Summary
The ability of nuclear receptors (NRs) to repress transcriptional responses to diverse signaling pathways is an essential aspect of their biological activities, but mechanisms determining the specificity and functional consequences of transrepression remain poorly understood. Here, we report the identification of nuclear receptor-specific mechanisms that mediate signal- and gene-specific repression of transcriptional responses initiated by engagement of toll-like receptors (TLR) 3, 4 and 9. The glucocorticoid receptor (GR) was found to repress a large set of functionally related inflammatory response genes by disrupting p65/interferon regulatory factor (IRF) complexes required for TLR4- or TLR9-dependent, but not TLR3-dependent, transcriptional activation. This mechanism enables the GR to differentially regulate pathogen-specific programs of gene expression. In contrast, PPARγ and LXR repressed overlapping but functionally distinct sets of genes by p65/IRF3-independent mechanisms. In concert with their utilization of distinct transrepression mechanisms, simultaneous activation of GR and PPARγ or GR and LXRs resulted in additive/synergistic inhibition of subsets of TLR4 responses in vivo and in vitro. These findings reveal a molecular basis for combinatorial control of homeostasis and immune responses by NRs and suggest new approaches for treatment of inflammatory diseases.
Introduction
Members of the nuclear receptor superfamily play diverse roles in the regulation of development, homeostasis and immune responses by both positively and negatively regulating gene expression (Chawla et al., 2001; Evans, 1988; Kastner et al., 1995; Mangelsdorf and Evans, 1995). The glucocorticoid receptor (GR) is prototypic of a subset of ligand-dependent nuclear receptors that integrate host immune responses with physiological circuits that are required for maintenance of necessary organ functions. The ability of GR to repress transcriptional responses to inflammatory signals is an essential component of its homeostatic functions and a primary mechanism by which natural and synthetic GR agonists exert anti-inflammatory effects in a variety of disease settings (Reichardt et al., 1998; Reichardt et al., 2001). Negative regulation of inflammatory responses is thought to result, at least in part, from the ability of GR to interfere by transrepression, with the activities of other signal-dependent transcription factors that include NF-κB and activator protein-1 (AP-1) family members (Caldenhoven et al., 1995; Jonat et al., 1990; Scheinman et al., 1995b; Schüle et al., 1990; Yang-Yen et al., 1990) and reviewed in (De Bosscher et al., 2003). Numerous models have been proposed for GR-mediated transrepression, including direct interactions with NF-κB components (Caldenhoven et al., 1995; Liden et al., 1997; Scheinman et al., 1995b), regulation of components of signal transduction pathways involved in NF-κB and AP-1 activation (Auphan et al., 1995; Caelles et al., 1997; Scheinman et al., 1995a), competition for essential coactivators (Kamei et al., 1996; Sheppard et al., 1998), alternative utilization of coactivators (Kassel et al., 2004; Rogatsky et al., 2001; Scheinman et al., 1995b), recruitment of corepressors (Nissen and Yamamoto, 2000), and modifications of core transcription factors (De Bosscher et al., 2000; Nissen and Yamamoto, 2000). However, most of these models have been developed based on analysis of limited sets of specific target genes, often in transient transfection assays, and general applicability to broad programs of gene expression activated during inflammatory responses have not been established.
Anti-inflammatory activities have also been documented in vivo and/or in vitro for several other members of the nuclear receptor family, including estrogen receptors (ERs) (McKay and Cidlowski, 1999), vitamin D receptors (VDRs) (Nagpal et al., 2001), peroxisome proliferator-activated receptors (PPARs) (Devchand et al., 1996; Jiang et al., 1998; Marx et al., 2000; Ricote et al., 1998; Staels et al., 1998) and LXRs (Castrillo et al., 2003; Joseph et al., 2004; Joseph et al., 2003). PPARs and LXRs are regulated by fatty acid and cholesterol metabolites, respectively, and were initially characterized as nuclear receptors that play critical roles in lipid homeostasis (Forman et al., 1995; Issemann and Green, 1990; Janowski et al., 1996; Kliewer et al., 1995). Emerging evidence suggests that their ability to counter-regulate inflammatory responses plays important roles in both immunity and metabolic control (Joseph et al., 2003; Ricote et al., 1998). For example, PPARγ is thought to improve insulin resistance by both positively and negatively regulating gene expression, with PPARγ agonists suppressing the expression of inflammatory genes in adipocytes and adipose tissue-associated macrophages that are correlated with impaired insulin signaling (Pittas et al., 2004; Weisberg et al., 2003; Xu et al., 2003). While PPARs and LXRs have also been documented to antagonize the actions of NF-κB and AP-1 transcription factors (Ricote et al., 1998), the underlying mechanisms remain poorly understood.
Here, we have used toll-like receptor (TLR) signaling as a model system to explore mechanisms by which different members of the nuclear receptor superfamily repress pro-inflammatory programs of gene expression. TLR3, TLR4 and TLR9 are members of the TLR family of cell surface receptors that regulate transcriptional programs involved in innate immunity by recognizing pathogen-associated molecular patterns (Janeway and Medzhitov, 2002). TLR3 is activated by double stranded RNA and plays an important role in innate immunity directed against dsRNA viral pathogens by coupling to TRIF-dependent signal transduction pathways (Alexopoulou et al., 2001; Yamamoto et al., 2002). TLR9 is activated by immunostimulatory DNA sequences and is essential for innate immunity directed against certain double-stranded DNA viruses by coupling to the MyD88 signaling adapter (Hacker et al., 2000; Hemmi et al., 2000; Krug et al., 2004; Lund et al., 2003). TLR4 is activated by lipopolysaccharide (LPS) and mediates innate immunity against gram-negative bacterial infections by coupling to both MyD88 and TRIF (Alexopoulou et al., 2001; Poltorak et al., 1998). MyD88 and TRIF use overlapping but distinct signal transduction pathways to regulate the expression and function of numerous transcription factors that drive inflammatory responses, including NF-κB, AP-1, and interferon regulatory factors (IRFs) (Akira and Takeda, 2004; Pitha, 2004). In the present studies, GR, PPARγ and LXR agonists were found to repress both common and nuclear receptor-specific subsets of TLR target genes through the use of nuclear receptor- and TLR-specific transrepression mechanisms. Combinations of agonists for GR, PPARγ and LXRs resulted in additive or synergistic inhibition of a subset of TLR4-target genes both in cultured macrophages and in vivo, consistent with the simultaneous targeting of these genes by distinct mechanisms. These findings suggest that nuclear receptors function in a combinatorial manner to coordinately regulate the evolution of host immune responses.
Results
Nuclear Receptors Inhibit Overlapping but Distinct Subsets of LPS-Inducible Genes
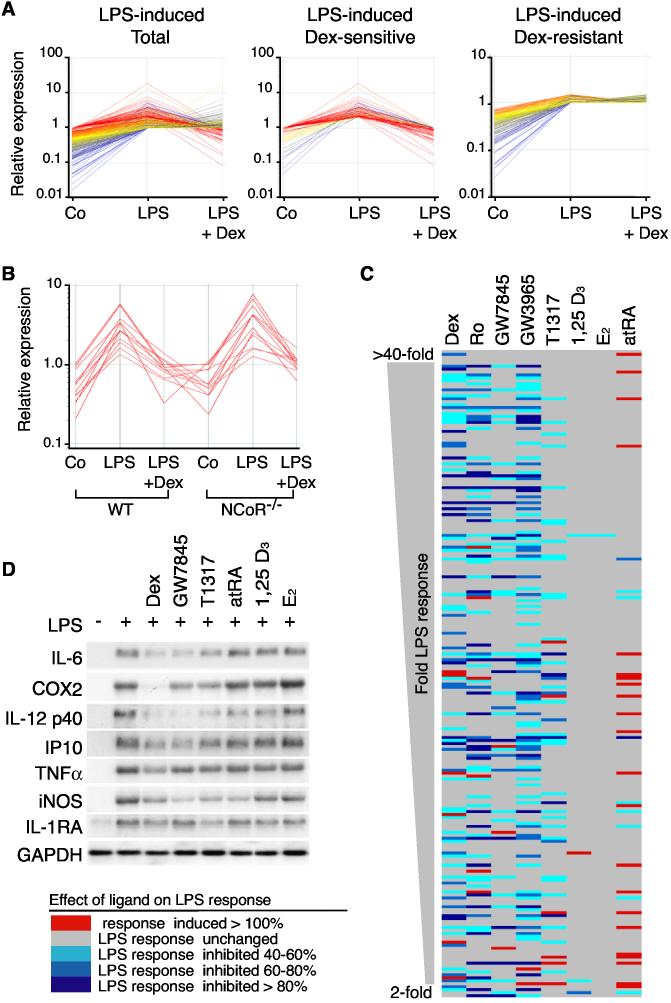
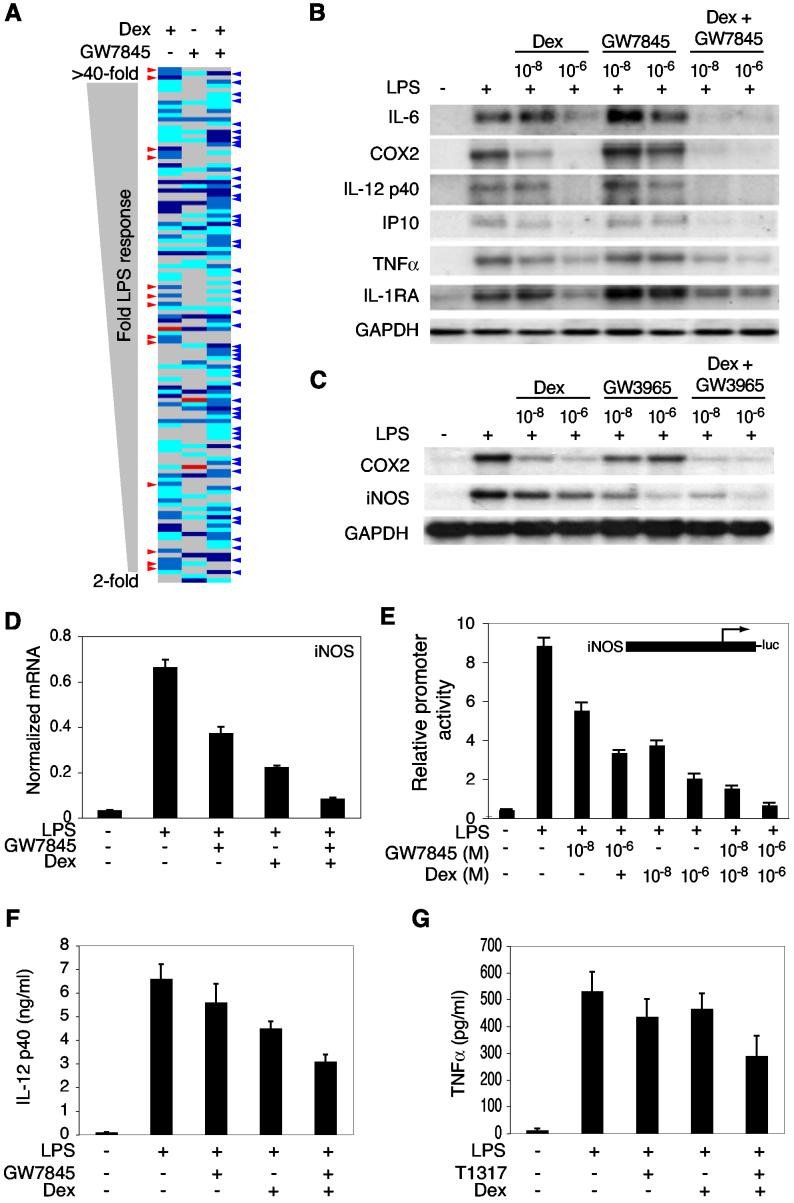
