Abstract
Nuclear spatial positioning plays an important role in the epigenetic regulation of eukaryotic gene expression. Here we show a role for nuclear spatial positioning in regulating episomal transgenes that are delivered by virus-like particles (VLPs). VLPs mediate the delivery of plasmid DNA (pDNA) to cell nuclei but lack viral factors involved in initiating and regulating transcription. By tracking single fluorescently labeled VLPs, coupled with luciferase reporter gene assays, we found that VLPs transported pDNA to cell nuclei efficiently but transgenes were immediately silenced by the cell. An investigation of the nuclear location of fluorescent VLPs revealed that the pDNAs were positioned next to centromeric heterochromatin. The activation of transcription by providing viral factors or inhibiting histone deacetylase activity resulted in the localization to euchromatin regions. Further, the activation of transcription induced the recruitment of PML nuclear bodies (PML-NBs) to the VLPs. This association did not play a role in regulating transgene expression, but PML protein was necessary for the inhibition of transgene expression with alpha interferon (IFN-α). These results support a model whereby cells can prevent foreign gene expression at two levels: by positioning transgenes next to centromeric heterochromatin or, if that is overcome, via the type I IFN response facilitated by PML-NB recruitment.
The epigenetic modulation of gene expression regulated by histone modifications, including acetylation, is fundamental to all aspects of cell growth and development (28). The nuclear location of genes can play a significant role in this regulation (2, 40). For instance, gene-dense chromosomes and active chromosomal genes are frequently found more centrally in the nucleus rather than at the nuclear periphery (6, 13, 54, 64) and the positioning of genes to centromeric heterochromatin is conserved evolutionarily and thought to facilitate silencing (8, 7, 14, 24). A role for promyelocytic leukemia nuclear bodies (PML-NBs) in transcriptional regulation has also been proposed, as transcription has been detected at the bodies or has been closely associated with them (4, 30, 34, 60). Further, a number of proteins that are involved in transcriptional regulation, including histone acetyltransferases (HATs), reside in or shuttle in and out of these structures (5, 34).
Recent studies have demonstrated the importance of epigenetic regulation immediately following the nuclear entry of DNA viruses. Histone deacetylase (HDAC) inhibitors, such as trichostatin A (TSA), which raise overall histone acetylation levels in cells (45), increase the infectivity of viruses, such as simian virus 40 (SV40), herpes simplex virus (HSV), and cytomegalovirus (43, 46, 58). Viral early gene products can also modulate HATs and HDACs. For example, the simian virus 40 (SV40) large T antigen (T-ag) upregulates the HAT activity of CREB-binding protein (CBP) (58) and HSV encodes several factors that disrupt repressor complexes or that associate with or phosphorylate HDACs (25, 35, 46). In addition, despite the uncertain cellular role of PML-NBs, the initial replication and transcription steps for many DNA viruses occur at these bodies, suggesting that the juxtaposition of incoming viral genomes with nuclear structures can influence infection (16). Viral factors are required to regulate these events, and the transcription of DNA containing either the SV40 or HSV-1 origin of replication (ori) sequences at this site depends on the presence of T-ag or ICP4 and ICP27 (55, 57).
Here we use mouse polyomavirus-like particles (VLPs) to explore the regulation of foreign DNA by epigenetic mechanisms. VLPs consist of protein nanospheres that are assembled from 360 molecules of the polyomavirus major coat protein VP1, stably associated with a single molecule of plasmid DNA (pDNA), but do not contain any other viral genes or their products (19, 52). VLPs mediate gene expression in a virus-like manner, requiring the viral receptor and the cellular microtubule network (33). Furthermore, the adsorption and internalization of VLPs into the cell cytoplasm appears indistinguishable from that of natural polyoma virions (1, 37). Thus, these particles retain sufficient information to deliver transgenes into the cell via pathways that are used by the natural virus. However, they lack the viral genes and regulatory DNA sequences that mediate the efficient initiation of viral gene expression and, as such, cannot carry out subsequent steps in the viral life cycle.
We demonstrate that VLP-delivered transgenes are efficiently silenced upon entering the nucleus and that expression can be activated by providing viral replication factors or inhibiting HDAC activity. The transgene expression status is related to the nuclear spatial positioning of VLPs. In nonexpressing cells, VLPs are positioned to centromeric heterochromatin in a pDNA-dependent manner, whereas in transcriptionally active cells, VLPs are repositioned to euchromatin regions and associate with PML-NBs even in the absence of viral replication factors. Further, we show that the regulation of transgene expression is unaffected by PML-NB disruption but PML protein is required for the inhibition of expression by alpha interferon (IFN-α). These results suggest that cells have developed specific mechanisms that can prevent expression from foreign genes as soon as they enter the nucleus.
MATERIALS AND METHODS
Cell culture and DNA constructs.
Mouse Swiss albino 3T3, cos7, and HeLa cells were maintained in Dulbecco's modified Eagle's medium with 5% fetal calf serum as previously described (33). Mouse primary embryo fibroblasts from pml−/− or wild-type mice were cultured as described above but in 10% fetal calf serum to a maximum of eight passages. For luciferase assays, the luciferase marker pDNA pCIKLUX, without the SV40 ori (ori mutant) (21) or with the ori (ori+) were used. The latter pDNA was generated by inserting the SV40 noncoding region from nucleotides 5179 to 290 (18) into the NaeI site of pCIKLUX as a PCR-amplified fragment from pEGFP (BD Biosciences) by using primers 5′-TGAGGCGGAAAGAACCAGCTGTG-3′ and 5′-GCCGGAGAACCTGCGTGCAA-3′. pCIKLUX with the mouse polyomavirus ori was prepared similarly but by using PCR fragments amplified using primers 5′-TCGATGAGGTCTACTAG-3′ and 5′-GGTGGTGAGGCTGAAATG-3′ encoding the mouse polyomavirus noncoding region from nucleotides 4967 to 171 (51). pCMVLT2 (53) was used to the express polyomavirus large T antigen. pEGFP (BD Biosciences) was delivered to cells for the determination of transgene expression status in individual cells.
VLP preparation and delivery.
VP1 nanospheres were gradient purified from sonicated insect cells that were infected with recombinant baculovirus encoding polyomavirus VP1, associated with pDNA at a molar ratio of 5:1, and delivered to cells as described previously (33). Calcium phosphate precipitate (CaPi) transfections used standard protocols and similar quantities of pDNA per transfection as those used for the VLP treatment. For fluorescence microscopy, VP1 nanospheres were covalently coupled with succinimidyl-ester-linked Cy3 fluorophores prior to association with pDNA by using 20× (Fig. 2A and C) or 1× (Fig. 2B) the published concentration of Cy3 dye as described previously (23). Qualitatively similar results were obtained by using either 0.5 or 0.05 μg pDNA/5 × 104 cells (data not shown). Cy3 labeling did not significantly compromise VLP-mediated transgene expression (Table 1). When stated, pDNA was labeled with AlexaFluor-488 by using the Ulysis labeling kit (Molecular Probes). Combining Cy3 and AlexaFluor-488 labeling abolished VLP-mediated transgene expression. However, using CaPi, the labeled pDNA was able to express marker genes (Table 2), providing evidence that no free pDNA is present in the VLP-treated cell preparations. Cells were incubated in TSA (Sigma Chemical Company) (solubilized in dimethyl sulfoxide) for 24 h prior to harvesting for 3T3 and primary mouse fibroblast (300 nM), HeLa (600 nM), and cos7 (100 nM) cells or incubated for 24 h prior to VLP or CaPi treatment in IFN-α (Wellferon; 1,000 U/ml), maintained throughout the experiment.
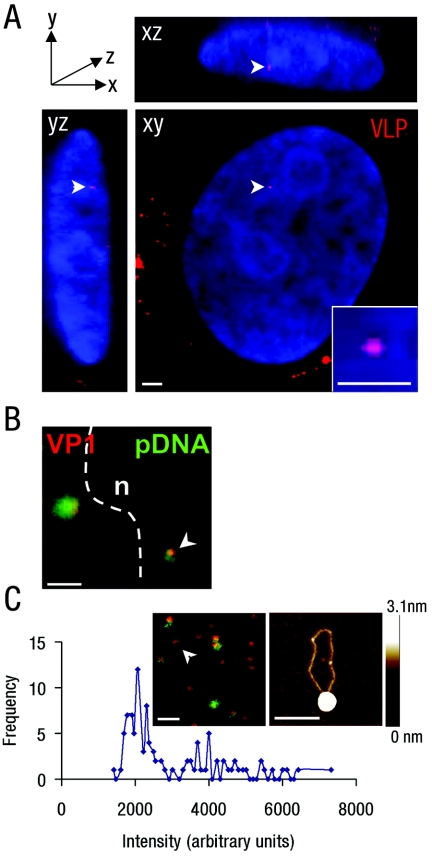
FIG. 2.
