Abstract
The Streptococcus pneumoniae capsular polysaccharides and pneumococcal surface protein A (PspA) are major determinants of virulence that are antigenically variable and capable of eliciting protective immune responses. By genetically switching the pspA genes of the capsule type 2 strain D39 and the capsule type 3 strain WU2, we showed that the different abilities of antibody to PspA to protect against these strains was not related to the PspA type expressed. Similarly, the level of specific antibody binding to PspA, other surface antigens, and surface-localized C3b did not depend on the PspA type but instead was correlated with the capsule type. The type 3 strain WU2 and an isogenic derivative of D39 that expresses the type 3 capsule bound nearly identical amounts of antibody to PspA and other surface antigens, and these amounts were less than one-half the amount observed with the type 2 parent strain D39. Expression of the type 3 capsule in D39 also reduced the amount of C3b deposited and its accessibility to antibody, resulting in a level intermediate between the levels observed with WU2 and D39. Despite these effects, the capsule type was not the determining factor in anti-PspA-mediated protection, as both D39 and its derivative expressing the type 3 capsule were more resistant to protection than WU2. The specific combination of PspA and capsule type also did not determine the level of protection. The capsule structure is thus a major determinant in accessibility of surface antigens to antibody, but certain strains appear to express other factors that can influence antibody-mediated protection.
Streptococcus pneumoniae is a major cause of pneumonia, otitis media, and meningitis, especially in the elderly, young children, and immunocompromised individuals. The capsular polysaccharide and pneumococcal surface protein A (PspA) are important determinants of virulence for S. pneumoniae. Both molecules are surface components that exhibit a high degree of variability among strains and are capable of eliciting protective immunity. For the capsule, 90 different serotypes have been identified (20), and immunity to one capsule type is not cross-protective against other types (28). Strains expressing certain capsular serotypes appear to be better able to cause disease than others, and only a small number of serotypes account for the majority of pneumococcal infections in humans (8, 12, 15). Levels of virulence in mice have been shown to vary with the capsule type expressed, although the genetic background also plays an important role (2, 25, 26, 41, 44). The capsule inhibits phagocytosis by blocking access to cell wall-localized C3b (7, 42). Differences in virulence may be related to the ability of the specific capsular polysaccharide structure to block the accessibility of antibodies and complement to surface components or to mask cell-wall bound C3b from recognition by phagocytic receptors (6, 7, 11, 42). Hostetter demonstrated that strains with different capsular types differ in the amount and site of bound C3b, as well as the C3b degradation products, potentially affecting opsonophagocytosis (23).
PspA occurs on all S. pneumoniae strains, and although variability exists, all PspAs appear to possess a signal peptide, an α-helical N-terminal region that contains most of the immunogenic epitopes and antigenic variability, a proline-rich domain, a choline binding domain important in anchoring the protein to the bacterial surface, and a short C-terminal tail (21, 30, 31, 35, 43). PspA reduces complement activation (40) and binds lactoferrin (16). Based on reactivity with a panel of monoclonal antibodies, PspAs were originally divided into 31 serotypes (9). Subsequent analyses of the sequences encoding the N-terminal halves of the proteins resulted in assignment of PspAs to three families and further subdivision into six clades (21). In general, PspAs from one serotype can elicit protection against strains representing different capsule and PspA types, although the levels of protection may differ between strains (4, 30, 32, 34). However, PspA-elicited protection against the capsule type 2 strain D39 is poor, even when immunization is with PspA obtained from D39 derivatives (4, 30, 32, 34). These same PspAs elicit protection against a wide variety of strains, including the capsule type 3 strain WU2 (4, 32, 34). The poor protection against D39 is consistent with the observation that PspA− mutants of D39 remain virulent, whereas the absence of PspA in WU2 results in avirulence (5, 36, 47; Abeyta and Yother, unpublished data). This finding suggests that the background of the invading strain may be an important determinant of its resistance to anti-PspA-mediated protection.
Because the studies described above were based on work with strains having different genetic backgrounds, it is not clear how, or if, the capsule and PspA serotypes influence the results. Here, we examined isogenic strains to determine the effects of alterations in PspA and capsule type on the levels of anti-PspA-mediated protection and accessibility of antibodies to surface components and bound C3b. We used the capsule type 2 strain D39 and the capsule type 3 strain WU2 for comparison because of the differences in PspA-elicited protection noted above. The type 2 capsule is a branched structure with subunits comprised of a linear backbone of →4)-β-d-Glcp-(1→3)-α-l-Rhap-(1→3)-α-l-Rhap-(1→3)-β-l-Rhap-(1→and a side chain of α-d-GlcpUA-(1→6)-α-d-Glcp linked α-(1→2) from the first rhamnose in the backbone to the side chain glucose (24). The type 3 capsule is a linear repeating unit of →3)-β-d-GlcpUA-(1→4)-β-d-Glcp-(−1→] (37). Based on a single difference in reactivity with a panel of monoclonal antibodies, the D39 and WU2 PspAs are classified in different serotypes (serotypes 25 and 1, respectively) (9). The pspA sequences of the D39 derivatives Rx1 and R6 have been reported previously and exhibit complete identity (22, 43). A comparison of the predicted N-terminal halves of the WU2 and Rx1 PspA sequences showed that despite a number of differences, the PspAs were closely related and were members of the same PspA clade (21). By independently altering the capsule and PspA types in D39 and WU2, we obtained additional insights into the contributions of these surface structures to the pathogenesis of S. pneumoniae.
MATERIALS AND METHODS
Bacterial strains and genetic manipulations.
