Abstract
Dynamic protein–protein interactions are involved in most physiological processes and, in particular, for the formation of multiprotein signaling complexes at transmembrane receptors, adapter proteins and effector molecules. Because the unregulated induction of signaling complexes has substantial clinical relevance, the investigation of these complexes is an active area of research. These studies strive to answer questions about the composition and function of multiprotein signaling complexes, along with the molecular mechanisms of their formation. In this review, the adapter protein, linker for activation of T cells (LAT), will be employed as a model to exemplify how signaling complexes are characterized using a range of techniques. The intensive investigation of LAT highlights how the systematic use of complementary techniques leads to an integrated understanding of the formation, composition and function of multiprotein signaling complexes that occur at receptors, adapter proteins and effector molecules.
Keywords: LAT, multiprotein complexes, signal transduction, T cells, T cell receptor
Reversible protein–protein interactions are a characteristic of most biochemical pathways. One process dependent on dynamic protein–protein interactions is the formation of multiprotein signaling complexes. These signaling complexes form at the cytoplasmic domain of transmembrane receptors, and at modular enzymes and nonenzymatic adapter proteins. Such complexes are vital for the activation and propagation of intracellular signals that regulate cellular function [1,2]. In fact, the uncontrolled activation of receptors and enzymes, which can lead to the inappropriate formation of signaling complexes, has been linked to many pathological conditions, including cancer, diabetes, and autoimmune and cardiovascular diseases. Formation of a signaling complex is mediated by inducible or constitutive interactions between discrete protein domains and specific motifs found on various signaling molecules [3-5]. Examples of these interaction domains and motifs include SH2 and PTB domains (which bind phosphorylated tyrosine residues) SH3 and WW domains (which constitutively associate with proline-rich domains) and PH domains (which interact with phosphorylated membrane lipids) [3-5]. Signaling proteins can contain multiple interaction domains and binding motifs, resulting in the formation of multiprotein signaling complexes that often have substantial specificity and a defined stoichiometry [6].
The study of multiprotein signaling complexes has raised several basic questions. What is the composition and stoichiometry of these signaling complexes? What is the molecular mechanism for the induction of these complexes? How does the formation of signaling complexes lead to the activation of downstream signaling pathways? Numerous techniques have been employed to address these questions. To truly obtain an integrated view of the cellular function of a signaling protein, it is optimal to use a panel of overlapping and complementary techniques, each having particular strengths and weaknesses. The purpose of this review is to show the benefits of a comprehensive examination of the formation of signaling complexes by multiple techniques. Our example is the characterization of the hematopoietic-specific adapter protein, linker for activation of T cells (LAT). The composition, formation and function of signaling complexes at LAT has been examined using a range of techniques, including the use of cellular and genetic structure/function studies, live cell imaging and the biophysical examination of purified LAT-interacting proteins (Table 1). These investigations of LAT will be summarized to illustrate that the use of multiple techniques can give a more complete characterization of a signaling molecule that nucleates multiprotein signaling complexes.
Table 1.
Summary of techniques used to analyze the induction of multiprotein complexes. Techniques used to analyze the formation, function and composition of multiprotein complexes are detailed. Also described is the information gained from these techniques and examples of the use of these techniques for the examination of linker for activation of T cells (LAT)-mediated multiprotein complexes. FRET, fluorescence resonance energy transfer.
| Technique | Information gained from the technique | LAT references |
|---|---|---|
| Sequence analysis | Identify potential interaction domains and motifs | [12,13] |
| Site directed mutagenesis | (1) Determine individual sites required for protein–protein interactions | [17,18,21- 23,26,27] |
| (2) Examine the formation and composition of in vivo protein complexes | ||
| (3) Investigate effects of complex formation on downstream signaling pathways | ||
| Mouse models with directed mutations | Characterize effects of multiprotein complex formation on cellular function | [33-41] |
| Confocal microscopy | (1) Visualize the co-localization of proteins to macromolecular structures in a cell | [42-46,48] |
| (2) Examine dynamic movement of signaling proteins and complexes | ||
| FRET microscopy | Quantify direct protein interactions in a cell | [51] |
| Electron microscopy | Obtain high-resolution images of protein localization and protein–protein interactions | [55] |
| Isothermal titration calorimetry | Measure thermodynamic constants, affinity and stoichiometry of a protein–protein interaction | [28] |
| Analytical ultracentrifugation | Characterize various multiprotein complexes in protein mixture | [28] |
| Yeast two-hybrid analysis | Determine potential binding partners | Not completed |
| Proteomic analysis | (1) Determine potential binding partners | Not completed |
| (2) Identify site-specific phosphorylation | Not completed |
Identification and cloning of LAT
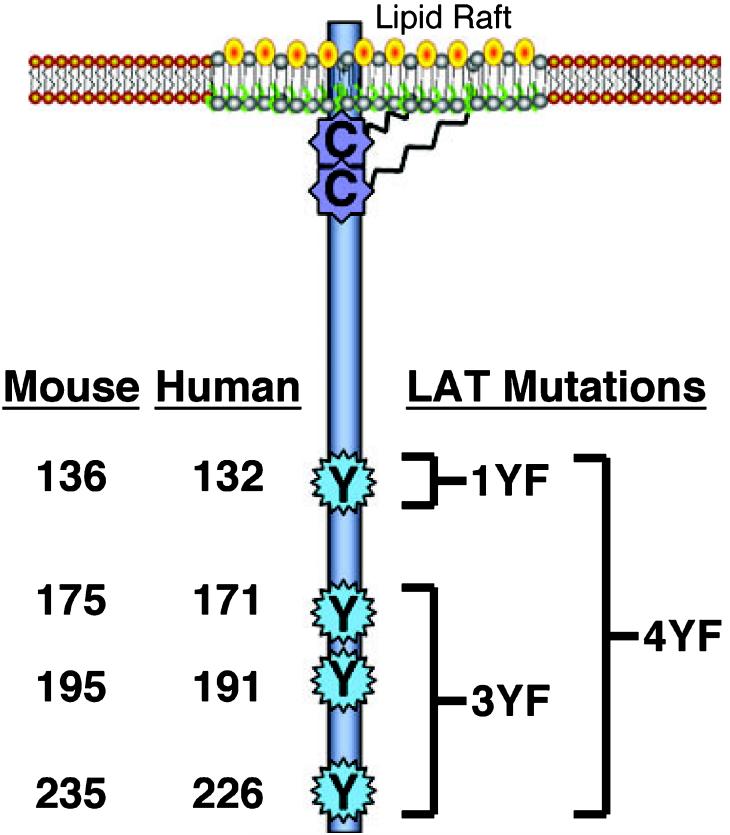
