Abstract
One of the major drawbacks limiting the use of synthetic peptide vaccines in genetically distinct populations is the fact that different epitopes are recognized by T cells from individuals displaying distinct major histocompatibility complex molecules. Immunization of mice with peptide (181-195) from the immunodominant 43 kDa glycoprotein of Paracoccidioides brasiliensis (gp43), the causative agent of Paracoccidioidomycosis (PCM), conferred protection against infectious challenge by the fungus. To identify immunodominant and potentially protective human T-cell epitopes in gp43, we used the TEPITOPE algorithm to select peptide sequences that would most likely bind multiple HLA-DR molecules and tested their recognition by T cells from sensitized individuals. The 5 most promiscuous peptides were selected from the gp43 sequence and the actual promiscuity of HLA binding was assessed by direct binding assays to 9 prevalent HLA-DR molecules. Synthetic peptides were tested in proliferation assays with peripheral blood mononuclear cells (PBMC) from PCM patients after chemotherapy and healthy controls. PBMC from 14 of 19 patients recognized at least one of the promiscuous peptides, whereas none of the healthy controls recognized the gp43 promiscuous peptides. Peptide gp43(180-194) was recognized by 53% of patients, whereas the other promiscuous gp43 peptides were recognized by 32% to 47% of patients. The frequency of peptide binding and peptide recognition correlated with the promiscuity of HLA-DR binding, as determined by TEPITOPE analysis. In silico prediction of promiscuous epitopes led to the identification of naturally immunodominant epitopes recognized by PBMC from a significant proportion of a genetically heterogeneous patient population exposed to P. brasiliensis. The combination of several such epitopes may increase the frequency of positive responses and allow the immunization of genetically distinct populations.
INTRODUCTION
Recent advances in peptide biochemistry and immunochemistry have led to the development of protective antimicrobial vaccines based on synthetic peptides and defined epitopes. Immunization with synthetic peptides has been widely used in animal models (1-3), and the relative ease, low cost of preparation, safety, prolonged shelf-life, and the ability to focus on defined epitopes (4-7) encouraged the use of synthetic peptide vaccines in animal (8-14) and human diseases (15,16).
The central event in the adaptive immune response to invasive microorganisms is the specific recognition of intracellularly processed foreign antigenic peptides bound to the peptide-binding region of the human leukocyte antigen (HLA) class II molecules on the surface of antigen-presenting cells by the T-cell receptor of CD4+ T cells. This is followed by activation, proliferation, and differentiation of specific CD4+ T cells to effector cells that are fully capable of interacting with other inflammatory cells and thus inducing specialized effector immune responses. Pathogen-induced activation of CD4+ T cells can generate memory T cells, which will be ready to respond more rapidly in case of a later contact with the same pathogen.
To induce protective immunity, epitopes contained in synthetic peptide vaccines must match epitopes naturally presented to the immune system during infection, be recognized by the entire human population targeted for vaccination, and induce an effector immune response capable of effectively eliminating the pathogen. Single epitope-based vaccines may, however, have some drawbacks. On the pathogen side, the monospecificity of the induced immune response favors the emergence of sequence mutants that can escape from the vaccine’s protective effect (7). On the host side, it is unlikely that T cells from large proportions of genetically distinct populations can recognize—and therefore be protected—by the same single peptide epitope. This is secondary to the wide polymorphism of HLA molecules that present antigenic peptides to T cells. Because the antigen-binding groove of each of the hundreds of allelic HLA molecules has distinct peptide-binding preferences, a distinct set of epitopes from a given protein antigen will be presented to T cells in each individual bearing a different HLA molecule. Additionally, some HLA molecules may not be able to bind at all to any of the peptides derived from a given protein (17,18).
Such interindividual variation of antigen recognition can be a problem for selecting immunogens for peptide vaccines, because they must contain immunodominant epitopes recognized by individuals with a wide range of different HLA molecules. The identification of single peptides that can bind to multiple HLA types, the so-called “promiscuous” epitopes, could lead to effective coverage of the human population by a peptide-based vaccine. Thus, the major challenge of a peptide-based vaccine is the identification of 1 or several “promiscuous” epitopes that could bind to many HLA alleles and thus cover close to 100% of a genetically diverse human population (19).
Until very recently, the search for immunodominant peptides relied on the direct testing of substantial numbers of overlapping peptides or peptide libraries. The identification of major histocompatibility complex (MHC)-binding motifs allowed the prediction of potential T cell epitopes (20,21), and such motifs were found to cluster in certain protein regions (22). The TEPITOPE algorithm, that predicts binding to 25 distinct HLA-DR molecules based on quantitative matrices established from HLA-DR binding assays (18,23), leads to the selection of high affinity-binding peptides, those with the highest chance of eliciting effective T-cell responses against immunogens (24). Additionally, TEPITOPE also allows detection of sequences predicted to bind to several HLA-DR molecules simultaneously, opening the possibility of selection of promiscuous T-cell epitopes. This approach has been used to successfully identify allele-specific and promiscuous T-cell epitopes (25-35). However, no study has addressed PBMC recognition of TEPITOPE-predicted promiscuous peptides in a genetically heterogeneous group previously exposed to an infectious agent.
PCM is a prevalent human systemic mycosis in Latin America where 90 million people live in endemic areas and almost 10 million may be infected with the fungus Paracoccidioides brasiliensis (36-38). Cellular immunity seems to be the major defense mechanism in both experimental and human PCM (39,40). Patients with active disease have high levels of anti-P. brasiliensis antibodies (41-45) as well as transient antigen-specific cell-mediated immunosupression (43), whereas conversion to positive delayed-type hypersensitivity (DTH) skin test and reduction in the antibody levels are useful parameters for successful chemotherapy and cure.
The immunodominant 43 kDa glycoprotein gp43 is the major diagnostic antigen from P. brasiliensis (46-48), being recognized by sera from patients with active PCM (49,50). Treated and healed PCM patients display positive DTH to P. brasiliensis gp43 (51). Gp43-immunized Balb/c (H-2d), A/Sn (H-2a), and C57bl/6 (H-2b) mice induce lymph node cells to proliferate and secrete interferon-γ and interleukin (IL)-2 but not IL-4, IL-5 or IL-10, when stimulated by the immunogen, indicating a T helper cell (Th)1 response (2). Epitope mapping of the entire gp43 identified peptide P10 (gp43[181-195]) as that carrying the immunodominant epitope in lymphocyte proliferation assays. Immunization of Balb/c mice with either purified gp43 or P10 was protective against subsequent intratracheal challenge by virulent P. brasiliensis (2). Furthermore, peptide P10 (gp43 [181-195]) does not elicit an antibody response, which is not protective, and may down-regulate the cellular immune response (2).
To identify the immunodominant epitopes in the gp43 of P. brasiliensis for immune response in humans, we used the TEPITOPE algorithm to select several different sequences that were predicted to bind to multiple HLA-DR molecules (promiscuous epitopes) and tested their binding to multiple HLA-DR molecules in peptide-binding assays and recognition in primary PBMC responses. Additionally, we assessed the specificity and sensitivity of the prediction by TEPITOPE, selecting peptides that were predicted not to bind to any of the HLA-DR molecules. Peptides were synthesized and tested in proliferation assays with PBMC from nonanergic treated and healed, genetically heterogeneous PCM patients. The antigen-induced primary in vitro PBMC proliferation assay detects the “central” memory T cells expanded by previous exposure to the pathogen (52). In the proliferation assay, one measures the DNA synthesis in lymphocytes after incubation with the antigen, which is presented by monocytic cells expressing MHC class II molecules. Presentation occurs after endocytosis and processing of proteins like gp43, or by direct binding of a synthetic peptide antigen to HLA-DR, -DP, or -DQ molecules on the cell surface of human antigen presenting cells. Peptide-induced assays with PBMC from sensitized individuals can lead to the identification of naturally presented epitopes. With the aid of the TEPITOPE algorithm, we successfully identified multiple promiscuous, naturally presented immunodominant CD4+ T cell epitopes in the gp43 glycoprotein from P. brasiliensis.
MATERIALS AND METHODS
Patients and Healthy Individuals
Heparin-treated venous blood for PBMC isolation and EDTA-treated venous blood for DNA extraction were obtained from clinically healed PCM patients represented by low or negative anti–P. brasiliensis antibody titers with positive cutaneous test for paracoccidiodin. Patients originally diagnosed after direct fungal identification on lesion samples underwent chemotherapy with either ketoconazole or itraconazole or a combination of trimethoprim with sulfadiazine or sulfamethoxazole prior to the current study. All patients were followed at the Department of Infectious and Parasitic Diseases, University Hospital, University of São Paulo Medical School. Healthy volunteers, paired by age and gender to the patients, were also tested. All subjects gave their written informed consent to participate in this study, which was approved by the Internal Review Board of the University of São Paulo Medical School.
