Abstract
The genome of enteropathogenic Escherichia coli (EPEC) encodes a global regulator, Ler (locus of enterocyte effacement [LEE]-encoded regulator), which activates expression of several polycistronic operons within the 35.6-kb LEE pathogenicity island, including the LEE2-LEE3 divergent operon pair containing overlapping −10 regions and the LEE5 (tir) operon. Ler is a predicted 15-kDa protein that exhibits amino acid similarity with the nucleoid protein H-NS. In order to study Ler-mediated activation of virulence operons in EPEC, we used a molecular approach to characterize the interactions of purified Ler protein with the upstream regulatory sequences of the LEE5 operon. We determined the cis-acting DNA sequences necessary for Ler binding at LEE5 by mobility shift and DNase I protection assays, demonstrating that Ler acts directly at LEE5 by binding sequences between positions −190 and −73 in relation to the transcriptional start site. Based on the molecular weight of Ler, the similarity to H-NS, and the extended region of protection observed in a DNase I footprint at LEE5, we hypothesized that multiple Ler proteins bind upstream of the LEE5 promoter to increase transcriptional activity from a distance. Using an hns deletion strain, we demonstrated that like the LEE2-LEE3 operon pair, H-NS represses LEE5 transcription. We describe a model in which Ler activates transcription at both divergent overlapping paired and single promoters by displacing H-NS, which results in the disruption of a repressing nucleoprotein complex.
Enteropathogenic E. coli (EPEC) is a leading cause of infant diarrhea in developing countries (35), and there is some indication that diarrhea caused by EPEC in the United States is underreported (5). EPEC is the prototype organism of a group of pathogenic bacteria that cause attaching and effacing (AE) intestinal lesions (25, 33, 35). A variety of gram-negative pathogens are capable of forming AE lesions, including enterohemorrhagic E. coli serotype O157:H7, which causes hemorrhagic colitis and hemolytic-uremic syndrome (52), Hafnia alvei, which causes diarrhea in children (2), the mouse pathogen Citrobacter rodentium, (43), and rabbit enteropathogenic E. coli, which causes diarrhea in rabbits (40).
All genes necessary for the AE phenotype in EPEC are encoded in a 35.6-kb pathogenicity island that contains 41 predicted open reading frames termed the locus of enterocyte effacement (LEE) (15, 19, 28, 35). The genetic organization of the EPEC LEE was determined previously by defining transcriptional units and mapping transcriptional start points (30). Components of the type III secretion system are transcribed from three polycistronic operons designated LEE1, LEE2, and LEE3, and the secreted Esp molecules are part of a fourth polycistronic operon designated LEE4. Genes involved in intimate attachment to the host cell, tir, cesT, and eae, were found to be transcribed from a fifth polycistronic operon termed LEE5 (tir) (30, 42).
Ler (LEE-encoded regulator) increases expression of at least four polycistronic operons found within the LEE (14, 30). In a cascade fashion, the EAF plasmid-encoded regulator Per modulates the expression of the Ler protein, which goes on to increase expression of most, if not all, of the genes necessary for the AE phenotype (30). Ler increases the transcription of divergent operons of the LEE possessing overlapping promoter regions (e.g., the LEE2-LEE3 promoter pair), as well as single nondivergent operons. Interestingly, Ler activates transcription from both LEE2 and LEE3 by binding over an extended region (positions −221 to −100) on only one side of the overlapping promoters, upstream of LEE2 (47). In addition, Ler activates the expression of espC, which encodes an enterotoxin (14) and is contained within a second pathogenicity island in EPEC (31), and thus Ler is considered to be a global regulator of virulence genes.
The predicted 15-kDa Ler protein exhibits significant amino acid similarity to the H-NS family of DNA-binding proteins, including 24% identity and 44% similarity to H-NS of Salmonella enterica serovar Typhimurium (47). The C-terminal domain of H-NS contains a conserved DNA-binding domain (4), whereas the N terminus contains a coiled-coil domain involved in protein oligomerization (27). Greater amino acid sequence similarity to H-NS was observed in the C terminus than in the N terminus. Base substitution in the putative oligomerization domain in the N terminus of Ler eliminated the ability of the protein to bind to DNA and the ability to activate expression of a LEE2-lacZ fusion, strongly suggesting that Ler must form oligomers in order to activate transcription (47).
H-NS is a 15-kDa, histone-like, nucleoid-associated protein that was originally described as a protein that compacts and alters the topology of DNA (20, 37). This protein is also a pleiotropic transcription factor, affecting the expression of approximately 5% of the genes in E. coli (3, 21). One commonality of the seemingly unrelated genes regulated by H-NS is that many respond to environmental stress conditions, such as osmotic shock and cold shock (13, 16, 26, 29, 50). H-NS was shown to influence the thermo-osmotic regulation of virulence genes in Shigella (38). H-NS is now known to regulate virulence gene expression in several gram-negative pathogens (16, 18, 41, 54).
H-NS forms homodimers or tetramers (48) and is known to form higher-order oligomeric structures in solution (46). It is now apparent that the H-NS protein binds to DNA possessing a particular conformation rather than to a specific consensus binding sequence (10, 12, 50). There is a large body of data that demonstrates that there is transcriptional repression by H-NS binding to promoter regions (1, 17, 26, 39, 53). Recent discoveries indicate that H-NS can hold RNA polymerase in an open complex while it forms a nucleoprotein structure that represses transcription at the rrnB P1 promoter (11, 44). Ler has been shown to relieve the transcriptional repression caused by H-NS at the LEE2 and LEE3 operons (6) and the transcriptional repression of the LEE5 operon caused by an unknown negative regulator (42).
In order to learn more about the molecular pathogenesis of EPEC, we used a biochemical approach to study the mechanism of Ler binding at the LEE5 operon. Ler is a key virulence gene regulator in EPEC, and a better understanding of the details of its DNA binding activity and the DNA sequences required for binding should provide insight into to how Ler activates transcription. The ability of the Ler protein to increase transcriptional activity from a distance at both divergent overlapping and nondivergent promoter sequences and the similarity to the H-NS family of proteins that, in most cases, repress transcription prompted us to study the mechanism of Ler binding at the molecular level.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phage.
