Abstract
Upregulation of HER2/ErbB2/Neu occurs in 15–30% of human breast cancers and correlates with poor prognosis. Identification of ErbB2/Neu transcriptional targets should facilitate development of novel therapeutic approaches. Development of breast cancer is a multistep process; thus, to identify the transcriptomes associated with different stages of progression of tumorigenesis, we compared expression profiles of mammary tumors and preneoplastic mammary tissue from MMTV-Neu transgenic mice to expression profiles of wild-type mammary glands using Affymetrix microarrays. We identified 324 candidate genes that were unique to ErbB2/Neu-induced tumors relative to normal mammary gland tissue from wild-type controls. Expression of a subset of these genes (82) was also changed in the preneoplastic mammary glands compared to wild-type controls, indicating that they may play a pivotal role during early events of ErbB2/Neu-initiated mammary tumorigenesis. Further analysis of the microarray data revealed that expression of several known transforming growth factor (TGF)-β target genes was altered, suggesting that the TGF-β signaling cascade is downregulated in ErbB2/Neu-induced tumors. Western blot analysis for TGF-β-Receptor-I/ALK5 and immunohistochemistry for TGF-β-Receptor-I/ALK5 and phosphorylated/activated Smad2 confirmed that the Smad-dependent TGF-β signaling cascade was inactive in these tumors. Although absent in most of the tumor, phosphorylated Smad2 was present in the periphery of tumors. Interestingly, presence of phosphorylated/activated Smad2 correlated with expression of Activin-Receptor-IB/ALK4, suggesting that although Smad-dependent TGF-β signaling is absent in ErbB2/Neu-induced tumors, Activin signaling may be active at the leading edge of these tumors. Cumulatively, these data indicate that the TGF-β pathway is intrinsically suppressed in ErbB2/Neu tumors via a mechanism involving loss of TGF-β-Receptor-I/ALK5.
Keywords: mammary cancer progression, gene expression profiling, ErbB2/Neu/HER2, transgenic mice, transforming growth factor-β
Introduction
Activation of the ErbB family of growth factor receptors and subsequent stimulation of their associated intracellular signaling pathways is a significant factor in the genesis of several human cancers (Salomon et al., 1995). Amplification of the ErbB2 (hereafter referred to as ErbB2/Neu) gene and consequent protein overexpression occurs in 15–30% of primary human breast tumors (Slamon et al., 1989, 1995). This overexpression is strongly associated with poor prognosis (Salomon et al., 1995), as well as resistance to endocrine and conventional chemotherapy (Wright et al., 1992). The oncogenic potential of ErbB2/Neu has been confirmed in the mammary epithelia of transgenic mice (Muller et al., 1988; Bouchard et al., 1989; Guy et al., 1992, 1996). Mice bearing the rat c-neu proto-oncogene under transcriptional control of the mouse mammary tumor virus (MMTV) promoter/enhancer (hereafter referred to as MMTV-Neu mice) stochastically develop focal mammary tumors and pulmonary metastases after a long latency (Guy et al., 1992). This mouse model serves as an excellent tool for deciphering the molecular pathways responsible for ErbB2/Neu-induced tumorigenesis and identifying novel targets for both chemoprevention and chemotherapy (Li et al., 1997; Boggio et al., 1998; Lenferink et al., 2000; Shepherd et al., 2001; Yu et al., 2001; Bulavin et al., 2004).
The ErbB2/Neu gene is located on human chromosome 17q12 and encodes a 185-kDa member of the ErbB family of cell surface receptor tyrosine kinases (Yamamoto et al., 1986). This family of growth factor receptors is comprised of four members: epidermal growth factor receptor (also termed ErbB1/HER1), ErbB2/Neu/HER2/p185, ErbB3/HER3, and ErbB4/HER4 (Hynes and Stern, 1994). Although ErbB2/Neu is an orphan receptor with no high-affinity ligand (Holbro et al., 2003), it is the preferred heterodimerization partner of the other ligand-activated family members (Tzahar et al., 1996; Graus-Porta et al., 1997). Consequent to ligand-induced formation of receptor heterodimers, each receptor subunit is activated by transphosphorylation. These phosphorylated residues serve as docking sites for a host of intracellular signaling molecules. Ultimately, these diverse signaling cascades converge on the nucleus to alter the cellular transcriptome, and the transcriptional targets of receptor activation mediate many of the physiological changes manifested by these receptors. These changes include regulation of cell growth, differentiation, motility, and death (Alroy and Yarden, 1997). As such, dysregulation of any component of the pathway such as the ErbB ligands, cell surface receptors, signaling molecules, or transcriptional targets has profound implications for both normal development and malignant transformation of multiple tissues.
Development of breast cancer is a multistep process, beginning with a benign stage and progressing through intermediate stages marked by hyperproliferation of breast epithelium and ultimately resulting in invasive carcinomas (Wellings and Jensen, 1973; Krishnamurthy and Sneige, 2002). Although partial transcriptomes have been developed for ErbB2/Neu tumors, these have been limited to association of the gene expression profile to a specific tumor type without assessment of preneoplastic changes associated with ErbB2/Neu expression. In contrast, characterizing the early events of ErbB2/Neu-induced tumorigenesis should provide insight into the molecular mechanisms of tumorigenesis. To identify progressive changes in the transcriptome that occur during the transition from normal mammary gland to an overt ErbB2/Neu-initiated tumor, the partial transcriptomes of preneoplastic mammary glands and tumors from MMTV-Neu mice were compared to the partial transcriptomes of glands from wild-type mice using the Affymetrix U74Av2 GeneChip arrays. Analysis of these data revealed a subset of genes that were altered in expression during the progression from preneoplastic mammary gland to overt tumors.
In addition to identifying a transcriptional profile associated with neoplastic progression, we found that several genes downregulated in ErbB2/Neu-induced tumors were known targets of the transforming growth factor (TGF)-β signaling pathway. This suggested that the TGF-β pathway might be suppressed in ErbB2/Neu-induced mammary tumors. Normally, the TGF-β signaling pathway is activated when a member of the TGF-β superfamily of ligands (TGF-β, Activin, bone morphogenetic protein, Nodal) induces formation of a heterotetrameric complex of two type II receptors and two type I receptors (Massague, 1998; Yue and Mulder, 2001). Signals are transduced from the cell surface to the nucleus by activated Smad complexes. In the canonical signaling pathway, the binding of TGF-β, Activin, or Nodal ligands to their selective receptors specifically activates the Smad2 and Smad3 receptor-activated Smads via phosphorylation. Smad2 and Smad3 then form heteromeric complexes with the common mediator-Smad, Smad4, to enable translocation to the nucleus and transcriptional regulation of numerous genes. Induction of expression of the inhibitory Smads, Smad6 and Smad7, serves as a negative feedback mechanism. In the mammary gland, the TGF-β pathway regulates normal ductal and alveolar development and remodeling during postlactational involution (Barcellos-Hoff and Ewan, 2000; Wakefield et al., 2000). The TGF-β pathway also has a paradoxical role in mammary tumorigenesis (Reiss, 1999; Derynck et al., 2001; Roberts and Wakefield, 2003; Tang et al., 2003). While TGF-β is growth suppressive during early tumorigenesis, it can promote malignancy and metastasis later during tumorigenic progression. Thus, evaluating the role of TGF-β during ErbB2/Neu tumorigenesis should yield significant insight into the processes of ErbB2/Neu tumor initiation and progression.
Results
Identification of an ErbB2/Neu mammary tumor molecular signature
No previous studies have examined early changes in mammary tissues that ultimately accumulate ErbB2/Neu-initiated tumors. To identify early changes associated with ErbB2/Neu expression, we utilized the MMTV-Neu mouse model of mammary cancer (Guy et al., 1992). We evaluated three different mammary tissues: wild-type, preneoplastic tissue that expresses ErbB2/Neu, and ErbB2/Neu-induced tumors. The latter two were collected from the same mice. The tissue dissection is illustrated in Figure 1a. To ensure the distinction between tumor and preneoplastic tissue, the tissue immediately bordering the tumor was discarded, while the tumor and more distal surrounding mammary tissue (hereafter referred to as adjacent ErbB2/Neu) were used in comparative analysis of gene expression profiles. We anticipated that the mammary gland tissue surrounding ErbB2/Neu-induced tumors would express the ErbB2/Neu transgene and be preneoplastic, but does not contain an overt tumor. Northern blot analysis confirmed that the MMTV-Neu transgene was indeed expressed in the adjacent ErbB2/Neu mammary glands (Figure 1b).
Figure 1.
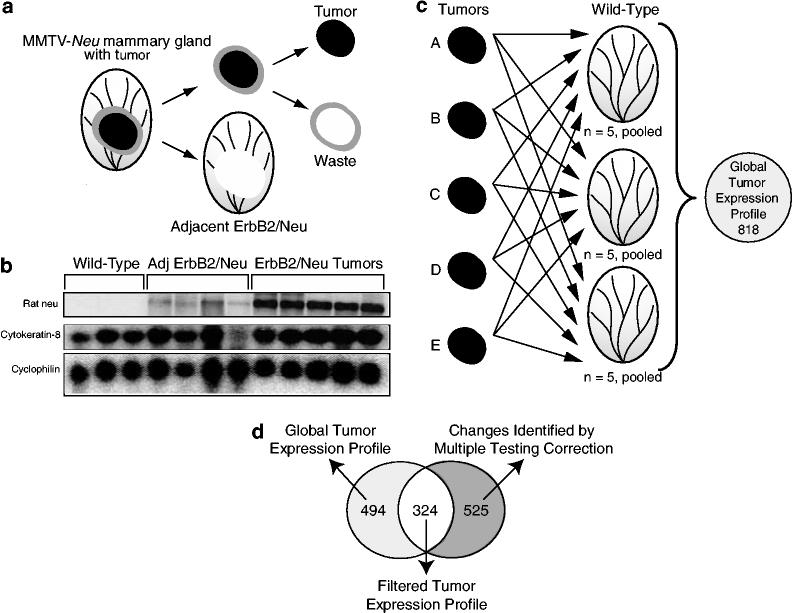
Identification of an ErbB2/Neu mammary tumor molecular signature. (a) Experimental design. Three mammary gland tissue types were isolated from mice for gene expression microarray analyses including: age-matched wild-type (Wild-Type), ErbB2/Neu tumor, and tissue adjacent to, but not including, the tumor. For the ErbB2/Neu-expressing tissues, the entire mammary gland was removed from the MMTV-Neu mouse. The tumor plus the 2 mm border of tissue immediately surrounding the tumor were cut out. This 2 mm bordering peritumoral tissue was removed and discarded. The adjacent ErbB2/Neu gland and the tumor were retained for analysis. (b) Northern blot analysis confirms ErbB2/Neu transgene expression in adjacent ErbB2/Neu tissue. In total, 20 μg of total RNA from microarray samples were separated by gel electrophoresis, transferred to nylon membrane, and then hybridized with a PCR-generated probe for Rat neu, cytokeratin-8 (epithelial cell marker), and cyclophilin (loading control). (c) Identification of genes with altered expression using the Affymetrix Change Call Algorithm. Samples were analysed by Affymetrix MGU74Av2 microarrays. The gene expression profile of each tumor (n = 5) was compared to the expression profile of each pooled wild-type sample (n = 3 profiles; five pooled tissues per profile). Only genes that were called changed in every comparison using the Affymetrix change call algorithm were retained. The intersection of these 15 comparisons (five tumors × three wild-type samples) contained 821 genes and ESTs. Removal of genes that were called absent on all of the microarrays reduced the list to 818 genes and ESTs. These genes represent the global tumor expression profile for ErbB2/Neu-induced mammary tumors. (d) Comparison of the Affymetrix subset of genes to the subset of genes acquired using Welch's approximate t-test with the Benjamini and Hochberg multiple testing correction. Statistical analysis of the entire 12 448 genes identified 849 genes that were statistically different in expression levels when comparing the ErbB2/Neu tumor partial transcriptomes to the wild-type partial transcriptomes. A total of 324 genes met both the Affymetrix algorithm and the statistical criteria, as indicated by the Venn diagram. These genes represent a filtered ErbB2/Neu tumor expression profile
Each of the three tissue types described above was analysed by gene expression profiling. To obtain a representative average of wild-type gene expression for baseline comparisons, RNA was isolated from 15 age-matched wild-type control mammary glands and then pooled into three groups of five samples, thus minimizing contributions due to interindividual variation. All samples including pooled, wild-type controls (n = 3), adjacent ErbB2/Neu glands (n = 4), and ErbB2/Neu tumors (n = 5) were analysed by Affymetrix U74Av2 microarrays containing 12 448 probe sets for known genes and ESTs. A subtractive approach was used to identify differentially expressed transcriptional targets in ErbB2/Neu tumors compared to wild-type tissue. Using Affymetrix Microarray Suite, data from each ErbB2/Neu tumor microarray analysis (n = 5) were compared to the data collected from each pooled wild-type sample (n = 3) resulting in 15 total comparisons (five tumors × three wild-type) (Figure 1c). Transcripts that were called ‘not changed’ in any comparison by the Affymetrix algorithm were eliminated from the gene list. The intersection of these 15 lists contained 821 genes. This list was further reduced to 818 genes by removing any genes that were called ‘absent’ in all analyses. The small reduction of genes by this second filter suggests that the first filtering step was sufficiently stringent to remove most genes whose expression remained unchanged when comparing tumors to wild-type samples. This list of 818 genes represents the global gene expression profile of ErbB2/Neu-induced tumors when compared to wild-type glands after interrogating the 12 488 gene set from Affymetrix and is displayed in Supplementary Table 1.