The bacterial strains used are listed in Table 1. S. pneumoniae strains were grown in Todd-Hewitt broth (Difco Laboratories, Detroit, Mich.) supplemented with 0.5% yeast extract (THY) or on blood agar base no. 2 (Difco) supplemented with 3% sheep blood in a candle jar at 37°C. Escherichia coli strains were grown in L-broth or on L-agar at 37°C. Erythromycin was used at a concentration of 0.3 μg/ml for S. pneumoniae and at a concentration 300 μg/ml for E. coli. Ampicillin was used at a concentration 50 μg/ml for E. coli. S. pneumoniae was transformed as previously described (19, 46).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmida | Derivation and propertiesb,c | Reference(s) |
|---|---|---|
| S. pneumoniae strains | ||
| D39 | Type 2 encapsulated; PspA+ (84 kDa) | 1, 33 |
| GH1003 | pGG108 × WU2; Emr; insertion downstream of pspA; PspA+ (92 kDa) | This study |
| JD611 | Nonencapsulated derivative of WU2; Cps3D−; PspA+ (92 kDa) | 10 |
| JD770 | Type 3 encapsulated derivative of WU2 containing insertion-duplication of cps3DSU; Emr; PspA+ (92 kDa) | 10 |
| JD803 | JD770 × D39; Emr; type 3 encapsulated; PspA+ (84 kDa) | 25 |
| JD804 | Same as JD803; independent derivative | 25 |
| JY1123 | pKSD300 × WU2; Emr; PspAtr (46 kDa) | 45 |
| MA101 | pJY4173 × D39; Emr; insertion downstream of pspA; PspA+ (84 kDa) | This study |
| MA139 | GH1003 × D39; Emr; PspA+ (92 kDa) | This study |
| MA158 | Same as MA139; independent derivative | This study |
| MA200 | WG44.1 × R36A; Emr; PspA− | This study |
| MA1024 | MA101 × WU2; Emr; PspA+ (84 kDa) | This study |
| MA1025 | Same as MA1024; independent derivative | This study |
| R36A | Nonencapsulated derivative of D39; PspA+ (84 kDa) | 1 |
| Rx1 | Avirulent derivative of D39; PspA+ (84 kDa) | 33, 39 |
| WG44.1 | PspA− derivative of Rx1 (ΔpspA); Emr | 36, 45 |
| WU2 | Type 3 encapsulated, virulent; PspA+ (92 kDa) | 3, 33 |
| Plasmids | ||
| pGG103 | pJY4163::1.8-kb Sau3AI-KpnI fragment containing 3′ region of Rx1/WU2 pspA | This study |
| pGG108 | pJY4163::1.2-kb SacI-KpnI fragment containing WU2 pspA 3′ region | This study |
| pGH160 | pUC18::1.2-kb SphI-SalI containing WU2 pspA 5′ region; encodes PspA46 | This study |
| pJY4163 | Vector for insertion-duplication mutagenesis; replicates in E. coli, not in S pneumoniae; Emr | 45 |
| pJY4173 | pJY4163 containing 3′ region of Rx1 pspA | 45 |
| pJY4306 | pUC18::1.0-kb HindIII-DraI fragment (Rx1 pspA′ bp 1-997); encodes PspA42 | 45 |
| pJY4310 | pUC18::HindIII-SacI (Rx1 pspA′ bp 1-790); encodes PspA30 | 45 |
| pKSD300 | pVA891::0.55-kb internal Sau3AI fragment of Rx1 pspA | 36, 43 |
| pRS102 | HindIII self-ligated fragment from JY1123 (in pVA891); C-terminal coding region of WU2 pspA; Emr | This study |
| pVA891 | Vector for insertion-duplication mutagenesis; replicates in E. coli, not in S. pneumoniae; Cmr (E. coli only); Emr | 29 |
Strains MA139, MA158, MA1024, MA1025, JD803, and JD804 are each the result of at least three backerosses with the parental recipients.
Crosses were done by transformation.
The apparent molecular masses of PspA (in parentheses) were determined by SDS-PAGE and Western immunoblotting. PspA+, full-length PpA; PspAtr, truncated PspA.
The C-terminal coding region of WU2 pspA was targeted and cloned essentially as described previously for Rx1 pspA (45). The WU2 derivative JY1123 was constructed by transformation and insertion of pKSD300, which harbors an internal 550-bp fragment from the 5′ half of Rx1 pspA in pVA891(Em) (36, 43, 45). Chromosomal DNA from JY1123 was digested with HindIII, self-ligated, and transformed into E. coli, with selection for erythromycin-resistant transformants. The resulting plasmid, pRS102, contained the C-terminus-encoding two-thirds of WU2 pspA and flanking DNA. A 1.8-kb BamHI/KpnI fragment containing the 3′ half of WU2 pspA was subcloned into pJY4163 to generate pGG103, and subclones of this plasmid were used to obtain the WU2 pspA sequence. Sequencing was performed by the UAB Automated Sequencing Core Facility (University of Alabama at Birmingham, Birmingham) or by the Sanger dideoxy method (38) by using a Sequenase 2.0 kit (Amersham Life Sciences, Arlington Heights, Ill.). Plasmid and chromosomal DNAs were isolated by using Qiagen columns (Qiagen, Chatsworth, Calif.).
For construction of D39 derivatives that expressed the WU2 PspA, D39 was transformed with chromosomal DNA from GH1003, a WU2 derivative carrying an erythromycin resistance marker downstream of pspA. Erythromycin-resistant transformants were selected and backcrossed three times with the parental recipient strain. Two independently obtained derivatives, MA139 and MA158, were used in the subsequent studies. A similar procedure was used to introduce D39 pspA into WU2. The donor for this construction was MA101, and the independent derivatives obtained were MA1024 and MA1025. Switching of the pspA sequences was confirmed by Southern and Western analyses, as described below.
Transcription analyses.
RNA was isolated from S. pneumoniae by using the FastPrep system (Bio 101). For Northern analyses, RNA was denatured, electrophoresed through a 1% agarose-2.2 M formaldehyde gel, transferred to a Duralon-UV membrane (Stratagene), hybridized, and developed by using the Genius system (Boehringer Mannheim Corp., Indianapolis, Ind.). DNA probes were made as described below for Southern analyses. The pspA probe was generated by incorporation of a labeled nucleotide during PCR amplification of the pJY4310 insert.
Primer extension was performed by a modification of a previously described method (27). Briefly, 20 μg of RNA was mixed with 5 ng of a 5′-biotinylated pspA primer (5′-TAGCGACGCTGGCTAGACTTG-3′) in hybridization buffer (200 mM Tris hydrochloride [pH 8.0], 1 M NaCl, 1 mM disodium EDTA) in a 10-μl (final volume) reaction mixture and heated at 100°C for 2 min and then at 40°C for 2 h. After the mixture was cooled to room temperature, 5 μl of 2× reverse transcriptase buffer (0.1 M Tris hydrochloride [pH 8.2], 12 mM MgCl2, 20 mM dithiothreitol, 0.4 mM dATP, 0.4 mM dTTP, 0.4 mM dGTP, 0.4 mM dCTP), 0.2 μl of actinomycin D (5-mg/ml stock), 10 U of RNasin, and 5 U of avian myeloblastosis virus reverse transcriptase (Promega) were added. After incubation at 42°C for 1 h, 10 μl of sequencing dye was added to stop the reaction, and the tubes were stored at −20°C. Four microliters of each reaction mixture was electrophoresed on a 5% polyacrylamide sequencing gel along with a sequencing ladder from pspA generated with the same primer by using pJY4306 and pGH160 as templates. Primer extension products were visualized by exposure to X-ray film after chemiluminescent detection with the Plex system (NEB).