The plasmids, strains, and phage used in this study are listed in Table 1. Strains were grown at 37°C with aeration in Luria-Bertani medium supplemented with ampicillin (100 μg/ml) or kanamycin (50 μg/ml). Single-copy chromosomal lacZ fusions were constructed by homologous recombination between λRS45 and plasmids containing LEE5 regulatory fragments cloned upstream of the promoterless lacZYA operon contained in pRS551 and were subsequently transduced into MC4100, selecting for kanamycin resistance as previously described (30, 45).
TABLE 1.
Bacterial strains, plasmids, and phage used in this study
| Strain, plasmid or phage | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Lab stocks |
| MC4100 | araD139 Δ(argF-lac) U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | 7 |
| HN4104 | MC4100 Δ(hns tdk adhE oppABCD)118 zch-506::Tn10 | 8 |
| KMRS551 | MC4100 φ(promoterless) lacZ | This study |
| KMTIR2 | MC4100 φLEE5-lacZ(−405 to +172) | This study |
| KMTIR3 | MC4100 φLEE5-lacZ(−303 to +172) | This study |
| KMTIR4 | MC4100 φLEE5-lacZ(−198 to +172) | This study |
| CRTIR5 | MC4100 φLEE5-lacZ (−75 to +172) | This study |
| JFHNTIR3 | HN4104 φLEE5-lacZ(−303 to +172) | This study |
| JFHNTIR4 | HN4104 φLEE5-lacZ(−198 to +172) | This study |
| JFHNTIR5 | HN4104 φLEE5-lacZ(−75 to +172) | This study |
| KHTIR3M | MC4100 φΔ(−51 to −47)LEE5-lacZ(−303 to 172) | This study |
| Plasmids | ||
| pBR322 | Cloning vector, bla | 49 |
| pBluescript-II KS | Cloning vector, bla | Stratagene |
| pVS45 | Minimal ler in pBADMycHis | 47 |
| pSE1100 | Minimal ler in pBR322, bla | 30 |
| pTHK113 | Minimal hns in pBR322, bla | 23 |
| pRS551 | (Promoterless) lacZ reporter fusion vector | 45 |
| pKMTIR2 | LEE5 (−405 to +172) in pRS551 | This study |
| pKMTIR3 | LEE5 (−303 to +172) in pRS551 | This study |
| pKMTIR4 | LEE5 (−198 to +172) in pRS551 | This study |
| pCRTIR5 | LEE5 (−75 to +172) in pRS551 | This study |
| pKHTIR3M | Δ(−51 to −47)LEE5 (−303 to 172) in pRS551 | This study |
| pBlueTIR3 | LEE5 (−303 to +172) in pBluescript | This study |
| pBlueTIR4 | LEE5 (−198 to +172) in pBluescript | This study |
| pBlueTIR5 | LEE5 (−75 to +172) in pBluescript | This study |
| pBlueTIR3M | Δ(−51 to −47) LEE5 (−303 to 172) in pBluescript | This study |
| pKH273 | LEE5 (−272 to −31) in pBluescript | This study |
| Phage λRS45 | Specialized transducing phage | 45 |
Generation of LEE5 regulatory fragments.
Specific LEE5 regulatory fragments were generated by PCR by using Pwo DNA polymerase, which contains proofreading activity (Boehringer Mannheim). The oligonucleotides used in the study are listed in Table 2 and were purchased from Invitrogen. PCRs were performed by using standard protocols. Restriction endonucleases and DNA ligase were obtained from New England Biolabs and used according to the manufacturer's instructions. The PCR-generated LEE5 regulatory fragments were gel isolated with a Qiaquick gel extraction kit obtained from Qiagen. DNA fragments were then cloned into pRS551 by using a Bio-Rad Micropulser electroporation apparatus to transform E. coli strains DH5α and MC4100. Clones containing plasmids with LEE5 regulatory sequences fused to lacZ were verified by restriction mapping, PCR, DNA sequence analysis, and β-galactosidase assays.
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence (5′ to 3′)a |
|---|---|
| TIR1 | CGCGGATCCGCGCCGTCTGTTTGTGAA |
| TIR2 | CCGGAATTCGGTAAAGGAGTGGATCCCA |
| TIR3 | CCGGAATTCAGTGATATCAAGGCTCTAA |
| TIR4 | CCGGAATTCAGGTCTCTATAGACGTTTAAA |
| TIR5 | CCGGAATTCGTTGGAAATACAGACATGCA |
| JFTIR1 | AATACAGACATGCATTTCTGTTATTTTGCTTG CATCAAAA |
| JFTIR3 | TTTTGATGCAAGCAAAATAACAGAAATGCAT GTCTGTATT |
| TF1 | CCGGAATTCTTGCAGACAATGTGCAGGAT |
| TF5 | CGCGGATCCATGCAAGCAAAATAACGCAC |
| BlueNot | GGCCGCTCTAGAACTAGTG |
Restriction sites used in cloning are underlined.
Enzymatic assays.
Lysogens of E. coli strain MC4100 containing single-copy chromosomal lacZ fusions were transformed with pSE1100 (minimal ler clone [30]), pTHK113 (minimal hns clone [23]), or pBR322 (control) and grown at 37°C in Luria-Bertani medium supplemented with the appropriate antibiotics with aeration to an absorbance at 600 nm of approximately 0.3 to 0.5 and then subjected to β-galactosidase assays performed by the method described by Miller (32). Assays were performed in triplicate, and values (in Miller units) were expressed as the mean ± one standard deviation. Statistical analysis was performed with raw data generated from β-galactosidase assays by using StatView (SAS Institute Inc.).
Ler protein purification.
His-tagged Ler protein was isolated from DH5α(pVS45) as described previously (47). The purified Ler protein migrated as a single species when it was separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and stained by Coomassie brilliant blue. The concentration of purified Ler protein, 50 μg/ml, was estimated by comparison with a known concentration of a standard protein having a similar molecular weight (lysozyme from Sigma).
DNA mobility assays.