A subset of genes was further delineated by additional statistical analyses. First, we applied the Welch's t-test with the Benjamini and Hochberg Multiple Testing Correction (False Discovery Rate of 5%) to all 12 448 probe sets represented on the microarrays. Comparison of the five tumors to three wild-type control samples resulted in the identification of 829 transcripts that had statistically different levels of expression in ErbB2/Neu tumor tissue compared to wild-type controls. We next evaluated the overlap in the two sets of genes identified by the Affymetrix algorithm (818 genes) or using the multiple testing correction (829 genes). In all, 324 genes were independently retained by both analytical approaches, thus delineating a filtered subset of transcripts with statistically significant alterations of gene expression (Figure 1d; Supplementary Table 2). Genes with a P-value <0.01 are shown in Table 1. At this level of significance, 23 genes were increased in tumors while 42 genes were decreased. To determine the reproducibility of the gene expression data, we analysed two additional independent tumors by microarray analysis and found high repeatability (93% confirmed in both tumors) as indicated by the ‘repeated observation’ column in Table 1. To further assess the accuracy of the microarray data through an independent approach, we analysed 87 of the 324 genes that were included in the filtered gene expression profile by real time reverse transcriptase (RT)–PCR. This analysis involved comparisons between three additional tumors as well as three more wild-type control samples. The direction of alteration of gene expression (increased/decreased) in tumors compared to three wild-type controls was confirmed for 73 of the 87 genes (88%). Only three genes (3%) were changed in opposite directions when comparing the microarray and real-time RT–PCR data. Overall, the data generated by real-time RT–PCR provided independent confirmation of the microarray results, suggesting that the method of analysis employed for evaluating the microarray data was sufficiently stringent to identify characteristic changes associated with ErbB2/Neu-induced mammary tumors.
Table 1.
A total of 65 genes consistently demonstrated significant (P<0.01) alterations of gene expression in ErbB2/Neu-induced mammary tumors relative to age-matched wild-type control mammary glands
| Gene name | Genbank accession | Affymetrix accession | Abs calla |
Fold changeb |
SOM Cl #c | Rep Obsd | RT–PCRe | |
|---|---|---|---|---|---|---|---|---|
| ANvsWT | TUvsWT | |||||||
| Increased in tumors | ||||||||
| Tumor protein D52-like 1 | AF004428 | 101446_at | A to P | 2.7 | 12.1 | 2 | +/+ | Y |
| Dual specificity phosphatase 6 | AI845584 | 93285_at | 3.1 | 10.4 | 2 | +/+ | ||
| Desmoglein 2 | AI152659 | 104480_at | 4.1 | 6.1 | 4 | +/+ | ||
| Inositol 1,4,5-triphosphate receptor 5 | AF031127 | 101441_i_at | 2.0 | 4.7 | 2 | +/+ | Y | |
| Cytidine monophospho-N-acetylneuraminic acid synthetase | AJ006215 | 98593_at | 1.7 | 4.7 | 2 | +/+ | ||
| p53 apoptosis effector related to Pmp22 | AI854029 | 97825_at | 1.9 | 4.0 | 2 | +/+ | ||
| Folate receptor 1 (adult) | AV035020 | 162302_f_at | A to P | 2.2 | 3.7 | 4 | +/+ | |
| Latexin | D88769 | 96065_at | 1.7 | 3.4 | 2 | +/+ | ||
| Hypothetical protein 6330505N24 | AI430272 | 104207_at | 1.5 | 3.0 | 2 | +/+ | ||
| RIKEN cDNA 1200015G06 gene | AI844370 | 93590_at | 2.3 | 2.8 | 2 | +/+ | ||
| ATPase, H+ transporting, V1 subunit A, isoform 1 | AW123765 | 95746_at | 1.2 | 2.6 | 2 | +/+ | ||
| DNA segment, Chr 3, UCLA 1 | AI843466 | 95708_at | 1.8 | 2.6 | 4 | +/+ | ||
| CD9 antigen | L08115 | 95661_at | 1.4 | 2.5 | +/+ | |||
| Insulin-like growth factor 2 receptor | U04710 | 95117_at | 1.3 | 2.5 | +/+ | Y | ||
| CASP2 and RIPK1 domain containing adaptor with death domain | AJ224738 | 102952_g_at | 1.2 | 2.4 | 2 | +/+ | N | |
| T-cell immunoglobulin and mucin domain containing 2 | AA795198 | 97335_at | 1.5 | 2.4 | 4 | +/– | ||
| Golgi phosphoprotein 3 | AW060175 | 160688_at | 1.4 | 2.3 | +/+ | |||
| Calcium-binding protein, intestinal | Y00884 | 95423_at | 1.5 | 2.2 | 2 | +/+ | ||
| Bcl2-associated athanogene 2 | W71352 | 160962_at | 1.6 | 2.2 | +/+ | |||
| Death-associated protein | AI196645 | 93842_at | 1.3 | 2.1 | +/+ | |||
| RAB18, member RAS oncogene family | L04966 | 94319_at | 1.2 | 1.8 | +/+ | 2 of 3 | ||
| WD-40-repeat-containing protein with a SOCS box 2 | AF033188 | 160296_at | 1.2 | 1.7 | +/+ | |||
| Interleukin 25 | AW045739 | 96318_at | 1.4 | 1.7 | +/+ | |||
| Decreased in tumors | ||||||||
| RIKEN cDNA 1620401E04 gene | AW125480 | 96900_at | 0.66 | 0.49 | 3 | +/+ | ||
| ATPase, Na+/K+ transporting, beta 3 polypeptide | U59761 | 99579_at | 0.75 | 0.48 | +/+ | |||
| l-3-hydroxyacyl-Coenzyme A dehydrogenase, short chain | D29639 | 95485_at | 0.71 | 0.45 | 3 | +/+ | ||
| Phytanoyl-CoA hydroxylase | AF023463 | 96608_at | 0.72 | 0.44 | 3 | +/+ | ||
| Amyloid beta (A4) precursor-like protein 2 | M97216 | 93498_s_at | 0.64 | 0.41 | 3 | +/+ | ||
| Transforming growth factor beta 1 induced transcript 4 | X62940 | 93728_at | 0.82 | 0.38 | 1 | –/+ | ||
| RIKEN cDNA 1110021N07 gene | AI846382 | 93983_at | P to A | 0.58 | 0.38 | 3 | +/+ | |
| Carnitine palmitoyltransferase 2 | U01170 | 95646_at | 0.64 | 0.36 | 3 | +/+ | ||
| Myristoylated alanine-rich protein kinase C substrate | M60474 | 96865_at | 0.90 | 0.35 | 3 | +/+ | ||
| Branched chain ketoacid dehydrogenase E1, beta polypeptide | L16992 | 102302_at | 0.59 | 0.33 | 3 | +/+ | ||
| RIKEN cDNA 4921515A04 gene | AI642098 | 104494_at | 0.53 | 0.33 | 3 | +/+ | ||
| Glycogenin 1 | AW049730 | 100597_at | 0.51 | 0.31 | 3 | +/+ | ||
| Acetyl-Coenzyme A acyltransferase 2 | AI849271 | 95064_at | 0.65 | 0.30 | 3 | +/+ | ||
| Phosphoglucomutase 2 | AI842432 | 104313_at | 0.52 | 0.29 | 3 | +/+ | ||
| RIKEN cDNA 1110001I14 gene | AW047554 | 104605_at | 0.45 | 0.26 | 3 | +/+ | ||
| 3-Ketoacyl-CoA thiolase B | AW012588 | 99571_at | P to A | 0.51 | 0.25 | 3 | +/+ | |
| RIKEN cDNA D330037A14 gene | AI854506 | 96206_at | P to A | 0.69 | 0.25 | 1 | +/+ | |
| RIKEN cDNA 1200015A22 gene | AI854863 | 98633_at | P to A | 0.44 | 0.24 | 3 | +/+ | |
| Kruppel-like factor 4 (gut) | U20344 | 99622_at | 0.69 | 0.23 | 3 | +/+ | Y | |
| Glycerol phosphate dehydrogenase 2, mitochondrial | D50430 | 98984_f_at | 0.57 | 0.20 | 3 | +/+ | ||
| Preproenkephalin 1 | M55181 | 94516_f_at | 0.56 | 0.20 | 3 | +/+ | ||
| Phosphodiesterase 8A | AF067806 | 160941_at | P to A | 0.82 | 0.19 | 1 | +/+ | Y |
| Actin, alpha 2, smooth muscle, aorta | X13297 | 93100_at | 0.87 | 0.18 | 1 | +/+ | ||
| Neuropilin | D50086 | 95016_at | 0.82 | 0.18 | 3 | +/+ | ||
| Serum deprivation response | AI839175 | 160373_i_at | 0.54 | 0.18 | 3 | +/+ | Y | |
| Procollagen, type VI, alpha 3 | AF064749 | 101110_at | 0.60 | 0.17 | 3 | +/+ | ||
| Glycerol-3-phosphate acyltransferase, mitochondrial | U11680 | 101867_at | 0.38 | 0.17 | 3 | +/+ | ||
| Hephaestin | AF082567 | 104194_at | 0.40 | 0.16 | 1 | +/+ | ||
| Small chemokine (C-C motif) ligand 11 | U77462 | 92742_at | P to A | 0.58 | 0.15 | 3 | +/+ | |
| Synaptonemal complex protein 3 | AW212131 | 93994_at | 0.54 | 0.15 | 3 | +/+ | ||
| UDP glycosyltransferase 1 family, polypeptide A6 | U16818 | 99580_s_at | P to A | 0.64 | 0.12 | 3 | +/+ | |
| Chemokine (C-X-C motif) ligand 12 | AV139913 | 162234_f_at | P to A | 0.58 | 0.12 | 3 | +/+ | |
| Aldehyde dehydrogenase family 1, subfamily A7 | U96401 | 94778_at | P to A | 0.54 | 0.10 | 3 | +/+ | |
| Sushi-repeat-containing protein | AB028049 | 103568_at | P to A | 0.68 | 0.10 | 1 | +/+ | Y |
| Dermatopontin | AA717826 | 96742_at | 0.68 | 0.09 | 3 | +/+ | Y | |
| Prostaglandin E synthase | AI060798 | 104406_at | P to A | 0.71 | 0.08 | 1 | +/+ | |
| Protease, serine, 11 (Igf binding) | AW125478 | 96920_at | 0.54 | 0.08 | 3 | +/+ | Y | |
| RIKEN cDNA 2610042L04 gene | AI853444 | 93568_i_at | P to A | 1.26 | 0.08 | 1 | +/+ | |
| Lumican | AF013262 | 93353_at | 0.90 | 0.07 | 1 | +/+ | ||
| Cytochrome P450, family 4, subfamily b, polypeptide 1 | AV376161 | 162044_f_at | P to A | 0.86 | 0.07 | 1 | +/+ | |
| Glutathione peroxidase 3 | U13705 | 101676_at | 0.33 | 0.04 | 3 | +/+ | ||
| Thrombospondin type 1 domain-containing gene | AB016768 | 98312_at | P to A | 1.04 | 0.03 | 1 | +/+ | |
Absolute call is the Affymetrix parameter that measures detection. P = present A = absent. ‘A to P’ indicates that a gene was called ‘absent’ in control and ‘present’ in tumor. If a gene changes from absent to present or vice versa, the fold change is irrelevant
Fold change was derived from the Affymetrix signal log ratio (SLR). AN = adjacent ErbB2/Neu, WT = wild-type control, and TU = tumor
‘SOM CL #’ = self-organizing map cluster number. Clusters are shown in Figure 3b
Repeated observation indicates whether the gene was confirmed by microarray data from two additional independent tumor samples. ‘+/+’ indicates confirmed by both tumors analysed and ‘+/–’ or ‘–/+’ by one tumor
Direction (increased/decreased) of expression confirmed by real-time RT–PCR. ‘Y’ = yes confirmed in three tumors, ‘2 of 3’ = confirmed in two of three tumors, and ‘N’ = not confirmed. If blank, then not assayed
We considered the possibility that a number of genes in the global tumor profile may be altered due to changes in cellularity when comparing tumors to wild-type tissue. To address this issue, we analysed the expression changes for cytokeratins 8 and 18, both of which are epithelial cell markers. These genes were upregulated in tumors by 1.8±0.8-fold. Thus, any changes in gene expression that are less than 3.4 (the median fold change + two standard deviations) may be due to changes in cellular content. Genes that fail to surpass this cutoff are denoted within the tables.