VLPs enter the nuclei of transgene-silent and transgene-active cells. (A) Three-dimensional reconstruction of sequential confocal sections of a cos7 cell nucleus in xy, xz, and yz planes, showing the sections containing a single nuclear Cy3-VLP (red; arrowhead). Nuclear DNA stained with DAPI (blue). Inset, digital zoom of single nuclear Cy3-VLP. Bars, 2 μm (main panel) and 1 μm (inset). (B) Overlay confocal images of Cy3-VLPs (red) and AlexaFluor-488-labeled pDNA (green), showing a single nuclear Cy3-VLP (arrowhead), nuclear boundary (dotted line), and nucleus (n). Control experiments with unlabeled pDNA gave a low-level diffuse signal corresponding to secondary antibody binding (data not shown). Bar, 2 μm. (C) VLPs labeled with Cy3 (on VP1 nanospheres; red) and AlexaFluor-488 (on pDNA; green) were dropped onto glass, and a single confocal section was captured (inset). Intensities of 100 individual Cy3 spots were measured and plotted according to fluorescence intensities. The intensity peak corresponding to a single VLP (approximately 2,000 arbitrary units as described previously) (23) and the fluorescence signal corresponding to a single Cy3-labeled VP1 nanosphere associated with a single AlexaFluor-488-labeled pDNA is indicated (inset of left panel, white arrowhead). Similar complexes were dropped onto mica and analyzed by AFM (inset right; height ranges are indicated by the bar to the right). Bars, 1 μm (inset left) and 200 nm (inset right).
TABLE 1.
Influence of Cy3-labeling and pDNA concentration on VLP-mediated reporter gene activitya
| pDNA concn (μg pDNA/5 × 104 cells) | Luciferase activity (RLU/mg of protein) at 24 h
|
||
|---|---|---|---|
| Unlabeled | 1× Cy3 label | 20× Cy3 label | |
| 0.5 | 146,993 | 259,861 | 125,683 |
| 0.05 | 26,626 | 77,237 | 35,847 |
Results are shown as luciferase activity at 24 h from cos7 cells treated with unlabeled or Cy3-labeled VLPs carrying pDNA with the ori.
TABLE 2.
Influence of AlexaFluor-488 pDNA labeling on VLP-delivered reporter gene activitya
| Treatment | Luciferase activity at 24 h
|
|
|---|---|---|
| Unlabeled pDNA | AlexaFluor-488-labeled pDNA | |
| Cy3-VLPs | 1,625,762 | 74 |
| CaPi | 159,278 | 17,930 |
Results are shown as luciferase activity at 24 h from cos7 cells treated with Cy3-VLPs or transfected using CaPi carrying either unlabeled or AlexaFluor-488-labeled pDNA with the ori (0.5 μg/5 × 104 cells).
Luciferase assays.
Cell lysates were assayed at 40 h post-VLP treatment by using a luciferase reporter kit according to the manufacturer's instructions (Promega, United Kingdom). Luciferase activity, given in relative light units (RLU)/mg total protein, was measured in a luminometer (TD-20/20; Turner Biosystems, CA) and standardized against recombinant luciferase (1 RLU is equivalent to 1 U), and the bicinchoninic acid assay (Sigma Chemical Company) was used for measuring total protein. All data are representative of three independent experiments performed in duplicate or triplicate.
Fluorescence microscopy.
Cells were harvested by formaldehyde fixation at 24 h post-Cy3-VLP treatment as described previously (23). Cells were washed with phosphate-buffered saline and permeabilized with 0.1% Triton X-100 in phosphate-buffered saline before incubation with anti-centromeric protein antisera (15), Sm human autoimmune sera, or rabbit anti-PML protein antisera (Santa Cruz, Calif.). After washing, cells were subsequently incubated with anti-human fluorescein isothiocyanate (Jackson ImmunoResearch Laboratories, Inc.) or anti-rabbit AlexaFluor-488-labeled or anti-rabbit AlexaFluor-633-labeled (Molecular Probes) secondary antiserum as appropriate and mounted on glass slides with Vectorshield (Vector Laboratories). The signal from AlexaFluor-488-labeled pDNA (Ulysis labeling kit; Molecular Probes) was amplified with rabbit anti-AlexaFluor-488 antibody (Molecular Probes) (see Fig. 2B, inset). Confocal images were collected sequentially at room temperature on a Leica TCS SP1 or SP2 confocal laser scanning microscope with a 100× oil immersion objective lens (numerical aperture, 1.40) by using argon UV (SP1 only), green argon (488 nm), red HeNe (548 nm for SP2 and 568 nm for SP1), and far red HeNe (633 nm) lasers with a pinhole equivalent to 1 Airy disk unit. Sequential 0.2442-μm sections were collected by using the Leica confocal software in the z axis (minimum 20/stack), and data were averaged from four scans per section. For multiple labeling, no “bleed-through” was detected between channels. Three-dimensional reconstructions were generated by using Volocity software (Improvisions). Metamorph (Universal Imaging Corp.) was used to generate overlaid confocal images. Nuclear VLPs were defined as discrete Cy3 signals in DAPI (4′,6′-diamidino-2-phenylindole), TOTO-3, or Hoechst 33342 staining regions (Molecular Probes). Hoechst 33342 (39) was used to identify centromeric heterochromatin in 3T3 cells, as AT-rich repeat regions are very abundant at this site in mouse cells. Intensity differences for Hoechst 33342 or Sm staining (as a marker for transcriptionally active regions) (59) were used to score Cy3-VLP nuclear localization in 3T3 cells as centromeric heterochromatin regions (colocalized with or next to Hoechst 33342-high/Sm-negative foci), intermediate (Hoechst 33342-medium/Sm-medium regions), or euchromatin (Hoechst 33342-negative/Sm-high foci) regions. In cos7 cells, centromeric heterochromatin is less AT-rich and, therefore, anticentromere human autoimmune sera recognizing the centromeric proteins CENP-A, -B, and -C and marking the location of pericentromeric heterochromatin was used (15). Colocalization and “next to” were defined as overlapping or adjacent pixels of fluorescence signal, respectively. The frequency of nuclear Cy3-VLPs at defined locations was scored for at least three independent experiments. The chi-square test (Minitab version 14) was used to establish that, for a given variable, there was no significant difference between experiments. Data were then pooled, and chi-square analysis was performed to determine whether the observed reproducible differences in nuclear location in the absence or presence of TSA or following the delivery of pDNA without or with ori were statistically significant.
Atomic force microscopy.
VP1 nanosphere-pDNA complexes, prepared as for cell experiments, were diluted in water to 60 ng/μl, dropped onto the surface of freshly cleaved mica, and allowed to adsorb for 1 min. The mica surface was then rinsed with water, and excess water was removed under a stream of nitrogen for 30 s. Silicon probe tips were used to scan the samples in tapping mode by using a MultiMode SPM and Nanoscope IIIa control system (Digital Instruments, Inc., Santa Barbara, CA).
Hirt extracts, Southern blotting, and PCR.
Low-molecular-weight DNA was isolated from transfected cells (10 μg pDNA/1 × 106) at 24 h according to the method of Hirt (26). Transfections were carried out at submaximal levels of pDNA to ensure that CaPi delivery was not saturating. Proteins were removed by phenol-chloroform extraction and DNA ethanol precipitated. One-sixth of each extract, which is equivalent to 2 μg low-molecular-weight nucleic acid, was then digested with restriction endonucleases in the presence of RNase A, and the DNA fragments were separated on a 1% agarose gel and transferred to nylon membrane by Southern blotting. pCIKLux was radiolabeled with [32P]dCTP by random hexamer priming, and the blot was probed using standard protocols.