The LEE5 regulatory fragments were subcloned into pBluescript by using E. coli strain DH5α. Plasmid DNA was isolated with a Qiagen plasmid midi kit (Qiagen). The purified plasmid DNA was cut with NotI restriction endonuclease, which left a 5′ overhang that was subsequently filled in by using [α-32P]dCTP (Amersham), dGTP, and Klenow DNA polymerase (Promega). The plasmid DNA was then cut with EcoRI, releasing the LEE5 regulatory fragments. Radiolabeled and nonlabeled DNA fragments were separated in a 6% polyacrylamide-1× Tris-borate-EDTA (TBE) gel and were isolated by soaking the gel slice in elution buffer (0.5 M ammonium acetate, 0.1% SDS, 1 mM EDTA) overnight and precipitating the DNA with ethanol. Ler binding reactions were performed by incubating 10 μg of poly(dI-dC) (U.S. Biochemicals) per ml (to prevent nonspecific binding), 8 μg of purified Ler protein per ml, and 20,000 cpm of probe DNA in binding buffer (10 mM Tris-HCl [pH 7.4], 1 mM EDTA, 50 μg of bovine serum albumin per ml, 5 mM NaCl, 50 mM KCl) for 30 min at room temperature. A 50-fold excess of an identical unlabeled fragment was used as a specific competitor for each fragment as a control. The binding reaction mixtures were electrophoresed in a 6% polyacrylamide-1× TBE gel at 4°C, dried at 80°C for 1 h, and exposed to Kodak X-OMAT film.
PCR-generated deletions.
Overlapping PCR was performed by using oligonucleotide primers which contained deletions of the LEE5 sequence corresponding to positions −51 to −47. Primers JFTIR1 and JFTIR3 contain corresponding five-base deletions and were used with primers TIR1 and TIR3, respectively, to independently amplify LEE5 sequences. Individual PCR products were amplified, gel isolated, mixed, and used as template DNA for a secondary overlapping PCR (22), which resulted in a product that contained the 5-bp deletion. The PCR product was cloned into pRS551 and sequenced to verify the deletion.
DNase I footprint analysis.
Ler binding reactions were performed as described above, except that various amounts of Ler protein, 70,000 cpm of radiolabeled probe DNA (both coding and noncoding strands individually from positions −272 to −31 of the LEE5 regulatory region), and 100 μg of poly(dI-dC) (U.S. Biochemicals) per ml were used. The coding and noncoding strands were labeled individually by cutting the plasmid, pKH273, with either EcoRI or NotI first, followed by filling in of the EcoRI site with [α-32P]dATP or of the NotI site with [α-32P]dCTP and the Klenow fragment of DNA polymerase. The polymerase was then heat inactivated by heating the preparation at 70°C for 15 min. Subsequently, the fragments were released from the plasmid by a final restriction with the alternate enzyme NotI or EcoRI and isolated as described above. After Ler binding, 5 mU of DNase I (Sigma), MgCl2 (final concentration, 5 mM), and CaCl2 (final concentration, 1 mM) were added and incubated at room temperature for 2 min. The reaction was terminated by addition of stop solution (200 mM NaCl, 2 mM EDTA, 1% SDS), and this was followed by phenol-chloroform extraction and ethanol precipitation with 7.5 M ammonium acetate (pH 7.5) and 1 μl of glycogen (Invitrogen). The footprint reaction mixtures were denatured at 75°C for 2 min prior to loading and then were electrophoresed in a denaturing 6% polyacrylamide-1× TBE gel, dried at 80°C for 1 h, and exposed to Kodak X-OMAT film. Sequencing reaction mixtures (Sequenase DNA sequencing kit, version 2; U.S. Biochemicals) with the TF1 and BlueNot primers were loaded adjacent to the footprint reaction mixtures to determine the positions of protected sequences.
RESULTS
Deletion analysis of LEE5 regulatory sequences.
In order to define LEE5 regulatory fragments that were appropriate for Ler binding studies, sequences upstream of the LEE5 promoter were deleted (Fig. 1A). PCR amplification of LEE5 sequences from positions −405, −303, −198, and −75 to position +172 relative to the transcriptional start site of the tir gene, which is the first gene of the LEE5 operon, were fused to a promoterless lacZ reporter gene in pRS551 (45). Individual fusions were allowed to recombine with λRS45 and inserted, as single copies, into the chromosome of MC4100 (see Materials and Methods). The resulting isogenic, single-copy fusion strains were transformed with pBR322 as a negative control or pSE1100, which encodes a minimal ler fragment in pBR322 (30). The β-galactosidase activities of these strains were assayed in the presence and absence of the Ler protein expressed in trans from plasmid pSE1100. Fusion strains KMTIR2 (positions −405 to +172), KMTIR3 (positions −303 to +172), and KMTIR4 (positions −198 to 172) were all induced approximately four- to fivefold in the presence of pSE1100 (Fig. 1B). Fusion strain CRTIR5 (positions −75 to +172) exhibited high levels of β-galactosidase activity independent of ler expression. These results indicated that Ler-mediated activation of the LEE5 operon required sequences between positions −198 and −75. The observation that high levels of β-galactosidase activity were detected independent of Ler in the CRTIR5 strain was consistent with the activity of a negative regulator that represses LEE5 expression and with the hypothesis that sequences necessary for repression also reside within the same region required for activation by Ler.
FIG. 1.
Deletion analysis of the LEE5 regulatory region. (A) Schematic representation of LEE5 fragments generated by PCR for Ler binding experiments. The strains used were K-12 derived and contained single-copy, chromosomal LEE5-lacZ fusions. The first gene of the LEE5 operon is tir. The termini of each fragment in relation to the LEE5 transcriptional start site (+1) are indicated (30, 42). (B) β-Galactosidase activities derived from the single-copy, chromosomal LEE5-lacZ fusion strains shown in panel A. Plasmid pSE1100 contains a minimal ler fragment cloned into pBR322 and was described previously (30). The error bars indicate one standard deviation. The values are the means for multiple independent assays performed in triplicate. Strain KMRS551 contained the single-copy, chromosomal, promoterless lacZ fusion in MC4100 as a negative control.
DNA mobility assays with purified Ler protein.