The adjacent ErbB2/Neu tissue has preneoplastic characteristics
Once we had characterized the molecular profile associated with ErbB2/Neu-induced mammary tumors, we turned our focus to identifying gene expression changes that occur early in the tumorigenic cascade. Mammary gland tissue surrounding the ErbB2/Neu tumors was used for this analysis. Several lines of evidence suggested that this tissue expresses the ErbB2/Neu transgene, exhibits active ErbB2/Neu signaling, and is preneoplastic. Histological examination of the mammary gland surrounding ErbB2/Neu-induced tumors demonstrated modest focal hyperplasia and distended ducts as reported previously (Boggio et al., 1998) (Figure 2a). Furthermore, immunohistochemical analysis of independent samples for phosphorylated ErbB2/Neu (Tyr-877) demonstrated activation of ErbB2/Neu signaling in adjacent ErbB2/Neu mammary tissue, whereas wild-type tissue lacked phospho-ErbB2/Neu immunoreactivity (Figure 2b). To determine whether this tissue harbored early transcriptional changes induced by active ErbB2/Neu signaling, the microarray data were inspected for known ErbB2/Neu targets. Ets variant 1 (ER81), Cyclin D1, and LIM Only-4 (LMO4) genes have previously been reported to be upregulated in ErbB2/Neu-induced mammary tumors or cell lines with activated ErbB2/Neu signaling (Lee et al., 2000; Shepherd et al., 2001; Wang et al., 2004). These genes appeared in the global ErbB2/Neu tumor molecular signature and were also upregulated in the adjacent ErbB2/Neu samples (Supplementary Table 1; Figure 2c). Furthermore, three previously characterized ErbB2/Neu tumor markers (FXYD3, WDNM1/EXPI, casein-k) (Morrison and Leder, 1994) were elevated in the adjacent ErbB2/Neu samples as well as in tumors (Figure 2c). Confirmation of active ErbB2/Neu signaling and expression of known tumor markers in combination with the gross appearance and histological morphology of the adjacent ErbB2/Neu mammary tissue indicates that this tissue is indeed preneoplastic and, therefore, should be useful in identifying changes in gene expression associated with early carcinogenic mechanisms.
Figure 2.
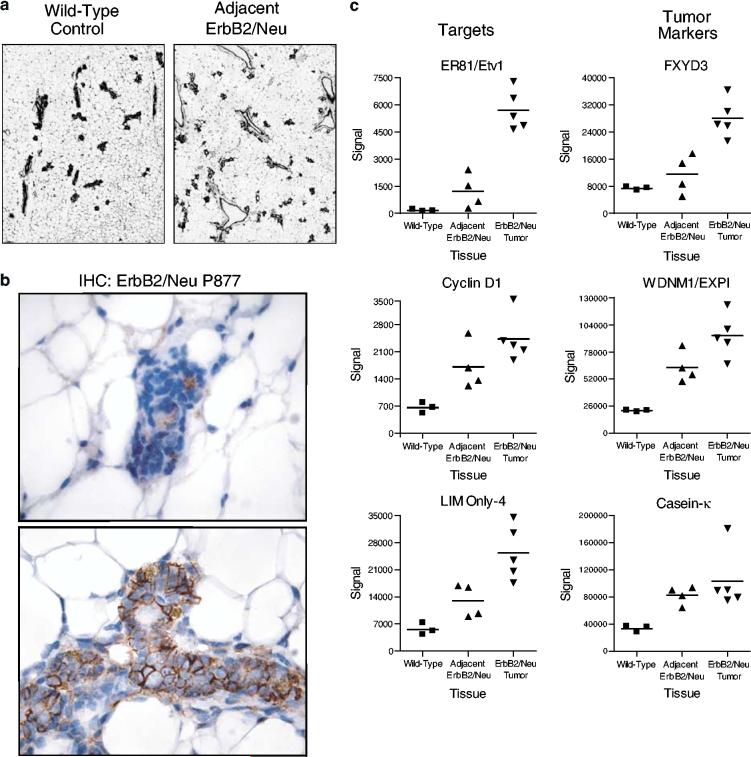
The adjacent ErbB2/Neu tissue has preneoplastic characteristics. (a) Mammary gland tissue adjacent to ErbB2/Neu-induced tumors displays focal hyperplasia. Representative sections of wild-type control mammary gland and tissue adjacent to an ErbB2/Neu-induced tumor are shown (H&E, × 40). (b) Adjacent ErbB2/Neu mammary epithelia demonstrate active ErbB2/Neu signaling. Representative sections ( × 400) of immunohistochemical staining for phosphorylated ErbB2/Neu (Tyr-877) in wild-type mammary gland tissue (top panel) and adjacent ErbB2/Neu tissue (bottom panel) are shown. Note the selective membrane staining in the adjacent ErbB2/Neu tissue. (c) Known targets of ErbB2/Neu and previously characterized ErbB2/Neu tumor markers are expressed in the adjacent ErbB2/Neu samples. The signal intensity from microarray data is depicted on the y-axis with the tissue type on the x-axis. The expression values for individual samples are represented with the mean value for each group being indicated by a horizontal line
The molecular profile of the adjacent ErbB2/Neu tissue is intermediate between the profiles of tumors and control mammary tissues
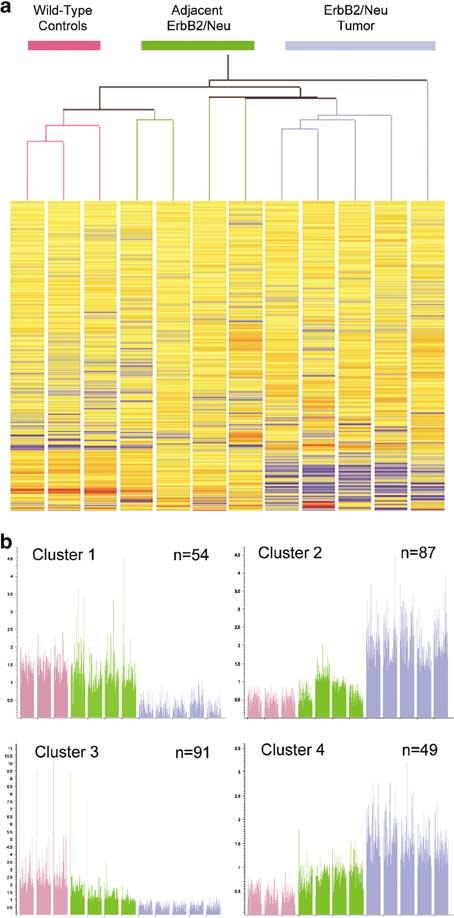
To determine whether subgroups of tissue types could be identified independently based solely on their gene expression profiles, hierarchical clustering analysis (Eisen et al., 1998) was applied to the entire set of transcripts (7976 genes) that were called ‘present’ or ‘marginal’ on at least one of the arrays from the three different tissue types (wild-type, adjacent ErbB2/Neu, and tumor (Figure 3a)). As expected, all tumor samples appeared on separate branches of the dendrogram from wild-type samples with one tumor appearing to be an extreme outlier of the tumor classification. Importantly, placement on the dendrogram did not correlate with somatic mutation of the transgene. Siegel et al. (1994) previously identified activating somatic deletion mutations of the Neu expression cassette in mammary tumors from MMTV-Neu mice. Although previously described as occurring in 65% of tumors in these mice, we found that only one of the seven tumors that were evaluated by the microarray analysis contained a somatic mutation of the transgene (data not shown), and this tumor is the center tumor in the dendrogram. In addition to segregating tumors from wild-type molecular signatures, this analysis revealed that the profiles from two of the adjacent ErbB2/Neu samples were more similar to the wild-type molecular profiles, whereas the other two adjacent ErbB2/Neu samples were more similar to the ErbB2/Neu tumors as indicated by their placement on the dendrogram. Hence, the adjacent ErbB2/Neu molecular profiles were intermediate between the wild-type and ErbB2/Neu tumor molecular profiles, supporting the concept that these samples and their molecular profiles reflect intermediate stages of tumorigenic progression.
Figure 3.
The molecular profile of the adjacent ErbB2/Neu tissue is intermediate between the profiles of tumors and control mammary tissues. (a) A dendrogram derived by two-way hierarchical clustering analysis of 7976 genes that were called ‘present’ or ‘marginal’ on at least one microarray is shown. Tissue types are grouped vertically, and genes are grouped horizontally. The arms of the tree are color coded by tissue type (pink = wild-type control (n = 3); green = adjacent ErbB2/Neu (n = 4); blue = ErbB2/Neu tumor (n = 5)). Expression levels are displayed as red = high, yellow = intermediate, and blue = low expression. (b) Self-organizing Map (SOM) analysis reveals progressive alterations of gene expression that correlate with tumorigenic stage. The y-axis represents the signal intensity derived from microarray analyses. For individual genes, values were normalized to the median value across all the samples and expression of individual genes is represented by a thin vertical line for each gene. Each bar represents a compression of all genes in an individual sample. The sample type is color coded (pink = wild-type control (n = 3); green = adjacent ErbB2/Neu (n = 4); blue = tumor (n = 5)). The clusters have been numbered from 1 to 4 (i.e. Cluster 1 to Cluster 4), with the total number of genes represented in each cluster indicated in the top right corner. Genes within individual clusters are identified in Tables 1 and 2 and in Supplementary Tables 1 and 2
Self-organizing Map (SOM) analysis reveals progressive alterations of gene expression that correlate with tumorigenic stage
Given the placement of tissue types on the hierarchical tree, we suspected that subsets of genes expressed in adjacent ErbB2/Neu samples would display intermediate behavior between wild-type glands and tumors. To assess this directly, we used SOM analysis (Tamayo et al., 1999) to classify the 324 significantly altered genes (i.e. filtered tumor signature) into descriptive patterns of expression. Using the Affymetrix Data Mining Tool, we empirically determined that four nodes generated informative, nonredundant, SOM clusters. SOM cluster 1 (Figure 3b; Table 1 and Supplementary Table 1) contains transcripts that are more highly expressed in wild-type and adjacent ErbB2/Neu mammary glands than in the ErbB2/Neu-induced tumors. SOM clusters 2, 3, and 4 (Figure 3b) reveal progressive patterns in which the expression level of genes in the adjacent ErbB2/Neu samples was intermediate between the wild-type samples and the ErbB2/Neu tumors, thus correlating with ErbB2/Neu transgene expression and neoplastic progression. We considered the possibility that the progressive nature of the changes of gene expression in the preneoplastic tissue was reflective of an experimental artifact due to contaminating tumor tissue in the adjacent ErbB2/Neu samples. The data in clusters 2 and 3 argue against this possibility. Expression of numerous genes in cluster 2 is high in tumors, yet the expression of many of these same genes is unchanged in the adjacent ErbB2/Neu tissues. Likewise, low expression of genes in tumors in cluster 3 should not significantly reduce expression in adjacent ErbB2/Neu tissues; however, these genes are indeed downregulated in adjacent ErbB2/Neu samples compared to wild-type controls. Neither of these clusters can be explained by minor tumor contaminants in adjacent ErbB2/Neu samples. Furthermore, we identified genes that were exclusively altered in expression in the adjacent ErbB2/Neu samples compared to both wild-type and tumors, thus confirming the unique nature of these tissue specimens (data not shown).