Gene expression profiling experiments were initially performed to identify LPS-inducible genes in primary macrophages that were sensitive to transrepression by the GR agonist dexamethasone (Dex) (Figure 1A). The observation that about half of the LPS-inducible genes were Dex-sensitive raised the questions of how GR discriminated between sensitive and resistant genes and whether these two classes of genes exert distinct biological functions. Recent findings indicate that NCoR corepressor complexes occupy a subset of NF-κB and AP-1 target genes under basal conditions and are cleared in a signal-dependent manner as a prerequisite to transcriptional activation (Ogawa et al., 2004; Perissi et al., 2004). Because some NCoR target genes were also subject to Dex-mediated repression, microarray experiments were performed using NCoR−/− macrophages to determine whether NCoR was required for Dex-sensitivity (Ogawa et al., 2004). Lack of NCoR had no impact on GR-mediated repression (Figure 1B), indicating the utilization of NCoR-independent mechanisms. To investigate whether different nuclear receptors repress a common set of LPS target genes, gene expression profiling experiments were performed using nuclear receptor-specific agonists for PPARγ (rosiglitazone (Ro) and GW7845), LXRα/β (GW3965 and T1317), VDR (1,25-(OH)2vitamin D3), ER (17β-estradiol; E2), and RARs (all-trans retinoic acid; atRA). These experiments demonstrated that each agonist exerted an overlapping but distinct impact on LPS-dependent gene expression (Figures 1C and 1D). GR, LXR and PPARγ agonists were the most potent inhibitors of the LPS response, with VDR-, ER- and RAR-specific agonists exerting relatively modest inhibitory effects under these conditions (Figure 1C). Dex, GW3965 and Ro inhibited expression of a larger number of genes than they activated (Figures 1C and data not shown). Sensitivity to repressive effects of GR, PPARγ or LXR agonists did not correlate with degree of responsiveness to LPS (Figure 1C), absolute expression levels, or a requirement for the p65 compoment of NF-κB (Figure S1 and data not shown). In concert, these experiments suggest that negative regulation of gene expression is a dominant biological function of GR, LXR and PPARγ in activated macrophages and indicate that these nuclear receptors utilize distinct molecular mechanisms to repress TLR4-dependent gene expression.
Figure 1.
Inhibition of LPS-Dependent Gene Expression by Nuclear Receptor Agonists (A) Relative expression of LPS-inducible genes in peritoneal macrophages under control conditions and after 6 h of LPS treatment in the absence or presence of 1 μM Dex. Left panel; genes induced by LPS >2-fold. Middle panel; genes induced by LPS >2-fold and inhibited by Dex more than 50% (Dex-sensitive). Right panel; genes induced by LPS >2-fold and resistant to Dex. Expression data was collected using Affymetrix U74A microarrays and represent results obtained from four independent experiments. (B) Dex-mediated transrepression of LPS-inducible genes in macrophages derived from fetal liver-derived macrophages of wild-type (WT) and NCoR−/− mice. The illustrated gene expression values are for the 12 most highly repressed genes in wild-type macrophages. Expression data was collected using Affymetrix U74A microarrays and represents results obtained from two independent experiments. (C) Effect of nuclear receptor agonists on responsiveness of 208 LPS-target genes in peritoneal macrophages. Genes are ordered based on magnitude of average LPS induction over 4 experiments from >40-fold at the top to 2-fold at the bottom. Genes in which the LPS response was not altered by agonist treatment are illustrated in gray. Red indicates ligand-dependent upregulation and blue ligand-dependent downregulation of the LPS response. The magnitude of the effect is indicated by the key at lower left. Expression data was collected using Affymetrix U74A microarrays and represents results obtained from a minimum of two independent experiments for each drug treatment. Data for Ro is from experiments performed under identical conditions (Welch et al., 2003). (D) Confirmation of negative regulation of LPS-target genes by Northern blotting. Macrophages were treated with LPS for 6 h in the presence of 1 μM concentrations of the indicated agonists.
p65/IRF3 Complexes Mediate Signal-specific Inhibition of Transcriptional Responses
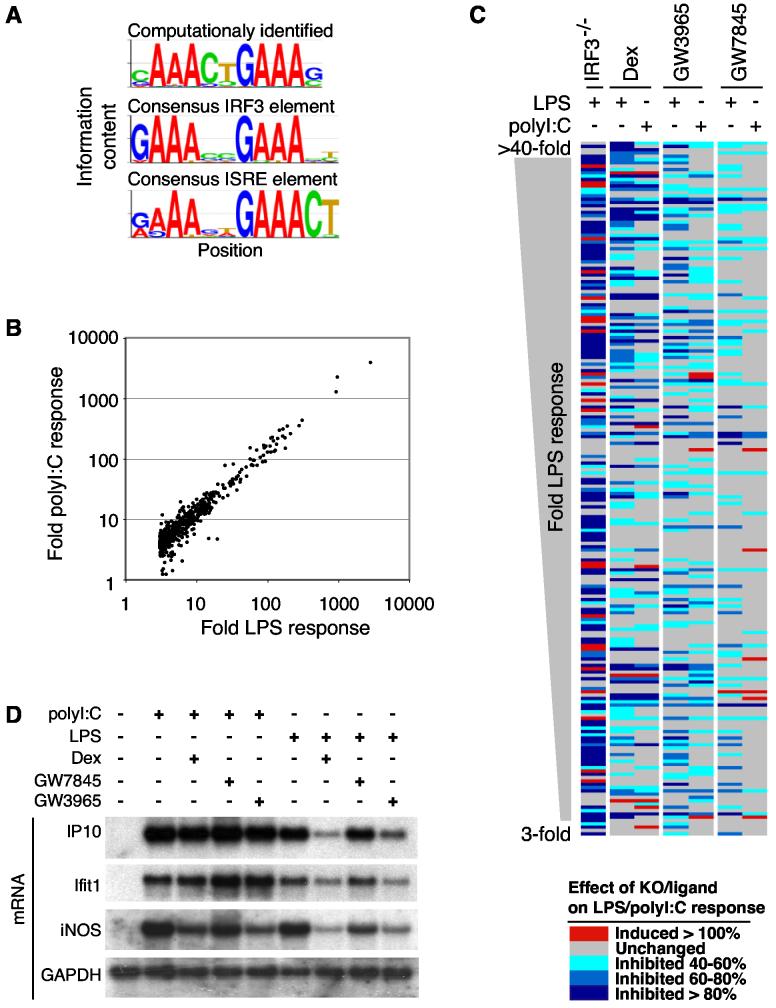
Computational motif discovery methods were used to search for potential transcriptional regulatory elements mediating LPS-dependent activation and nuclear receptor-mediated transrepression. The sequence cAAActGAAAg was identified as the most highly significant motif enriched in the promoter sequences of LPS target genes (Figure 2A). This motif is nearly identical to consensus IRF3 and interferon (IFN)-sensitive response element (ISRE) sequences recognized by IRF3 and the type I IFN-inducible ISGF3 complex (Juang et al., 1998; Wathelet et al., 1998; Yoneyama et al., 1998). Recent findings indicate that binding of poly I:C to TLR3 and LPS to TLR4, respectively, activates IRF3 and induces ISRE-mediated gene activation (Fitzgerald et al., 2003; Juang et al., 1998; Navarro and David, 1999; Pitha, 2004; Wathelet et al., 1998; Yoneyama et al., 1998). We therefore performed gene expression profiling experiments to identify IRF3-target genes by comparing LPS responses of wild-type and IRF3−/− macrophages (Dang et al., 2004; Sato et al., 2000). In parallel, expression profiling experiments were performed to evaluate the impact of GR, PPARγ and LXR agonists on transcriptional responses of macrophages to LPS and poly I:C (Figure 2C). Although TLR3 and TLR4 differ with respect to use of MyD88 as an adapter protein for signal transduction (Akira and Takeda, 2004; Pitha, 2004), the qualitative and quantitative pattern of genes induced more than 3-fold by LPS and poly I:C at the 6 h time point were very similar (Figure 2B). Of the 543 genes induced more than 3-fold by LPS in wild-type macrophages, 159 genes exhibited a 90% or more loss of activation in IRF3−/− macrophages and an additional 160 genes exhibited at least a 40% reduction in LPS activation (Figure 2C, column 1). A complete listing of expression data and response element sequences is provided as Table S1.
Figure 2.
Differential Repression of TLR Responses by GR, PPARγ, and LXRs (A) Identification of IRF3-binding sites as highly enriched sequence motifs in the promoters of LPS-inducible genes. The top sequence logo is representative of the most significant motif present in the promoters of genes that are LPS-inducible and not present in promoters of non-LPS-inducible genes. This motif was found de novo without previous knowledge of known transcription factor binding sites. The sequence logos representing the known consensus IRF3-binding and ISRE motifs are shown for comparison. (B) A scatter plot illustrating fold responses to poly I:C compared to genes activated at least 3-fold by LPS. Expression data was collected using Codelink Uniset Mouse 1 microarrays and is representative of results obtained from four independent experiments. (C) Effect of IRF3-deficiency and GR (Dex), LXR (GW3965) or PPARγ (GW7845) agonists on transcriptional responses to LPS and poly I:C. The panel illustrates the 208 most highly induced LPS-responsive genes. Sensitivity of each gene to loss of IRF3 (column 1) or treatment with Dex, GW3965 and GW7845 is color coded as indicated in the key at the bottom. Expression data was collected using Amersham Codelink Mouse Uniset 1 microarrays and represents results from two independent experiments for each ligand. (D) Confirmation of signal-specific repression of LPS- and poly I:C-inducible genes by GR, LXR and PPARγ-specific agonists. Macrophages were treated with LPS or poly I:C for 6 h in the presence of 1 μM concentrations of the indicated agonists.