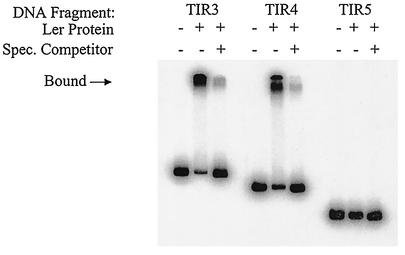
To identify sequences necessary for Ler binding, we performed mobility shift experiments with purified Ler protein (47) and radiolabeled DNA fragments corresponding to the LEE5 regulatory sequences used in the deletion analysis (Fig. 1A). As expected, DNA fragments corresponding to TIR3 and TIR4 exhibited pronounced shifts in mobility when purified Ler protein was added to the binding reaction mixture compared to the results obtained with an identical reaction mixture without the Ler protein (Fig. 2). DNA fragment TIR5, which showed no Ler-dependent activation (Fig. 1B), also exhibited no shift in mobility in the presence of purified Ler protein (Fig. 2). This observation clearly demonstrated that Ler did not bind to sequences from positions −75 to +172. These results were consistent with the fusion data in that Ler-mediated transcriptional activation was observed with the same regulatory fragments that also exhibited shifts in mobility due to Ler binding.
FIG. 2.
Mobility shift assay of LEE5 regulatory fragments. Purified Ler protein (8 μg/ml) was incubated with the radiolabeled LEE5 regulatory fragments shown in Fig. 1A and 10 μg of nonspecific competitor DNA [poly(dI-dC)] per ml. The specific competitor DNA (Spec. Competitor) consisted of a 50-fold excess of the identical fragment that was not labeled and added to the binding reaction mixture. Unbound species and bound species (indicated by the arrow) were separated in a 6% nondenaturing polyacrylamide gel and visualized by autoradiography.
To examine the specificity of Ler binding, we added nonspecific competitor DNA [unlabeled poly(dI-dC)] at a concentration of 10 μg/ml to each binding reaction mixture to minimize nonspecific interactions. In addition, a 50-fold excess of unlabeled specific competitor DNA (identical to the labeled probe DNA) was added to the reaction mixtures (Fig. 2), which resulted in specific competition. Under these conditions, the binding of Ler protein to radiolabeled TIR3 and TIR4 fragments was severely reduced (Fig. 2). We therefore concluded that the Ler protein bound specifically to sequences upstream of position −75 and acted directly to increase transcription at LEE5.
Deletion of sequences between the regulatory region and the promoter affected repression but not activation of the LEE5 operon.
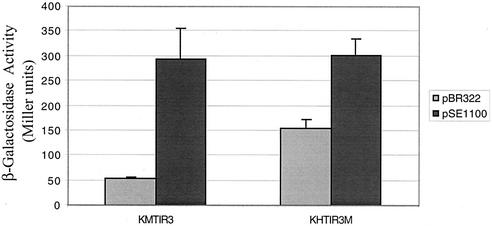
Since Ler bound at a distance from the LEE5 promoter and also the LEE2 promoter (47), we hypothesized that Ler may influence transcription by altering DNA topology. By deleting 5 bp at positions −51 to −47, we altered the spacing of the LEE5 regulatory region (positions −198 to −75) in relation to the promoter by one half-helical turn of DNA without altering the sequences required for repression or Ler-mediated activation. Strains with single-copy lacZ fusions, encoding the wild-type TIR3 regulatory fragment or the TIR3M (five-base deletion) regulatory fragment, were assayed for Ler-dependent β-galactosidase activity as described above. Strains KMTIR3 and KHTIR3M transformed with pSE1100 exhibited Ler-mediated activation of the same magnitude, ∼300 Miller units (Fig. 3). Strain KHTIR3M transformed with pBR322, as a control, exhibited threefold-higher β-galactosidase activity (155 Miller units) than strain KMTIR3 (53 Miller units) also transformed with pBR322 (P < 0.0001). These results indicated that repression of the LEE5 operon was compromised by the 5-bp deletion.
FIG. 3.
β-Galactosidase activity derived from a LEE5-lacZ fusion containing a 5-bp deletion centered at position −49. Wild-type TIR3 and TIR3M (5-bp deletion) regulatory fragments were fused to lacZ, and β-galactosidase activities were assayed. The error bars indicate one standard deviation. Construct pSE1100 described in the legend to Fig. 1B encodes a minimal ler clone that constitutively expresses Ler protein (30). The values are means for two experiments performed in triplicate.
Mobility shift experiments were performed with the TIR3M fragment and purified Ler protein. The deletion fragment TIR3M exhibited shifts in mobility in the presence of Ler protein that were identical to those exhibited by the wild-type TIR3 regulatory fragment, indicating that the five-base deletion had no effect on Ler binding (data not shown).
H-NS-dependent repression of LEE5.
As indicated previously, H-NS is involved in repression of the LEE2 and LEE3 operons (6). However, because the identity of the negative-acting factor for the LEE5 operon remained unclear, we wished to determine whether H-NS affected the expression of LEE5 as well. Therefore, we assayed β-galactosidase activities derived from LEE5-lacZ single-copy fusions in HN4104 strains having a deletion of hns and the isogenic parent strain, MC4100. The TIR3 and TIR4 fusion strains which contained all known regulatory sequences and from which hns was deleted exhibited β-galactosidase activities that were approximately fourfold greater than the β-galactosidase activities of isogenic strains with wild-type hns (60 versus 250 Miller units) (Table 3). Providing H-NS expressed from the minimal hns gene on a plasmid restored transcriptional activity to low basal levels (67 to 96 Miller units) in the TIR3 and TIR4 fusion strains from which hns was deleted. Under the conditions tested, maximal LEE5 expression was observed in the absence of H-NS and in the presence of Ler; under these conditions the β-galactosidase activities reached 1,400 to 2,000 Miller units. β-Galactosidase activities were similarly high for all of the strains containing the TIR5 fragment fused to lacZ, from which all currently known positive and negative regulatory sequences were deleted (Table 3).
TABLE 3.