For the PCR-based assay, DNA was isolated from 4 × 104 cells at 24 h post-VLP addition by using InstaGene chelate mix (Roche, Lewes, United Kingdom) according to the manufacturer's instructions. Column purification using the High Pure PCR purification kit (Roche) removed the InstaGene chelate mix, as this inhibits restriction endonuclease digestion. Isolated DNAs were then titrated for pDNA concentration against known pDNA template concentrations by PCR amplification with plasmid-specific primers spanning a region containing MboI and DpnI restriction endonuclease sites (5′-GTAACCACCACACCCGCCGCG-3′ and 5′-AGGAGAGCGCACGAGGGAGCTTCCAGGGG-3′). DNA amplification was performed in a 50-μl reaction mix of 0.5 U of Taq polymerase (Promega), 50 pmol of each primer, 125 μM of each deoxynucleoside triphosphate, and 1.5 mM MgCl2. PCRs were run for 25 cycles of denaturation at 95°C for 1 min, with annealing at 64°C for 1 min and elongation at 72°C for 3 min. Finally, a volume equivalent to 10 picograms of isolated pDNA was subjected to mock or DpnI or MboI restriction endonuclease digestion followed by PCR amplification as described above. PCR products were fractionated on a 1.2% agarose gel containing 5 ng/ml ethidium bromide. Note: DpnI treatment digests input pDNA, leading to specific amplification of pDNA replicated within the transfected cells. MboI treatment digests DNA replicated within mammalian cells, resulting in specific amplification of input pDNA.
Nucleosome assay.
Nuclei were prepared from HeLa cells (1 × 106) 24 h after transfection with VLPs or CaPi carrying pCIKLux with ori by centrifugation, as described previously (10). Following lysis in nonionic detergent, nuclei were exposed to increasing concentrations of micrococcal nuclease (0 to 100 units/1 × 105 nuclei) for 5 min at 37°C. Sodium dodecyl sulfate was added to a final concentration of 2%, and products were loaded directly onto a 1.8% agarose gel followed by subsequent staining with ethidium bromide. Fragments were transferred to nylon membrane and probed for pDNA-specific sequences as described above. Control experiments were carried out with untransfected HeLa cell nuclei mixed with pDNA (0.1 μg per digest).
RESULTS
VLP-delivered transgenes are silenced following nuclear entry but can be activated by viral replication factors or TSA.
In most cell lines, transgene expression from VLP-delivered pDNA was barely detectable (as shown for two commonly used cell lines, cos7 and 3T3) (Fig. 1A, white bars) even though transfection with calcium phosphate precipitate (CaPi) typically yielded luciferase expression of over 104 RLU/mg total protein for most cell lines tested and over 106 RLU/mg total protein for cos7 cells. When viral replication factors were added to the system, VLP-mediated transgene expression was activated in all cell lines tested (illustrated for cos7 cells expressing T-ag treated with pDNA containing the SV40 ori [Fig. 1A and see below]). Alternatively, expression was activated when cells were cultured with HDAC inhibitors, such as TSA (Fig. 1B, gray bars).
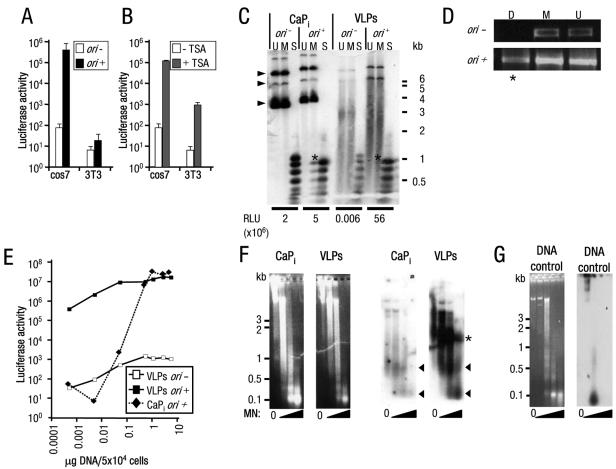
FIG. 1.
High-level, copy number-independent VLP-delivered transgene expression activated by viral factors or TSA. (A and B) Luciferase activity (RLU/mg total protein) from (A) cos7 or 3T3 cells treated with VLPs carrying pDNA without (−) or with (+) ori and from (B) cos7 or 3T3 cells treated with VLPs carrying pDNA without ori, cultured without or with TSA. Note: mock VLP-treated cells cultured with TSA resulted in less than 100 RLU/mg total protein (data not shown); y axis is a log scale. Error bars indicate the standard errors. (C) Southern blot analysis of low-molecular-weight DNA Hirt extracts (26) from cos7 cells transfected using CaPi or treated with VLPs carrying pDNA without (−) or with (+) ori. Restriction endonuclease digests of DNA extracts (U, mock-digested DNA; M, MboI [inhibited by dam methylation, digests only DNA replicated in mammalian cells]; or S, Sau3A [digests all DNA]) demonstrated pDNA replication products present in CaPi-transfected but not VLP-treated cells (indicated by asterisks). The positions of migration of form I, II, and III DNAs are indicated by arrowheads. Considerable degradation is observed for the VLP-delivered pDNA, presumably due to the majority of the plasmid being unprotected from nuclease attack in the cell (52). Luciferase activity (RLU/mg total protein) from an aliquot of each transfected cell culture taken prior to Hirt extraction is given below the appropriate tracks. Positions of migration of molecular weight standard markers are shown to the right of the gel. (D) PCR analyses of low-molecular-weight DNA extracts from cos7 cells treated with VLPs as described for panel C. Plasmid-specific PCR amplification was performed after digestion of the extracts with the restriction endonucleases DpnI (D, inhibited by mammalian methylation; PCR amplification detects mammalian replicated pDNA only; indicated by asterisks) or MboI (M, inhibited by dam methylation; PCR amplification detects bacterial input pDNA only) or mock-digested DNA (U). (E) Luciferase activity from cos7 cells treated with increasing amounts of VLPs or CaPi (equivalent to 0.0005 to 5 μg pDNA/5 × 104 cells) carrying pDNA without (−) or with (+) ori. Note: both axes are a log scale. (F) Micrococcal nuclease assays of HeLa cell nuclei transfected with CaPi or treated with VLPs. Following electrophoresis of micrococcal nuclease (MN)-treated samples, the agarose gels were stained with ethidium bromide (left panels) and then subjected to Southern blot analysis (right panels). Consistent with previous observations (29), atypical nucleosome ladders for CaPi-transfected pDNA were detected, represented by protected pDNA fragments of approximately 0.1 and 0.6 kb (arrowheads). Similar ladders were detected for VLP-delivered pDNA. A further band of VLP-protected pDNA fragments (52) of 1.2 to 1.5 kb was also detected (asterisks). (G) Untransfected HeLa cell nuclei with pDNA added to isolated nuclei prior to micrococcal nuclease digestion and then analyzed as described above for panel F. Left panel, ethidium bromide stain; right panel, Southern blot analysis. Positions of migration of molecular weight standard markers are shown to the left of the gels.
The most obvious reasons for the viral replication factors generating expression could be that T-ag either increased the copy number by driving ori-dependent pDNA replication (22) or enhanced the transport of the pDNA to the nucleus (41), thus providing more templates for transcription. However, it seems unlikely that TSA, by simply inhibiting HDACs, would be able to reproduce the combined effect of T-ag and ori. We therefore considered an alternative explanation, which was that each method directly activated expression of the transgene by epigenetic means rather than enhancing copy number or delivery. We present several lines of evidence in support of this notion. First, our data suggest that expression is not proportional to copy number. The amount of replicated ori-containing pDNA was used as an indicator of the number of copies of pDNA delivered to the nucleus of cos7 cells that were treated with VLPs or transfected with CaPi. These data were then compared with luciferase expression for each culture (Fig. 1C). In this experiment, the Southern blotting of Hirt-extracted low-molecular-weight DNA showed that replication of pDNA with ori in VLP-treated cells was far lower than that in control cells transfected using CaPi, as judged by the signal intensity for pDNA cleaved with the mammalian methylase-sensitive enzyme MboI (lanes M). Low-level replication was, however, detected by PCR for VLP-delivered pDNA; this is consistent with very low copy number delivery (Fig. 1D). Since the amount of replication of VLP-delivered pDNA was less than that observed for CaPi delivery yet luciferase expression from the same cultures was approximately 10-fold higher (55.8 × 106 versus 4.9 × 106 RLU/mg total protein), this suggests that expression is not proportional to copy number. Second, expression from VLP-delivered pDNA was barely detectable in the absence of viral replication factors, irrespective of the number of VLPs added per cell, yet in the presence of replication factors, significant levels of expression could be obtained by adding as few as 103 VLPs per cell (equivalent to 0.0005 μg pDNA/5 × 104 cells) (Fig. 1E). Third, the increased expression in the presence of TSA is not due to enhanced delivery since this reagent, added at any time before or following VLP treatment (even two weeks later), resulted in similar activations of the transgene (data not shown). Fourth, nucleosome assembly of VLP-delivered pDNA could be demonstrated, which is consistent with a role for epigenetic effects in the regulation of the pDNA-encoded genes (Fig. 1F). Nuclei from CaPi-transfected or VLP-treated cells were incubated with micrococcal nuclease, which cleaves DNA between nucleosomes, to generate a ladder of protected fragments on agarose gels (3). pDNA-specific sequences delivered by either CaPi or VLPs migrated as two fragments (Fig. 1F); this was similar to nucleosome ladders that were previously observed for transfected pDNAs (29). Untransfected pDNA was totally degraded (Fig. 1G), demonstrating that the DNA sequences were not inherently resistant to nuclease attack. Thus, VLP-delivered pDNA is assembled into nucleosomes, consistent with a role for TSA or replication in facilitating transcription by nucleosome displacement or reorganization. In summary, there appear to be two distinct expression states for VLP-delivered transgenes: a silent state (transgene silent) which occurs by default immediately following the delivery of pDNA to the nucleus and a specifically activated state (transgene active) that is obtained by adding viral replication factors or TSA to the system.