As in the case of LPS-dependent gene expression, GR, PPARγ and LXR agonists repressed both common and nuclear receptor-specific targets of poly I:C-inducible genes (Figure 2C). However, despite the overall similarity in the sets of genes that were transcriptionally activated by LPS and poly I:C, the patterns of GR, PPARγ and LXR-mediated transrepression were significantly different, indicating that nuclear receptor transrepression is regulated in a signal-specific manner. For example, a substantial number of genes that were Dex-sensitive when activated by LPS became Dex-resistant when activated by poly I:C, illustrated for IP10 and Ifit1 in Figure 2D.
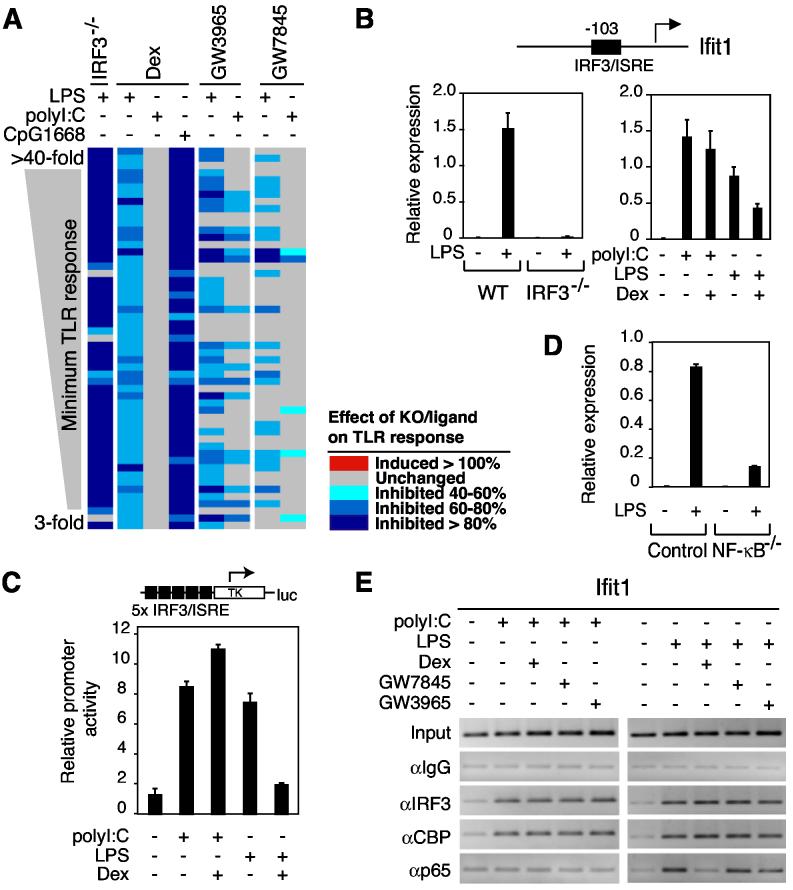
Unexpectedly, nearly all of the highly inducible LPS- and poly I:C-target genes that were Dex-sensitive when activated by LPS but Dex-resistant when activated by poly I:C were also highly dependent on IRF3 for LPS induction (Figure 3A and data not shown). The promoters for many of the genes exhibiting this pattern of expression contained proximal IRF3/ISRE sequences, exemplified by Ifit1 (Figure 3B). In contrast, while PPARγ and LXR agonists also inhibited a significant number of IRF3-dependent genes, the signal-specific pattern of sensitivity and resistance differed (Figures 2C and 3A). This pattern therefore suggested a mechanistic link between IRF3 and signal-specific transrepression by GR. To test this hypothesis, we evaluated the ability of GR to repress transcriptional activation of an artificial promoter constructed to exclusively contain ISRE elements, which was activated by LPS and poly I:C (Figure 3C). Significantly, the induction of the ISRE-dependent reporter was strongly inhibited by Dex when LPS was used as a stimulus, but not when poly I:C was used as a stimulus (Figure 3C).
Figure 3.
Signal-specific Repression by GR Correlates with a Requirement for IRF3 for Transcriptional Activation (A) Transcriptional responses of 54 genes highly induced by LPS, poly I:C and CpG1668, exhibiting sensitivity to Dex when activated by LPS (column 2) and resistance to Dex when activated by poly I:C (column 3). The dependence of the LPS response on IRF3 is indicated in the first column and the effect of CpG1668 (1 μM) is indicated column 4. Effects of IRF3-deficiency or nuclear receptor agonists on LPS, CpG1668 or poly I:C responses are color coded as in Figure 2C. (B) Promoter structure and expression profile of Ifit1 in response to LPS, poly I:C and Dex in wild-type (WT) and IRF3−/− peritoneal macrophages. (C) An ISRE-specific promoter exhibits LPS-specific repression by Dex. U373 cells were transfected with a 5xISRE-Luc reporter plasmid. Cells were treated with the indicated combinations of LPS (100 ng/ml), poly I:C (50 μg/ml) and Dex, and analyzed for luciferase activity 18 h later. (D) Expression of Ifit1 in response to LPS (1 μg/ml) in control and NF-κB−/− fetal liver-derived macrophages. (E) p65 recruitment to the proximal promoter region of Ifit1 is specifically induced by LPS and inhibited by activation of GR. Primary macrophages were treated with LPS (100 ng/ml), poly I:C (50 μg/ml), and the indicated agonists for GR, PPARγ and LXRs for 1 h. ChIP assays were performed with antibodies against IRF3, CBP, p65 and control IgG, respectively. Immunoprecipitated DNA was analyzed by PCR using primers specific for the promoter.
While TLR3 and TLR4 signaling both lead to activation of IRF3, recent studies suggest that p65/RelA functions as an essential coactivator of IRF3 in the case of TLR4 signaling, but not in case of TLR3 signaling (Wietek et al., 2003). The p65 requirement for LPS induction of an ISRE-dependent gene was confirmed by experiments demonstrating markedly impaired activation of the Ifit1 gene in NF-κB-deficient macrophages (Figure 3D). These observations raised the possibility that GR inhibited IRF3-target genes in response to TLR4 signaling, but not TLR3 signaling, by targeting the p65 requirement. We therefore characterized the composition of activation complexes bound to the proximal promoter region of Ifit1 in primary macrophages by chromatin immunoprecipitation (ChIP) assay. These experiments demonstrated that IRF3 and CBP were prominently recruited to the ISRE-containing promoter in response to both LPS and poly I:C (Figure 3E). In contrast, p65 was specifically recruited to the ISRE in response to LPS, but not in response to poly I:C. Interestingly, the recruitment of p65 to the ISRE in response to LPS was largely inhibited by Dex but not by PPARγ or LXR agonists (Figure 3E). Similar findings were obtained using ChIP to evaluate the composition of activation complexes bound to the artificial promoter construct exclusively containing ISRE elements (Figure S2).
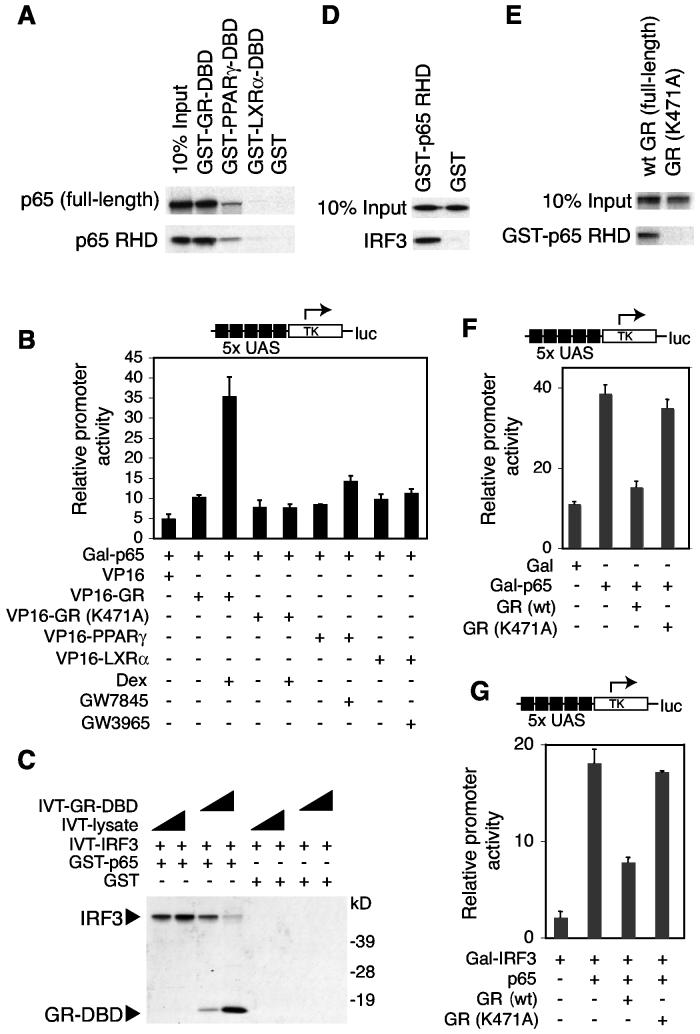
Previous studies have suggested that nuclear receptor interactions with p65 are involved in transrepression (Liden et al., 1997; Scheinman et al., 1995b), but how this interaction could account for receptor-, signal- and gene-specific repression has not been established. We therefore evaluated the possibility that GR inhibited LPS induction of the ISRE promoter through direct interactions with p65. In vitro interaction assays confirmed that the GR-DBD strongly interacted with both full-length p65 and further narrowed this interaction to the N-terminal Rel-homology domain (RHD) (Figure 4A). In contrast, the PPARγ-DBD and the LXRα-DBD exhibited minimal interaction with p65 in vitro (Figure 4A). These results were confirmed by mammalian two-hybrid assays using a Gal4DBD-p65 fusion protein as bait and VP16 fusions with full-length GR, PPARγ or LXRα as preys (Figure 4B).
Figure 4.