Peptide Selection
The amino acid sequence of P. brasiliensis gp43 glycoprotein (Genbank accession number AY005437) was scanned by a TEPITOPE algorithm that can predict, after scanning all 9-mer windows starting with a hydrophobic residue on a protein sequence, those sequences that have a potential ability to bind to 1 or more of 25 different HLA-DR molecules by use of 25 virtual matrices that cover most of the HLA class II peptide binding specificities in the Caucasian population (18). Briefly, the algorithm incorporates, for each of the 25 HLA-DR molecules, a matrix of values for each amino acid residue at each position, p2 to p9. Position- and residue-specific matrix values were assembled from empirical HLA-DR peptide binding assays (53). The algorithm provides a score—the algebraic sum of the matrix values for each peptide position—to each of the 9-mer windows along the scanned sequence. Nonamers attaining a score above the threshold for a given HLA-DR molecule (for example, a 3% threshold selects sequences with HLA-binding scores equal to or higher than those of the 3% sequences with highest scores in the TEPITOPE database) are selected by the software. The algorithm also allows the selection of sequences predicted to bind simultaneously and thus, promiscuously, to several HLA-DR molecules (23). Peptides predicted to bind to the largest number of HLA molecules at the highest threshold (1%) in addition to displaying binding to an increased number of distinct HLA-DR molecules (3% threshold) were selected (Table 1). We synthesized peptides that correspond to an inner nonamer core selected by TEPITOPE as the HLA-binding motif with flanking amino acids added at both N- and C-terminal ends, to increase the efficiency of in vitro peptide presentation to CD4+ T cells (23).
Table 1.
Synthetic peptides from gp43 of P. brasiliensis used in this study
| Numbera | Peptide | Sequence |
|---|---|---|
| 1 | gp43 (45-59) | IGGWLLLEPWISPSV-NH2 |
| 2 | gp43 (94-108) | TEDDFKNIAAAGLNHV-NH2 |
| 3 | gp43 (106-120) | LNHVRIPIGYWAVNP-NH2 |
| 4 | gp43 (283-298) | IDQHVKLACSLPHGRL-NH2 |
| 5 | gp43 (180-194) | KQTLIAIHTLAIRYA-NH2 |
| 6 | gp43 (183-197) | LIAIHTLAIRYANRT-NH2 |
| 7 | gp43 (179-199) | IKQTLIAIHTLAIRYANRTDV-NH2 |
| 8 | gp43 (181-195)b | QTLIAIHTLAIRYAN-NH2 |
| 9 | gp43 (118-132) | VNPIEGEPYVQGQLD-NH2 |
| 10 | gp43 (161-175) | GHRGAINWQHGDTIK-NH2 |
| 11 | gp43 (244-258) | DASLPPRTWNGFLAP-NH2 |
| 12 | gp43 (259-271) | KTYKNVYIDTYHN-NH2 |
| 13 | gp43 (347-360) | SKGSSSELSAQQKK-NH2 |
1-5: selected peptides that were predicted to bind to a large number of HLA-DR molecules. 6-8: peptides neighboring peptide gp43(181-195), which is immunodominant and conferred protection in mice infected intratracheally with a virulent strain of P. brasiliensis. 9-13: peptides predicted not to bind to any of the 25 HLA-DR molecules.
Corresponds to the P10 peptide of Taborda and others (2).
Synthetic Peptides
All peptides (see Table 1) derived from gp43 antigen were synthesized by solid phase technology using 9-fluorenylmethoxycarbonyl (Fmoc) strategy (54) on an automated benchtop simultaneous multiple solid-phase peptide synthesizer PSSM8 (Shimadzu, Tokyo, Japan) with Fmoc protected amino acid residues and TGR resin (Novabiochem, San Diego, CA, USA). Therefore, all peptides were obtained with the C-terminal carboxyl group in amide form. All peptides were deprotected and cleaved from the resins by treatment with K reagent composed by 80% trifluoroacetic acid, 2.5% triisopropylsilane, 2.5% ethanedithiol, 5.0% anisol, 5.0% water, and 5.0% phenol (55). The resulting peptides were analyzed by reverse-phase high performance liquid chromatography (Shimadzu, Tokyo, Japan) on a C18 column eluted at 1 mL/min using 5% to 95% gradient of acetonitrile 90% in 0.1% TFA over 30 min. Peptide quality was assessed by Matrix Assisted Laser Desorption and Ionization—Time of Flight instrument (Micromass, Manchester, UK) using a-cyano-4 hydroxy cinnamic acid as the matrix.
Proliferation Assay
PBMC were isolated from peripheral blood by density gradient centrifugation (d = 1.077). Cells were incubated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% normal human serum, 2 mM l-glutamine, 1 mM sodium pyruvate, 50 μg/mL gentamicin, and 10 mM HEPES buffer, in triplicate 96-well U-bottom culture plates (105 cells/well; final volume 0.2 mL) with gp43 (1 and 10 μg/mL), and synthetic peptides (0.1; 1.0 and 10.0 μM). Plates containing phytohaemagglutinin (PHA; 2.5 mg/mL) and complete culture medium were used as positive and negative controls, respectively. In some assays, we tested the proliferative responses of whole or CD4+ T-cell–depleted PBMC from patients to gp43 and derived peptides. Plates were incubated in 5% CO2 at 37 °C for 5 d, and cultures were pulsed with 1 μCi/well (H3+)-thymidine (Amersham, Buckinghamshire, UK) for the next 18 h (H3+)-thymidine incorporation was determined with a Betaplate β counter (Wallac, Turku, Finland). Data are represented as mean counts per min (cpm) of triplicate cultures and the stimulation index (SI) defined as mean cpm response value with antigen/mean cpm of culture medium control. SI values ≥ 2.0 were considered positive.
CD4+ T Lymphocyte Depletion and Flow Cytometry Analysis
CD4+ T cells were depleted from PBMC with Dynabeads M-450 (Dynal, Oslo, Norway) with shaking at 4 °C for 30 min and washed with 0.1% phosphate buffered saline–bovine serum albumin. The efficiency of T-CD4+ depletion was evaluated by flow cytometry analysis with a FACScan (Becton Dickinson, San Jose, CA, USA). Antibody conjugates used included anti–CD4-PE, anti–CD8-FITC, and anti–CD3-FITC (Dakopatts, Glostrup, Denmark), with anti-β2 microglobulin-FITC (56) and anti–HBS-FITC (57) used as positive and negative controls, respectively, as previously described (58). Purity of cell populations is expressed as percentage of stained cells.
HLA Class II Typing
DNA was extracted alternatively by DTAB/CTAB (59) or salting out (60) methods and HLA-DR typing was performed by low resolution PCR-SSP (61).
Immunoblotting
Heparin-treated plasma from PCM patients or healthy controls was tested for anti-gp43 IgG responses. Briefly, purified P. brasiliensis gp43 (2) was resolved in 10% acrylamide minigels (15 mA, constant amperage) using a Hoefer mini-VE Electrophoresis Unit (Pharmacia Biotech, Uppsala, Sweden), and electroblotted onto nitrocellulose membranes. Blots were blocked for 2 h at room temperature with 5% nonfat dry milk in TBS-Tween (Tris-buffered saline; 10 mM Tris, pH 7.5; 100 mM NaCl; 0.1% Tween-20) prior to incubation for 2 h at room temperature with plasma from PCM patients and healthy individuals diluted at 1:40. After serum incubation, membranes were extensively washed with several changes of TBS-Tween-5% milk and then probed for 2 h at room temperature with anti-human IgG conjugate with alkaline phosphatase at a dilution of 1:1000. Membranes were washed with several changes of T-TBS and processed for detection by o-phenylenediamine substrate. In all blots, bands corresponding to the protein of interest were identified by reference to molecular weight protein standards (markers) run in parallel and were scanned using a conventional scanner.
ELISA Analysis
Anti-gp43 antibodies were measured using ELISA with gp43 in the solid phase. Briefly, Corning polypropylene 96-well microtiter ELISA plates (Corning, New York, USA) were sensitized with gp43 (12.5 ng/well), overnight at 4 °C. After blocking with 2% bovine serum albumin, serum samples (diluted 1:100 in phosphate-buffered saline) from 16 PCM patients and 6 healthy individuals were incubated for 2 h at 37 °C in duplicate wells. The reaction was developed using an anti-human IgG-peroxidase conjugated with o-phenylenediamine as chromogenic substrate. The absorbance was subsequently measured at 490 nm.
Class II Peptide-Binding Assays
HLA class II molecules were purified from Epstein-Barr virus transformed homozygous B lymphoblastoid cell lines or transfected fibroblasts by affinity chromatography, as previously described (14). Peptide binding assays were performed by incubating purified human class II molecules (5 to 500 nM) with various concentrations of unlabeled peptide inhibitors and 1 to 10 nM 125I-radiolabeled probe peptides for 48 h in phosphate buffered saline containing 0.05% to 0.15% Nonidet P-40 in the presence of a protease inhibitor cocktail (4,14). Class II peptide complexes were separated from free peptide by gel filtration on TSK200 columns (part number 16215; Tosoh Biosciences LLC, Montgomeryville, PA, USA) and the fraction of bound peptide calculated. Alternatively, the percent of MHC bound radioactivity was determined by capturing MHC/peptide complexes on LB3.1 antibody coated Lumitrac 600 plates (Greiner Bio-one, Fricken-hausen, Germany), and determining bound cpm using the Top-Count (Packard Instrument Co., Meriden, CT, USA) microscintillation counter.