The first observation of the molecule that would later be named as LAT, was as a 36–38 kDa protein that was tyrosine phosphorylated upon T cell receptor (TCR) activation [7]. This molecule was dubbed pp36/38 and several groups went on to demonstrate that it interacted with the SH2 domains of PLC-γ1, Grb2 and the p85 subunit of phosphatidylinositol (PI) 3-kinase after TCR activation [8-11]. Although pp36/38 was first observed in 1990, it proved exceedingly difficult to isolate and identify. Not until 1998 was pp36/38 isolated in large-scale purifications from activated Jurkat T cells and thymocytes [12,13]. When cloned and sequenced, human LAT was found to be a 233 amino acid protein whose expression is restricted to hematopoietic cell lineages, including T cells, pro B cells, mast cells, natural killer cells, megakaryocytes and platelets [12-16]. The mouse and rat versions of LAT were also cloned and shown to comprise 242 and 241 amino acids, respectively, and to have 65–70% identity with human LAT [12,13]. Structurally, LAT contains a short predicted extracellular region of four amino acids, a single transmembrane-spanning region and long intracellular region with no apparent intrinsic enzymatic activity (Fig. 1) [12]. It is a member of the family of class III transmembrane proteins that lacks a signal sequence [12]. The intracellular domain of LAT contains nine conserved tyrosines, with the five most distal tyrosines – 127, 132, 171, 191 and 226 of the human sequence – rapidly phosphorylated upon TCR activation, creating potential sites for SH2 domain-mediated interactions (Fig. 1) [17]. The kinase(s) directly responsible for the phosphorylation of these sites is still controversial. Early studies suggested that ZAP-70, a kinase rapidly activated after TCR stimulation, is responsible for the in vivo phosphorylation of individual LAT tyrosines [12,18]. However, two other tyrosine kinases, Itk and Lck, activated upon TCR stimulation, have been shown to phosphorylate LAT peptides in vitro. Whether these kinases are involved in the in vivo phosphorylation of LAT is still unknown.
Fig. 1.
Structure and membrane localization of the linker for activation of T cells (LAT). LAT contains a short extracellular region, a single transmembrane domain and an intracellular region with no apparent enzymatic activity. Although the transmembrane domain is sufficient for membrane localization, the palmitoylation of cysteine residues near the plasma membrane anchors LAT in defined membrane domains called lipid rafts. The intracellular region of LAT contains multiple conserved tyrosines that are phosphorylated upon receptor activation. The last four tyrosines (the amino acid numbers for human and mouse LAT are shown here) are required for LAT function. Importantly, several mutant forms of LAT that have been used are defined here.
Structure/function studies
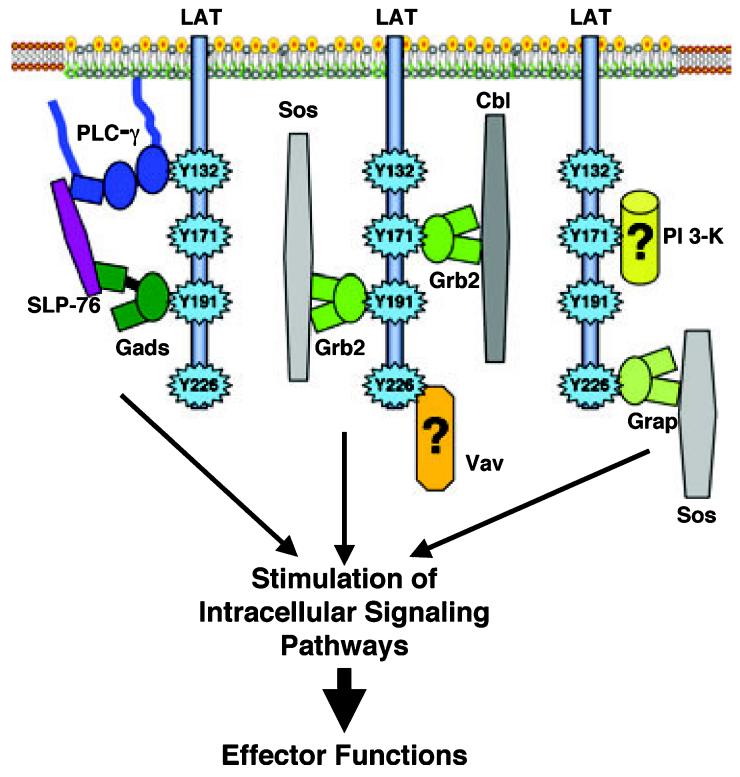
Initial analysis of the intracellular region of LAT, along with the early characterization of pp36/38, suggested that LAT functions as a classic adapter protein by facilitating the inducible formation of multiprotein signaling complexes. In order to test this hypothesis, structure/function investigations were carried out to determine which individual LAT tyrosines, when phosphorylated, were required for the direct and indirect binding of various signaling molecules. Early sequence analysis of motifs surrounding four distal LAT tyrosines provided insight into the potential binding partners for individual phosphorylated LAT tyrosines [12,13]. LAT tyrosines 171 (YVNV), 191 (YVNV) and 226 (YENL), when phosphorylated, are found in the sequence context of consensus-binding sites (pYXNX) for Grb2, a ubiquitously expressed SH2 and SH3 domain-containing adapter protein [19]. Similarly, LAT tyrosine 132 (YLVV), when phosphorylated, is within a consensus-binding site (pYLXV) for PLC-γ1, an SH2 and SH3 domain-containing signaling protein important for Ca2+ influx and protein kinase C activation [20]. These potential interactions were confirmed in several structure/function studies using the LAT-deficient Jurkat T-cell line, JCaM 2.5 [21]. In these studies, the JCaM 2.5 cell line was transfected with various mutant forms of LAT, and the association of LAT with individual signaling proteins was then assessed by immunoprecipitation. When examined in the mutant JCaM 2.5 cell lines, LAT tyrosines 171, 191 and 226 were shown to be vital for the interaction of LAT with Grb2 and a related molecule, Grap (Fig. 2) [18,22,23]. The interaction of phosphorylated LAT with these molecules was mediated by the central SH2 domains of Grb2 and Grap. The SH2 domain of both of these proteins is flanked by two SH3 domains that constitutively bind several ligands, including Sos, a guanine nucleotide exchange factor for Ras, and two E3 ubiquitin ligases, c-Cbl and Cbl-b [15]. In fact, LAT tyrosines 171, 191 and 226 were shown to be crucial for the indirect binding of LAT to Sos and Cbl-b (Fig. 2) [18]. Interestingly, two Grb2-binding sites, in any combination, were needed for the stable association of LAT with Grb2 [17], indicating that the binding of LAT to multiple Grb2 proteins may be required for the interaction between these molecules. In addition to Grb2 and Grap, T cells express a third Grb2-like molecule, called Gads, that contains a central SH2 domain flanked by two SH3 domains [15]. As determined by immunoprecipitation, Gads principally binds phosphorylated LAT tyrosine 191 and also shows some binding to phosphorylated LAT tyrosine 171, but fails to interact with phosphorylated LAT tyrosine 226 (Fig. 2) [17,22], in subtle contrast to what was found for Grb2 and Grap. In confirmation of this finding, LAT tyrosines 171 and 191 are vital for the binding of LAT with SLP-76, a high affinity SH3 domain ligand for Gads (Fig. 2) [22].