GR Specifically Inhibits the Interaction of p65 with IRF3 (A) GR-DBD preferentially interacts with the p65 RHD. GST pull-down assays were performed using the indicated GST-NR-DBD fusion proteins and in vitro translated full-length p65 or p65 RHD, respectively. (B) Wild-type GR preferentially interacts with p65 in vivo in a ligand-dependent manner. The mammalian two-hybrid assay was performed in RAW264.7 cells using Gal-p65 as bait and the indicated VP16-nuclear receptor fusion proteins as preys in the presence and absence of agonists. (C) GR-DBD inhibits interaction of IRF3 with p65. GST pull-down assays were performed using GST-p65 and increasing amounts of in vitro translated full-length IRF3 and/or GR-DBD as indicated. (D) IRF3 interacts with the p65 RHD in vitro. GST pull-down assays were performed using GSTp65 RHD and in vitro translated full-length IRF3. (E) GR-DBD mutant GRK471A is unable to interact with the p65 RHD in vitro. GST pull-down assays were performed using GST-p65 RHD and in vitro translated full-length GR and GRK477A, respectively. (F) Inhibition of Gal-p65 transactivation by liganded wild-type GR but not by GRK477A. Endogenous GR expression in mouse RAW264.7 cells was knocked down by pretreatment with a GR-specific siRNA for 48 h. Cells were then transfected with expression vectors for Gal4 (Gal), Gal4-p65 (Gal-p65), wild-type human GR or GRK477A as indicated in the presence of Dex. Luciferase activity was analyzed 24 h later. (G) Coactivation of Gal-IRF3 by p65 is inhibited by liganded wild-type GR but not by GRK477A. Endogenous GR expression in RAW264.7 cells was knocked down by pretreatment with a GR-specific siRNA for 48 h prior to transfection with expression vectors for human wild-type GR or GRK477A and treatment with Dex as indicated.
We next evaluated whether IRF3 interacted directly with p65 and whether this interaction was influenced by GR. In vitro binding assays demonstrated that a glutathione-S-transferase (GST)-p65 fusion protein interacted with IRF3 (Figure 4C). Addition of increasing amounts of GR-DBD to the binding reaction led to decreased IRF3 interaction with p65 and a concomitant increase in the binding of the GR-DBD (Figure 4C). Furthermore, IRF3 interacted with the RHD of p65 (Figure 4D). These findings suggest that GR and IRF3 compete for the same binding site and that GR preferentially interacts with p65.
To determine whether the interaction of GR with p65 was relevant to its repression function, we evaluated a GR mutant in which lysine 471 in the second zinc finger of the DBD was changed to alanine (GRK471A) based on a previous report that a corresponding mutant is defective for inhibition of p65 activity (Liden et al., 1997). In contrast to wild-type GR, GRK471A exhibited little interaction with p65 in vitro or in vivo in mammalian two-hybrid assays (Figures 4B and 4E), and GRK471A lacked inhibitory activity against Gal-p65 mediated transactivation (Figure 4F). Consistent with these in vitro findings, wild-type GR was able to inhibit p65-dependent transactivation of a Gal-IRF3 fusion gene, while GRK471A was not (Figure 4G), suggesting that direct interaction of GR and p65 is required for GR-mediated transrepression of IRF3. Taken together, these findings support a model in which the requirement of ISRE-containing genes for p65 as a coactivator following LPS activation, but not poly I:C stimulation, accounts for the LPS-specific sensitivity of these genes to transrepression by GR.
IRF3/p65 Complexes Mediate Gene-specific Inhibition of Transcriptional Responses
In addition to the IRF3-binding motif, κB elements were also highly enriched in promoter regions of LPS-inducible genes (Table S1). However, the presence of these sequences did not correlate with signal or gene-specific patterns of regulation. NF-κB binding sites were identified in promoters of genes that were sensitive to nuclear receptor agonists following activation by LPS or poly I:C (e.g., iNOS, Figure 2D), in promoters of genes that were nuclear receptor-sensitive following LPS but not poly I:C activation (e.g., Clic4, Figure 5A and data not shown), and in promoters of genes that were nuclear receptor-resistant regardless of the signal (e.g., Nfkbia and Gro1, Figure 5A and data not shown). In each case, transcriptional responses to LPS required p65 (Figure S1). The recent finding that a subset of NF-κB sites appear to determine the utilization of IRF3 as a coactivator of p65 (Leung et al., 2004) suggested the possibility that this might be a basis for gene-specific sensitivity to repression by GR. To examine this, we chose the Scyb9 and Clic4 genes because they were highly IRF3-dependent, Dex-sensitive NF-κB target genes that did not contain ISRE motifs within their proximal promoter or distal upstream regions (Figures 5A and 5B). The Nfkbia and Gro1 genes were chosen for comparison because they were highly induced by LPS in an IRF3-independent manner, were Dex-resistant, contained well-characterized κB elements and lacked proximal or distal ISRE elements (Figures 5A and 5B). ChIP experiments revealed that p65 was recruited to each of these genes in response to LPS, as expected (Figure 5C). IRF3 was recruited to the proximal promoter regions of Scyb9 and Clic4 in response to LPS, but not to the Nfkbia or Gro1 promoters, consistent with the requirement of Scyb9 and Clic4 for IRF3 for activation and confirming a gene-specific recruitment of IRF3 to a subset of p65-target genes. Significantly, treatment with Dex had no effect on the recruitment of p65 to any of these four target genes but inhibited the recruitment of IRF3 to the Scyb9 and Clic4 promoters, coincident with ligand-dependent recruitment of GR to these promoters (Figure 5C).
Figure 5.
Utilization of IRF3 as a Coactivator of p65 Determines Gene-specific Sensitivity (A) Location of NF-κB sites in proximal promoter regions of Scyb9, Clic4, Nfkbia and Gro1, and their expression profiles in response to LPS in wild-type (WT) and IRF3−/− peritoneal macrophages. (B) Expression profiles of Scyb9, Clic4, Nfkbia and Gro1 in response to LPS and Dex in wild-type peritoneal macrophages. (C) Recruitment of IRF3 to the proximal promoter regions of Scyb9 and Clic4, but not Nfkbia or Gro1, in response to LPS. Recruitment of IRF3 was largely inhibited by Dex. Proximal regions of Scyb9, Clic4, Nfkbia and Gro1 promoters that includes an NF-κB site were analyzed by ChIP assays using the indicated antibodies. Crosslinking was performed 1 h after treatment with LPS and Dex. (D) Expression profiles of IP10, Clic4 and Nfkbia in response to LPS and Dex in control (WT) and MyD88−/− peritoneal macrophages. Gene expression was determined by real-time quantitative PCR.
MyD88 Dictates GR-sensitivity of IRF3/7-dependent Gene Expression
Because TLR3 and TLR4 activate IRF3 and NF-κB through the TRIF-dependent pathway, while TLR4, in addition, activates NF-κB and MAP kinases via the MyD88-dependent pathway, these observations raised the possibility that glucocorticoid sensitivity was dictated by the utilization of the MyD88-dependent pathway. To initially test this hypothesis, we determined the profile of dexamethasone-sensitive genes in macrophages treated with immunostimulatory DNA (CpG1668) to activate TLR9, which exclusively couples to the MyD88-dependent pathway (Hacker et al., 2000; Krug et al., 2004). As in the case of polyI:C-stimulated cells, the overall profile of transcriptional activation induced by CpG1668 was very similar to that induced by LPS. Remarkably, of the genes that were highly induced by all three TLR agonists and were Dex-resistant when activated by polyI:C but Dex-sensitive when activated by LPS, 100% were also Dex-sensitive when activated by CpG1668 (Figure 3A). Furthermore, the quantitative extent of Dex-mediated repression was more pronounced following TLR9 stimulation than TLR4 stimulation in nearly every case (Figure 3A). TLR9 primarily induces ISRE gene expression through IRF7 (Kawai et al., 2004), suggesting that the mechanism of GR-mediated repression established for IRF3 extends to IRF7-dependent gene expression. To determine whether Dex-sensitivity of the response to LPS requires signaling through the MyD88 pathway, experiments were performed in MyD88−/− macrophages (Adachi et al., 1998). In these cells, LPS activation is entirely TRIF-dependent (Akira and Takeda, 2004; Kawai et al., 2001). Remarkably, IRF3-dependent genes that were Dex-sensitive in wild-type cells became Dex-resistant in MyD88−/− cells, regardless of whether they contained ISRE or κB elements (Figure 5D). These findings suggest that signaling through the MyD88-dependent pathway specifies Dex-sensitivity of this set of genes.
GR, PPARγ and LXR Function in a Combinatorial Manner to Inhibit LPS Responses
The observation that GR, PPARγ and LXR agonists repressed overlapping but distinct sets of LPS-target genes by p65-dependent and p65-independent mechanisms raised the possibility that they might exert combinatorial effects on inflammatory responses. To test this hypothesis, gene expression profiling experiments were performed to characterize LPS responses in the presence or absence of combinations of saturating concentrations of GR, PPARγ and LXR agonists. The results of this analysis for the combination of Dex and the PPARγ agonist GW7845 are illustrated in Figure 6A, restricted to the subset of genes transrepressed by at least one agonist. While several examples were observed in which nuclear receptor-specific inhibitory effects of one agonist were reversed by addition of the second agonist (red arrows), the major impact of the combination of agonists was to increase the strength of inhibition of a subset of LPS target genes (blue arrows). These results were confirmed by additional experiments that examined the concentration dependence of combinatorial interactions by Northern blot analysis and quantitative PCR analysis of representative target genes (Figures 6B and 6D). Low concentrations of Dex and GW7845 (10 nM) that exerted relatively little repressive effects when used individually could act synergistically in combination (Figure 6B). Parallel studies of combinations of Dex and GW3965 also demonstrated additive or synergistic effects on LPS-target genes (Figure 6C and data not shown).
Figure 6.
GR and PPARγ Function in a Combinatorial Manner to Inhibit LPS Responses (A) Combinatorial interactions between GR and PPARγ agonists at a genome-wide level. Peritoneal macrophages were stimulated with LPS in the absence or presence of Dex alone, GW7845 alone, or the combination of Dex plus GW7845. Each agonist was used at 1 μM. The panel illustrates LPS-target genes exhibiting a > 40% reduction of the LPS response in the presence of at least one agonist. Effects of agonists on the LPS response are color coded according to the legend in Figure 1C. Red arrows indicate genes in which one agonist abolished strong inhibitory effects of the other agonist. Blue arrows indicate genes in which the combination of Dex and GW7845 resulted in stronger inhibition of the LPS response than either agonist alone. Expression data was collected using Affymetrix U74A microarrays and represents results obtained from two independent experiments. (B) Confirmation of combinatorial effect of GR and PPARγ agonists on regulation of LPS-target genes by Northern blotting. Macrophages were treated with LPS for 6 h in the presence of the indicated concentrations (10 nM, 1 μM) of agonists. (C) Confirmation of combinatorial effect of GR and LXR agonists on regulation of LPS-target genes by Northern blotting. (D) Confirmation of combinatorial effect of GR and PPARγ agonists (1 μM) on iNOS expression by real-time quantitative PCR. (E) Combinatorial interactions between GR and PPARγ at the promoter levels. RAW 264.7 cells were transfected with a luciferase reporter plasmid under transcriptional control of the iNOS promoter, PPARγ and RXRα expression plasmids. Cells were treated with the indicated combinations of LPS (100 ng/ml), Dex (10 nM, 1 μM) and GW7845 (10 nM, 1 μM), analyzed for luciferase activity 24 h later. (F) In vivo effects of combinations of GR and PPARγ agonists on the response to intraperitoneal injection of LPS. Six C57BL6 mice were pretreated with the indicated combinations of GW7845 (1 mg/kg) and Dex (1 mg/kg) for 7 days, injected intraperitoneally with LPS (1 mg) and circulating levels of IL-12 p40 were measured by ELISA 8 h later. (G) In vivo effects of combinations of GR and LXR agonists on the response to intraperitoneal injection of LPS. Six C57BL6 mice were pretreated with the indicated combinations of Dex (1 mg/kg) and T1317 (10 mg/kg) for 7 days, injected intraperitoneally with LPS (1 mg) and circulating levels of TNFα were measured by ELISA 6 h later.