The radiolabeled probe peptides utilized were HA Y307-319 (sequence YPKYVKQNTLKLAT; DRB1*0101), an analog of TT Y828-843 (sequence YATFFIKANSKFIGITE; DRB5*0101, DRB1*1101, DRB1*0701), MBP Y85-100 (sequence PVVHF-FKNIVTPRTPPY; DRB1*1501, DRB1*0401, DRB1*0405), and an analog of TT 830-843 (sequence QYIKANAKFIGITE; DRB1*1302).
Statistical Analysis
Comparison of SI and absorbance values between clinical groups was performed with nonparametric Mann-Whitney’s U test. Correlations between TEPITOPE prediction, binding assays, and proliferation assays were performed with the nonparametric Spearman correlation test with 95% confidence intervals.
RESULTS
Anti-gp43 IgG Antibody Responses in Patients and Controls
Nineteen patients with PCM (21 to 75 y old) and 6 healthy controls (30 to 59 y old) were studied. All tested PCM patients, but none of the control individuals, displayed IgG antibodies against P. brasiliensis gp43 on immunoblotting assay (Figure 1). The median absorbance of anti gp43 IgG detected by ELISA was 0.850 in patients (Abs490 = 0.224 to 1.711) and 0.059 in healthy individuals (Abs490 = 0.027 to 0.176) (P < 0.0002).
Figure 1.
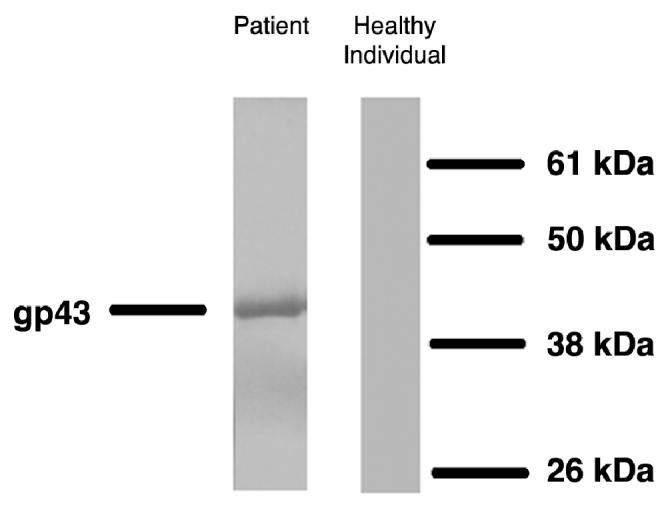
Western blotting of PCM patients’ sera and a healthy individual against gp43 of P. brasiliensis. Serum dilution 1:40; gp43 concentration used: 2.5 μg/mL.
Peptide Selection, Binding Analysis, and T Lymphocyte Response to P. brasiliensis gp43 and Synthetic Peptides
The entire sequence of P. brasiliensis gp43 (416 residues) was scanned by the TEPITOPE algorithm at 3% threshold, which led to the identification of regions predicted to bind with high affinity to several different HLA-DR molecules (TEPITOPE promiscuous region scanning profile for gp43 in Figure 2). Peptides predicted to bind to the largest number of HLA molecules at the highest threshold (1%) in addition to displaying binding to an increased number of distinct HLA-DR molecules (3% threshold) were selected (see Table 1): gp43(45-59), gp43(94-108), gp43(106-120), gp43(283-298), and gp43(180-194) that bound to 18, 12, 5, 15, and 21 HLA-DR molecules, respectively. The gp43(180-194) peptide was predicted to be the most promiscuous ligand, binding to 84% (21/25) of the HLA-DR molecules at 3% and 36% (12/25) of the HLA-DR molecules at 1% threshold (data not shown).
Figure 2.
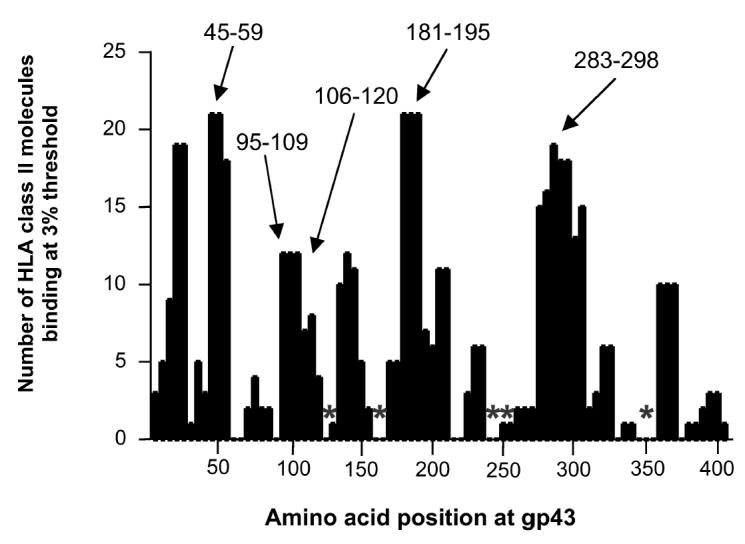
Scanning of gp43 of P. brasiliensis by TEPITOPE algorithm at 3% threshold. Five peptides marked with arrows, predicted to bind to at least 10 of the 25 HLA-DR molecules included in the algorithm, were selected as promiscuous epitopes. Peptides marked with asterisks were predicted not to bind significantly to any HLA-DR molecules and were used as peptide controls.
Binding assays of these peptides with the 9 most prevalent HLA-DR molecules in the general population, showed that gp43(180-194) bound to 100% (9/9) of the HLA-DR molecules followed by gp43(283-298), gp43(94-108), gp43(45-59), and gp43(106-120) that bound to 89% (8/9), 56% (5/9), 33% (3/9), and 22% (2/9) of the tested molecules, respectively (Table 2).
Table 2.
Peptide binding analysis of the 5 peptides selected by TEPITOPE with 9 most prevalent HLA-DR molecules in the populationa
| Binding capacity/IC50% (nM)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptides | DR1 DRB1 *0101 | DR3 DRB1 *0301 | DR4w4 DRB1 *0401 | DR4w15 DRB1 *0405 | DR7 DRB1 *0701 | DR5w11 DRB1 *1101 | DR6w19 DRB1 *1302 | DR2w2 B1 DRB1 *1501 | DR2w2 B2 DRB5 *0101 | Molecules bound | Freqb |
| gp43(45-59) | 0.87 | – | 1337 | 122 | 1795 | 1349 | 40767 | 3.8 | 1509 | 3 | 33% |
| gp43(95-108) | 0.49 | – | 6449 | 241 | 51 | 17790 | 80 | 7662 | 11 | 5 | 56% |
| gp43(106-121) | 25 | – | 5775 | 5248 | 2347 | 1852 | 33333 | 31 | 5466 | 2 | 22% |
| gp43(180-194) | 0.27 | 93 | 37 | 33 | 8.8 | 4.8 | 2.7 | 6.3 | 2.6 | 9 | 100% |
| gp43(283-298) | 470 | 3427 | 135 | 59 | 92 | 106 | 1275 | 506 | 23 | 7 | 89% |
The results are expressed in IC50% (nM). Peptide binding assays were performed by incubating purified human class II molecules (5 to 500 nM) with various concentrations of unlabeled test peptides and 125I-radiolabeled probe peptides. Bold numbers = significant affinity threshold below 1000; dash (–) means IC50% > 50000.
Freq = frequency.
From the PBMC samples of the 19 patients tested, 14 responded to 1 to 5 peptides, whereas 5 failed to respond to any peptide (Table 3); PBMC from healthy controls who did not respond to the gp43 failed to respond to any of the peptides (Table 4). The distribution of HLA-DR molecules predicted by TEPITOPE to bind to the gp43 peptides was similar between the peptide responder and non-responder PCM patient groups (Table 5). We observed that depletion of CD4+ T cells from PBMC from patients p24 and p25 completely abrogated the proliferative response against native gp43 protein and all gp43-derived peptides tested (data not shown). Interestingly enough, PBMC samples from the PCM patients that failed to proliferate to gp43 peptides also showed significantly lower proliferative responses against purified gp43 (median SI = 9.8) than gp43 peptide-reactive PCM patients (median SI = 78.3; P = 0.0026). However, median PHA responses did not differ significantly between groups.
Table 3.