To further delineate the transcriptome alterations associated with ErbB2/Neu neoplastic progression, we identified a subset of genes that were consistently changed when comparing wild-type tissue to the two adjacent ErbB2/Neu tissues that shared the most commonalities with the tumor molecular profiles. In total, 388 genes were consistently altered in expression in these two adjacent ErbB2/Neu samples compared to the wild-type control samples as indicated by the Affymetrix ‘change’ parameter (six comparisons: two adjacent ErbB2/Neu × three wild-type controls). Of these 388 genes, 230 were contained in the global ErbB2/Neu tumor molecular profile (Supplementary Table 1) and 82 were included in the filtered ErbB2/Neu tumor molecular signature (Supplementary Table 2). The data for these 82 genes are presented in Table 2.
Table 2.
Identification of genes whose expression changes during the transition from wild-type to preneoplastic mammary tissue and is retained in overt tumors
| Gene name | Genbank accession | Affymetrix accession | Abs calla |
Fold changeb |
SOM Cl #c | P-valued | Rep Obse | |
|---|---|---|---|---|---|---|---|---|
| AN vs WT | TU vs WT | |||||||
| Increased in adjacent erbB2/neu samples | ||||||||
| Nephronectin | AA592182 | 103721_at | A to P | 6.8 | 49.9 | 2 | 0.0419 | +/+ |
| Fatty acid-Coenzyme A ligase, long chain 4 | AA619207 | 102381_at | 5.7 | 19.4 | 2 | 0.0210 | +/+ | |
| ets variant gene 1 | L10426 | 92927_at | A to P | 5.5 | 34.1 | 2 | 0.0187 | +/+ |
| Downstream of tyrosine kinase 1 | U78818 | 102896_at | A to P | 4.9 | 8.5 | 2 | 0.0208 | +/+ |
| RIKEN cDNA 1600029D21 gene | AI121305 | 97413_at | 4.2 | 8.6 | 4 | 0.0403 | +/+ | |
| Desmoglein 2 | AI152659 | 104480_at | 4.1 | 6.1 | 4 | 0.0049 | +/+ | |
| est | C77278 | 97165_r_at | A to P | 3.1 | 3.9 | 4 | 0.0236 | +/+ |
| Potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 | AF042487 | 102198_at | 3.0 | 10.1 | 2 | 0.0306 | +/+ | |
| RIKEN cDNA 1700051C09 gene | AV260411 | 161227_r_at | 2.9 | 5.2 | 4 | 0.0252 | +/+ | |
| Extracellular proteinase inhibitor | X93037 | 103051_at | 2.8 | 3.6 | 0.0352 | +/+ | ||
| Lipocalin 2 | X81627 | 160564_at | 2.7 | 6.9 | 0.0111 | +/+ | ||
| Homer homolog 2 (Drosophila) | AF093259 | 160695_i_at | A to P | 2.6 | 7.2 | 4 | 0.0276 | +/+ |
| Kangai 1 (suppression of tumorigenicity 6, prostate) | D14883 | 99584_at | 2.3 | 3.1 | 4 | 0.0133 | +/+ | |
| Stromal cell-derived factor receptor 2 | D50464 | 103421_at | 2.3 | 3.1 | 4 | 0.0218 | +/+ | |
| Inhibitor of DNA binding 2 | AF077861 | 93013_at | 2.1 | 3.7 | 4 | 0.0462 | +/+ | |
| UDP-N-acetyl-alpha-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 | AV055653 | 162313_f_at | 2.0 | 5.1 | 4 | 0.0295 | +/+ | |
| S-phase kinase-associated protein 1A | Z47088 | 99607_at | 2.0 | 3.1 | 4 | 0.0248 | +/+ | |
| Tumor protein D52 | U44426 | 160249_at | 1.8 | 2.9 | 4 | 0.0356 | +/+ | |
| Ribonucleotide reductase M2 | M14223 | 102001_at | 1.8 | 2.9 | 4 | 0.0130 | +/+ | |
| WW domain-binding protein 5 | U92454 | 100522_s_at | 1.7 | 2.8 | 4 | 0.0201 | +/+ | |
| RIKEN cDNA 3110003A17 gene | AA833425 | 96135_at | 1.7 | 2.3 | 4 | 0.0161 | +/+ | |
| RIKEN cDNA 2310009N05 gene | AW061073 | 160801_at | 1.6 | 3.1 | 2 | 0.0189 | +/+ | |
| Checkpoint kinase 1 homolog (Saccharomyces pombe) | AF016583 | 103064_at | 1.4 | 2.0 | 4 | 0.0356 | –/+ | |
| RIKEN cDNA 2610042L04 gene | AI853444 | 93568_i_at | 1.3 | 0.1 | 1 | 0.0056 | +/+ | |
| Decreased in adjacent erbB2/neu samples | ||||||||
| RIKEN cDNA 1110020P15 gene | AW046239 | 94078_at | 0.8 | 0.6 | 3 | 0.0193 | +/+ | |
| ATPase, Na+/K+ transporting, beta 3 polypeptide | U59761 | 99579_at | 0.8 | 0.5 | 0.0097 | +/+ | ||
| Dehydrogenase/reductase (SDR family) member 7 | AW120882 | 95620_at | 0.7 | 0.5 | 0.0289 | +/+ | ||
| NADH dehydrogenase (ubiquinone) flavo-protein 1 | AI846127 | 96267_at | 0.7 | 0.5 | 0.0234 | +/+ | ||
| RIKEN cDNA 1620401E04 gene | AW125480 | 96900_at | 0.7 | 0.5 | 3 | 0.0085 | +/+ | |
| RIKEN cDNA 1300003D03 gene | AA871791 | 102052_at | 0.7 | 0.4 | 3 | 0.0407 | +/+ | |
| Acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) | AI849271 | 95064_at | 0.7 | 0.3 | 3 | 0.0050 | +/+ | |
| Amyloid beta (A4) precursor-like protein 2 | M97216 | 93498_s_at | 0.6 | 0.4 | 3 | 0.0085 | +/+ | |
| Carnitine palmitoyltransferase 2 | U01170 | 95646_at | 0.6 | 0.4 | 3 | 0.0083 | +/+ | |
| RIKEN cDNA 1110015E22 gene | AW045753 | 104217_at | 0.6 | 0.1 | 3 | 0.0387 | –/– | |
| Insulin-like growth factor-binding protein 6 | X81584 | 103904_at | 0.6 | 0.0 | 1 | 0.0230 | +/+ | |
| NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1 | AI849803 | 96909_at | 0.6 | 0.5 | 3 | 0.0155 | +/+ | |
| MAP kinase-interacting serine/threonine kinase 2 | AI845732 | 101007_at | 0.6 | 0.3 | 3 | 0.0499 | +/+ | |
| Branched chain aminotransferase 2, mitochondrial | AF031467 | 100443_at | 0.6 | 0.3 | 3 | 0.0403 | +/+ | |
| Pyruvate dehydrogenase E1 alpha 1 | M76727 | 98102_at | 0.6 | 0.4 | 3 | 0.0339 | +/+ | |
| Chemokine (C-X-C motif) ligand 12 | L12030 | 100112_at | 0.6 | 0.3 | 3 | 0.0208 | +/+ | |
| Immunoglobulin joining chain | M90766 | 102372_at | 0.6 | 0.1 | 3 | 0.0470 | +/+ | |
| Niemann pick type C2 | AB021289 | 160344_at | 0.6 | 0.5 | 0.0189 | +/+ | ||
| Procollagen, type VI, alpha 3 | AF064749 | 101110_at | 0.6 | 0.2 | 3 | 0.0088 | +/+ | |
| Branched chain ketoacid dehydrogenase E1, beta polypeptide | L16992 | 102302_at | 0.6 | 0.3 | 3 | 0.0014 | +/+ | |
| Uridine monophosphate kinase | L31783 | 94381_at | 0.6 | 0.5 | 3 | 0.0100 | +/+ | |
| Protein phosphatase 2, regulatory subunit B (B56), alpha isoform | AI956230 | 93826_at | 0.6 | 0.2 | 3 | 0.0409 | +/+ | |
| Nuclear receptor subfamily 1, group H, member 3 | AF085745 | 104381_at | 0.6 | 0.1 | 3 | 0.0390 | +/+ | |
| Aminolevulinate, delta-, dehydratase | X13752 | 101044_at | 0.6 | 0.3 | 3 | 0.0375 | +/+ | |
| RIKEN cDNA 1110032O19 gene | AI853294 | 97386_at | 0.6 | 0.3 | 3 | 0.0100 | +/+ | |
| Glycerol phosphate dehydrogenase 2, mitochondrial | D50430 | 98984_f_at | 0.6 | 0.2 | 3 | 0.0001 | +/+ | |
| Glutathione transferase zeta 1 (maleylacetoacetate isomerase) | AW060750 | 160350_at | 0.6 | 0.1 | 3 | 0.0301 | +/+ | |
| Preproenkephalin 1 | M55181 | 94516_f_at | 0.6 | 0.2 | 3 | 0.0098 | +/+ | |
| Sterol carrier protein 2-pseudogene | X87685 | 95787_s_at | 0.6 | 0.4 | 3 | 0.0480 | –/– | |
| 2,4-dienoyl CoA reductase 1, mitochondrial | AI844846 | 160711_at | 0.6 | 0.2 | 3 | 0.0480 | +/+ | |
| Synaptonemal complex protein 3 | AW212131 | 93994_at | 0.5 | 0.2 | 3 | 0.0069 | +/+ | |
| Aldehyde dehydrogenase family 1, subfamily A7 | U96401 | 94778_at | 0.5 | 0.1 | 3 | 0.0050 | +/+ | |
| Protease, serine, 11 (Igf binding) | AW125478 | 96920_at | 0.5 | 0.1 | 3 | 0.0001 | +/+ | |
| RIKEN cDNA 4921515A04 gene | AI642098 | 104494_at | 0.5 | 0.3 | 3 | 0.0083 | +/+ | |
| RAB34, member of RAS oncogene family | AI835712 | 160317_at | 0.5 | 0.3 | 3 | 0.0345 | +/+ | |
| Eucaryotic translation initiation factor 4E-binding protein 1 | U28656 | 100636_at | 0.5 | 0.4 | 3 | 0.0178 | +/+ | |
| Phosphoglucomutase 2 | AI842432 | 104313_at | 0.5 | 0.3 | 3 | 0.0062 | +/+ | |
| 3-ketoacyl-CoA thiolase B | AW012588 | 99571_at | 0.5 | 0.3 | 3 | 0.0060 | +/+ | |
| Glycogenin 1 | AW049730 | 100597_at | 0.5 | 0.3 | 3 | 0.0049 | +/+ | |
| Electron transferring flavoprotein, beta polypeptide | AW046273 | 96947_at | 0.5 | 0.2 | 3 | 0.0445 | +/+ | |
| Fat-specific gene 27 | M61737 | 102016_at | 0.5 | 0.0 | 3 | 0.0382 | +/+ | |
| CD36 antigen | L23108 | 93332_at | 0.5 | 0.2 | 3 | 0.0306 | +/+ | |
| Sialyltransferase 10 (alpha-2,3-sialyltransferase VI) | AI153959 | 102208_at | 0.5 | 0.2 | 3 | 0.0415 | +/+ | |
| Methylcrotonoyl-Coenzyme A carboxylase 1 (alpha) | AW123316 | 94940_at | 0.5 | 0.3 | 3 | 0.0350 | +/+ | |
| Glucan (1,4-alpha-), branching enzyme 1 | AW210370 | 96803_at | 0.5 | 0.3 | 3 | 0.0257 | +/+ | |
| Lipin 1 | AI846934 | 98892_at | 0.5 | 0.2 | 3 | 0.0219 | +/+ | |
| Diacylglycerol O-acyltransferase 1 Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme | AF078752 | 104371_at | 0.5 | 0.1 | 3 | 0.0378 | +/+ | |
| A hydratase (trifunctional protein), beta subunit | AW122615 | 96913_at | 0.5 | 0.2 | 3 | 0.0365 | +/+ | |
| RIKEN cDNA 1110001I14 gene | AW047554 | 104605_at | 0.5 | 0.3 | 3 | 0.0013 | +/+ | |
| RIKEN cDNA 1200015A22 gene | AI854863 | 98633_at | 0.4 | 0.2 | 3 | 0.0085 | +/+ | |
| Guanine nucleotide-binding protein, alpha inhibiting 1 | AI153412 | 104412_at | 0.4 | 0.0 | 3 | 0.0364 | +/+ | |
| Glycerol-3-phosphate acyltransferase, mitochondrial | U11680 | 101867_at | 0.4 | 0.2 | 3 | 0.0028 | +/+ | |
| Protein tyrosine phosphatase, receptor type, B | X58289 | 92289_at | P to A | 0.4 | 0.0 | 3 | 0.0308 | +/+ |
| Carboxylesterase 3 | AW226939 | 101538_i_at | 0.4 | 0.0 | 3 | 0.0189 | +/+ | |
| Growth hormone receptor | U15012 | 99108_s_at | 0.4 | 0.1 | 3 | 0.0317 | +/+ | |
| Glutathione peroxidase 3 | U13705 | 101676_at | 0.3 | 0.0 | 3 | 0.0001 | +/+ | |
| Adrenergic receptor, beta 3 | X72862 | 92537_g_at | 0.3 | 0.0 | 3 | 0.0410 | +/+ | |
Absolute call is the Affymetrix parameter that measures detection. P = present A = absent. ‘A to P’ indicates that a gene was called ‘absent’ in control and ‘present’ in adjacent ErbB2/Neu tissue. If a gene changes from absent to present or vice versa, then the fold change is irrelevant
Fold change was derived from the Affymetrix signal log ratio (SLR). AN = adjacent ErbB2/Neu, WT = wild-type control, and TU = tumor
SOM CL # = self-organizing map cluster number. Clusters are shown in Figure 3b
P-value was calculated for average tumor vs average control using Welch's t-test with the Benjamini and Hochberg Multiple Testing Correction (false discovery rate of 5%)
Repeated observation indicates whether the gene was confirmed by microarray data from two additional independent tumor samples. ‘+/+’ indicates confirmed by both tumors analysed, ‘+/–’ or ‘–/+’ by one tumor, and ‘–/–’ by neither
While comparisons of tumors to wild-type tissues can lead to important discoveries regarding the unique molecular signature of tumors, these studies are complicated by the difference in cellularity that exist between tumors and normal glands. We addressed this by standardizing all data to the changes observed in epithelial-specific markers, cytokeratins 8 and 18. The comparison of adjacent ErbB2/Neu to wild-type control tissue described herein is not affected by this limitation because these tissues, on average, are similar in cellular composition. In comparing adjacent ErbB2/Neu microarray data to wild-type control data, we found that fat-specific markers were only slightly decreased (average 30% reduction; data not shown) while cytokeratins were marginally increased (average 10% increase; Supplementary Table 1). Furthermore, Western blot analysis for expression of E-cadherin, another epithelial cell marker, provided further corroboration that the wild-type control and adjacent ErbB2/Neu mammary glands have comparable epithelial cell content (Figure 4). These data support the supposition that comparisons between adjacent ErbB2/Neu samples and wild-type tissues can reveal early gene expression changes that are due to overexpression of ErbB2/Neu and not due to large changes in cellular composition. In conclusion, the progressive changes in expression of these 82 genes correlate with tumorigenic stage, suggesting that a subset of this population of genes may play a pivotal role in tumor promotion or progression.