Fig. 2.
Linker for activation of T cells (LAT)-mediated multiprotein signaling complexes. The phosphorylation of LAT on the distal four tyrosines results in the formation of several multiprotein signaling complexes with distinct composition and stoichiometry. These include complexes that contain either Grb2 or Grap and a reported interaction between PLC-γ1 and the Gads–SLP-76 complex. Vav and the p85 subunit of phosphatidylinositol (PI) 3-kinase may also interact with LAT, but whether these proteins directly or indirectly associate with LAT is still controversial. The result of the formation of LAT-mediated complexes is the activation of signaling pathways and the induction of effector functions.
These structure/function studies have also examined the binding of LAT to other effector molecules important for intracellular signaling. Phosphorylated LAT tyrosine 132 was demonstrated to be the principal direct binding site for PLC-γ1, an interaction mediated by the N-terminal SH2 domain of PLC-γ1 (Fig. 2) [18,22,24-26]. Interestingly, along with its direct binding to LAT tyrosine 132, PLC-γ1 also requires the presence of two or more of LAT tyrosines 171, 191 and 226 for a stable in vivo interaction with LAT [17,22]. This suggests that PLC-γ1 requires direct associations with both LAT tyrosine 132 and other proteins simultaneously bound to LAT tyrosines 171, 191 and 226 for a stable interaction with LAT. This hypothesis was seemingly confirmed when PLC-γ1 was reported to interact, via an SH3 domain-mediated interaction, with the Gads–SLP-76 complex when these proteins are all bound to LAT (Fig. 2) [27]. However, recent reports have shown that the interaction between PLC-γ1 and SLP-76 has a surprisingly weak affinity for an SH3 domain-mediated interaction [28] and that the interaction of PLC-γ1 and SLP-76 is not required for the cellular function of PLC-γ1 [29]. Although PLC-γ1 probably interacts with the Gads–SLP-76 complex when these proteins are bound to LAT, it is still an open question as to whether this or other interactions are required for the stable binding of LAT to PLC-γ1.
Along with PLC-γ1, the direct binding of LAT to Vav and to the p85 subunit of PI 3-kinase has also been suggested. When examined using far western blotting, Vav appeared to directly associate with LAT tyrosines 171, 191 and 226 (Fig. 2) [18]. Similarly, the p85 subunit of PI 3-kinase was observed to bind directly to LAT, primarily via LAT tyrosine 171 (Fig. 2) [18]. These findings were surprising in that LAT does not contain the apparent consensus-binding sequences for either Vav (pYMXX) or the p85 subunit of PI 3-kinase (pYMXM) [12,19,20]. Further experiments are needed to determine whether Vav and the p85 subunit of PI 3-kinase directly or indirectly associate with LAT.
These structure/function studies have also characterized how the phosphorylation of specific LAT tyrosines links to the activation of intracellular signaling. In LAT-deficient JCaM 2.5 cells, proximal kinases, such as ZAP-70, Lck and Fyn, are still active, but there is little signaling downstream of LAT [21]. This results in severe defects in TCR-induced Ca2+ influx and in the activation of mitogen-actived protein (MAP) kinases and the transcription factors, AP-1 and NF-AT [21]. When these activation events were investigated in JCaM 2.5 cells reconstituted with wild-type and mutated LAT, LAT tyrosine 132 alone was not sufficient for Ca2+ influx but the presence of tyrosines 132, 171 and 191 was required for relatively normal Ca2+ influx [17,26]. As stated above, this result may reflect the requirement for both the direct binding to LAT tyrosine 132, and the indirect interaction via the Gads–SLP-76 complex, for the stable interaction between PLC-γ1 and LAT [17,22,27]. Interestingly, the presence of only LAT tyrosines 171, 191 and 226 was not sufficient for TCR-mediated MAP kinase activation [17]. This indicates that recruitment of the Grb2–Sos complex to the plasma membrane, via its association with LAT at these sites, is not sufficient for MAP kinase activation, in contrast to other previously described receptor systems [30]. Instead, the presence of LAT tyrosines 132, 171 and 191 on a single LAT protein was required for full activation of MAP kinases [17,22,26]. These LAT tyrosines are needed for the recruitment of PLC-γ1 to LAT, suggesting that the activation of PLC-γ1 is crucial for the TCR-mediated stimulation of MAP kinase activity, an effect apparently mediated by RasGRP, a Ca2+ and diacylglycerol-sensitive guanine nucleotide exchange factor [31,32]. In confirmation of the role of these LAT tyrosines on Ca2+ influx and the activation of MAP kinases, either mutation of LAT tyrosine 132 or LAT tyrosines 171, 191 and 226 resulted in the inhibition of the Ca2+ and MAP kinase-sensitive transcription factors, NF-AT and AP-1 [18,22,26]. Together, this indicates that the formation of multiprotein signaling complexes at LAT is crucial for linking LAT phosphorylation to the stimulation of intracellular signaling pathways. Yet, exactly which complexes are required for the activation of specific pathways is still not completely understood.
Together, these structure/function studies have given detailed information on the binding of specific SH2 domain-containing signaling proteins to individual phosphorylated LAT tyrosines. They have also provided substantial information on the composition of the multiprotein signaling complexes that occur at LAT and how these complexes facilitate the direct and indirect association of signaling proteins to LAT. These studies have also begun to connect LAT-mediated complexes to the activation of specific intracellular signaling pathways. The characterization of LAT by these structure/function studies highlights how these experiments are an important and vital first step for the investigation of multiprotein signaling complexes.
