Abstract
Interleukin-4 (IL-4)–mediated pro-oxidative and pro-inflammatory vascular environments have been implicated in the pathogenesis of atherosclerosis. The cellular and molecular regulatory mechanisms underlying this process, however, are not fully understood. In the present study, we employed GeneChip microarray analysis to investigate global gene expression patterns in human vascular endothelial cells after treatment with IL-4. Our results showed that mRNA levels of a total of 106 genes were significantly up-regulated and 41 genes significantly down-regulated with more than a 2-fold change. The majority of these genes are critically involved in the regulation of inflammatory responses, apoptosis, signal transduction, transcription factors, and metabolism; functions of the remaining genes are unknown. The changes in gene expression of selected genes related to inflammatory reactions, such as vascular cell adhesion molecule-1 (VCAM-1), E-selectin, monocyte chemoattractant protein-1 (MCP-1), and interleukin-6 (IL-6), were verified by quantitative real-time reverse transcriptase–polymerase chain reaction (RT-PCR) and enzyme-linked immunosorbent assay (ELISA) analyses. IL-4 treatment also significantly increased the adherence of inflammatory cells to endothelial cell monolayers in a dose-dependent manner. These results may help determine the molecular mechanisms of action of IL-4 in human vascular endothelium. In addition, a better understanding of IL-4–induced vascular injury at the level of gene expression could lead to the identification of new therapeutic strategies for atherosclerosis.
INTRODUCTION
Inflammatory responses elicited by a variety of stimuli in the vascular endothelium have been implicated in the development of cardiovascular disease. It is now widely believed that atherosclerosis is an inflammatory disease of the vessel wall, and inflammatory reactions in endothelial cells are primarily regulated through the production of inflammatory mediators and their close interactions (1). In fact, enhanced expressions of adhesion molecules, chemokines, and pro-inflammatory cytokines in vascular endothelial cells facilitate recruiting and adhering of inflammatory cells, such as lymphocytes and monocytes/macrophages, into the vessel wall, and thus stimulate transendothelial migration, which can be considered an early atherogenic process (2–5). These studies strongly support the idea that an inflammatory environment in the vascular endothelium is critical for the initiation and development of atherosclerosis.
Interleukin-4 (IL-4) is a pleiotropic immunomodulatory cytokine secreted by T-helper 2 lymphocytes, eosinophils, and mast cells (6,7). IL-4 is present at high levels in tissues of patients with chronic inflammatory diseases, where it may play a critical role in the disease progression. Indeed, elevated levels of IL-4 were detected in atherosclerotic lesions (8). Additionally, a growing body of evidence indicates that IL-4 may play a role in atherogenesis through induction of inflammatory responses, such as up-regulation of vascular cell adhesion molecule-1 (VCAM-1) (9,10) and monocyte chemoattractant protein-1 (MCP-1) (11,12). IL-4 may also be considered as a pro-oxidative cytokine, which can increase the oxidative potential of target cells (10,13,14).
It has been proposed that the IL-4–mediated overexpression of inflammatory mediators is regulated at the transcriptional level through activation of a variety of redox-responsive transcription factors. For example, we have shown that IL-4–induced oxidative stress up-regulates the expression of VCAM-1 and MCP-1 genes via activation of Sp-1 and signal transducers and activators of transcription, respectively (10,12). Although previous studies have established the potential role of IL-4 in the development of cardiovascular disease, the cellular and molecular regulatory mechanisms underlying this process are not fully understood. In the present study, GeneChip microarray analysis was conducted to investigate the global gene expression changes of IL-4–treated human vascular endothelial cells using the Affymetrix GeneChip® Human Genome U133A Arrays. In addition, we also employed the quantitative real-time reverse transcriptase–polymerase chain reaction (RT-PCR) and enzyme-linked immunosorbent assay (ELISA) to confirm changes in the levels of expression of selective genes of interest. We found that IL-4 significantly regulates the expression of genes known to be involved in inflammation, apoptosis, signal transduction, and transcription factors.
MATERIALS AND METHODS
Cell Cultures
Human umbilical vein endothelial cells (HUVEC) were isolated as described previously (15). HUVEC were cultured in enriched M199 medium supplemented with 20% fetal calf serum, 1% each of penicillin/streptomycin, glutamine, and antibiotic-antimycotic, heparin (300 μg/mL; Gibco BRL, Grand Island, NY, USA), HEPES (6 mg/mL; Sigma Chemical, St. Louis, MO, USA), and endothelial cell growth supplement (40 μg/mL; Collaborative Research, Bedford, MA, USA) in 5% CO2 at 37 ºC. Cells were determined to be endothelial by their cobblestone morphology and uptake of fluorescent-labeled acetylated LDL (1,1′-dioctadecyl-3,3,3′3′-tetramethylindocarbocyanine perchlorate; Molecular Probes Inc., Eugene, OR, USA). HUVEC from passage 2 were used in all experiments. The human monocytic leukemia cells (THP-1) were purchased from American Type Culture Collection (Manassas, VA, USA) and used to study cell adhesion assay. THP-1 cells were cultured in suspension in RPMI 1640 medium supplemented with 10% fetal calf serum, 25 mM glucose, 10 mM HEPES, 1.0 mM sodium pyruvate, 50 μM 2-mercaptoethanol, and 1% each of penicillin/streptomycin in 5% CO2 at 37 ºC.
GeneChip Microarray Analysis
Microarray gene expression analysis was performed using the Affymetrix GeneChip System with Human Genome U133A Arrays (Affymetrix Inc, Santa Clara, CA, USA).
RNA isolation and GeneChip microarray processing.
HUVEC were either untreated or treated with 10 ng/mL of IL-4 for 4 h. Total RNA was isolated and purified using RNeasy Mini Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s protocols. The labeling of RNA samples, human GeneChip (HG-U133A) hybridization, and array scanning were carried out as described earlier (16,17) and according to the Affymetrix GeneChip Expression Analysis Technical Manual. Briefly, an average yield of 40 μg of biotin-labeled cRNA target was obtained from 5 μg of total RNA from each sample, of which 20 μg of cRNA was applied to 1 gene chip. The hybridization was run overnight in a rotating oven (Affymetrix GeneChip Hybridization Oven 640) at 45 °C. The chips were then washed and stained on a fluidics station (Affymetrix GeneChip Fluidics Station 400), and scanned at a resolution of 3 μm in a confocal scanner (Affymetrix GeneArray Scanner).
Microarray data analysis.
