Abstract
The chromodomain (CD) of the Drosophila Polycomb protein exhibits preferential binding affinity for histone H3 when trimethylated at lysine 27. Here we have investigated the five mouse Polycomb homologs known as Cbx2, Cbx4, Cbx6, Cbx7, and Cbx8. Despite a high degree of conservation, the Cbx chromodomains display significant differences in binding preferences. Not all CDs bind preferentially to K27me3; rather, some display affinity towards both histone H3 trimethylated at K9 and H3K27me3, and one CD prefers K9me3. Cbx7, in particular, displays strong affinity for both H3K9me3 and H3K27me3 and is developmentally regulated in its association with chromatin. Cbx7 associates with facultative heterochromatin and, more specifically, is enriched on the inactive X chromosome. Finally, we find that, in vitro, the chromodomain of Cbx7 can bind RNA and that, in vivo, the interaction of Cbx7 with chromatin, and the inactive X chromosome in particular, depends partly on its association with RNA. We propose that the capacity of this mouse Polycomb homolog to associate with the inactive X chromosome, or any other region of chromatin, depends not only on its chromodomain but also on the combination of histone modifications and RNA molecules present at its target sites.
During the development of multicellular organisms, highly orchestrated networks of gene regulators dictate gene expression patterns, such as those of the homeobox (hox) genes. The Polycomb (PcG) and Trithorax (TrxG) group proteins maintain repression or activation of target genes, respectively, and allow for “cellular memory” throughout subsequent cell divisions and development (30). The founding member of the PcG genes is Drosophila melanogaster Polycomb (Pc), mutations in which result in body segment transformations. Pc is encoded by a single gene in Drosophila, while the mouse homologs have expanded into five family members known as Chromobox 2 (Cbx2) (mPc1 or M33), Cbx4 (mPc2), Cbx6, Cbx7, and Cbx8 (mPc3) (34). Importantly, these proteins contain a highly conserved N-terminal chromodomain (CD), a module first identified in the Drosophila proteins heterochromatin protein 1 (HP1) and Pc (24). The CD is found in a wide range of chromatin-associated proteins, most with transcriptionally repressive functions. The CD binds to methylated histones: the CD of Drosophila HP1 binds histone H3K9me2 and me3, while that of Pc specifically binds K27me3 on H3 (2, 9, 16, 18). Besides methyl-lysine binding, several reports have also suggested that certain CDs bind nucleic acids (1, 5).
Consistent with gene silencing functions, PcG proteins have also been implicated in X inactivation, whereby one of the two female X chromosomes is inactivated to provide gene dosage between the sexes. The polycomb repressive complex 2 (PRC2) containing Eed and E(z) is responsible for trimethylating H3 at K27. This mark, likely acting in concert with other repressive methyl marks (see below), is critical for the early stages of silencing the inactive X chromosome (Xi) (26, 32) and is thought to facilitate the recruitment of a second complex, PRC1. Some PRC1 components, including polyhomeotic 1 (Phc1) and Phc2, Bmi1, and Cbx2, have recently been shown to localize to the Xi (27), although it is unclear whether they are recruited directly by K27me3. The noncoding Xist transcript, which coats the X chromosome in cis and triggers X inactivation during early development, may also have a role in recruiting both PRC2 and PRC1 proteins to chromatin (7, 26, 27), as inducible Xist transgenes result in the rapid appearance of PRC2 and PRC1 proteins on the chromosome. Other histone modifications that are enriched on the Xi include H3K9me2 (4, 13) and H4K20me1 (17), although the binding effectors that “read” these marks and the enzyme complexes that “write” these marks on the Xi have yet to be identified. In particular, any participation of these histone modifications in the recruitment of Polycomb group proteins has not been examined.
In the present study, we examine the binding affinities of all five mouse Pc-like Cbx CDs for mono-, di-, and trimethylated K9 and K27 on the histone tail of H3 as well as mono-, di-, and trimethylated K20 on histone H4. Interestingly, we find that unlike Drosophila Pc, some of the mammalian Pc-like CDs bind to K9me3 as well as K27me3 (Cbx2 and Cbx7), Cbx4 prefers K9me3, and Cbx6 and Cbx8 do not bind significantly to either modification under our assay conditions. Furthermore, we demonstrate that all Cbx proteins, except Cbx4, localize to the Xi during female mouse embryonic stem (ES) cell differentiation and that the global association of Cbx7 with chromatin is developmentally regulated. Finally, we show that although histone methyl marks mediate binding of the Pc-like proteins, RNA is also an important component for the association of Cbx proteins with chromatin. Collectively, these studies demonstrate that related chromatin-binding motifs exhibit differences in binding characteristics that likely translate into distinct biological readouts and highlight the need to carefully analyze the differences between mammalian and fly chromodomain proteins.
MATERIALS AND METHODS
Recombinant proteins and peptide synthesis.
Cbx CDs (amino acids [aa] 1 to 62) and full-length Cbx7 were cloned into pGEX-6P-1 (Amersham); glutathione S-transferase (GST) fusion proteins were produced in Escherichia coli BL21. Bacterial lysates were purified over glutathione Sepharose 4B (Amersham) as recommended by the manufacturer. 6× His-tagged human HP1β and Drosophila Pc proteins were a gift of W. Fischle (aa 15 to 72 and aa 15 to 77, respectively, both cloned into pET-11a; Promega). Point mutations of caging aromatic residues in Cbx7 were created using QuikChange site-directed mutagenesis (Stratagene). Peptides used for fluorescence polarization were synthesized at the Rockefeller University Proteomics Resource Center corresponding to H3 1-15, H3 19-35, and H4 12-27 (unmodified, me1, me2, and me3 at H3K9, K27, and H4K20). 5-Carboxyfluorescein was coupled directly to the N terminus. Biotinylated peptides for pull-down assays were synthesized at either Baylor College of Medicine Protein Chemistry Facility or Rockefeller University Proteomics Resource Center.
Fluorescence polarization.
