Abstract
The frequency at which the genes responsible for capsule biosynthesis occurred in field isolates of Streptococcus uberis was determined. Of the two genotypes detected (hasABC and hasC), the capsular genotype (hasABC) was more common. This genotype was present at a higher frequency in a population isolated from mastitis cases than in a population isolated from cattle bedding. The virulence of a mutant strain of S. uberis (TRF0-6) that lacked the ability to produce a hyaluronic acid capsule due to an insertion within its single copy of hasA (P. N. Ward, T. R. Field, W. G. F. Ditcham, E. Maguin, and J. A. Leigh, Infect. Immun. 69:392-399, 2001) was compared to that of the capsular parental strain (0140J). Strains TRF0-6 and 0140J infected all mammary gland quarters following experimental challenge. The wild type and the mutant induced overt signs of disease in four out of four and in six out of eight mammary gland quarters, respectively. Both the wild type and the hasA mutant were resistant to killing by bovine neutrophils following cultivation in bovine milk. The ability to withstand the bactericidal action of neutrophils following growth in milk was therefore independent of the capsule and coincided with the ability of supernatants from such cultures to prevent the bactericidal action of neutrophils. This investigation revealed that, in the absence of the capsule, S. uberis is able to withstand the bactericidal effect of bovine neutrophils and induce mastitis in dairy cows.
Bovine mastitis, inflammation of the udder, usually arises as a result of intra-mammary gland infection by bacteria (8). Over 135 infectious agents have been associated with clinically apparent episodes of mastitis (27), but the vast majority of episodes are due to infection with one of five bacterial species. Those most commonly implicated are Streptococcus uberis, S. dysgalactiae, S. agalactiae, Escherichia coli, and Staphylococcus aureus (16). Mastitis is one of the most common infectious diseases of dairy cattle. In the United Kingdom, it occurs at a frequency of between 35 and 40 cases per 100 cows per year and at an annual cost of about £170,000,000 (17). The manifestation of disease can range from visible abnormalities in the milk (protein aggregates or clots) to the production of a secretion that is composed solely of aggregated protein in a serous fluid and often results in pain and swelling in the affected gland.
Milk from the uninfected gland contains leukocytes, including macrophages, neutrophils, and lymphocytes, typically at <150,000 cells/ml. Infection usually results in an inflammatory response which leads to an increase in the number of cells, primarily due to the influx of neutrophils from the peripheral circulation (23). Milk from clinically infected quarters usually contains in excess of 2,000,000 cells/ml; >90% of these are neutrophils (22). The inflammatory reaction and the increase in the number of neutrophils result in a lower rate of milk production and a gross deterioration of the quality of the secretion.
Phagocytosis and killing of bacteria by neutrophils constitute a major defense mechanism of the lactating bovine mammary gland. This process is responsible for controlling infections caused by S. aureus (24) and eliminating infections caused by E. coli (13). The role of phagocytic cells in the control of infection by S. uberis appears less clear (10). However, it was demonstrated that a capsular strain of S. uberis (0140J) was more capable of establishing an infection in the lactating gland than a noncapsular strain (EF20), which was readily eliminated (14, 20).
Experimental infection of the mammary gland with virulent strains of S. uberis results in the appearance of large numbers of neutrophils in the interstitial tissues and secretion (10, 25). However, these do not reduce the number of S. uberis organisms present in the secretion (10). Microscopic examination of infected tissues showed that bacteria detected in the secretion were found only rarely in association with phagocytic cells, and in no instance were these identified as neutrophils (25).
The ability of S. uberis to resist the bactericidal action of neutrophils has been reproduced in vitro by growth of this organism in media containing casein-derived peptides (18) or milk whey produced following the action of rennin on skim milk (1). The ability to alter the resistance of this organism to phagocytosis in vitro, by altering growth conditions, has enabled a comparison of individual strains that exhibit phagocytosis-resistant and -susceptible phenotypes. In these experiments, the production of a hyaluronic acid capsule correlated with the ability to resist phagocytosis by neutrophils (18). Removal of the capsule and inhibition of its reformation with hyaluronidase resulted in a significant reduction in the ability of S. uberis to resist phagocytosis by neutrophils in vitro (18). Capsulation in S. uberis was shown to be dependent on the presence of at least two genes (26) by the production of mutants in which the synthesis of hyaluronic acid was blocked by lesions in either hasA (hyaluronate synthase) or hasC (UDP-glucose pyrophosphorylase). Furthermore, this study showed in vitro that the capsule is a significant barrier to the efficient uptake and killing of S. uberis by bovine neutrophils (26).
Current evidence would therefore indicate that the resistance of S. uberis to phagocytosis and killing by bovine neutrophils is mediated through a hyaluronic acid capsule. In this communication, we report the first use of a genetically altered strain of S. uberis (TRF0-6) to determine the role of the capsule in vivo in an experimental model of infection in the target species.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. uberis strain 0140J was obtained from a case of clinical bovine mastitis, and strain C198 was obtained from a dairy cow with a subclinical intra-mammary gland infection. Strain TRF0-6, a noncapsular (hasA) mutant derivative of strain 0140J, was produced and characterized as described by Ward et al. (26). E. coli strain P4 was originally isolated from a case of clinical bovine mastitis in the United Kingdom. Strains are currently kept within the culture collection of the Institute for Animal Health. They were routinely cultured at 37°C in Todd-Hewitt broth or a chemically defined medium (18) to which enzymatically produced casein hydrolysate had been added to a final concentration of 1% (wt/vol) (CDM-CH). In addition, S. uberis strains were grown in raw skim milk at 37°C. Milk was collected aseptically from uninfected healthy animals within the dairy herd of the Institute for Animal Health, and skim milk was removed from the upper fat layer following centrifugation at 1,000 × g for 10 min at 4°C.