Figure 4.
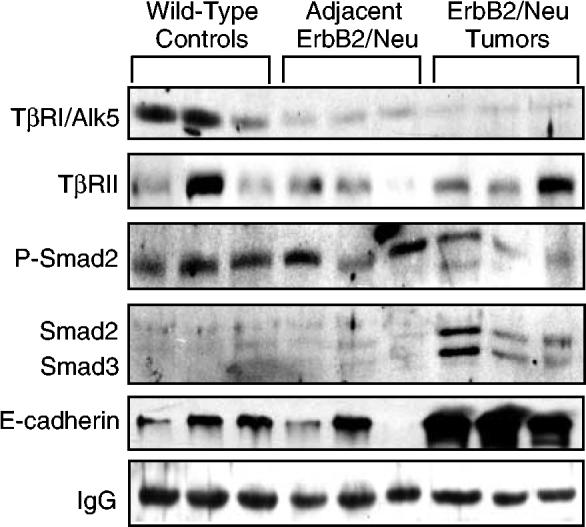
Western blot analysis confirms alterations in components of the TGF-β pathway in ErbB2/Neu-induced mammary tumors. Whole-cell lysate (150 μg) from mammary gland tissue was resolved by SDS–PAGE and transferred to PVDF membrane. Membranes were sequentially evaluated for the presence/absence of several TGF-β pathway components. E-cadherin was included as a marker for epithelial cell content and the IgG band represents a loading control
The molecular profile of ErbB2/Neu tumors suggests that these tumors may have lost functional TGF-b signaling
During analysis of the genes contained in the filtered ErbB2/Neu tumor molecular signature, we noted that several of the genes that were decreased in tumors were known TGF-β-inducible genes. Included in this list were TGF-β-1-induced transcript 4, TGF-β-induced-microtubule-associated protein 4, dermatopontin, matrix metalloproteinase 3, serine protease 11 (IGF binding), gap junction membrane channel protein alpha 1, zinc-finger homeobox 1a, and others (Verrecchia et al., 2001; Pimentel et al., 2002; Chambers et al., 2003). Although tumor cells and surrounding stroma often produce abundant TGF-β ligands (Reiss, 1999), the altered expression of these TGF-β targets lead us to hypothesize that the TGF-β pathway might be downregulated in ErbB2/Neu-induced tumors. Unfortunately, none of the canonical TGF-β intracellular signaling pathway members satisfied the criteria required to be included in the ErbB2/Neu tumor expression profile (i.e. TGF-β1, TGF-β2, TGF-β3, TβRI, TβRII, Smad2/3, Smad4, or Smad7). To directly determine whether the TGF-β pathway is perturbed in ErbB2/Neu-induced tumors, we generated Western blots with independent tumor, preneoplastic mammary gland, and wild-type samples to examine protein expression levels of multiple components of the TGF-β signaling pathway. We evaluated expression of TGF-β receptors (TβRI/Alk5 and TβRII) and Smad2/3 (total and phosphorylated). Western blot analyses revealed that TβRI/Alk5 protein levels were decreased in the adjacent ErbB2/Neu mammary glands and tumors relative to wild-type controls, whereas total Smad2/3 levels were increased in tumors compared to wild-type controls and adjacent ErbB2/Neu mammary glands (Figure 4). Phosphorylated Smad2 was unchanged to marginally decreased in tumors relative to wild-type controls. These results lead us to examine the most definitive indicator of activation of the Smad-dependent TGF-β signaling pathway: activation/phosphorylation of Smad2 within individual cells in the various tissue samples.
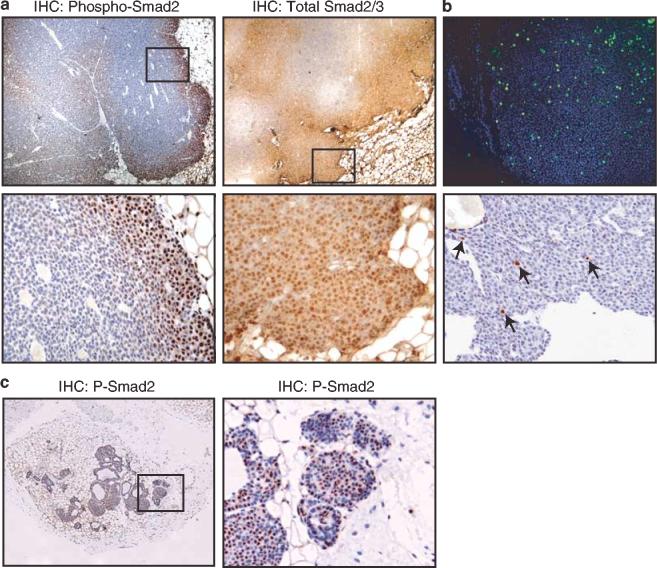
Smad2 is inactive throughout ErbB2/Neu-induced mammary tumors except at the tumor/stroma interface
Immunohistochemical evaluation of nuclear, phosphorylated Smad2 revealed that Smad2 is inactive in the majority of the cells of an ErbB2/Neu-induced tumor. Any activated Smad2 that was translocated to the nucleus was confined to cells in the periphery of these tumors, in areas of invagination by stromal tissue (Figure 5a), and in small lobes of tumors (data not shown). Immunohistochemical evaluation of total Smad2/3 demonstrated increased nuclear staining in the periphery of the tumors and mostly cytoplasmic staining in the rest of the tumor with some regions of negative staining (Figure 5a). These data indicate that the Smad signaling pathway is quiescent in the majority of an ErbB2/Neu-induced mammary tumor.
Figure 5.
Smad2 is inactive throughout ErbB2/Neu-induced mammary tumors except at the tumor/stroma interface. (a) Immunohistochemical analysis of phosphorylated and total Smad2 in ErbB2/Neu-induced tumors. The top panels are representative immunohistochemical analyses for phosphorylated Smad2 and total Smad2/3 on ErbB2/Neu tumor sections ( × 40). The bottom panels are a higher magnification ( × 200) of the boxed area from the panels above. Note the selective nuclear staining (brown) in the enlarged sections, identifying cells with activated Smad2. Sections were counterstained with Gill's hematoxylin (blue). Seven independent tumors from ErbB2/Neu mice were examined. These tumors were comparable to the size of tumors that were evaluated by microarray analyses and utilized for subsequent immunohistochemical analysis. (b) Assessment of DNA synthesis and apoptosis reveals cycling cells throughout ErbB2/Neu-induced mammary tumors. The top panel ( × 100) is representative staining for BrdU (green). Nuclei were counterstained with DAPI (blue). The bottom panel ( × 200) is representative in situ analysis for apoptosis. The brown cells (indicated by arrows) are apoptotic cells. (c) Smad2 is activated throughout small ErbB2/Neu tumors. The left panel is a representative immunohistochemical analysis for phosphorylated Smad2 on sections of small ErbB2/Neu tumors ( × 40). The right panel is a higher magnification ( × 200) of the boxed area from the panel on the left. Note the selective nuclear staining (brown) in the enlarged section, identifying cells with activated Smad2
We considered the possibility that the peripheral staining pattern of Smad2 was reflective of nonviable cells in the center of these tumors. The cells lacking Smad2 activation appeared healthy and non-necrotic. To further evaluate cell viability, proliferative indices including bromodeoxyuridine (BrdU) incorporation during DNA synthesis (Figure 5b), phosphorylated-histone 3 expression (data not shown), and Ki67 expression (data not shown) were evaluated. Additionally, apoptotic cells were assessed by detection of fragmented DNA and morphological characteristics (Figure 5b). These data revealed a uniform distribution of proliferating and apoptotic cells throughout the tumors, indicating that there are healthy cycling cells throughout these tumors, both in the periphery and in the center of the tumor. Thus, lack of phospho-Smad2 staining in the center of tumors is not due to loss of cell viability.
The pattern of Smad2 activation in the periphery of ErbB2/Neu-induced tumors suggested that a unique tumor microenvironment induced by proximity to the stromal compartment may lead to activation of Smad2. If so, smaller tumors with greater accessibility to the stroma may have uniform activation of Smad2. Immunohistochemical analysis for phosphorylated Smad2 in smaller tumors revealed activated Smad2 throughout the tumor (Figure 5c). Together, these data suggest that the majority of epithelial cells comprising an ErbB2/Neu-induced mammary tumor have lost Smad signaling and that accessibility to the stromal microenvironment prevents this loss.
Immunohistochemical analysis demonstrates heterogeneous activation of the ErbB2/Neu receptor in ErbB2/Neu tumor sections
To determine whether differential activation of Smad2 in the periphery vs the center of the tumor correlated with differences in activation of ErbB2/Neu signaling, ErbB2/Neu receptor phosphorylation on positions 877 or 1248 was evaluated using immunohistochemistry. As expected, the ErbB2/Neu receptor was expressed throughout most of the tumor, whereas the activated/phosphorylated receptor demonstrated positive staining in the periphery of these tumors and heterogeneous staining in the center of the tumors with interspersed regions of both positive and negative staining (Figure 6). The patterns of staining for the different phosphorylated tyrosine residues (Tyr877 and Tyr1248) were consistently very similar. Generally, phosphorylated ErbB2/Neu staining overlapped with regions of positive staining for phosphorylated Smad2 at the periphery of the tumors as well as regions of negative staining in the center of the tumor, suggesting that activated Smad2 can coexist with active ErbB2/Neu signaling.