Fluorescence polarization assays were performed essentially as described previously (9). All mouse Pc CDs were analyzed using buffer containing 20 mM imidazole, 25 mM NaCl, and 2 mM dithiothreitol (DTT), and mHP1α was assayed using a buffer containing 50 mM Na2HPO4, pH 7, 25 mM NaCl, 1 mM MgCl2, 2 mM DTT. Data were obtained with a Hidex Chameleon plate reader.
Peptide pull-down assays.
Peptide pull-down assays were performed as described previously (35): 5 μg of recombinant protein plus 20-fold excess of bovine serum albumin (internal control) was incubated in assay buffer (150 mM KCl, 20 mM HEPES, and 0.2% Triton X-100), and beads were washed six times in assay buffer.
Far-Western blotting.
Far-Western blottings were performed essentially as described previously (23) with acid extracted LF2 histones and recombinant H3 (Upstate Biotechnologies, Inc.). Recombinant GST proteins were incubated at 0.3 μg/ml, and peptide competitions were performed at 10 μg/ml.
Chromatin fractionation, mononucleosomal IP, and RNase treatment.
Mouse Cbx7 was cloned into pCMV-Tag 4A (Stratagene), generating a C-terminal flag fusion. A stable Cbx7-293 cell line was created. Biochemical fractionation and microccocal nuclease digestion were performed as described previously (21, 35). Chromatin samples were treated with MNase for 6 min at 37°C to produce mononucleosomal DNA, and extracts were used for immunoprecipitation (IP). Extracts expressing the Cbx7-flag and 293 cells were incubated with α-flag beads (M2 affinity gel; Sigma) for 4 h at 4°C and washed 5× with 1× Tris-buffered saline. Cbx7-flag protein and bound histones were peptide eluted overnight (Sigma) and immunoblotted. Chromatin from female ES cells was prepared at day 6 of differentiation as described above, resuspended in low-salt buffer (10 mM HEPES, 10 mM KCl, and 1.5 mM MgCl2), and treated with RNase A at 0, 2, or 5 μg per 100 μl of chromatin. Samples were incubated at 37°C for 10 min, and chromatin was run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis for immunoblots.
Antibodies.
Antibodies used include monoclonal α-Flag (M2, Sigma); Upstate Biotechnologies polyclonal histone antibodies H3K9me2 (07-441), H3K9me3 (07-442), H3K27me3 (07-449), H3K4me3 (07-473), H4K20me1 (07-440), and H4K20me3 (07-463); monoclonal α-Hp1β (Chemicon); α-GST for far Western blotting (Amersham); monoclonal α-green fluorescent protein (GFP) (Roche); monoclonal α-Eed (gift of A. Otte); α-WDR5 (gift of W. Herr); and monoclonal α-H3K27me3 for immunofluorescence (gift of D. Reinberg).
Cell culture and differentiation.
Female ES cell line LF2 (a gift from A. Smith) was grown and differentiated as previously described (31).
Cbx cDNA fusions.
Cbx cDNAs were cloned with BglII/SalI into EGFP-N1 (BD Biosciences), producing C-terminal enhanced GFP (EGFP) fusion proteins. Cbx2 cDNA was a gift of M. Narita. Cbx4 cDNA was cloned from female ES cell line HP310 by reverse transcription-PCR; cDNAs for Cbx6 and Cbx8 were purchased from Open Biosystems; Cbx7 cDNA was previously described (11). All Cbx cDNAs were verified. Point mutations in Cbx7-EGFP were created as for recombinant proteins. The Cbx4 CD swap into Cbx7 was made by staggered PCR by incorporation of overlapping oligonucleotides, and the chimeric cDNA was cloned into EGFP-N1.
Immunofluorescence and RNase treatment.
LF2 cells were transiently transfected at day 2 of differentiation with Lipofectamine 2000 (Invitrogen), and on day 3 cells were stained as previously described (6). Line histograms were made with processing software MetaMorph Offline. RNase treatments on living LF2 cells were performed as described previously (23). In brief, 24 h posttransfection, cells were incubated on ice for 10 min in PNS (20 mM PIPES, pH 6.8, 200 mM NaCl, 600 mM sucrose)-0.3% Triton X-100. Cells were incubated for 1 h on ice in PNS with or without RNase A (1 mg/ml) and fixed in 4% paraformaldehyde. Immunofluorescence was performed as described above.
GST overlay cell assays.
GST overlay assays were performed as previously described (23). Polyclonal α-GST antibody was a gift of A. E. L. Marjou.
RNA gel shifts.
RNA was in vitro transcribed with T7 Megascript (Ambion) and [32P]UTP. Fragments (100 and 500 nucleotides [nt]) of cyclin E were used as a template for single- and double-stranded RNAs. RNAs were gel purified. Cyclin E DNA was labeled with [γ-32P]ATP using T4 polynucleotide kinase. Increasing amounts of protein (10 to 500 pmol) were incubated in buffer containing 20 mM HEPES, 100 mM KCl, 2 mM EDTA, 0.01% NP-40, 1 mM DTT, 3 μg tRNA for RNA gel shifts (Ambion), 1 μg poly(dI-dC) for DNA gel shifts (Amersham), RNasin for RNA gel shifts (Promega), and 10,000 cpm of RNA or DNA in a total reaction volume of 20 μl. Reactions were incubated on ice for 30 min; a 5% native 0.5× Tris-borate-EDTA gel was prerun at room temperature; and gels were run at 4°C for 3 h at 250 V and imaged with a FLA-5000 phosphorimager (Fujifilm). For denaturation experiments, proteins were heated at 95°C for 10 min and assayed as described above.
