Abstract
Background
Wegener granulomatosis (WG) belongs to the heterogeneous group of systemic vasculitides. The multifactorial pathophysiology of WG is supposedly caused by yet unknown environmental influence(s) on the basis of genetic predisposition. The presence of anti-neutrophil cytoplasmic antibodies (ANCA) in the plasma of patients and genetic involvement of the human leukocyte antigen system reflect an autoimmune background of the disease. Strong associations were revealed with WG by markers located in the major histocompatibility complex class II (MHC II) region in the vicinity of human leukocyte antigen (HLA)-DPB1 and the retinoid X receptor B (RXRB) loci. In order to define the involvement of the 6p21.3 region in WG in more detail this previous population-based association study was expanded here to the respective 3.6 megabase encompassing this region on chromosome 6. The RXRB gene was analysed as well as a splice-site variation of the butyrophilin-like (BTNL2) gene which is also located within the respective region. The latter polymorphism has been evaluated here as it appears as a HLA independent susceptibility factor in another granulomatous disorder, sarcoidosis.
Methods
150–180 German WG patients and a corresponding cohort of healthy controls (n = 100–261) were used in a two-step study. A panel of 94 microsatellites was designed for the initial step using a DNA pooling approach. Markers with significantly differing allele frequencies between patient and control pools were individually genotyped. The RXRB gene was analysed for single strand conformation polymorphisms (SSCP) and restriction fragment length polymorphisms (RFLP). The splice-site polymorphism in the BTNL2 gene was also investigated by RFLP analysis.
Results
A previously investigated microsatellite (#1.0.3.7, Santa Cruz genome browser (UCSC) May 2004 Freeze localisation: chr6:31257596-34999883), which was used as a positive control, remained associated throughout the whole two-step approach. Yet, no additional evidence for association of other microsatellite markers was found in the entire investigated region. Analysis of the RXRB gene located in the WG associated region revealed associations of two variations (rs10548957 pallelic = 0.02 and rs6531 pallelic = 5.20 × 10-5, OR = 1.88). Several alleles of markers located between HLA-DPB1, SNP rs6531 and microsatellite 1.0.3.7 showed linkage disequilibrium with r2 values exceeding 0.10. Significant differences were not demonstrable for the sarcoidosis associated splice-site variation (rs2076530 pallelic = 0.80) in our WG cohort.
Conclusion
Since a microsatellite flanking the RXRB gene and two intragenic polymorphisms are associated significantly with WG on chromosome 6p21.3, further investigations should be focussed on extensive fine-mapping in this region by densely mapping with additional markers such as SNPs. This strategy may reveal even deeper insights into the genetic contributions of the respective region for the pathogenesis of WG.
Background
Wegener granulomatosis (WG) is a granulomatous disorder belonging to the heterogeneous group of systemic vasculitides (SV). A common feature of SV is the inflammation of the endothelium [1,2]. SV are classified according to the size of affected vessels and the type of auto-antibodies, namely anti-neutrophil cytoplasmic antibodies (ANCAs), which are used for differential diagnosis [3,4].
WG has an annual incidence of 5–10/million individuals in Caucasians [5]. The pathophysiology of WG still remains largely unknown with a supposedly multifactorial basis [6,7]. Presence of ANCA in the plasma of ~90% of WG patients reflects autoimmune background of the disease. In WG patients ANCAs are mostly directed against proteinase 3 (PRTN3), presented in primary azurophil granules of polymorph nuclear neutrophils (PMN) and lysosomes of monocytes [8,9]. After cytokine priming of PMN, PRTN3 translocates to the cell surface where ANCAs can bind and activate PMN resulting in a respiratory burst and release of proteolytic enzymes [10]. This may then lead to a self sustaining inflammatory process.
Several candidate genes such as PRTN3, α1-antitrypsine, adhesion molecule CD18 or interleukin 1 and its receptor have been investigated for WG association [11-15]. In addition, there is genetic evidence that the human leukocyte antigen (HLA) system is involved in WG development [16-19]. Yet, mostly these studies showed exclusively spurious WG associations. Recently, an extended association screen (EAS) revealed strong WG association of a microsatellite marker (UCSC May 2004 Freeze chr6:31257596-34999883) located in major histocompatibility complex class II (MHCII) region in the immediate vicinity of the HLA-DPB1 and retinoid X receptor β (RXRB) genes in two German populations [20]. Genotyping 19 alleles of the HLA-DPB1 gene with increased frequency of the HLA-DPB1*0401 allele supported the evidence that this region harbours at least one genetic factor for WG. But further association mapping encompassing a region of ~280 kb with additional microsatellite and SNP markers did not allow to decide between several alternatives. Either this region harbours one major locus for WG, encompasses two susceptibility loci or the association of specific marker alleles is due to linkage with a causative locus at some distance. In this context, the complex linkage disequilibrium (LD) patterns are crucial with areas of densely packed recombination hot spots as well as large LD blocks on chromosome 6p thus further complicating to define associations in this region [21,22]. In a recent study on the granulomatous disorder sarcoidosis, a fine mapping approach using SNPs led to the definition of an HLA independent genetic factor, BTNL2 [23]. This study underlines the necessity of investigations on larger regions encompassing defined associations in order to evaluate aforementioned considerations on extended LD on chromosome 6, i.e. how far an association can extend, or HLA independent risk factors. Here, we expanded association mapping to a 3.6 mb region encompassing the WG associated 6p21.3 as well as adjacent regions with additional microsatellites. This study was carried out in a two-step population based design using pooled DNA for the initial scan and, subsequently individual genotyping of markers that differed statistically significantly between WG patients and controls.
Furthermore, we focussed on two candidate genes on chromosome 6, RXRB and butyrophilin-like 2 (BTNL2) located in the vicinity or even in the respective WG-associated region [20]. RXRB presents as a good candidate as the protein binds to hormone nuclear receptors such as the vitamin D receptor (VDR) [24]. VDR and its ligand calcitriol have widespread influence on immuno-regulatory systems [25-28]. Thereby, RXRB variation may be involved in the pathogenesis of WG. In order to find functionally relevant polymorphisms the entire coding region and ~593 bp of the promoter were screened for variations. Furthermore, a recently described splice-site polymorphism of the BTNL2 gene has been reported to represent a HLA independent risk factor for another granulomatous disorder affecting the lung, namely sarcoidosis [23]. BTNL2 is located on 6p21.3 in the vicinity of the HLA-DRB1 locus. On the basis of sharing traits in WG and sarcoidosis such as granulomas and pulmonary affections, the functionally relevant splice-site polymorphism was genotyped in our cohorts.