To determine whether combinatorial effects of GR, PPARγ and LXR agonists acted at the promoter level, iNOS promoter activity was evaluated in RAW264.7 cells. The iNOS promoter was chosen for this analysis because the endogenous iNOS gene was subject to combinatorial inhibition by GR, PPARγ, and LXR agonists, its transcriptional activation requires binding sites for NF-κB (Lowenstein et al., 1993), and maximum responses to LPS required IRF3 (data not shown). As shown in Figure 6E, GR and PPARγ agonists inhibited iNOS promoter activity in a dose-dependent manner. When cells were treated with the combination of Dex and GW7845, at least additive effects with respect to inhibition of LPS response were observed at both saturating and non-saturating concentrations of ligands (Figure 6E). Similar results were observed for the combination of GR and LXR agonists (data not shown).
Although GR-mediated repression did not require NCoR (Figure 1B), we recently found that the ability of PPARγ to repress LPS activation of the iNOS promoter required NCoR (G.P., C.K.G., unpublished). Consistent with this, knockdown of NCoR expression with specific siRNAs had no effect on transrepression by Dex, but significantly impaired the ability of PPARγ and LXR agonists to repress the iNOS promoter (Figure S3). These findings indicate that at least two distinct, receptor-specific mechanisms are utilized by GR and PPARγ/LXR to repress LPS activation of the iNOS promoter, providing a potential explanation for synergistic repression when GR and PPARγ or LXR agonists are used in combination.
To investigate whether combinatorial interactions between PPARγ agonists and Dex observed in primary macrophages would also occur in an in vivo model system, we evaluated the IL-12 p40 subunit, as this was synergistically repressed in primary macrophages by GW7845 and Dex, but not by the combination of Dex and T1317. Consistent with these findings, treatment of mice with the combination of GW7845 (1 mg/kg/day) and Dex (1 mg/kg/day) prior to injection with LPS resulted in significantly greater inhibition of circulating IL-12 p40 than either agonist alone (Figure 6F). For the combination of GR and LXR agonists, we evaluated TNFα, based on synergistic inhibition by Dex and T1317 in primary macrophages. Treatment of mice with the combination of Dex (1 mg/kg/day) and T1317 (10 mg/kg/day) resulted in significantly greater inhibition of circulating tumor necrosis factor α (TNFα) levels following injection with LPS than observed following either agonist alone (Figure 6G).
Biological Functions Associated with Nuclear Receptor-sensitive and Nuclear Receptor-resistant TLR-target Genes
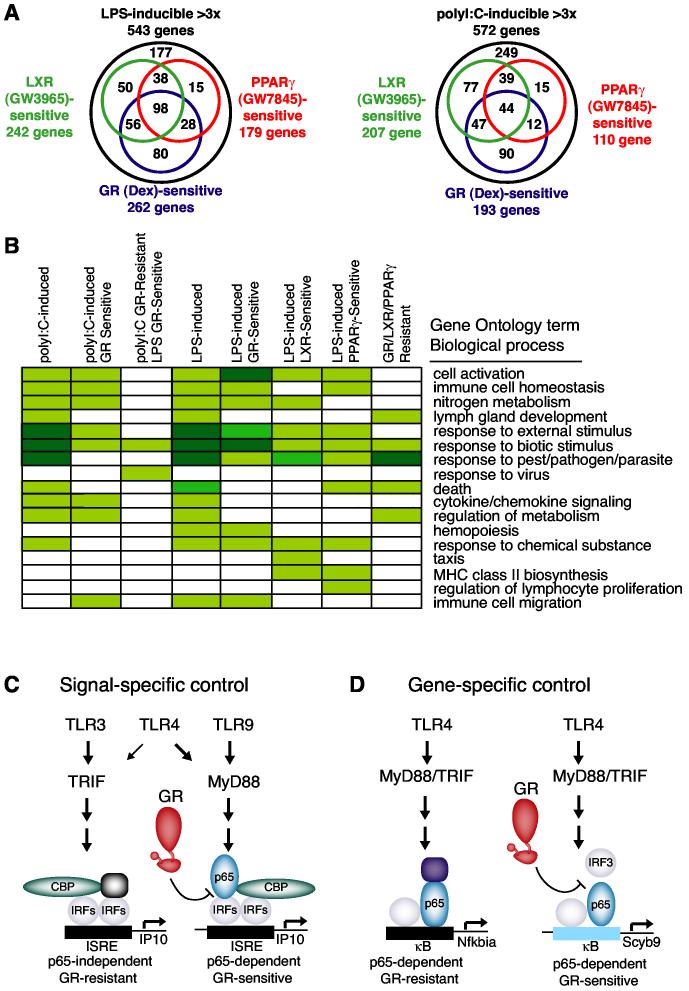
The identification of distinct subsets of TLR3- and TLR4-target genes exhibiting nuclear receptor-sensitive or nuclear receptor-resistant profiles raises the question of whether these genes participate in distinct biological processes. Venn diagrams illustrating the overlapping and distinct subsets of genes subject to transrepression by GR, PPAR and LXR following macrophage activation by LPS and poly I:C are illustrated in Figure 7A. To investigate the potential meaning of these findings at a biological level, statistical analysis of the functional annotations associated with specific sets of differentially regulated genes was performed using annotations provided by the Gene Ontology (GO) database (Gene Ontology Consortium, 2001). A subset of this analysis is presented in Figure 7B, illustrating major categories including immune cell homeostasis, response to virus, cytokine, chemokine signaling, etc. Although the effort to assign functional annotations to all mammalian gene products is at an early stage of development, the results of this analysis suggests significant functional differences in the sets of nuclear receptor-sensitive and nuclear receptor-resistant LPS-target genes. For example, the set of genes that was activated by LPS and resistant to GR, PPARγ and LXR agonists was enriched for functional annotations linked to metabolism (Figure 7B). In addition, transcriptional activation of the core components of the NF-κB pathway by LPS or poly I:C was almost completely resistant to repression by all three nuclear receptor agonists (Figure S4B). We also placed data for GR-mediated repression of LPS- and polyI:C-inducible genes on KEGG pathway maps (Kanehisa, 1996) and provide an example for the TLR signaling pathway in Figure S5. This figure indicates that components of the TLR signaling pathway in addition to NF-κB factors are Dex resistant, while TLR-activated chemokines and cytokines exhibit differential patterns of sensitivity that relate to proinflammatory effects and chemotaxis.
Figure 7.
Receptor-, Signal- and Gene-specific Counter-regulation of Inflammatory Responses by GR, PPARγ and LXRs (A) Venn diagram indicating sensitivity of LPS-responsive genes to GR, PPARγ and LXR-specific agonists. Nuclear receptor sensitivity was defined as >40% repression of the LPS response in a minimum of two independent microarray experiments (left). Venn diagram indicating sensitivity of poly I:C-responsive genes to GR, PPARγ and LXR-specific agonists. Nuclear receptor sensitivity was defined as >40% repression of the poly I:C response in a minimum of two independent microarray experiments (right). Venn diagrams are derived from the complete data set used to generate Figures 2B and 2C. (B) Representative functional annotations corresponding to Biological Process terms derived from the Gene Ontology database that were significantly enriched in the sets of LPS- and poly I:C-responsive genes. p represents the probability of obtaining the indicated number n genes within the category by chance determined as previously described (Ogawa et al., 2004). Color coding corresponds to the following p values; [unk]; p less than 0.01, [unk]; p less than 0.0001, [unk]; p less than 10-6. (C) Model for signal-specific GR-mediated transrepression, determined by utilization of p65 as an obligate TLR4-specific co-activator of IRF3. IRF-mediated activation of ISRE-containing genes by TLR4 and TLR9 through MyD88-pathway requires that p65 function as a signal-specific co-activator. The p65/IRF interaction is disrupted by liganded GR, resulting in transrepression. TLR3-specific activation of IRF3 through the TRIF pathway is p65-independent, and hence GR-resistant. (D) Model for gene-specific GR-mediated transrepression, determined by utilization of IRF3 as an obligate promoter-specific co-activator of NF-κB. The IRF3/p65 interaction is disrupted by liganded GR, providing an explanation for promoter-specific inhibition of the LPS response.
GR, PPARγ and LXR regulated functionally overlapping sets of genes, but also targeted genes in functionally related groups in a nuclear receptor-specific manner. For example, the list of repressed genes with functional annotations linked to hemopoiesis by the Gene Ontology consortium reached statistical significance for Dex, but not LXR or PPARγ agonists (Figure 7B). Repressive actions of each nuclear receptor ligand on specific genes involved in immune cell migration, differentiation and activation are illustrated in Figure S4B. Overall, Dex inhibited a larger number of genes involved in immune cell activation to a greater extent than LXR or PPARγ agonists, which may explain in part why LXR and PPARγ agonists are not as effective as Dex in acute models of inflammation. Significant differences in effects of the three receptor-specific agonists on chemokine gene expression were observed, suggesting that each receptor may play a context-specific role in regulating recruitment of specific immune cells to sites of inflammation.
Discussion
Signal-specific, Gene-specific and Nuclear Receptor-specific Transrepression
The present studies have used a combination of gene expression profiling and molecular analysis to investigate nuclear receptor-specific and combinatorial mechanisms of transrepression by nuclear receptors. These observations extend the spectrum of nuclear receptor- and promoter-specific inhibition of signal-dependent gene expression, demonstrating that GR, PPARγ and LXR repress overlapping but distinct subsets of inflammatory response genes, consistent with the large number of mechanisms that have been proposed for negative regulation by nuclear receptors (De Bosscher et al., 2003). Although evidence supporting global inactivation of NF-κB activity by GR and ER has been reported in other cell types (Auphan et al., 1995; Ghisletti et al., 2005; Scheinman et al., 1995a), the present studies indicate that GR, LXRs and PPARγ target only a subset of the TLR-inducible NF-κB-dependent genes in macrophages. These findings indicate that the mechanisms of transrepression in this cell type must operate in a promoter-specific manner.