Southern blot analysis.
Chromosomal DNA was digested with HindIII and KpnI, electrophoresed through 0.8% agarose, and transferred to Duralon-UV membrane by using a Possiblot pressure blotter (Stratagene, La Jolla, Calif.). The pspA-specific probe was generated by PCR by using primers specific for the pspA leader sequence and 3′ transcription termination site (5′-GTCTCAGCCTACTGTTGT-3′ and 5′-GGGGTACCTAAAGAATACG-3′, respectively) (43). Taq polymerase (Fisher Scientific, Pittsburgh, Pa.) was used for PCR amplification. The probe was labeled with digoxigenin-dUTP, and blots were developed by using the DIG/Genius system (Boehringer Mannheim).
Western immunoblot analyses of PspA.
S. pneumoniae samples were prepared as previously described (45). Equivalent amounts of all samples (based on cell number) were electrophoresed in sodium dodecyl sulfate (SDS)-10% polyacrylamide gels. Western immunoblotting and processing were performed as previously described (45). The PspA-specific monoclonal antibody Xi126 was provided by David Briles (University of Alabama at Birmingham).
Generation of PspA-specific polyclonal antiserum and protection studies.
BALB/cByJ mice, obtained from Jackson Laboratory, Bar Harbor, Maine, were immunized by subcutaneous injection with 1 μg of PspA purified from strain R36A as previously described (48) and emulsified in complete Freund's adjuvant. Control serum was obtained by immunization with an equivalent fraction from the PspA− R36A derivative MA200. After 14 days, the mice were boosted by intraperitoneal injection by using the same amount of antigen without adjuvant. Seven days later, blood was collected, and the serum was pooled and stored at −85°C. The serum was examined for reactivity with PspA in a Western immunoblot, as well as by an indirect enzyme-linked immunosorbent assay (ELISA), as previously described (34).
In protection studies, 8- to 10-week-old BALB/cByJ mice were passively immunized by intraperitoneal injection with 20 μl of PspA-specific polyclonal serum or control serum diluted in 200 μl of sterile lactated Ringer's solution. After 1 h, the mice were challenged by intravenous injection with 107 CFU of pneumococci. Survival was monitored for 21 days, and the results were compared by using Fisher's exact test.
Antibody binding and complement assays.
Indirect ELISAs to detect binding of antibodies to surface antigens were performed essentially as described previously (18). S. pneumoniae cultures were grown to mid-exponential phase (∼3 × 108 CFU/ml) in THY, heat killed by incubation at 65°C for 20 min, and centrifuged (14,000 × g, 20 min). The pellets were washed once and resuspended in the original culture volume of phosphate-buffered saline (PBS) (140 mM NaCl, 3 mM KCl, 5 mM Na2HPO4, 2 mM KH2PO4; pH 7.4). The volumes were then adjusted so that the optical densities (OD600) of all samples were equivalent. Microtiter plates were coated overnight at 4°C with 50-μl portions of samples for use in ELISAs. The PspA-specific polyclonal antiserum described above (diluted 1:1,000) was used to detect PspA. For detection of other surface antigens, a polyclonal antibody specific for serotype 19 (typing serum obtained from Statens Seruminstitut, Copenhagen, Denmark; diluted 1:5,000) was used. Because whole cells are used as immunogens for this antiserum and because of the poor immunogenicity of the type 19 capsule, the majority of the antibodies are reactive with noncapsular antigens (18). The antiserum can thus be used to detect general exposure of surface antigens to antibody. The results of the antibody binding assays were normalized to the binding observed with JD611, a nonencapsulated derivative of the type 3 strain WU2. The anti-type 19 polyclonal antiserum reacts with JD611 at antiserum dilutions of >2 × 105 (18).
The amount of type 3 capsule on the cell surface does not affect binding of the bacteria to a microtiter plate (18). Likewise, strains expressing type 2 and type 3 capsules bound equivalently to microtiter plates. Two methods were used to determine the latter result. First, heat-killed bacteria (∼3 × 108 CFU/ml) were fluorescein labeled (13) by incubation in a 0.05 M sodium carbonate-0.1 M NaCl buffer containing 1 mg of fluorescein (Sigma Chemical Co., St. Louis, Mo.) per ml for 30 min at room temperature. The suspensions were centrifuged, washed, and resuspended in PBS; serial dilutions were then coated on microtiter plates. Following incubation overnight at 4°C, the plates were washed, and the percentage of bacteria bound to each plate was determined by dividing the amount of fluorescence bound by the total fluorescence in the coating sample. Fluorescence was determined by using a Victor2 multilabel counter (Perkin-Elmer) at excitation and emission wavelengths of 485 and 535 nm, respectively. Comparable levels of binding were observed for WU2, D39, and JD803. As a second test, the binding assays were carried out in suspension. Live and heat-killed bacteria were incubated with antisera (as described above) and live bacteria were incubated with complement (as described below). Samples were then centrifuged, washed, and resuspended in buffer containing the secondary antibody. Following this incubation, samples were centrifuged, washed, and resuspended in buffer containing the detection substrate. The results were normalized to the OD595 of the final suspension. The results were comparable to those obtained by using bacteria bound to microtiter plates.