The gene expression levels of samples were analyzed using the Affymetrix Microarray Suite software according to the manufacturer’s recommendation. All data presented in Tables 1 and 2 show the mean fold change in gene expression from 3 independent experiments in IL-4–treated HUVEC compared with untreated control cell cultures. All genes presented were significantly changed (P < 0.05) and the mean fold minimum change chosen for presentation was 2.0.
Table 1.
Up-regulation of specific gene expression in human vascular endothelial cells treated with interleukin-4
| Gene symbol | NCBI accession nr | Fold change | P value | Description |
|---|---|---|---|---|
| Adhesion molecules | ||||
| VCAM1 | NM_001078 | 21.7 | <0.001 | Vascular cell adhesion molecule 1 |
| CSPG2 | D32039 | 5.4 | 0.006 | Chondroitin sulfate proteoglycan 2 |
| SELE | NM_000450 | 4.3 | <0.001 | E-selectin |
| AIM1 | U83115 | 4.1 | <0.001 | Absent in melanoma 1 |
| AGC1 | X17406 | 3.2 | 0.002 | Aggrecan 1 |
| PCDH7 | NM_002589 | 2.7 | <0.001 | BH-protocadherin |
| FCN3 | NM_003665 | 2.2 | 0.023 | Ficolin (collage/fibrinogen domain) 3 |
| FAT | NM_005245 | 2.0 | 0.007 | FAT tumor suppressor homolog 1 |
| Chemokines and cytokines | ||||
| CCL2 | S69738 | 8.2 | <0.001 | Monocyte chemoattractant protein-1 |
| IL6 | NM_000600 | 2.0 | 0.003 | Interleukin-6 |
| Apoptosis | ||||
| PAWR | NM_002583 | 2.6 | 0.001 | PRKC, apoptosis, WT1, regulator |
| CASP3 | NM_004346 | 2.5 | 0.008 | Caspase 3 |
| CASP2 | AF314174 | 2.0 | 0.028 | Caspase 2 |
| Signal transduction | ||||
| PMCH | NM_002674 | 331.2 | <0.001 | Promelanin–concentrating hormone |
| PIK3CG | AF327656 | 9.9 | 0.013 | Phosphoinositide-3-kinase, catalytic, γ |
| SOCS1 | AB005043 | 5.8 | <0.001 | Suppressor of cytokine signaling 1 |
| LIFR | NM_002310 | 5.7 | <0.001 | Leukemia inhibitory factor receptor |
| BMP4 | D30751 | 3.4 | <0.001 | Bone morphogenetic protein 4 |
| CSF2RB | AV756141 | 3.2 | <0.001 | Colony stimulating factor 2 receptor β |
| KIAA0551 | AF172268 | 3.2 | <0.001 | Traf2 and NCK interacting kinase |
| RGS2 | NM_002923 | 3.1 | <0.001 | Regulator of G-protein signaling 2 |
| INHBA | M13436 | 3.0 | <0.001 | Inhibin βA |
| MET | BG170541 | 2.7 | 0.003 | Hepatocyte growth factor receptor |
| RICS | NM_014715 | 2.3 | 0.045 | Rho GTPase-activating protein |
| H11 | AF133207 | 2.2 | 0.002 | Protein kinase H11 |
| BMP2 | NM_001200 | 2.2 | <0.001 | Bone morphogenetic protein 2 |
| GUCY1B3 | W93728 | 2.1 | 0.010 | Guanylate cyclase 1, soluble, β3 |
| EXT1 | NM_000127 | 2.0 | <0.001 | Exostoses (multiple) 1 |
| ARL7 | NM_005737 | 2.0 | 0.005 | ADP-ribosylation factor-like 7 |
| Transcription factors | ||||
| CREM | D14826 | 8.5 | 0.004 | cAMP responsive element modulator |
| PKNOX2 | AK023792 | 6.2 | 0.001 | PBX/knotted 1 homeobox 2 |
| MAD | NM_002357 | 5.4 | <0.001 | MAX dimerization protein 1 |
| FOXC1 | AU145890 | 4.6 | 0.011 | Forkhead box C1 |
| CITED2 | NM_006079 | 4.2 | <0.001 | Cbp/p300-interacting transactivator |
| IRLB | BE268538 | 4.1 | <0.001 | c-Myc promoter-binding protein |
| CEBPB | AL564683 | 2.9 | <0.001 | CCAAT/enhancer binding protein β |
| POU4F1 | NM_006237 | 2.6 | 0.005 | POU domain, class 4, transcription factor 1 |
| GATA6 | D87811 | 2.3 | 0.018 | GATA binding protein 6 |
| KIAA0146 | NM_005195 | 2.3 | <0.001 | CCAAT enhancer binding protein (CEBP) |
| SSRP2 | NM_012446 | 2.2 | 0.003 | Single-stranded DNA binding protein 2 |
| ELL2 | NM_012081 | 2.1 | 0.022 | ELL-related RNA polymerase II, elongation factor |
| KLF4 | BF514079 | 2.1 | 0.011 | Kruppel-like factor 4 |
| TRAP95 | NM_005481 | 2.0 | 0.038 | Thyroid hormone receptor-associated protein, 95-kD subunit |
| CART1 | NM_006982 | 2.0 | 0.024 | Cartilage paired-class homeoprotein 1 |
| TOX | AI961231 | 2.0 | 0.009 | Thymus high mobility group box protein |
| Others | ||||
| MCHP | S64288 | 204.2 | <0.001 | Melanin-concentrating hormone precursor |
| HS3ST1 | BF000296 | 10.7 | 0.002 | Heparan sulfate 3-O-sulfotransferase 1 |
| CDC45L | NM_003504 | 7.4 | 0.003 | CDC45 cell division cycle 45-like |