Functional studies in mice
The structure/function studies of LAT gave initial insights into the link between the signaling complexes formed at individual LAT tyrosines and the activation of intracellular signaling pathways. However, these studies were performed using Jurkat T cells, which although are an excellent model system for examining early TCR-mediated signaling, cannot be used to address the effects of LAT mutations on T-cell differentiation and all aspects of normal T-cell function. Therefore, to elucidate the role that LAT-mediated signaling complexes play in the differentiation and function of various immune cells, LAT-deficient mice, and mice with the wild-type LAT sequence replaced with mutated versions of LAT, were produced. Mutant mice with the last four tyrosines of LAT mutated to phenylalanine (4YF; Fig. 1) were phenotypically indistinguishable from LAT-deficient mice, with severely reduced numbers of all mature T-cell subsets caused by an early block in T-cell differentiation [33,34]. This suggested that the formation of signaling complexes induced by LAT phosphorylation are absolutely necessary for normal T-cell differentiation. Interestingly, mice with a tyrosine to phenylalanine mutation of mouse LAT tyrosine 136 (1YF; Fig. 1) (i.e. the tyro-sine homologous to human LAT tyrosine 132), developed a polyclonal lymphoproliferative disease by 8 weeks of age and later showed hallmarks of autoimmune disease [35,36]. T cells from these mice had decreased PLC-γ1 activation, resulting in severely reduced levels of TCR-induced Ca2+ influx and NF-AT activation compared with wild-type littermates [35]. However, these mice had relatively normal levels of MAP kinase activation compared with wild-type littermates [35], in contrast to the LAT 1YF reconstituted JCaM 2.5 cells [22,26]. Similarly to LAT 1YF mice, mice containing tyrosine to phenylalanine mutations in the distal three tyrosines of LAT (3YF; Fig. 1), which leads to a loss of Grb2, Gads and Grap binding, had abnormal expansion of a specific subset of T cells, leading to a lymphoproliferative disease [37]. The function of LAT-mediated signaling complexes in the activation and differentiation of B cells and mast cells has also been examined in cell lines derived from LAT-deficient mice. In B-cell lines retrovirally reconstituted with various forms of LAT, the last four intracellular tyrosines, especially LAT tyrosine 136, appeared important for the ability of LAT to facilitate early B-cell differentiation and suppress the mitogenic potential of these B-cell lines [38]. In retrovirally reconstituted bone marrow-derived mast cells, cells with LAT 1YF or 4YF mutations had severe defects in FcεR1 receptor-mediated signaling, degranulation and cytokine release, similar to those seen in LAT-deficient mice [39-41]. Together, these data suggested that the ability of phosphorylated LAT to form signaling complexes was crucial for the differentiation and function of multiple immune cell types.
The studies of immune cells derived from various mutant mice have shown that the formation of LAT-mediated signaling complexes play a complex role in the differentiation and homeostasis of T-cell populations, the maturation of B cells and the activation of mast cells by the FcεR1 receptor. These investigations have given unique insight into the functional consequences of complex formation at LAT that could not be observed using established cell lines. In total, these studies have highlighted that subtle mutations in LAT, leading to alterations in the formation of multiprotein signaling complexes, have profound deleterious effects on the differentiation and function of T cells, B cells and mast cells.
Imaging studies
Cellular imaging is a highly informative method that is becoming widely used, not only to qualitatively examine the cellular localization of individual signaling proteins, but also to quantitatively investigate protein–protein interactions. The recruitment and localization of LAT and LAT-binding proteins to the sites of receptor activation has been extensively characterized by both confocal and electron microscopy. Using confocal microscopy, several groups have shown that LAT is quickly recruited to the contact site between a Jurkat T cell and a staphylococcal enterotoxin E (SEE)-labeled Raji B cell [42] or an anti-TCR-coated bead [43-45]. However, owing to the awkward geometry of these interactions and poor time synchronization, these studies were not able to provide high-resolution, dynamic images of LAT localization upon TCR activation. In order to obtain this information, a method was developed to image T-cell activation on a planar surface [46,47]. In this method, glass coverslips were coated with TCR stimulatory antibodies, and Jurkat T cells expressing labeled signaling molecules were activated by dropping these cells onto the stimulatory coverslips [46,47]. Using this method, TCR components and the protein tyrosine kinase, ZAP-70, were observed to localize to punctate clusters that formed immediately upon contact and were coincident with sites of tight interactions between the Jurkat T cell and the coverslip [48]. These punctate clusters are similar to clusters of ZAP-70 seen within 2 min of the interaction between T cells and antigen-presenting cells, suggesting that they are the physiologically relevant proto-synapses that are induced early after TCR stimulation [49,50]. Interestingly, the sites of TCR and ZAP-70 clustering co-localized with punctate clusters of LAT and multiple known LAT-interacting proteins, such as Grb2, Gads, c-Cbl, SLP-76, WASp and Nck [48,51]. The recruitment of LAT to the sites of TCR and ZAP-70 clustering occurred within 30 s of the T cell–coverslip contact, and these clusters were reported dissipate within 150 s of T-cell activation [48]. Thus, these clusters contain dynamic multiprotein signaling complexes, many of which are mediated by phosphorylated LAT.
Although informative, the qualitative methods used to detect proteins in these imaging studies can only determine whether two proteins are co-localized to the same macromolecular structure, but cannot show whether these proteins are interacting directly, as would occur in signaling complexes. However, a recent study, using fluorescence resonance energy transfer (FRET) microscopy, has examined whether LAT directly or indirectly associates with various signaling molecules [51]. FRET is a biophysical method that measures the transfer of energy from an excited donor fluorophore directly to an acceptor fluorophore, leading to an increased fluorescence emission of the acceptor and a quenching of the emission fluorescence of the donor [52-54]. For FRET to occur, the donor and acceptor must have a sufficient spectral overlap, a favorable orientation and a separation of 1–10 nm [52]. Because of the ability of FRET to measure only close interactions, it is a valuable approach for assessment and measurement of protein–protein interactions in living cells [52-54]. Using this technique, measurable but low FRET was detected between LAT and both SLP-76 and Nck, suggesting, as shown previously, that these molecules closely, but indirectly, associate with LAT [51]. In contrast, SLP-76 demonstrated substantial FRET with Nck, indicating that SLP-76 binds directly to this protein [51]. In the future, FRET analysis will prove to be a powerful tool for quantifying the interactions of intracellular signaling molecules that have been suggested by numerous biochemical studies to occur upon TCR activation.
