
A notion that processing of box H/ACA intronic RNAs is independent from their intronic location implies that recognition of box H/ACA signal sequences by H/ACA RNP proteins and selection of the host pre-mRNA splice sites by the splicing machinery are independent molecular events. Therefore, in principle, assembly of H/ACA RNPs could occur before, during, or after splicing of the host pre-mRNA. To define the timing of intronic box H/ACA RNA selection, we took advantage of two observations. We had earlier noticed that the second intron of the human globin gene contains a cryptic 5′ splice site at G976 that can be activated upon providing a 3′ splicing signal sequence (T. Kiss, unpublished data) (Fig. 5A). A scrutiny of human box H/ACA RNA sequences revealed that the H-box motif of human U92 scaRNA and its preceding nucleotides show a striking homology to the consensus sequence at the 3′ splice sites of human U2-type introns (Fig. 5A). In other words, the H-box region of U92 has the capacity to bind H/ACA RNP proteins and also to recruit splicing factors. In theory, when located upstream of the G976 cryptic 5′ splice site of the globin pre-mRNA, the U92 RNA might be either processed to mature U92 scaRNA or, alternatively, its 3′ half could be spliced to the second exon (E2) of globin as part of a newly included exon (Ex) depending on whether box H/ACA RNA recognition or splice site selection occurs first.
FIG. 5.
Processing of globin-b pre-mRNA carrying the U92 box H/ACA scaRNA. (A) Schematic structure of the 3′-terminal part of the globin-b-U92 transcript. Dashed lines indicate alternative splicing patterns. Position of exon x (Ex) (boxed) and sequences at the cryptic 5′ and 3′ splice sites are shown. A potential branch point (BP) is underlined, and the box H motif of U92 is shaded. The consensus sequence at human U2-type 3′ splice sites is shown (8). Arrows represent primers used for RT-PCR. Numbering is from the 5′ end of mature U92 RNA or from the transcription initiation site of the human β-globin gene. (B) Mutations introduced into the H or ACA box of U92. Altered nucleotides are shown by lowercase letters, and the expected consequences on U92 excision and Ex exon inclusion are listed. (C) Analysis of RNA processing by RT-PCR. RNAs obtained from mouse L929 cells nontransfected (N) or transfected with the pCMV-globin-b expression construct carrying the wild-type (WT) or mutant (m1 to m4) U92 RNA genes were analyzed by RT-PCR. Lane C represents control RT-PCR performed with Escherichia coli tRNA. The amplified fragments were separated on a 2% agarose gel, cloned, and subjected to sequence analysis. Sequencing gels representing the E2-Ex and Ex-E3 junction regions are shown. (D) RNase A/T1 mapping. RNAs from mouse cells expressing (T) globin-b pre-mRNAs carrying wild-type (WT) or mutant (m1 to m4) U92 RNAs were mapped with sequence-specific RNA probes spanning the entire globin primary transcripts (see Fig. 3A). Control mappings with nontransfected mouse (N) or HeLa (H) cellular RNAs or E. coli tRNA (C) are shown.
To test this assumption, the human U92 gene was inserted into the b site of pCMV-globin-b upstream of the G976 cryptic 5′ splice site. Upon transient expression in mouse L929 cells, processing of the globin-b-U92 pre-mRNA was assayed by RT-PCR (Fig. 5C). By using primers specific for the second and third exon of the globin pre-mRNA, two amplified DNA fragments of about 100 and 210 bp were obtained (lane 3). Cloning and sequencing of the amplified DNAs revealed that the shorter fragment corresponded to the correctly spliced globin mRNA, while the longer product represented an alternatively spliced version of the globin mRNA in which a 106-nt-long extra exon, Ex, was inserted between E2 and E3. As predicted, the “box H” cryptic 3′ splice site of U92 and the intronic G976 cryptic 5′ splice site were utilized for inclusion of Ex (Fig. 5C, lower panel). RNase A/T1 mapping confirmed the inclusion of Ex into the globin mRNA and also revealed that the U92 scaRNA was also processed from a fraction of the transiently expressed globin-b-U92 RNA (Fig. 5D, lane 4). These results demonstrate that the box H region of U92 in the context of the globin-b pre-mRNA sequence can function as a 3′ splicing acceptor site. On the other hand, the observed coaccumulation of mature U92 scaRNA and the alternatively spliced globin mRNA suggested that binding of H/ACA RNP proteins and recruitment of splicing factors to the box H region of U92 are parallel and, most probably, competitive molecular events.