| SIAT8A | L32867 | 6.5 | <0.001 | Sialyltransferase 8A |
| TMOD1 | NM_003275 | 5.9 | 0.004 | Tropomodulin 1 |
| MTHFR | AJ249275 | 5.7 | 0.009 | 5,10–Methylenetetrahydrofolate reductase |
| ENPP1 | NM_006208 | 5.2 | 0.004 | Ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| LOX | NM_002317 | 5.1 | <0.001 | Lysyl oxidase |
| AMIGO4 | AC004010 | 4.2 | <0.001 | Amphoterin induced gene 2 |
| SULF1 | AW043713 | 4.6 | <0.001 | Sulfatase 1 |
| GJA5 | NM_005266 | 4.3 | <0.001 | Gap junction protein (Connexin 40) |
| ARK5 | NM_014840 | 3.9 | <0.001 | KIAA0537 gene product |
| FKBP5 | NM_004117 | 3.8 | 0.002 | FK506 binding protein 5 |
| LRRTM2 | NM_015564 | 3.8 | 0.017 | Leucine-rich repeat transmembrane neuronal 2 protein |
| COVA1 | NM_006375 | 3.6 | 0.014 | Cytosolic ovarian carcinoma antigen 1 |
| DACT1 | NM_016651 | 3.6 | <0.001 | Dapper homolog 1, antagonist of β-catenin |
| DMD | NM_004010 | 3.6 | <0.001 | Dystrophin |
| LRRN3 | AI221950 | 3.5 | 0.006 | Leucine rich repeat neuronal 3 |
| PTX3 | NM_002852 | 3.5 | <0.001 | Pentaxin-related gene |
| CLCN4 | AA071195 | 3.5 | 0.006 | Chloride channel 4 |
| SLC38A1 | NM_030674 | 3.5 | <0.001 | Amino acid transporter system A1 |
| FLJ11743 | NM_024527 | 3.5 | 0.011 | Hypothetical protein FLJ11743 |
| OSPBL11 | NM_022776 | 3.4 | <0.001 | Oxysterol binding protein-like 11 |
| CH25H | NM_003956 | 3.2 | 0.026 | Cholesterol 25-hydroxylase |
| FBN1 | AI264196 | 2.8 | 0.003 | Fibrillin 1 (Marfan syndrome) |
| ELAVL2 | NM_004432 | 2.7 | <0.001 | ELAV (embryonic lethal, abnormal vision)-like 2 |
| CPT2 | M58581 | 2.7 | 0.020 | Carnitine palmitoyltransferase II |
| NNMT | NM_006169 | 2.5 | <0.001 | Nicotinamide N-methyltransferase |
| UP | NM_003364 | 2.5 | <0.001 | Uridine phosphorylase |
| KCNK3 | NM_002246 | 2.5 | 0.022 | Potassium channel, subfamily K, member 3 |
| DAAM1 | AK021890 | 2.5 | <0.001 | Dishevelled associated activator of morphogenesis 1 |
| SLC22A4 | NM_003059 | 2.4 | <0.001 | Solute carrier family 22, member 4 |
| PSCD1 | NM_004762 | 2.4 | <0.001 | Cytohesin 1 |
| JAG1 | NM_000214 | 2.4 | 0.024 | Jagged 1 (Alagille syndrome) |
| EGLN3 | NM_022073 | 2.4 | 0.007 | Egl 9 homolog 3 |
| TPK1 | NM_022445 | 2.3 | 0.005 | Thiamin pyrophosphokinase 1 |
| LOC169611 | AL050002 | 2.3 | 0.004 | Hypothetical protein LOC169611 |
| PVRL3 | AA129716 | 2.3 | 0.004 | Poliovirus receptor-related 3 |
| DOK5 | AL050069 | 2.2 | 0.002 | Docking protein 5 |
| MRF2 | BG285011 | 2.2 | <0.001 | Modulator recognition factor 2 |
| DNAJC3 | NM_006260 | 2.2 | 0.034 | DnaJ (Hsp40) homolog, subfamily C, 3 |
| CPR8 | AK022459 | 2.2 | 0.005 | Cell cycle progression 8 protein |
| SIAT1 | AV695711 | 2.1 | 0.002 | Sialyltransferase 1 |
| RDX | NM_002906 | 2.1 | 0.024 | Radixin |
| CYP1B1 | NM_000104 | 2.0 | 0.010 | Cytochrome P450 1B1 |
| ALDH1A2 | NM_003888 | 2.0 | 0.002 | Aldehyde dehydrogenase1A2 |
| GCNT1 | NM_001490 | 2.0 | <0.001 | β-1,6-N-acetylglucosaminyltransferase |
| Unknown | ||||
| DKK2 | NM_014421 | 26.5 | <0.001 | Dickkopf homolog 2 |
| T12479 | AW029169 | 8.5 | <0.001 | Hypothetical protein DKFZp 564N1362.1 |
| FLJ10713 | NM_018189 | 5.0 | <0.001 | Hypothetical protein FLJ10713 |
| KIAA0977 | NM_014900 | 3.8 | <0.001 | KIAA0977 protein |
| LOC51334 | NM_016644 | 2.8 | 0.010 | Mesenchymal stem cell protein DSC54 |
| CGI-115 | NM_016052 | 2.6 | <0.001 | CGI-115 protein |
| C13orf7 | NM_024546 | 2.4 | <0.001 | Chromosome 13 open reading frame 7 |
| FLJ20378 | AI336206 | 2.2 | <0.001 | Hypothetical protein FLJ20378 |
| SCA1 | NM_000332 | 2.2 | 0.012 | Spinocerebellar ataxia 1 |
| FLJ90005 | W27419 | 2.1 | <0.001 | Hypothetical protein FLJ90005 |
| 24739 | AF070571 | 2.1 | 0.011 | Homo sapiens clone 24739 |
| FJX1 | NM_014344 | 2.1 | 0.007 | Four jointed box 1 |
| FLJ10901 | NM_018265 | 2.0 | 0.020 | Hypothetical protein FLJ10901 |
| HRASLS | NM_020386 | 2.0 | 0.002 | HRAS-like suppressor |
| Housekeeping genes | ||||
| ACTB | Hs.426930 | 1.0 | 0.694 | β-Actin |
| GAPDH | M33197 | 1.0 | 0.680 | Glyceraldehyde-3-phosphate dehydrogenase |
Table 2.