Analysis of S. uberis for the distribution of has-related genes in field isolates by multiplex PCR.
The distribution of two of the three has genes (hasA and hasB) in S. uberis (26) was investigated by multiplex PCR, and the distribution of hasC (26) was investigated by single-gene PCR. The strain collection kept at the Cattle Health Laboratory (Broerup, Denmark) consisted of samples from 19 Danish dairy herds; 77 strains were isolated from cases of clinical mastitis in lactating cattle, and 103 strains were isolated from bedding materials. All isolates were cultivated on blood agar containing esculin and putatively identified as S. uberis following analysis by using the API-20 STREP system (bioMerieux).
Confirmation of the species-level identification was achieved by multiplex PCR for the simultaneous identification of S. uberis 23S rRNA gene sequences for hasA and hasB. The PCR included 0.8× PCR buffer (Applied Biosystems); 200 μM each dATP, dCTP, dGTP, and dTTP (Applied Biosystems); primers (0.075 to 0.3 μM each) (Table 1); 2.5 μM MgCl2; 1.25 U of Taq DNA polymerase (Applied Biosystems); genomic DNA template (5 μl); and autoclaved ultrafiltered water to a final volume of 50 μl. Oligonucleotide primer pairs were designed with Gene Fisher software (7) on the basis of DNA sequences for the genes hasA and hasB (GenBank accession no. AJ242946) and hasC (GenBank accession no. AJ400707). For the identification of S. uberis, oligonucleotide primers designed for a specific part of the 23S rRNA gene were used (9). All oligonucleotide primers were obtained from a commercial source (DNA-technology, Aarhus, Denmark). The concentrations of the primers used in the PCR as well as the expected sizes of the four different amplicons are shown in Table 1.
TABLE 1.
Oligonucleotide primer sequences and their applications, individual primer concentrations, and predicted PCR product sizes
| Designation | Sequence | Application | Primer concn (μM) | Size of PCR product (bp) |
|---|---|---|---|---|
| hasA-for | 5′-GAAAGGTCTGATGCTGATG | Amplification of hasA DNA | 0.1 | 319 |
| hasA-rev | 5′-TCATCCCCTATGCTTACAG | 0.1 | ||
| hasB-for | 5′-TCTAGACGCCGATCAAGC | Amplification of hasB DNA | 0.1 | 532 |
| hasB-rev | 5′-TGAATTCCTATGCGTCGATC | 0.1 | ||
| hasC-for | 5′-TGCTTGGTGACGATTTGATG | Amplification of hasC DNA | 0.3 | 225 |
| hasC-rev | 5′-GTCCAATGATAGCAAGGTCAC | 0.3 | ||
| Ub-23S-I | 5′-CGTATTTAAAATTGACTTTAGCC | Amplification of S. uberis DNA | 0.075 | 451 |
| Ub-23-S-II | 5′-AATTTCTCGCTACCCAC | 0.075 |
For DNA template preparation, bacteria were grown on blood agar plates for 18 h at 37°C and subsequently overnight in brain heart infusion broth (Difco) containing 1% (wt/vol) glucose. Genomic DNA was isolated from 2 ml of an overnight culture by using an Easy-DNA kit (Invitrogen) according to the manufacturer's guidelines. Amplification of DNA was performed with a thermal cycler (PTC 200; MJ Research, Inc.) by using the following procedure: initial denaturation at 95°C for 5 min; denaturation at 95°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 2 min for 28 cycles; and a final extension at 72°C for 5 min. The PCR products were separated by electrophoresis on 1.5% SeaKem GTG agarose gels (BMA, Rockland, Maine) in 0.1 M Tris-borate-0.001 M EDTA (pH 8.3) buffer at room temperature by using a 7-V/cm voltage gradient. The gels were stained with ethidium bromide, and the results were visualized with Bio-print (Vilber Lourmat). Both the multiplex PCR and the single-gene PCR were validated with a bovine strain of S. uberis, 0140J, previously shown to contain all three has genes (26). The odds ratio for the presence of the hasABC genotype in clinical versus environmental isolates was calculated.
Experimental challenge with S. uberis strains 0140J and TRF0-6.
Six dairy cows in their first lactation were each challenged in two mammary gland quarters by infusion of between 600 and 1,000 CFU of S. uberis suspended in 1 ml of pyrogen-free saline (Sigma). Two animals were challenged with S. uberis strain 0140J, and four animals were challenged with strain TRF0-6. Animals were challenged in two groups of three (one with 0140J and two with TRF0-6 in each group). Animals were housed in contained experimental facilities and milked twice daily at 0700 and 1600. Each animal was milked by using a separate unit to ensure no cross contamination between animals. Samples were collected at each milking for the enumeration of bacteria and somatic cells as described previously (5, 6). Animals were assessed clinically at each milking, and those in which predetermined criteria for clinical end points (clotted and discolored milk and/or swollen or painful udder quarters) had been reached were treated with proprietary branded antibiotics.
RFLP of chromosomal DNA.
Bacterial colonies isolated following experimental challenge were transferred to Todd-Hewitt broth and grown at 37°C for 18 h. Chromosomal DNA was isolated (26) and subjected to restriction fragment length polymorphism (RFLP) analysis (12). The RFLP profiles were compared to those obtained following a similar analysis of cultures of the challenge strains TRF0-6 and 0140J.