High-resolution electron microscopy has been used to examine the localization of LAT in mast cells both before and after FcεRI activation. In these studies, the membrane localization of LAT and other signaling proteins in stimulated and unstimulated mast cells was imaged by electron microscopy [55]. In resting mast cells, LAT was localized to small membrane clusters that usually contained fewer than 10 LAT molecules [55]. Upon FcεRI activation, LAT coalesced into larger clusters, often containing 100–150 LAT molecules, that did not appear to co-localize with the FcεRI receptor [55]. These large LAT clusters did, however, co-localize with PLC-γ1 and partially co-localized with the p85 subunit of PI-3 kinase [55]. This study provided high-resolution images of LAT localization upon receptor activation, confirming the clustering of LAT with other intracellular signaling molecules that was observed by confocal microscopy.
The use of cellular imaging techniques has proven to be a highly informative method to examine the localization and function of LAT in activated T cells and mast cells. In particular, these studies have revealed that upon both TCR and FcεRI activation, proximal signaling molecules, including ZAP-70, LAT and SLP-76, are recruited to punctate clusters similar to those seen early after T cell–antigen-presenting cell contacts. The presence of these clusters is an important observation for understanding LAT function and could only be easily observed and characterized using microscopic techniques. As observed by confocal and FRET microscopy, the punctate signaling clusters multiple known proteins that interact with LAT, suggesting that these clusters are partially composed of LAT-mediated multiprotein signaling complexes. Yet, it is still unknown the exact role that LAT-mediated signaling complexes play in the formation and regulation of these punctate clusters. In the future, high-resolution, quantitative methods, such as electron microscopy and FRET, will be combined with standard confocal microscopy to provide greater insight into the formation, composition and function of the multiprotein complexes that occur at LAT.
Biophysical studies
Recently, state-of-the-art biophysical methods have been used to examine the in vitro formation of LAT complexes. In these studies, the association of purified LAT-binding proteins, including Grb2, Gads, SLP-76 and PLC-γ1, with each other and with synthesized LAT peptides, has been examined using multiple complementary biophysical methods. These approaches offer the opportunity to probe the basic mechanism of protein–protein interactions in great detail. First, the affinity of the LAT-binding proteins, PLC-γ1, Grb2 and Gads, for synthesized phosphopeptides that contain phosphorylated LAT tyrosines 132, 171, 191 and 226, was assessed using isothermal titration calorimetry [28], a method that allows for the simultaneous measurement of the affinity, binding stoichiometry and thermodynamic constants [56-59]. This study was performed to characterize the properties that drive the binding of individual signaling proteins to specific LAT tyrosines. To this end, the preferential in vivo binding of Grb2 to phosphorylated LAT tyrosines 171, 191 and 226 appeared to be driven primarily by substantial differences in affinity for these sites compared with phosphorylated LAT tyrosine 132 [28]. In contrast, the specific in vivo interaction of Gads with phosphorylated LAT tyrosines 171 and 191 appeared to be driven by a combination of affinity preferences (the explanation for the lack of in vivo binding of Gads to phosphorylated LAT tyrosine 132) and the formation of multiprotein signaling complexes (the reason for the lack of detectable in vivo association of Gads with phosphorylated LAT tyrosine 226) [28]. Finally, the in vivo association of PLC-γ1 with LAT tyrosine 132 was principally driven by the formation of multiprotein complexes and not by substantially increased affinity of PLC-γ1 for LAT tyrosine 132 compared with other LAT tyrosines [28]. These experiments have shown that forces which drive the binding specificity of SH2 domain-containing proteins for individual LAT tyrosines are complicated, with the interaction of each signaling protein driven by a different combination of affinity preferences and complex formation.
Along with examining binding specificity, the binding between PLC-γ1 and the Gads–SLP-76 complex, which has been reported to occur at LAT [27], has also been examined using multiple biophysical techniques. As measured by isothermal titration calorimetry and fluorescence polarization, the affinity of Gads for both the short 10 amino acid core-binding motif and the complete proline-rich region of SLP-76 was extremely strong and, in fact, was one of the strongest reported SH3 domain-mediated interactions [28,60]. In contrast, the affinity of PLC-γ1 for SLP-76 was extremely weak and probably does not occur unaided in a cellular context [28]. Interestingly, it appeared that the proline-rich region of SLP-76 underwent a substantial change in secondary structure upon binding both Gads and PLC-γ1 [28,61]. This suggested that the prestructuring of SLP-76 by a high-affinity interaction with Gads could increase the affinity of SLP-76 for PLC-γ1. This possibility was examined using sedimentation velocity analytical ultracentrifugation (SV-AUC). SV-AUC follows the sedimentation of proteins in solution under a centrifugal field, allowing for the characterization of the thermodynamic and hydrodynamic properties of the proteins [62,63]. It is an excellent method for characterizing the multiprotein complexes formed in a mixture of proteins. As assessed by SV-AUC, PLC-γ1 appeared to have substantially stronger binding to the Gads–SLP-76 complex than to SLP-76 alone [28]. Together, this implies that the interaction of PLC-γ1 with the Gads–SLP-76 complex, although occurring at a low level in unstimulated cells, is probably stabilized when all the proteins are bound to LAT (Fig. 2).
The biophysical examination of the LAT complex has provided a number of interesting and novel observations. These studies are uniquely able to define the properties that drive the substantial binding specificity of SH2 domain-containing proteins to individual LAT tyrosines. They are also able to quantitatively examine the multiprotein complexes that form at LAT, providing insights into the formation and function of these complexes that could not be observed using other methods. These experimental techniques hold great promise for quantitatively characterizing the composition, stoichiometry and specificity of the multiprotein complexes occurring at a single LAT molecule.
Conclusions
Our understanding of LAT has progressed from the identification of a 36–38 kDa phosphorylated protein that binds several intracellular signaling proteins to the realization that LAT is a nucleating site for multiprotein signaling complexes which are vital for the differentiation and function of T cells, B cells and mast cells. This knowledge comes from using many different, yet complementary, techniques that have led to an integrated understanding of the formation, composition and function of LAT-mediated signaling complexes (Table 1). But even with all that is known about these complexes, there is still more that needs to be examined. The composition of the multiprotein complexes that occur at LAT, along with the individual LAT tyrosines that mediate these interactions, must be examined using a combination of proteomic, yeast two-hybrid and structure/function studies. The mechanism of how subtle LAT mutations, which lead to the disruption of specific signaling complexes, can result in alterations in T-cell homeostasis and the induction of autoimmune disease, needs to be addressed. The dynamic formation of LAT-induced signaling complexes needs to be further examined using confocal and electron microscopy, and the individual protein– protein interactions that occur in these complexes need to be quantitatively measured using FRET microscopy. Finally, the affinity, binding specificity and stoichiometry of the multiprotein signaling complexes that occur at LAT need to be quantitatively characterized using biophysical methodology to provide a better understanding of the molecular mechanisms of complex formation at a single LAT molecule. In total, the investigation of the multiprotein signaling complexes that form at LAT is an excellent example of how to approach the study of a signaling protein with adapter function. The systematic use of multiple complementary techniques, each providing a different viewpoint, is the optimal way to gain a complete and detailed understanding of the physiological function of signaling proteins that nucleate multiprotein complexes.