Proliferative response of PBMC from PCM patients to 5 gp43 peptides selected by TEPITOPE programa
| gp43 (45-59)c |
gp43 (94-108)c |
gp43 (106-120)c |
gp43 (283-298)c |
gp43 (180-194)c |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patientsb | 0.1 | 1.0 | 10.0 | 0.1 | 1.0 | 10.0 | 0.1 | 1.0 | 10.0 | 0.1 | 1.0 | 10.0 | 0.1 | 1.0 | 10.0 | gp43 (1 μg) | gp43 (10 μg) | PHA |
| p1 | 3.2 | 17.5 | 14.9 | – | 2.6 | 2.7 | 4.5 | 3.9 | 4.5 | 3.0 | 2.5 | 8.4 | 5.0 | 12.0 | 8.1 | 210.6 | 189.4 | 2368.6 |
| p2 | – | – | – | 2.8 | 5.2 | 3.9 | 2.0 | 2.1 | – | 2.8 | 8.3 | 2.9 | 3.2 | 5.3 | 4.3 | 26.9 | 48.9 | 16.9 |
| p3 | 5.7 | 3.8 | – | 3.4 | 4.6 | – | 2.9 | 2.3 | – | 3.3 | 2.9 | – | 3.1 | 9.2 | – | 5.0 | 13.3 | 243.0 |
| p4 | 2.0 | – | – | – | 2.1 | – | 2.0 | – | – | – | 2.1 | 3.0 | 2.6 | 3.5 | 2.2 | 40.8 | 61.5 | 1181.7 |
| p5 | – | – | 6.8 | – | 2.4 | 2.6 | – | – | – | – | – | – | – | 2.3 | 3.5 | 25.6 | 81.5 | 121.6 |
| p6 | 4.2 | 4.5 | – | – | – | – | – | – | – | – | – | – | 10.6 | 4.5 | – | 88.3 | 123.1 | 710.7 |
| p7 | – | – | – | 3.9 | – | – | 12.0 | 2.6 | – | – | – | – | – | – | – | 19.4 | 31.7 | 761.3 |
| p8 | – | – | – | – | – | 2.0 | – | – | – | – | – | – | – | 2.3 | 6.8 | 30.6 | 185.8 | 977.9 |
| p9 | – | – | – | – | – | – | – | – | – | – | 2.2 | 5.4 | – | – | – | 23.4 | 51.7 | 293.1 |
| p10 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 2.6 | 11.6 | 102.7 | 261.6 |
| p11 | – | – | – | – | 2.2 | – | – | – | 2.3 | – | – | – | – | 3.4 | – | 29.8 | 71.3 | 340.1 |
| p12 | 2.0 | – | – | – | – | – | – | – | – | – | – | – | – | 2.8 | – | 29.8 | 99.5 | 803.8 |
| p13 | – | – | – | – | – | – | – | – | – | – | – | 2.1 | – | – | – | 3.8 | 22.0 | 236.7 |
| p14 | – | – | 3.4 | – | – | – | – | – | – | – | – | – | – | – | – | 8.8 | 13.9 | 2205.8 |
| p15 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 19.9 | 6.9 | 261.4 |
| p16 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 3.2 | 7.2 | 53.4 |
| p17 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 15.0 | 15.5 | 162.4 |
| p18 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 2.3 | 7.0 | 106.5 |
| p19 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 18.1 | 12.6 | 740.0 |
Only positive stimulation index values (SI ≥ 2) are shown. Dash (–) corresponds to SI < 2.
p = patients.
PBMC (105 cells/well) were stimulated with peptides at 0.1, 1.0, and 10.0 μM as described.
Table 4.
Proliferation response of PBMC from healthy individuals to 5 gp43 peptides selected by TEPITOPE programa
| gp43 (45-59)c |
gp43 (94-108 c |
gp43 (106-120)c |
gp43 (283-298)c |
gp43 (180-194)c |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Controlsb | 0.1 | 1.0 | 10.0 | 0.1 | 1.0 | 10.0 | 0.1 | 1.0 | 10.0 | 0.1 | 1.0 | 10.0 | 0.1 | 1.0 | 10.0 | gp43 1 μg | PHA |
| c1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 234.1 |
| c2 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 439.2 |
| c3 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 232.1 |
| c4 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 216.4 |
| c5 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 353.2 |
| c6 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 545.3 |
Only positive stimulation index values (SI ≥ 2) are shown. Dash (–) corresponds to SI < 2.
c = healthy control individual.
PBMC (105 cells/well) were stimulated with peptides at 0.1, 1.0, and 10.0 μM as described.
Table 5.
HLA-DR molecules carried by responders and nonresponders to each gp43 peptide selected by TEPITOPEa
| HLA-DR
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 7 | 8 | 9 | 10 | 11 | 13 | 14 | 15 | 16 | 17 | Blank | Responseb | ||
| gp43(45-59) | Responder | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 2 | 2 | 10/11 | |||
| Nonresponder | 1 | 4 | 1 | 2 | 2 | |||||||||
| gp43(94-108) | Responder | 2 | 1 | 2 | 1 | 1 | 4 | 1 | 1 | 2 | 1 | 10/11 | ||
| Nonresponder | 3 | 1 | 3 | 1 | 1 | 2 | 1 | |||||||
| gp43(106-120) | Responder | 2 | 2 | 1 | 1 | 3 | 2 | 1 | 7/11 | |||||
| Nonresponder | 4 | 1 | 3 | 1 | 1 | 2 | 2 | 1 | ||||||
| gp43(180-194) | Responder | 2 | 3 | 2 | 1 | 4 | 1 | 2 | 3 | 10/11 | ||||
| Nonresponder | 3 | 1 | 1 | 2 | ||||||||||
| Gp43(283-298) | Responder | 2 | 1 | 1 | 3 | 1 | 3 | 1 | 7/11 | |||||
| Nonresponder | 4 | 1 | 2 | 4 | 1 | 2 | 1 | 1 | ||||||
Results are among the 14 PCM patients that responded to at least 1 peptide.
Response = number of HLA-DR molecules associated with response.
Considering positive responses to any of the 3 peptide concentrations tested, gp43(180-194) was the most frequently recognized peptide (53%), followed by gp43(94-108), gp43(45-59), gp43(283-298), and gp43(106-120) at frequencies of 42%, 37%, 32%, and 32%, respectively. We observed that the frequency of responders to each peptide was commensurate with the promiscuity of HLA-DR binding predicted by TEPITOPE (Figure 3). Moreover, we observed that the frequency of peptide responses accurately predicted by TEPITOPE, taking into account the HLA-DR molecules carried by each patient, varied from 40% to 80% of all observed responses. On the other hand, the proportion of observed responses not predicted by TEPITOPE varied from 20% to 60% (see Table 3 and data not shown).
Figure 3.
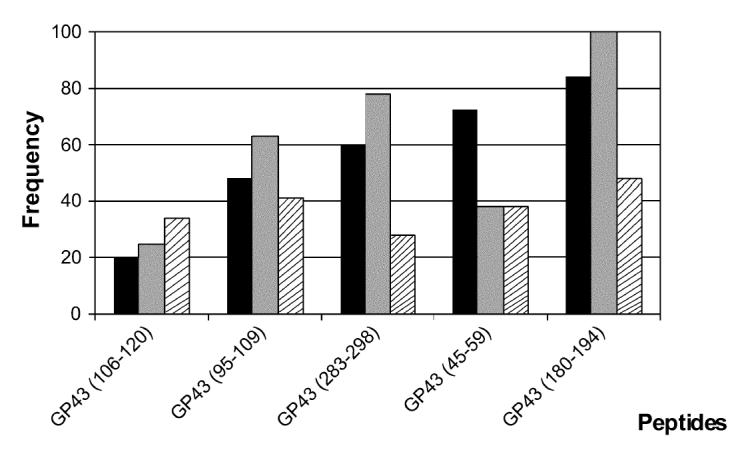
Predicted promiscuity of peptides for binding with high affinity to different HLA-DR molecules at 3% threshold (▪); frequency of peptides binding to 9 different HLA-DR molecules (□); frequency of responders to peptides tested in proliferation assay with PBMC of 19 treated and healed nonanergic PCM patients (▧).
Furthermore, we observed that for most of the TEPITOPE-selected peptides, at least 50% of the responder patients recognized the peptide at the lowest concentration (0.1 mM): peptides gp43(106-120) by 5 of 6 responding patients, gp43(45-59) by 5 of 7, gp43(180-194) by 5 of 10, gp43(283-298) by 3 of 6, and gp43(94-108) by 3 of 8, consistent with high avidity T-cell recognition. Peptide gp43(180-194) was the peptide with the highest average SI at 0.1 mM concentration (4.9) followed by gp43(106-120), gp43(45-59), gp43(94-108), and gp43(283-298) with 4.7, 3.4, 3.3, and 3.0 average SI, respectively. Analysis of the HLA-DR profile from patients that responded to any peptide indicated that a wide diversity of HLA-DR molecules was represented among the responders for each peptide and few HLA-DR molecules were only expressed among non-responders (see Table 5).