PCR amplification of the hasA gene sequence in isolates recovered following challenge.
Bacterial colonies isolated following enumeration of bacteria from milk samples obtained following experimental challenge were transferred to Todd-Hewitt broth and grown at 37°C for 18 h. A sample (5 μl) from each culture was heated at 100°C for 5 min and used as a source of template DNA for the amplification of hasA by PCR with the procedures and primers described previously (26). The amplification products were compared to those obtained from cultures of each of the challenge strains, 0140J and TRF0-6.
Measurement of the bactericidal action of bovine neutrophils.
The bactericidal action of bovine neutrophils against various bacteria was determined. In order to compare the data obtained here with those obtained previously, strains of S. uberis were subjected to an assay similar to that used in earlier studies (18, 20, 26). Briefly, 0.5 ml of a washed bacterial suspension (106 CFU/ml) in phosphate-buffered saline (PBS) was mixed with 1.0 ml of a suspension of bovine neutrophils (107 cells/ml) and 1.5 ml of aseptically collected bovine skim milk. The mixtures were rolled at 120 rpm for 180 min on a Coulter roller (Coulter Electronics) at 37°C. Samples (100 μl) were removed, and the total number of viable bacteria was determined. The data obtained were compared to those obtained from a similarly incubated bacterial suspension to which the same volume of PBS but no neutrophils had been added.
This assay was also adapted to determine the ability of neutrophils to function in the milk in which S. uberis strains 0140J and TRF0-6 had been cultivated. Bacterial cells were removed from a milk culture (24 h, 37°C) by centrifugation (15,000 × g, 15 min), and the milk was sterilized by filtration (0.2-μm-pore-size filter). The culture filtrate (1.5 ml) was mixed with 100 μl of peripheral blood neutrophils (108 cells/ml) and 50 μl of E. coli strain P4 (108 CFU/ml). The mixtures were rolled at 120 rpm for 180 min on a Coulter roller at 37°C. Samples (100 μl) were removed, and the total number of viable bacteria was determined. The data obtained were compared to those obtained from assays in which filtered milk that had been incubated at either 37 or 4°C for 24 h in the absence of bacteria was used in place of the milk culture filtrate. Each assay was also controlled by comparison to identical assays from which neutrophils were omitted.
In all experiments, at least three independent assays were conducted in triplicate and bacterial numbers were estimated following dilution of samples in isotonic saline and incubation of 50 μl of each dilution on blood agar containing 1% (wt/vol) esculin at 37°C for 18 h.
Determination of the ability of S. uberis in milk cultures to withstand the bactericidal action of bovine neutrophils.
The ability of S. uberis strains 0140J and TRF0-6 to resist the bactericidal action of neutrophils following growth in bovine milk was assessed. Bacteria were cultured in bovine skim milk at 37°C for 24 h, and 2.4 ml of this culture was mixed with 100 μl of a suspension of isolated peripheral blood neutrophils (108 cells/ml of PBS). The mixtures were rolled at 120 rpm for 180 min on a Coulter roller at 37°C. Samples (100 μl) were removed, and the total number of viable bacteria was determined. The data obtained were compared to those obtained from a similarly incubated bacterial culture to which the same volume of PBS but no neutrophils had been added. The number of bacteria was determined by dilution of samples in isotonic saline and direct plating as described above. This assay was conducted in triplicate on three separate occasions.
RESULTS
Distribution of has genes in isolates of S. uberis obtained from cases of clinical bovine mastitis and cattle bedding materials.
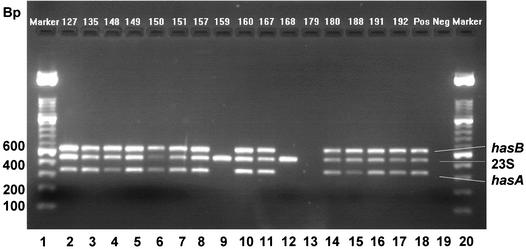
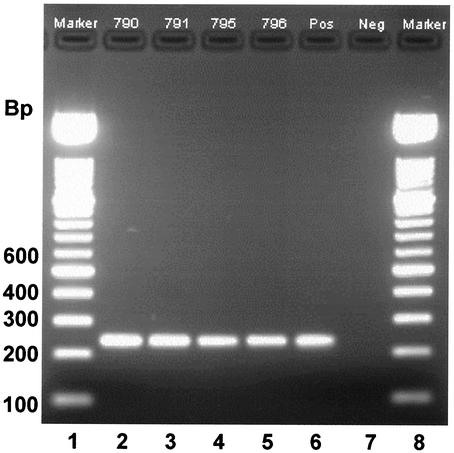
A survey by multiplex PCR (Fig. 1) and single-gene PCR (Fig. 2) of the presence of capsule genes in isolates of S. uberis from cases of clinical bovine mastitis and from environmental sources within 19 Danish dairy herds revealed that 180 isolates of S. uberis corresponded to two genotypes. Simultaneous amplification of hasA, hasB, and species-specific sequences of 23S rRNA genes allowed confirmation of the species identity of isolates and determination of the presence or absence of hasA and hasB (illustrative data from such an amplification are provided in Fig. 1). Simultaneous amplification of hasC was not reliable; however, single-gene PCR revealed that hasC was present in all isolates of S. uberis (illustrative data from such an amplification are provided in Fig. 2). In each amplification reaction, S. uberis strain 0140J, which is known to contain hasAB and C (26), was included as a positive control, and reactions conducted in the absence of template DNA served as negative controls (Fig. 1 and 2).
FIG. 1.