RESULTS AND DISCUSSION
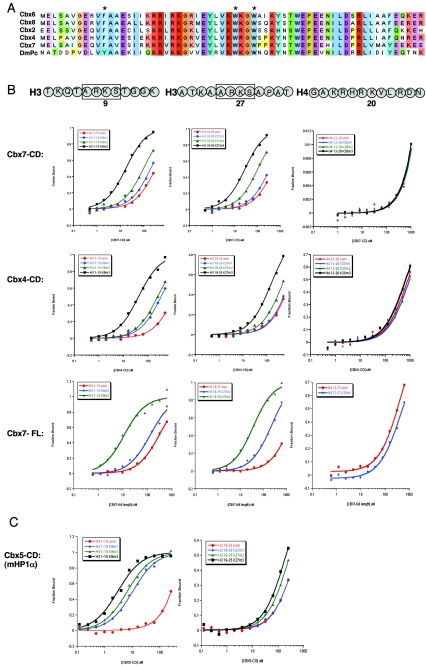
The Drosophila Polycomb CD binds preferentially to histone H3 trimethylated at lysine 27 (9, 22). We set out to test whether or not this tenet holds true for its mammalian counterparts. Recombinant protein for the CDs of mouse Cbx2, Cbx4, Cbx6, Cbx7, and Cbx8 as well as full-length Cbx7 was bacterially expressed and purified. The CDs of these Pc-like proteins are remarkably conserved at the amino acid sequence level (Fig. 1A), making it plausible that all would behave similarly in their ability to bind an H3K27me3 peptide. Surprisingly, however, the Pc-like Cbx CDs do not behave like Drosophila Pc, as determined by two independent assays. The first assay used was a quantitative biophysical approach, fluorescence polarization (FP), in which recombinant CDs were incubated with fluorescently labeled peptides that mimic histone tails. Unmodified and modified peptides (me1, me2, and me3) of H3K9 and H3K27 as well as H4K20me1, H4K20me2, and H4K20me3 were tested with each CD in addition to full-length Cbx7. These marks were chosen because they represent well-characterized repressive histone modifications associated with either constitutive (e.g., K9me3 on pericentromeres) or facultative (e.g., K9me2, K27me3, and K20me1 on the Xi) heterochromatin (12, 25). Representative binding curves are shown for the CDs of the Pc homologs Cbx4, Cbx7, and full-length Cbx7 (Fig. 1B). All FP data collected are summarized in Fig. 1D. Interestingly, Cbx2 and Cbx7 bind both to K9me3 and K27me3, with Cbx7 having a higher affinity for these modifications (Fig. 1B and D). Cbx7 has a dissociation constant in the low-micromolar range for both K9me3 and K27me3 (Fig. 1D), similar to those of Drosophila HP1 for K9me3 and Pc for K27me3 (9), and full-length Cbx7 behaves identically to its CD in this assay (Fig. 1B). Notably, Cbx4 was the only Pc-like CD to have significant preference for K9me3 (Fig. 1B and D), thus functionally representing an HP1-like protein in this regard. Moreover, although K9 and K27 are both embedded in an ARKS motif in H3, Cbx4 can clearly distinguish between these methyl marks, while Cbx7 binds both (Fig. 1B, see histone sequences). In order to ensure that our FP analysis was consistent with published results, we examined mouse Cbx5 (HP1α) for its binding affinity to H3K9 and H3K27 methylated peptides (Fig. 1C and D). Indeed, we find that our binding constants are reflective of the published work on the mammalian (and Drosophila) HP1 proteins (9, 10).
FIG. 1.
Analysis of Pc-like Cbx CD binding affinities for trimethylated H3K9 and H3K27. (A) ClustalW alignment of the five mouse Pc-like CDs (aa 1 to 62) and the Drosophila Pc CD (aa 16 to 78). The asterisks represent the caging aromatic residues that mediate the histone methyl-lysine interaction. Note the high degree of conservation among these family members. (B) Fluorescence polarization of Cbx7 (top) and Cbx4 (middle) to histone tail peptides, including the me1, me2, and me3 states on residues K9 and K27 of H3, and K20me1, K20me2, and K20me3 of H4, respectively. Full-length Cbx7 (bottom) was tested against all of the above except for the monomethylated forms of H3K9 and H3K27, H4K20me2 and H4K20me3, and behaves identically to its CD for peptides tested. Histone tail sequences are represented above; note the ARKS motifs of H3K9 and H3K27. See panel D for actual peptides used. (C) Fluorescence polarization of Cbx5 (mouse HP1α) to H3K9 and H3K27 histone tail peptides in the me1, me2, and me3 states. (D) Dissociation constants (Kd, in micromolars) for Cbx2, Cbx4, Cbx6, Cbx7, and Cbx8, as well as Cbx5 (mHP1α), with each series of methylated peptides for each backbone shown. Low-micromolar binding constants are highlighted in red. The asterisk depicts weak binding of Cbx7 for H3K9me2 (a mark that is enriched on the Xi). Values represent averages ± standard deviations for at least three independent experiments in all cases (except for Cbx5 with certain peptides). ND, not determined. (E) Peptide pull-down assays. (Left) All CDs were examined for binding to unmodified and trimethylated peptides representing H3K9, H3K27, and H4K20. Results support those obtained by FP (D). (Right) Cbx7 CD, Cbx7 caging aromatic point mutant F11A, and human HP1β CD recombinant proteins were tested for the ability to bind unmodified and me1, me2, and me3 peptides of H3K9 and K27. Note the trimethyl specificity of Cbx7. (Right, bottom) GST and full-length Cbx7 were examined for binding to unmodified and trimethylated biotinylated peptides of H3K9 and H3K27. Full-length Cbx7 behaves the same as its CD alone. GST does not bind any peptide, as expected. un, unmodified.
None of the Polycomb Cbx CDs bind to the mono-, di-, or trimethylated forms of H4K20 (Fig. 1B and D) or, as expected, to trimethylated H3K36 (a transcriptional activation mark; data not shown). Unexpectedly, however, neither Cbx6 nor Cbx8 binds significantly to K27me3 or to any other modification tested (Fig. 1D; FP data not shown), suggesting that these CDs may bind to another methylation site not yet identified or tested; perhaps, for example, a mark that is embedded in an ARKS-like motif of another histone or nonhistone protein. Furthermore, they may require a factor for methyl-histone binding that is not present in these recombinant assays.