To confirm the above conclusion and to further investigate the relationship of U92 processing and globin pre-mRNA splicing, mutations were introduced into the H or ACA motif of U92 (Fig. 5B). Previous functional analysis of the H box (consensus AnAnnA) of human H/ACA snoRNAs revealed that replacement of the evolutionarily conserved A3 residue for a G is fatal to snoRNA accumulation (21). Consistent with this, an A3-to-G3 transition fully abolished the processing of the mutant U92-m2 RNA from the globin-b pre-mRNA (Fig. 5D, lane 8). This mutation also abolished accumulation of the wild-type globin mRNA and only the alternatively spliced, Ex-containing longer globin mRNA was detectable (Fig. 5C, lane 5). This demonstrates that recognition of the “box H” cryptic 3′ splice site and inclusion of Ex occurs with 100% efficacy if box H/ACA RNP proteins cannot bind to the H box of U92. As was highly predictable, replacement of the G2 residue for a C fully devastated accumulation of the alternatively spliced version of globin mRNA (Fig. 5C, lane 7; see also D, lane 12). More importantly, abolishment of Ex inclusion doubled the accumulation of the mutant U92-m4 snoRNA compared to the wild-type snoRNA (compare lanes 4 and 12). This provides further support to the notion that splice site selection and H/ACA snoRNA recognition are competitive events.
In great accordance with previous results (21), conversion of the A3 residue in the H box of U92 to a pyrimidine (U) strongly inhibited, but did not fully abolish, accumulation of U92-m3, and alteration of the ACA box of U92 fully abolished processing of U92-m1 (Fig. 5D, lanes 6 and 10). However, to some surprise, processing of the wild-type globin mRNA was not severely inhibited or fully abolished by the U92-m3 and U92-m1 mutations, respectively (Fig. 5C, lanes 4 and 6). A simple explanation for this observation is that box H/ACA RNP proteins still can loosely bind to the structural core motif of U92-m1 and U92-m3, even in the absence of fully functional H or ACA box motifs, and they partially prevent the recruitment of splicing factors. Nevertheless, the above results demonstrate that recognition of the U92 box H/ACA intronic RNA and reading of splicing signals in the globin-b-U92 primary transcript are competitive events. Since splicing of pre-mRNAs already occurs during transcription elongation (40), we conclude that recognition of box H/ACA intronic RNAs is a cotranscriptional event that occurs on the nascent host pre-mRNA parallel with spliceosome assembly.
Dyskerin interacts with pre-mRNA transcripts hosting U64 intronic snoRNA.
We next investigated whether H/ACA snoRNP proteins could interact with nascent pre-mRNA transcripts hosting a box H/ACA intronic snoRNA. Mouse L929 cells were transfected with the pCMV-RPS2 expression construct (see Fig. 2A). Cellular extracts were prepared and dyskerin (Nap57) and Gar1 H/ACA proteins were precipitated with anti-NAP57 and anti-GAR1 antibodies, respectively (Fig. 6). Coimmunoprecipitation of nascent RPS2 pre-mRNA transcripts was monitored with RT-PCR by utilization of PCR primers that were specific for unspliced pre-mRNA. Precipitation of endogenous H/ACA snoRNPs was verified by Northern blot analysis performed with a deoxyoligonucleotide probe specific for mouse U17 H/ACA snoRNA. The anti-NAP57 antibody precipitated mouse U17 as well as the transiently expressed RPS2 pre-mRNA (lane 4). The anti-GAR1 antibody failed to precipitate the RPS2 pre-mRNA (lane 5), although it recognized U17 snoRNP. Finally, a control antibody specific for fibrillarin box C/D snoRNP protein did not recognize U17 and RPS2 but precipitated the mouse U75 box C/D snoRNA (lane 3). These results demonstrate that dyskerin, an essential central component of box H/ACA snoRNPs (25, 59), binds to intronic box H/ACA snoRNAs already in the newly synthesized host pre-mRNA. We also propose that Gar1, another H/ACA snoRNP protein that is dispensable for snoRNA stability, binds at a later stage of intronic H/ACA snoRNP biogenesis (3, 11, 59).
FIG. 6.
