Abstract
The oxysterol binding protein (OSBP)-related proteins (ORPs) are conserved from yeast to man 1,2 and are implicated in regulation of sterol pathways 3,4 and in signal transduction 5. The structure of the full-length yeast ORP Osh4 was determined at 1.5–1.9 Å resolution in complexes with ergosterol, cholesterol, and 7-, 20-, and 25-hydroxycholesterol. A single sterol molecule binds in a hydrophobic tunnel in a manner consistent with a transport function for ORPs. The entrance is blocked by a flexible N-terminal lid and surrounded by functionally critical basic residues. The structure of the open state of a lid-truncated form of Osh4 was determined at 2.5 Å resolution. Structural analysis and limited proteolysis show that sterol binding closes the lid and stabilizes a conformation favoring transport across aqueous barriers and transmitting signals. The unliganded structure exposes potential phospholipid-binding sites that are positioned for membrane docking and sterol exchange. Based on these observations we propose a model in which sterol and membrane binding promote reciprocal conformational changes that facilitate a sterol transfer and signaling cycle.
OSBP was first discovered 6,7 as a cytosolic receptor for oxysterols that downregulate cholesterol synthesis 8. The cloning of OSBP1 led to the identification of a large family of OSBP-related proteins, the ORPs, with 7 members in S. cerevisiase and 12 in H. sapiens2. ORPs are essential for life in eukaryotes. The deletion of all 7 ORPs leads to severe defects in sterol and lipid distribution and endocytosis in yeast 3,4, and OSBP is essential for embryonic development in mice (M. Brown, personal communication). All ORPs contain a core OSBP-related domain (ORD), and many also contain pleckstrin homology (PH) domains, transmembrane regions, endoplasmic reticulum (ER)-targeting FFAT motifs, GOLD domains, and/or ankyrin repeats 2. These additional domains localize ORPs by binding to phosphoinositides 9, the ER protein VAP10, and other targeting signals. The localization of ORPs is dynamic. Oxysterol binding changes the subcellular localization of certain ORPs from the cytosol to the Golgi or ER 9,11. ORPs can bind lipids other than oxysterols, including phosphoinositides and phosphatidic acid 12,13. OSBP is a cholesterol sensing regulator of two protein phosphatases, a PTPPBS family member, and Ser/Thr phosphatase PP2A5.
The structure of full-length Osh4 was determined by multiple isomorphous replacement and refined to a free R-factor of 23 % at 1.5 Å resolution (Supplementary Fig. 1; Supplementary Table 1). Osh4 is built around a central antiparallel β-sheet of 19 strands (residues 115–293) which form a nearly complete β-barrel (Fig. 1a, Supplementary Fig. 2). The strands of the barrel are pitched at ~45 degrees to its axis. The barrel has structural similarity to the large β-barrels of various bacterial outer membrane transporters 14 as scored by Dali 15 (Supplementary Fig. 3). There is no PH domain within the ORD13. A tunnel with largely hydrophobic walls runs through center of the barrel in a location that corresponds to the pore in bacterial outer membrane transporters (Fig. 1b). As a soluble β-barrel protein with a hydrophilic exterior and a hydrophobic tunnel in the barrel center, Osh4 resembles an inside-out porin. Residues 1–29 form a lid that covers the tunnel opening (Fig. 1c, d). The N-terminal domain following the lid (residues 30–117) consists of a two-stranded β-sheet and three α-helices that form a 50 Å long antiparallel bundle, which runs the entire length of the barrel. The portion of the bundle distal to the lid fills the center of the barrel and thereby plugs the far end of the tunnel. The proximal portion of the bundle forms one wall of the tunnel, replacing the missing strands of the barrel. There is a large C-terminal region (residues 308–434) following the barrel. The exterior surface around the lid of the tunnel contains a half-dozen highly conserved basic residues (Fig. 1c; Supplementary Fig. 4).
Figure 1. Structure of Osh4.
a. The overall structure of Osh4. The N-terminal lid (1–29) is red, the central helices (30–116) orange, the β-barrel (117–307) green, and the C-terminal sub-domain (308–434) cyan. b. Surface Osh4: basic residues are blue, acidic red, hydrophobic green, and neutral polar white. Waters are shown as red spheres. c. Strictly conserved residues are colored cyan. The N-terminal lid is shown in ribbons and colored red. d. Solvent-accessible conserved residues are clustered around the tunnel entrance. e. Residues that bind 25-HC are colored green. f. Recognition of 25-HC 25-hydroxyl group. The ɛ-amino group of Lys336 has two conformations in all complex crystal structures, with the left-hand conformation (closest to Lys 109) predominating. g. Superposition of five sterols in the binding site. 7-HC is colored gray, 20-HC cyan, 25-HC red, cholesterol green and ergosterol blue. Hydroxyl groups in the sterols are shown in spheres. Hydrogen bonds are shown in dashed lines.