The second assay used to test binding preferences was a peptide pull-down approach, in which biotinylated histone tail peptides conjugated to avidin beads were used as bait for the recombinant proteins. Results of this assay lend strong support to the relative binding constants determined by FP: Cbx7 binds preferentially to both K9me3 and K27me3, whereas Cbx4 prefers K9me3 (Fig. 1E, left panel). Peptide pull-down assays for Cbx2, Cbx6, and Cbx8 also strongly support the FP results (Fig. 1E, left panel). Moreover, none of the CDs bind to H4K20me3, a lysine residue not found in ARKS motif and a mark of pericentric heterochromatin (Fig. 1E, left panel). Notably, a Cbx7 methyl-lysine caging aromatic residue (found to be structurally essential in an HP1 [15]) point mutant (F11A) fails to bind either K9me3 or K27me3, demonstrating that a functional CD is critical for binding (Fig. 1E, right panel). Human HP1β was used as a control for K9me binding (Fig. 1E, right panel), and the mouse HP1 homologs show similar results (data not shown). Full-length Cbx7 was also tested in this assay and behaves like its CD (Fig. 1E, bottom right).
We next focused our attention on Cbx7, as it showed the strongest affinity for both K27me3 and K9me3 and is thus somewhat divergent from Drosophila Pc. Interestingly, Cbx7 has recently been shown to extend the life span of mammalian primary cells by prolonging senescence (11). Moreover, mutations in critical residues of the CD inhibited the ability of Cbx7 to extend the life span (11). We further investigated the role of Cbx7 in the context of chromatin and, more specifically, tested its association with facultative heterochromatin and coupled histone modifications.
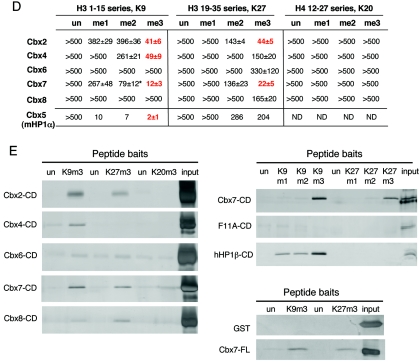
In order to test the binding preferences of Cbx7 for histone H3 methyl marks in the context of endogenous histones, far Western blotting assays were performed. Using recombinant GST-Cbx7, we demonstrate that Cbx7 has specificity for K9me3 and K27me3, as it was competed from histone H3 derived from female ES cells with these peptides but not the unmodified peptides (Fig. 2A). Although the exact mechanism by which Cbx7 binds the nucleosome is not fully understood, this finding supports the peptide binding results described above. As expected, Cbx7 did not bind recombinant H3, which is devoid of modifications. Moreover, GST alone and the caging point mutations (F11A and W35A) all failed to recognize endogenously modified H3 (Fig. 2A).
FIG. 2.
Cbx7-associated histone modifications. (A) Far Western blotting assay. The GST-Cbx7 CD binds to female ES cell (LF2) histone H3 through K9me3 and K27me3, demonstrated by peptide competitions (lanes 1 to 6); Cbx7 CD does not bind to recombinant H3 (lane 2); GST alone and caging aromatic mutants fail to associate (lanes 7 to 9); Ponceau for equal loading of total histones (bottom). (B) IPs of mononucleosomal extracts prepared from Cbx7-flag stable 293 cells versus control cells. Inputs in both cases show presence of all histone modifications; Cbx7-flag associated histones (IP) are marked predominantly by facultative heterochromatic modifications, including H3K9me2, H3K27me3, and H4K20me1. un, unmodified.
In order to determine the histone modification profile of Cbx7-associated chromatin, we took advantage of a biochemical fractionation scheme to isolate chromatin (21, 35). Chromatin from a 293-derived Cbx7-flag stable cell line and control cells was digested into mononucleosomes, and anti-flag immunoprecipitations were performed. As expected, input from control cells and Cbx7-stable cells contained all histone modifications tested (Fig. 2B). However, histones were only precipitated from Cbx7-flag cells and were modified with H3K27me3, H3K9me2, H3K9me3, and H4K20me1 (Fig. 2B). These marks are characteristic of facultative heterochromatin, in particular the Xi (see below). Ubiquitylation of H2A by the E3 ligases Ring1A and Ring1B has also been linked with the Xi (7, 8, 33), and Cbx7 also specifically associated with histones bearing this epigenetic modification (data not shown). Interestingly, Cbx7 did not associate with either H4K20me3, a mark of pericentric heterochromatin, or, as expected, with H3K4me3, a mark of actively transcribing genes (Fig. 2B).
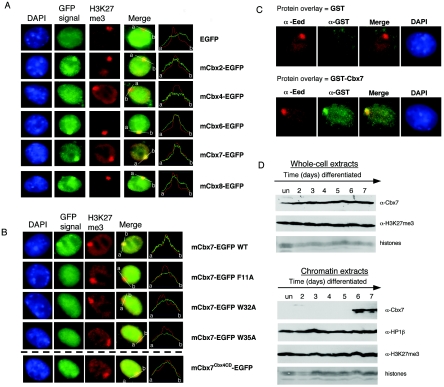
We next tested the association of Cbx proteins with facultative heterochromatin in vivo, in particular their association with the Xi. Various epigenetic mechanisms are thought to be critical for maintaining the silent state of the Xi, including K27 trimethylation. Due to their role in developmentally regulated repression in Drosophila, the PcG proteins were proposed to be involved in maintaining the silent X (30). Recently, members of PRC1 have been shown to associate with the Xi (7, 8, 14, 27). A key question that we sought to address is whether any of the mammalian Pc proteins could be the effectors that bind K27me3 on the Xi in order to recruit PRC-like complexes. Thus, we created EGFP fusion proteins for Cbx2, Cbx4, Cbx6, Cbx7, and Cbx8 to assess their association with the Xi in 3-day-differentiated female ES cells, a point at which K27me3 is at its peak on the Xi. Cbx2, Cbx6, Cbx7, and Cbx8 do associate with the Xi (Fig. 3A; see also the line histograms). On the other hand, Cbx4, which preferentially binds K9me3, is not enriched on the Xi (Fig. 3A) or on pericentric heterochromatic regions (4′,6′-diamidino-2-phenylindole [DAPI]-dense chromocenters; data not shown), indicating that K9me3 binding affinity alone is not driving recruitment. Cbx7, which has the highest affinity for H3K27me3 in vitro, also shows the most striking enrichment on the Xi, suggesting a recruitment mechanism by this mark. We further performed protein overlay assays whereby GST and GST-Cbx7 (full-length) recombinant proteins were incubated with cells at day 3 of differentiation, followed by GST antibody staining. GST-Cbx7, but not GST, is enriched on the Xi, confirming our GFP localization studies (Fig. 3C).