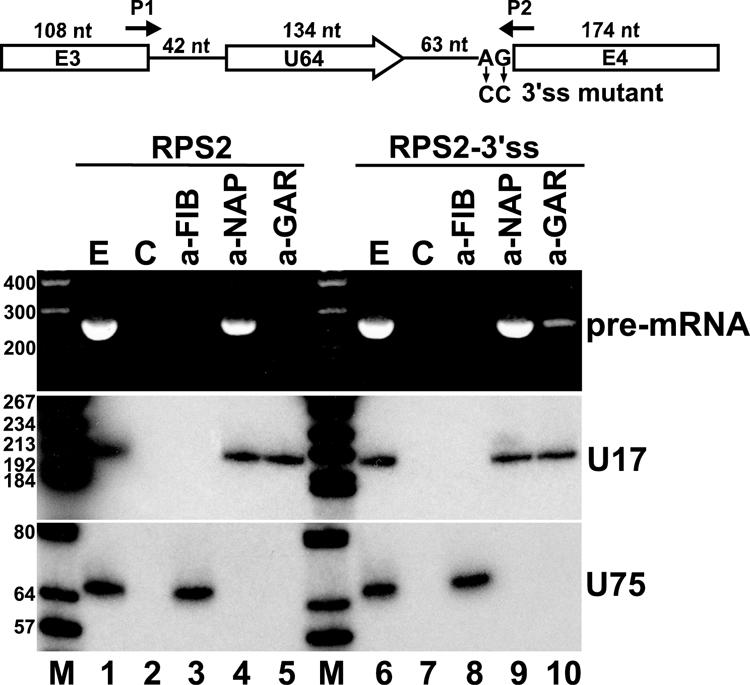
Coimmunoprecipitation of RPS2 pre-mRNA with dyskerin. A schematic structure of transiently expressed human RPS2 pre-mRNA hosting U64 snoRNA is shown. Altered nucleotides in RPS2-3′ss mutant pre-mRNA are shown. Arrows P1 and P2 represent deoxyoligonucleotide primers used for PCRs. For other details, see the legend to Fig. 2A. Cellular extracts prepared from mouse L929 cells transfected with pCMV-RPS2 or pCMV-RPS2-3′ss were treated with protein A-agarose-bound antifibrillarin (a-FIB), antidyskerin (a-NAP), or anti-GAR1 (a-GAR) antibody. RNAs recovered from the extracts (E) and protein A agarose beads either containing or lacking (C) antibodies were analyzed by RT-PCR (upper panel) or Northern blot analysis (lower panels). The amplified RT-PCR fragments were analyzed on 2% agarose gel. For Northern blot analysis, RNAs were separated on a 6% sequencing gel, electrotransferred onto a nylon membrane, and probed with terminally labeled U17- and U75-specific deoxyoligonucleotides. Lanes M represent size markers in base pairs (upper panel) or in nucleotides (lower panels).
RNase protection experiments demonstrated that exons 3 and 4 of the transiently expressed RPS2 pre-mRNA were correctly spliced in mouse cells (Fig. 2B and data not shown). To abolish splicing of RPS2 and to prevent binding of splicing factors, we destroyed the 3′ splice site signal sequence in the pCMV-RPS2 expression construct (Fig. 6). Upon transient expression in mouse cells, as predicted, the mutant RPS2-3′ss pre-mRNA was not spliced (data not shown). Immunoprecipitation with anti-NAP57 followed by RT-PCR analysis, however, revealed that dyskerin was successfully recruited by the unspliced RPS2-3ss pre-mRNA (lane 9), providing further support to the conclusion that recognition of intronic box H/ACA snoRNAs and binding of H/ACA core proteins does not depend on spliceosome assembly. Interestingly, in contrast to the wild-type RPS2 transcript (lane 5), the mutant RPS2-3′ss pre-mRNA showed a weak but reproducible interaction with Gar1 protein (lane 10). In fact, the RPS2-3′ss transcript was processed to mature U64 snoRNA with very low efficiency (data not shown), indicating that efficient splicing of host pre-mRNA, although not essential for H/ACA snoRNP assembly, facilitates snoRNA production through providing entry sites for processing exonucleases (36). In summary, the above results further corroborated the conclusion that box H/ACA central core proteins are recruited to the newly synthesized host pre-mRNA in a splicing-independent manner shortly after or already during transcription elongation.
Correct expression of U64 snoRNP depends on polymerase II transcription.