Acknowledgements
We thank Dr Connie Sommers for helpful discussions. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Abbreviations
- FRET
fluorescence resonance energy transfer
- LAT
linker for activation of T cells
- MAP
kinase, mitogen-actived protein kinase
- PI
phosphatidylinositol
- TCR
T cell receptor
References
- 1.Vondriska TM, Pass JM, Ping P. Scaffold proteins and assembly of multiprotein signaling complexes. J Mol Cell Cardiol. 2004;37:391–397. doi: 10.1016/j.yjmcc.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 2.Burack WR, Cheng AM, Shaw AS. Scaffolds, adaptors and linkers of TCR signaling: theory and practice. Curr Opin Immunol. 2002;14:312–316. doi: 10.1016/s0952-7915(02)00347-3. [DOI] [PubMed] [Google Scholar]
- 3.Mayer BJ. Protein–protein interactions in signaling cascades. Mol Biotechnol. 1999;13:201–213. doi: 10.1385/MB:13:3:201. [DOI] [PubMed] [Google Scholar]
- 4.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 5.Sudol M. From Src homology domains to other signaling modules: proposal of the ‘protein recognition code’. Oncogene. 1998;17:1469–1474. doi: 10.1038/sj.onc.1202182. [DOI] [PubMed] [Google Scholar]
- 6.Tomlinson MG, Lin J, Weiss A. Lymphocytes with a complex: adapter proteins in antigen receptor signaling. Immunol Today. 2000;21:584–591. doi: 10.1016/s0167-5699(00)01716-3. [DOI] [PubMed] [Google Scholar]
- 7.June CH, Fletcher MC, Ledbetter JA, Samelson LE. Increases in tyrosine phosphorylation are detectable before phospholipase C activation after T cell receptor stimulation. J Immunol. 1990;144:1591–1599. [PubMed] [Google Scholar]
- 8.Gilliland LK, Schieven GL, Norris NA, Kanner SB, Aruffo A, Ledbetter JA. Lymphocyte lineage-restricted tyrosine-phosphorylated proteins that bind PLC gamma 1 SH2 domains. J Biol Chem. 1992;267:13610–13616. [PubMed] [Google Scholar]
- 9.Buday L, Egan SE, Rodriguez Viciana P, Cantrell DA, Downward J. A complex of Grb2 adaptor protein, Sos exchange factor, and a 36-kDa membrane-bound tyrosine phosphoprotein is implicated in ras activation in T cells. J Biol Chem. 1994;269:9019–9023. [PubMed] [Google Scholar]
- 10.Sieh M, Bolen JB, Weiss A. CD45 specifically modulates binding of Lck to a phosphopeptide encompassing the negative regulatory tyrosine of Lck. Embo J. 1993;12:315–321. doi: 10.1002/j.1460-2075.1993.tb05659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukazawa T, Reedquist KA, Panchamoorthy G, Soltoff S, Trub T, Druker B, Cantley L, Shoelson SE, Band H. T cell activation-dependent association between the p85 subunit of the phosphatidylinositol 3-kinase and Grb2/phospholipase C-gamma 1-binding phosphotyrosyl protein pp36/38. J Biol Chem. 1995;270:20177–20182. doi: 10.1074/jbc.270.34.20177. [DOI] [PubMed] [Google Scholar]
- 12.Zhang W, Sloan-Lancaster J, Kitchen J, Trible RP, Samelson LE. LAT: the ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell. 1998;92:83–92. doi: 10.1016/s0092-8674(00)80901-0. [DOI] [PubMed] [Google Scholar]
- 13.Weber JR, Orstavik S, Torgersen KM, Danbolt NC, Berg SF, Ryan JC, Tasken K, Imboden JB, Vaage JT. Molecular cloning of the cDNA encoding pp36, a tyrosine-phosphorylated adaptor protein selectively expressed by T cells and natural killer cells. J Exp Med. 1998;187:1157–1161. doi: 10.1084/jem.187.7.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su YW, Jumaa H. LAT links the pre-BCR to calcium signaling. Immunity. 2003;19:295–305. doi: 10.1016/s1074-7613(03)00202-4. [DOI] [PubMed] [Google Scholar]
- 15.Samelson LE. Signal transduction mediated by the T cell antigen receptor: the role of adapter proteins. Annu Rev Immunol. 2002;20:371–394. doi: 10.1146/annurev.immunol.20.092601.111357. [DOI] [PubMed] [Google Scholar]
- 16.Wange RL. LAT, the linker for activation of T cells: a bridge between T cell-specific and general signaling pathways. Sci STKE RE1. 2000 doi: 10.1126/stke.2000.63.re1. [DOI] [PubMed] [Google Scholar]
- 17.Zhu M, Janssen E, Zhang W. Minimal requirement of tyrosine residues on linker for activation of T cells in T cell activation and thymocyte development. J Immunol. 2003;170:325–333. doi: 10.4049/jimmunol.170.1.325. [DOI] [PubMed] [Google Scholar]
- 18.Paz PE, Wang S, Clarke H, Lu X, Stokoe D, Abo A. Mapping the ZAP-70 phosphorylation sites on LAT (linker for activation of T cells) required for recruitment and activation of signalling proteins in T cells. Biochem J. 2001;356:461–471. doi: 10.1042/0264-6021:3560461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Songyang Z, Shoelson SE, McGlade J, Olivier P, Pawson T, Bustelo XR, Barbacid M, Sabe H, Hanafusa H, Yi T, et al. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol Cell Biol. 1994;14:2777–2785. doi: 10.1128/mcb.14.4.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ, et al. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 21.Finco TS, Kadlecek T, Zhang W, Samelson LE, Weiss A. LAT is required for TCR-mediated activation of PLCgamma1 and the Ras pathway. Immunity. 1998;9:617–626. doi: 10.1016/s1074-7613(00)80659-7. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W, Trible RP, Zhu M, Liu SK, McGlade CJ, Samelson LE. Association of Grb2, Gads, and phospholipase C-gamma 1 with phosphorylated LAT tyrosine residues. Effect of LAT tyrosine mutations on T cell antigen receptor-mediated signaling. J Biol Chem. 2000;275:23355–23361. doi: 10.1074/jbc.M000404200. [DOI] [PubMed] [Google Scholar]