Complement assays were performed by a modification of the method of Gordon et al. (14). For opsonization, serum from BALB/cByJ or CBA/N mice was pooled and stored at −85°C. Bacteria were grown in THY to a concentration of ∼3 × 108 CFU/ml. Cells in 0.5-ml samples were harvested by centrifugation, washed once in gelatin Veronal buffer (Sigma), and resuspended in 100 μl of gelatin Veronal buffer. Serum was added to the bacterial suspension at a level that was 10% of the final volume, and opsonization was carried out for 30 min at 37°C. Bacteria were then washed three times in PBS and resuspended in 100 μl of PBS, and 50 μl of a bacterial suspension containing ∼2 × 107 CFU of bacteria was added in duplicate to 96-well microtiter plates (Falcon; Becton Dickinson, Oxnard, Calif.). The wells were coated overnight at 4°C, rinsed three times with PBS containing 0.05% Tween 20, and blocked for 1 h at room temperature with 1% bovine serum albumin in PBS. Samples were incubated at room temperature for 1 h with 50 μl of peroxidase-conjugated goat antiserum to C3 (anti-C3-HRP; Cappel Research Products, ICN Biomedicals, Costa Mesa, Calif.) diluted 1:500 in 1% bovine serum albumin in PBS. After three washes with PBS containing 0.05% Tween, color development with 2,2′-azino-di-(3-ethylbenzthiazolinesulfonic acid) (ABTS) peroxidase substrate (Fisher) and H2O2 was measured at OD415. Nonspecific binding was determined by incubation of bacteria in heat-inactivated (30 min, 56°C) serum. The amounts of bound C3 were calculated relative to the amount of a normal mouse standard consisting of serum amyloid P component (Calbiochem, La Jolla, Calif.) coated on ELISA wells and processed as described above. For determination of the total C3 bound, 20-μl suspensions containing bacteria opsonized as described above were separated by SDS-polyacrylamide gel electrophoresis (PAGE) (4 to 15% polyacrylamide gradient gels) and transferred to nitrocellulose. C3 was detected by using anti-C3-HRP diluted 1:1,000 and development with SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, Ill.). The ratios of total C3 binding were determined by densitometric analysis of the immunoblot. For both the accessibility and total C3 binding assays, comparable results were obtained with BALB/cByJ and CBA/N mouse sera. The latter serum contains an X-linked immunodeficiency that results in poor antibody responses to polysaccharide antigens and low levels of intrinsic antibody to the phosphocholine component of the pneumococcal cell wall (3).
In studies examining the effect of antibody on opsonization, bacteria (∼2 × 107 CFU) were incubated with heat-inactivated (30 min, 56°C) PspA-specific polyclonal serum or control serum diluted 1/10 in 100 μl (final volume) of PBS. After incubation for 30 min on ice, the suspension was centrifuged, and cells were washed three times with PBS prior to opsonization as described above.
Nucleotide sequence accession numbers.
The WU2 and Rx1 pspA nucleotide sequences have been deposited in the GenBank database under accession numbers AF516671 and M74122, respectively.
RESULTS
Characterization of PspA.
Analysis of the C-terminal half of the predicted WU2 PspA sequence revealed a small number of differences with the sequence of PspA from the D39 derivative Rx1. In both, the proline-rich domain contains two segments, proline A and proline B, separated by a short spacer sequence. Both proline regions are larger in WU2 than in Rx1 (Fig. 1). The choline-binding domain of Rx1 PspA contains 10 nearly identical repeats of 20 amino acids each. The 9th and 10th repeats differ slightly from the other repeats, and an additional amino acid is present in the 9th repeat (Fig. 1) (43). This same pattern occurs in WU2 PspA. Additionally, repeats 7 through 10 in WU2 PspA are effectively a duplication of repeats 3 through 6 (Fig. 1). The C-terminal tail in both Rx1 PspA and WU2 PspA is 17 amino acids long, and only two residues differ (Fig. 1).
FIG. 1.
Comparison of the predicted sequences of the C-terminal halves of the Rx1 and WU2 PspAs. The sequence begins at the proline A domain (amino acid 289 of the mature Rx1 PspA) and continues to the end of the protein. The individual sequences are contiguous throughout, with spacing added for clarity of alignment. Amino acids in the WU2 sequence that are identical to those in the Rx1 sequence are indicated by dots.
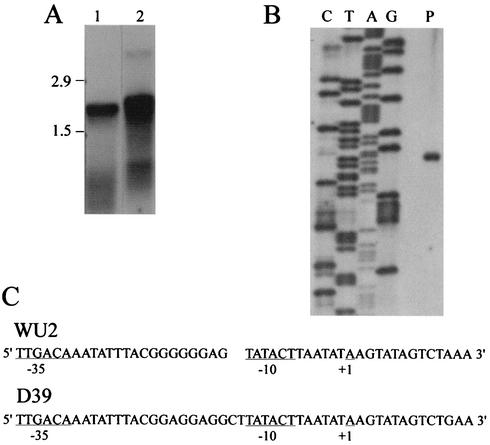
To aid in genetically switching the D39 and WU2 pspA sequences, an erythromycin resistance marker was integrated downstream of the putative pspA transcription terminator (43), as described below. To determine whether the insertions might cause polar effects on downstream genes, we performed Northern and primer extension analyses to determine the sizes of the transcripts containing pspA. Northern blotting revealed pspA-specific transcripts of approximately 1.9 and 2.0 kb for Rx1 and WU2, respectively (Fig. 2A). These sizes are consistent with those predicted for monocistronic transcripts and reflect the slightly larger size of WU2 PspA (see below). As previously reported for WU2 (17), the amount of the pspA transcript was constant from early exponential phase through early stationary phase (data not shown). Using primer extension analysis, we located the site of transcription initiation for the WU2 (Fig. 2B) and D39 (data not shown) pspA genes. For both genes, initiation occurred 7 nucleotides downstream of the −10 site (at an A residue), thus confirming the location of the putative promoter sequence (Fig. 2C).
FIG. 2.
Transcription of pspA. (A) Northern analysis. Lanes were loaded with 20 μg of RNA and probed with a pspA-specific DNA probe. Lane 1, Rx1; lane 2, WU2. (B) Primer extension analysis of WU2. Nucleotides are indicated above the lanes. The ladder reads 5′ to 3′ from bottom to top. The initiating nucleotide was confirmed by spiking the A sequencing reaction mixture with the primer extension reaction mixture. Lane P contained the primer extension product. (C) Nucleotide sequences of the WU2 and D39 pspA promoter regions. The transcription initiation site is indicated (+1). The D39 sequence contains two additional nucleotides in the spacer region between the −35 and −10 sequences. A gap was therefore introduced into the WU2 sequence for clarity of alignment.
Effect of alterations in PspA and capsular serotype on anti-PspA-mediated protection.