Down-regulation of specific gene expression in human vascular endothelial cells treated with interleukin-4
| Gene symbol | NCBI accession nr | Fold change | P value | Description |
|---|---|---|---|---|
| Adhesion molecules | ||||
| ITGA2 | NM_002203 | −3.6 | <0.001 | Integrin α2 |
| Cytokines, chemokines, and receptors | ||||
| IL8 | NM_000584 | −5.6 | <0.001 | Interleukin-8 |
| IL7R | NM_002185 | −3.4 | 0.005 | Interleukin-7 receptor |
| CXCL2 | M57731 | −2.2 | 0.001 | Chemokine (C-X-C motif) ligand 2 |
| Growth factors and receptors | ||||
| GAS1 | NM_002048 | −14.0 | 0.007 | Growth arrest-specific 1 |
| NDRG4 | AV724216 | −2.1 | 0.011 | NDRG family member 4 |
| EGFR | AW157070 | −2.1 | 0.006 | Epidermal growth factor receptor |
| PDGFB | NM_002608 | −2.0 | 0.029 | Platelet-derived growth factor β polypeptide |
| Signal transduction | ||||
| KIT | NM_000222 | −9.0 | <0.001 | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| ITPKB | NM_002221 | −2.6 | 0.001 | Inositol 1,4,5-triphophate 3-kinase B |
| RAB11B | AL575337 | −2.2 | 0.016 | RAB11B, member RAS oncogene family |
| TNS | AL046979 | −2.0 | 0.036 | Tensin |
| Transcription factors | ||||
| KLF15 | NM_014079 | −2.7 | 0.004 | Kruppel-like factor 15 |
| SNAPC4 | AK023513 | −2.2 | 0.002 | Small nuclear RNA activating complex, polypeptide 4, 190 kDa |
| NPAS2 | AW000928 | −2.1 | <0.001 | Neuronal PAS domain protein 2 |
| HOXB6 | NM_018952 | −2.0 | 0.011 | Homeo box B6 |
| Others | ||||
| GJA4 | NM_002060 | −4.8 | 0.006 | Gap junction protein (connexin 37) |
| HIP14 | AF161412 | −2.9 | 0.019 | Huntingtin interacting protein 14 |
| KCNN2 | NM_021614 | −2.8 | <0.001 | Potassium intermediate/small conductance calcium-activated channel N2 |
| AMN | NM_030943 | −2.6 | 0.029 | Amnionless homolog |
| NFNG | AI760053 | −2.6 | 0.023 | Manic fringe homolog |
| CLTB | X81637 | −2.5 | 0.032 | Clathrin light chain b gene |
| CHST2 | NM_004267 | −2.2 | <0.001 | Carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 |
| FLJ12800 | NM_022903 | −2.2 | 0.030 | Hypothetical protein FLJ12800 |
| CLDN18 | BE551219 | −2.2 | 0.046 | Claudin 18 |
| RASGRP3 | NM_015376 | −2.1 | 0.003 | RAS guanine releasing protein 3 |
| CYP2A6 | NM_000762 | −2.1 | <0.001 | Cytochrome P450 2A6 |
| TRPV5 | NM_019841 | −2.1 | 0.008 | Trasient receptor potential cation channel, subfamily V, member 5 |
| CAV3 | NM_001234 | −2.0 | 0.021 | Caveolin 3 |
| BPAG1 | BG253119 | −2.0 | 0.031 | Bullous pemphigoid antigen 1 |
| WIZ | AL390184 | −2.0 | 0.009 | Widely interspaced zinc finger motifs |
| Unknown | ||||
| CHI3L1 | AJ251847 | −4.3 | 0.005 | CHI3L1 gene for cartilage glycoprotein-39 |
| R29124_1 | BF110434 | −3.3 | 0.004 | Hypothetical protein R29124_1 |
| FLJ11983 | AK022045 | −2.9 | 0.004 | FLJ11983 fis, clone HEMBA1001337 |
| PRO1598 | NM_018503 | −2.7 | 0.022 | Hypothetical protein PRO1598 |
| FLJ10002 | AK000864 | −2.6 | 0.007 | FLJ10002 fis, clone HEMBA1000046 |
| FLJ23497 | NM_025089 | −2.6 | 0.044 | Hypothetical protein FLJ23497 |
| KIAA0795 | NM_025010 | −2.5 | 0.013 | KIAA0795 protein |
| HL14 | M14087 | −2.4 | 0.049 | HL14 gene encoding β-galactoside-binding lectin |
| FLJ20378 | AI734156 | −2.2 | 0.033 | Hypothetical protein FLJ20378 |
| LOC92346 | AL035295 | −2.0 | 0.040 | PAC 106H8, similar to Dynamin |
Real-time Reverse Transcriptase–Polymerase Chain Reaction (RT-PCR)
Quantitative real-time RT-PCR, also known as fluorescence-based kinetic RT-PCR, was employed to confirm specific gene expression changes detected by the GeneChip analysis. The fluorogenic 5′ nuclease assay technology using TaqMan® probes was used to ensure specificity and sensitivity. Total RNA was isolated and purified using RNeasy Mini Kit (Qiagen) according to the protocol of the manufacturer. Then, 1 μg of total RNA was reverse-transcribed at 25 ºC for 15 min, 42 ºC for 45 min, and 99 ºC for 5 min in 20 μL of 5 mM MgCl2, 10 mM Tris-HCl, pH 9.0, 50 mM KCl, 0.1% Triton X-100, 1 mM dNTP, 1 unit/μL of recombinant RNasin ribonuclease inhibitor, 15 units/μg of AMV reverse transcriptase, and 0.5 μg of random hexamers. For quantitative PCR, amplifications of individual genes were performed on ABI PRISM® 7000 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) using TaqMan® Universal PCR Master Mix, gene-specific TaqMan PCR probes and primers, and a standard thermal cycler protocol (50 ºC for 2 min before the 1st cycle, 95 ºC for 15 s, and 60 ºC for 1 min, repeated 45 times). For specific probes and primers of PCR amplifications, Assay-on-Demand™ Products for human VCAM-1 and E-selectin, and TaqMan Pre-Developed Assay Reagents for human MCP-1, IL-6, and β-actin, were obtained from Applied Biosystems. The threshold cycle (CT), which indicates the fractional cycle number at which the amount of amplified target gene reaches a fixed threshold, from each well was determined using ABI Prism 7000 SDS software. Relative quantification, which represents the change in gene expression from real-time quantitative PCR experiments between IL-4–treated group and untreated control group, was calculated by the comparative CT method as described earlier (18,19). The data were analyzed using equation 2–ΔΔCT, where ΔΔCT = [CT of target gene – CT of housekeeping gene]treated group – [CT of target gene – CT of housekeeping gene]untreated control group. For the treated samples, evaluation of 2–ΔΔCT represents the fold change in gene expression, normalized to a housekeeping gene (β-actin) and relative to the untreated control.