Effect of hns deletion on the expression of LEE5-lacZ fusionsa
| Relevant genotypeb | Plasmid | β-Galactosidase activityc |
|---|---|---|
| MC4100 ΦTIR3-lacZ | 61 (3) | |
| MC4100 ΦTIR3-lacZ | pBR322 | 64 (3) |
| MC4100 ΦTIR3-lacZ | pSE1100 (ler+) | 307 (36) |
| MC4100 ΦTIR3-lacZ | pTHK113 (hns+) | 48 (6) |
| HN4104 ΦTIR3-lacZ (Δhns) | 225 (15) | |
| HN4104 ΦTIR3-lacZ (Δhns) | pBR322 | 252 (23) |
| HN4104 ΦTIR3-lacZ (Δhns) | pSE1100 (ler+) | 2,074 (79) |
| HN4104 ΦTIR3-lacZ (Δhns) | pTHK113 (hns+) | 67 (12) |
| MC4100 ΦTIR4-lacZ | 77 (4) | |
| MC4100 ΦTIR4-lacZ | pBR322 | 60 (5) |
| MC4100 ΦTIR4-lacZ | pSE1100 (ler+) | 581 (52) |
| MC4100 ΦTIR4-lacZ | pTHK113 (hns+) | 39 (6) |
| HN4104 ΦTIR4-lacZ (Δhns) | 243 (14) | |
| HN4104 ΦTIR4-lacZ (Δhns) | pBR322 | 318 (20) |
| HN4104 ΦTIR4-lacZ (Δhns) | pSEI100 (ler+) | 1,566 (41) |
| HN4104 ΦTIR4-lacZ (Δhns) | pTHK113 (hns+) | 96 (21) |
| MC4100 ΦTIR5-lacZ | 1,405 (307) | |
| MC4100 ΦTIR5-lacZ | pBR322 | 2,319 (91) |
| MC4100 ΦTIR5-lacZ | pSE1100 (ler+) | 1,432 (111) |
| MC4100 ΦTIR5-lacZ | pTHK113 (hns+) | 1,188 (37) |
| HN4104 ΦTIR5-lacZ (Δhns) | 1,850 (498) | |
| HN4104 ΦTIR5-lacZ (Δhns) | pBR322 | 2,176 (124) |
| HN4104 ΦTIR5-lacZ (Δhns) | pSE1100 (ler+) | 2,040 (69) |
| HN4104 ΦTIR5-lacZ (Δhns) | pTHK113 (hns+) | 1,520 (151) |
Overnight cultures of each strain were diluted 1:500 into fresh Luria-Bertani medium containing the appropriate antibiotics. Strains were grown to an optical density at 600 nm of 0.3 to 0.5 and assayed as described in Materials and Methods.
Strain genotypes are shown in Table 1.
β-Galactosidase activity is expressed in Miller units, and the standard error is indicated in parentheses. Assays were performed in triplicate with at least two independent cultures.
To ensure that H-NS activity was indeed absent from the HN4104 hns deletion strain used for our assays, phenotypes associated with this strain were confirmed prior to and after transduction of the LEE5-lacZ fusions into the strains. As expected, HN4104 and its derivatives were able to ferment salicin, presumably by expression of the cryptic bgl operon, which is known to be expressed in the absence of H-NS (34). Thus, we concluded that like the LEE2-LEE3 operon pair, H-NS was involved in the negative regulation of LEE5.
DNase I footprint analysis of purified Ler bound to LEE5 regulatory sequences.
A 241-bp fragment (positions −272 to −31) containing the LEE5 regulatory region and flanking sequences was radiolabeled in separate reactions on both the coding and noncoding strands and subjected to DNase I digestion in the presence and absence of purified Ler protein. Ler protein protected approximately 117 bp of DNA (positions −190 to −73) from digestion by DNase I (Fig. 4). Because of the low predicted molecular mass of Ler protein (15 kDa), multiple Ler proteins would be required to protect such an extended region of DNA (>100 bp). Consistently, Ler appeared to exhibit cooperative binding. At low concentrations of Ler protein (0 to 0.15 μg of Ler protein per ml), little or no protection was observed, yet when the protein level reached 0.5 μg/ml Ler protein protected an extended region of DNA. One striking feature of these footprints was that Ler protein protected a region of DNA rich in AT (A+T content, >80%).
FIG. 4.
DNase I footprint of Ler protein on the upstream LEE5 regulatory region. Concentrations of Ler protein (in micrograms per milliliter) are indicated at the top. (A) DNase I protection of the LEE5 coding strand. The Ler protein protected a region of DNA from DNase I digestion from positions −190 to −73, which is represented by an open bar. A DNase I-hypersensitive site was observed at position −111 on the coding strand and is indicated by an arrow. (B) DNase I protection of the LEE5 noncoding strand. Ler protein protected a region of DNA from positions −190 to −79, which is represented by an open bar. At higher concentrations of Ler protein, the regions of protection extended to positions −60 and −221 on the coding and noncoding strands, respectively, as indicated by shaded bars.
At high concentrations of Ler protein protection extended slightly further downstream on the coding strand (position −60) and slightly further upstream on the noncoding strand (position −221). This most likely represented weak binding and was observed only at high concentrations of Ler protein. Thus, similar to binding upstream of the LEE2 promoter (from positions −221 to −100 [47]), Ler bound between positions −190 and −73 in relation to the LEE5 transcriptional start site. A Ler-dependent DNase I-hypersensitive site was observed at position −111 on the coding strand. We concluded from this finding that the binding of Ler protein to the LEE5 regulatory region induced a structural change in the DNA that resulted in increased cleavage, specifically at position −111, by DNase I.
DISCUSSION
We demonstrated that Ler acts directly to increase transcription of the LEE5 operon. As determined by DNase I footprint analyses, purified Ler protein bound to an approximately 117-bp region of DNA (positions −190 to −73) upstream of the LEE5 transcriptional start site. As determined by a mobility shift assay, Ler protein specifically bound to sequences between positions −198 and −75, which is consistent with the lacZ fusion data (Fig. 1), with which we independently identified and confirmed a regulatory region (42) required for both H-NS-dependent repression and Ler-mediated derepression. One apparent conflict in our data was that as determined by a mobility shift assay, the TIR5 regulatory fragment (positions −75 to +172) did not bind Ler, whereas a DNase I protection assay showed that there was protection to the −60 position on the coding strand (Fig. 4A). This might be explained by the observation that Ler did not protect sequences downstream of the −79 position on the noncoding strand, and thus the binary complex (TIR5 DNA and Ler protein) was most likely too unstable to remain intact in the mobility shift assay.