Methods
Patients
German WG patients were clinically diagnosed according to international standards by applying the 1990 classification criteria of the American College of Rheumatology [29] and the definitions of the 1994 Chapel Hill Consensus Conference [3]. All patients have defined ANCA-status (npositive = 151; nnegative = 30). WG was biopsy-proven in the German reference centre for vasculitis (Department of Pathology, University of Schleswig-Holstein Campus Luebeck, Germany) by 2 different observers. Control groups comprised healthy adults from northern Germany and the Ruhr area (Germany). The institutional Ethics Committee of Bochum Ruhr-University approved this study. Informed consent was obtained from all individuals.
Pooling of DNA
DNA pooling was performed as described before [20]. Briefly, DNA concentrations were measured spectrophotometrically in 4 steps, and samples were diluted accordingly. The final DNA concentration in pools was 50 ng/μl. In order to control for artefacts during pooling, 150 ANCA-positive patients were divided into 3 sub-pools and 100 controls from the northern German cohort into 2 sub-pools, respectively.
Microsatellite marker panel
A panel of 94 markers was designed encompassing the WG associated 6p21.32 and flanking regions (annotated by Santa Cruz genome browser (UCSC): chro6:31255920-34938760, May 2004 freeze, see [30]). Microsatellites were chosen utilizing the simple-repeat option of UCSC. The average distance between microsatellite markers was 41 kb (standard deviation ~30 kb) with five gaps of distances ≥100 kb (<152 kb). Most of the microsatellites harboured dinucleotide repeats (68%), the remaining comprised tri-, tetra- or penta-nucleotide repeat motifs. Further information about the microsatellites is available as additional file 1 (website: [31]). No marker revealed significantly differing 'intra sub-pool' distributions as reflected by the p value (see below).
PCR
For fragment analysis we used fluorescence 5'FAM labelled, tailed oligonucleotide added to the 5'-part of the sequence specific primer as described before [20]. Primers were designed with the Primer Express 2.0 Software (Applied Biosystems, Foster City, USA). Melting temperature was set to 55°C. Three primers were used for PCR: tailed forward primer (tailed F), reverse primer and labelled primer (labelled F) corresponding to the 5'-tail sequence of tailed F. Reaction mix consisted of 1 × PCR buffer (Qiagen GmbH, Hilden, Germany), 1.5 pmol labeled F, 0.2 mM each dNTP, 3 mM MgCl2, 0.2 pmol tailed F, 1.5 pmol reverse primer, 0.25 U Qiagen Hot Start Taq (Qiagen) and 50 ng DNA. PCR was performed in a Biometra T-Gradient Thermoblock (Biometra, Göttingen, Germany) using the following characteristics: activation step at 95°C for 15 min; 35 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 1 min and extension at 72°C for 1 min; and a final extension at 72°C for 10 min.
Initial screen
Electrophoresis and data analysis were performed on a 48-well ABI377 slab-gel system using ABI Genotyper software as described before [20]. Data analysis generated marker-specific allele image profiles (AIPs) representing distributions of alleles within each sub-pool.
Statistical analysis
AIPs of WG pools were compared with those of the control cohort. Peak heights were cumulated to 100% (representing 100 alleles in each pool, respectively). Afterwards allele frequencies (corresponding to peaks height) were calculated according to the estimated total allele count. Association was tested for each marker by comparison of the AIPs from WG-DNA pools with those of controls by contingency tables using a significance level of p = 0.05. In order to focus the statistics on major effects, alleles with a frequency of less than 5% were summarized to one allele. Next, frequency distributions were compared by means of contingency tables as mentioned above. The resulting p values were corrected for multiple testing using QValue software [32,33] with a 'cut off' of 5%. Nevertheless, non-corrected p values were simply ranked according to their evidence for association in order to select markers for further studies. In addition, intra-population differences were tested by comparing WG pools with each other as well as both control pools.
Individual genotyping
In order to exclude false positive significances, patients and controls were individually genotyped for respective markers. Genotyping was performed on the Beckman Coulter CEQ8000 8-capillary system using 'Fragment Analysis Module' software (Beckman Coulter, Inc., Fullerton, USA). Reaction mix contained 1 × PCR buffer (Qiagen), 0.75 pmol fluorescent labelled F, 0.2 mM each dNTP, 3 mM MgCl2, 0.2 pmol tailed F, 1.5 pmol reverse primer, 0.25 U Qiagen Hot Start Taq (Qiagen) and 50 ng DNA. Parameters for amplification were used as described above. Allele and genotype frequency of WG patients and controls were compared by χ2-testing. Additionally, Hardy-Weinberg equilibrium was tested using Genepop software [20].
Screening of the coding and a 593 bp promoter region in the RXRB gene
The coding and promoter regions (593 bp) of the RXRB gene were initially screened by single strand conformation polymorphism (SSCP) with 48 patient (ANCA-positive) and 48 control DNAs. The RXRB sequence was downloaded from UCSC database (NM021976, July 2003 Freeze). Oligonucleotides were designed using Primer Express 2.0 Software (Applied Biosystems) with an optimal annealing temperature of 57°C (see table 1). PCR was performed using Qiagen HotStar Taq (Qiagen) with an initial denaturation at 95°C for 15 min, 35 cycles of 94°C for 30 sec -57°C for 30 sec -72°C for 30 sec and a final extension at 72°C for 10 min. PCR conditions were chosen as recommended by Qiagen. Fragments were labelled with α-dATP (exons 2–10; Hartmann Bioanalytics, Braunschweig, Germany) and α-dCTP (exon 1a and 1b; Hartmann Bioanalytics). SSCP were analysed on 6% and 5% polyacrylamide gels with either 5% or 10% glycerol. The gels were run at room temperature at 35 W. Amplificates containing obvious band shifts were sequenced using dye terminator cycle sequencing on an ABI377 automatic sequencer (Applied Biosystems). Statistical comparisons of allele frequencies and genotypes were based on the χ2 test. In addition, Hardy-Weinberg equilibrium has been tested for all SNPs.