Enzyme-linked Immunosorbent Assay (ELISA)
Cell surface expression levels of adhesion molecules such as VCAM-1 and E-selectin were quantified by ELISA Development kits (R&D Systems, Minneapolis, MN, USA) according to the manufacturer ’s procedure, with modifications. Briefly, HUVEC monolayers were incubated with either anti-human VACM-1 or E-selectin monoclonal antibody (2.5 μg/mL) for 1 h at 37 °C. The cells were then incubated with biotinylated goat antimouse IgG antibody (1:1,000 dilution) for 1 h at 37 °C. After washing the wells thoroughly, the working dilution of Streptavidin-HRP was added to each well and incubated for 20 min at room temperature. The cells were incubated with HRP Substrate Solution for 20 min at room temperature with subsequent addition of Stop Solution. After color development, absorbance from each well was measured by a microtiter plate reader at 450 nm to 570 nm.
The protein levels of human MCP-1 and IL-6 in cell culture supernatants were determined using Human MCP-1 Immunoassay and Human IL-6 Immunoassay kits (R&D Systems) according to the protocol of the manufacturer, respectively. This assay employs the quantitative sandwich enzyme immunoassay technique using a murine monoclonal antibody against human MCP-1 or IL-6, and a polyclonal secondary antibody conjugated with horseradish peroxidase. The minimum detectable concentration of MCP-1 and IL-6 was less than 5.0 and 0.70 pg/mL, respectively.
Cell Adhesion Assay
Adhesion studies were performed with the human monocytic leukemia cell line, THP-1, as previously described (20) with modifications (21). Briefly, HUVEC were grown to confluence on 24-well plates and exposed to IL-4 for 8 and 24 h. Prior to the cell-cell adhesion assay, the HUVEC monolayers were washed twice with Hank’s Balanced Salt Solution (HBSS) and then washed with M199 medium containing 10% fetal bovine serum. Calcein acetoxymethyl ester (calcein AM; Calbiochem, La Jolla, CA, USA) was employed to label THP-1 cells. The fluorescence labeling of THP-1 cells was achieved by incubating cells (2.5 × 105 cells/mL) with 5 μg/mL of calcein AM. After loading of calcein AM for 20 min at 37 °C, the cells were washed 3 times with HBSS, and then washed with M199 medium containing 10% fetal bovine serum. The calcein AM–labeled THP-1 cells were added onto the HUVEC monolayers and incubated for 20 min at 37 °C. The non-adherent THP-1 cells were removed from monolayers by washing each well 3 times with HBSS. The fluorescence intensity was measured by a fluorescence plate reader using excitation of 490 nm and emission of 517 nm.
Statistical Analysis
Routine statistical analysis of data was completed using Sigma-Stat 2.03 (SPSS, Chicago, IL, USA). Statistical probability of P < 0.05 was considered significant.
RESULTS
IL-4 Up-regulates Adhesion of Leukocytes to Human Vascular Endothelial Cell Monolayers
The adherence of human acute monocytic leukemia cells, THP-1, to HUVEC monolayers was determined to verify the functional integrity of inflammatory mediators up-regulated by human vascular endothelial cells after stimulation with IL-4. Following an 8-to 24-h incubation with IL-4 doses ranging from 0.1 to 10 ng/mL, endothelial cell function was significantly and dose-dependently altered as assessed by changes in THP-1 adherence to the HUVEC monolayer (Figure 1). Hence, endothelial cells were exposed to 10 ng/mL of IL-4 for 4 h and changes in gene expression were assessed using microarray analysis.
Figure 1.
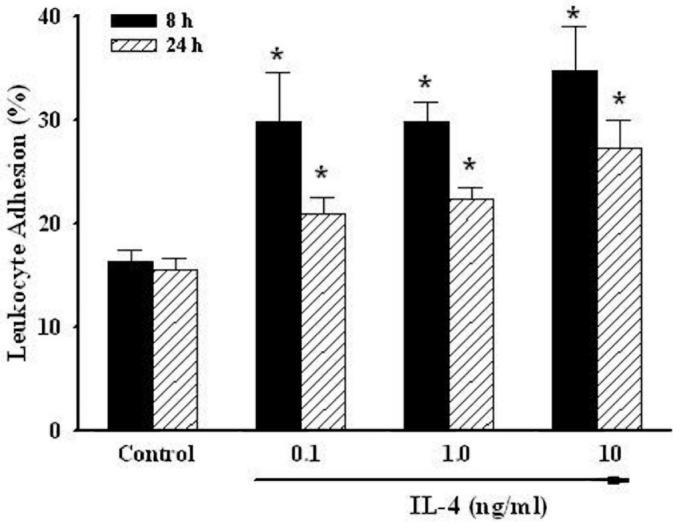
IL-4 up-regulates the adhesion of leukocytes to human vascular endothelial cell monolayers. HUVEC were either untreated or treated with the indicated concentrations of IL-4 (0.1, 1.0, and 10 ng/mL) for up to 24 h. The adherence of calcein AM–labeled THP-1 cells was measured by fluorescent microplate reader using excitation of 490 nm and emission of 517 nm. Data are means ± SD of 4 determinations. *Statistically significant compared with the control group (P < 0.05).
Identification of Global Gene Expression Changes in IL-4–Treated Human Vascular Endothelial Cells
The gene expression profile of human vascular endothelial cells treated with IL-4 was assessed using microarray technology with the Affymetrix GeneChip Human Genome U133A Arrays, which contain more than 22000 human genes. As shown in Table 1, 106 genes were significantly up-regulated at the mRNA level with more than a 2-fold change in HUVEC after treatment with IL-4 for 4 h. Classification by function revealed that IL-4 treatment up-regulated genes mainly responsible for inflammatory reactions, apoptosis, signal transduction, and transcription factors. Among these, mRNA levels of the inflammatory mediators, such as adhesion molecules (VCAM-1 and E-selectin), chemokine (MCP-1), and pro-inflammatory cytokine (IL-6), were markedly and significantly induced, suggesting that IL-4 can play a crucial role in the pro-inflammatory pathways in human vascular endothelium. The expression of 2 housekeeping genes, β-actin and glyceraldehyde-3-phosphate dehydrogenase, was not affected with IL-4 treatment. In addition, exposure of HUVEC to IL-4 resulted in a significant down-regulation of 41 genes by at least 2-fold factor, as compared with untreated control cell cultures (Table 2).