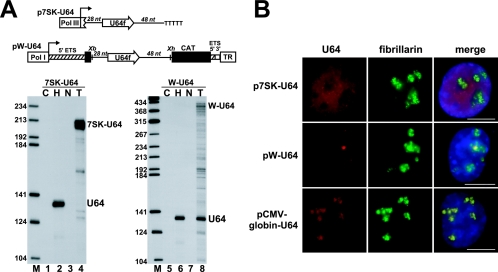
After having demonstrated that selection of intronic H/ACA RNA sequences is independent from pre-mRNA splicing, we tested whether H/ACA RNAs can be processed from precursor RNAs synthesized by RNA pol I and pol III. The U64f fragment carrying the human U64 snoRNA gene with 28 bp upstream and 48 bp downstream flanking sequences was inserted into the pW mouse ribosomal minigene construct (24) or fused to the promoter and terminator regions of the RNA pol III-specific 7SK snRNA gene (39, 47) (Fig. 7A). The pW-U64 and p7SK-U64 expression constructs were transfected into mouse L929 and COS7 cells, respectively, and processing of U64 was assayed by RNase mapping. The RNA pol III-synthesized 7SK-U64 transcript stably accumulated in COS7 cells, but it was not processed into mature U64 snoRNA, indicating that pol III transcripts cannot serve as substrates for box H/ACA snoRNA processing (lane 4). Fluorescent in situ hybridization with a human U64-specific oligonucleotide probe revealed that the transiently expressed 7SK-U64 RNA accumulated in the nucleoplasm, and it was rather excluded from the nucleolus where mature U64 was expected to accumulate (Fig. 6B).
FIG. 7.
Correct expression of U64 snoRNP depends on pol II transcription. (A) Processing of U64 RNA from pol I and pol III transcripts. Schematic structures of the pol III-specific p7SK-U64 and the pol I-specific pW-U64 expression constructs are shown. The human intronic U64 snoRNA gene, together with 28-bp upstream and 48-bp downstream flanking sequences, was fused to the promoter (Pol III) and terminator (TTTTT) of the human 7SK snRNA gene or inserted into the XbaI (Xb) and XhoI (Xh) sites of the pW ribosomal minigene (24). The mouse pol I promoter (Pol I) and terminator (TR), fragments derived from the 5′ (hatched boxes) and 3′ (open box) external transcribed spacers (ETS) of the mouse rRNA gene, and a fragment of the chloramphenicol acetyltransferase (CAT) gene are shown. RNAs extracted from HeLa (H) cells, transfected (T) or nontransfected (N) COS7 cells (lanes 3 and 4), or mouse L929 (lanes 7 and 8) cells were analyzed by RNase A/T1 mapping with sequence-specific probes as indicated above the lanes. Lane C represents control mapping with Escherichia coli tRNA. (B) In situ localization. COS7 cells transfected with the pCMV-globin-U64 or p7SK-U64 and mouse L929 transfected with the pW-U64 expression constructs were hybridized with a fluorescent oligonucleotide probe specific for human U64. The nucleolus was visualized by staining with an anti-fibrillarin antibody. Nuclear DNA was stained with DAPI (blue). Bar, 10 μm.
RNase A/T1 mapping revealed that U64 was correctly, although with low efficiency, processed from the W-U64 transcript (Fig. 7A, lane 8). Since pol I-mediated synthesis of the W-U64 RNA was expected to occur in the nucleolus (22, 24, 46), it was not surprising that the mature U64 snoRNA and the unprocessed or partially processed W-U64 RNAs accumulated in the nucleolus (Fig. 7B). Immunoprecipitation with an antibody against Gar1p, a late binding H/ACA protein (3, 11, 59), demonstrated that the excised U64 snoRNA was assembled into H/ACA snoRNP (data not shown). However, in situ hybridization also revealed that the U64 snoRNA processed from the W-U64 pol I transcript in the nucleolus had an aberrant localization pattern concentrated in small dot-like regions of the nucleolus (Fig. 7B). In contrast, the control U64 snoRNA excised from the second intron of the RNA pol II-synthesized globin pre-mRNA showed the characteristic distribution of snoRNAs: it coaccumulated with fibrillarin in the fibrillar compartment of the nucleolus (Fig. 7B). We assume that in the nucleolus, a fraction of the W-U64 pol I transcript and H/ACA RNP proteins can assemble into precursor snoRNP (3, 11, 59) before nucleases, maybe the ones responsible for regular processing of intronic snoRNAs, degrade the flanking sequences. However, default processing from a nucleolar pol I transcript cannot support correct intranucleolar localization of box H/ACA snoRNPs. Hence, we conclude that efficient and correct expression of human box H/ACA snoRNPs requires cotranscription with RNA pol II-synthesized pre-mRNAs.