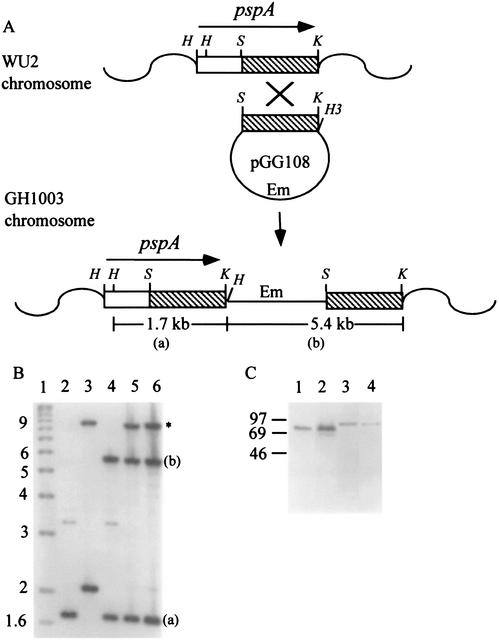
Derivatives of the capsule type 2 strain D39 and the capsule type 3 strain WU2 in which the pspA sequences were exchanged were constructed by transforming the recipient strain with donor chromosomal DNA carrying an insertion downstream of pspA, located at a KpnI site approximately 50 bp downstream of the pspA transcription terminator. Following selection for the erythromycin resistance marker carried on the insertion and three additional backcrosses, replacement of the recipient pspA with pspA of the donor was confirmed by Southern analyses, in which differences in restriction fragment sizes between the two pspA sequences were apparent. The procedure used for construction of D39 derivatives carrying WU2 pspA and the results of the subsequent Southern blot analysis are shown in Fig. 3A and B. A similar procedure and confirmation analysis were used for construction of WU2 derivatives carrying D39 pspA (data not shown). The D39 and WU2 PspAs differ in size, and this difference in the derivatives was confirmed by Western immunoblot analyses (Fig. 3C). Because D39 and WU2 differ in the time required to kill BALB/cByJ mice following intravenous infection (25), we examined the virulence of the derivatives administered by this route. None of the derivatives differed from the corresponding parent strain, indicating that the insertions downstream of pspA did not affect virulence and the differences in virulence between D39 and WU2 are not attributable to PspA (data not shown).
FIG. 3.
Construction and characterization of derivatives expressing altered PspA serotypes. (A) Construction of WU2 derivatives carrying an erythromycin resistance insertion downstream of pspA. WU2 was transformed with pGG108, which contains the 3′ 1.2 kb (▧) of WU2 pspA. Recombination resulted in insertion of the plasmid immediately downstream of pspA, with duplication of the 1.2-kb fragment (GH1003 chromosome). Chromosomal DNA from this strain was used to transform WU2 pspA into D39, with selection for erythromycin resistance following recombination in homologous sequences flanking pspA. The positions of HindIII (H), SacI (S), and KpnI (K) restriction sites are indicated. (a) and (b) indicate restriction fragments shown in the Southern blot in panel B. (B) Southern blot analysis. HindIII-KpnI-digested chromosomal DNA was hybridized with a full-length pspA-specific probe, as described in Materials and Methods. The complete D39 pspA gene (lane 3) is present on a single 2.1-kb HindIII-KpnI fragment (43). The 8.9-kb band (indicated by an asterisk) in lanes 3, 5, and 6 results from hybridization of the probe with the closely related pspC gene, which is present in D39 but not WU2 (35). WU2 pspA (lane 2) contains an internal HindIII site, as shown in panel A. Not shown is the 350-bp HindIII fragment, which was detected in WU2 and all derivatives containing WU2 pspA. Lane 1, kilobase ladder; lane 2, WU2; lane 3, D39; lane 4, GH1003 (WU2 with insertion downstream of pspA); lane 5, MA139 (D39 with WU2 pspA); lane 6, MA158 (D39 with WU2 pspA). (C) Western immunoblot analysis of cell lysates from derivatives expressing an altered PspA serotype. Blots were reacted with the PspA-specific monoclonal antibody Xi126. The positions of molecular mass standards (in kilodaltons) are indicated on the left. Lane 1, D39; lane 2, MA1025 (WU2 derivative expressing D39 PspA); lane 3, WU2; lane 4, MA139 (D39 derivative expressing WU2 PspA).
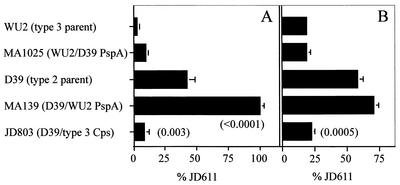
To examine the effect of alterations in PspA type on the protective ability of antibody to PspA, mice passively immunized with PspA-specific or control serum were challenged intravenously with the derivatives or their parents. The PspA-specific antiserum was obtained by using PspA isolated from R36A, a nonencapsulated derivative of D39. The control serum was obtained by immunizing mice with a similarly isolated fraction from a PspA-negative derivative of R36A. As shown in Table 2, mice challenged with the type 3 strain WU2 or its derivative expressing D39 PspA (MA1025) were fully protected from fatal infection. When mice were challenged with the type 2 strain D39 or its derivatives expressing WU2 PspA (MA139 and MA158), however, there was no significant difference in survival between mice that received PspA-specific antibody and mice that did not. Thus, the specific PspA type is not responsible for the differences in anti-PspA-mediated protection.
TABLE 2.
Protection of BALB/cByJ mice passively immunized with anti-PspA serum
| Challenge strain | Capsule serotype | PspA serotype | No. of mice alive/ no. totalb
|
Pc | |
|---|---|---|---|---|---|
| Without antibody | With antibody | ||||
| WU2 | 3 | 1 | 0/12 | 12/12 | <0.0001 |
| MA1025 | 3 | 25 | 0/5 | 5/5 | 0.0079 |
| D39 | 2 | 25 | 4/14 | 6/14 | NS |
| MA139/MA158a | 2 | 1 | 3/14 | 6/14 | NS |
| JD803/JD804a | 3 | 25 | 6/14 | 7/14 | NS |
Data was combined for the two independently obtained derivatives, which did not differ from each other.
Mice were infected intravenously with 107 CFU 1 h after intraperitoneal immunization with anti-PspA polyclonal antiserum (with antibody) or a control antiserum lacking PspA-specific antibody (without antibody).
Calculated by using Fisher's exact test. NS, not significant.
To test the effect of alteration in capsule type on anti-PspA-mediated protection, we used the D39 derivatives JD803 and JD804, in which the type 2 capsule locus was replaced with the WU2 type 3 capsule locus. The amount of type 3 capsule produced by these strains is equivalent to the amount produced by WU2, but the virulence remains the same as that of D39 (25). These derivatives were as resistant to anti-PspA-mediated protection as D39 was (Table 2). Thus, capsule type alone does not determine the level of protection. In addition, the WU2 derivative expressing D39 PspA (MA1025) and the D39 type 3 derivatives (JD803 and JD804) had identical PspA and capsule serotypes yet had very different levels of resistance to anti-PspA-mediated protection. Thus, the specific combination of PspA and capsule type of the invading strain also does not determine resistance to protection. The effect of expressing the type 2 capsule in the type 3 background was not tested in these assays because such derivatives produce only small amounts of type 2 capsule and exhibit severely reduced virulence (unpublished data).