Our data clearly demonstrated that H-NS is involved in the negative regulation of the LEE5 operon (Table 3). The observation that under the conditions tested, maximal activity was achieved only in the presence of Ler and in the absence of H-NS may suggest that an additional protein(s) acts negatively at LEE5. An alternate explanation for the high transcriptional activity observed in the presence of Ler and in the absence of H-NS is that greater Ler activation of LEE5 requires environmental conditions specific to conditions inside the human gut. Whether H-NS acts directly or indirectly through control of expression of an alternate negative regulator to repress LEE5 transcription remains to be determined.
There is an extensive body of work which suggests that the global regulator H-NS is a non-sequence-specific DNA-binding protein yet specifically recognizes structural features of DNA (10, 12, 50). When we aligned the upstream LEE5 and LEE2 (47) regulatory sequences to which Ler bound, we found no significant sequence similarity that allowed us to identify a Ler consensus binding sequence (data not shown). These sequences were, however, similar lengths, approximately 120 bp, and the A+T contents were high, approximately 80%. AT-rich DNA, particularly poly(A) tracts, are known to form curves (24). Based on these observations, the molecular mass of Ler (15 kDa), the amino acid sequence similarity to H-NS, the extended protection from DNase I digestion, and the likelihood that multiple Ler proteins bind upstream of LEE5 and LEE2 (47), we propose that like H-NS, Ler recognizes DNA structural motifs instead of specific nucleotide sequences.
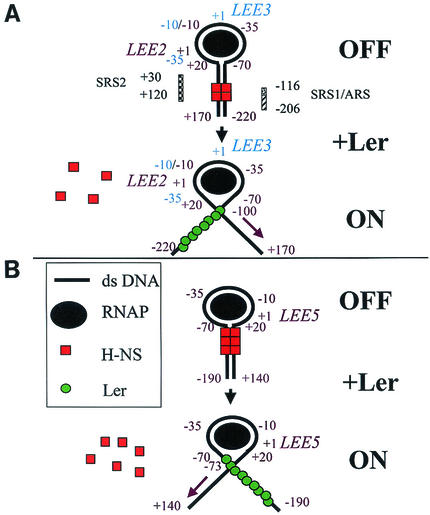
Our current model of Ler function is that multiple Ler proteins bind to the upstream regulatory regions of both LEE5 and LEE2, disrupting a nucleoprotein complex responsible for the repression of the LEE5, as well as both the LEE2 and LEE3 operons (Fig. 5). It has been proposed that in the presence of H-NS, LEE2-LEE3 transcription is repressed by the binding of H-NS to the silencing regulatory sequences, identified previously by genetic analysis (6). Thus, repression requires both upstream and downstream silencing regions (Fig. 5A). By using scanning force microscopy, it was recently demonstrated that H-NS represses the rrnB P1 promoter by forming a collar-like structure wrapping RNA polymerase in an open complex (11), and some evidence suggests that this type of repression may be a general phenomenon (9). Thus, our model includes RNA polymerase in the nucleoprotein complex, which is disrupted by the binding of Ler to the LEE2 upstream region, allowing transcription of LEE2. It is also possible, however, that RNA polymerase is not part of the repressing nucleoprotein complex and that H-NS simply occludes RNA polymerase binding. We propose that transcription of the LEE3 operon is also increased due to the absence of the repressing nucleoprotein complex.
FIG. 5.
Model for Ler action at LEE virulence operons. See the text for details. (A) LEE2-LEE3 divergent operon pair. (B) LEE5 operon. Assignment of the region of DNA contact with RNA polymerase (positions −70 to 20) was adapted from the three-step model of initiation by E. coli RNA polymerase previously described (9). Silencer regulatory sequences (SRS1 and SRS2), which were proposed to bind H-NS, and the putative attenuator regulatory sequence (ARS) (6) are shown. LEE2 and LEE5 operon regulatory positions, in relation to the starts of transcription (+1), are indicated by purple type, while LEE3 positions are indicated by blue type. Operon labels are adjacent to their cognate transcriptional start sites. The purple diagonal arrows indicate the directions of transcription for LEE2 and LEE5. ds DNA, double-stranded DNA; RNAP, RNA polymerase.
As shown in Fig. 5B, LEE5 is also negatively regulated by H-NS. It is reasonable to propose that LEE5 transcription is inhibited by the formation of a nucleoprotein complex similar to that proposed for the LEE2-LEE3 operon pair. Ler binding to the upstream region of LEE5 disrupts the H-NS-dependent nucleoprotein complex, allowing an increase in LEE5 transcription. Thus, our model begins to explain how Ler can bind on only one side of divergent operon pairs to increase transcriptional activity at both promoters, as well as single nondivergent operons which are repressed by the same protein, H-NS.
At LEE5, the upstream regulatory region (between positions −198 and −75) is required for both H-NS-dependent repression and Ler-mediated activation. KHTIR3M, the strain containing the 5-bp deletion fragment fused to lacZ, which showed significant derepression with no alteration of Ler-mediated activation or Ler binding, provides a unique opportunity to study potential topology constraints required for the nucleoprotein complex. If the five-base deletion exhibits derepression due to the one half-helical turn of DNA difference between the regulatory region and the putative downstream regulatory region of LEE5, one could then predict the transcriptional activity of multiple deletion derivatives.
H-NS is a modulator of expression of environmentally regulated genes, particularly those that respond to temperature, osmolarity, and pH (for a review see reference 3), and has been demonstrated to control the expression of virulence-associated genes in several genera, including Shigella (8, 51) and Vibrio (36, 54). The proU operon of E. coli, which is activated in response to osmotic stress, is negatively regulated by H-NS binding to sites located both upstream and downstream of the promoter (26), similar to what has been proposed for the LEE2-LEE3 operon pair (6) and LEE5. Experiments performed with a proU′-lacZ transcriptional fusion showed that Ler can neither substitute for nor exert a dominant negative effective on H-NS, demonstrating that Ler is distinct from H-NS and suggesting that Ler does not interact directly with H-NS (14). These data, combined with our observation that Ler does not derepress the expression of the cryptic bgl operon in the presence of H-NS, demonstrate that Ler is a specific regulator of virulence genes rather than a general antagonist of H-NS. Thus, we concluded that the specific regulator Ler is part of a complex regulatory network controlling the expression of EPEC virulence genes in response to environmental signals inside the human gut.