Multiplex PCR amplification of hasA, hasB, and 23S rRNA genes from isolates of S. uberis. Lanes 1 and 20, 100-bp molecular size markers (DNA molecular weight marker XIV; Roche). Lanes 2 to 17, PCR products from isolates obtained from both subclinically and clinically infected mammary gland quarters. Lane 18, positive control (strain 0140J). Lane 19, negative control (DNA template free).
FIG. 2.
PCR amplification of hasC gene from isolates of S. uberis. Lanes 1 and 8, 100-bp molecular size markers (DNA molecular weight marker XIV). Lanes 2 to 5, PCR products from isolates obtained from bedding samples. Lane 6, positive control (strain 0140J). Lane 7, negative control (DNA template free). Strains 790, 791, and 795 (lanes 2 to 4, respectively) were shown by multiplex PCR to contain both hasA and hasB.
The predominant genotype in both the disease-associated and the environmental populations was hasABC (Table 2), and statistical analysis revealed that this genotype was overrepresented in the clinical versus the environmental isolates (odds ratio, 15; 95% confidence interval). The remaining genotype (hasC) lacked both hasA and hasB (e.g., strains 159 and 168; Fig. 1, lanes 9 and 12, respectively).
TABLE 2.
Distribution and frequency of has genes in environmental and bovine milk S. uberis isolates
| has gene(s) present | No. (%) of S. uberis isolates in the following clinical samples:
|
||
|---|---|---|---|
| Bedding | Clinical | Total | |
| hasC | 17 (16.5) | 1 (1.3) | 18 (10) |
| hasABC | 86 (83.5) | 76 (98.7) | 162 (90) |
A previous analysis of mutants of S. uberis revealed that the presence of both hasA and hasC is essential for the production of a hyaluronic acid capsule (26). Consequently, isolates with the hasC genotype, lacking hasA, are unable to produce a hyaluronic acid capsule. Given the overrepresentation of isolates carrying the hasAB gene cluster (26) in clinical disease and in light of the data supporting the role of the capsule in determining the resistance of S. uberis to killing by bovine neutrophils (1, 2, 18, 20, 26), an investigation of the role of the capsule in vivo was undertaken.
Experimental challenge with S. uberis strains 0140J and TRF0-6.
The virulence of a well-characterized strain of S. uberis, 0140J, and that of its mutant derivative, TRF0-6, which lacks the ability to produce hyaluronic acid due to insertional disruption of its single copy of hasA (26), were compared in a well-established experimental model (5, 6, 10, 14, 15, 19) for bovine mastitis.
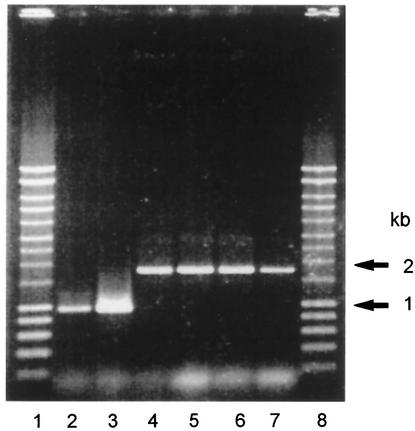
S. uberis was isolated in pure cultures from milk obtained from the first milking postchallenge in all animals. In each sample, the bacteria were shown to be of the correct strain lineage by RFLP analysis (data not shown) and genotype by PCR amplification of the appropriately sized products (950 bp for 0140J and 1,750 bp for TRF0-6) from the hasA gene (Fig. 3).
FIG. 3.
PCR amplification of hasA and hasA::ISS1 from bacteria isolated from milk within 24 h of challenge with strain 0140J or TRF0-6. Lanes 1 and 8, molecular size markers. Lanes 2 to 7, PCR products from hasA of bacteria isolated from milk of animals challenged with either strain 0140J (lanes 2 and 3) or strain TRF0-6 (lanes 4 to 7).
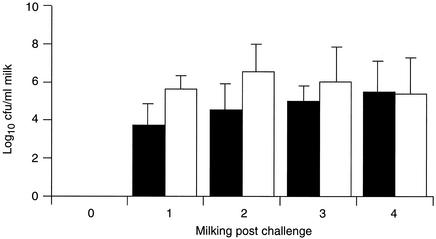
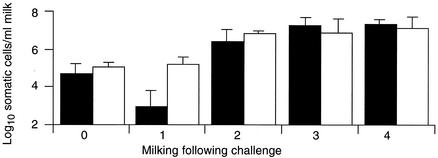
All four quarters on two animals challenged with strain 0140J shed bacteria (Fig. 4), exhibited inflammatory responses (measured by an increase in the somatic cell count) (Fig. 5), and showed signs of disease (discolored milk containing clots, accompanied by swollen and painful udder quarters). All challenged quarters in which clinical signs were apparent were treated with antibiotics after the fourth milking postchallenge.
FIG. 4.
Bacterial recovery following challenge. Data are the geometric means of the number of bacteria obtained from the milk of animals challenged with either strain 0140J (solid bars; n = 4) or strain TRF0-6 (open bars; n = 8). The standard error is shown as a line above each mean.
FIG. 5.
Inflammatory response following challenge. Data are the geometric means of the number of somatic cells obtained from the milk of animals challenged with either strain 0140J (solid bars; n = 4) or strain TRF0-6 (open bars; n = 8). The standard error is shown as a line above each mean.