In addition, we selected 5 gp43 peptides that were predicted not to bind at 3% threshold to any HLA-DR molecule: gp43(118-132), gp43(161-175), gp43(244-258), gp43(259-271), and gp43(347-360) (marked with asterisks in Figure 2 and shown as peptides 9 to 13 in Table 1). The binding assays of these peptides showed that peptides gp43(244-258) and gp43(347-360) failed to bind to any of the 9 HLA-DR molecules tested, peptide gp43(259-271) bound to 11% (1/9) and peptides gp43(118-132) and gp43(161-175) showed significant binding to 33% (3/9) of the HLA-DR molecules tested (Table 6). Four of 5 peptides were recognized by 10% to 20% of the 10 patients tested in PBMC proliferation assay, however, peptide gp43(161-175) was recognized by 5 of 10 patients tested (Table 7), a frequency of recognition similar to TEPITOPE-selected immunodominant peptide gp43(180-194) (see Table 3).
Table 6.
Peptide binding analysis with 9 most prevalent HLA-DR molecules with 5 peptides predicted by TEPITOPE not to bind promiscuouslya
| Binding capacity/IC50% (nM)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptides | DR1 DRB1 *0101 | DR3 DRB1 *0301 | DR4w4 DRB1 *0401 | DR4w15 DRB1 *0405 | DR7 DRB1 *0701 | DR5w11 DRB1 *1101 | DR6w19 DRB1 *1302 | DR2w2 B1 DRB1 *1501 | DR2w2 B2 DRB5 *0101 | Bound molecules | Freqb |
| gp43(118-132) | 3952 | 4420 | 1885 | 4728 | 767 | 724 | 171 | 1432 | 1228 | 3 | 33% |
| gp43(161-175) | 68 | – | 8540 | 7163 | 9787 | 12477 | 165 | 405 | 1690 | 3 | 33% |
| gp43(244-258) | 4401 | – | – | – | 21540 | – | 16254 | 5170 | 22074 | 0 | 0% |
| gp43(259-271) | 20724 | 5044 | 16989 | 1854 | 7530 | 16257 | 5621 | 429 | 19765 | 1 | 11% |
| gp43(347-360) | – | – | – | 21038 | – | 12983 | 22876 | – | 7758 | 0 | 33% |
The results are expressed in IC50% (nM). Peptide binding assays performed as described in Table 2. Bold numbers = significant affinity threshold below 1000. Dash (–) means IC50% > 30000.
Freq = frequency.
Table 7.
Proliferation response of PBMC from 10 PCM patients to 5 gp43 peptides selected by TEPITOPE program not predicted to bind to any of the HLA-DR moleculesa
| gp43 (118-132)c |
gp43 (161-175)c |
gp43 (244-258)c |
gp43 (259-271)c |
gp43 (347-360)c |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patientsb | 0.1 | 1.0 | 10.0 | 0.1 | 1.0 | 10.0 | 0.1 | 1.0 | 10.0 | 0.1 | 1.0 | 10.0 | 0.1 | 1.0 | 10.0 | gp43 10 μg | PHA |
| p2 | 5.4 | 4.5 | 2.8 | 2.0 | – | 2.0 | – | 2.0 | – | – | – | – | – | – | – | 43.5 | 111.4 |
| p3 | – | – | – | 2.4 | – | – | – | – | – | – | – | – | – | – | – | 57.6 | 288.6 |
| p4 | – | – | – | 10.5 | 10.2 | 7.1 | – | – | – | – | – | – | 3.0 | – | – | 51.3 | 294.8 |
| p10 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 14.3 | 216.6 |
| p11 | – | 2.4 | 2.0 | 6.9 | 6.8 | 6.9 | – | – | – | – | – | – | – | – | – | 26.7 | 130.9 |
| p12 | – | – | – | – | – | – | – | – | – | 3.0 | 5.3 | 4.8 | – | – | – | 28.4 | 421.1 |
| p20 | – | – | – | – | 2.4 | 2.8 | – | – | – | – | – | – | – | – | – | 95.5 | 521.7 |
| p21 | – | – | – | – | – | – | – | – | – | – | – | 2.0 | – | – | – | 346.1 | 409.3 |
| p22 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 84.9 | 452.7 |
| p23 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 5.2 | 114.1 |
Only positive stimulation index values (SI = 2) are shown. Dash (–) corresponds to SI < 2.
p = patient.
PBMC (105 cells/well) were stimulated by peptides at 0.1, 1.0, and 10.0 μM as described.
T-cell Recognition of Epitopes Neighboring the Immunodominant gp43(180-194)
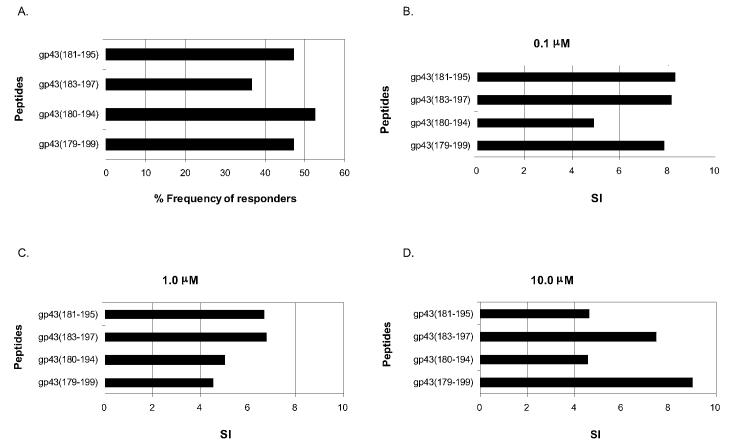
Peptide gp43(180-194), the most frequently recognized sequence (see Figure 1, Table 3), contains 14 of 15 residues of the P10 peptide shown to be immunodominant and protective in mice (2). TEPITOPE analysis at 3% threshold of the region 180-200, which comprises the immunodominant epitopes, showed the presence of 2 additional nonamer core sequences 186-194 (IHTLAIRYA) and 191-199 (IRYANRTDV), in addition to the major HLA-binding nonamer core sequence 183-191 (LIAIHT-LAI), predicted to bind to 5 of 25, 6 of 25, and 20 of 25 HLA-DR molecules, respectively. To finely map the peptide epitopes within the immunodominant region, we synthesized and tested for HLA-DR binding assays as well as proliferation of PBMC from PCM patients and healthy controls 3 additional peptides positioned in different registers in relation to the nonamer core sequences contained within the positions 180-200 sequence of gp43. They were gp43(181-195), equivalent to peptide P10 (2), gp43(183-197), the center piece of which had the 183-191(LIAI-HTLAI) nonamer core sequence, and a 21-mer containing both nonamers and additional flanking residues, gp43(179-199) (see Table 1). The binding assays of these peptides with 9 different HLA-DR molecules showed that all peptides bound to 89% to 100% of the HLA-DR molecules similarly to the peptide gp43(180-194) (see Table 8). Also, these peptides were tested in proliferation assays of PBMC from 19 patients and 6 healthy individuals. Taking into account responses to any peptide concentration, gp43(181-195) and the 21-mer peptide gp43(179-199) were recognized by 47% and peptide gp43(183-195) by 37% of the patients tested (Figure 4A), compared with 53% of gp43(180-194), whereas none of the gp43-negative healthy individuals responded to any of the peptides (data not shown). We observed that peptides gp43(181-195), gp43(183-197), and gp43(179-199) were recognized at the lowest concentration (0.1 mM) by 56%, 57%, and 67% of peptide responders, respectively, similar to the 50% of the original peptide gp43(180-194). Average SI values of peptides gp43(181-195), gp43(183-197), and gp43(179-199) at 0.1 mM were 8.3, 8.2, and 7.9, respectively, higher than the 4.9 SI of gp43(180-194) (Figures 4B to 4D).
Table 8.
Peptide binding analysis of peptides neighboring gp43(180-195) with 9 most prevalent HLA-DRa
| Binding capacity/IC50% (nM)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptides | DR1 DRB1 *0101 | DR3 DRB1 *0301 | DR4w4 DRB1 *0401 | DR4w15 DRB1 *0405 | DR7 DRB1 *0701 | DR5w11 DRB1 *1101 | DR6w19 DRB1 *1302 | DR2w2 B1 DRB1 *1501 | DR2w2 B2 DRB5 *0101 | Bound Molecules | Freqb |
| gp43(183-197) | 13 | 1447 | 464 | 190 | 11 | 5.1 | 146 | 59 | 51 | 8 | 89% |
| gp43(179-199) | 1.8 | 116 | 92 | 11 | 18 | 12 | 9.1 | 5.9 | 6.2 | 9 | 100% |
| gp43(181-195) | 0.080 | 736 | 68 | 23 | 2.0 | 11 | 1.3 | 4.9 | 1.0 | 9 | 100% |
| gp43(180-194) | 0.27 | 93 | 37 | 33 | 8.8 | 4.8 | 2.7 | 6.3 | 2.6 | 9 | 100% |
The results are expressed in IC50% (nM). Peptide binding assays were performed as described in Table 2. Bold numbers = significant affinity threshold below 1000.