Acknowledgments
We thank James B. Kaper for an initial gift of purified Ler protein, Bianca Colonna for the gift of the hns deletion strain HN4104, and Tom Kawula for the gift of the minimal hns clone pTHK113. We thank Vanessa Sperandio and Arthur Glasfeld for helpful comments on the manuscript and Peter J. Russell and Maryanne McClellan for critical discussions. We also thank The Vollum Institute at Oregon Health Sciences University for performing the DNA sequence analysis.
This study was supported by grant R15 AI47802 awarded to J.L.M. by the National Institutes of Health and by generous start-up funds provided by Reed College, in addition to National Science Foundation and Howard Hughes Medical Institute awards to Reed College for undergraduate research.
Editor: V. J. DiRita
REFERENCES
- 1.Afflerbach, H., O. Schroder, and R. Wagner. 1999. Conformational changes of the upstream DNA mediated by H-NS and FIS regulate E. coli rrnB P1 promoter activity. J. Mol. Biol. 286:339-353. [DOI] [PubMed] [Google Scholar]
- 2.Albert, M. J., S. M. Faruque, M. Ansaruzzaman, M. M. Islam, K. Haider, K. Alam, I. Kabir, and R. Robins-Browne. 1992. Sharing of virulence-associated properties at the phenotypic and genetic levels between enteropathogenic Escherichia coli and Hafnia alvei. J. Med. Microbiol. 37:310-314. [DOI] [PubMed] [Google Scholar]
- 3.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 4.Bertin, P., N. Benhabiles, E. Krin, C. Laurent-Winter, C. Tendeng, E. Turlin, A. Thomas, A. Danchin, and R. Brasseur. 1999. The structural and functional organization of H-NS-like proteins is evolutionarily conserved in gram-negative bacteria. Mol. Microbiol. 31:319-329. [DOI] [PubMed] [Google Scholar]
- 5.Bokete, T. N., T. S. Whittam, R. A. Wilson, C. R. Clausen, C. M. O'Callahan, S. L. Moseley, T. R. Fritsche, and P. I. Tarr. 1997. Genetic and phenotypic analysis of Escherichia coli with enteropathogenic characteristics isolated from Seattle children. J. Infect. Dis. 175:1382-1389. [DOI] [PubMed] [Google Scholar]
- 6.Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664-678. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 8.Colonna, B., M. Casalino, P. A. Fradiani, C. Zagaglia, S. Naitza, L. Leoni, G. Prosseda, A. Coppo, P. Ghelardini, and M. Nicoletti. 1995. H-NS regulation of virulence gene expression in enteroinvasive Escherichia coli harboring the virulence plasmid integrated into the host chromosome. J. Bacteriol. 177:4703-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coulombe, B., and Z. F. Burton. 1999. DNA bending and wrapping around RNA polymerase: a “revolutionary” model describing transcriptional mechanisms. Microbiol. Mol. Biol. Rev. 63:457-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dame, R. T., C. Wyman, and N. Goosen. 2001. Structural basis for preferential binding of H-NS to curved DNA. Biochimie 83:231-234. [DOI] [PubMed] [Google Scholar]
- 11.Dame, R. T., C. Wyman, R. Wurm, R. Wagner, and N. Goosen. 2002. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J. Biol. Chem. 277:2146-2150. [DOI] [PubMed] [Google Scholar]
- 12.Dersch, P., K. Schmidt, and E. Bremer. 1993. Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth-phase control and autoregulation. Mol. Microbiol. 8:875-889. [DOI] [PubMed] [Google Scholar]
- 13.Donato, G. M., M. J. Lelivelt, and T. H. Kawula. 1997. Promoter-specific repression of fimB expression by the Escherichia coli nucleoid-associated protein H-NS. J. Bacteriol. 179:6618-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliot, S. J., V. Sperandio, J. A. Giron, J. L. Mellies, L. A. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliot, S. J., L. A. Wainwright, T. McDaniel, B. Macnamara, M. Konnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 16.Falconi, M., B. Colonna, G. Prosseda, G. Micheli, and C. O. Gualerzi. 1998. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 17:7033-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falconi, M., N. P. Higgins, R. Spurio, C. L. Pon, and C. O. Gualerzi. 1993. Expression of the gene encoding the major bacterial nucleoid protein H-NS is subject to transcriptional auto-repression. Mol. Microbiol. 10:273-282. [DOI] [PubMed] [Google Scholar]
- 18.Falconi, M., G. Prosseda, M. Giangrossi, E. Beghetto, and B. Colonna. 2001. Involvement of FIS in the H-NS-mediated regulation of virF gene of Shigella and enteroinvasive Escherichia coli. Mol. Microbiol. 42:439-452. [DOI] [PubMed] [Google Scholar]
- 19.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 20.Higgins, C. F., J. C. Hinton, C. S. Hulton, T. Owen-Hughes, G. D. Pavitt, and A. Seirafi. 1990. Protein H1: a role for chromatin structure in the regulation of bacterial gene expression and virulence? Mol. Microbiol. 4:2007-2012. [DOI] [PubMed] [Google Scholar]
- 21.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J. P. Le Caer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40:20-36. [DOI] [PubMed] [Google Scholar]
- 22.Innis, M. A., D. H. Gelfand, J. J. Sninsky, and T. J. White. 1990. PCR protocols. Academic Press, San Diego, Calif.