All eight quarters on four animals challenged with strain TRF0-6 shed bacteria (Fig. 4) and exhibited inflammatory responses (Fig. 5); in six quarters, there were overt signs of disease indistinguishable from those produced in response to infection with strain 0140J. These quarters required treatment with antibiotics after the fourth (two animals) or fifth (one animal) milking postchallenge. The remaining two quarters, both on the same animal, did not show clinical signs of disease. However, both continued to shed bacteria up to the 10th milking postchallenge, despite the presence of over 106 neutrophils per ml of milk. Infection in these mammary gland quarters was resolved only at the end of the experiment by treatment with antibiotics.
Inflammatory responses (Fig. 5) and bacterial recoveries (Fig. 4) measured in both groups of animals were similar and were therefore not dependent on the challenge strain.
Resistance to the bactericidal action of bovine neutrophils.
The failure of animals to clear the noncapsular mutant following experimental challenge was in marked contrast to the interaction between this strain and bovine neutrophils reported previously in studies conducted in vitro (26).
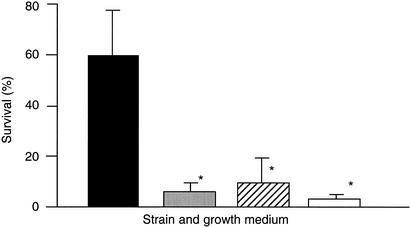
In order to confirm the earlier data, both strains were grown in CDM-CH, known to induce capsule expression in the wild-type strain (18). Washed bacterial suspensions were subjected to a bactericidal assay similar to that used previously (18, 20, 26). The data obtained here were similar to those obtained before and confirmed that the noncapsular mutant was considerably less able to resist the bactericidal action of neutrophils than the capsular parental strain in such an assay (Fig. 6). These data were not consistent with the observations in vivo, where both strains appeared to survive at similar levels in the presence of neutrophils.
FIG. 6.
Survival of S. uberis 0140J or TRF0-6 obtained from cultures grown in CDM-CH or raw bovine milk. Data are means (n = 9) of the percent survival of bacteria in the presence of neutrophils compared to that in equivalent assays in the absence of neutrophils. The standard error is shown as a line above each mean. Strains 0140J and TRF0-6 were grown in either CDM-CH (solid and open bars, respectively) or raw skim milk (shaded and hatched bars, respectively) and washed prior to incubation with bovine neutrophils. Survival that was significantly different (P < 0.001) from the survival of 0140J grown in CDM-CH is indicated by asterisks.
In order to more closely mimic the situation in vivo, each strain was grown in raw skim milk, and neutrophils or an equivalent volume of PBS was added directly to each culture. Both the wild type and the noncapsular mutant survived similarly in the presence or absence of neutrophils, and the numbers of bacteria remained constant throughout the experiment. These results indicate that the capsule may not be the sole effector of resistance to neutrophils in this bacterium following the growth of the bacterium in bovine milk.
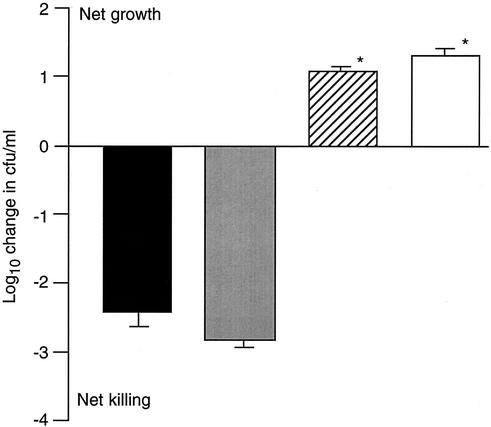
To investigate whether the additional hypothesized antiphagocytic component was cell associated and/or released into milk during bacterial growth, two further experiments were conducted. In the first experiment, strains 0140J and TRF0-6 were grown in milk, collected, and washed, and their ability to withstand the bactericidal action of bovine neutrophils was compared to that of washed suspensions of the same strains following growth in CDM-CH (Fig. 6). The two strains extracted from milk cultures survived at levels similar to each other and to the level of the noncapsular mutant grown in CDM-CH. Each of these cultures was significantly (P < 0.001) less able to resist the bactericidal action of neutrophils than the parental strain cultured in CDM-CH (Fig. 6). In the second experiment, the effect of filtrates from milk cultures of either 0140J or TRF0-6 on the bactericidal action of bovine neutrophils was determined. Bovine neutrophils were added to milk culture filtrates, and a washed suspension of E. coli (strain P4) was added to the mixture as a reporter of bactericidal action. Filtered milk that had not been used to culture bacteria but that had been incubated at either 4 or 37°C was used as a control medium. Similarly, the viability of the reporter in each medium was determined in the absence of neutrophils. Milk that had not been used to culture S. uberis supported the bactericidal action of neutrophils, and milk that did not contain neutrophils supported the growth of E. coli. However, milk in which either 0140J or TRF0-6 had been cultured was significantly (P < 0.001) less able to support the bactericidal action of neutrophils (Fig. 7).
FIG. 7.
Effect of S. uberis milk culture filtrates on killing of E. coli by bovine neutrophils. Data are means (n = 9) of the change in the number of E. coli organisms detected in milk in the presence of neutrophils. The standard error is shown as a line above or below each mean. Prior to use in the assays, milk was incubated for 24 h at 4°C (solid bar) or 37°C (shaded bar) in the absence of bacteria or was used to culture (24 h, 37°C) either strain 0140J (hatched bar) or strain TRF0-6 (open bar). Data that were significantly different (P < 0.001) from those obtained in milk incubated at either 4 or 37°C in the absence of bacteria are indicated by asterisks. In the absence of neutrophils, each sample supported a similar increase in the number of E. coli organisms of approximately log10 2.0 CFU/ml.