Figure 6.

Immunohistochemical analysis demonstrates heterogeneous activation of the ErbB2/Neu receptor in ErbB2/Neu tumor sections. The top panels ( × 40) are representative ErbB2/Neu tumor sections that were incubated with antibodies to total ErbB2/Neu and phosphorylated ErbB2/Neu (P-877 and P-1248). The bottom panels ( × 400) are higher magnifications of the boxed areas in the top panels. Note the selective membrane staining (brown). Sections were counterstained with Gill's hematoxylin (blue)
Immunohistochemical assessment reveals loss of detectable TGF-β-Receptor-I in adjacent ErbB2/Neu mammary gland and ErbB2/Neu tumor epithelia
Phosphorylated Smad2 observed in the outer rim of ErbB2/Neu tumors could be due to restricted TGF-β signaling in this area or induction by other members of TGF-β superfamily capable of activating Smad2 such as Activin (Massague, 1998). The Western blot analysis (Figure 4) suggested that the type I TGF-β receptor/Alk5 (TβRI) might be greatly reduced or lost in ErbB2/Neu tumors. To determine whether expression of TβRI correlated with regions of Smad2 activation, we examined the cellular localization of TβRI by immunohistochemistry. Corroborating the Western blot data, staining for TβRI was mostly negative in epithelial cells of both the adjacent ErbB2/Neu tissue and in ErbB2/Neu tumors but positive in wild-type epithelial cells (Figure 7). These data indicate that expression of TβRI, and hence, TGF-β signaling is suppressed in adjacent ErbB2/Neu tissue and throughout ErbB2/Neu tumors.
Figure 7.
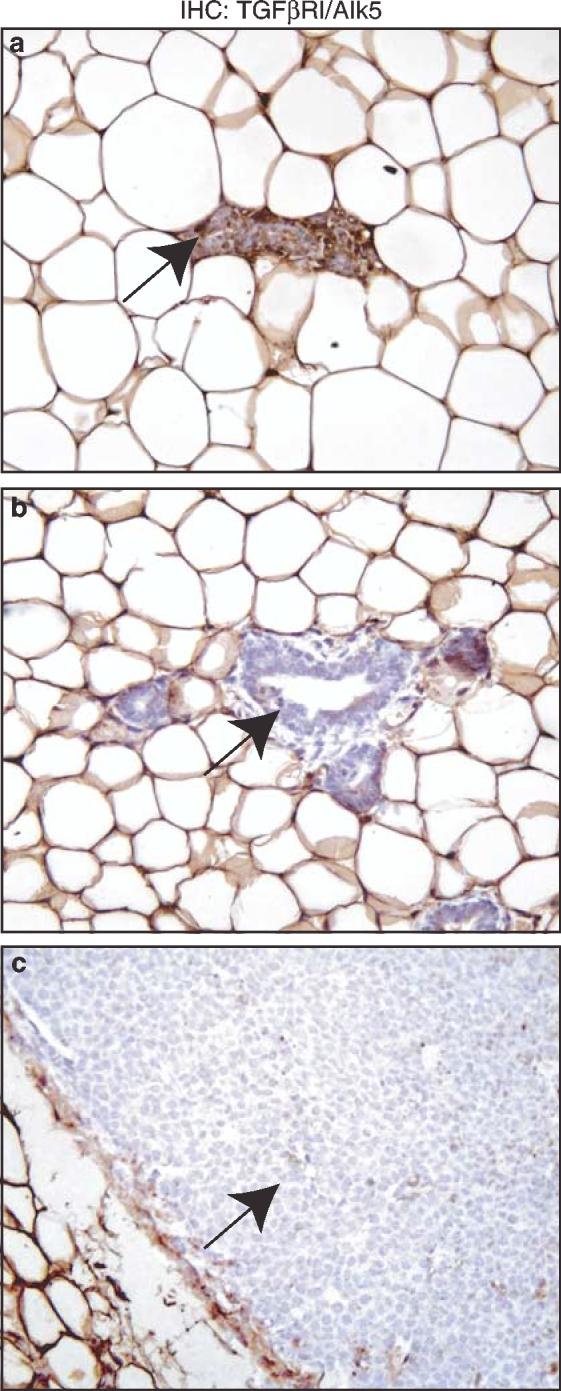
Immunohistochemical assessment reveals loss of detectable TGF-β-Receptor-I in adjacent ErbB2/Neu mammary gland and ErbB2/Neu tumor epithelia. The panels contain representative immunohistochemical staining for TGF-β-Receptor-I/ALK5 in wild-type control tissue sections (a, × 200), adjacent ErbB2/Neu tissues (b, × 200), and the ErbB2/Neu tumor tissues (c, × 200). Note the brown epithelial cells (arrow) in wild-type glands and the loss of this staining in epithelial cells in adjacent glands and tumors (arrows)
Activin-Receptor-IB expression correlates with active Smad2 signaling
Loss of TβRI in the entire tumor was surprising since active Smad2 was observed in the periphery of the tumor. Smad2 can also be activated by Activin ligand binding with Activin receptors, thus we examined cellular localization of the type I Activin receptor/Alk4 (ActRIB) by immunohistochemistry. Interestingly, Act-RIB staining occurred in the periphery of the tumor sections and in the adjacent ErbB2/Neu tissue (Figure 8), indicating that Activin signaling may be responsible for the activation of Smad2 at the tumor/stroma interface of ErbB2/Neu tumors.
Figure 8.
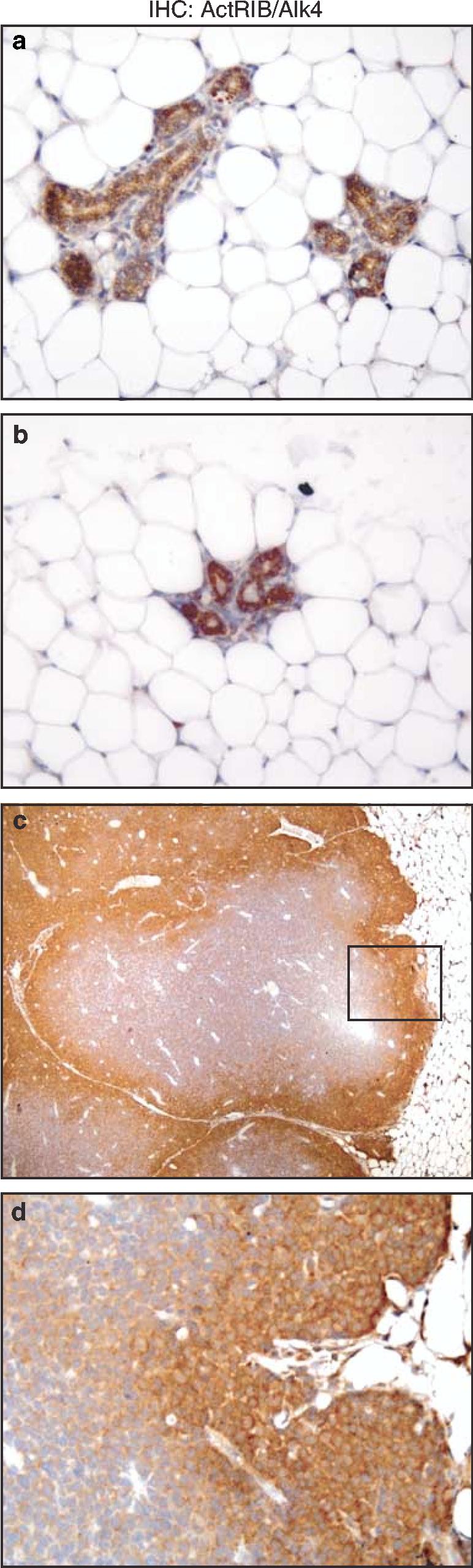
Activin-Receptor-IB expression correlates with active Smad2 signaling. The panels contain representative immunohistochemical staining for Activin-Receptor-IB on wild-type mammary glands (a, × 200), adjacent ErbB2/Neu mammary glands (b, 200), and ErbB2/Neu tumor sections (c, × 40). The bottom panel is a higher magnification (d, × 200) of the boxed area from the ErbB2/Neu tumor panel above. Sections were counterstained with Gill's hematoxylin (blue)
Discussion
In this study, we identified a core set of genes that are progressively altered in expression during ErbB2/Neu-induced tumorigenesis in an in vivo model of breast cancer. Further characterization of these genes should provide insight into the tumorigenic events associated with ErbB2/Neu overexpression and thus a more complete understanding of the underlying biological mechanisms of the ErbB2/Neu tumorigenic cascade. Owing to the importance of ErbB2/Neu overexpression in human breast cancers, several groups have previously used expression profiling to identify genes associated with ErbB2/Neu expression in breast tumors and cell lines (Perou et al., 2000; Kauraniemi et al., 2001, 2004; Desai et al., 2002; Wilson et al., 2002; Andrechek et al., 2003; Dressman et al., 2003; Kumar-Sinha et al., 2003; Mackay et al., 2003; Bertucci et al., 2004). Importantly, none have examined the expression profiles of preneoplastic tissue to identify alterations of gene expression that are associated with tumor progression in an in vivo model. Indeed, Green and co-workers (Desai et al., 2002) suggested that comparative expression profiling of various developmental stages of oncogene-induced tumors would greatly increase our understanding of the progression of genetic changes associated with tumorigenesis. Accordingly, the core set of 82 genes that were described in this report whose expression is associated with ErbB2/Neu-initiated cancer progression may provide new diagnostic and predictive markers, as well as new chemotherapeutic or chemoprevention targets.
Golub and co-workers have previously identified a 17 gene signature associated with human breast cancer metatastasis (Ramaswamy et al., 2003). Of the metastasis genes that generated informative data in our analysis, 33% were contained within the global tumor profile (Supplementary Table 1). Furthermore, 30% of the genes encoding prognosis discriminator proteins for human breast cancer (Jacquemier et al., 2005) were also identified by our analysis of ErbB2/Neu tumors. These data support the utility of this mouse model in identifying genes that may be important in human breast tumorigenesis. Several of the 82 genes (Etv1, Eif4ebp1, Ghr, Id2, Kai1, Tpd52) that were progressively altered during ErbB2/Neu-induced tumorigenesis are already known to be associated with human breast cancer, highlighting the relevance of this model in evaluating alterations of the transcriptome that are associated with progression of tumorigenesis. Interestingly in previous studies, there were only a small number of genes that were common to all of the human breast tumor expression profiling experiments that identified ErbB2/Neu tumor molecular signatures, and these genes are contained in the ErbB2/Neu amplicon (Perou et al., 2000; Kauraniemi et al., 2001; Wilson et al., 2002; Dressman et al., 2003; Bertucci et al., 2004). The analytical approaches and perhaps the heterogeneity of human ErbB2/Neu-positive breast tumors in these experiments seem to preclude identification of universal downstream targets of ErbB2/Neu signaling. This further supports the use of the simplified ErbB2/Neu mouse model for identifying candidate genes for further analysis.
In comparing the global gene expression profile defined by our microarray analysis with previously published expression profiling data associated with ErbB2/Neu overexpression (Desai et al., 2002; Wilson et al., 2002; Andrechek et al., 2003; Kumar-Sinha et al., 2003; Mackay et al., 2003; Bertucci et al., 2004; Kauraniemi et al., 2004), we found only small subsets of overlap between the current data and these other expression profiles, and none of the genes were consistent among all of the data sets. There are several variations among the approaches used that may account for these discrepancies. These variations include: microarray formats with differential representation of the genome, analytical and statistical approaches, experimental paradigms, and starting material (i.e. cell lines vs tissue samples). Not surprisingly, our gene expression profile shared the most commonalities with the MMTV-Neu-tumor signature identified by Desai et al. (2002). Despite the use of different microarray platforms and the collection of tumors at different time points, 70 of the 324 genes identified herein as ‘signature genes’ were present in the list of genes reported by Desai et al. Only four of these genes were changed in opposite directions (i.e. down- vs upregulated), suggesting strong concordance between the two tumor analyses. Thus, the study presented herein both confirms the data reported by Desai et al. (2002), and extends this work by evaluating a different set of genes whose expression changes in tumors compared to wild-type tissue and examining the transcripts associated with preneoplastic changes initiated by ErbB2/Neu expression.