- 23.Perez-Villar JJ, Whitney GS, Sitnick MT, Dunn RJ, Venkatesan S, O'Day K, Schieven GL, Lin TA, Kanner SB. Phosphorylation of the linker for activation of T-cells by Itk promotes recruitment of Vav. Biochemistry. 2002;41:10732–10740. doi: 10.1021/bi025554o. [DOI] [PubMed] [Google Scholar]
- 24.Irvin BJ, Williams BL, Nilson AE, Maynor HO, Abraham RT. Pleiotropic contributions of phospholipase C-gamma1 (PLC-gamma1) to T-cell antigen receptor-mediated signaling: reconstitution studies of a PLC-gamma1-deficient Jurkat T-cell line. Mol Cell Biol. 2000;20:9149–9161. doi: 10.1128/mcb.20.24.9149-9161.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stoica B, DeBell KE, Graham L, Rellahan BL, Alava MA, Laborda J, Bonvini E. The amino-terminal Src homology 2 domain of phospholipase C gamma 1 is essential for TCR-induced tyrosine phosphorylation of phospholipase C gamma 1. J Immunol. 1998;160:1059–1066. [PubMed] [Google Scholar]
- 26.Lin J, Weiss A. Identification of the minimal tyrosine residues required for Linker for Activation of T cell function. J Biol Chem. 2001;276:29588–29595. doi: 10.1074/jbc.M102221200. [DOI] [PubMed] [Google Scholar]
- 27.Yablonski D, Kadlecek T, Weiss A. Identification of a phospholipase C-gamma1 (PLC-gamma1) SH3 domain-binding site in SLP-76 required for T-cell receptor-mediated activation of PLC-gamma1 and NFAT. Mol Cell Biol. 2001;21:4208–4218. doi: 10.1128/MCB.21.13.4208-4218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houtman JC, Higashimoto Y, Dimasi N, Cho S, Yamaguchi H, Bowden B, Regan C, Malchiodi EL, Mariuzza R, Schuck P, et al. Binding specificity of multiprotein signaling complexes is determined by both cooperative interactions and affinity preferences. Biochemistry. 2004;43:4170–4178. doi: 10.1021/bi0357311. [DOI] [PubMed] [Google Scholar]
- 29.Gonen R, Beach D, Ainey C, Yablonski D. T cell receptor-induced activation of phospholipase C-gamma1 depends on a sequence-independent function of the P-I region of SLP-76. J Biol Chem. 2005;280:8364–8370. doi: 10.1074/jbc.M409437200. [DOI] [PubMed] [Google Scholar]
- 30.Tari AM, Lopez-Berestein G. GRB2: a pivotal protein in signal transduction. Semin Oncol. 2001;28:142–147. doi: 10.1016/s0093-7754(01)90291-x. [DOI] [PubMed] [Google Scholar]
- 31.Dower NA, Stang SL, Bottorff DA, Ebinu JO, Dickie P, Ostergaard HL, Stone JC. RasGRP is essential for mouse thymocyte differentiation and TCR signaling. Nat Immunol. 2000;1:317–321. doi: 10.1038/79766. [DOI] [PubMed] [Google Scholar]
- 32.Ebinu JO, Stang SL, Teixeira C, Bottorff DA, Hooton J, Blumberg PM, Barry M, Bleakley RC, Ostergaard HL, Stone JC. RasGRP links T-cell receptor signaling to Ras. Blood. 2000;95:3199–3203. [PubMed] [Google Scholar]
- 33.Sommers CL, Menon RK, Grinberg A, Zhang W, Samelson LE, Love PE. Knock-in mutation of the distal four tyrosines of Linker for Activation of T cells blocks murine T cell development. J Exp Med. 2001;194:135–142. doi: 10.1084/jem.194.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W, Sommers CL, Burshtyn DN, Stebbins CC, DeJarnette JB, Trible RP, Grinberg A, Tsay HC, Jacobs HM, Kessler CM, et al. Essential role of LAT in T cell development. Immunity. 1999;10:323–332. doi: 10.1016/s1074-7613(00)80032-1. [DOI] [PubMed] [Google Scholar]
- 35.Sommers CL, Park CS, Lee J, Feng C, Fuller CL, Grinberg A, Hildebrand JA, Lacana E, Menon RK, Shores EW, et al. A LAT mutation that inhibits T cell development yet induces lymphoproliferation. Science. 2002;296:2040–2043. doi: 10.1126/science.1069066. [DOI] [PubMed] [Google Scholar]
- 36.Aguado E, Richelme S, Nunez-Cruz S, Miazek A, Mura AM, Richelme M, Guo XJ, Sainty D, He HT, Malissen B, et al. Induction of T helper type 2 immunity by a point mutation in the LAT adaptor. Science. 2002;296:2036–2040. doi: 10.1126/science.1069057. [DOI] [PubMed] [Google Scholar]
- 37.Nunez-Cruz S, Aguado E, Richelme S, Chetaille B, Mura AM, Richelme M, Pouyet L, Jouvin-Marche E, Xerri L, Malissen B, et al. LAT regulates gamma/delta T cell homeostasis and differentiation. Nat Immunol. 2003;4:999–1008. doi: 10.1038/ni977. [DOI] [PubMed] [Google Scholar]
- 38.Su YW, Herzog S, Lotz M, Feldhahn N, Muschen M, Jumaa H. The molecular requirements for LAT-mediated differentiation and the role of LAT in limiting pre-B cell expansion. Eur J Immunol. 2004;34:3614–3622. doi: 10.1002/eji.200425445. [DOI] [PubMed] [Google Scholar]
- 39.Saitoh S, Arudchandran R, Manetz TS, Zhang W, Sommers CL, Love PE, Rivera J, Samelson LE. LAT is essential for Fc (epsilon) RI-mediated mast cell activation. Immunity. 2000;12:525–535. doi: 10.1016/s1074-7613(00)80204-6. [DOI] [PubMed] [Google Scholar]
- 40.Saitoh S, Odom S, Gomez G, Sommers CL, Young HA, Rivera J, Samelson LE. The four distal tyrosines are required for LAT-dependent signaling in FcepsilonRI-mediated mast cell activation. J Exp Med. 2003;198:831–843. doi: 10.1084/jem.20030574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malbec O, Malissen M, Isnardi I, Lesourne R, Mura AM, Fridman WH, Malissen B, Daeron M. Linker for Activation of T cells integrates positive and negative signaling in mast cells. J Immunol. 2004;173:5086–5094. doi: 10.4049/jimmunol.173.8.5086. [DOI] [PubMed] [Google Scholar]