The observation that a significant set of genes that were sensitive to nuclear receptor-dependent repression when activated through TLR4 became resistant to repression when activated through TLR3 also indicates that transrepression programs mediated by GR, PPARγ and LXRs are regulated in a signal-specific manner. The finding of a highly significant correlation between IFR3-dependent transcriptional activation and signal-dependent transrepression by glucocorticoid receptor (GR) led to the discovery that GR effectively disrupts the formation of an IRF3/p65 activator/coactivator complex that is required for activation of ISRE-containing promoters by TLR4-but not TLR3-dependent signaling (Figures 3E and 4C). Of the 262 genes scored as being LPS-inducible and Dex-sensitive in these studies, at least 85 genes fit with the hypothesis that disruption of IRF3/p65 complexes is a quantitatively important component of the transrepression mechanism. Taken together with the results of studies in MyD88−/− macrophages (Figure 5D) and the patterns of gene expression following TLR9 activation (Figure 3A), these findings support a unifying model in which TLR signaling through MyD88 specifies glucocorticoid sensitivity of IRF-dependent genes through the utilization of IRF/p65 complexes (Figure 7C). As IRF7 is involved in TLR9-MyD88-dependent gene activation (Kawai et al., 2004), these results imply that the mechanism of GR-mediated repression operates through both IRF3 and IRF7. These findings thus reveal the molecular basis for an example of signal-specific transrepression that is utilized by a large group of functionally inter-related genes. Distinct regions of GR appear to be involved in mediating repression of AP-1 target genes (Bladh et al., 2005) and it will be of interest to explore signal-specific modulation of transrepression in response to other proinflammatory cytokines that induce AP-1 and STAT transcription factors, such as TNFα and IFNβ.
Conversely, the ability of IRF3 to function as an essential coactivator of p65 on a subset of NF-κB target genes provides an explanation for how transrepression by GR can be achieved in a gene-specific manner (Figure 7D). NF-κB target genes that are resistant to GR-mediated transrepression are predicted to utilize other classes of coactivators, such as Bcl3 (Leung et al., 2004), that may prevent the interaction of GR with DNA-bound NF-κB. Consistent with this, ChIP experiments demonstrated recruitment of GR to the Dex-sensitive Scyb9 and Clic4 promoters. A significant number of Dex-sensitive NF-κB target genes are not IRF3-dependent, indicating a requirement for additional mechanisms. Among the 100 most LPS and polyI:C-inducible genes, all of the genes that were Dex-sensitive following polyI:C stimulation were also Dex-sensitive following activation by LPS (Figure S4A). Similarly, among the 100 most polyI:C and CpG1668-inducible genes, all of the genes that were Dex-sensitive following polyI:C stimulation were also Dex-sensitive following activation by CpG1668 (Figure S4A). These results suggest that a common set of mechanisms is used for Dex repression of this subset of TLR-inducible genes. We anticipate that these mechanisms will include gene-specific recruitment of coactivators other than IRF3 that are also sensitive to competition with GR.
In contrast to GR, transrepression mediated by PPARγ and LXRs did not seem to involve p65. This observation is consistent with the overlapping but distinct sets of genes that were found to be sensitive to repression by PPARγ and LXRs and implies that distinct targeting mechanisms are employed by these nuclear receptors. We and others recently reported that a subset of genes induced by pro-inflammatory signals are occupied in the basal state by NCoR/SMRT corepressor complexes that act as transcriptional checkpoints to maintain these genes in a transcriptionally silent state (Hoberg et al., 2004; Ogawa et al., 2004; Perissi et al., 2004). Signal-dependent transcriptional activation of these genes requires clearance of NCoR/SMRT complexes by an ubiquitin/proteosome-dependent step as a prerequisite to transcriptional activation. Recent studies suggest that PPARγ represses LPS induction of the iNOS gene by preventing the clearance of the NCoR corepressor complex (G.P., C.K.G., unpublished). The convergence of NCoR-dependent and NCoR-independent transrepression mechanisms on the iNOS promoter support the hypothesis that synergistic repression of LPS target genes by GR and PPARγ is achieved by the combinatorial utilization of distinct, nuclear receptor-specific mechanisms.
Physiological Implications for Cellular Responses to Bacterial and Viral Pathogens
The observation that TLR-responsive genes exhibit different sensitivities to repression by nuclear receptors suggests that they play distinct biological roles in determining cellular responses to infection and other inflammatory processes. By specifically targeting p65/IRF3 complexes, GR is able to discriminate signals initiated by TLRs that either do or do not couple to the MyD88 signaling pathway, providing a biological rationale for the context-specific utilization of these complexes. For example, we predict that the antiviral program of TLR3–should be resistant to GR-mediated repression. In contrast, the antiviral program elicited by activation of TLR9, which is involved in innate responses to herpes simplex virus (HSV) (Krug et al., 2004; Lund et al., 2003), should be GR-sensitive. This prediction is in fact consistent with clinical experience and provides a potential molecular explanation for why steroid use is contraindicated in the presence of HSV-infected retinitis, encephalitis, and uveitis.
In addition to defining combinatorial control of inflammatory responses at the promoter level, these studies also suggest a higher level of regulation that serves to integrate both local and systemic signaling pathways. In the case of GR, the endogenous corticosteroid ligands are classic endocrine hormones, produced in the adrenal cortex in response to systemic physiological circuits (i.e., the hypothalamic-pituitary-adrenal axis). Circulating corticosteroids diffuse into cells and bind to GR in virtually all tissues with sub-nanomolar affinities, providing coordinate regulation of gene expression at the whole body level. In contrast, the adopted orphan receptors such as PPARγ and LXRs are activated by metabolites of fatty acids and cholesterol, respectively, that are produced locally within the cell and bind with relatively low affinity (Forman et al., 1997; Forman et al., 1995; Kliewer et al., 1995; Kliewer et al., 1997; Nagy et al., 1998). In addition, the expression of PPARγ and LXRα and the production of regulatory ligands are determined by local cytokines and other regulatory systems (Ricote et al., 1998). These results also suggest that different combinations of NR ligands will exert different biases with respect to evolution of inflammatory responses. Taken together, these observations suggest a model in which GR and PPARγ/LXR integrate systemic and local regulatory signals so as to coordinate transcriptional responses to infection throughout the body. In support of this, LXRs have recently been shown to play important roles in protection against bacterial infection (Joseph et al., 2004; Valledor et al., 2004).
Clinical Implications
Nuclear receptors are important targets of drugs used in a variety of human disease settings. In many cases, the ability to achieve desirable therapeutic effects with a natural or synthetic nuclear receptor agonist is limited by undesirable or unacceptable side effects. For example, glucocorticoids are potent anti-inflammatory drugs, but can cause or exacerbate hypertension, diabetes, obesity, and dyslipidemia. Emerging information on the ability of selective modulators of nuclear receptors to alter the specificity of coactivator and corepressor recruitment raises new possibilities for the development of novel pharmaceutical agents (Smith and O'Malley, 2004; Wagner et al., 2003). The present studies suggest an alternative, and potentially complementary, strategy to leverage desirable therapeutic effects while minimizing side effects. Using chronic, steroid-dependent inflammatory diseases as an example, it is possible that anti-inflammatory actions of synthetic glucocorticoids could be achieved at lower doses with fewer side effects by simultaneous administration of PPARγ or LXR agonists. The present findings may also influence decisions to use glucocorticoids or other classes of nuclear receptor agonists in the setting of viral or bacterial infection. Lack of efficacy of glucocorticoids in preventing some inflammatory complications of viral infections such as viral myocarditis (Vallejo and Mann, 2003), may potentially be explained by the viral activation of GR-resistant signaling pathways. Because PPARγ and LXR agonists exerted repressive effects on some of the GR-resistant inflammatory mediators induced by TLR3, it is possible that they might have therapeutic value in these settings.
Taken together, we propose that the GR, PPARs and LXRs function in a combinatorial manner to integrate systemic and local signals that control inflammatory gene expression and the evolution of innate and acquired immune responses. Further elucidation of these combinatorial mechanisms may provide further insights into how nuclear receptors control signal-activated transcription and lead to new strategies for treatment of inflammatory diseases.
Experimental Procedures
Cell Culture
Thioglycollate-elicited macrophages were isolated by peritoneal lavage 3 days following peritoneal injection of 2.5 ml 3% thioglycollate (DIFCO). Cells were plated in RPMI medium 1640 and 10% fetal bovine serum, washed after 5 h the medium was removed and cells were fed with fresh medium containing 0.5% fetal bovine serum. LPS (Sigma) was used at a concentration of 100 ng/ml. Fetal liver-derived macrophages generated from E14.5 embryo liver were plated and cultured in RPMI with 10% fetal bovine serum plus L-cell media for 7 days as described (Ogawa et al., 2004). Fetal liver-derived macrophages of TNF−/−cRel−/− (control) and TNF−/−cRel−/−RelA−/− (NF-κB−/−) were obtained by mating TNF−/−cRel−/−RelA+/− mice and by genotyping the embryos.
Expression Array Profiling
Cells were lysed with Trizol (Invitrogen) and total RNA was purified using RNeasy columns (Qiagen). cRNA was generated from 10 μg total RNA using Superscript (Invitrogen) and the High Yield RNA transcription labeling kit (Enzo). Fragmented cRNA was hybridized to Affymetrix Mu11 or Codelink mouse Uniset 1 microarrays according to manufacture's instructions. Data was analyzed with Microarray Suite (Affymetrix), GeneSpring (Silicongenetics) and in-house software developed as described (Ogawa et al., 2004; Sasik et al., 2002).
Computational Analysis
Proximal promoter regions were extracted for each gene represented on the microarray using the May 2004 mouse genome assembly with the method as described (Halees et al., 2003). Analysis was restricted to the region 1 kb upstream of the transcription start site. In cases were several possible alternative promoters may be present, analysis was focused on the most 5' transcription start site. Motif discovery was performed using a comparative algorithm previously described (Segal et al., 2002). Promoters were initially divided into two sets: those that were up regulated by LPS and those that were present on the array but did not change in response to LPS. An exhaustive search for all n-mers (6 < n < 12) was performed and each n-mer was scored for its enrichment in the promoters up regulated by LPS using the hypergeometric distribution. The top 500 n-mers with a p-value less than 0.01 were then clustered together and used to create position specific probability matrices. The matrices were then further optimized to discriminate between the LPS responsive and non-responsive genes by the methods as described (Segal et al., 2002).
Transient Transfection and Reporter Studies
Transient transfections were performed as described using Superfect reagent (Qiagen) (Ogawa et al., 2004; Ricote et al., 1998). RAW 264.7 cells were transfected with iNOS promoter-luciferase, pCMX-PPARγ and pCMX-RXRα plasmids. pRL-TK, a renilla luciferase reporter plasmid was also co-transfected as an internal control (Promega). GR, PPAR and LXR agonists were used at the indicated concentrations in 0.5% fetal bovine serum and cells were harvested 24 h later for analysis of luciferase activity. Double-stranded, short interfering RNAs (siRNA) were synthesized by Dharmacon Research (Lafayette) and were transfected for 48 h prior to activation with ligands and LPS induction as previously described (Perissi et al., 2004). Data are represented as mean ± SD.