VLPs deliver an average of two pDNA molecules to transgene-silent or transgene-active cell nuclei.
To confirm that transgene delivery to the nucleus is unaffected by treatments that activated transgene expression, we asked whether the delivery of VLPs could be detected in the nuclei of transgene-silent and transgene-active cells. VP1 nanospheres were coupled covalently to the fluorescent dye Cy3 and complexed with pDNA to generate fluorescently labeled VLPs (Cy3-VLPs). Cells were then treated with Cy3-VLPs and examined by confocal microscopy. Three-dimensional reconstructions of sequential confocal sections showed only small numbers of Cy3-related fluorescent spots in each nucleus and only in cells that were treated with Cy3-VLPs (Fig. 2A; control data not shown). In 3T3 cells, these VLP-related spots were detected in a quarter to a half of treated cells, with one or two VLPs on average per nucleus, irrespective of the expression status of the culture (Table 3). In cos7 cells, VLPs were detected in more nuclei (up to 68%), with an average of two per nucleus, irrespective of expression status (Table 3). No nuclei were found with more than eight VLPs. Further, if Cy3-labeled nanospheres were associated with pDNA conjugated to AlexaFluor-488, pDNA-specific fluorescence was associated with nuclear Cy3 spots and little or no free signal was detected (Fig. 2B). As spot fluorescence intensities of nuclear Cy3-VLPs were similar to those of single Cy3-VLPs spread on glass (23) (Fig. 2C shows an example of calculating intensities for single VLPs associated with AlexaFluor-488-labeled pDNA), each nuclear spot most likely represents a single VLP. The associated pDNA fluorescence most probably represents no more than one molecule of pDNA, as electron microscopy (52) and atomic force microscopy (AFM) studies (Fig. 2C) of the VLP protein-DNA complexes show that each VP1 nanosphere interacts with only one pDNA molecule. Although the AlexaFluor-488 and Cy3 fluorescence do not always overlap completely, only 1 to 2 kb of pDNA is closely associated with the protein nanospheres (52). AFM analysis of VLPs (Fig. 2C) revealed a considerable proportion of pDNA trailing several hundred nanometers from the VP1 nanosphere, providing one possible explanation for the offset in the fluorescence signals. Thus, VLPs deliver a similar and small number of pDNA molecules to both transgene-silent and transgene-active cell nuclei. Further, since the addition of TSA could activate transgene expression even after the time at which the above data were collected (for example, at 24 h or later, following VLP addition), expression-competent pDNA must have been present in a significant proportion of the transgene-silent cells. This confirms that, in the absence of specific activation, VLP-delivered transgenes are effectively silenced within the nucleus.
TABLE 3.
Number of VLPs delivered per cell nucleus in the absence or presence of viral replication factors or TSAa
| Parameter | Value for:
|
||||
|---|---|---|---|---|---|
| 3T3 cells
|
cos7 cells
|
||||
| ori mutant without TSA (n = 121; silent)b | ori mutant with TSA (n = 123; active) | ori mutant without TSA (n = 78; silent) | ori mutant with TSA (n = 31; active) | ori† without TSA (n = 64; active) | |
| No. of nuclei containing the following no. of VLPs | |||||
| 0 | 72 | 89 | 42 | 10 | 25 |
| 1 | 26 | 22 | 21 | 8 | 19 |
| 2 | 13 | 9 | 11 | 4 | 12 |
| 3 | 7 | 2 | 2 | 3 | 2 |
| 4 | 2 | 0 | 1 | 3 | 2 |
| 5 | 1 | 1 | 1 | 3 | 2 |
| 6 | 0 | 0 | 0 | 0 | 1 |
| 7 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 0 | 0 | 0 | 1 |
| % of nuclei with VLPs | 40.5 | 27.6 | 46.2 | 67.7 | 60.9 |
| Avg. no. of VLPs/nucleusc | 1.75 | 1.5 | 1.58 | 2.48 | 2.07 |
Number of Cy3-fluorescent VLPs counted (as described in the legend for Fig. 2) in 3T3 or cos7 cells with VLPs carrying either ori-mutant or ori+ pDNA and incubated with or without TSA.
The expression status was determined by luciferase assays of parallel cultures and is indicated as active or silent. n, total number of nuclei counted per cell line and treatment.
Calculated for VLP-positive nuclei only.
VLPs adopt different nuclear locations in transgene-silent or transgene-active cells.
Using the ability to switch between expression states, we asked whether the nuclear location of VLPs differed between transgene-silent and transgene-active cells and whether or not there is any difference in location between the two methods of activation (i.e., T-ag/ori or TSA). The nuclear periphery and centromeric heterochromatin have been associated with low gene activity (as described above). Cy3-VLPs did not appear to localize preferentially to the periphery in transgene-silent cells (data not shown), so their location with respect to centromeric heterochromatin was examined. For cos7 cells, the location of pericentromeric heterochromatin was determined by immunofluorescence with autoimmune sera against human centromere proteins (Fig. 3A and K). In transgene-silent cos7 nuclei, 63% of Cy3-VLPs were found next to this site. In mouse cells, the A/T nucleotide content of centromeric heterochromatin results in intense staining with the DNA dye Hoechst 33342, whereas euchromatin regions lack staining (39). Therefore, Hoechst 33342 was used to distinguish the location of Cy3-VLPs in 3T3 cells, as it allowed the scoring of VLP locations with respect to both centromeric heterochromatin and euchromatin. In this case, 54% of VLPs were next to centromeric heterochromatin, although some Cy3-VLPs (15%) were also localized to euchromatin regions (Fig. 3B and L). By contrast, in transgene-active cos7 cells treated with Cy3-VLPs carrying pDNA with ori, only 13% of Cy3-VLPs were next to centromeric heterochromatin (Fig. 3C and K). Similarly, when 3T3 cells were cultured with TSA, the proportion of Cy3-VLPs that was associated with centromeric heterochromatin was reduced significantly, from 54% to 27% (Fig. 3D and L). Further, in Hoechst 33342-stained 3T3 cells, the Cy3-VLPs not only moved away from centromeric heterochromatin upon the addition of TSA but also mostly (54%) repositioned to euchromatin regions (Fig. 3D and L).
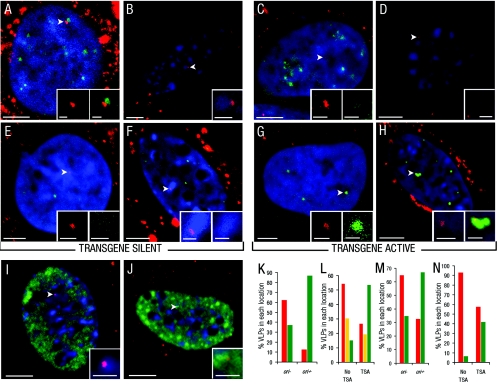
FIG. 3.