DISCUSSION
The use of a mutant derivative of S. uberis 0140J that was unable to produce a capsule due an insertional lesion within its single hyaluronic acid synthase gene (26) has enabled the role of the capsule in the pathogenesis of bovine mastitis to be investigated in the target species for the first time. In contrast to data from in vitro studies indicating the importance of the capsule in resisting uptake and killing by phagocytes (1, 2, 18, 20, 26) and the undoubted role of neutrophils in protecting the bovine mammary gland from bacterial infection (13, 24), the present study indicates that the virulence of the mutant strain was similar to that of the parental strain.
Following challenge, infection was established in all quarters challenged with the wild-type strain and the hasA mutant. Of these, 100 and 75%, respectively, showed overt clinical signs of disease. In compliance with legislation in the United Kingdom on the use of experimental animals, each animal showing clinical signs of disease was treated with antibiotics. In no instance were bacteria eliminated from the mammary gland in the absence of antibiotic therapy, indicating that intact hasA and consequently the hyaluronic acid capsule were not required for infection. Furthermore, the influx of neutrophils into the mammary gland did not result in a decrease in the number of bacteria present in the secretion, indicating that the absence of the capsule did not noticeably alter the resistance of the bacteria to the bactericidal action of bovine neutrophils in vivo. These results are consistent with the results of previous studies on infection of the bovine mammary gland with wild-type S. uberis (5, 6, 10, 14, 15, 19) but are in contrast to results obtained with other mastitis pathogens, such as E. coli (13) and Staphylococcus aureus (24), where an influx of neutrophils corresponds to a decrease in bacterial numbers and even elimination of the infecting organisms.
This infection model has been used on a number of occasions by various researchers (5, 6, 10, 14, 15, 19) and typically yields clinical disease in approximately 90% of challenges with strain 0140J. Failure to generate clinical disease in two quarters (both on the same animal) out of eight quarters challenged with the hasA mutant may indicate a minor reduction in the virulence of this strain. However, in light of the importance of neutrophils in controlling intra-mammary gland infection, this result did not reflect the requirement for the capsule hypothesized from earlier studies conducted in vitro (1, 2, 18, 20, 26). Given the similarity of the responses of animals challenged with the mutant strain and animals challenged with the parental strain in both this study and previous studies (5, 6, 10, 14, 15, 19), it is considered unlikely that the absence of clinical mastitis in one animal was due solely to the absence of a capsule on the challenge strain. In the animal that did not show overt signs of disease, the mutant strain persisted in the presence of a potentially overwhelming neutrophil response (more than 10 neutrophils per CFU) in both quarters for 120 h (the duration of the experiment), indicating that even in this animal, the mutant strain was able to withstand the bactericidal action of incoming neutrophils.
A comparison of the abilities of two field strains (0140J and EF20) to withstand the bactericidal action of neutrophils in vitro, to produce a capsule (20), and to infect the lactating mammary gland (15) led to the conclusion that these observations were linked. Strain EF20 was considered to have reduced virulence due to the absence of a capsule and a consequent reduction in its resistance to the bactericidal action of neutrophils. In this study and in a previous investigation (26), strain TRF0-6 was also shown to be unable to resist the bactericidal action of neutrophils during bactericidal assays similar to those used previously with strain EF20. Strain TRF0-6 was also shown to be unable to produce detectable levels of hyaluronic acid due a lesion within its single copy of hasA (26). In this study, it was shown clearly that strain TRF0-6 was able to infect the bovine mammary gland (eight of eight challenges) and cause clinical mastitis (six of eight challenges). These results are in contrast to those of a previous study, carried out with the same infection model, which showed that strain EF20 was able to infect only 2 out of 18 challenged quarters; in comparison, in the same experiment, strain 0140J caused clinical disease in 16 out of 18 challenged quarters (15). Therefore, although the present investigation confirmed that the capsule is able to confer resistance to the bactericidal action of neutrophils under some assay conditions in vitro, it also leads to the conclusion that such assays do not accurately reflect all of the possible interactions between bacteria and neutrophils in vivo. These data also cast doubt on the original interpretation that strain EF20 showed reduced virulence, compared to strain 0140J, due to the absence of a capsule (15, 20) and imply that another, unidentified difference relating to virulence exists between these two field strains.
In the field, S. uberis infections of the bovine mammary gland arise predominantly as a result of challenge at the teat end from bacteria present in environmental reservoirs, such as bedding materials. If the presence of the capsule is of greater significance to the pathogenesis of disease than to environmental survival, then it is likely that the expression of the capsule will be an overrepresented phenotypic characteristic in a collection of isolates obtained from cases of mastitis. For S. uberis, it has been demonstrated that the presence of both hasA and hasC is required for capsule production (26); consequently, of the two genotypes of S. uberis detected in this study, only hasABC can be predicted to have the genetic capability for capsule production. This genotype occurred at a higher frequency in the isolates associated with disease, suggesting that the capsule is required for some aspects of intra-mammary gland infection and pathogenesis. It is considered unlikely that the minor perturbation of virulence that we observed in one out of the four animals challenged with the noncapsular mutant would account for the difference in the distribution of potentially capsular and noncapsular genotypes. With the infection model used in the present investigation, bacteria are placed beyond the teat canal, into the cistern of the mammary gland; therefore, this model does not allow investigation of all of the possible aspects of infection in the field. As a result, it can be hypothesized that during infection in the field, the capsule may play a role in teat end colonization and/or teat canal penetration.