Table 1.
Oligonucleotides and restriction enzymes used for SSCP analyses of the exonic and 593 bp promoter regions of the RXRB gene.
| Exon | Oligonucleotide sense/antisense | PCR product (bp) | Restriction enzyme; length of fragments (bp) |
| 01a | CGGTATCCCTACTCTCAGCCA/GCGGGATCCAGCCAGG | 164 | - |
| 01b | GGTGCGAAAAGAAATGCA/CACTGGCTCGCCTGCC | 202 | - |
| 02 | CTCTTGTGTGTCTGTGTGCCT/GGTTAGAGGATTGGAAGGTCA | 403 | AvaII 176/227 |
| 03 | GACTTCCCTATTTCCCCCAT/TCTCGGAGAAGAGGAGGCT | 207 | - |
| 04 | TGAAGGTGTCTCCATGCAAC/AGAAGGGCATGTGGTCTAAGA | 275 | BsmFI 126/149 |
| 05 | CTGATGAGGCCGTAAGGATA/GTGGATTGACCCCAACACT | 280 | Eco24I 130/150 |
| 06 | TCCATAACTCTTACCCCCGT/CTGGGTACGCAAGGTAAGG | 255 | MboII 102/153 |
| 07 | CCTGATTTCTGGCTCCTGAC/CCTCTATCTACATGCCAGCCT | 209 | - |
| 08 | GCTGAGCTGTGACCTTTGAGT/TCTCGGAGAAGAGGAGGCT | 199 | - |
| 09 | CTCCTCTCTTCCCTGCCAT/CCTCATAGCACTCCCCACC | 199 | - |
| 10 | CTCAATCCCCTTCTCCCAC/AGTGTGAGAAGCACCACGTC | 209 | - |
| Promoter (593 bp) | |||
| Sys1 | GGGCTTAATTCGACCCAA/GCATTTCTTTTCGCACC | 213 | - |
| Sys2 | GCTAACAGGCCGGAGGAGA/GGATTGATCGGAGGATTAGCT | 212 | - |
| Sys3 | CTCTCCTTTCCCGGTTTG/TCGTCTAGTTGGAAACCGAG | 234 | - |
| Sys4 | CTCTTTATCCCGAACCACCT/TCTCGCGGGATCTAAAGG | 221 | - |
| Sys5 | CATCACGCTGACCAGAGG/TGGTGAAAGATTAGTGTCCCA | 214 | - |
Genotyping of rs6531
The sequence variation rs6531 was genotyped in 151 ANCA-positive patients and 201 controls by RFLP analysis. A restriction site for DdeI (NewEnglandBiolabs, Ipswich, USA) was generated by using the tailed mismatch oligonucleotide, mmantisense-CATCGCTGATTCGCACATCATCAATGGATCGGTCTGA. PCR was performed by 'the tailed primer' method using three oligonulceotides: the mmantisene-primer, an oligonucleotide corresponding to the tail and the forward oligonucleotide sense-CCGATCTTTAGTGACCCCAGT. Restriction with DdeI results in a 226 bp fragment (T allele) and in case of the C allele in 2 fragments of 192 and 34 bp, respectively. Electrophoresis was done on a 3% agarose gel containing ethidium bromide. The results were analysed with the χ2 test and Hardy-Weinberg equilibrium was controlled.
Genotyping of the Val95Ala polymorphism in the RXRB gene
The previously reported the Val95Ala polymorphism [34] was genotyped by RFLP analysis in 94 patients and 90 controls. Fragments were amplified using oligonucleotides Val95Ala sense-CGGTGGGGTATTAGAGAATT and Val95Ala antisense-CCCATGGAAGAACTGATGACTGG. PCR was performed with Qiagen HotStar Taq (Qiagen) as described above with an annealing temperature of 55–60°C and different MgCl2-concentrations generating a 300bp fragment. The 2 alleles (Val, T allele; Ala, C allele) are detected as 232 bp and 193 bp bands, respectively.
Electrophoresis was done in ethidium bromide containing 2% agarose gels. In order to control the results of the RFLP analysis, DNAs of 15 patients and 20 controls were sequenced as described above. Association was verified with the χ2 test and Hardy-Weinberg equilibrium.
Genotyping the functionally relevant BTNL2 variation rs2076530
Primer extension analysis was used to genotype the truncating splice site mutation in 180 patients and 261 controls. Two primers were designed for amplification, forward TCCAGATACTCAGTGCCAGA, reverse: TTGTCCAGGAACTAGCATATT. PCR conditions were: 1 × PCR buffer (Qiagen), 0.2 mM each dNTP, 0.2 pmol forward primer, 0.2 pmol reverse primer, 0.25 U Qiagen Hot Start Taq (Qiagen) and 50 ng DNA. The reaction was performed with initial denaturation at 95°C for 15 min, 35 cycles of 94°C for 30 sec -60°C for 30 sec -72°C for 30 sec and a final extension at 72°C for 10 min The product was purified with magnetic beads system (AMPure; ABgene, Epsom, UK). A primer extension reaction was performed with the interrogation primer: GCCCAGTTTGGATCTGAAGGTGGTA using Beckmann CEQ™ SNP-Primer Extension Kit (Beckman Coulter) under recommended conditions. Electrophoresis and data analysis was performed using Beckman Coulter CEQ8000. Statistical analysis was based on the χ2 tests, and allele frequencies were tested for Hardy-Weinberg equilibrium.
LD analyses
We calculated pair-wise LD between HLA-DPB1 alleles (genotypes obtained from a previous study [20], see additional file 2, website: [35]), rs6531 and the significantly associated microsatellite #1.0.3.7 in our control and patient cohorts. Microsatellite, SNP and HLA-DPB1 alleles with frequencies below 0.05 were excluded. For pairwise LD calculation we presumed each microsatellite allele to be biallelic, i.e. one distinct allele against all others pooled. The same procedure was applied for the most frequent HLA-DPB1 alleles *0401, *0402, *0201 and *0301. The strength of LD between all three markers was quantified by D' and r2 values. LD analyses were calculated by Haploview 3.2 [36].