DISCUSSION
In mammalian cells, a large number of small regulatory RNAs, including box C/D and H/ACA modification guide RNAs and many microRNAs, are processed from pre-mRNA introns. The functional relationship between processing of intron-encoded RNAs and splicing of host pre-mRNAs is poorly understood. Previous works revealed that removal and debranching of the host introns provide the entry sites for exonucleases responsible for processing the majority of intronic box C/D and H/ACA snoRNAs (36, 48). Recently, a more intimate role of the spliceosome played in the assembly and processing of intron-encoded box C/D snoRNAs has been discovered (27, 28). In vitro processing experiments demonstrated that recruitment of the 15.5-kDa core protein to the K-turn motif of box C/D snoRNAs is dependent on splicing factors specific to the C1 complex, and consequently a proper location relative to the branch point is essential for efficient snoRNA processing. In this study, we have investigated the in vivo processing of box H/ACA RNAs that represent another major class of human intronic ncRNAs. Several lines of evidence support the conclusion that assembly of box H/ACA snoRNPs, in contrast to that of box C/D snoRNPs, is independent of pre-mRNA splicing and that the sole function of the splicing machinery is in liberation of the box H/ACA RNA-containing host intron for exonucleolytic processing.
In contrast to intronic box C/D snoRNA genes which are typically located 60 to 90 bp upstream of the 3′ splice site (28), the human H/ACA RNA genes show no preferential localization close to the 3′ end of their host introns (Fig. 1) (51). While an average human intron is about 3,400 bp (41), we found that the average length of box H/ACA RNA-containing introns is about 7,400 bp. The functional significance of this observation, if any, remains uncertain, especially since some box H/ACA RNAs are processed from very short introns. Nevertheless, the findings that human box H/ACA RNA genes preferentially reside within longer than average introns and that they localize randomly relative to the ends of the host intron strongly support the notion that recognition of intronic box H/ACA RNAs is independent of binding of the splicing machinery. Indeed, in vivo processing experiments demonstrated that expression of the human U64 box H/ACA snoRNA is not affected by its intronic position relative to the branch point (Fig. 2) and that the U17, U19, and U64 H/ACA snoRNAs flanked with short natural sequences are equally efficiently processed from the 5′-terminal, the middle, or the 3′-terminal region of the second intron of the human β-globin pre-mRNA (Fig. 4). Overall, these results strongly argue against a synergy between human box H/ACA RNA processing and pre-mRNA splicing.
In vivo processing of U17, U19, and U64 snoRNAs from the globin pre-mRNA also revealed that natural intronic flanking sequences can stimulate the efficient processing of box H/ACA RNAs (Fig. 3 and 4). Previous studies showed that formation of external intronic stem structures can facilitate the expression of box C/D snoRNAs, probably through safeguarding the accurate folding of the K-turn core motif (13, 58). The role of intronic flanking sequences played in H/ACA RNA processing is unclear. Formation of an external stem is evident for only a few H/ACA intronic RNAs, but in most cases, computer folding failed to reveal an apparent intronic stem or other common RNA structure (unpublished data). We assume that the naturally occurring intronic flanking nucleotides represent harmless sequences which do not interfere with the correct folding of the imbedded intronic H/ACA RNA. For example, intronic sequences capable of forming base-pairing interactions with snoRNA sequences would hamper snoRNA folding and recognition by box H/ACA proteins or by other auxiliary factors involved in snoRNP assembly.
To determine exactly when selection of box H/ACA intronic RNAs occurs during pre-mRNA processing in living cells, we expressed a modified globin pre-mRNA in which the same intronic nucleotides could be recognized either as the box H motif of the U92 intronic RNA or as the 3′ splice site signal of an alternatively spliced intron (Fig. 5). Employing such an artificial snoRNA processing versus a pre-mRNA splicing competition system demonstrated that recognition of H/ACA intronic snoRNA sequences and selection of the 3′ splice site occur concurrently on the native pre-mRNA. Since all mRNA-processing reactions, including splicing, occur cotranscriptionally (reviewed in references 43 and 49), we conclude that selection of intronic box H/ACA RNAs already takes place during synthesis of the host pre-mRNA. This notion was supported by the finding that dyskerin, a central component of box H/ACA snoRNPs, specifically associates with nascent pre-mRNAs carrying the U64 H/ACA intronic snoRNA (Fig. 6). Moreover, demonstration that dyskerin can bind to a mutant U64 host pre-mRNA that lacks a functional 3′ splice site further supported the conclusion that selection of H/ACA intronic snoRNAs occurs in a splicing-independent manner.
In yeast, the Nhp2p and Cbf5p (the yeast equivalent of dyskerin) H/ACA snoRNP proteins as well as Naf1p, a trans-acting protein factor essential for yeast box H/ACA snoRNP assembly (16, 19, 65), can be cross-linked to the coding region of actively transcribed yeast H/ACA snoRNA genes (4, 64). Since Naf1p can interact with the H/ACA core proteins Cbf5p and Nhp2p as well as with the phosphorylated carboxy-terminal domain (CTD) of RNA pol II (19, 29, 31), and since it copurifies with the pol II factors Spt16p, Sub1p, and Tfg1p (64), it may play a central role in recruitment of H/ACA core proteins to RNA pol II and to the newly synthesized snoRNA (4, 19, 64). Although the majority of yeast box H/ACA snoRNAs are transcribed from independent genes, we can envisage that cotranscriptional assembly of box H/ACA snoRNPs is an evolutionarily conserved phenomenon.