FIG. 3.
Cbx proteins associate with the inactive X. (A) All Cbx-EGFP fusion proteins, except Cbx4, localize to the Xi in 3-day-differentiated female ES cells. The Xi is visualized by K27me3 staining (red). Fluorescence intensity plots (white line from a to b) across the nucleus and including the K27me3 signal (red) on the Xi illustrate the enrichment for, or lack of, each of the Cbx-GFP fusion proteins (green). (B) Point mutations of the caging aromatic residues in the Cbx7 CD (F11A, W32A, and W35A) disrupt localization to the Xi. When the CD of Cbx7 is replaced by the CD of Cbx4 (mCbx7Cbx4CD-EGFP), the chimeric protein no longer strongly associates with the Xi, although it is not disrupted completely (bottom panel). Note that the K27me3 peak coincides with a Cbx7-GFP peak for the wild-type (WT) and Cbx7Cbx4CD proteins but not for the three mutated versions. (C) GST overlay assays were performed on 3-day-differentiated female mouse ES cells. Protein was detected on paraformaldehyde-fixed cells using α-GST antibody (green). The Xi was visualized by α-Eed antibody (red). GST protein does not associate with the Xi (top), while full-length GST-Cbx7 is enriched on the Xi (bottom). It should be noted that although these experiments support our EGFP-fusion results, they were somewhat variable in their efficiency. (D) Female ES cells were tested throughout differentiation (up to day 7) for Cbx7 association with chromatin (un, undifferentiated). Cbx7 and K27me3 are expressed throughout the differentiation process (top, whole-cell extracts); however, Cbx7 specifically associates with chromatin on day 6 and onward (compare to HP1β and K27me3 in chromatin extracts at the bottom of the panel).
Point mutations of the caging aromatic residues in the CD of Cbx7-EGFP were found to abrogate its localization to the Xi, underlining the importance of the chromodomain in the association of Cbx7 with the Xi (Fig. 3B). Furthermore, the enrichment of Cbx7 on the Xi supports its association with the histone modifications shown in Fig. 2B, as K20me1, but not K20me3, is a predominant mark on the Xi in mouse ES cells. Intriguingly, Cbx6 and Cbx8 do not bind significantly to K27me3 in vitro (or any other modification tested), suggesting that this mark does not play a major role in the recruitment of these particular proteins to the Xi. Thus, it remains to be determined if another histone methylation (or other) mark on the Xi is responsible for the recruitment of Cbx6 and Cbx8. It is also a formal possibility that these Cbx proteins are recruited to their chromatin-binding sites in a histone methylation-independent manner.
To test whether a K27me3-binding CD is necessary for Xi localization, we replaced the CD of Cbx7 with the CD of Cbx4 (an H3K9me3-binding CD), thus creating a chimeric Pc protein (Cbx7Cbx4CD). This “swapped” protein was no longer enriched on the Xi, although its association was not completely disrupted (see the line histograms at the bottom of Fig. 3B). This suggests that although a K27me-binding CD is important for localization of Cbx7 to the Xi, other recruitment mechanisms may be involved, including interacting proteins and/or RNA interactions (see below). Consistent with this result, the Cbx7 CD alone fused to EGFP failed to localize to the Xi (as did those of Cbx6 and Cbx8; data not shown).
The lack of immunofluorescence-compatible Cbx antibodies has made it difficult to assess the kinetics of their association with the Xi through ES cell differentiation. Thus, we surveyed the association of Cbx7 with chromatin on a global level. Chromatin was isolated from female ES cells across differentiation, and the association of Cbx7 with bulk chromatin was assayed by immunoblot analyses. Interestingly, although Cbx7 is present throughout differentiation (whole-cell extracts in Fig. 3D), the protein only associates with chromatin at later stages of differentiation typically observed first at day 6 (chromatin extracts in Fig. 3D) through day 10 of differentiation (data not shown). While these data represent a global chromatin association, we note that the histone variant macroH2A first associates with the Xi at a similar stage (12). It is striking to compare the chromatin association of Cbx7 with that of HP1β and the H3K27me3 modification, both of which remain globally constant across differentiation (Fig. 3D). The mechanism underlying this regulation is unknown but suggests a potentially important role for Cbx7 in development. For example, the temporal coincidence of Cbx7 and macroH2A recruitment to chromatin indicates a potential mechanistic link. It should be noted that although Cbx7 associates with the Xi at day 3 of differentiation in transient transfection assays (Fig. 3A) and endogenous Cbx7 associates with chromatin on day 6, the latter represents a global association and not an Xi-specific one.
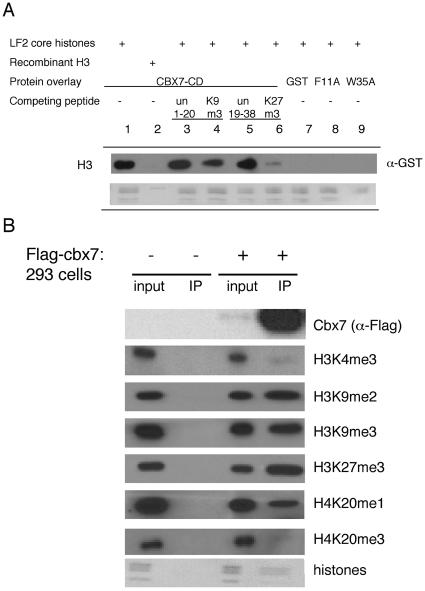
In mammals containing more than one X chromosome, the X inactivation process is initiated during early development by the noncoding RNA Xist, which coats the X chromosome chosen for silencing. The PRC1 and PRC2 complexes, as well as the H3K27me3, H3K9me2, and H4K20me1 histone modifications, become rapidly enriched on the Xist RNA-coated chromosome. Subsequently, the incorporation of macroH2A and DNA (CpG) methylation is observed. It is believed that together these epigenetic marks assist in maintaining this silenced state (12, 14, 27). Given that the Xist RNA is essential for silencing (12) and may have a role in recruiting PRC proteins, we tested whether the recombinant Cbx CDs were able to bind RNA in vitro. Furthermore, numerous PcG- and CD-containing proteins have been demonstrated to bind RNA (3). Interestingly, when the CDs of mouse HP1α (Cbx5) and Cbx7 (Pc-like) were directly compared in an electromobility shift assay with a 500-nt single-stranded RNA, only Cbx7 bound to the RNA (Fig. 4A). All Pc-like CDs were then assayed, and with the exception of Cbx2, they were able to bind RNA (Fig. 4B). Although the Cbx2 CD does not bind RNA in this assay, it is also the only Cbx that contains an AT-hook motif (34), which lies C terminal to the CD (and thus is not present in our CD construct) and may represent a nucleic acid binding module for this Cbx. The CDs of mouse HP1β (Cbx1) and HP1α (Cbx5) were unable to bind RNA (Fig. 4A and B), consistent with previous reports (23). The hinge region of HP1α, which lies between the CD and the chromoshadow domain (the latter is not present in Pc-like proteins), has been shown to be required for RNA binding (23).