Results
Initial association mapping
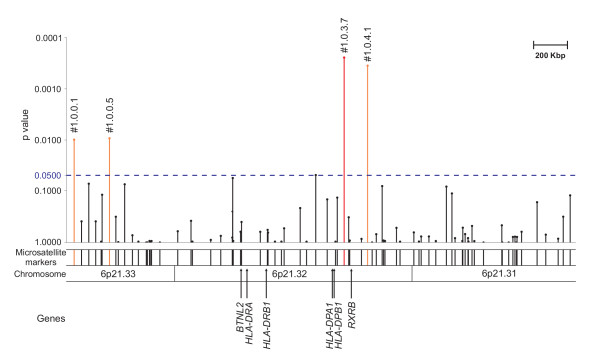
Eighty four markers were analysed in an initial scan (after exclusion of non-polymorphic and recurrently artefact-producing microsatellites). Thereof, five microsatellites revealed significant differences in allele distribution between WG patients and controls (see table 2). Correction for multiple comparisons with QValue rejected significance for all results. Nonetheless, markers were simply ranked according to their p values. Thereafter, four markers were selected for individual genotyping (see figure 1). One microsatellite showed only marginal significance (#1.0.3.4) after summation of rare alleles (<5%) and thus was excluded from further investigations. Individual genotyping confirmed exclusively one marker (adjacent to RXRB), the positive control, as statistically significantly different: #1.0.3.7 (p = 3 × 10-4, see table 2).
Table 2.
P values for microsatellite markers after the initial step and individual genotyping.
| p values | |||
| Code of marker | Number of alleles | after analysis with pooled DNA | after summation of rare alleles (<5%) |
| #1.0.0.1 | 5 | 0.02 | 0.01 |
| #1.0.0.5 | 12 | 0.03 | 0.01 |
| #1.0.3.4 | 3 | 0.04 | 0.05 |
| #1.0.3.7* | 5 | 2 × 10-4 | 3 × 10-4 |
| #1.0.4.1 | 6 | 1 × 10-4 | 4 × 10-4 |
| p values | |||
| Code of marker | after analyses of each single allele (most significant allele) | after individual genotyping (pc value) | after correction by Q-value |
| #1.0.0.1 | 0.03 | n.s. | n.s. |
| #1.0.0.5 | >0.05 | n.s. | n.s. |
| #1.0.3.4 | 0.03 | n.d. | n.d. |
| #1.0.3.7* | < 1 × 10-4 | 4 × 10-5 (pc = 3 × 10-4; c = 5) | n.s. |
| #1.0.4.1 | 3 × 10-3 | n.s. | n.s. |
n.s. = not significant; n.d. = not determined; * = positive control
Figure 1.
Map of the investigated HLA region with p values of the markers used in the initial screen. Figure shows the investigated fragment of chromosome 6 comprising the previously defined WG associated region encompassing the HLA-DPB1 and RXRB genes. Vertical lines represent studied microsatellites. The lengths/heights of these lines correspond to p values after analyses in the initial screen with pooled DNA. The 4 markers, which were genotyped individually, are labelled. The p values of markers #1.0.0.1, #1.0.0.5 and #1.0.4.1 (pool analysis) could not be replicated by genotyping individually. Drawn in red is the marker (#1.0.3.7) that remained significantly different after individual genotyping. This marker has been used as a positive control (see text). For further information of fine localisation of each investigated marker and inter-marker distances see additional file 1.
Analyses of the RXRB gene
SSCP analyses in the RXRB gene did not reveal any variations in exons 1–5, 8–10 and the 593 bp promoter region. Similarly, the Val95Ala polymorphism was neither detectable by RFLP nor by sequence analyses in our patient and control cohorts. Yet, the analysis of the PCR product comprising exon 6 revealed a band shift. Subsequent sequencing showed an intronic CT-INDEL polymorphism which has been reported before (rs10548957). Statistical analysis of the allele frequencies revealed a p value of 0.02 with a minor allele frequency of 0.07 in the control cohort (n = 47) and total absence in the patients (n = 43). In exon 7 a single nucleotide polymorphism (SNP) (rs6531) was detected using SSCP and sequencing. Comparison of allele frequencies of patients (n = 151) vs. controls (n = 201) revealed significant p values (see table 3 for detailed analyses). Additionally 28 ANCA-negative patients were genotyped for this SNP and compared to either the ANCA-positive patients or the control cohort. There is a significant deviation between the ANCA-negative and -positive patients regarding this SNP. The ANCA-negative cohort did not differ from the control group.
Table 3.
Genotyping of rs6531 in the RXRB gene.
| Genotypes/phenotypes/alleles | Patients ANCA+ (n = 151) | Patients ANCA- (n = 36) | Controls (n = 201) | |
| CC | 36 | 15 | 92 | |
| CT | 87 | 19 | 88 | |
| TT | 28 | 2 | 21 | |
| CC+CT (vs. TT) | 123 | 34 | 180 | |
| TT+CT (vs. CC) | 115 | 21 | 109 | |
| C | 159 | 49 | 272 | |
| T | 143 | 23 | 130 | |
| ANCA+ against ANCA- patients | ANCA+ patients against controls | |||
| Genotypes/phenotypes/alleles | ||||
| P value | OR (95 % C.I.) | P value | OR (95 % C.I.) | |
| CC | 0.03 | 2.3 × 10-5 | ||
| CT | 0.10 | 1.91 (0.87 to 4.16)* | 1.6 × 10-4 | 2.53 (1.55 to 4.11)* |
| TT | 0.02 | 5.83 (1.23 to 27.65)* | 0.03 | 5.89 (3.01 to 11.53)* |
| CC+CT (vs. TT) | 0.07 | 0.26 (0.06 to 1.14) | 0.03 | 0.51 (0.29 to 0.94) |
| TT+CT (vs. CC) | 0.03 | 2.26 (1.17 to 4.88) | 2.3 × 10-5 | 2.70 (1.69 to 4.30) |
| C | 1.0 × 10-2 | 5.2 × 10-5 | ||
| T | 1.91 (1.11 to 3.30) | 1.88 (1.38 to 2.56) | ||
OR = odds ratio, C.I. = confidence interval.