Verification of Microarray Analysis Using Real-Time RT-PCR and ELISA
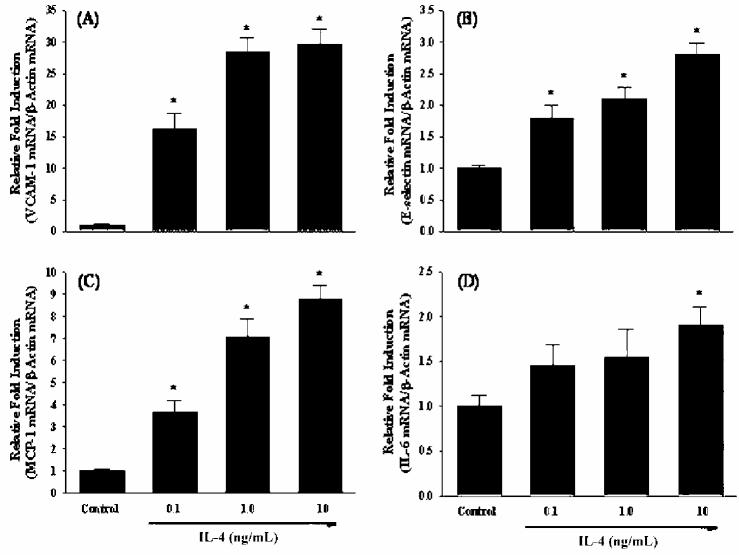
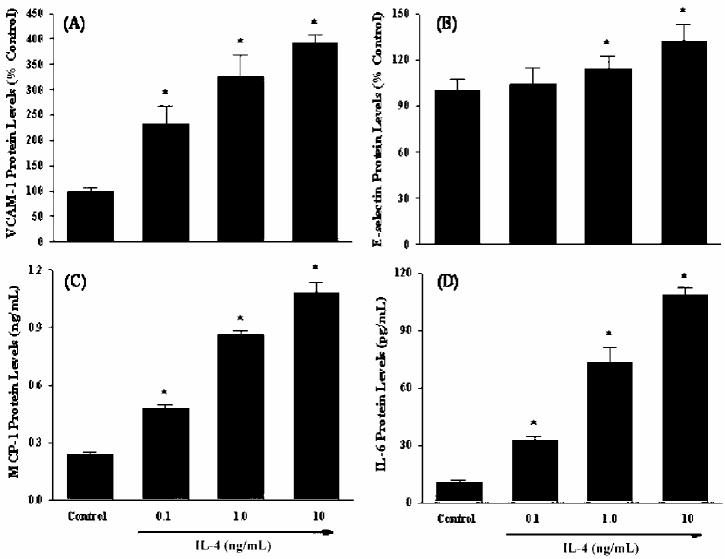
To validate the changes in gene expression of IL-4-treated human vascular endothelial cells observed in microarray analysis, we performed quantitative real-time RT-PCR for several target genes that were up-regulated in HUVEC treated with IL-4. In the present study, we selected 4 inflammatory genes encoding VCAM-1, E-selectin, MCP-1, and IL-6. Real-time RT-PCR showed that increasing concentrations of IL-4 dramatically induced mRNA expression of adhesion molecules, such as VCAM-1 and E-selectin (Figures 2A and 2B). A significant and dose-dependent induction of chemokine MCP-1 gene was also observed in HUVEC treated with IL-4 (see Figure 2C). Additionally, IL-4 treatment markedly up-regulates gene expression of pro-inflammatory cytokine IL-6 (see Figure 2D). These results confirm that up-regulation of selected genes identified by microarray analysis correlates with mRNA expression measured by real-time RT-PCR. In parallel with gene expression analyses, a series of ELISA was conducted to determine whether IL-4–induced increases in mRNA levels could translate to elevated protein expression. Consistent with the data on gene expression, treatment with IL-4 resulted in a significant and dose-dependent up-regulation of protein expression of VCAM-1, E-selectin, MCP-1, and IL-6 (Figures 3A to 3D).
Figure 2.
IL-4 up-regulates the mRNA expression of inflammatory mediators in human vascular endothelial cells. HUVEC were either untreated or treated with the indicated concentrations of IL-4 (0.1, 1.0, and 10 ng/mL) for 4 h. The mRNA levels of VCAM-1 (A), E-selectin (B), MCP-1 (C), and IL-6 (D) were determined by real-time RT-PCR as described in Materials and Methods. Data are means ± SE of 4 determinations. *Statistically significant compared with the control group (P < 0.05).
Figure 3.
IL-4 up-regulates the protein expression of inflammatory mediators in human vascular endothelial cells. HUVEC were either untreated or treated with the indicated concentrations of IL-4 (0.1, 1.0, and 10 ng/mL) for 12 h (VCAM-1), 6 h (E-selectin), or 16 h (MCP-1 and IL-6). The protein levels of VCAM-1 (A), E-selectin (B), MCP-1 (C), and IL-6 (D) were measured by ELISA as described in Materials and Methods. Data are means ± SD of 4 determinations. *Statistically significant compared with the control group (P < 0.05).
DISCUSSION
Microarray analysis is one of the most advanced and emerging molecular biological technologies, and it has been widely adopted for analyzing the global gene expression profiles in vivo and in vitro (22,23). Recent studies have demonstrated the potential of this technology for investigating molecular pathophysiological mechanisms involved in a variety of human diseases. In fact, microarray technology has been used as a novel experimental approach to analyze alterations in gene expression in cancer (24), atherosclerosis (25), stroke (26), Alzheimer’s disease (27), HIV infection (28), schizophrenia (29), and muscular dystrophy (30).
In the present study, we performed microarray analysis using the Affymetrix GeneChip Human Genome U133A Arrays to further understand transcriptional regulatory mechanisms of action of IL-4 in human vascular endothelium. Our results revealed that mRNA levels of a total of 106 genes were significantly up-regulated and 41 genes significantly down-regulated with more than a 2-fold change in HUVEC treated with IL-4 compared with the control cell cultures (see Tables 1 and 2). Interestingly, many of IL-4–up-regulated genes are involved in inflammatory reactions, which are critical to initiate and promote early stage of atheroge-nesis. Previous studies from our group and others have demonstrated that IL-4–induced oxidative stress can produce a pro-inflammatory vascular environment through up-regulation of inflammatory genes, such as adhesion molecules, chemokines, and cytokines (9–12,31,32). The present data, showing significant up-regulation of VCAM-1, E-selectin, MCP-1, and IL-6, strongly support that IL-4 is a key mediator to induce pro-oxidative and pro-inflammatory pathways in human vascular endothelium.