The role of the hyaluronic acid capsule of S. pyogenes (Lancefield group A streptococci) has been investigated in vitro and in vivo, in mouse models of colonization and disease. While the majority of the studies indicated a role for the capsule, the manifestation of its effects in vitro differed according to the presence of serum, whole blood (21), or fibrinogen (3). In these situations, the consequence of the absence of the capsule was less marked due to the presence of an alternate antiphagocytic mechanism, namely, M protein. It would appear that with S. uberis, infection of the bovine mammary gland is a particular and specialized circumstance in which the hyaluronic acid capsule appears to have little relevance for resistance to the action of phagocytic cells during the pathogenesis of infection.
The present investigation led to the hypothesis that a factor or factors other than the capsule must be able to effect the resistance of S. uberis to killing by bovine neutrophils. This hypothesis was substantiated following the demonstration that neutrophils added directly to milk-grown cultures were unable to kill either the wild type or the noncapsular mutant. This observation was in agreement with those obtained in vivo, where neither strain was cleared or even reduced in number following the influx of neutrophils. This observation was not attributable to the induction of a factor associated with bacterial cells, as washed suspensions of either strain grown in this medium were unable to withstand the bactericidal action of bovine neutrophils effectively. However, cell-free filtrates from milk-grown cultures of either strain were shown to be unable to support the bactericidal action of neutrophils, suggesting that the resistance of S. uberis to neutrophil-mediated killing may be effected through an extracellular factor.
As the bacterium used as a reporter of bactericidal action, E. coli, and S. uberis are not immunologically cross-reactive and as the opsonization of either bacterial species can be effected by antibody alone (4, 11, 18, 28), it is considered unlikely that the inability of milk culture filtrates to support phagocytosis was due to the specific depletion of opsonin. Furthermore, nonspecific depletion of immunoglobulin by S. uberis is not considered likely, as S. uberis does not produce any detectable antibody binding proteins and binds antibody only in an antigen-dependent manner (18).
An extracellular factor(s) capable of inhibiting neutrophils has been reported for S. uberis 0140J (4), and other strains of S. uberis have been shown to exert an inhibitory effect against bovine macrophages (2). It is possible, therefore, that material released or secreted by either 0140J or its noncapsular mutant derivative, TRF0-6, was capable of inhibiting phagocytic killing by bovine neutrophils. A previous interpretation of these observations (2, 4), in light of data from in vitro studies supporting the role of the capsule, led to the conclusion that this effect was exerted only in concert with the capsule. However, the data presented in this communication have separated the presence of the capsule and an inhibitory effect, thus demonstrating their functional and genetic independence.
In conclusion, many of the isolates from cases of bovine mastitis possessed a hyaluronic acid capsule, implying a role for this structure. The data presented in this communication suggested that the role of the capsule with respect to the pathogenesis of bovine mastitis is limited but implied that the capsule may exert an effect at the level of penetration of the mammary gland. Furthermore, these data alluded to the possibility that following entry into the bovine mammary gland, S. uberis may be able to produce an environment in which the bactericidal effect exerted by the influx of neutrophils is inhibited. Even in the absence of a capsule, this effect appears to be sufficient to permit persistence of the infection to a level at which it results in the induction of clinical mastitis in the target species. Given previous reports (2, 4), this finding may reflect the production of an activity capable of inhibiting neutrophil function. However, the nature and precise mode of action of this inhibitory activity are yet to be established.
Acknowledgments
We acknowledge the valuable contribution of Zoe Penrose for assistance in the maintenance, handling, and day-to-day care of the dairy cows during the challenge experiments. We are grateful to numerous colleagues for helpful comments during the compilation of the manuscript.
The authors from the Institute for Animal Health acknowledge the financial support of the Ministry of Agriculture, Fisheries, and Food (now the Department for the Environment, Food, and Rural Affairs, United Kingdom).
Editor: V. J. DiRita
REFERENCES
- 1.Almeida, R. A., and S. P. Oliver. 1993. Growth curve, capsule expression and characterization of the capsular material of selected strains of Streptococcus uberis. Zentbl. Veterinaermed. 40:697-706. [DOI] [PubMed] [Google Scholar]
- 2.Almeida, R. A., and S. P. Oliver. 1993. Antiphagocytic effect of the capsule of Streptococcus uberis. Zentbl. Veterinaermed. 40:707-714. [DOI] [PubMed] [Google Scholar]
- 3.Dale, J. B., R. G. Washburn, M. B. Marques, and M. R. Wessels. 1996. Hyaluronate capsule and surface M protein in resistance to opsonization of group A streptococci. Infect. Immun. 64:1495-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Field, T. R., P. M. Norton, A. P. Bland, and J. A. Leigh. 1997. Changes in bovine neutrophils induced by the capsule of Streptococcus uberis. Adv. Exp. Med. Biol. 418:957-960. [DOI] [PubMed] [Google Scholar]
- 5.Finch, J. M., A. W. Hill, T. R. Field, and J. A. Leigh. 1994. Local vaccination with killed Streptococcus uberis protects the bovine mammary gland from experimental infection following intramammary challenge with the same strain. Infect. Immun. 62:3599-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finch, J. M., A. Winter, A. Walton, and J. A. Leigh. 1997. Further studies on the efficacy of a live vaccine against mastitis caused by Streptococcus uberis. Vaccine 15:1138-1143. [DOI] [PubMed] [Google Scholar]
- 7.Giegerich, R., F. Meyer, and C. Sleichermacher. 1996. Gene Fisher—software support for the detection of postulated genes, p. 68-77. ISMB-96. In D. J. States, P. Agarwal, T. Gaasterland, L. Hunter, and R. F. Smith (ed.), Proceedings of the Fourth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif. [PubMed]
- 8.Guidry, A. J. 1985. Mastitis, p. 237-262. In B. L. Larson (ed.), Lactation. Iowa State University Press, Ames, Iowa.