*Odds ratios for the CT and TT genotypes are contrasted against the CC genotype. No evidence for statistical difference between the ANCA-negative WG patients and the control cohort was revealed, when calculating all off above-used comparisons.
LD analyses
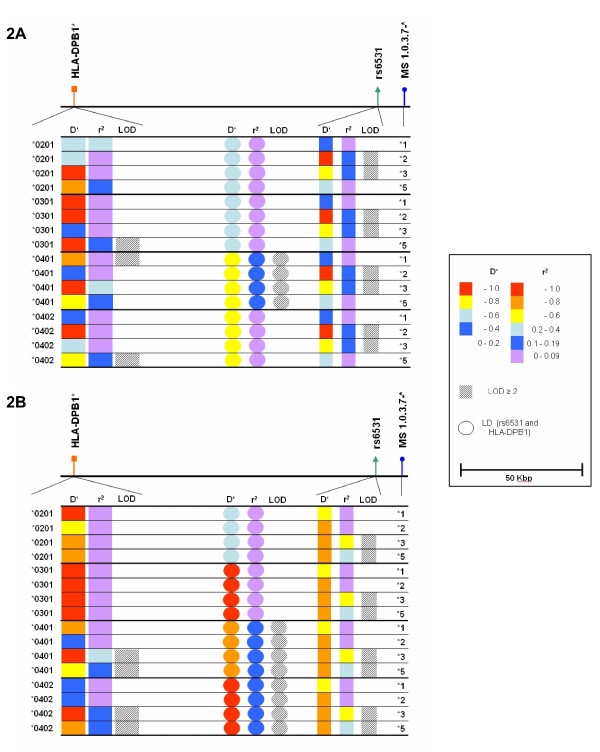
Several alleles between HLA-DPB1, rs6531 and microsatellite marker #1.0.3.7 showed LD with r2 values exceeding 0.10 (for detailed description see figure 2A and 2B). For example, strong LD exists between HLA-DPB1*0401 and microsatellite allele *3 as well as the rs6531 polymorphism in the patient and control cohorts.
Figure 2.
Graphical overview of LD analyses with frequent HLA-DPB1 alleles, the rs6531 polymorphism in the RXRB gene and alleles of the significantly WG-associated #1.0.3.7 microsatellite marker. Pair-wise LD evaluated by D' and r2 between markers in the previously found WG associated chromosome 6 region for controls (2A) and patients (2B). Vertical lines with circle, triangle and square represent the relative localisation of each tested marker (HLA-DPB1, RXRB rs6531 and microsatellite marker #1.0.3.7, respectively). D' and r2 values are indicated by colour coding. Logarithm of odds (LOD) values equal to or exceeding 2 are represented by dashed rectangles, circles or squares. Rectangles are related to LDs between the microsatellite #1.0.3.7 and HLA-DPB1 alleles. Circles represent LD values between HLA-DPB1 and the SNP. Squares show LDs between the microsatellite alleles and rs6531 SNP. The investigated alleles of the microsatellite markers are shown at the left hand side of the figure, whereas the different designations of the HLA-DPB1 alleles are depicted at the right hand side. MS: microsatellite.
Analysis of the functionally relevant BTNL2 variation
Genotyping of rs2076530 polymorphism in 180 patients and 261 controls (Northern Germany and Ruhr area populations) did not reveal any statistically significant difference in allele, phenotype and genotype frequencies between all WG patients and healthy individuals (see table 4). In addition, when dividing the patient cohort into ANCA+ and ANCA-patients, significant differences were not detected between the respective patient and control cohorts (data not shown).
Table 4.
Genotyping of rs2076530 in the BNTL2 gene.
| Genotypes/phenotypes/alleles | Patients *1 (n = 180) | Controls (n = 261) | P value |
| GG | 68 | 79 | 0.27 |
| GA | 76 | 135 | 0.14 |
| AA | 36 | 47 | 0.94 |
| GG+GA | 144 | 214 | 0.84 |
| AA+GA | 112 | 182 | 0.19 |
| G | 212 | 293 | 0.80 |
| A | 148 | 229 |
*1 ANCA positive (n = 150) and ANCA negative (n = 30) patients. No statistically significant difference between the ANCA positive and negative patients was evident.
Discussion
The approach using pooled DNA and allele image profiles for microsatellite based association studies was used to investigate groups of candidate genes for different multifactorial disorders revealing novel predisposition factors for respective diseases [20,37]. For these studies markers were chosen located intra- or juxtagenically to distinct genes. Here, we used a population based association study approach with microsatellites and DNA pooling for scanning a ~3.6 mb region on 6p21.3. This chromosomal segment includes the previously characterized, WG associated region [20]. Several other studies have revealed associations of HLA genes with WG and other vasculitides (e.g. [17,19,38-41]). Yet, most of these results could not be replicated. Considering the highly variable HLA complex, the strong positive selection effects on this region in combination with the relatively limited number of investigated patients, definitive conclusions are truly challenging. By using our EAS approach we searched for further loci for WG or whether the identified HLA-DPB1/RXRB associations are due to linkage with other factors located more distantly on 6p21.3 comprised in the genomic region investigated here. Yet, we could neither reveal any evidence for an additional locus nor for an expanded, significantly associated 'HLA-DPB1/RXRB' region, respectively. In our study, the only marker (#1.0.3.7) which remained significantly associated has been reported before [20]. This marker is located in the immediate vicinity of the RXRB gene and ~125 kb centromeric to HLA-DPB1. Hence we calculated LD of the microsatellite, HLA-DPB1 alleles (with data from a previous study [20]; see additional file 2) and the significantly differing, intragenic rs6531 RXRB SNP. These analyses e.g. confirmed results of Rajisbaum et al. [42] which revealed significant LD between polymorphisms in RXRB and HLA-DPB1. In order to definitively investigate this very locus, a more detailed approach with a dense map of additional markers (preferential SNPs) appears mandatory.