FIG. 4.
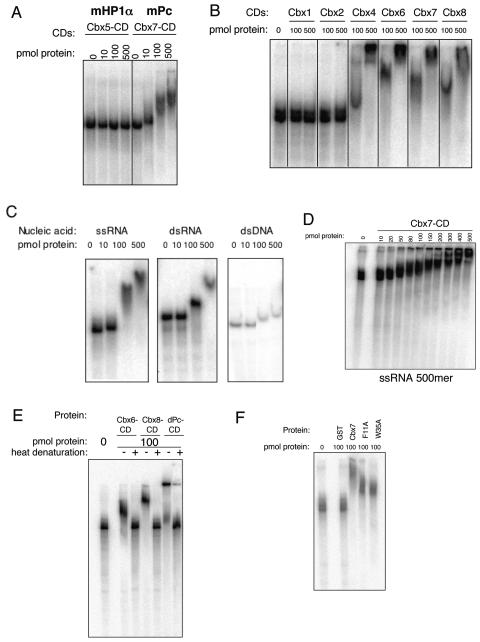
Pc-like Cbx CDs bind RNA. Recombinant CDs were tested for RNA binding by gel shift assays. (A) The Cbx5 (mouse HP1α) CD was compared to Cbx7 (mPc) for RNA binding with a 500-nt single stranded RNA (ssRNA), and only the latter demonstrates a shift. (B) Cbx4, Cbx6, Cbx7, and Cbx8 CDs interact with RNA, while Cbx1 (mHP1β) and Cbx2 do not. (C) Cbx7 binds to both single-stranded RNA (ssRNA) and double-stranded RNA (dsRNA). Cbx7 binds dsDNA with only minor affinity. (D) Gel shift analysis of the Cbx7 CD with ssRNA (500mer) was performed by extensive titration of protein in order to determine a dissociation constant, which is approximately 100 μM. (E) Heat denaturation of Cbx6, Cbx8, and dPc CD proteins disrupts the interaction with RNA (+, heat denaturation). (F) Point mutations in the caging aromatic residues of the Cbx7 CD do not abrogate RNA binding. Recombinant GST was used as a negative control.
The ability to bind RNA was not sequence specific, as the Cbx CDs bound to various RNAs tested, including Xist sequences (data not shown). Similar results have been reported for other RNA-binding CDs (1, 23), although we cannot rule out the possibility that the Cbx7 CD has sequence specificity in vivo. Cbx7 appears to bind single-stranded RNA with higher affinity than double-stranded RNA and to double-stranded DNA with minor affinity (Fig. 4C). Furthermore, the affinity of Cbx7 for single-stranded RNA is approximately 100 μM, as estimated by gel shift analysis of extensively titrated protein (Fig. 4D). The GST tag of Cbx7 does not facilitate binding, as GST protein alone does not bind to RNA (Fig. 4F). Moreover, Cbx7 with its GST tag removed binds RNA in an identical manner to GST-Cbx7 CD, as does full-length Cbx7 (data not shown).
In order to assess whether the secondary structure of the CD is important for the RNA interaction, the CDs of Cbx6, Cbx8, and Drosophila Pc (which behaves similarly to its mammalian homologs; Fig. 4E and data not shown) were heat denatured followed by gel shift analysis (Fig. 4E). In all cases, the CDs no longer bound the RNA when denatured, suggestive that an intact secondary structure is required for binding. Furthermore, point mutations of the caging aromatic residues of the Cbx7 CD do not completely abrogate RNA binding, suggesting that the region of the CD responsible for the RNA interaction may be distinct from that which binds the methyl-lysine residue (Fig. 4F). This finding suggests that the CD module could participate in targeting Cbx proteins to chromatin in at least two ways, via histone methylation and via RNA.