The expression profiling of ErbB2/Neu tumors suggested that the TGF-β pathway may be altered in these tumors. The lack of Smad2 activation in the majority of cells within these tumors was accompanied by reduction of TβRI expression, suggesting that ErbB2/Neu induction of tumors results in loss of at least one component of the TGF-β signaling pathway. Recently, another group reported that ErbB2/Neu can collaborate with ER81 to upregulate the expression of the inhibitory Smad7 in a breast cancer cell line (Dowdy et al., 2003), suggesting a direct mechanism for regulation of TGF-β signaling by the ErbB2/Neu pathway. Cumulatively, these data suggest that there are several possible mechanisms for suppression of the TGF-β pathway during ErbB2/Neu tumorigenesis. Whether loss of TβRI is due to active repression by ErbB2/Neu or an indirect mechanism remains unknown.
Three recent studies have evaluated the impact of genetically manipulating the TGF-β pathway on ErbB2/Neu-induced tumorigenesis in mice (Yang et al., 2002; Muraoka et al., 2003; Siegel et al., 2003). All three studies reported that the TGF-β pathway promotes metastasis of ErbB2/Neu-induced mammary tumors; however, discrepancies were reported regarding primary tumor latency. Whereas overexpression of secreted TβRII or active TGF-β1 had no impact on primary tumor latency, forced expression of constitutively active TβRI in the same cells that express ErbB2/Neu delayed tumor development. In addition, multiple studies also found that forced activation of the TGF-β signaling pathway decreases the proliferative rate of ErbB2/Neu-induced tumors (Muraoka et al., 2003; Siegel et al., 2003). These data are consistent with the results reported herein indicating that intrinsic loss of TβRI provides a growth advantage for ErbB2/Neu-induced mammary tumorigenesis. It is important to note that all of the bitransgenic mouse studies discussed above focused on assessing the ability of the TGF-β pathway to alter ErbB2/Neu-induced tumorigenesis under conditions in which both pathways are exogenously regulated without evaluating whether ErbB2/Neu-induced tumors undergo spontaneous alterations in TGF-β signaling. The study presented herein is the first to our knowledge to show that ErbB2/Neu-induced mammary tumors normally lose the Smad-dependent TGF-β signaling pathway resulting from loss of TβRI.
The presence of phosphorylated Smad2 in areas of the tumor that were in close proximity to stromal tissue occurred in the apparent absence of TbRI. This suggested that another member the TGF-β superfamily might be responsible for Smad2 activation. The correlation of immunohistochemical data for phosphorylated/activated Smad2 and Activin-Receptor-IB/Alk4 suggests that ErbB2/Neu and Activin signaling can coexist within ErbB2/Neu-induced tumors, but this coexistence is limited to the subset of tumor cells that are in close apposition to the stroma. Determining the role of Activin signaling and the mechanism for suppression of TGF-β signaling during the progression of ErbB2/Neu-initiated mammary tumorigenesis will be interesting questions to pursue in future studies.
Materials and methods
Materials
Radiolabeled nucleotides were purchased from Perkin Elmer Life Sciences (Boston, MA, USA). All chemicals were purchased from Sigma (St Louis, MO, USA). Antibodies were purchased from companies as follows: phospho-specific Smad2 (#3101), phospho-specific ErbB2/Neu (Tyr877; #2241), ErbB2/Neu (#2242) from Cell Signaling Technology, Inc. (Beverly, MA, USA); Smad3 (FL-425), TGF-β-Receptor I (sc-398), and TGF-β-Receptor II (H-567) from Santa Cruz Biotechnology (Santa Cruz, CA, USA); phospho-specific ErbB2/Neu (Tyr1248, AB-18 PN2A) from NeoMarkers (Fremont, CA, USA); ActivinRIB (AF222) from R&D Systems (Minneapolis, MN, USA); and anti-BrdU (Beckton Dickinson, San Jose, CA, USA; #347580). Secondary antibodies include: horseradish peroxidase-conjugated goat anti-mouse and anti-rabbit (Santa Cruz Biotechnology) and fluorescein conjugated-goat anti-mouse (Jackson ImmunoResearch, West Grove, PA, USA; #115-095-003).
Transgenic mice
All mice were housed in microisolator-plus units under pathogen-free conditions. Food and water were provided ad libitum and a 12 h light/dark cycle was maintained. Mice (FVB/N-TgN(MMTV-neu)202Mul) containing the rat proto-oncogene c-neu transgene targeted to mammary epithelium by the MMTV-LTR promoter (Guy et al., 1992) were purchased from Jackson Laboratories (Bar Harbor, ME, USA) and bred to generate a colony of MMTV-Neu and nontransgenic, wild-type control mice. Transgenic mice were genotyped by PCR with primers specific to the Neu transgene: forward: 5′ CGCAACCCACATCAGGCC 3′ and reverse: 5′ TTCCTGCAGCAGCCTACGC 3′. Nulliparous mice were palpated weekly to detect tumors. At 1–3 weeks after initial tumor detection, mice were killed by asphyxiation in a CO2 chamber, cardiac blood was collected, mammary tissues were removed, and other organs were examined for metastases. All animal studies were approved by the Case Western Reserve University Institutional Animal Care and Use Committee.
Tissue isolation
Tumor and surrounding mammary gland were removed 1–3 weeks after tumor detection (average tumor latency = 37 weeks) and placed in RNAlater (Ambion, Austin, TX, USA) to prevent degradation of RNA. The adjacent, grossly nontumorigenic mammary gland (adjacent ErbB2/Neu) was isolated, leaving a perimeter of approximately 2 mm of normal tissue encasing the tumor (Figure 1a). This small border of tissue surrounding the tumor was then removed and discarded. This resulted in two tissue samples: (1) tumors and (2) adjacent tissue that was at least 2 mm from the tumor and thus contained no overt tumor tissue. The adjacent ErbB2/Neu mammary gland and tumor tissue were frozen on dry ice and stored at -80°C. For wild-type controls, thoracic mammary glands were removed from 15 age-matched, nulliparous wild-type animals (age 31 or 41 weeks). RNA isolated from each gland was pooled into three groups, each representing five different wild-type animals.
Microarray analysis
All data and detailed protocols have been submitted to Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) according to Minimum Information About a Microarray Experiment (MIAME) guidelines (Brazma et al., 2001). Total RNA was extracted from tissues using the TRIzol method (Invitrogen, Carlsbad, CA, USA). Using a 10 μg total RNA template and a custom primer containing both T7 primer sequence and oligo(dT)24 (Genset Corp, La Jolla, CA, USA), cDNA was synthesized (Superscript Choice System, Invitrogen, Carlsbad, CA, USA). In vitro transcription (Enzo Diagnostics, Inc., Farmingdale, NY, USA) was performed with Bio-CTP and Bio-UTP to produce biotin-labeled anti-sense cRNA. Biotinylated cRNA was purified using RNeasy spin columns (Qiagen, Inc., Valencia, CA, USA) and delivered to the Gene Expression Array Core Facility at Case Western Reserve University (http://www.geacf.net/). Biotinylated cRNA (10 μg) was fragmented and spiked with procaryotic control RNAs followed by overnight hybridization at 451C to the Affymetrix (Santa Clara, CA, USA) Murine U74Av2 GeneChip Array. After washing, arrays were stained with streptavidin–phycoerythrin and scanned using an Agilent Gene Array scanner 2000 driven by the Affymetrix Micro-Array Suite 5.0. The scanner PMT was set to the wide dynamic range setting.
Computational analyses were performed with Microarray Suite (v.5.0, Affymetrix), Data Mining Tool (DMT v.3.0, Affymetrix), MicroDB (v.3.0, Affymetrix), and GeneSpring (v.6.0, Silicon Genetics) software. Graphs representing values for individual genes were generated using GraphPad Prism version 4.00 for Windows, GraphPad Software (San Diego, CA, USA; www.graphpad.com). Scanned images were analysed using Affymetrix MAS 5.0. Four columns of data were given particular attention when assessing the results including: ‘signal’, ‘detection’, ‘signal log ratio’, and ‘change’. The microarray data generated from analysis of each tumor (n = 5) sample was compared to the microarray data generated from analysis of each pooled age-matched, wild-type control sample (n = 3) as illustrated in Figure 1c. Probes that received an Affymetrix change call of ‘increased’, ‘decreased’, ‘marginally increased’, or ‘marginally decreased’ were retained for further analysis. Probes that were not called ‘present’ or ‘marginal’ by Affymetrix detection call on at least one GeneChip were removed. Application of Welch's approximate t-test with the Benjamini and Hochberg multiple testing correction defined a set of probes with a false discovery rate of 5% (P<0.05).
Histology
Following fixation in 4% (w/v) paraformaldehyde/PBS, one inguinal mammary gland and isolated tumors from each mouse were paraffin-embedded, cut into 5-μm sections, and then stained with hematoxylin and eosin.
Northern blot analysis
Following isolation of total RNA according to the TRIzol reagent protocol (Invitrogen, Carlsbad, CA, USA), 20 μg of total RNA was separated by electrophoresis on a 1% denaturing agarose gel, transferred to Hybond-N + Nylon membrane (Amersham Pharmacia Biotech, Visscataway, NJ, USA) with a Turbo Blotter (Schleicher and Scheull, Keene, NJ, USA), and hybridized to a denatured, double-stranded DNA probe in QuikHyb solution (Stratagene, Cedar Creek, TX, USA) according to the recommended protocol. Double-stranded DNA templates for probes were generated by PCR with the neu transgene primers described above and transgenic mouse genomic DNA template. Probes were radiolabeled with a-32P-labeled dCTP by random priming (DECAprime II, Ambion, Austin, TX, USA).
Real-time RT–PCR
Applied Biosystem 7900HT Gene Expression Micro Fluidic Cards, configuration 9, were designed and purchased from Applied Biosystems (Foster City, CA, USA). Each of these 384 well cards contained 95 gene targets including controls with four replicates per target. To serve as negative controls, four wells were left empty. Total RNA was extracted from tissues using the TRIzol method (Invitrogen, Carlsbad, CA, USA). RNA was treated to remove potential contaminating DNA according to the protocol from the DNA-free kit (Ambion, Austin, TX, USA) and delivered to the Gene Expression Array Core Facility at CWRU. cDNA was generated using 3 μg of RNA in a 100 μl reaction volume in accordance with the High Capacity cDNA Archive Kit protocol (Applied Biosystems, Foster City, CA, USA). Data were evaluated using ABI Prism SDS 2.2 software (Applied Biosystems, Foster City, CA, USA). The following adjustable analysis settings were used: automatic threshold cycle (CT), automatic outlier removal, and relative quantification (RQ) min/max confidence 95%. All data were calibrated relative to an endogenous control, glyceraldehyde-3-phosphate dehydrogenase. Nine independent tissue samples were evaluated (three different samples per experimental group (tumors, adjacent glands, or wild-type controls)).
Immunoblotting
Whole tissues were homogenized in nondenaturing protein lysis buffer (20 mm Tris-HCl, pH 7.5, 1% Triton X-100, 100 mm NaCl, 40 mm NaF, 1 mm EDTA, 1 mm EGTA) with additional phosphatase and protease inhibitors (1 mm Na3VO4, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 mm PMSF). Following lysis on ice for 30 min, homogenates were clarified by centrifugation (12 000 g) for 10 min at 41C. The supernatant was retained as the whole-cell lysate for subsequent Western blot analysis. Proteins were quantified by Bradford Protein Assay (Biorad, Hercules, CA, USA).
Whole-cell lysate (150 μg) was resolved by discontinuous SDS–PAGE, and proteins were transferred to PVDF membrane in Towbin Buffer (192 mg/l glycine, 25 mg/l Trisma Base, 10–20% (vol/vol) methanol). SDS–PAGE gels were stained with Coomassie blue stain to confirm complete transfer. Membranes were blocked with 5% (w/v) skim milk in PBS with 0.05% (v/v) Tween-20 (PBS-T) (1 h, RT). Membranes were incubated with primary antibody in 5% (w/v) bovine serum albumin (overnight, 4°C) in PBS-T plus sodium azide (0.02% (v/v) followed by incubation with appropriate secondary antibody conjugated to horseradish peroxidase in 5% skim milk in PBS-T (1 h, RT). Bound antibodies were detected by chemiluminescence (Lumiglo, Cell Signaling Technology, Inc., Beverly, MA, USA).