- 42.Bonello G, Blanchard N, Montoya MC, Aguado E, Langlet C, He HT, Nunez-Cruz S, Malissen M, Sanchez-Madrid F, Olive D, et al. Dynamic recruitment of the adaptor protein LAT: LAT exists in two distinct intracellular pools and controls its own recruitment. J Cell Sci. 2004;117:1009–1016. doi: 10.1242/jcs.00968. [DOI] [PubMed] [Google Scholar]
- 43.Hartgroves LC, Lin J, Langen H, Zech T, Weiss A, Harder T. Synergistic assembly of Linker for Activation of T cells signaling protein complexes in T cell plasma membrane domains. J Biol Chem. 2003;278:20389–20394. doi: 10.1074/jbc.M301212200. [DOI] [PubMed] [Google Scholar]
- 44.Harder T, Kuhn M. Selective accumulation of raft-associated membrane protein LAT in T cell receptor signaling assemblies. J Cell Biol. 2000;151:199–208. doi: 10.1083/jcb.151.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanimura N, Nagafuku M, Minaki Y, Umeda Y, Hayashi F, Sakakura J, Kato A, Liddicoat DR, Ogata M, Hamaoka T, et al. Dynamic changes in the mobility of LAT in aggregated lipid rafts upon T cell activation. J Cell Biol. 2003;160:125–135. doi: 10.1083/jcb.200207096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bunnell SC, Kapoor V, Trible RP, Zhang W, Samelson LE. Dynamic actin polymerization drives T cell receptor-induced spreading: a role for the signal transduction adaptor LAT. Immunity. 2001;14:315–329. doi: 10.1016/s1074-7613(01)00112-1. [DOI] [PubMed] [Google Scholar]
- 47.Bunnell SC, Barr VA, Fuller CL, Samelson LE. High-resolution multicolor imaging of dynamic signaling complexes in T cells stimulated by planar substrates. Sci STKE PL8. 2003 doi: 10.1126/stke.2003.177.pl8. [DOI] [PubMed] [Google Scholar]
- 48.Bunnell SC, Hong DI, Kardon JR, Yamazaki T, McGlade CJ, Barr VA, Samelson LE. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J Cell Biol. 2002;158:1263–1275. doi: 10.1083/jcb.200203043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freiberg BA, Kupfer H, Maslanik W, Delli J, Kappler J, Zaller DM, Kupfer A. Staging and resetting T cell activation in SMACs. Nat Immunol. 2002;3:911–917. doi: 10.1038/ni836. [DOI] [PubMed] [Google Scholar]
- 50.Lee KH, Dinner AR, Tu C, Campi G, Raychaudhuri S, Varma R, Sims TN, Burack WR, Wu H, Wang J, et al. The immunological synapse balances T cell receptor signaling and degradation. Science. 2003;302:1218–1222. doi: 10.1126/science.1086507. [DOI] [PubMed] [Google Scholar]
- 51.Barda-Saad M, Braiman A, Titerence R, Bunnell SC, Barr VA, Samelson LE. Dynamic molecular interactions linking the T cell antigen receptor to the actin cytoskeleton. Nat Immunol. 2005;6:80–89. doi: 10.1038/ni1143. [DOI] [PubMed] [Google Scholar]
- 52.Wallrabe H, Periasamy A. Imaging protein molecules using FRET and FLIM microscopy. Curr Opin Biotechnol. 2005;16:19–27. doi: 10.1016/j.copbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 53.Pollok BA, Heim R. Using GFP in FRET-based applications. Trends Cell Biol. 1999;9:57–60. doi: 10.1016/s0962-8924(98)01434-2. [DOI] [PubMed] [Google Scholar]
- 54.Gordon GW, Berry G, Liang XH, Levine B, Herman B. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys J. 1998;74:2702–2713. doi: 10.1016/S0006-3495(98)77976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson BS, Pfeiffer JR, Surviladze Z, Gaudet EA, Oliver JM. High resolution mapping of mast cell membranes reveals primary and secondary domains of Fc (epsilon) RI and LAT. J Cell Biol. 2001;154:645–658. doi: 10.1083/jcb.200104049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 57.Leavitt S, Freire E. Direct measurement of protein binding energetics by isothermal titration calorimetry. Curr Opin Struct Biol. 2001;11:560–566. doi: 10.1016/s0959-440x(00)00248-7. [DOI] [PubMed] [Google Scholar]
- 58.Ladbury JE, Chowdhry BZ. Sensing the heat: the application of isothermal titration calorimetry to thermodynamic studies of biomolecular interactions. Chem Biol. 1996;3:791–801. doi: 10.1016/s1074-5521(96)90063-0. [DOI] [PubMed] [Google Scholar]
- 59.Velazquez-Campoy A, Leavitt SA, Freire E. Characterization of protein–protein interactions by isothermal titration calorimetry. Methods Mol Biol. 2004;261:35–54. doi: 10.1385/1-59259-762-9:035. [DOI] [PubMed] [Google Scholar]
- 60.Berry DM, Nash P, Liu SK, Pawson T, McGlade CJ. A high-affinity Arg-X–X-Lys SH3 binding motif confers specificity for the interaction between Gads and SLP-76 in T cell signaling. Curr Biol. 2002;12:1336–1341. doi: 10.1016/s0960-9822(02)01038-2. [DOI] [PubMed] [Google Scholar]
- 61.Liu Q, Berry D, Nash P, Pawson T, McGlade CJ, Li SS. Structural basis for specific binding of the Gads SH3 domain to an RxxK motif-containing SLP-76 peptide: a novel mode of peptide recognition. Mol Cell. 2003;11:471–481. doi: 10.1016/s1097-2765(03)00046-7. [DOI] [PubMed] [Google Scholar]
- 62.Harding SE, Winzor DJ. Sedimentation velocity analytical ultracentrifugation. In: Harding SE, Chowdhry BZ, editors. Protein–Ligand Interactions: Hydrodynamics and Calorimtry. Oxford University Press; Oxford: 2001. pp. 75–103. [Google Scholar]
- 63.Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 2002;11:2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]