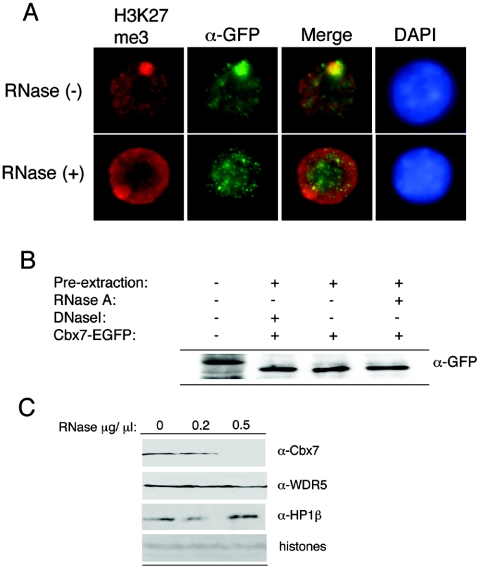
To test this hypothesis more directly, we examined whether RNA is an important factor for the localization of Cbx7 to the Xi in vivo. Treatment of ES cells at day 3 of differentiation with RNase A, which degrades single-stranded RNA, resulted in a substantial loss of Cbx7 from the Xi (Fig. 5A). As expected, the Xist RNA signal was abolished by this RNase treatment based on RNA-fluorescent in situ hybridization analysis (data not shown), while K27me3 on the Xi was not affected (Fig. 5A). Moreover, the level and integrity of the Cbx7-GFP protein and the global level of other proteins were not affected by the RNase treatment based on Western and Coomassie staining, respectively (Fig. 5B and data not shown), and immunofluorescence analysis of other nuclear proteins (such as lamin B1) did not reveal any disruption following RNase treatment (data not shown). Finally, we also examined whether RNA is involved in the ability of Cbx7 to associate with chromatin on a global level. Chromatin from 6-day-differentiated female ES cells was treated with RNase. Here we observed that the ability of Cbx7 to associate with chromatin was also RNA dependent (Fig. 5C). In contrast, neither WDR5, an effector of H3K4me2 (35), nor HP1β was affected by RNase treatment (Fig. 5C). Similar RNase studies performed in mouse cells resulted in depletion of HP1α from pericentric regions; however, the total amount of HP1α detected by immunoblot was unaffected (20). While the HP1 isoforms may differ, our results suggest that HP1 proteins may be displaced from distinct chromosomal regions by RNase and yet not be affected on a global chromatin level. Although our results do not permit the conclusion that Cbx7 interacts with chromatin through RNA directly, the ability of the CD to bind RNA in vitro suggests that this interaction may indeed be involved in its recruitment to target loci, including the Xi. Our data further suggest that the association of Cbx7 with chromatin, and the Xi in particular, is at least partially dependent on RNA, the nature of which is currently under investigation.
FIG. 5.
Cbx7 chromatin association is RNA dependent. (A) RNase treatment of 3-day-differentiated ES cells strongly diminished the accumulation of Cbx7-EGFP on the Xi. The Xi is visualized by K27me3 staining (red), and the signal of Cbx7-EGFP was enhanced using α-GFP antibody (green). (B) Degradation of Cbx7 is not observed after RNase treatment (or DNase I treatment), as detected by Western blot with GFP antibody. (C) Cbx7 is depleted from 6-day-differentiated ES cell chromatin by RNase treatment (top), while WDR5 and HP1β are not affected. See histones for equal loading (bottom).
Chromodomains are well-recognized methyl-lysine docking modules that are thought to bring about downstream effects that ultimately affect the structure of chromatin. Although the mechanism of chromatin compaction through the recruitment of repressive binding proteins such as HP1 and Pc is still unclear, a paradigm has been established whereby HP1 binds to H3K9me2 and H3K9me3, and H3K27me3 is bound by Pc (9). However, our investigation of the five mammalian Pc proteins and their distinct binding preferences suggests that this paradigm is not so straightforward. In support, Ringrose and colleagues have demonstrated that Drosophila Pc staining on polytene chromosomes overlaps with both H3K9me3 and H3K27me3 at PcG target genes and that both trimethylated H3K9 and K27 peptides compete for Pc from the chromosomes (29). While the biological readout of the Cbx binding preferences remains unclear, it is likely that these proteins interact with different PRC-like complexes. For example, Cbx2 is a component of the PRC1 complex, which includes Bmi1, Ring1, and Phc (30). Cbx7, however, has been shown to interact with Ring1 but not Bmi1 (11). Thus, the mammalian Pc proteins not only seem to show differential methylated histone binding preferences but also exist in distinct complexes that are likely to be dynamically regulated throughout development.
The stably silenced state of the Xi is in part regulated by epigenetic modifications, including K27me3, and the effector proteins that bind them. Based on our findings with Cbx7, we conclude that although the H3K27me3 mark seems to be important for the recruitment of Cbx7 via its CD, other components, including RNAs, must also be required for its recruitment and maintenance on the Xi. Indeed, recent reports have described the recruitment of several PRC1 proteins to the Xi, which may be part of one or multiple complexes (7, 8, 14, 27). It will be interesting to identify Cbx7-interacting proteins at different stages of development in order to understand the mechanism(s) by which Cbx7 associates with the inactive X chromosome, and with chromatin in general, during ES cell differentiation. For example, in addition to representing a potential mechanism by which Ring1 is able to ubiquitylate H2A on the Xi, the association of Cbx7 with chromatin during ES cell differentiation may be linked to the rather sudden appearance of macroH2A on the Xi (28). Finally, the role of RNA in Cbx7 chromatin association is of particular interest in light of recent evidence in fission yeast, flies, and mammals suggesting that noncoding RNAs can impact the chromatin template (3, 19). Moreover, TAP-tag purification of Cbx7 from human cells has demonstrated that this Pc protein interacts with an RNA-helicase, suggesting the involvement of RNA in Cbx7-mediated repression (J. Nicholls and J. Gil, unpublished observations). The ability of particular CDs to potentially bind both methylated histone tails and RNA suggests that a cooperative binding mechanism may mediate the enrichment of particular CD-containing proteins in chromatin. Future functional and structural studies will be required to determine the nature of this potential synergy, particularly in the case of Cbx7 and Xist RNA in the context of the inactive X chromosome.
Acknowledgments
We are grateful to members of the Allis laboratory for helpful discussions, in particular, W. Fischle and S. Hake. We greatly appreciate W. Fischle for purified HP1 and Pc proteins, Y. Wang for the initial work on this project, R. Diaz for help with peptide purification, and L. Cooper for assistance with protein purification. We thank W. Herr, M. Narita, A. Otte, A. E. L. Marjou, D. Reinberg and A. Smith for reagents and C. Maison for helpful discussions.
This work is funded by the Centre National de la Recherche Scientifique (ATIPE), the Curie Institute (PIC program), the “Epigenome” European Network of Excellence, and the Human Frontier Science Program to E.H., as well as the MRC (J.G.), the NSF (E.B.), and NIH grant GM53512 (C.D.A.).