Freq = frequency.
Figure 4.
A: Frequency of responders to P10 peptide (gp43 [181-195]) and its neighboring peptides gp43(183-197), gp43(180-194), and gp43(179-199). B–D: Average SI of responders to peptides P10 and its neighboring peptides at 0.1 μM (B), 1.0 μM (C), and 10.0 μM (D)
DISCUSSION
In silico analysis of P. brasiliensis gp43 glycoprotein in search of promiscuous peptides binding to different HLA-DR alleles led us to the identification of several immunodominant epitopes recognized by primary CD4+ T cells from treated and healed PCM patients. Furthermore, we observed that the frequency of peptide binding and recognition was commensurate with the promiscuity of HLA binding as determined by TEPITOPE analysis.
The observation that sera from all treated and healed PCM patients, but none of the healthy individuals, had a significant IgG antibody response against gp43 (see Figure 1 and ELISA results, data not shown) showed that a T-helper anti-gp43 response was induced among PCM patients. This is in agreement with the fact that T-cell response against gp43 and its peptides in treated-healed PCM patients is restricted to the CD4+ T-cell subset (data not shown).
The fact that all TEPITOPE-selected gp43 peptides were recognized by a significant proportion of PCM patients tested (see Table 3) indicates that the algorithm was able to successfully identify promiscuous epitopes in gp43. Furthermore, the observation that the frequency of peptide binding to multiple HLA-DR molecules and the frequency of responders to each peptide was commensurate with the promiscuity of HLA-DR binding predicted by TEPITOPE (see Figure 3) indicates that the algorithm can correlate with actual peptide binding results and identify promiscuous epitopes in a semi-quantitative manner. Analyzed as a whole, the 5 TEPITOPE selected peptides showed significant binding in 27 of 45 assays (60%), and 37 of 95 proliferation assays (39%), whereas the 5 peptides predicted not to bind only showed binding in 7 of 45 assays (16%) and 11 of 50 proliferation assays (22%), emphasizing the accuracy of the algorithm for predicting promiscuous HLA-DR binding. The observation that all TEPITOPE-selected peptides were recognized by 32% to 53% of PCM patients, while 4 of 5 peptides not predicted to bind to any HLA-DR molecules were recognized by 20% or less patients, emphasizes the usefulness of the algorithm. This can be observed in patients p3 and p4, whose PBMC recognized 5 of 5 TEPITOPE-selected peptides, but only 1 and 2 of 5 peptides that were predicted not to bind to HLA-DR molecules, respectively. Additionally, there was significant correlation between the promiscuity of TEPITOPE prediction and HLA-DR binding assay (P < 0.001, r2 = 0.78), or proliferation assay (P = 0.01, r2 = 0.48); significant correlation of promiscuity was also observed between HLA-DR binding assay versus proliferation assay (P = 0.001, r2 = 0.60). However, the observation that peptide gp43(161-175)—predicted not to bind to any of the HLA-DR molecules—bound to 33% of the HLA-DR molecules tested and was recognized by 50% of patients indicates that TEPITOPE prediction misses some immunodominant epitopes. By virtue of the built-in strategy of the TEPITOPE algorithm, it is only able to identify HLA-DR binding sequences starting with hydrophobic residues (17,23). This means that it was not built to identify every single potential epitope, but to be able to reliably identify those with this structural feature. One has to bear in mind that epitopes predicted by TEPITOPE not to bind to any of the 25 build in HLA-DR molecules may still be presented by other HLA-DR, -DP, or -DQ molecules.
The observation that a wide diversity of HLA-DR molecules was associated with recognition of each peptide (see Table 5) suggests that peptides predicted to be promiscuous by TEPITOPE were indeed capable of being presented by multiple HLA-DR molecules. In addition, the frequency of allelic variation in the HLA-DR distribution of the patients tested is similar to the spectrum of the Brazilian mixed population; thus, the lack of some particular haplotypes reflects the low frequency of the same haplotypes in the Brazilian population (62). The slight differences of frequency of DR1, DR10, and DR16 observed between the patients tested and the Brazilian population can be attributed to the low number of individuals tested. The observation that TEPITOPE-selected promiscuous peptides could often be recognized by individuals carrying HLA molecules to which they were predicted not to bind (data not shown) may indicate that peptides were even more promiscuous than predicted. It is known that the HLA-DR binding motifs from different HLA molecules are relatively similar (20,22,63), and it is conceivable that epitopes predicted to bind to several HLA-DR molecules included in the TEPITOPE repertoire may also bind to other HLA class II molecules. Recently we observed that a peptide from Plasmodium vivax antigen identified as promiscuous by TEPITOPE was recognized by mice bearing distinct H-2 molecules (data not shown). This is in agreement with the observation that peptides derived from the herpes simplex virus selected with the TEPITOPE algorithm to bind to multiple HLA-DR molecules were capable to eliciting potent T-cell responses in several MHC-disparate strains of mice (34). On the other hand, the lack of response to a given peptide by some individuals whose HLA profile is predicted by TEPI-TOPE to respond underlines the fact that TEPITOPE predicts HLA binding but not necessarily T-cell recognition. The observation that PCM patients sharing the same HLA-DR molecules (DR7, DR13: p8, p10, and p14) recognized different peptides, suggests that the individual-specific peptide recognition may also depend on the T-cell receptor repertoire.
The fact that PBMC from some patients failed to respond to the promiscuous gp43 peptides tested was probably related to the significantly lower stimulation indexes observed in the same patients when their PBMC were stimulated with gp43 (see Table 3). This may suggest that these patients had comparatively weak T-cell responses against gp43, which could be secondary to incomplete recovery from the nonresponsiveness/anergy observed during active disease (43).
A fine analysis of the immunodominant region of gp43 (positions 180-200) indicated that the 3 additional peptides were significantly antigenic, although not more than the original TEPITOPE-predicted gp43 (180-194). The fact that their overall frequency of recognition was lower, but the relative recognition at 0.1 μM was more frequent and attaining higher SI values than gp43 (180-194) indicated that the presence of additional HLA binding registers may not increase promiscuity but can affect the avidity of peptide recognition. Importantly, this study demonstrated that peptide P10, which is immunodominant and protective in murine models of P. brasiliensis infection (2), is also an immunodominant and promiscuous epitope in PCM patients.
The analysis of responsiveness and avidity among TEPITOPE-selected promiscuous peptides showed that the peptides gp43(180-194) and neighbors, including P10, are the most immunodominant and promiscuous epitopes of gp43, being recognized by more than 50% of all tested patients followed by TEPITOPE-nonselected peptide gp43(161-175). Panigada and others (30) also identified TEPITOPE-derived peptides capable of being recognized by peptide-primed T-cell lines from PBMC of Mycobacterium tuberculosis-infected patients (30). Given the fact that in vitro immunization can elicit antigen-specific T-cell lines even in naive donors, the reported epitope may not have been naturally processed upon infection (64). The fact that we showed primary PBMC recognition of TEPITOPE-derived promiscuous gp43 peptides among PCM patients is evidence that such epitopes were actually presented during infection, thus being prime candidates for vaccinal epitopes, because immunity triggered by such epitopes generated memory responses that can be boosted by contact with the same epitope or infectious agent. Recent data have suggested that single epitope-based vaccines are not powerful enough to induce full protective immunity. The combination of multiple B-cell and T-cell epitopes and conjugates as a pool or as a multiepitope polypeptide was shown to increase the immunogenicity (65,66). In the present study, we observed that 74% of the patients recognized the combination of the 5 TEPITOPE selected promiscuous gp43 peptides, gp43(180-194), gp43(45-59), gp43(283-298), gp43(94-108), and gp43(106-120). Preliminary results in a different set of PCM patients indicated that the response to the 5 pooled peptides reached 75% (data not shown). It is conceivable that inclusion of additional TEPITOPE-derived peptides to the above mentioned peptide combination may cover close to 100% of a genetically distinct population.
Acknowledgments
We would like to thank Dr Maria Lúcia Marin who helped us with determining the HLA of patients, Dr Simone Gonçalves da Fonseca and Renata Cristina Ferreira who helped with FACS acquisition, and Washington Robert da Silva for technical assistance. This study was supported by grant 00-08404-3 and LKI was supported by fellowship grant 99/15319-6 from São Paulo State Science Funding Agency (FAPESP). ECN is the recipient of productivity grant 520533/97-6 from the Brazilian National Research Council (CNPq). AS and JS are supported by National Institutes of Health contracts N01-AI-95362 and HHSN266200400006C.