Analysis of antibody binding to PspA and surface antigens.
A possible explanation for the results described above is that antibody binding to PspA varies with the background strain in a manner that is not dependent on capsule type. We therefore compared binding of the polyclonal PspA-specific antiserum to strains D39 and WU2. In contrast to what might have been expected from the protection results, a higher level of binding was observed with D39 than with WU2 (Fig. 4A). Expression of D39 PspA in WU2 (MA1025) resulted in binding similar to that observed for the parent strain WU2, but the D39 derivative expressing WU2 PspA (MA139) bound more than twice as much antibody as the parent strain D39 (Fig. 4A). For the D39 derivative expressing the type 3 capsule (JD803), binding was similar to that observed with the WU2 (type 3) background strains.
FIG. 4.
Reactivity with polyclonal antisera to PspA and surface antigens. The data are the averages for at least three ELISA experiments in which plates were coated with whole cells and reacted with either PspA-specific antiserum (A) or antiserum reactive with noncapsular surface antigens (B). For MA1025, MA139, and JD803, the results for two independently obtained derivatives, which did not differ from each other, were combined. Binding was calculated relative to the binding of the capsule-negative strain JD611, which was defined as 100%. The results were compared by using Student's t test. P values for MA139 and JD803 are shown in parentheses for comparisons with D39. MA1025 and JD803 were not significantly different from WU2. The difference in antibody binding for D39 and WU2 was significant in both assays (P = 0.003 and P = 0.0009 for panels A and B, respectively).
The overall accessibility of the pneumococcal surface to antibody was assessed by using a polyclonal antiserum that reacted with surface antigens. The binding of this antiserum was generally similar to the binding observed with the PspA-specific antiserum; i.e., regardless of the genetic background, strains expressing the type 3 capsule bound less antibody than strains expressing the type 2 capsule (Fig. 4B). Switching the PspA types did not significantly affect the binding. Taken together, the binding results suggest that accessibility of PspA and the entire pneumococcal cell surface is dependent upon the capsule type, but differences in accessibility between strains do not account for differences in anti-PspA protection levels.
Effects of PspA serotype, capsular serotype, and PspA-specific antibody on complement binding.
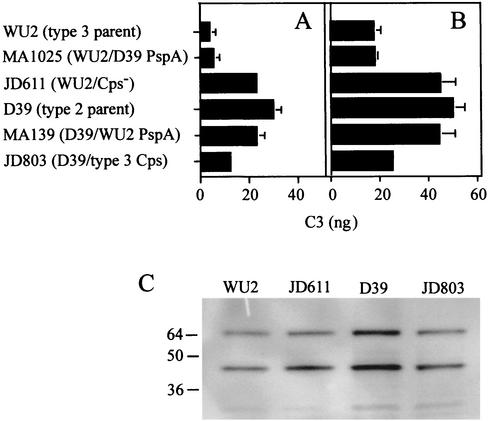
Levels of accessible surface-bound C3b (and its degradation products) resulting from opsonization in mouse serum were measured by using C3-specific antiserum. More C3b was detectable on D39 than on WU2, and switching the WU2 and D39 PspAs did not alter these results (Fig. 5A). Alteration of the capsule type did have an influence, and the amount of detectable C3b on the D39 derivative expressing the type 3 capsule was intermediate between the amounts observed for two parents (Fig. 5A). The nonencapsulated D39 derivative R36A bound approximately 3.5-fold more anti-C3 than the WU2 nonencapsulated derivative JD611, indicating the importance of the genetic background in complement activation and/or deposition (data not shown). Incubation with PspA-specific antiserum enhanced complement fixation by all of the strains (Fig. 5B). Again, however, less C3b was detectable on strains expressing the type 3 capsule than on strains expressing the type 2 capsule.
FIG. 5.
Detection of complement bound to D39, WU2, and derivatives. (A and B) C3 bound to bacteria after opsonization in BALB/cByJ serum, detected by ELISA, as described in Materials and Methods. Comparable results were obtained following opsonization in CBA/N serum. The data are the averages for at least three experiments. For MA1025, MA139, and JD803, the data for two independently obtained derivatives were combined. The results were compared by using Student's t test. (A) Accessible surface-bound C3 after incubation in serum. MA1025 and MA139 were not significantly different from parent strains WU2 and D39, respectively. JD803 was significantly different from both WU2 and D39 (P = 0.02 and P = 0.002, respectively). WU2 and D39 were significantly different (P = 0.006). (B) Accessible surface-bound C3 after opsonization with the PspA-specific polyclonal antiserum followed by incubation in BALB/cByJ serum. JD803 was significantly different from D39 (P = 0.007) but not from WU2. D39 and WU2 were significantly different (P = 0.004). (C) Total bound C3. Equivalent numbers of bacteria opsonized in CBA/N serum were separated by SDS-PAGE and immmunoblotted by using peroxidase-conjugated anti-C3. Comparable results were obtained following opsonization in BALB/cByJ serum. The bands represent the C3b β-chain (65 kDa) and iC3b α-chain (46 kDa) fragments (40).
To determine whether the amount of C3b accessible to antibody was a reflection of the amount of C3b deposited, opsonized bacteria were examined by using SDS-PAGE and immunoblotting with the C3-specific antiserum. Consistent with the accessibility assay, more C3b products were present on D39 than on the other strains (Fig. 5C). However, the differences in accessibility were not completely accounted for by differences in total C3b binding (i.e., for WU2, JD611, D39, and JD803, the relative levels of total C3b binding were approximately 1:1.5:2.1:1.4, whereas those of accessible C3b were approximately 1:6.1:7.7:3.2). The capsule, its structure, and the genetic background thus appear to influence both the total amount of complement deposited and its accessibility to antibodies.
DISCUSSION
The reasons for differences in virulence between S. pneumoniae strains are likely multifactorial. We previously observed that the effect of expression of the type 3 capsule on the virulence of recipients with different genetic backgrounds was unpredictable and could result in avirulence, enhanced virulence, or no change in virulence (25). Specific combinations of factors therefore appear to play an important role. Expression of the type 3 capsule in D39 did not alter the virulence of this strain (25), but as shown here, it decreased the ability of antibodies to bind surface components, including PspA and surface-localized C3b, and affected the amount of C3b deposited on the cell. Whether this effect is due solely to the alteration in capsule structure or is dependent on other factors present in the D39 background is not known. However, antibody binding to surface components of the D39 type 3 derivatives was observed at levels almost identical to the levels observed for WU2 and its derivatives that expressed the type 3 capsule, suggesting that the capsule structure itself is the major determinant of surface exposure.