Sterols bind to Osh4 in the central tunnel of the β-barrel in a head down orientation. The 3-hydroxyl is buried at the bottom of the tunnel and the side-chain touches the inner surface of the lid (Fig. 1e). Although Osh4 has a novel fold, the burial of its ligands in a central hydrophobic tunnel is reminiscent of the structures of other lipid binding and transport proteins. The closest analogy is to the cholesterol-binding steroidogenic acute regulatory protein (StAR) transport (START) domain proteins MLN64 16 and StarD4 17, the phosphatidylcholine (PC) binding START domain protein PC-TP 18, and the mammalian phosphatidylinositol transfer proteins 19–21. In these structures the ligands are completely sequestered from solution. The yeast Sec14 phospholipid transfer protein 22 and its relatives 23,24, the GM2 activator protein 25, the cholesterol-binding proteins NPC226 and Scp2 27,28, and the glycolipid transfer protein GLTP 29 have similar ligand-binding tunnels, although some of these are open at one end 22,25,29. The structural analogy to other lipid transporters suggested to us that Osh4 might be an ergosterol transporter, and that other ORPs might also transport sterols or other lipids.
The most striking aspects of oxysterol recognition by Osh4 are the absence of direct hydrogen bonds between sterol hydroxyl groups and side-chains of conserved amino acids, and the prominence of water-mediated interactions. The 3-hydroxyl of cholesterol, ergosterol, and oxysterols binds to two water molecules and to the side-chain of Gln 96. This Gln is part of a hydrated cluster of polar side-chains at the bottom of the tunnel (Fig. 1e, g). Trp 46, Tyr 97, Asn 165, and Gln 181 comprise the remainder of the polar cluster at the tunnel bottom, and form water-mediated interactions with the 3-hydroxyl. The 20- and 25-hydroxyl groups of the respective oxysterols interact with more or less ordered water molecules but not directly with the protein. The 7-hydroxyl of 7-hydroxycholesterol (7-HC) has no apparent hydrogen bonds to either protein or waters. The 25-hydroxyl is linked via two water molecules to the conserved Lys-109 side-chain, but other oxysterol hydroxyls are linked to less well-ordered waters. The lack of direct protein interaction with oxysterol hydroxyls in Osh4 contrasts sharply to the specific recognition of 24(S), 25 epoxycholesterol by a His-Trp pair in LXR 30, a specific effector of a subset of oxysterols.
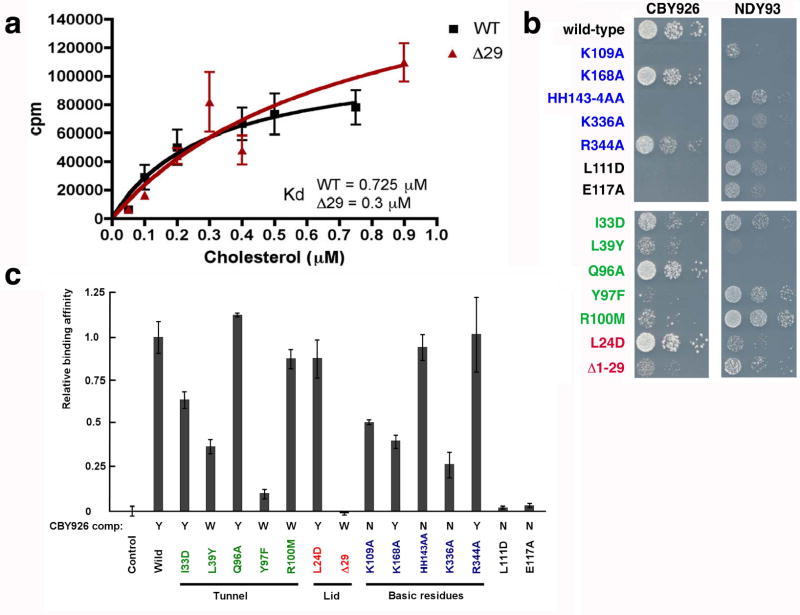
The absence of direct interactions between hydroxyl groups and the protein is difficult to reconcile with the concept that Osh4 is a specific effector of oxysterol signaling. We examined the relative affinities of Osh4 for cholesterol and oxysterols. Cholesterol binds to Osh4 with a Kd of 300 nM (Fig. 2a), as compared to a Kd of 55 nM for the tightest-binding oxysterol, 25-HC (Supplementary Fig. 5). Given the modest difference in affinities, and given that cholesterol is far more abundant than oxysterols in mammalian cells, we reasoned that cholesterol might be a physiological ligand for ORPs. We report elsewhere tests of this hypothesis that show Osh4 transports ergosterol, the yeast counterpart of cholesterol (Raychaudhuri, Im, Hurley & Prinz, in preparation).