REFERENCES
- 1.Muller GM, Shapira M, Arnon R. Anti-influenza response achieved by immunization with a synthetic conjugate. Proc Natl Acad Sci USA. 1982;79:569–73. doi: 10.1073/pnas.79.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taborda CP, Juliano MA, Puccia R, Franco M, Travassos LR. Mapping of the T-cell epitope in the major 43-kilodalton glycoprotein of Paracoccidioides brasiliensis which induces a Th-1 response protective against fungal infection in BALB/c mice. Infect Immun. 1998;66:786–93. doi: 10.1128/iai.66.2.786-793.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith DJ, King WF, Barnes LA, Peacock Z, Taubman MA. Immunogenicity and protective immunity induced by synthetic peptides associated with putative immunodominant regions of Streptococcus mutans glucan-binding protein B. Infect. Immun. 2003;71:1179–84. doi: 10.1128/IAI.71.3.1179-1184.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Southwood S, et al. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160:3363–73. [PubMed] [Google Scholar]
- 5.Hunziker IP, et al. Perspectives: toward a peptide-based vaccine against hepatitis C virus. Mol Immunol. 2001;38:475–84. doi: 10.1016/s0161-5890(01)00083-9. [DOI] [PubMed] [Google Scholar]
- 6.Tsuji M, Zavala F. Peptide-based subunit vaccines against pre-erythrocytic stages of malaria parasites. Mol Immunol. 2001;38:433–42. doi: 10.1016/s0161-5890(01)00079-7. [DOI] [PubMed] [Google Scholar]
- 7.Ertl HC, Xiang Z. Novel vaccine approaches. J Immunol. 1996;156:3579–82. [PubMed] [Google Scholar]
- 8.DiMarchi R, Brooke G, Gale C, Cracknell V, Doel T, Mowat N. Protection of cattle against foot-and-mouth disease by a synthetic peptide. Science. 1986;232:639–41. doi: 10.1126/science.3008333. [DOI] [PubMed] [Google Scholar]
- 9.Casal JI, et al. Peptide vaccine against canine parvovirus: identification of two neutralization subsites in the N terminus of VP2 and optimization of the amino acid sequence. J Virol. 1995;69:7274–7. doi: 10.1128/jvi.69.11.7274-7277.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taboga O, et al. A large-scale evaluation of peptide vaccines against foot-and-mouth disease: lack of solid protection in cattle and isolation of escape mutants. J Virol. 1997;71:2606–14. doi: 10.1128/jvi.71.4.2606-2614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nargi F, et al. Protection of swine from foot-and-mouth disease with one dose of an all-D retro peptide. Vaccine. 1999;17:2888–93. doi: 10.1016/s0264-410x(99)00127-9. [DOI] [PubMed] [Google Scholar]
- 12.Van Regenmortel MH, Guichard G, Benkirane N, Briand JP, Muller S, Brown F. The potential of retro-inverso peptides as synthetic vaccines. Dev Biol Stand. 1998;92:139–43. [PubMed] [Google Scholar]
- 13.Dong XN, Wei K, Liu ZQ, Chen YH. Candidate peptide vaccine induced protection against classical swine fever virus. Vaccine. 2002;21:167–73. doi: 10.1016/s0264-410x(02)00466-8. [DOI] [PubMed] [Google Scholar]
- 14.Sidney J, Southwood S, Oseroff C, del Guercio MF, Sette A, Grey HM. The measurement of MHC/peptide interactions by gel infiltration. Curr. Protocols Immunol. 1998:18.3.1–18.3.19. doi: 10.1002/0471142735.im1803s31. [DOI] [PubMed] [Google Scholar]
- 15.Patarroyo ME, et al. A synthetic vaccine protects humans against challenge with asexual blood stages of Plasmodium falciparum malaria. Nature. 1988;332:158–61. doi: 10.1038/332158a0. [DOI] [PubMed] [Google Scholar]
- 16.Smith JW, et al. Adjuvant immunization of HLA-A2-positive melanoma patients with a modified gp100 peptide induces peptide-specific CD8+ T-cell responses. J Clin Oncol. 2003;21:1562–73. doi: 10.1200/JCO.2003.09.020. [DOI] [PubMed] [Google Scholar]
- 17.Hammer J, Sturniolo T, Sinigaglia F. HLA class II peptide binding specificity and autoimmunity. Adv Immunol. 1997;66:67–100. doi: 10.1016/s0065-2776(08)60596-9. [DOI] [PubMed] [Google Scholar]
- 18.Sturniolo T, et al. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat Biotechnol. 1999;17:555–61. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- 19.Cunha-Neto E. MHC-restricted antigen presentation and recognition: constraints on gene, recombinant and peptide vaccines in humans. Braz J Med Biol Res. 1999;32:199–205. doi: 10.1590/s0100-879x1999000200008. [DOI] [PubMed] [Google Scholar]
- 20.Rammensee HG. Chemistry of peptides associated with MHC class I and class II molecules. Curr Opin Immunol. 1995;7:85–96. doi: 10.1016/0952-7915(95)80033-6. [DOI] [PubMed] [Google Scholar]
- 21.Rammensee HG, Friede T, Stevanoviic S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 22.Meister GE, Roberts CG, Berzofsky JA, De Groot AS. Two novel T-cell epitope prediction algorithms based on MHC-binding motifs; comparison of predicted and published epitopes from Mycobacterium tuberculosis and HIV protein sequences. Vaccine. 1995;13:581–91. doi: 10.1016/0264-410x(94)00014-e. [DOI] [PubMed] [Google Scholar]
- 23.Bian H, Reidhaar-Olson JF, Hammer J. The use of bioinformatics for identifying class II-restricted T-cell epitopes. Methods. 2003;29:299–309. doi: 10.1016/s1046-2023(02)00352-3. [DOI] [PubMed] [Google Scholar]
- 24.Schroers R, Huang XF, Hammer J, Zhang J, Chen SY. Identification of HLA DR7-restricted epitopes from human telomerase reverse transcriptase recognized by CD4+ T-helper cells. Cancer Res. 2002;62:2600–5. [PubMed] [Google Scholar]
- 25.de Lalla C, Sturniolo T, Abbruzzese L, Hammer J, Sidoli A, Sinigaglia F, Panina-Bordignon P. Cutting edge: identification of novel T cell epitopes in Lol p5a by computational prediction. J Immunol. 1999;163:1725–9. [PubMed] [Google Scholar]
- 26.Stassar MJ, Raddrizzani L, Hammer J, Zoller M. T-helper cell-response to MHC class II-binding peptides of the renal cell carcinoma-associated antigen RAGE-1. Immunobiology. 2001;203:743–55. doi: 10.1016/S0171-2985(01)80003-6. [DOI] [PubMed] [Google Scholar]
- 27.Cochlovius B, Stassar M, Christ O, Raddrizzani L, Hammer J, Mytilineos I, Zoller M. In vitro and in vivo induction of a Th cell response toward peptides of the melanoma-associated glycoprotein 100 protein selected by the TEPITOPE program. J Immunol. 2000;165:4731–41. doi: 10.4049/jimmunol.165.8.4731. [DOI] [PubMed] [Google Scholar]
- 28.Manici S, et al. Melanoma cells present a MAGE-3 epitope to CD4(+) cytotoxic T cells in association with histocompatibility leukocyte antigen DR11. J Exp Med. 1999;189:871–6. doi: 10.1084/jem.189.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Consogno G, et al. Identification of immunodominant regions among promiscuous HLA-DR-restricted CD4+ T-cell epitopes on the tumor antigen MAGE-3. Blood. 2003;101:1038–44. doi: 10.1182/blood-2002-03-0933. [DOI] [PubMed] [Google Scholar]
- 30.Panigada M, et al. Identification of a promiscuous T-cell epitope in Mycobacterium tuberculosis Mce proteins. Infect Immun. 2002;70:79–85. doi: 10.1128/IAI.70.1.79-85.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruger S, Schroers R, Rooney CM, Gahn B, Chen SY. Identification of a naturally processed HLA-DR-restricted T-helper epitope in Epstein-Barr virus nuclear antigen type 1. J Immunother. 2003;26:212–21. doi: 10.1097/00002371-200305000-00005. [DOI] [PubMed] [Google Scholar]
- 32.Shen L, Schroers R, Hammer J, Huang XF, Chen SY. (2003) Identification of a MHC class-II restricted epitope in carcinoembryonic antigen [online]. Cancer Immunol. Immunother 18. [DOI] [PMC free article] [PubMed]
- 33.Schroers R, et al. Identification of MHC class II-restricted T-cell epitopes in prostate-specific membrane antigen. Clin Cancer Res. 2003;9:3260–71. [PubMed] [Google Scholar]
- 34.BenMohamed L, et al. Identification of novel immunodominant CD4+ Th1-type T-cell peptide epitopes from herpes simplex virus glycoprotein D that confer protective immunity. J Virol. 2003;77:9463–73. doi: 10.1128/JVI.77.17.9463-9473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campi G, et al. CD4(+) T cells from healthy subjects and colon cancer patients recognize a carcinoembryonic antigen-specific immunodominant epitope. Cancer Res. 2003;63:8481–6. [PubMed] [Google Scholar]
- 36.Brummer E, Castaneda E, Restrepo A. Paracoccidioidomycosis: an update. Clin Microbiol Rev. 1993;6:89–117. doi: 10.1128/cmr.6.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shikanai-Yasuda MA. (1996) Paracoccioidomycosis. In: Oxford Textbook of Medicine Weatherall DJ, Ledinghan JGG, Warrell DA (eds.) Oxford Medical Publications, Oxford University Press, Oxford, UK. pp. 814–24.