REFERENCES
- 1.Akhtar, A., and P. B. Becker. 2000. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell 5:367-375. [DOI] [PubMed] [Google Scholar]
- 2.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, E., and C. D. Allis. 2005. RNA meets chromatin. Genes Dev. 19:1635-1655. [DOI] [PubMed] [Google Scholar]
- 4.Boggs, B. A., P. Cheung, E. Heard, D. L. Spector, A. C. Chinault, and C. D. Allis. 2002. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 30:73-76. [DOI] [PubMed] [Google Scholar]
- 5.Bouazoune, K., A. Mitterweger, G. Langst, A. Imhof, A. Akhtar, P. B. Becker, and A. Brehm. 2002. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J. 21:2430-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaumeil, J., I. Okamoto, M. Guggiari, and E. Heard. 2002. Integrated kinetics of X chromosome inactivation in differentiating embryonic stem cells. Cytogenet. Genome Res. 99:75-84. [DOI] [PubMed] [Google Scholar]
- 7.de Napoles, M., J. E. Mermoud, R. Wakao, Y. A. Tang, M. Endoh, R. Appanah, T. B. Nesterova, J. Silva, A. P. Otte, M. Vidal, H. Koseki, and N. Brockdorff. 2004. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 7:663-676. [DOI] [PubMed] [Google Scholar]
- 8.Fang, J., T. Chen, B. Chadwick, E. Li, and Y. Zhang. 2004. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J. Biol. Chem. 279:52812-52815. [DOI] [PubMed] [Google Scholar]
- 9.Fischle, W., Y. Wang, S. A. Jacobs, Y. Kim, C. D. Allis, and S. Khorasanizadeh. 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17:1870-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischle, W., B. S. Tseng, H. L. Dormann, B. M. Ueberheide, B. A. Garcia, J. Shabanowitz, D. F. Hunt, H. Funabiki, and C. D. Allis. 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438:1116-1122. [DOI] [PubMed] [Google Scholar]
- 11.Gil, J., D. Bernard, D. Martinez, and D. Beach. 2004. Polycomb CBX7 has a unifying role in cellular lifespan. Nat. Cell Biol. 6:67-72. [DOI] [PubMed] [Google Scholar]
- 12.Heard, E. 2004. Recent advances in X-chromosome inactivation. Curr. Opin. Cell Biol. 16:247-255. [DOI] [PubMed] [Google Scholar]
- 13.Heard, E., C. Rougeulle, D. Arnaud, P. Avner, C. D. Allis, and D. L. Spector. 2001. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell 107:727-738. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez-Munoz, I., A. H. Lund, P. van der Stoop, E. Boutsma, I. Muijrers, E. Verhoeven, D. A. Nusinow, B. Panning, Y. Marahrens, and M. van Lohuizen. 2005. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc. Natl. Acad. Sci. USA 102:7635-7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobs, S. A., and S. Khorasanizadeh. 2002. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295:2080-2083. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs, S. A., S. D. Taverna, Y. Zhang, S. D. Briggs, J. Li, J. C. Eissenberg, C. D. Allis, and S. Khorasanizadeh. 2001. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 20:5232-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohlmaier, A., F. Savarese, M. Lachner, J. Martens, T. Jenuwein, and A. Wutz. 2004. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2:E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 19.Maison, C., and G. Almouzni. 2004. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell. Biol. 5:296-304. [DOI] [PubMed] [Google Scholar]
- 20.Maison, C., D. Bailly, A. H. Peters, J. P. Quivy, D. Roche, A. Taddei, M. Lachner, T. Jenuwein, and G. Almouzni. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30:329-334. [DOI] [PubMed] [Google Scholar]
- 21.Mendez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Min, J., Y. Zhang, and R. M. Xu. 2003. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 17:1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muchardt, C., M. Guilleme, J. S. Seeler, D. Trouche, A. Dejean, and M. Yaniv. 2002. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 3:975-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paro, R., and D. S. Hogness. 1991. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc. Natl. Acad. Sci. USA 88:263-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters, A. H., and D. Schubeler. 2005. Methylation of histones: playing memory with DNA. Curr. Opin. Cell Biol. 17:230-238. [DOI] [PubMed] [Google Scholar]
- 26.Plath, K., J. Fang, S. K. Mlynarczyk-Evans, R. Cao, K. A. Worringer, H. Wang, C. C. de la Cruz, A. P. Otte, B. Panning, and Y. Zhang. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300:131-135. [DOI] [PubMed] [Google Scholar]
- 27.Plath, K., D. Talbot, K. M. Hamer, A. P. Otte, T. P. Yang, R. Jaenisch, and B. Panning. 2004. Developmentally regulated alterations in Polycomb repressive complex 1 proteins on the inactive X chromosome. J. Cell Biol. 167:1025-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasmussen, T. P., M. A. Mastrangelo, A. Eden, J. R. Pehrson, and R. Jaenisch. 2000. Dynamic relocalization of histone MacroH2A1 from centrosomes to inactive X chromosomes during X inactivation. J. Cell Biol. 150:1189-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ringrose, L., H. Ehret, and R. Paro. 2004. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol. Cell 16:641-653. [DOI] [PubMed] [Google Scholar]
- 30.Ringrose, L., and R. Paro. 2004. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 38:413-443. [DOI] [PubMed] [Google Scholar]
- 31.Rougeulle, C., J. Chaumeil, K. Sarma, C. D. Allis, D. Reinberg, P. Avner, and E. Heard. 2004. Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol. Cell. Biol. 24:5475-5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silva, J., W. Mak, I. Zvetkova, R. Appanah, T. B. Nesterova, Z. Webster, A. H. Peters, T. Jenuwein, A. P. Otte, and N. Brockdorff. 2003. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev. Cell. 4:481-495. [DOI] [PubMed] [Google Scholar]
- 33.Smith, K. P., M. Byron, C. M. Clemson, and J. B. Lawrence. 2004. Ubiquitinated proteins including uH2A on the human and mouse inactive X chromosome: enrichment in gene rich bands. Chromosoma 113:324-335. [DOI] [PubMed] [Google Scholar]
- 34.Tajul-Arifin, K., R. Teasdale, T. Ravasi, D. A. Hume, and J. S. Mattick. 2003. Identification and analysis of chromodomain-containing proteins encoded in the mouse transcriptome. Genome Res. 13:1416-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wysocka, J., T. Swigut, T. A. Milne, Y. Dou, X. Zhang, A. L. Burlingame, R. G. Roeder, A. H. Brivanlou, and C. D. Allis. 2005. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121:859-872. [DOI] [PubMed] [Google Scholar]