Since mammalian introns are very unstable after removal, it seems reasonable to assume that in order to maximize the efficiency of intronic RNA expression, the H/ACA core proteins and putative auxiliary factors are actively recruited to the nascent host pre-mRNA. This idea is fuelled by the observation that binding of H/ACA proteins can efficiently compete with recruitment of splicing factors that are in fact actively loaded on the pre-mRNA during transcription elongation (Fig. 5). The existence of a mechanism stimulating H/ACA precursor RNP assembly on pre-mRNAs is further supported by our results that correct and efficient expression of human H/ACA RNPs is dependent on RNA pol II transcription (Fig. 6). Since Naf1p is an evolutionarily conserved protein, it might function also in the active recruitment mammalian box H/ACA RNP proteins and other auxiliary factors to nascent pre-mRNAs hosting intronic box H/ACA RNAs (19).
In conclusion, the results presented in this paper indicate that recognition of human intron-encoded box H/ACA RNA sequences occurs cotranscriptionally and in parallel with splice site selection. However, assembly of H/ACA precursor RNPs, in contrast to that of box C/D snoRNPs (27, 28), is mechanistically independent from splicing of the host pre-mRNA. Instead, efficient processing and correct nucleolar localization of human intronic box H/ACA snoRNPs depends on RNA pol II transcription, supporting the idea that factors associated with pol II facilitate the assembly of box H/ACA snoRNPs both in humans and yeast (4, 64). In the future, identification of the trans-acting factors mediating cotranscriptional deposition of box H/ACA core proteins on nascent intronic H/ACA RNA sequences will be a challenging task.
Acknowledgments
We are grateful to U. T. Meier (Albert Einstein College of Medicine, New York, N.Y.), W. Filipowicz (Friedrich Miescher Institut, Basel, Switzerland), and J. A. Steitz (Yale University School of Medicine, New Haven, Conn.) for providing us with anti-NAP57, anti-GAR1, and antifibrillarin antibodies, respectively.
P.R. and X.D. were funded by la Fondation pour la Recherche Médicale. A.M.K. was supported by a short-term EMBO fellowship, a Hungarian State Eötvös fellowship, and l’Association pour la Recherche contre le Cancer. Our work was supported by grants from la Ligue Nationale contre le Cancer and la Fondation pour la Recherche Médicale.
REFERENCES
- 1.Ambros, V. 2004. The functions of animal microRNAs. Nature 431:350-355. [DOI] [PubMed] [Google Scholar]
- 2.Bachellerie, J. P., J. Cavaillé, and A. Hüttenhofer. 2002. The expanding snoRNA world. Biochimie 84:775-790. [DOI] [PubMed] [Google Scholar]
- 3.Baker, D. L., O. A. Youssef, M. I. Chastkofsky, D. A. Dy, R. M. Terns, and M. P. Terns. 2005. RNA-guided RNA modification: functional organization of the archaeal H/ACA RNP. Genes Dev. 19:1238-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballarino, M., M. Morlando, F. Pagano, A. Fatica, and I. Bozzoni. 2005. The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 25:5396-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein, E., and C. D. Allis. 2005. RNA meets chromatin. Genes Dev. 19:1635-1655. [DOI] [PubMed] [Google Scholar]
- 7.Bortolin, M. L., P. Ganot, and T. Kiss. 1999. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J. 18:457-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burge, C., T. Tuschl, and P. Sharp. 1999. Splicing of precursors to mRNAs by the spliceosomes, p. 525-560. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N. Y.