- 38.Restrepo A, McEwen JG, Castaneda E. The habitat of Paracoccidioides brasiliensis: how far from solving the riddle? Med Mycol. 2001;39:233–41. doi: 10.1080/mmy.39.3.233.241. [DOI] [PubMed] [Google Scholar]
- 39.Mota NG, et al. Correlation between cell-mediated immunity and clinical forms of paracoccidioidomycosis. Trans R Soc Trop Med Hyg. 1985;79:765–772. doi: 10.1016/0035-9203(85)90112-9. [DOI] [PubMed] [Google Scholar]
- 40.Calich VL, Singer-Vermes LM, Russo M, Vaz CA, Burger E. (1994) Immunogenetics in Paracoccidioidomycosis. In: Paracoccidioidomycosis Franco M, Lacaz CS, Restrepo A, and Del Negro GM (eds.) CRC Press, Boca Raton, FL. pp. 151–73.
- 41.Biagioni L, Souza MJ, Chamma LG, Mendes RP, Marques SA, Mota NG, Franco M. Serology of paracoccidioidomycosis. II Correlation between class-specific antibodies and clinical forms of the disease. Trans R Soc Trop Med Hyg. 1984;78:617–21. doi: 10.1016/0035-9203(84)90220-7. [DOI] [PubMed] [Google Scholar]
- 42.Camargo ZP, Unterkircher C, Travassos LR. Identification of antigenic polypeptides of Paracoccidioides brasiliensis by immunoblotting. J Med Vet Mycol. 1989;27:407–12. [PubMed] [Google Scholar]
- 43.Benard G, Mendes-Giannini MJ, Juvenale M, Miranda ET, Duarte AJ. Immunosuppression in paracoccidioidomycosis: T-cell hyporesponsiveness to two Paracoccidioides brasiliensis glycoproteins that elicit strong humoral immune response. J Infect Dis. 1997;175:1263–7. doi: 10.1086/593694. [DOI] [PubMed] [Google Scholar]
- 44.Del Negro GM, et al. Evaluation of tests for antibody response in the follow-up of patients with acute and chronic forms of paracoccidioidomycosis. J Med Microbiol. 2000;49:37–46. doi: 10.1099/0022-1317-49-1-37. [DOI] [PubMed] [Google Scholar]
- 45.Souza AR, et al. Anti-idiotypic antibodies in patients with different clinical forms of paracoccidioidomycosis. Clin Diagn Lab Immunol. 2000;7:175–81. doi: 10.1128/cdli.7.2.175-181.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puccia R, Schenkman S, Gorin PA, Travassos LR. Exocellular components of Paracoccidioides brasiliensis: identification of a specific antigen. Infect Immun. 1986;53:199–206. doi: 10.1128/iai.53.1.199-206.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puccia R, Travassos LR. 43-kilodalton glycoprotein from Paracoccidioides brasiliensis: immunochemical reactions with sera from patients with paracoccidioidomycosis, histoplasmosis, or Jorge Lobo’s disease. J Clin Microbiol. 1991;29:1610–5. doi: 10.1128/jcm.29.8.1610-1615.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Travassos LR, Taborda CP, Iwai LK, Cunha-Neto E, Puccia R. (2004) The gp43 from Paracoccidioides brasiliensis: A major diagnostic antigen and vaccine candidate. In: The Mycota XII. Human Fungal Pathogens Domer JE, Kobayashi GS (eds.) Springer-Verlag, Berlin-Heidelberg, pp. 279–96.
- 49.De Camargo Z, Unterkircher C, Campoy SP, Travassos LR. Production of Paracoccidioides brasiliensis exoantigens for immunodiffusion tests. J Clin Microbiol. 1988;26:2147–51. doi: 10.1128/jcm.26.10.2147-2151.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taborda CP, Camargo ZP. Diagnosis of paracoccidioidomycosis by passive haemagglutination assay of antibody using a purified and specific antigen-gp43. J Med Vet Mycol. 1993;31:155–60. doi: 10.1080/02681219380000171. [DOI] [PubMed] [Google Scholar]
- 51.Saraiva EC, Altemani A, Franco MF, Unterkircher CS, Camargo ZP. Paracoccidioides brasiliensis-gp43 used as paracoccidioidin. J Med Vet Mycol. 1996;34:155–61. doi: 10.1080/02681219680000261. [DOI] [PubMed] [Google Scholar]
- 52.Geginat J, Lanzavecchia A, Sallusto F. Proliferation and differentiation potential of human CD8+ memory T-cell subsets in response to antigen or homeostatic cytokines. Blood. 2003;100(11):4260–6. doi: 10.1182/blood-2002-11-3577. [DOI] [PubMed] [Google Scholar]
- 53.Hammer J, et al. High-affinity binding of short peptides to major histocompatibility complex class II molecules by anchor combinations. Proc Natl Acad Sci U S A. 1994;91:4456–60. doi: 10.1073/pnas.91.10.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Atherton E, Sheppard RC. (1989) Solid Phase Peptide Synthesis: A Practical Approach IRL Press, Oxford, UK. 152 p.
- 55.King DS, Fields CG, Fields GB. A cleavage method which minimizes side reactions following Fmoc solid phase peptide synthesis. Int J Pept Protein Res. 1990;36:255–66. doi: 10.1111/j.1399-3011.1990.tb00976.x. [DOI] [PubMed] [Google Scholar]
- 56.Crevat D, Kalil J, Rosa F, Fellous M. Presence of 2 different epitopes on the human beta 2-microglobulin defined by monoclonal antibodies. Ann Immunol (Paris) 1983;134C:31–41. [PubMed] [Google Scholar]
- 57.Kalil J, Crevat D, Fellous M, Drouet J, Courouce AM, Ropars C. Production of monoclonal antibodies against HBs. Ann Immunol (Paris) 1981;132C:319–26. [PubMed] [Google Scholar]
- 58.Guilherme L, et al. T-cell reactivity against streptococcal antigens in the periphery mirrors reactivity of heart-infiltrating T lymphocytes in rheumatic heart disease patients. Infect Immun. 2001;69:5345–51. doi: 10.1128/IAI.69.9.5345-5351.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gustincich S, Manfioletti G, Del Sal G, Schneider C, Carninci P. A fast method for high-quality genomic DNA extraction from whole human blood. Biotechniques. 1991;11:298–300. 302. [PubMed] [Google Scholar]
- 60.Bignon JD, Fernandez-Vina MA. (1997) Protocols of the 12th International Histocompatibility Workshop for typing of HLA class II alleles by DNA amplification by the polymerase chain reaction (PCR) and hybridation with sequence specific oligonucleotide probes (SSOP). In: Genetic Diversity of HLA Functional and Medical Implication Fauchet R, Charron D (eds.) Paris, France, EDK Medical and Scientific International Publisher, pp. 584–95.
- 61.Olerup O, Zetterquist H. HLA-DR typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours: an alternative to serological DR typing in clinical practice including donor-recipient matching in cadaveric transplantation. Tissue Antigens. 1992;39:225–35. doi: 10.1111/j.1399-0039.1992.tb01940.x. [DOI] [PubMed] [Google Scholar]
- 62.Goldberg AC, Marin ML, Chiarella J, Rosales C, Kalil J. (1997) Brazil normal. In: HLA 1997 Gjertson DW, Terasaki PI (eds.) UCLA Tissue Typing Laboratory, American Society for Histocompatibility and Immunogenetics, Los Angeles, CA. 330 p.
- 63.Hammer J. New methods to predict MHC-binding sequences within protein antigens. Curr Opin Immunol. 1995;7:263–9. doi: 10.1016/0952-7915(95)80013-1. [DOI] [PubMed] [Google Scholar]
- 64.Sinigaglia F, Romagnoli P, Guttinger M, Takacs B, Pink JR. Selection of T-cell epitopes and vaccine engineering. Methods Enzymol. 1991;203:370–86. doi: 10.1016/0076-6879(91)03021-8. [DOI] [PubMed] [Google Scholar]
- 65.Meloen RH, Langeveld JP, Schaaper WM, Slootstra JW. Synthetic peptide vaccines: unexpected fulfillment of discarded hope? Biologicals. 2001;29:233–6. doi: 10.1006/biol.2001.0298. [DOI] [PubMed] [Google Scholar]
- 66.Alexander J, et al. A decaepitope polypeptide primes for multiple CD8+ IFN-gamma and Th lymphocyte responses: evaluation of multiepitope polypeptides as a mode for vaccine delivery. J Immunol. 2002;168:6189–98. doi: 10.4049/jimmunol.168.12.6189. [DOI] [PubMed] [Google Scholar]