Location of VLPs with respect to centromeric heterochromatin, euchromatin regions, and PML-NBs. (A to J) Location of Cy3-VLPs relative to centromeric heterochromatin or euchromatin regions or PML-NBs in cos7 cells treated with Cy3-VLPs carrying pDNA without (transgene silent) (A and E) or with ori (transgene active) (C and G) or in 3T3 cells cultured without (transgene silent) (B and F) or with TSA (transgene active) (D and H). The top panels show representative overlay images of single confocal sections with nuclear Cy3-VLPs (red; arrowheads; determined as described in the legend for Fig. 2A) relative to immunostaining for centromeric proteins in cos7 cells (green) with DAPI-stained nuclei (blue) (A and C) or Hoechst 33342-stained nuclear regions in 3T3 cells (blue) (B and D). The middle panels show representative overlay images of single confocal sections with nuclear Cy3-VLPs (red; arrowheads) relative to immunostaining for PML protein (green) with DAPI-stained nuclei (blue) in cos7 cells (E and G) or Hoechst 33342-stained regions (blue) in 3T3 cells (F and H). Note: the signals for immunostained PML protein in panels E and F are less intense than for those in panels G and H as in the former, PML-NBs are not in the same sections as the VLPs. The bottom panels show representative overlay images of single confocal section with nuclear Cy3-VLPs (red; arrowheads) relative to immunostaining for Sm antigen (green)- and Hoechst 33342-stained nuclear regions (blue) in 3T3 cells cultured without (I) or with (J) TSA. Insets show digital zooms of Cy3-VLPs with corresponding cellular structure. (K through N) Graphical representation of the percentage of VLPs in each nuclear location in cos7 cells without (−) or with (+) ori or 3T3 cells without or with TSA, with respect to location next to centromeric heterochromatin or euchromatin regions (red and green bars, respectively, in panel K); centromeric heterochromatin, euchromatin regions, or intermediate regions (red, green or yellow bars, respectively, in panel L); or away from or next to PML-NBs (red and green bars, respectively, in panels M and N). Using chi-squared analysis (described in Material and Methods) the observed difference in Cy3-VLP nuclear location following treatment with TSA or delivery of ori for each panel was statistically significant (K through N) (P < 0.001). Bars, 5 μm (main panels) and 1 μm (insets).
Sm antigens comprise a family of factors that are involved in splicing RNA, and their locations are reported to correlate with sites of transcriptional activity (31, 38, 49). Therefore, the localization of VLPs was also compared with that of the Sm antigen by immunofluorescence. Consistent with the results for staining for centromeric heterochromatin and euchromatin, VLPs in transgene-silent cells were more frequently localized in areas of low Sm antigen immunostaining, and in transgene-active cells, they were mostly in high-Sm-antigen-staining areas (Fig. 3I and J, respectively). Thus, the repositioning of VLPs upon activation of transgene expression resulted in transfer from areas of low to high transcriptional activity.
As cos7 cells express T-ag constitutively, it was not possible to determine whether incoming VLPs carrying pDNA with ori initially localized to centromeric heterochromatin and were then repositioned or whether they were targeted directly to euchromatin regions. However, for 3T3 cells, which are transgene silent until TSA is added (Fig. 1B shows an example of activation by TSA added at 24 h following VLP treatment; also see Materials and Methods), Cy3-VLPs must initially localize to centromeric heterochromatin and then be repositioned to euchromatin regions.
Since the localization of the VLPs to centromeric heterochromatin appeared very specific, we next explored the question of which component of VLPs might be responsible by determining the nuclear location of fluorescent VP1 nanospheres in the absence of complexation with pDNA. These particles entered transgene-silent 3T3 cell nuclei with a frequency similar to that of VLPs (i.e., in 33.3% of treated cells). However, they did not tend to localize with centromeric heterochromatin since in only 11% of the cells were Cy3-VP1 nanospheres found next to these structures compared to 71% of the cells for VLPs with pDNA. These data indicate that nuclear positioning depends on the pDNA rather than the VP1 nanospheres.
To summarize these results, the default pathway for VLP-delivered pDNA results in very efficient transgene silencing accompanied by pDNA-dependent deposition at the relatively transcriptionally inactive centromeric heterochromatin. Conversely, the activation of expression by viral replication factors or HDAC inhibition correlates with the repositioning of the transgenes to the more transcriptionally active euchromatin regions.
VLPs colocalize with PML-NBs in transgene-active cells.
The addition of T-ag and ori, which activate VLP-mediated expression, is known to be sufficient to localize the transcription of transfected pDNA to PML-NBs (55). Therefore, we examined whether these viral replication factors localized Cy3-VLPs to PML-NBs. In transgene-silent cos7 cells, 35% of Cy3-VLPs carrying pDNA lacking ori were found next to PML-NBs (Fig. 3E and M), whereas in 3T3 cells in the absence of both T-ag and ori, only 7% of Cy3-VLPs were near these structures (Fig. 3F and N). Conversely, as predicted for cos7 cells in the presence of both T-ag and ori, the majority of VLPs (66%) were associated with PML-NBs (Fig. 3G and M). Unexpectedly, TSA treatment of 3T3 cells also led to a considerable increase in Cy3-VLPs next to PML-NBs (from 7 to 42%) (Fig. 3H and N). This was surprising as it is thought that, for SV40, replication of the viral DNA is required to localize transcription to this site (55). Our data, however, show that VLPs per se have little affinity for PML-NBs but that, when transgene expression is activated by TSA and VLPs are repositioned to euchromatin regions, PML-NBs associate with VLPs even in the absence of SV40 T-ag and ori. Thus, the colocalization of VLP-delivered transgenes with PML-NBs is a consequence of transgene expression rather than the presence of specific viral replication factors.
Association of VLPs with PML-NBs plays a role in posttranscriptional events.
Since the association of PML-NBs with VLPs was linked to transgene expression, we asked whether PML-NBs were necessary for transgene expression. To address this, mouse embryo fibroblasts from pml−/− mice, in which PML-NBs are disorganized, were treated with VLPs. Under transgene-silent or transgene-active conditions (induced by TSA), transgene expression in pml−/− mouse fibroblasts was similar to that of their wild-type counterparts (Fig. 4A). Thus, the PML protein itself is not required for transgene silencing or activation.
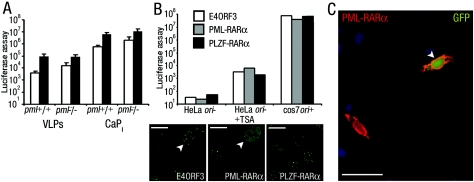
FIG. 4.
Disruption of PML-NBs does not affect VLP-delivered transgene expression status. (A) Luciferase activity from primary embryo fibroblasts isolated from wild-type (pml+/+) or pml−/− mice that were treated with VLPs or transfected using calcium phosphate precipitate (CaPi) cultured without (open bars) or with TSA (closed bars). Error bars indicate the standard errors. (B) Top, Luciferase activity from cells (as indicated) pretransfected with PML-NB-disrupting gene E4ORF3 or PML-RARα or with nondisrupting control gene PLZF-RARα. Cells were cultured without or with TSA as indicated or in the presence of viral factors (cos7 ori+). Bottom, PML-NB disruption was confirmed by immunofluorescence for PML protein (transfected cells are indicated with arrowheads), and overexpression of RARα fusion proteins was confirmed by immunofluorescence for RARα (data not shown). Bar, 10 μm. (C) Fluorescence image of HeLa cells pretransfected with PML-RARα and then treated with VLPs carrying pDNA encoding the EGFP protein and cultured with TSA. A cell overexpressing PML-RARα (red; detected by immunostaining for RARα) and expressing EGFP (green) is indicated by the arrowhead. Cell nuclei were stained with DAPI (blue). Bar, 20 μm.
Next, PML-NBs were transiently disrupted by the overexpression of a viral gene or cellular promyelocytic leukemia protein-retinoic acid receptor (RAR) gene fusion product (E4ORF3 or PML-RARα) (Fig. 4B) (9, 62). Approximately 50% of the transiently transfected cells expressed E4ORF3, PML-RARα, or a control gene fusion between the promyelocytic leukemia zinc finger protein and retinoic acid receptor (PLZF-RARα) (32). If PML-NB integrity is required for transgene silencing, disruption of PML-NBs in 50% of cells would result in a readily detectable activation of VLP-mediated expression. This was not observed (Fig. 4B) and leads us to conclude that PML-NBs are not involved in regulating transgene silencing. These experiments also suggest that PML-NBs do not play a role in transgene activation, although the loss in transgene activation following PML-NB disruption in 50% of cells would equate to an only twofold decrease in expression, which may not be reliably detected. If PML-NB integrity is necessary for transgene activation, it should not be possible to detect VLP-delivered transgene-expressing cells with disrupted PML-NBs. Therefore, PML-RARα- and PLZF-RARα-expressing cells were compared on an individual basis for VLP-mediated enhanced green fluorescent protein (EGFP) expression under transgene-activating conditions (induced by TSA). EGFP expression was readily detected in cells transiently expressing PML-RARα (Fig. 4C). Of the cells that expressed the fusion proteins, 21% of those expressing PML-RARα (i.e., with disrupted PML-NBs) and 24.5% of those expressing PLZF-RARα (as control) also expressed VLP-delivered EGFP transgenes. These results confirm that the disruption of PML-NBs does not reduce the potential for transgene activation. Thus, PML-NB integrity is not necessary for the silencing or activation of VLP-delivered transgenes.