To verify the alterations in gene expression observed in microarray analysis, as well as to further explore the potential role of IL-4 in inflammatory pathways in human vascular endothelium, the present study focused on a set of genes related to inflammatory reactions such as VCAM-1, E-selectin, MCP-1, and IL-6. VCAM-1 is expressed primarily on endothelial cells and mediates cell-cell interactions via binding to its integrin counter receptor, very late antigen-4, which may be involved in the recruitment of mononuclear leukocytes to the vascular lesions in early atherosclerosis (33). We and others have shown that IL-4 up-regulates VCAM-1 expression in vascular endothelial cells through antioxidant-sensitive mechanisms (10,31,32). In agreement with previous studies, a marked and significant increase in mRNA and protein expression of VCAM-1 was observed in IL-4–treated HUVEC by real-time RT-PCR and ELISA, respectively (see Figures 2A and 3A).
Another adhesion molecule studied in the present study was E-selectin. E-selectin is present exclusively on the surface of endothelial cells and plays a key role in mediating early leukocyte-endothelial interactions such as initial attachment and rolling during an inflammatory response. It is well documented that E-selectin is up-regulated at the transcriptional level following exposure to a series of pro-inflammatory mediators, such as IL-1β, TNF-α, and lipopolysaccharide (34). In contrast, it has been proposed that treatment of endothelial cells with IL-4 suppresses IL-1β– or TNF-α–stimulated E-selectin gene transcription (35,36). Direct effects of IL-4 on E-selectin expression in human vascular endothelial cells, however, remain unclear. In the present study, we provide new evidence to indicate that IL-4 could directly up-regulate mRNA and protein expression of E-selectin in HUVEC (see Figures 2B and 3B). These results suggest that E-selectin may play an important role in IL-4–mediated inflammatory pathways in vascular endothelium.
Among a variety of chemokines and inflammatory cytokines, MCP-1 and IL-6 are of critical significance in the early stages of atherosclerosis. MCP-1 is secreted by a variety of cell types, including vascular endothelial cells, and promotes the recruitment of inflammatory cells and their migration throughout the vascular endothelium that are thought to be critical early pathological events in atherogenesis (37,38). Consistent with previous experiments (11,12), the present study showed that IL-4 treatment resulted in up-regulation of mRNA and protein expression of MCP-1 in human vascular endothelial cells (see Figures 2C and 3C).
IL-6, a multifunctional pro-inflammatory cytokine, plays a major role in inflammatory responses in vascular endothelium and has also been implicated in the pathogenesis of atherosclerosis (1,39). Although recent evidence indicates that IL-4 synergistically amplifies the TNF-α–, IL-1β–, or LPS-induced production of IL-6 protein in HUVEC (40), the molecular basis for the induction of this cytokine by IL-4 has not been elucidated. Therefore, our results, showing that IL-4 significantly induced the expression of IL-6 mRNA and increased IL-6 production in HUVEC (Figures 2D and 3D), appear to be the first to document the stimulatory effect of IL-4 on IL-6 gene expression in human vascular endothelial cells.
In conclusion, the present study provides the first quantitative large-scale gene expression analysis of IL-4–stimulated human vascular endothelial cells. We identified 147 differentially regulated genes that are responsible for the regulation of inflammatory responses, apoptosis, signal transduction, transcription factors, metabolism, and several unknown functions. Because IL-4 is involved in the early stages of atherogenesis, these results could contribute to a deeper understanding of fundamental insights of pathophysiological mechanisms involved in atherosclerosis at the level of gene expression and provide a foundation for development of therapeutic strategies for vascular diseases.
Acknowledgments
This work was supported by grants from the American Heart Association, Ohio Valley Affiliate, NIEHS/NIH (ES07380), and the University of Kentucky Microarray Facility Program.
REFERENCES
- 1.Ross R. Atherosclerosis is an inflammatory disease. Am. Heart. J. 1999;138:S419–20. doi: 10.1016/s0002-8703(99)70266-8. [DOI] [PubMed] [Google Scholar]
- 2.Davies MJ, et al. The expression of the adhesion molecules ICAM-1, VCAM-1, PECAM, and E-selectin in human atherosclerosis. J. Pathol. 1993;171:223–9. doi: 10.1002/path.1711710311. [DOI] [PubMed] [Google Scholar]
- 3.Cotran RS, Mayadas-Norton T. Endothelial adhesion molecules in health and disease. Pathol. Biol. 1998;46:164–70. [PubMed] [Google Scholar]
- 4.Reape TJ, Groot PH. Chemokines and atherosclerosis. Atherosclerosis. 1999;147:213–25. doi: 10.1016/s0021-9150(99)00346-9. [DOI] [PubMed] [Google Scholar]
- 5.Rus HG, Niculescu F, Vlaicu R. Tumor necrosis factor-alpha in human arterial wall with atherosclerosis. Atherosclerosis. 1991;89:247–54. doi: 10.1016/0021-9150(91)90066-c. [DOI] [PubMed] [Google Scholar]
- 6.Paul WE. Interleukin-4: A prototypic immunoregulatory lymphokine. Blood. 1991;77:1859–70. [PubMed] [Google Scholar]
- 7.Rocken M, Racke M, Shevach EM. IL-4-induced immune deviation as antigen-specific therapy for inflammatory autoimmune disease. Immunol. Today. 1991;17:225–31. doi: 10.1016/0167-5699(96)80556-1. [DOI] [PubMed] [Google Scholar]
- 8.Sasaguri T, et al. A role for interleukin 4 in production of matrix metalloproteinase 1 by human aortic smooth muscle cells. Atherosclerosis. 1998;138:247–53. doi: 10.1016/s0021-9150(97)00296-7. [DOI] [PubMed] [Google Scholar]
- 9.Galea P, Chartier A, Lebranchu Y. Increased lymphocyte adhesion to allogeneic endothelial cells by interleukin-4 (IL-4) Transplant Proc. 1991;23:243–44. [PubMed] [Google Scholar]