GST Pull-down Assays
GST pull-down assays were carried out as described previously (Li et al., 2000). GST fusion proteins were produced as crude bacterial lysates and immobilized on glutathione agarose beads. p65, IRF3, full-length GR, GR(K471A), and GR-DBD proteins were translated in vitro using 35S-labeled methionine and TnT-coupled reticulocyte lysate system (Promega).
ChIP Assay
ChIP assay was conducted as previously (Baek et al., 2002; Perissi et al., 2004). Briefly, macrophages were fixed with 1% formaldehyde and cross-linked adducts were resuspended and sonicated resulting in DNA fragments of 300–900 bp. Protein-bound, immunoprecipitated DNA was reverse cross-linked at 65°C overnight and then purified using PCR purification Kit (Qiagen). 4 ul from a 50 ul DNA extraction were used for PCR amplification. Anti-IRF3 (Zymed), anti-CBP (Santa Cruz), anti-p65 (Santa Cruz), and anti-GR (Santa Cruz) antibodies were used in immunoprecipitation experiments.
RNA Analysis
RNA analysis by Northern blotting was performed as described (Ogawa et al., 2000). Five μg total RNA were separated by gel electrophoresis and transferred to nylon (Supercharge, Scheicher & Schuell). Prior to hybridization, membranes were UV cross-linked (Stratagene) and stained with Methylene Blue (Molecular Research Center, Cincinnati). Probes were generated by RT-PCR followed by random priming labeling (Invitrogen) and hybridization with QuickHyb (Stratagene). Real-time quantitative PCR (SYBRgreen) analysis was performed on an Applied Biosystems 7300 Real-Time PCR system.
LPS-Induced Endotoxin Shock and In Vivo Studies
Combinatorial effects of GR, PPARγ and LXR agonists in antagonism of LPS responses in vivo were evaluated by measuring TNFα and IL-12 p40 levels 6 and 8 hs after intraperitoneal injection of LPS at 1 mg/mouse, respectively. We initially performed a series of pilot studies to establish dosing schedules of Dex that would result in less than 50% inhibition of the LPS-induced TNFα and IL-12 p40 responses. Mice were orally dosed daily with Dex for 7 days and were intraperitoneally injected with LPS (1 mg/mouse). Blood was collected after LPS-stimulation, and analyzed for cytokine levels by ELISA. Based on the results of these pilot studies, two independent studies evaluating effects of combinations of PPARγ agonists and LXR agonists with Dex at the dosing levels indicated in Figure 6. At a minimum, six mice were used for each experimental condition.
Acknowledgements
We are grateful to U.C. San Diego BIOGEM Core Facility for assistance with microarray experiments, to S. Akira for MyD88−/− mice, to T. Taniguchi for IRF3−/− mice, to B. Beutler for CpG1668, and to T. Willson (GlaxoSmithKline) for GW7845. We thank A. Z. Howarth for figure preparation. M.G.R. is an investigator with the Howard Hughes Medical Institute. These studies were supported in part by NIH grants CA52599 and GM069338 and a grant from the Stanford Reynolds Center to C.K.G. These studies were supported in part by NIH grant and the Sandler Program for Asthma Research to M.G.R.
References
- Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, Akira S. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- Akira S, Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Auphan N, DiDonato JA, Rosette C, Helmberg A, Karin M. Immunosuppression by glucocorticoids: inhibition of NF-κB activity through induction of IκB synthesis. Science. 1995;270:286–290. doi: 10.1126/science.270.5234.286. [DOI] [PubMed] [Google Scholar]
- Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-κB and β-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- Bladh LG, Liden J, Dahlman-Wright K, Reimers M, Nilsson S, Okret S. Identification of endogenous glucocorticoid repressed genes differentially regulated by a glucocorticoid receptor mutant able to separate between nuclear factor-κB and activator protein-1 repression. Mol. Pharmacol. 2005;67:815–826. doi: 10.1124/mol.104.005801. [DOI] [PubMed] [Google Scholar]
- Caelles C, Gonzalez-Sancho JM, Munoz A. Nuclear hormone receptor antagonism with AP-1 by inhibition of the JNK pathway. Genes Dev. 1997;11:3351–3364. doi: 10.1101/gad.11.24.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldenhoven E, Liden J, Wissnik S, Van de Stoipe A, Raaijmakers J, Koenderman L, Okret S, Gustafsson JÅ, Van der Sagg PT. Negative cross-talk between RelA and the glucocorticoid receptor: a possible mechanism for the antiinflammatory action of glucocorticoids. Mol. Endocrinol. 1995;9:401–412. doi: 10.1210/mend.9.4.7659084. [DOI] [PubMed] [Google Scholar]
- Castrillo A, Joseph SB, Vaidya SA, Haberland M, Fogelman AM, Cheng G, Tontonoz P. Crosstalk between LXR and Toll-like receptor signaling mediates bacterial and viral antagonism of cholesterol metabolism. Mol. Cell. 2003;12:805–816. doi: 10.1016/s1097-2765(03)00384-8. [DOI] [PubMed] [Google Scholar]
- Chawla A, Boisvert WA, Lee CH, Laffitte BA, Barak Y, Joseph SB, Liao D, Nagy L, Edwards PA, Curtiss LK, et al. A PPARγ-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol. Cell. 2001;7:161–171. doi: 10.1016/s1097-2765(01)00164-2. [DOI] [PubMed] [Google Scholar]
- Dang O, Navarro L, Anderson K, David M. Cutting edge: anthrax lethal toxin inhibits activation of IFN-regulatory factor 3 by lipopolysaccharide. J. Immunol. 2004;172:747–751. doi: 10.4049/jimmunol.172.2.747. [DOI] [PubMed] [Google Scholar]
- De Bosscher K, Vanden Berghe W, Haegeman G. The interplay between the glucocorticoid receptor and nuclear factor-κB or activator protein-1: molecular mechanisms for gene repression. Endocr. Rev. 2003;24:488–522. doi: 10.1210/er.2002-0006. [DOI] [PubMed] [Google Scholar]
- De Bosscher K, Vanden Berghe W, Vermeulen L, Plaisance S, Boone E, Haegeman G. Glucocorticoids repress NF-κB-driven genes by disturbing the interaction of p65 with the basal transcription machinery, irrespective of coactivator levels in the cell. Proc. Natl. Acad. Sci. USA. 2000;97:3919–3924. doi: 10.1073/pnas.97.8.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devchand PR, Keller H, Peters JM, Vazquez M, Gonzalez FJ, Wahli W. The PPARα-leukotriene B4 pathway to inflammation control. Nature. 1996;384:39–43. doi: 10.1038/384039a0. [DOI] [PubMed] [Google Scholar]
- Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald KA, Rowe DC, Barnes BJ, Caffrey DR, Visintin A, Latz E, Monks B, Pitha PM, Golenbock DT. LPS-TLR4 signaling to IRF-3/7 and NF-κB involves the toll adapters TRAM and TRIF. J. Exp. Med. 2003;198:1043–1055. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proc. Natl. Acad. Sci. USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman BM, Tontonoz P, Chen J, Brun RP, Spiegelman BM, Evans RM. 15-deoxy-Δ12,14 prostaglandin J2 is a ligand for the adipocyte determination factor PPARγ. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- Gene ontology consortium Creating the gene ontology resource: design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Meda C, Maggi A, Vegeto E. 17β-estradiol inhibits inflammatory gene expression by controlling NF-κB intracellular localization. Mol. Cell. Biol. 2005;25:2957–2968. doi: 10.1128/MCB.25.8.2957-2968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker H, Vabulas RM, Takeuchi O, Hoshino K, Akira S, Wagner H. Immune cell activation by bacterial CpG-DNA through myeloid differentiation marker 88 and tumor necrosis factor receptor-associated factor (TRAF)6. J. Exp. Med. 2000;192:595–600. doi: 10.1084/jem.192.4.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halees AS, Leyfer D, Weng Z. PromoSer: A large-scale mammalian promoter and transcription start site identification service. Nucleic Acids Res. 2003;31:3554–3559. doi: 10.1093/nar/gkg549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Hoberg JE, Yeung F, Mayo MW. SMRT derepression by the IκB kinase a; A prerequisite to NF-κB transcription and survival. Mol. Cell. 2004;16:245–255. doi: 10.1016/j.molcel.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- Janeway CA, Jr., Medzhitov R. Innate immune recognition. Annu. Rev. Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor LXRα. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- Jiang C, Ting AT, Seed B. PPARγ agonists inhibit production of monocyte inflammatory cytokines. Nature. 1998;391:82–86. doi: 10.1038/34184. [DOI] [PubMed] [Google Scholar]
- Jonat C, Rahmsdorf HJ, Park KK, Ponta H, Herrlich P. Anti-tumor promotion and antiinflammation: down-modulation of AP-1 (Fos/Jun) activity by glucocorticoid hormone. Cell. 1990;62:1189–1204. doi: 10.1016/0092-8674(90)90395-u. [DOI] [PubMed] [Google Scholar]
- Joseph SB, Bradley MN, Castrillo A, Bruhn KW, Mak PA, Pei L, Hogenesch J, O'Connell R M, Cheng G, Saez E, et al. LXR-dependent gene expression is important for macrophage survival and the innate immune response. Cell. 2004;119:299–309. doi: 10.1016/j.cell.2004.09.032. [DOI] [PubMed] [Google Scholar]
- Joseph SB, Castrillo A, Laffitte BA, Mangelsdorf DJ, Tontonoz P. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat. Med. 2003;9:213–219. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- Juang YT, Lowther W, Kellum M, Au WC, Lin R, Hiscott J, Pitha PM. Primary activation of interferon α and interferon β gene transcription by interferon regulatory factor 3. Proc. Natl. Acad. Sci. USA. 1998;95:9837–9842. doi: 10.1073/pnas.95.17.9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin SC, Heyman RA, Rose DW, Glass CK, Rosenfeld MG. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- Kanehisa M. Toward pathway engineering: a new database of genetic and molecular pathways. Science & Technology Japan. 1996;59:34–38. [Google Scholar]