Despite the binding of higher levels of antibody to PspA and surface-localized C3b by D39, this strain is much more resistant to anti-PspA-mediated protection than WU2. Neither the capsule type nor the PspA type appeared to be solely responsible for the resistance to protection, suggesting that strains such as D39 possess other factors that can impede antibody-mediated protection. Possibly, the C3b deposited on the D39 surface is localized at a site that is poorly accessible to phagocytic C3b receptors despite being available for antibody binding. Alternatively, the results could reflect differences between the in vitro assay and the in vivo situation. Whereas the virulence of PspA-negative mutants of WU2 is significantly reduced, mutants of D39 remain highly virulent (5, 36, 47; Abeyta and Yother, unpublished). PspA thus appears to be less important in D39 virulence, and reduced levels of PspA or reduced exposure on the surface during animal infections could explain both the minimal effect of mutations on virulence and the poor protection seen with antibody to PspA. Conversely, in vivo alterations in expression of capsule or other factors in the WU2 background may account for its sensitivity to protection.
Both capsule and PspA play important roles in resistance to complement. The capsule blocks access to cell wall-bound C3b (7, 42; this study) and also appears to affect the amount of C3b deposited. PspA likewise reduces the amount of complement bound (40). Alteration of the PspA types examined in this study did not affect the function of PspA or the amount of C3b detected on the cell surface, suggesting that the functional domains may be conserved between PspA types. In contrast, the type 3 capsule was more effective than the type 2 capsule at blocking antibody access to surface antigens and surface-localized C3b. The structure of the capsular polysaccharide thus appears to play a major role in its ability to block accessibility to the pneumococcal surface.
Acknowledgments
This study was supported by Public Health Service grants AI28457, F31 AG05699, T32 HL07553, and T32 AI07041 from the National Institutes of Health.
We thank Chuck Turnbough for advice regarding the primer extension analyses.
Editor: E. I. Tuomanen
REFERENCES
- 1.Avery, O. T., C. M. MacLeod, and M. McCarty. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 79:137-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briles, D. E., M. J. Crain, B. M. Gray, C. Forman, and J. Yother. 1992. Strong association between capsular type and virulence for mice among human isolates of Streptococcus pneumoniae. Infect. Immun 60:111-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briles, D. E., M. Nahm, K. Schoroer, J. Davie, P. Baker, J. Kearney, and R. Barletta. 1981. Antiphosphocholine antibodies found in normal mouse serum are protective against intravenous infection with type 3 Streptococcus pneumoniae. J. Exp. Med. 153:694-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briles, D. E., R. C. Tart, E. Swaitlo, J. P. Dillard, P. Smith, K. A. Benton, B. A. Ralph, A. Brooks-Walter, M. J. Crain, S. K. Hollingshead, and L. S. McDaniel. 1998. Pneumococcal diversity: considerations for new vaccine strategies with emphasis on pneumococcal surface protein A (PspA). Clin. Microbiol. Rev. 11:645-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briles, D. E., J. Yother, and L. S. McDaniel. 1988. Role of pneumococcal surface protein A in the virulence of Streptococcus pneumoniae. Rev. Infect. Dis. 10:S372-S374. [DOI] [PubMed] [Google Scholar]
- 6.Brown, E. J., S. W. Hosea, C. H. Hammer, C. G. Burch, and M. M. Frank. 1982. A quantitative analysis of the interactions of antipneumococcal antibody and complement in experimental pneumococcal bacteremia. J. Clin. Investig. 69:85-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, E. J., K. A. Joiner, R. M. Cole, and M. Berger. 1983. Localization of complement component 3 on Streptococcus pneumoniae: anti-capsular antibody causes complement deposition on the pneumococcal capsule. Infect. Immun 39:403-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler, J. C., R. F. Breiman, H. B. Lipman, J. Hofmann, and R. R. Facklam. 1995. Serotype distribution of Streptococcus pneumoniae infections among preschool children in the United States, 1978-1994: implications for development of a conjugate vaccine. J. Infect. Dis. 171:885-889. [DOI] [PubMed] [Google Scholar]
- 9.Crain, M. J., W. D. Waltman II, J. S. Turner, J. Yother, D. K. Talkington, L. S. McDaniel, B. M. Gray, and D. E. Briles. 1990. Pneumococcal surface protein A (PspA) is serologically variable and is expressed by all clinically important capsular serotypes of Streptococcus pneumoniae. Infect. Immun. 58:3293-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dillard, J. P., and J. Yother. 1994. Genetic and molecular characterization of capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 3. Mol. Microbiol. 12:959-972. [DOI] [PubMed] [Google Scholar]
- 11.Fine, D. P. 1975. Pneumococcal type-associated variability in alternate complement pathway activation. Infect. Immun. 12:772-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finland, M., and M. Barnes. 1977. Changes in occurrence of capsular serotypes of Streptococcus pneumoniae at Boston City Hospital during selected years between 1935 and 1974. J. Clin. Microbiol. 5:154-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geelen, S., C. Bhattacharyya, and E. Tuomanen. 1993. The cell wall mediates pneumococcal attachment to and cytopahtology in human endothelial cells. Infect. Immun. 61:1538-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon, D. L., J. Rice, J. J. Finley-Jones, P. J. McDonald, and M. K. Hostetter. 1988. Analysis of C3 deposition and degradation on bacterial surfaces after opsonization. J. Infect. Dis. 157:697-704. [DOI] [PubMed] [Google Scholar]
- 15.Gray, B. G., and J. H. C. Dillon. 1986. Clinical and epidemiologic studies of pneumococcal infection in children. Pediatr. Infect. Dis. 5:201-207. [DOI] [PubMed] [Google Scholar]