- 10.Lee YW, Kühn H, Hennig B, Neish AS, Toborek M. IL-4-induced oxidative stress upregulates VCAM-1 gene expression in human endothelial cells. J. Mol. Cell. Cardiol. 2001;33:83–94. doi: 10.1006/jmcc.2000.1278. [DOI] [PubMed] [Google Scholar]
- 11.Rollins BJ, Pober JS. Interleukin-4 induces the synthesis and secretion of MCP-1/JE by human endothelial cells. Am. J. Pathol. 1991;138:1315–9. [PMC free article] [PubMed] [Google Scholar]
- 12.Lee YW, Hennig B, Toborek M. Redox-regulated mechanisms of IL-4-induced MCP-1 expression in human vascular endothelial cells. Am. J. Physiol. Heart Circ. Physiol. 2003;284:H185–92. doi: 10.1152/ajpheart.00524.2002. [DOI] [PubMed] [Google Scholar]
- 13.Brinckmann R, et al. Regulation of 15-lipoxygenase expression in lung epithelial cells by interleukin-4. Biochem. J. 1996;318:305–12. doi: 10.1042/bj3180305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee YW, et al. Interleukin 4 induces transcription of the 15-lipoxygenase I gene in human endothelial cells. J. Lipid Res. 2001;42:783–91. [PubMed] [Google Scholar]
- 15.Toborek M, Lee YW, Kaiser S, Hennig B. Measurement of inflammatory properties of fatty acids in human endothelial cells. Methods Enzymol. 2002;352:198–219. doi: 10.1016/s0076-6879(02)52020-6. [DOI] [PubMed] [Google Scholar]
- 16.Blalock EM, et al. Gene microarrays in hippocampal aging: Statistical profiling identifies novel processes correlated with cognitive impairment. J. Neurosci. 2003;23:3807–19. doi: 10.1523/JNEUROSCI.23-09-03807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blalock EM, et al. Incipient Alzheimer’s disease: Microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2173–8. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Deng X, Li H, Tang YW. Cytokine expression in respiratory syncytial virus-infected mice as measured by quantitative reverse-transcriptase PCR. J. Virol. Methods. 2003;107:141–6. doi: 10.1016/s0166-0934(02)00211-2. [DOI] [PubMed] [Google Scholar]
- 20.Braut-Boucher F, et al. A non-isotopic, highly sensitive, fluorimetric, cell-cell adhesion microplate assay using calcein AM-labeled lymphocytes. J. Immunol. Methods. 1995;178:41–51. doi: 10.1016/0022-1759(94)00239-s. [DOI] [PubMed] [Google Scholar]
- 21.Choi W, et al. PCB 104-induced proinflammatory reactions in human vascular endothelial cells: Relationship to cancer metastasis and atherogenesis. Toxicol. Sci. 2003;75:47–56. doi: 10.1093/toxsci/kfg149. [DOI] [PubMed] [Google Scholar]
- 22.Watson A, Mazumder A, Stewart M, Balasubramanian S. Technology for microarray analysis of gene expression. Curr. Opin. Biotechnol. 1998;9:609–14. doi: 10.1016/s0958-1669(98)80138-9. [DOI] [PubMed] [Google Scholar]
- 23.Schulze A, Downward J. Navigating gene expression using microarrays—a technology review. Nature Cell Biol. 2001;3:E190–5. doi: 10.1038/35087138. [DOI] [PubMed] [Google Scholar]
- 24.Golub TR, et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–7. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 25.Hiltunen MO, et al. Changes in gene expression in atherosclerotic plaques analyzed using DNA array. Atherosclerosis. 2002;165:23–32. doi: 10.1016/s0021-9150(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 26.Bowler RP, et al. A catalytic antioxidant (AEOL 10150) attenuates expression of inflammatory genes in stroke. Free Radic. Biol. Med. 2002;33:947–61. doi: 10.1016/s0891-5849(02)01008-0. [DOI] [PubMed] [Google Scholar]
- 27.Ginsberg SD, Hemby SE, Lee VM, Eberwine JH, Trojanowski JQ. Expression profile of transcripts in Alzheimer’s disease tangle-bearing CA1 neurons. Ann. Neurol. 2000;48:77–87. [PubMed] [Google Scholar]
- 28.Geiss GK, et al. Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology. 2000;266:8–16. doi: 10.1006/viro.1999.0044. [DOI] [PubMed] [Google Scholar]
- 29.Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 30.Chen YW, Zhao P, Borup R, Hoffman EP. Expression profiling in the muscular dystrophies: Identification of novel aspects of molecular pathophysiology. J. Cell Biol. 2000;151:1321–36. doi: 10.1083/jcb.151.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masinovsky B, Urdal D, Gallatin WM. IL-4 acts synergistically with IL-1β to promote lymphocyte adhesion to microvascular endothelium by induction of vascular cell adhesion molecule-1. J. Immunol. 1990;145:2886–95. [PubMed] [Google Scholar]
- 32.Blease K, Seybold J, Adcock IM, Hellewell PG, Burke-Gaffney A. Interleukin-4 and lipopolysaccharide synergize to induce vascular cell adhesion molecule-1 expression in human lung microvascular endothelial cells. Am. J. Respir. Cell Mol. Biol. 1998;18:620–30. doi: 10.1165/ajrcmb.18.5.3052. [DOI] [PubMed] [Google Scholar]
- 33.Cybulsky M, Gimbrone M. Endothelial expression of a mononuclear leukocyte adhesion molecule during atherogenesis. Science. 1991;251:788–91. doi: 10.1126/science.1990440. [DOI] [PubMed] [Google Scholar]
- 34.Whelan J. Selectin synthesis and inflammation. Trends Biochem. Sci. 1996;21:65–9. [PubMed] [Google Scholar]
- 35.Bennett BL, Cruz R, Lacson RG, Manning AM. Interleukin-4 suppression of tumor necrosis factor α-stimulated E-selectin gene transcription is mediated by STAT6 antagonism of NF-κB. J. Biol. Chem. 1997;272:10212–9. doi: 10.1074/jbc.272.15.10212. [DOI] [PubMed] [Google Scholar]
- 36.Thornhill MH, Haskard DO. IL-4 regulates endothelial cell activation by IL-1, tumor necrosis factor, or IFN-γ. J. Immunol. 1990;145:865–72. [PubMed] [Google Scholar]
- 37.Gu L, Tseng SC, Rollins BJ. Monocyte chemoattractant protein-1. Chem. Immunol. 1999;72:7–29. doi: 10.1159/000058723. [DOI] [PubMed] [Google Scholar]
- 38.Rollins BJ. Chemokines. Blood. 1997;90:909–28. [PubMed] [Google Scholar]
- 39.Libby P, Ridker PM, Maseri A. Inflammation and atherosclerosis. Circulation. 2002;105:1135–43. doi: 10.1161/hc0902.104353. [DOI] [PubMed] [Google Scholar]
- 40.Chen CC, Manning AM. TGF-β1, IL-10 and IL-4 differentially modulate the cytokine-induced expression of IL-6 and IL-8 in human endothelial cells. Cytokine. 1996;8:58–65. doi: 10.1006/cyto.1995.0008. [DOI] [PubMed] [Google Scholar]