- Kassel O, Schneider S, Heilbock C, Litfin M, Gottlicher M, Herrlich P. A nuclear isoform of the focal adhesion LIM-domain protein Trip6 integrates activating and repressing signals at AP-1- and NF-κB-regulated promoters. Genes Dev. 2004;18:2518–2528. doi: 10.1101/gad.322404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner P, Mark M, Chambon P. Nonsteroid nuclear receptors: what are genetic studies telling us about their role in real life? Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- Kawai T, Sato S, Ishii KJ, Coban C, Hemmi H, Yamamoto M, Terai K, Matsuda M, Inoue J, Uematsu S, et al. Interferon α induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 2004;5:1061–1068. doi: 10.1038/ni1118. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takeuchi O, Fujita T, Inoue J, Muhlradt PF, Sato S, Hoshino K, Akira S. Lipopolysaccharide stimulates the MyD88-independent pathway and results in activation of IFN-regulatory factor 3 and the expression of a subset of lipopolysaccharide-inducible genes. J. Immunol. 2001;167:5887–5894. doi: 10.4049/jimmunol.167.10.5887. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Lenhard JM, Willson TM, Patel I, Morris DC, Lehmann JM. A prostaglandin J2 metabolite binds peroxisome proliferator-activated receptorγ and promotes adipocyte differentiation. Cell. 1995;83:813–819. doi: 10.1016/0092-8674(95)90194-9. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Sundseth SS, Jones SA, Brown PJ, Wisely GB, Koble CS, Devchand P, Wahli W, Willson TM, Lenhard JM, Lehmann JM. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and γ. Proc. Natl. Acad. Sci. USA. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug A, Luker GD, Barchet W, Leib DA, Akira S, Colonna M. Herpes simplex virus type 1 activates murine natural interferon-producing cells through toll-like receptor 9. Blood. 2004;103:1433–1437. doi: 10.1182/blood-2003-08-2674. [DOI] [PubMed] [Google Scholar]
- Leung TH, Hoffmann A, Baltimore D. One nucleotide in a κB site can determine cofactor specificity for NF-κB dimers. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Li M, Pascual G, Glass C. Peroxisome proliferator-activated receptor y-dependent repression of the inducible nitric oxide synthase gene. Mol. Cell. Biol. 2000;20:4699–4707. doi: 10.1128/mcb.20.13.4699-4707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liden J, Franck D, Rafter I, Guftafson JÅ, Okret S. A new function for the C-terminal zinc finger of the glucocorticoid receptor; regression of RelA transactivation. J. Biol. Chem. 1997;272:21467–21472. doi: 10.1074/jbc.272.34.21467. [DOI] [PubMed] [Google Scholar]
- Lowenstein CJ, Alley EW, Raval P, Snowman AM, Snyder SH, Russell SW, Murphy WJ. Macrophage nitric oxide synthase gene: two upstream regions mediate induction by interferonγ and lipopolysaccharide. Proc. Natl. Acad. Sci. USA. 1993;90:9730–9734. doi: 10.1073/pnas.90.20.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J. Exp. Med. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Marx N, Mach F, Sauty A, Leung JH, Sarafi MN, Ransohoff RM, Libby P, Plutzky J, Luster AD. Peroxisome proliferator-activated receptor-γ activators inhibit IFNγ-induced expression of the T cell-active CXC chemokines IP-10, Mig, and I-TAC in human endothelial cells. J. Immunol. 2000;164:6503–6508. doi: 10.4049/jimmunol.164.12.6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay LI, Cidlowski JA. Molecular control of immune/inflammatory responses: interactions between nuclear factor-κB and steroid receptor-signaling pathways. Endocr. Rev. 1999;20:435–459. doi: 10.1210/edrv.20.4.0375. [DOI] [PubMed] [Google Scholar]
- Nagpal S, Lu J, Boehm MF. Vitamin D analogs: mechanism of action and therapeutic applications. Curr. Med. Chem. 2001;8:1661–1679. doi: 10.2174/0929867013371950. [DOI] [PubMed] [Google Scholar]
- Nagy L, Tontonoz P, Alvarez JG, Chen H, Evans RM. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARγ. Cell. 1998;93:229–240. doi: 10.1016/s0092-8674(00)81574-3. [DOI] [PubMed] [Google Scholar]
- Navarro L, David M. p38-dependent activation of interferon regulatory factor 3 by lipopolysaccharide. J. Biol. Chem. 1999;274:35535–35538. doi: 10.1074/jbc.274.50.35535. [DOI] [PubMed] [Google Scholar]
- Nissen RM, Yamamoto KR. The glucocorticoid receptor inhibits NF-κB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 2000;14:2314–2329. doi: 10.1101/gad.827900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Fujita M, Ishii Y, Tsurukami H, Hirabayashi M, Ikeda K, Orimo A, Hosoi T, Ueda M, Nakamura T, et al. Impaired estrogen sensitivity in bone by inhibiting both estrogen receptor α and β pathways. J. Biol. Chem. 2000;275:21372–21379. doi: 10.1074/jbc.M909675199. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Lozach J, Jepsen K, Sawka-Verhelle D, Perissi V, Sasik R, Rose DW, Johnson RS, Rosenfeld MG, Glass CK. An NCoR transcriptional checkpoint controlling AP-1-dependent gene networks required for macrophage activation. Proc. Natl. Acad. Sci. USA. 2004;101:14461–14466. doi: 10.1073/pnas.0405786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- Pitha PM. Unexpected similarities in cellular responses to bacterial and viral invasion. Proc. Natl. Acad. Sci. USA. 2004;101:695–696. doi: 10.1073/pnas.0307303101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittas AG, Joseph NA, Greenberg AS. Adipocytokines and insulin resistance. J. Clin. Endocrinol. Metab. 2004;89:447–452. doi: 10.1210/jc.2003-031005. [DOI] [PubMed] [Google Scholar]
- Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, Schütz G. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- Reichardt HM, Tuckermann JP, Gottlicher M, Vujic M, Weih F, Angel P, Herrlich P, Schutz G. Repression of inflammatory responses in the absence of DNA binding by the glucocorticoid receptor. EMBO J. 2001;20:7168–7173. doi: 10.1093/emboj/20.24.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricote M, Li AC, Willson TM, Kelly CJ, Glass CK. The peroxisome proliferator-activated receptor-γ is a negative regulator of macrophage activation. Nature. 1998;391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- Rogatsky I, Zarember KA, Yamamoto KR. Factor recruitment and TIF2/GRIP1 corepressor activity at a collagenase- 3 response element that mediates regulation by phorbol esters and hormones. EMBO J. 2001;20:6071–6083. doi: 10.1093/emboj/20.21.6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasik R, Calvo E, Corbeil J. Statistical analysis of high-density oligonucleotide arrays: a multiplicative noise model. Bioinformatics. 2002;18:1633–1640. doi: 10.1093/bioinformatics/18.12.1633. [DOI] [PubMed] [Google Scholar]
- Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-α/β gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- Scheinman RI, Cogswell PC, Lofquist AK, Baldwin AS., Jr. Role of transcriptional activation of IκB α in mediation of immunosuppression by glucocorticoids. Science. 1995a;270:283–286. doi: 10.1126/science.270.5234.283. [DOI] [PubMed] [Google Scholar]
- Scheinman RI, Gualberto A, Jewell CM, Cidlowski JA, Baldwin AS., Jr. Characterization of mechanisms involved in transrepression of NF-κB by activated glucocorticoid receptors. Mol. Cell. Biol. 1995b;15:943–953. doi: 10.1128/mcb.15.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüle R, Rangarajan P, Kliewer S, Ransone LJ, Bolado J, Yang N, Verma IM, Evans RM. Functional antagonism between oncoprotein c-Jun and the glucocorticoid receptor. Cell. 1990;62:1217–1226. doi: 10.1016/0092-8674(90)90397-w. [DOI] [PubMed] [Google Scholar]
- Segal E, Barash Y, Simon I, Friedman N, Koller D. Proc. 6th Inter. Conf. on Research in Computational Molecular Biology (RECOMB) Washington, DC: 2002. From promoter sequence to expression: a probabilistic framework. [Google Scholar]
- Sheppard KA, Phelps KM, Williams AJ, Thanos D, Glass CK, Rosenfeld MG, Gerritsen ME, Collins T. Nuclear integration of glucocorticoid receptor and nuclear factor-κB signaling by CREB-binding protein and steroid receptor coactivator-1. J. Biol. Chem. 1998;273:29291–29294. doi: 10.1074/jbc.273.45.29291. [DOI] [PubMed] [Google Scholar]
- Smith CL, O'Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr. Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- Staels B, Koenig W, Habib A, Merval R, Lebret M, Torra IP, Delerive P, Fadel A, Chinetti G, Fruchart JC, et al. Activation of human aortic smooth-muscle cells is inhibited by PPARα but not by PPARγ activators. Nature. 1998;393:790–793. doi: 10.1038/31701. [DOI] [PubMed] [Google Scholar]
- Valledor AF, Hsu LC, Ogawa S, Sawka-Verhelle D, Karin M, Glass CK. Activation of liver X receptors and retinoid X receptors prevents bacterial-induced macrophage apoptosis. Proc. Natl. Acad. Sci. USA. 2004;101:17813–17818. doi: 10.1073/pnas.0407749101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo J, Mann DL. Antiinflammatory therapy in myocarditis. Curr. Opin. Cardiol. 2003;18:189–193. doi: 10.1097/00001573-200305000-00003. [DOI] [PubMed] [Google Scholar]
- Wagner BL, Valledor AF, Shao G, Daige CL, Bischoff ED, Petrowski M, Jepsen K, Baek SH, Heyman RA, Rosenfeld MG, et al. Promoter-specific roles for liver X receptor/corepressor complexes in the regulation of ABCA1 and SREBP1 gene expression. Mol. Cell. Biol. 2003;23:5780–5789. doi: 10.1128/MCB.23.16.5780-5789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol. Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JS, Ricote M, Akiyama TE, Gonzalez FJ, Glass CK. PPARγ and PPARδ negatively regulate specific subsets of lipopolysaccharide and IFNγ target genes in macrophages. Proc. Natl. Acad. Sci. USA. 2003;100:6712–6717. doi: 10.1073/pnas.1031789100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wietek C, Miggin SM, Jefferies CA, O'Neill LA. Interferon regulatory factor-3-mediated activation of the interferon-sensitive response element by Toll-like receptor (TLR) 4 but not TLR3 requires the p65 subunit of NF-κB. J. Biol. Chem. 2003;278:50923–50931. doi: 10.1074/jbc.M308135200. [DOI] [PubMed] [Google Scholar]
- Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, Chen H. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Mori K, Hoshino K, Takeuchi O, Takeda K, Akira S. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-β promoter in the Toll-like receptor signaling. J. Immunol. 2002;169:6668–6672. doi: 10.4049/jimmunol.169.12.6668. [DOI] [PubMed] [Google Scholar]
- Yang-Yen H-F, Chambard J-C, Sun Y-L, Smeal T, Schmidt TJ, Drouin J, Karin M. Transcriptional interference between c-Jun and the glucocorticoid receptor: mutual inhibition of DNA binding due to direct protein-protein interaction. Cell. 1990;62:1205–1215. doi: 10.1016/0092-8674(90)90396-v. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]