It remains possible, that loci of small effects escaped detection in our microsatellite-based association mapping – a procedure known to ascertain strong associations. Such deficiency might be due to artefacts in the pooling procedure and analyses with pooled DNA such as length-dependent amplification of short microsatellite marker alleles or even the presence of null-alleles. In addition, the slab-gel system approach might reflect a further hindrance in this subtle procedure, since it is not 100% consistent. Altogether such shortcomings could result in false allele frequencies thus ignoring truly positive low-effect loci, for which extremely precise data are necessary. A further explanation for the negative results for most markers is that our panel may not represent the whole region completely. Hence the possibility remains that we missed causative alleles due to lack of LD between a distinct marker allele and the respective susceptibility factor. More detailed information on haplotype block structures and future definition of LD between multiallelic microsatellite markers may facilitate even more far-fetched interpretations of our analyses. Nevertheless, our data indicate that the here investigated region might harbour only one (major) locus associated with WG as represented by the control marker in the vicinity of RXRB (~5 kb) showing a positive result. By applying QValue correction tests, the significance of this marker would have been rejected. Similar controversies were presented recently in a study for narcolepsy utilizing EAS using correction according to Bonferroni [37]. Hence, further analyses on this region were performed regardless of the correction for multiple comparisons.
In addition to the indirect association study with microsatellites, we screened the entire coding (including the splice-sites) and the promoter region of RXRB in order to seek putative functionally relevant variations. In these analyses two polymorphisms were evident: an intronic CT-INDEL (rs10548957) variation revealing only a marginal association with WG, whereas the synonymous SNP (rs6531) in exon 7 was strongly associated (table 3). Nevertheless, this latter SNP does not affect known functional sequences influencing splicing and or regulation of gene expression. Therefore, the functional consequences of both polymorphisms are not immediately apparent. Both variations, however, confirmed the previously defined predisposing and protective haplotypes [20]. In this study only part of the potential promoter region was investigated. Therefore, the expression of RXRB may be influenced by polymorphisms/mutations in more distant promoter regions. Interestingly, the SNP in exon 7 might reflect differences between ANCA-negative and -positive patients. Whereas this polymorphism is associated in the ANCA-positive WG group, there is no statistical difference between the controls and ANCA-negative patient group. Although the amount of ANCA-negative patients in our study is rather small and therefore the statistics are almost underpowered, this result gives a further hint that both phenotypes may differ in their genetic background confirming recent studies, e.g. on the intracellular tyrosine phosphates (PTPN22) gene [43].
Although our initial mapping approach did not reveal an association in further regions in the HLA complex, we genotyped the previously found functionally relevant variation in the BTNL2 gene, which is associated with sarcoidosis and may present a predisposing/protective variation with low effect size [23]. Recently, several studies proposed common genetic factors for diseases characterised by autoimmune phenomena such as polymorphisms in the aforementioned PTPN22 gene or the cytotoxic T lymphocyte associated protein (CTLA4; see [44,45]). Considering common features of sarcoidosis and WG (e.g. autoimmune background, granulomata in the lung), variations within BTNL2 may represent risk factors for WG. Yet, in our study we did not find evidence for involvement of BTNL2 in the pathogenesis of WG, neither in the initial screen (both microsatellites flanking BTNL2 at ~25 kb: #1.0.2.5 and #1.0.2.6 showing p = 0.95 and p = 0.63, respectively) nor in individual genotyping with the functional rs2076530 polymorphism reported before (p = 0.42, table 4). As a remote possibility, different variations (of low effect size) in the BTNL2 gene may have predisposing/protective nature in the pathogenesis of WG due to allelic heterogeneity.
Conclusion
Based on our largely negative association data in context with the defined WG associated region on 6p21.3, we submit that only a single region of ~280 kb exists that is responsible for WG predisposition in the HLA complex. In case several loci of low effect size would exist, novel alternative methodological approaches are necessary to validate their association with WG on chromosome 6.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
PS carried out the practical laboratory work, performed primary data analyses and drafted the manuscript. PJ initiated this study, prepared the initial version of the manuscript and performed statistical analyses. EC and WLG were responsible for the clinical characterisation of the entire patient cohort. JTE encouraged the critical approach, rewrote and finalized the paper. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Microsatellite marker panel, additional data.
HLA-DPB1 and microsatellite #1.0.3.7 genotyping information.
Acknowledgments
Acknowledgements
Many thanks to Amer Denis Akkad for technical and laboratory support.
This work was supported by a grant from the German Bundesministerium für Bildung und Forschung (Rheumanet, 01 GI 9954). Supported by the DFG grant Schu 1308/1-3, the Kompetenznetzwerk Rheuma and Forschungsschwerpunkt "Koerpereigene Infektabwehr", Medical University of Luebeck.
Contributor Information
Paweł Szyld, Email: szyld@list.pl.
Peter Jagiello, Email: pjag@gmx.de.
Elena Csernok, Email: csernok@rheuma-zentrum.de.
Wolfgang L Gross, Email: gross@rheuma-zentrum.de.
Joerg T Epplen, Email: joerg.t.epplen@rub.de.