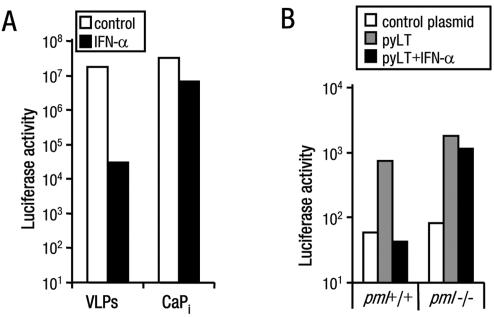
Cells lacking PML protein are not able to mount a successful IFN response against viral infection (11, 48). Therefore, the effect of IFN on VLP-delivered transgene expression was explored in cells with either intact or disorganized PML-NBs. First, the sensitivity of VLPs to IFN-α was determined. Consistent with the virus-like delivery of pDNA to cells by VLPs (33), treatment of cos7 cells with IFN-α resulted in a 1,000-fold reduction in replication factor-driven transgene expression. Under similar conditions, the expression from CaPi-delivered pDNA was reduced less than 10-fold (Fig. 5A). These data show that VLPs activate the IFN-α antiviral pathways (20) to a level sufficient to sensitize the cells to exogenously added IFN.
FIG. 5.
PML protein is necessary for inhibition of VLP-mediated expression by IFN-α. (A) Luciferase activity from cos7 cells that were treated with VLPs or transfected using CaPi carrying pDNA with ori cultured without or with IFN-α. (B) Luciferase activity from primary embryo fibroblasts isolated from wild-type (pml+/+) or pml−/− mice that were pretransfected with a control plasmid (pEGFP; open bars) or pCMVLT2 carrying the polyomavirus large T antigen (pyLT) gene and then treated with VLPs carrying pDNA with the polyomavirus ori cultured without (open and gray bars) or with IFN-α (closed bars). Data are representative examples of at least three independent experiments.
We next asked whether IFN-α would be similarly effective in pml−/− cells. Wild-type or pml−/− mouse cells were pretransfected with the mouse polyomavirus large T antigen (as the SV40 T-ag is not able to interact efficiently with the mouse DNA polymerase complex) (42). Cells were then treated with VLPs carrying pDNA with the polyomavirus ori. In both cell types, VLP-mediated transgene expression was detected and, although the levels of expression were low, they were reproducibly higher than the levels for cells that were pretransfected with a control pDNA (Fig. 5B). If cells were then cultured with IFN-α, expression was inhibited in the wild-type cells but little effect was observed in the pml−/− cells. Taken together, these data show that PML-NBs are not necessary for silencing or activating VLP-mediated expression but they may play a role in mediating the IFN antiviral response.
DISCUSSION
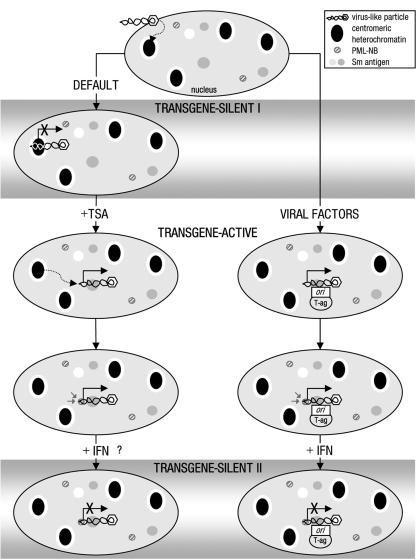
In this report, we describe a method for tracking single VLP-delivered pDNA molecules in cell nuclei. Using this technique, we have established a model for the earliest events following entry of foreign DNA delivered by VLPs into a mammalian cell nucleus (Fig. 6). VLP-delivered transgenes are immediately silenced following nuclear entry, and this correlates with the initial localization of VLPs to centromeric heterochromatin in a pDNA-dependent manner (transgene silent I). We suggest that this represents a default cellular defense pathway for recognizing and inactivating foreign DNA, which VLPs, lacking the majority of viral components, are unable to counteract. In the presence of the HDAC inhibitor TSA, the repositioning of transgene pDNA from centromeric heterochromatin to Sm antigen-rich euchromatin regions and activation of transcription occur (transgene active). Alternatively, in the presence of viral factors (T-ag and ori), VLPs are also localized to Sm antigen-rich regions and transcribe. In both cases, the activation of expression leads to an association with PML-NBs which may occur de novo, as has been observed for HSV-1 (17). However, rather than being required for regulating expression per se, we propose that the main function of PML-NBs' recruitment is to facilitate a second phase of cellular defense against viral attack via the IFN-α pathway (transgene silent II).
FIG. 6.
Model for a two-phase cellular defense against VLP-delivered pDNA. Hypothetical stages in the response of cells to invasion by foreign genes are given. The sequence of events is derived from the data presented, with the exception of the order of events for activation and repositioning of transgenes in the presence of TSA, since it is not known which occurs first. The locations and relative positions of VLP and cellular components shown are schematic representations derived from fluorescence data. Transgene expression status is based on luciferase assay data. Inhibition of HDAC activity can attenuate the IFN antiviral pathway (44); therefore, the effect of IFN-α treatment on TSA-mediated transgene activation has not been determined.
Association of viral genomes with centromeric heterochromatin has not been observed for DNA viruses, but this is likely to be due to the evolution of viral strategies to overcome this silencing mechanism. For example, polyomavirus genomes are assembled into nucleosomes with acetylated histones prior to packaging into the virion (12, 50). This may provide sufficient transcriptional activation for the expression of replication factors that are required to establish infection. Alternatively, factors packaged in the virion particle, for instance those of the more complex herpesviruses, may contribute to preventing deposition at centromeric heterochromatin. It is noteworthy in this context that an early electron microscopy study observed the deposition of oncoretroviral genomes at centromeric heterochromatin (36).
The presence of specific viral factors or the HDAC inhibitor TSA can trigger the activation of transcription. The viral factor that was used in this study, T-ag, is a multifunctional protein, so distinguishing which functions are relevant in activating expression is problematic. The fact that HDAC inhibition can produce a similar switch from transgene silent to transgene active states suggests that T-ag binding to the HAT p300/CBP is relevant (58). This could be a direct action on the chromatin-like structure of the transgenes via histone acetylation, as although the pDNA remains episomal (19) and is bacterial in origin, it is assembled into nucleosomes within 24 h following delivery (see Fig. 1F). However, for efficient T-ag-mediated activation, the pDNA also has to carry ori. These findings suggest three possible mechanisms for overcoming silencing by viral factors. First, T-ag binding to ori could recruit activated p300/CBP to the pDNA, increasing histone acetylation at the transgene promoter, facilitating recruitment of transcription factors, or decreasing the activity of corepressor complexes. Second, T-ag binding to ori could tether the pDNA at, or transport it to, euchromatin regions. Or, finally, T-ag binding to ori could initiate pDNA replication, leading to the unwinding of the structure and allowing access to transcription factors. Regarding this latter point, we found a clear requirement for both T-ag and ori for transcriptional activation, but also for active DNA polymerase (data not shown). Slightly higher basal levels of transgene expression are reproducibly observed in the presence of T-ag (i.e., in cos7 relative to 3T3 cells with VLPs carrying pDNA without ori), suggesting that T-ag can potentiate, but is not sufficient for, transgene activation. Thus, our data currently point to a model where the predominant event for activating transgene expression is modification of the chromatin structure, induced either by the initiation of pDNA replication by T-ag/ori or by the acetylation of histones by the action of TSA. However, we cannot rule out a small but significant contribution from the p300/CBP-activating properties of T-ag. Future experiments will help to resolve this issue and determine whether changes in the chromatin structure alone or acetylation of nonhistone proteins are involved in physically relocating the episomes from one site to another.