- 16.Hammerschmidt, S., G. Bethe, P. H. Remane, and G. S. Chhatwal. 1999. Identification of pneumococcal surface protein A as a lactoferrin-binding protein of Streptococcus pneumoniae. Infect. Immun. 67:1683-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardy, G. G., M. J. Caimano, and J. Yother. 2000. Capsule biosynthesis and basic metabolism in Streptococcus pneumoniae are linked through the cellular phosphoglucomutase. J. Bacteriol. 182:1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy, G. G., A. D. Magee, C. L. Ventura, M. J. Caimano, and J. Yother. 2001. Essential role for cellular phosphoglucomutase in virulence of type 3 Streptococcus pneumoniae. Infect. Immun. 69:2309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Havarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henrichsen, J. 1995. Six newly recognized types of Streptococcus pneumoniae. J. Clin. Microbiol. 33:2759-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollingshead, S. K., R. Becker, and D. E. Briles. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun. 68:5889-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hostetter, M. K. 1986. Serotypic variations among virulent pneumococci in deposition and degradation of covalently bound C3b: implications for phagocytosis and antibody production. J. Infect. Dis. 153:682-693. [DOI] [PubMed] [Google Scholar]
- 24.Jansson, P. E., B. Lindberg, M. Anderson, U. Lindquist, and J. Henrichsen. 1988. Structural studies of the capsular polysaccharide from Streptococcus pneumoniae type 2, a reinvestigation. Carbohydr. Res. 182:111-117. [DOI] [PubMed] [Google Scholar]
- 25.Kelly, T., J. P. Dillard, and J. Yother. 1994. Effect of genetic switching of capsular type on virulence of Streptococcus pneumoniae. Infect. Immun. 62:1813-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knecht, J. C., G. Schiffman, and R. Austrian. 1970. Some biological properties of pneumococcus type 37 and the chemistry of its capsular polysaccharide. J. Exp. Med. 132:475-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, J., and C. L. Turnbough, Jr. 1994. Effects of transcriptional start site sequence and position on nucleotide-sensitive selection of alternative start sites at the pyrC promoter in Escherichia coli. J. Bacteriol. 176:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacLeod, C. M., R. G. Hodges, M. Heildeberger, and W. G. Bernhard. 1945. Prevention of pneumococcal pneumonia by immunization with specific capsular polysaccharides. J. Exp. Med. 82:445-465. [PMC free article] [PubMed] [Google Scholar]
- 29.Macrina, F. L., R. P. Evans, J. A. Tobian, D. L. Hartley, D. B. Clewell, and K. R. Jones. 1983. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene 25:145-150. [DOI] [PubMed] [Google Scholar]
- 30.McDaniel, L. S., D. O. McDaniel, S. K. Hollingshead, and D. E. Briles. 1998. Comparison of the PspA sequence from Streptococcus pneumoniae EF5668 to the previously identified PspA sequence from strain Rx1 and the ability of PspA from EF5668 to elicit protection against pneumococci of different capsular types. Infect. Immun. 66:4748-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDaniel, L. S., B. A. Ralph, D. O. McDaniel, and D. E. Briles. 1994. Localization of protection-eliciting epitopes on PspA of Streptococcus pneumoniae between amino acid residues 192 and 260. Microb. Pathog. 17:323-337. [DOI] [PubMed] [Google Scholar]
- 32.McDaniel, L. S., G. Scott, J. F. Kearney, and D. E. Briles. 1984. Monoclonal antibodies against protease sensitive pneumococcal antigens can protect mice from fatal infection with Streptococcus pneumoniae. J. Exp. Med. 160:386-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDaniel, L. S., G. Scott, K. Widenhofer, J. Carroll, and D. E. Briles. 1986. Analysis of a surface protein of Streptococcus pneumoniae recognized by protective monoclonal antibodies. Microb. Pathog. 1:519-531. [DOI] [PubMed] [Google Scholar]
- 34.McDaniel, L. S., J. S. Sheffield, P. Delucchi, and D. E. Briles. 1991. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular type. Infect. Immun. 59:222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDaniel, L. S., J. S. Sheffield, E. Swaitlo, J. Yother, M. J. Crain, and D. E. Briles. 1992. Molecular localization of variable and conserved regions of pspA, and identification of additional pspA homologous sequences in Streptococcus pneumoniae. Microb. Pathog. 13:261-269. [DOI] [PubMed] [Google Scholar]
- 36.McDaniel, L. S., J. Yother, M. Vijayakumar, L. McGarry, W. R. Guild, and D. E. Briles. 1987. Use of insertional inactivation to facilitate studies of biological properties of pneumococcal surface protein A (PspA). J. Exp. Med. 165:381-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reeves, R. E., and W. F. Goebel. 1941. Chemoimmunological studies on the soluble specific substance of pneumococcus. V. The structure of the type III polysaccharide. J. Biol. Chem. 139:511-519. [Google Scholar]
- 38.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shoemaker, N. B., and W. R. Guild. 1974. Destruction of low efficiency markers is a slow process occurring at a heteroduplex stage of transformation. Mol. Gen. Genet. 128:283-290. [DOI] [PubMed] [Google Scholar]
- 40.Tu, A.-H. T., R. L. Fulgham, M. A. McCrory, D. E. Briles, and A. J. Szalai. 1999. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect. Immun. 67:4720-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walter, A. W., V. H. Guerin, M. W. Beattie, H. Y. Cotler, and H. B. Bucca. 1941. Extension of the separation of types among pneumococci: description of 17 types in addition to types 1 to 32 (Cooper). J. Immunol. 41:279. [Google Scholar]
- 42.Winkelstein, J. A., A. S. Abramovitz, and A. Tomasz. 1980. Activation of C3 via the alternative complement pathway results in fixation of C3b to the pneumococcal cell wall. J. Immunol. 124:2502-2506. [PubMed] [Google Scholar]
- 43.Yother, J., and D. E. Briles. 1992. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J. Bacteriol. 174:601-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yother, J., C. Forman, B. M. Gray, and D. E. Briles. 1982. Protection of mice from infections with Streptococcus pneumoniae by anti-phosphocholine antibody. Infect. Immun. 36:184-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yother, J., G. L. Handsome, and D. E. Briles. 1992. Truncated forms of PspA that are secreted from Streptococcus pneumoniae and their use in functional studies and cloning of the pspA gene. J. Bacteriol. 174:610-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yother, J., L. S. McDaniel, and D. E. Briles. 1986. Transformation of encapsulated Streptococcus pneumoniae. J. Bacteriol. 168:1463-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yother, J., L. S. McDaniel, M. J. Crain, D. F. Talkington, and D. E. Briles. 1991. Pneumococcal surface protein A: structural analysis and biological significance, p. 88-91. In G. M. Dunny, P. P. Cleary, and L. L. McKay (ed.), Genetics and molecular biology of streptococci, lactococci, and enterococci. American Society for Microbiology, Washington, D.C.
- 48.Yother, J., and J. M. White. 1994. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J. Bacteriol. 176:2976-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]