- 9.Caffarelli, E., M. Arese, B. Santoro, P. Fragapane, and I. Bozzoni. 1994. In vitro study of processing of the intron-encoded U16 small nucleolar RNA in Xenopus laevis. Mol. Cell. Biol. 14:2966-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caffarelli, E., A. Fatica, S. Prislei, E. De Gregorio, P. Fragapane, and I. Bozzoni. 1996. Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 15:1121-1131. [PMC free article] [PubMed] [Google Scholar]
- 11.Charpentier, B., S. Muller, and C. Branlant. 2005. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res. 33:3133-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darzacq, X., B. E. Jády, C. Verheggen, A. M. Kiss, E. Bertrand, and T. Kiss. 2002. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 21:2746-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darzacq, X., and T. Kiss. 2000. Processing of intron-encoded box C/D small nucleolar RNAs lacking a 5′,3′-terminal stem structure. Mol. Cell. Biol. 20:4522-4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datta, A. K. 1995. Efficient amplification using ‘megaprimer’ by asymmetric polymerase chain reaction. Nucleic Acids Res. 23:4530-4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decatur, W. A., and M. J. Fournier. 2003. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 278:695-698. [DOI] [PubMed] [Google Scholar]
- 16.Dez, C., J. Noaillac-Depeyre, M. Caizergues-Ferrer, and Y. Henry. 2002. Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol. Cell. Biol. 22:7053-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dragon, F., V. Pogacic, and W. Filipowicz. 2000. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 20:3037-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egloff, S., E. Van Herreweghe, and T. Kiss. 2006. Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol. Cell. Biol. 26:630-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fatica, A., M. Dlakic, and D. Tollervey. 2002. Naf1p is a box H/ACA snoRNP assembly factor. RNA 8:1502-1514. [PMC free article] [PubMed] [Google Scholar]
- 20.Filipowicz, W., and V. Pogacic. 2002. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 14:319-327. [DOI] [PubMed] [Google Scholar]
- 21.Ganot, P., M. Caizergues-Ferrer, and T. Kiss. 1997. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 11:941-956. [DOI] [PubMed] [Google Scholar]
- 22.Ganot, P., B. E. Jády, M. L. Bortolin, X. Darzacq, and T. Kiss. 1999. Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol. Cell. Biol. 19:6906-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodall, G. J., K. Wiebauer, and W. Filipowicz. 1990. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 181:148-161. [DOI] [PubMed] [Google Scholar]
- 24.Hadjiolova, K. V., A. Normann, J. Cavaille, E. Soupene, S. Mazan, A. A. Hadjiolov, and J. P. Bachellerie. 1994. Processing of truncated mouse or human rRNA transcribed from ribosomal minigenes transfected into mouse cells. Mol. Cell. Biol. 14:4044-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamma, T., S. L. Reichow, G. Varani, and A. R. Ferre-D'Amare. 2005. The Cbf5-Nop10 complex is a molecular bracket that organizes box H/ACA RNPs. Nat. Struct. Mol. Biol. 12:1101-1107. [DOI] [PubMed] [Google Scholar]
- 26.Hernandez, N. 2001. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J. Biol. Chem. 276:26733-26736. [DOI] [PubMed] [Google Scholar]
- 27.Hirose, T., M. D. Shu, and J. A. Steitz. 2003. Splicing-dependent and -independent modes of assembly for intron-encoded box C/D snoRNPs in mammalian cells. Mol. Cell 12:113-123. [DOI] [PubMed] [Google Scholar]
- 28.Hirose, T., and J. A. Steitz. 2001. Position within the host intron is critical for efficient processing of box C/D snoRNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 98:12914-12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 30.Hüttenhofer, A., P. Schattner, and N. Polacek. 2005. Non-coding RNAs: hope or hype? Trends Genet. 21:289-297. [DOI] [PubMed] [Google Scholar]
- 31.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jády, B. E., X. Darzacq, K. E. Tucker, A. G. Matera, E. Bertrand, and T. Kiss. 2003. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal bodies following import from the cytoplasm. EMBO J. 22:1878-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiss, A. M., B. E. Jády, E. Bertrand, and T. Kiss. 2004. Human box H/ACA pseudouridylation guide RNA machinery. Mol. Cell. Biol. 24:5797-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiss, T. 2004. Biogenesis of small nuclear RNPs. J. Cell Sci. 117:5949-5951. [DOI] [PubMed] [Google Scholar]
- 35.Kiss, T. 2001. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 20:3617-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiss, T., and W. Filipowicz. 1995. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 9:1411-1424. [DOI] [PubMed] [Google Scholar]
- 37.Kiss, T., and W. Filipowicz. 1993. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO J. 12:2913-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiss, T., and B. E. Jády. 2004. Functional characterization of 2′-O-methylation and pseudouridylation guide RNAs. Methods Mol. Biol. 265:393-408. [DOI] [PubMed] [Google Scholar]