- 23.Kawula, T. H., and P. E. Orndorff. 1991. Rapid site-specific DNA inversion in Escherichia coli mutants lacking the histone-like protein H-NS. J. Bacteriol. 173:4116-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koo, H. S., H. M. Wu, and D. M. Crothers. 1986. DNA bending at adenine thymine tracts. Nature 320:501-506. [DOI] [PubMed] [Google Scholar]
- 25.Levine, M. M., E. J. Bergquist, D. R. Nalin, D. H. Waterman, R. B. Hornick, C. R. Young, and S. Sotman. 1978. Escherichia coli strains that cause diarrhea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:1119-1122. [DOI] [PubMed] [Google Scholar]
- 26.Lucht, J. M., P. Dersch, B. Kempf, and E. Bremer. 1994. Interactions of the nucleoid-associated DNA-binding protein H-NS with the regulatory region of the osmotically controlled proU operon of Escherichia coli. J. Biol. Chem. 269:6578-6588. [PubMed] [Google Scholar]
- 27.Lupas, A. 1996. Coiled coils: new structures and new functions. Trends Biochem. Sci. 21:375-382. [PubMed] [Google Scholar]
- 28.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 29.Mellies, J., R. Brems, and M. Villarejo. 1994. The Escherichia coli proU promoter element and its contribution to osmotically signaled transcription activation. J. Bacteriol. 176:3638-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 31.Mellies, J. L., F. Navarro-Garcia, I. Okeke, J. Frederickson, J. P. Nataro, and J. B. Kaper. 2001. espC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. Infect. Immun. 69:315-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 33.Moon, H. W., S. C. Whipp, R. A. Argnezio, M. M. Levine, and R. A. Gianella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukerji, M., and S. Mahadevan. 1997. Characterization of the negative elements involved in silencing the bgl operon of Escherichia coli: possible roles for DNA gyrase, H-NS, and CRP-cAMP in regulation. Mol. Microbiol. 24:617-627. [DOI] [PubMed] [Google Scholar]
- 35.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nye, M. B., J. D. Pfau, K. Skorupski, and R. K. Taylor. 2000. Vibrio cholerae H-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade. J. Bacteriol. 182:4295-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pon, C. L., R. A. Calogero, and C. O. Gualerzi. 1988. Identification, cloning, nucleotide sequence and chromosomal map location of hns, the structural gene for Escherichia coli DNA-binding protein H-NS. Mol. Gen. Genet. 212:199-202. [DOI] [PubMed] [Google Scholar]
- 38.Porter, M. E., and C. J. Dorman. 1994. A role for H-NS in the thermo-osmotic regulation of virulence gene expression in Shigella flexneri. J. Bacteriol. 176:4187-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prosseda, G., P. A. Fradiani, M. Di Lorenzo, M. Falconi, G. Micheli, M. Casalino, M. Nicoletti, and B. Colonna. 1998. A role for H-NS in the regulation of the virF gene of Shigella and enteroinvasive Escherichia coli. Res. Microbiol. 149:15-25. [DOI] [PubMed] [Google Scholar]
- 40.Robins-Browne, R. M., A. M. Tokhi, L. M. Adams, V. Bennett-Wood, A. V. Moisidis, E. O. Krejany, and L. E. O'Gorman. 1994. Adherence characteristics of attaching and effacing strains of Escherichia coli from rabbits. Infect. Immun. 62:1584-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowe, S., N. Hodson, G. Griffiths, and I. S. Roberts. 2000. Regulation of the Escherichia coli K5 capsule gene cluster: evidence for the roles of H-NS, BipA, and integration host factor in regulation of group 2 capsule gene clusters in pathogenic E. coli. J. Bacteriol. 182:2741-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez-SanMartin, C., V. H. Bustamante, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of the orf19 gene and the tir-cesT-eae operon of enteropathogenic Escherichia coli. J. Bacteriol. 183:2823-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schauer, D. B., and S. Falkow. 1993. Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infect. Immun. 61:2486-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schroder, O., and R. Wagner. 2000. The bacterial DNA-binding protein H-NS represses ribosomal RNA transcription by trapping RNA polymerase in the initiation complex. J. Mol. Biol. 298:737-748. [DOI] [PubMed] [Google Scholar]
- 45.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 46.Smyth, C. P., T. Lundback, D. Renzoni, G. Siligardi, R. Beavil, M. Layton, J. M. Sidebotham, J. C. Hinton, P. C. Driscoll, C. F. Higgins, and J. E. Ladbury. 2000. Oligomerization of the chromatin-structuring protein H-NS. Mol. Microbiol. 36:962-972. [DOI] [PubMed] [Google Scholar]
- 47.Sperandio, V., J. L. Mellies, R. M. Delahay, G. Frankel, J. A. Crawford, W. Nguyen, and J. B. Kaper. 2000. Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Mol. Microbiol. 38:781-793. [DOI] [PubMed] [Google Scholar]
- 48.Spurio, R., M. Falconi, A. Brandi, C. L. Pon, and C. O. Gualerzi. 1997. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 16:1795-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sutcliffe, J. G. 1979. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb. Symp. Quant. Biol. 43:77-90. [DOI] [PubMed] [Google Scholar]
- 50.Tippner, D., H. Afflerbach, C. Bradaczek, and R. Wagner. 1994. Evidence for a regulatory function of the histone-like Escherichia coli protein H-NS in ribosomal RNA synthesis. Mol. Microbiol. 11:589-604. [DOI] [PubMed] [Google Scholar]
- 51.Tobe, T., M. Yoshikawa, T. Mizuno, and C. Sasakawa. 1993. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by virF and repression by H-NS. J. Bacteriol. 175:6142-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tzipori, S., I. K. Wachsmuth, C. Chapman, R. Birden, J. Brittingham, C. Jackson, and J. Hogg. 1986. The pathogenesis of hemorrhagic colitis caused by Escherichia coli O157:H7 in gnotobiotic piglets. J. Infect. Dis. 154:712-716. [DOI] [PubMed] [Google Scholar]
- 53.Westermark, M., J. Oscarsson, Y. Mizunoe, J. Urbonaviciene, and B. E. Uhlin. 2000. Silencing and activation of ClyA cytotoxin expression in Escherichia coli. J. Bacteriol. 182:6347-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, R. R., and V. J. DiRita. 2002. Regulation of gene expression in Vibrio cholerae by ToxT involves both antirepression and RNA polymerase stimulation. Mol. Microbiol. 43:119-134. [DOI] [PubMed] [Google Scholar]