Immunohistochemistry
Immunohistochemistry was performed utilizing the Dako Envision Plus HRP kit (DakoCytomation, Carpinteria, CA, USA) with minor modifications, except for ActivinRIB for which the Vectastain ABC kit was used (Vector Laboratories; Burlingame, CA, USA). Briefly, mammary gland sections were deparaffinized in xylene and rehydrated in 100 and 95% ethanol. Methods for antigen retrieval included: boiling in 10 mm citrate buffer (pH 6.0) for 10 min (PSmad2, ActRIB), boiling in 10 mm citrate buffer (pH 6.0) 20 min (Total Smad2/3), 1 min boil followed by subboil in 1mm EDTA (pH 8.0) for 15 min (P-ErbB2-877, P-ErbB2-1248, ErbB2), and 5 min trypsin digestion (Sigma tablets, TbRI). Sections were blocked with the blocking buffer included in the kit along with 15 μl/ml normal serum and then incubated with primary antibody (overnight, 4°C). Primary antibodies were diluted 1: 100 except for PSmad2 (1: 200) and ActRIB (1: 25). Following incubation with secondary antibody included in the Dako or Vectastain kit, bound antibody was detected by DAB reaction. Sections were counterstained with Gill's hematoxylin #3 (Polysciences, Inc., Washington, PA, USA), dehydrated, cleared, and mounted in Permount. A control (blocking buffer without primary antibody) was performed for each tissue stained. Seven mammary glands with tumors from seven individual transgenic animals were analysed.
Apoptotic cells were identified using the ApopTag Plus Peroxidase In Situ Apoptosis Detection Kit (Chemicon; Temecula, CA, USA). For assessment of cells undergoing DNA synthesis, mice received an injection (0.1 mg/g body weight) of BrdU 2 h before being killed. Immunohistochemistry for BrdU was carried out as described previously (Milliken et al., 2002).
Supplementary Material
Acknowledgements
We extend special thanks to Kristen Lozada for her dedicated technical support and Erin Milliken for intellectual discussions. Tumor histology was performed by the Histology Core Facility of the CASE Comprehensive Cancer Center. Microarray work was supported by the Gene Expression Array Core Facility of the CASE Comprehensive Cancer Center (P30CA43703). RAK was supported by National Institutes of Health Grant (RO1-CA90398). MDL was supported by a Breast Cancer Research Program (BCRP) Predoctoral Traineeship Award (DAMD17-03-1-0302) and the National Institutes of Health Molecular Therapeutics Training Program (GM08803A1).
References
- Alroy I, Yarden Y. FEBS Lett. 1997;410:83, 86. doi: 10.1016/s0014-5793(97)00412-2. [DOI] [PubMed] [Google Scholar]
- Andrechek ER, Laing MA, Girgis-Gabardo AA, Siegel PM, Cardiff RD, Muller WJ. Cancer Res. 2003;63:4920–4926. [PubMed] [Google Scholar]
- Barcellos-Hoff MH, Ewan KB. Breast Cancer Res. 2000;2:92–99. doi: 10.1186/bcr40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertucci F, Borie N, Ginestier C, Groulet A, Charafe-Jauffret E, Adelaide J, Geneix J, Bachelart L, Finetti P, Koki A, Hermitte F, Hassoun J, Debono S, Viens P, Fert V, Jacquemier J, Birnbaum D. Oncogene. 2004;23:2564–2575. doi: 10.1038/sj.onc.1207361. [DOI] [PubMed] [Google Scholar]
- Boggio K, Nicoletti G, Di Carlo E, Cavallo F, Landuzzi L, Melani C, Giovarelli M, Rossi I, Nanni P, De Giovanni C, Bouchard P, Wolf S, Modesti A, Musiani P, Lollini PL, Colombo MP, Forni G. J. Exp. Med. 1998;188:589–596. doi: 10.1084/jem.188.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard L, Lamarre L, Tremblay PJ, Jolicoeur P. Cell. 1989;57:931–936. doi: 10.1016/0092-8674(89)90331-0. [DOI] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Nat. Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Bulavin DV, Phillips C, Nannenga B, Timofeev O, Donehower LA, Anderson CW, Appella E, Fornace AJ., Jr Nat. Genet. 2004;36:343–350. doi: 10.1038/ng1317. [DOI] [PubMed] [Google Scholar]
- Chambers RC, Leoni P, Kaminski N, Laurent GJ, Heller RA. Am. J. Pathol. 2003;162:533–546. doi: 10.1016/s0002-9440(10)63847-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Akhurst RJ, Balmain A. Nat. Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- Desai KV, Xiao N, Wang W, Gangi L, Greene J, Powell JI, Dickson R, Furth P, Hunter K, Kucherlapati R, Simon R, Liu ET, Green JE. Proc. Natl. Acad. Sci. USA. 2002;99:6967. doi: 10.1073/pnas.102172399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdy SC, Mariani A, Janknecht R. J. Biol. Chem. 2003;278:44377–44384. doi: 10.1074/jbc.M307202200. [DOI] [PubMed] [Google Scholar]
- Dressman MA, Baras A, Malinowski R, Alvis LB, Kwon I, Walz TM, Polymeropoulos MH. Cancer Res. 2003;63:2194. [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Proc. Natl. Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graus-Porta D, Beerli RR, Daly JM, Hynes NE. EMBO J. 1997;16:1647–1655. doi: 10.1093/emboj/16.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CT, Cardiff RD, Muller WJ. J. Biol. Chem. 1996;271:7673–7678. doi: 10.1074/jbc.271.13.7673. [DOI] [PubMed] [Google Scholar]
- Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Proc. Natl. Acad. Sci. USA. 1992;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbro T, Civenni G, Hynes NE. Exp. Cell Res. 2003;284:99–110. doi: 10.1016/s0014-4827(02)00099-x. [DOI] [PubMed] [Google Scholar]
- Hynes NE, Stern DF. Biochim. Biophys. Acta. 1994;1198:165–184. doi: 10.1016/0304-419x(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Jacquemier J, Ginestier C, Rougemont J, Bardou VJ, Charafe-Jauffret E, Geneix J, Adelaide J, Koki A, Houvenaeghel G, Hassoun J, Maraninchi D, Viens P, Birnbaum D, Bertucci F. Cancer Res. 2005;65:767–779. [PubMed] [Google Scholar]
- Kauraniemi P, Barlund M, Monni O, Kallioniemi A. Cancer Res. 2001;61:8235–8240. [PubMed] [Google Scholar]
- Kauraniemi P, Hautaniemi S, Autio R, Astola J, Monni O, Elkahloun A, Kallioniemi A. Oncogene. 2004;23:1010–1013. doi: 10.1038/sj.onc.1207200. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S, Sneige N. Adv. Anat. Pathol. 2002;9:185–197. doi: 10.1097/00125480-200205000-00002. [DOI] [PubMed] [Google Scholar]
- Kumar-Sinha C, Ignatoski KW, Lippman ME, Ethier SP, Chinnaiyan AM. Cancer Res. 2003;63:132–139. [PubMed] [Google Scholar]
- Lee RJ, Albanese C, Fu M, D'Amico M, Lin B, Watanabe G, Haines GK, Siegel PM, Hung MC, Yarden Y, Horowitz JM, Muller WJ, Pestell RG. Mol. Cell. Biol. 2000;20:672–683. doi: 10.1128/mcb.20.2.672-683.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenferink AE, Simpson JF, Shawver LK, Coffey RJ, Forbes JT, Arteaga CL. Proc. Natl. Acad. Sci. USA. 2000;97:9609–9614. doi: 10.1073/pnas.160564197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Rosen JM, McMenamin-Balano J, Muller WJ, Perkins AS. Mol. Cell. Biol. 1997;17:3155–3163. doi: 10.1128/mcb.17.6.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay A, Jones C, Dexter T, Silva RL, Bulmer K, Jones A, Simpson P, Harris RA, Jat PS, Neville AM, Reis LF, Lakhani SR, O'Hare MJ. Oncogene. 2003;22:2680–2688. doi: 10.1038/sj.onc.1206349. [DOI] [PubMed] [Google Scholar]
- Massague J. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Milliken EL, Ameduri RK, Landis MD, Behrooz A, Abdul-Karim FW, Keri RA. Endocrinology. 2002;143:3671–3680. doi: 10.1210/en.2002-220228. [DOI] [PubMed] [Google Scholar]
- Morrison BW, Leder P. Oncogene. 1994;9:3417–3426. [PubMed] [Google Scholar]
- Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P. Cell. 1988;54:105–115. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- Muraoka RS, Koh Y, Roebuck LR, Sanders ME, Brantley-Sieders D, Gorska AE, Moses HL, Arteaga CL. Mol. Cell. Biol. 2003;23:8691–8703. doi: 10.1128/MCB.23.23.8691-8703.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de RM, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Pimentel RC, Yamada KA, Kleber AG, Saffitz JE. Circ. Res. 2002;90:671–677. doi: 10.1161/01.res.0000014823.75393.4d. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Ross KN, Lander ES, Golub TR. Nat. Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- Reiss M. Microbes Infect. 1999;1:1327–1347. doi: 10.1016/s1286-4579(99)00251-8. [DOI] [PubMed] [Google Scholar]
- Roberts AB, Wakefield LM. Proc. Natl. Acad. Sci. USA. 2003;100:8621–8623. doi: 10.1073/pnas.1633291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon DS, Brandt R, Ciardiello F, Normanno N. Crit. Rev. Oncol. Hematol. 1995;19:183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- Shepherd TG, Kockeritz L, Szrajber MR, Muller WJ, Hassell JA. Curr. Biol. 2001;11:1739–1748. doi: 10.1016/s0960-9822(01)00536-x. [DOI] [PubMed] [Google Scholar]
- Siegel PM, Dankort DL, Hardy WR, Muller WJ. Mol. Cell. Biol. 1994;14:7068–7077. doi: 10.1128/mcb.14.11.7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel PM, Shu W, Cardiff RD, Muller WJ, Massague J. Proc. Natl. Acad. Sci. USA. 2003;100:8430–8435. doi: 10.1073/pnas.0932636100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, Press MF. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Proc. Natl. Acad. Sci. USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B, Vu M, Booker T, Santner SJ, Miller FR, Anver MR, Wakefield LM. J. Clin. Invest. 2003;112:1116–1124. doi: 10.1172/JCI18899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzahar E, Waterman H, Chen X, Levkowitz G, Karunagaran D, Lavi S, Ratzkin BJ, Yarden Y. Mol. Cell. Biol. 1996;16:5276–5287. doi: 10.1128/mcb.16.10.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrecchia F, Chu ML, Mauviel A. J Biol. Chem. 2001;276:17058–17062. doi: 10.1074/jbc.M100754200. [DOI] [PubMed] [Google Scholar]
- Wakefield LM, Yang YA, Dukhanina O. Breast Cancer Res. 2000;2:100–106. doi: 10.1186/bcr41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Kudryavtseva E, Ch'en IL, McCormick J, Sugihara TM, Ruiz R, Andersen B. Oncogene. 2004;23:1507–1513. doi: 10.1038/sj.onc.1207288. [DOI] [PubMed] [Google Scholar]
- Wellings SR, Jensen HM. J. Natl. Cancer Inst. 1973;50:1111–1118. doi: 10.1093/jnci/50.5.1111. [DOI] [PubMed] [Google Scholar]
- Wilson KS, Roberts H, Leek R, Harris AL, Geradts J. Am. J. Pathol. 2002;161:1171. doi: 10.1016/S0002-9440(10)64394-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C, Nicholson S, Angus B, Sainsbury JR, Farndon J, Cairns J, Harris AL, Horne CH. Br. J. Cancer. 1992;65:118–121. doi: 10.1038/bjc.1992.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Ikawa S, Akiyama T, Semba K, Nomura N, Miyajima N, Saito T, Toyoshima K. Nature. 1986;319:230–234. doi: 10.1038/319230a0. [DOI] [PubMed] [Google Scholar]
- Yang Y, Dukhanina O, Tang B, Mamura M, Letterio JJ, MacGregor J, Patel SC, Khozin S, Liu Z, Green J, Anver MR, Merlino G, Wakefield LM. J. Clin. Invest. 2002;109:1607–1615. doi: 10.1172/JCI15333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Geng Y, Sicinski P. Nature. 2001;411:1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- Yue J, Mulder KM. Pharmacol. Ther. 2001;91:1–34. doi: 10.1016/s0163-7258(01)00143-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.