In euchromatin regions, VLPs are mostly found associated with PML-NBs. Although these bodies are found localized to sites of early transcription and replication for many viruses (16), this is the first time, to our knowledge, that transcription from delivered transgenes has been suggested to be the trigger for association with PML-NBs. The observation that PML-NBs associate with transcribing genes but are not needed to activate expression has been made for other viral genomes and chromosomal genes, but it was concluded that viral proteins acted as PML-NB retention signals (57) or that PML-NBs acted as a depot for transcription factors (61). Further, in pml−/− mouse cells lacking the PML protein, where the bodies are disorganized, we found no evidence for a requirement for PML protein. Similar observations have been made for infection by viruses (11, 56), but these experiments cannot exclude the possibility that the pml-deficient cells have compensated for the loss of the gene and have a sub-PML-NB structure that is functionally equivalent. However, we rule out a need for intact PML-NBs for the regulation of VLP-delivered genes, as transient disruption with viral factors or cellular oncogenes also had little effect. Thus, these results argue against a pivotal role for PML-NBs in facilitating viral DNA replication and transcription. Instead, the bodies appear to be involved in defending the cell from viral attack, as pml−/− cells can no longer support inhibition by IFN of VLP-mediated expression. A similar need for PML protein has also been demonstrated for the inhibition of viral replication by type I interferons for a number of viruses (11, 48). Therefore, since PML-NBs inhibit VLP-mediated expression by IFN and associate with VLPs in transgene-active cells even in the absence of viral replication factors, we propose that transcription from the foreign DNA is the trigger for PML-NBs recruitment, ready for the cells to mount the IFN response.
In conclusion, we have developed a model to describe the first interactions of foreign DNA with cellular nuclear architecture. Our data demonstrate that cells can efficiently assimilate foreign genes, even those encoded on purified bacterially expressed pDNA episomal molecules, into a form that is not just assembled into nucleosomes but can be recognized and silenced by sophisticated epigenetic means. Furthermore, we show that the pDNA is the major determinant for this epigenetic regulation. The facts that TSA can increase transgene expression for a number of nonviral delivery systems (27, 47, 63) and TSA induced a 350-fold increase in transgene expression from tissue culture cells treated with naked pDNA (unpublished results) most likely reflect that silencing by positioning to centromeric heterochromatin is not restricted to VLP delivery. These findings predict that the inactivation of viral factors that overcome targeting of their genomes to centromeric heterochromatin would provide a viable antiviral strategy. Further, the localization of pDNA to centromeric heterochromatin may be a primary barrier of transgene expression to be surmounted if effective gene therapy is to be developed. Finally, future studies to define factors that are involved in the repositioning of VLPs will help to advance the understanding of the mechanisms underlying the chromatin remodeling and nuclear spatial repositioning of cellular genes.
Acknowledgments
We thank A. Pombo, M. Merkenschlager, K. Linton, P. Hobson, and K. Brown for helpful discussions; W. Earnshaw and A. Pombo for gifts of autoimmune sera against centromeric proteins or Sm; P. Pandolfi for pml−/− and pml+/+ mouse fibroblasts; K. Leppard, P. Freemont, and A. Zelent for E4ORF3, PML-RARα, and PLZF-RARα expressing pDNAs; and A. Porter for the Wellferon. We also thank M. Elliott and N. Bennett for performing the AFM and F. Ramirez for performing chi-squared analysis.
We are grateful to the Medical Research Council United Kingdom and the Hammersmith Hospitals Trust Research Committee (grant no. 20223) for financial support.
REFERENCES
- 1.An, K., A. Q. Paulsen, M. B. Tilley, and R. A. Consigli. 2000. Use of electron microscopic and immunogold labeling techniques to determine polyomavirus recombinant VP1 capsid-like particles entry into mouse 3T6 cell nucleus. J. Virol. Methods 90:91-97. [DOI] [PubMed] [Google Scholar]
- 2.Baxter, J., M. Merkenschlager, and A. G. Fisher. 2002. Nuclear organisation and gene expression. Curr. Opin. Cell Biol. 14:372-376. [DOI] [PubMed] [Google Scholar]
- 3.Belmont, A. S., S. Dietzel, A. C. Nye, Y. G. Strukov, and T Tumbar. 1999. Large-scale chromatin structure and function. Curr. Opin. Cell Biol. 11:307-311. [DOI] [PubMed] [Google Scholar]
- 4.Boisvert, F. M., M. J. Hendzel, and D. P. Bazett-Jones. 2000. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J. Cell Biol. 148:283-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boisvert, F. M., M. J. Kruhlak, A. K. Box, M. J. Hendzel, and D. P. Bazett-Jones. 2001. The transcription coactivator CBP is a dynamic component of the promyelocytic leukemia nuclear body. J. Cell Biol. 152:1099-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle, S., S. Gilchrist, J. M. Bridger, N. L. Mahy, J. A. Ellis, and W. A. Bickmore. 2001. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 10:211-219. [DOI] [PubMed] [Google Scholar]
- 7.Brown, K. E., S. S. Guest, S. T. Smale, K. Hahm, M. Merkenschlager, and A. G. Fisher. 1997. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 91:845-854. [DOI] [PubMed] [Google Scholar]
- 8.Brown, K. E., J. Baxter, D. Graf, M. Merkenschlager, and A. G. Fisher. 1999. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell 3:207-217. [DOI] [PubMed] [Google Scholar]
- 9.Carvalho, T., J. S. Seeler, K. Ohman, P. Jordan, U. Pettersson, G. Akusjarvi, M. Carmo-Fonseca, and A. Dejean. 1995. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 131:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cereghini, S., and M. Yaniv. 1984. Assembly of transfected DNA into chromatin: structural changes in the origin-promoter-enhancer region upon replication. EMBO J. 3:1243-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chee, A. V., P. Lopez, P. P. Pandolfi, and B. Roizman. 2003. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J. Virol. 77:7101-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, Y. H., J. P. MacGregor, D. A. Goldstein, and M. R. Hall. 1979. Histone modifications in simian virus 40 and in nucleoprotein complexes containing supercoiled viral DNA. J. Virol. 30:218-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Croft, J. A., J. M. Bridger, S. Boyle, P. Perry, P. Teague, and W. A. Bickmore. 1999. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 145:1119-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duraisingh, M. T., T. S. Voss, A. J. Marty, M. F. Duffy, R. T. Good, J. K. Thompson, L. H. Freitas-Junior, A. Scherf, B. S. Crabb, and A. F. Cowman. 2005. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell 121:13-24. [DOI] [PubMed] [Google Scholar]
- 15.Earnshaw, W. C., and N. Rothfield. 1985. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91:313-321. [DOI] [PubMed] [Google Scholar]
- 16.Everett, R. D. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266-7273. [DOI] [PubMed] [Google Scholar]
- 17.Everett, R. D., and J. Murray. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 79:5078-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiers, W., R. Contreras, G. Haegemann, R. Rogiers, A. Van de Voorde, H. Van Heuverswyn, J. Van Herreweghe, G. Volckaert, and M. Ysebaert. 1978. Complete nucleotide sequence of SV40 DNA. Nature 273:113-120. [DOI] [PubMed] [Google Scholar]
- 19.Forstova, J., N. Krauzewicz, V. Sandig, J. Elliott, Z. Palkova, M. Strauss, and B. E. Griffin. 1995. Polyoma virus pseudocapsids as efficient carriers of heterologous DNA into mammalian cells. Hum. Gene Ther. 6:297-306. [DOI] [PubMed] [Google Scholar]
- 20.Foster, G. R. 1997. Interferons in host defense. Semin. Liver Dis. 17:287-295. [DOI] [PubMed] [Google Scholar]
- 21.Gill, D. R., S. E. Smyth, C. A. Goddard, I. A. Pringle, C. F. Higgins, W. H. Colledge, and S. C. Hyde. 2001. Increased persistence of lung gene expression using plasmids containing the ubiquitin C or elongation factor 1 alpha promoter. Gene Ther. 8:1539-1546. [DOI] [PubMed] [Google Scholar]
- 22.Gluzman, Y. 1981. SV40-transformed simian cells support the replication of early SV40 mutants. Cell 23:175-182. [DOI] [PubMed] [Google Scholar]
- 23.Gorelik, J., A. Shevchuk, M. Ramalho, M. Elliott, C. Lei, C. F. Higgins, M. J. Lab, D. Klenerman, N. Krauzewicz, and Y. Korchev. 2002. Scanning surface confocal microscopy for simultaneous topographical and fluorescence imaging: application to single virus-like particle entry into a cell. Proc. Natl. Acad. Sci. USA 99:16018-16023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grogan, J. L., M. Mohrs, B. Harmon, D. A. Lacy, J. W. Sedat, and R. M. Locksley. 2001. Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity 14:205-215. [DOI] [PubMed] [Google Scholar]