- 39.Kruger, W., and B. J. Benecke. 1987. Structural and functional analysis of a human 7 S K RNA gene. J. Mol. Biol. 195:31-41. [DOI] [PubMed] [Google Scholar]
- 40.Lacadie, S. A., and M. Rosbash. 2005. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol. Cell 19:65-75. [DOI] [PubMed] [Google Scholar]
- 41.Lander, E. S. e. a. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 42.Laneve, P., F. Altieri, M. E. Fiori, A. Scaloni, I. Bozzoni, and E. Caffarelli. 2003. Purification, cloning, and characterization of XendoU, a novel endoribonuclease involved in processing of intron-encoded small nucleolar RNAs in Xenopus laevis. J. Biol. Chem. 278:13026-13032. [DOI] [PubMed] [Google Scholar]
- 43.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 44.Mattick, J. S., and I. V. Makunin. 2005. Small regulatory RNAs in mammals. Hum Mol. Genet. 14(Spec No. 1):R121-R132. [DOI] [PubMed] [Google Scholar]
- 45.Maxwell, E. S., and M. J. Fournier. 1995. The small nucleolar RNAs. Annu. Rev. Biochem. 64:897-934. [DOI] [PubMed] [Google Scholar]
- 46.Moss, T., and V. Y. Stefanovsky. 1995. Promotion and regulation of ribosomal transcription in eukaryotes by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol. 50:25-66. [DOI] [PubMed] [Google Scholar]
- 47.Murphy, S., C. Di Liegro, and M. Melli. 1987. The in vitro transcription of the 7SK RNA gene by RNA polymerase III is dependent only on the presence of an upstream promoter. Cell 51:81-87. [DOI] [PubMed] [Google Scholar]
- 48.Ooi, S. L., D. A. Samarsky, M. J. Fournier, and J. D. Boeke. 1998. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of a precursor snoRNA. RNA 4:1096-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 50.Reimer, G., K. M. Pollard, C. A. Penning, R. L. Ochs, M. A. Lischwe, H. Busch, and E. M. Tan. 1987. Monoclonal autoantibody from a (New Zealand black x New Zealand white) F1 mouse and some human scleroderma sera target an Mr 34,000 nucleolar protein of the U3 RNP particle. Arthritis Rheum. 30:793-800. [DOI] [PubMed] [Google Scholar]
- 51.Schattner, P., S. Barberan-Soler, and T. M. Lowe. 2006. A computational screen for mammalian pseudouridylation guide H/ACA RNAs. RNA 12:15-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Selden, R. F. 1992. Transfection using DEAE-dextran. John Wiley and Sons, New York, N. Y.
- 53.Terns, M. P., and R. M. Terns. 2002. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 10:17-39. [PMC free article] [PubMed] [Google Scholar]
- 54.Tollervey, D., and T. Kiss. 1997. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 9:337-342. [DOI] [PubMed] [Google Scholar]
- 55.Tyc, K., and J. A. Steitz. 1989. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 8:3113-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tycowski, K. T., M. D. Shu, and J. A. Steitz. 1993. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev. 7:1176-1190. [DOI] [PubMed] [Google Scholar]
- 57.Veres, G., R. A. Gibbs, S. E. Scherer, and C. T. Caskey. 1987. The molecular basis of the sparse fur mouse mutation. Science 237:415-417. [DOI] [PubMed] [Google Scholar]
- 58.Villa, T., F. Ceradini, and I. Bozzoni. 2000. Identification of a novel element required for processing of intron-encoded box C/D small nucleolar RNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:1311-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, C., and U. T. Meier. 2004. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 23:1857-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watkins, N. J., A. Dickmanns, and R. Luhrmann. 2002. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 22:8342-8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watkins, N. J., R. D. Leverette, L. Xia, M. T. Andrews, and E. S. Maxwell. 1996. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA 2:118-133. [PMC free article] [PubMed] [Google Scholar]
- 62.Weinstein, L. B., and J. A. Steitz. 1999. Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol. 11:378-384. [DOI] [PubMed] [Google Scholar]
- 63.Xia, L., N. J. Watkins, and E. S. Maxwell. 1997. Identification of specific nucleotide sequences and structural elements required for intronic U14 snoRNA processing. RNA 3:17-26. [PMC free article] [PubMed] [Google Scholar]
- 64.Yang, P. K., C. Hoareau, C. Froment, B. Monsarrat, Y. Henry, and G.Chanfreau. 2005. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol. Cell. Biol. 25:3295-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang, P. K., G. Rotondo, T. Porras, P. Legrain, and G. Chanfreau. 2002. The Shq1p.Naf1p complex is required for box H/ACA small nucleolar ribonucleoprotein particle biogenesis. J. Biol. Chem. 277:45235-45242. [DOI] [PubMed] [Google Scholar]
- 66.Yu, Y. T., E. C. Scharl, S. C. M., and J. A. Steitz. 1999. The growing world of small nuclear ribonucleoproteins, p. 487-524. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.