- 9.Hassan, A. A., I. U. Khan, A. Abdulmawjood, and C. Lammler. 2001. Evaluation of PCR methods for rapid identification and differentiation of Streptococcus uberis and Streptococcus parauberis. J. Clin. Microbiol. 39:1618-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill, A. W., J. M. Finch, J. A. Leigh, and T. R. Field. 1994. Immune modification of the pathogenesis of Streptococcus uberis mastitis in the dairy cow. FEMS Immunol. Med. Microbiol. 8:109-118. [DOI] [PubMed] [Google Scholar]
- 11.Hill, A. W., D. J. S. Heneghan, T. R. Field, and M. R. Williams. 1983. Increase in specific opsonic activity in bovine milk following experimental Escherichia coli mastitis. Res. Vet. Sci. 35:222-226. [PubMed] [Google Scholar]
- 12.Hill, A. W., and J. A. Leigh. 1989. DNA fingerprinting of Streptococcus uberis: a useful tool for epidemiology of bovine mastitis. Epidemiol. Infect. 103:165-171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill, A. W., A. L. Shears, and K. G. Hibbit. 1978. The elimination of serum resistant Escherichia coli from experimentally infected mammary glands of healthy dairy cows. Res. Vet. Sci. 25:89-93. [PubMed] [Google Scholar]
- 14.Hill, A. W. 1988. Protective effect of previous intramammary infections with Streptococcus uberis against subsequent clinical mastitis in the cow. Res. Vet. Sci. 44:386-391. [PubMed] [Google Scholar]
- 15.Hill, A. W. 1988. Pathogenicity of two strains of Streptococcus uberis infused into the lactating and non-lactating mammary gland. Res. Vet. Sci. 45:400-404. [PubMed] [Google Scholar]
- 16.Hillerton, J. E., M. F. S. Shearn, R. M. Teverson, S.Langridge, and J. M. Booth. 1993. Effect of pre-milking teat dipping on clinical mastitis on dairy farms in England. J. Dairy Res. 60:31-41. [DOI] [PubMed] [Google Scholar]
- 17.Kossaibati, M. A., and R. J. Esslemont. 1997. The costs of production diseases in dairy herds in England. Vet. J. 154:41-51. [DOI] [PubMed] [Google Scholar]
- 18.Leigh, J. A., and T. R. Field. 1994. Streptococcus uberis resists phagocytosis despite the presence of bound immunoglobulin. Infect. Immun. 62:1854-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leigh, J. A., J. M. Finch, T. R. Field, N. C. Real, A. Winter, A. W. Walton, and S. M. Hodgkinson. 1999. Vaccination with the plasminogen activator from Streptococcus uberis induces an inhibitory response and protects against experimental infection in the dairy cow. Vaccine 17:851-857. [DOI] [PubMed] [Google Scholar]
- 20.Leigh, J. A., T. R. Field, and M. R. Williams. 1990. Two strains of Streptococcus uberis, of differing ability to cause clinical mastitis, differ in their ability to resist some host defence factors. Res. Vet. Sci. 49:85-87. [PubMed] [Google Scholar]
- 21.Moses, A. E., M. R. Wessels, K. Zalcman, S. Alberti, S. Natanson-Yaron, T. Menes, and E. Hanski. 1997. Relative contributions of hyaluronic acid capsule and M protein to virulence in a mucoid strain of the group A Streptococcus. Infect. Immun. 65:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paape, M. J., W. P. Wergin, A. J. Guidry, and W. D. Schultze. 1981. Phagocytic defense of the ruminant mammary gland. Adv. Exp. Med. Biol. 137:555-578. [PubMed] [Google Scholar]
- 23.Persson, K., I. Larsson, and C. Hallen-Sandgren. 1993. Effects of certain inflammatory mediators on bovine neutrophil migration in vivo and in vitro. Vet. Immunol. Immunopathol. 37:1464-1466. [DOI] [PubMed] [Google Scholar]
- 24.Schalm, O. W., J. Lasmanis, and N. C. Jain. 1976. Conversion of chronic staphylococcal mastitis to acute gangrenous mastitis after neutropenia in blood and bone marrow produced by an equine anti-bovine leucocyte. Am. J. Vet. Res. 37:885-890. [PubMed] [Google Scholar]
- 25.Thomas, L. H., W. Haider, A. W. Hill, and R. S. Cook. 1994. Pathologic findings of experimentally induced Streptococcus uberis infection in the mammary gland of cows. Am. J. Vet. Res. 55:1723-1728. [PubMed] [Google Scholar]
- 26.Ward, P. N., T. R. Field, W. G. F. Ditcham, E. Maguin, and J. A. Leigh. 2001. Identification and disruption of two discrete loci encoding hyaluronic acid capsule biosynthesis genes hasA, hasB, and hasC in Streptococcus uberis. Infect. Immun. 69:392-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watts, J. L. 1988. Etiologic agents of bovine mastitis. Vet. Microbiol. 16:41-66. [DOI] [PubMed] [Google Scholar]
- 28.Williams, M. R., and A. W. Hill. 1982. A role for IgM in the in vitro opsonisation of Staphylococcus aureus and Escherichia coli by bovine polymorphonuclear leukocytes. Res. Vet. Sci. 33:47-53. [PubMed] [Google Scholar]