References
- Savage CO, Cooke SP. The role of the endothelium in systemic vasculitis. J Autoimmun. 1993;6:237–249. doi: 10.1006/jaut.1993.1021. [DOI] [PubMed] [Google Scholar]
- Watts RA, Scott DG. Vasculitis. Baillieres Clin Rheumatol. 1995;9:529–554. doi: 10.1016/S0950-3579(05)80257-2. [DOI] [PubMed] [Google Scholar]
- Jennette JC, Falk RJ, Andrassy K, Bacon PA, Churg J, Gross WL, Hagen EC, Hoffman GS, Hunder GG, Kallenberg CG, Mccluskey RT, Sinico RA, Rees AJ, Van Es LA, Waldherr R, Wiik A. Nomenclature of systemic vasculitides. Proposal of an international consensus conference. Arthritis Rheum. 1994;37:187–192. doi: 10.1002/art.1780370206. [DOI] [PubMed] [Google Scholar]
- Falk RJ, Jennette JC. Thoughts about the classification of small vessel vasculitis. J Nephrol. 2004;17 Suppl 8:S3–9. [PubMed] [Google Scholar]
- Watts RA, Scott DG. Epidemiology of the vasculitides. Curr Opin Rheumatol. 2003;15:11–16. doi: 10.1097/00002281-200301000-00003. [DOI] [PubMed] [Google Scholar]
- Miterski B, Epplen JT, Gencik M. On the genetic contribution to selected multifactorial diseases with autoimmune characteristics. Cell Mol Biol (Noisy-le-grand) 2002;48:331–341. [PubMed] [Google Scholar]
- Hellmich B, Epplen JT, Gross WL. Are ANCA-associated vasculitides inherited? Clin Exp Rheumatol. 2004;22:S1–2. [PubMed] [Google Scholar]
- Ludemann J, Utecht B, Gross WL. Anti-neutrophil cytoplasm antibodies in Wegener's granulomatosis recognize an elastinolytic enzyme. J Exp Med. 1990;171:357–362. doi: 10.1084/jem.171.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specks U. ANCA subsets: influence on disease phenotype. Cleve Clin J Med. 2002;69 Suppl 2:SII56–9. doi: 10.3949/ccjm.69.suppl_2.sii56. [DOI] [PubMed] [Google Scholar]
- van der Geld YM, Limburg PC, Kallenberg CG. Proteinase 3, Wegener's autoantigen: from gene to antigen. J Leukoc Biol. 2001;69:177–190. [PubMed] [Google Scholar]
- Esnault VL, Testa A, Audrain M, Roge C, Hamidou M, Barrier JH, Sesboue R, Martin JP, Lesavre P. Alpha 1-antitrypsin genetic polymorphism in ANCA-positive systemic vasculitis. Kidney Int. 1993;43:1329–1332. doi: 10.1038/ki.1993.186. [DOI] [PubMed] [Google Scholar]
- Gencik M, Meller S, Borgmann S, Fricke H. Proteinase 3 gene polymorphisms and Wegener's granulomatosis. Kidney Int. 2000;58:2473–2477. doi: 10.1046/j.1523-1755.2000.00430.x. [DOI] [PubMed] [Google Scholar]
- Huang D, Zhou Y, Hoffman GS. Pathogenesis: immunogenetic factors. Best Pract Res Clin Rheumatol. 2001;15:239–258. doi: 10.1053/berh.2001.0142. [DOI] [PubMed] [Google Scholar]
- Meller S, Jagiello P, Borgmann S, Fricke H, Epplen JT, Gencik M. Novel SNPs in the CD18 gene validate the association with MPO-ANCA+ vasculitis. Genes Immun. 2001;2:269–272. doi: 10.1038/sj.gene.6363781. [DOI] [PubMed] [Google Scholar]
- Borgmann S, Endisch G, Hacker UT, Song BS, Fricke H. Proinflammatory genotype of interleukin-1 and interleukin-1 receptor antagonist is associated with ESRD in proteinase 3-ANCA vasculitis patients. Am J Kidney Dis. 2003;41:933–942. doi: 10.1016/S0272-6386(03)00190-2. [DOI] [PubMed] [Google Scholar]
- Spencer SJ, Burns A, Gaskin G, Pusey CD, Rees AJ. HLA class II specificities in vasculitis with antibodies to neutrophil cytoplasmic antigens. Kidney Int. 1992;41:1059–1063. doi: 10.1038/ki.1992.161. [DOI] [PubMed] [Google Scholar]
- Cotch MF, Fauci AS, Hoffman GS. HLA typing in patients with Wegener granulomatosis. Ann Intern Med. 1995;122:635. doi: 10.7326/0003-4819-122-8-199504150-00029. [DOI] [PubMed] [Google Scholar]
- Hagen EC, Stegeman CA, D'Amaro J, Schreuder GM, Lems SP, Tervaert JW, de Jong GM, Hene RJ, Kallenberg CG, Daha M, van der Woude FJ. Decreased frequency of HLA-DR13DR6 in Wegener's granulomatosis. Kidney Int. 1995;48:801–805. doi: 10.1038/ki.1995.353. [DOI] [PubMed] [Google Scholar]
- Griffith ME, Pusey CD. HLA genes in ANCA-associated vasculitides. Exp Clin Immunogenet. 1997;14:196–205. [PubMed] [Google Scholar]
- Jagiello P, Gencik M, Arning L, Wieczorek S, Kunstmann E, Csernok E, Gross WL, Epplen JT. New genomic region for Wegener's granulomatosis as revealed by an extended association screen with 202 apoptosis-related genes. Hum Genet. 2004;114:468–477. doi: 10.1007/s00439-004-1092-z. [DOI] [PubMed] [Google Scholar]
- Jongeneel CV, Briant L, Udalova IA, Sevin A, Nedospasov SA, Cambon-Thomsen A. Extensive genetic polymorphism in the human tumor necrosis factor region and relation to extended HLA haplotypes. Proc Natl Acad Sci U S A. 1991;88:9717–9721. doi: 10.1073/pnas.88.21.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys AJ, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- Valentonyte R, Hampe J, Huse K, Rosenstiel P, Albrecht M, Stenzel A, Nagy M, Gaede KI, Franke A, Haesler R, Koch A, Lengauer T, Seegert D, Reiling N, Ehlers S, Schwinger E, Platzer M, Krawczak M, Muller-Quernheim J, Schurmann M, Schreiber S. Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat Genet. 2005;37:357–364. doi: 10.1038/ng1519. [DOI] [PubMed] [Google Scholar]
- Yu VC, Delsert C, Andersen B, Holloway JM, Devary OV, Naar AM, Kim SY, Boutin JM, Glass CK, Rosenfeld MG. RXR beta: a coregulator that enhances binding of retinoic acid, thyroid hormone, and vitamin D receptors to their cognate response elements. Cell. 1991;67:1251–1266. doi: 10.1016/0092-8674(91)90301-E. [DOI] [PubMed] [Google Scholar]