- 25.Gu, H., Y. Liang, G. Mandel, and B. Roizman. 2005. Components of the REST/CoREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proc. Natl. Acad. Sci. USA 102:7571-7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 27.Ishiguro, K., and A. C. Sartorelli. 2004. Activation of transiently transfected reporter genes in 3T3 Swiss cells by the inducers of differentiation/apoptosis-dimethylsulfoxide, hexamethylene bisacetamide and trichostatin A. Eur. J. Biochem. 271:2379-2390. [DOI] [PubMed] [Google Scholar]
- 28.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 29.Jeong, S., and A. Stein. 1994. Micrococcal nuclease digestion of nuclei reveals extended nucleosome ladders having anomalous DNA lengths for chromatin assembled on non-replicating plasmids in transfected cells. Nucleic Acids Res. 22:370-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiesslich, A., A. von Mikecz, and P. Hemmerich. 2002. Cell cycle-dependent association of PML bodies with sites of active transcription in nuclei of mammalian cells. J. Struct. Biol. 140:167-179. [DOI] [PubMed] [Google Scholar]
- 31.Kim, E., L. Du, D. B. Bregman, and S. L. Warren. 1997. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J. Cell Biol. 136:19-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koken, M. H., A. Reid, F. Quignon, M. K. Chelbi-Alix, J. M. Davies, J. H. Kabarowski, J. Zhu, S. Dong, S. Chen, Z. Chen, C. C. Tan, J. Licht, S. Waxman, H. de The, and A. Zelent. 1997. Leukemia-associated retinoic acid receptor alpha fusion partners, PML and PLZF, heterodimerize and colocalize to nuclear bodies. Proc. Natl. Acad. Sci. USA 94:10255-10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krauzewicz, N., J. Stokrova, C. Jenkins, M. Elliott, C. F. Higgins, and B. E. Griffin. 2000. Virus-like gene transfer into cells mediated by polyoma virus pseudocapsids. Gene Ther. 7:2122-2131. [DOI] [PubMed] [Google Scholar]
- 34.LaMorte, V. J., J. A. Dyck, R. L. Ochs, and R. M. Evans. 1998. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc. Natl. Acad. Sci. USA 95:4991-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lomonte, P., J. Thomas, P. Texier, C. Caron, S. Khochbin, and A. L. Epstein. 2004. Functional interaction between class II histone deacetylases and ICP0 of herpes simplex virus type 1. J. Virol. 78:6744-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loni, M. C., and M. Green. 1975. Virus-specific DNA sequences in mouse and rat cells transformed by the Harvey and Moloney murine sarcoma viruses detected by in situ hybridization. Virology 63:40-47. [DOI] [PubMed] [Google Scholar]
- 37.Mannova, P., and J. Forstova. 2003. Mouse polyomavirus utilizes recycling endosomes for a traffic pathway independent of COPI vesicle transport. J. Virol. 77:1672-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Misteli, T., and D. L. Spector. 1999. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell 3:697-705. [DOI] [PubMed] [Google Scholar]
- 39.Misteli, T., A. Gunjan, R. Hock, M. Bustin, and D. T. Brown. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature 408:877-881. [DOI] [PubMed] [Google Scholar]
- 40.Misteli, T. 2004. Spatial positioning; a new dimension in genome function. Cell 119:153-156. [DOI] [PubMed] [Google Scholar]
- 41.Munkonge, F. M., D. A. Dean, E. Hillery, U. Griesenbach, and E. W. Alton. 2003. Emerging significance of plasmid DNA nuclear import in gene therapy. Adv. Drug Deliv. Rev. 55:749-760. [DOI] [PubMed] [Google Scholar]
- 42.Murakami, Y., C. R. Wobbe, L. Weissbach, F. B. Dean, and J. Hurwitz. 1986. Role of DNA polymerase alpha and DNA primase in simian virus 40 DNA replication in vitro. Proc. Natl. Acad. Sci. USA 83:2869-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy, J. C., W. Fischle, E. Verdin, and J. H. Sinclair. 2002. Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 21:1112-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nusinzon, I., and C. M. Horvath. 2003. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc. Natl. Acad. Sci. USA 100:14742-14747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peterson, C. L., and M. A. Laniel. 2004. Histones and histone modifications. Curr. Biol. 14:R546-551. [DOI] [PubMed] [Google Scholar]
- 46.Poon, A. P., Y. Liang, and B. Roizman. 2003. Herpes simplex virus 1 gene expression is accelerated by inhibitors of histone deacetylases in rabbit skin cells infected with a mutant carrying a cDNA copy of the infected-cell protein no. 0. J. Virol. 77:12671-12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reeves, R., C. M. Gorman, and B. Howard. 1985. Minichromosome assembly of non-integrated plasmid DNA transfected into mammalian cells. Nucleic Acids Res. 13:3599-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Regad, T., A. Saib, V. Lallemand-Breitenbach, P. P. Pandolfi, H. de The, and M. K. Chelbi-Alix. 2001. PML mediates the interferon-induced antiviral state against a complex retrovirus via its association with the viral transactivator. EMBO J. 20:3495-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richler, C., G. Ast, R. Goitein, J. Wahrman, R. Sperling, and J. Sperling. 1994. Splicing components are excluded from the transcriptionally inactive XY body in male meiotic nuclei. Mol. Biol. Cell 5:1341-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaffhausen, B. S., and T. L. Benjamin. 1976. Deficiency in histone acetylation in nontransforming host range mutants of polyoma virus. Proc. Natl. Acad. Sci. USA 73:1092-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soeda, E., J. R. Arrand, N. Smolar, J. E. Walsh, and B. E. Griffin. 1980. Coding potential and regulatory signals of the polyoma virus genome. Nature 283:445-453. [DOI] [PubMed] [Google Scholar]
- 52.Stokrova, J., Z. Palkova, L. Fischer, Z. Richterova, J. Korb, B. E. Griffin, and J. Forstova. 1999. Interactions of heterologous DNA with polyomavirus major structural protein, VP1. FEBS Lett. 445:119-125. [DOI] [PubMed] [Google Scholar]
- 53.Strauss, M., S. Hering, L. Lubbe, and B. E. Griffin. 1990. Immortalization and transformation of human fibroblasts by regulated expression of polyoma virus T antigens. Oncogene 5:1223-1229. [PubMed] [Google Scholar]
- 54.Tanabe, H., F. A. Habermann, I. Solovei, M. Cremer, and T. Cremer. 2002. Non-random radial arrangements of interphase chromosome territories: evolutionary considerations and functional implications. Mutat. Res. 504:37-45. [DOI] [PubMed] [Google Scholar]
- 55.Tang, Q., P. Bell, P. Tegtmeyer, and G. G. Maul. 2000. Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J. Virol. 74:9694-9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang, Q., and G. G. Maul. 2003. Mouse cytomegalovirus immediate-early protein 1 binds with host cell repressors to relieve suppressive effects on viral transcription and replication during lytic infection. J. Virol. 77:1357-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang, Q., L. Li, A. M. Ishov, V. Revol, A. L. Epstein, and G. G. Maul. 2003. Determination of minimum herpes simplex virus type 1 components necessary to localize transcriptionally active DNA to ND10. J. Virol. 77:5821-5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valls, E., X. de la Cruz, and M. A. Martinez-Balbas. 2003. The SV40 T antigen modulates CBP histone acetyltransferase activity. Nucleic Acids Res. 31:3114-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vautier, D., D. Besombes, D. Chassoux, F. Aubry, and P. Debey. 1994. Redistribution of nuclear antigens linked to cell proliferation and RNA processing in mouse oocytes and early embryos. Mol. Reprod. Dev. 38:119-130. [DOI] [PubMed] [Google Scholar]
- 60.von Mikecz, A., S. Zhang, M. Montminy, E. M. Tan, and P. Hemmerich. 2000. CREB-binding protein (CBP)/p300 and RNA polymerase II colocalize in transcriptionally active domains in the nucleus. J. Cell Biol. 150:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, J., C. Shiels, P. Sasieni, P. J. Wu, S. A. Islam, P. S. Freemont, and D. Sheer. 2004. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 164:515-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weis, K., S. Rambaud, C. Lavau, J. Jansen, T. Carvalho, M. Carmo-Fonseca, A. Lamond, and A. Dejean. 1994. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell 76:345-356. [DOI] [PubMed] [Google Scholar]
- 63.Yamano, T., K. Ura, R. Morishita, H. Nakajima, M. Monden, and Y. Kaneda. 2000. Amplification of transgene expression in vitro and in vivo using a novel inhibitor of histone deacetylase. Mol. Ther. 1:574-580. [DOI] [PubMed] [Google Scholar]
- 64.Zink, D., M. D. Amaral, A. Englmann, S. Lang, L. A. Clarke, C. Rudolph, F. Alt, K. Luther, C. Braz, N. Sadoni, J. Rosenecker, and D. Schindelhauer. 2004. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J. Cell Biol. 166:815-825. [DOI] [PMC free article] [PubMed] [Google Scholar]