- Mathieu C, Waer M, Laureys J, Rutgeerts O, Bouillon R. Prevention of autoimmune diabetes in NOD mice by 1,25 dihydroxyvitamin D3. Diabetologia. 1994;37:552–558. doi: 10.1007/BF00403372. [DOI] [PubMed] [Google Scholar]
- Adorini L. Immunomodulatory effects of vitamin D receptor ligands in autoimmune diseases. Int Immunopharmacol. 2002;2:1017–1028. doi: 10.1016/S1567-5769(02)00049-8. [DOI] [PubMed] [Google Scholar]
- Adorini L. Tolerogenic dendritic cells induced by vitamin D receptor ligands enhance regulatory T cells inhibiting autoimmune diabetes. Ann N Y Acad Sci. 2003;987:258–261. doi: 10.1111/j.1749-6632.2003.tb06057.x. [DOI] [PubMed] [Google Scholar]
- Adorini L, Penna G, Giarratana N, Roncari A, Amuchastegui S, Daniel KC, Uskokovic M. Dendritic cells as key targets for immunomodulation by Vitamin D receptor ligands. J Steroid Biochem Mol Biol. 2004;89-90:437–441. doi: 10.1016/j.jsbmb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Leavitt RY, Fauci AS, Bloch DA, Michel BA, Hunder GG, Arend WP, Calabrese LH, Fries JF, Lie JT, Lightfoot RWJ, Masi AT, Mcshane DJ, Mills JA, Stevens MB, Wallace SL, Zvaifler NJ. The American College of Rheumatology 1990 criteria for the classification of Wegener's granulomatosis. Arthritis Rheum. 1990;33:1101–1107. doi: 10.1002/art.1780330807. [DOI] [PubMed] [Google Scholar]
- Santa Cruz genome browser (UCSC) http://genome.ucsc.edu
- Microsatellite marker panel additional data. http://www.ruhr-uni-bochum.de/mhg/additional_file_1.pdf
- Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society, Series B. 2002;64:479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- Storey. JD; Tibshirani,R Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro H, Okubo Y, Ohtsuki T, Yamakawa-Kobayashi K, Arinami T. Mutation analysis of the retinoid X receptor beta, nuclear-related receptor 1, and peroxisome proliferator-activated receptor alpha genes in schizophrenia and alcohol dependence: possible haplotype association of nuclear-related receptor 1 gene to alcohol dependence. Am J Med Genet. 2002;114:15–23. doi: 10.1002/ajmg.1620. [DOI] [PubMed] [Google Scholar]
- HLA-DPB1 and microsatellite #1.0.3.7 genotyping information. http://www.ruhr-uni-bochum.de/mhg/additional_file_2.pdf
- Haploview software http://www.broad.mit.edu/personal/jcbarret/haploview/
- Wieczorek S, Jagiello P, Arning L, Dahmen N, Epplen JT. Screening for candidate gene regions in narcolepsy using a microsatellite based approach and pooled DNA. J Mol Med. 2004;82:696–705. doi: 10.1007/s00109-004-0569-5. [DOI] [PubMed] [Google Scholar]
- Katz P, Alling DW, Haynes BF, Fauci AS. Association of Wegener's granulomatosis with HLA-B8. Clin Immunol Immunopathol. 1979;14:268–270. doi: 10.1016/0090-1229(79)90150-8. [DOI] [PubMed] [Google Scholar]
- Elkon KB, Sutherland DC, Rees AJ, Hughes GR, Batchelor JR. HLA antigen frequencies in systemic vasculitis: increase in HLA-DR2 in Wegener's granulomatosis. Arthritis Rheum. 1983;26:102–105. doi: 10.1002/art.1780260118. [DOI] [PubMed] [Google Scholar]
- Papiha SS, Murty GE, Ad'Hia A, Mains BT, Venning M. Association of Wegener's granulomatosis with HLA antigens and other genetic markers. Ann Rheum Dis. 1992;51:246–248. doi: 10.1136/ard.51.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gencik M, Borgmann S, Zahn R, Albert E, Sitter T, Epplen JT, Fricke H. Immunogenetic risk factors for anti-neutrophil cytoplasmic antibody (ANCA)-associated systemic vasculitis. Clin Exp Immunol. 1999;117:412–417. doi: 10.1046/j.1365-2249.1999.00969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajsbaum R, Fici D, Fraser PA, Flores-Villanueva PO, Awdeh ZL. Polymorphism of the human retinoid X receptor beta and linkage disequilibrium with HLA-DPB1. Tissue Antigens. 2001;58:24–29. doi: 10.1034/j.1399-0039.2001.580104.x. [DOI] [PubMed] [Google Scholar]
- Jagiello P, Aries P, Arning L, Wagenleiter SE, Csernok E, Hellmich B, Gross WL, Epplen JT. The PTPN22 620W allele is a risk factor for Wegener's granulomatosis. Arthritis Rheum. 2005;52:4039–4043. doi: 10.1002/art.21487. [DOI] [PubMed] [Google Scholar]
- Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KM, Smith AN, Di Genova G, Herr MH, Dahlman I, Payne F, Smyth D, Lowe C, Twells RC, Howlett S, Healy B, Nutland S, Rance HE, Everett V, Smink LJ, Lam AC, Cordell HJ, Walker NM, Bordin C, Hulme J, Motzo C, Cucca F, Hess JF, Metzker ML, Rogers J, Gregory S, Allahabadia A, Nithiyananthan R, Tuomilehto-Wolf E, Tuomilehto J, Bingley P, Gillespie KM, Undlien DE, Ronningen KS, Guja C, Ionescu-Tirgoviste C, Savage DA, Maxwell AP, Carson DJ, Patterson CC, Franklyn JA, Clayton DG, Peterson LB, Wicker LS, Todd JA, Gough SC. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- Criswell LA, Pfeiffer KA, Lum RF, Gonzales B, Novitzke J, Kern M, Moser KL, Begovich AB, Carlton VE, Li W, Lee AT, Ortmann W, Behrens TW, Gregersen PK. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet. 2005;76:561–571. doi: 10.1086/429096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Microsatellite marker panel, additional data.
HLA-DPB1 and microsatellite #1.0.3.7 genotyping information.