Figure 2. Mutational analysis of binding and function.
a. Binding curves for wild-type and lid truncation mutant using low-stringency washing protocol. Each data point shown is the average of two measurements. b. Plasmids encoding Osh4 mutants were introduced into CBY926 (4) and NDY93. The strains were grown at permissive temperature (23°C) and dilution series were incubated at 37°C. All experiments were repeated at least 3 times with the same results. Expression of mutants in vivo was quantitated by immunoblotting (Supplementary Fig. 6). c. CPM measured for labeled cholesterol bound at a concentration of 0.15 μM to Osh4 mutants were scaled to wild type (100 %) using high-stringency washing protocol. The standard error from three measurements is shown by the bars.
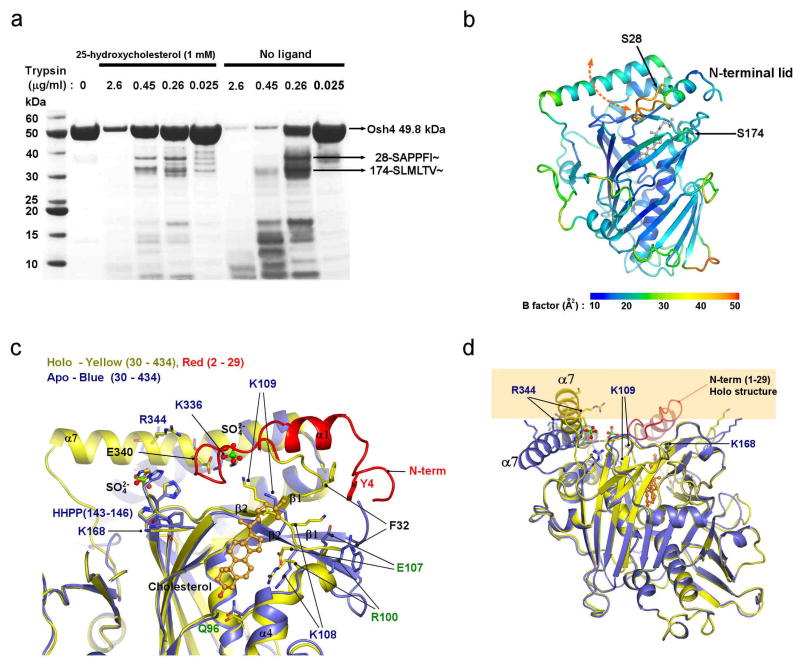
In the bound conformation of Osh4, sterol ligands are inaccessible from the outside. Therefore, a conformational change is required for the uptake and release of ligands. The lid has some of the highest B-values (Fig. 3b) in the structure, suggestive of flexibility. Following limited proteolysis by trypsin in the absence of ligand, Osh4 is rapidly converted to a stable fragment beginning at residue 28 (Fig. 3a), which corresponds to the end of the lid region (Fig. 3b). Binding of 25-HC stabilizes this region against proteolysis (Fig. 3a). Sterol ligands stabilize the closed conformation of the lid via direct van der Waals interactions with Trp 10, Phe 13, and the highly conserved residues Leu 24, and Leu 27 (Fig. 1f, g). We were unable to crystallize full-length Osh4 in the absence of ligand, which we attribute to the flexibility of the lid region in the unliganded state. We engineered a lidless Osh4 in which residues 1–29 were deleted and the flexible surface loop 236–240 was replaced by an ectopic dipeptide sequence. Mutation of loop 236–240 does not affect cholesterol binding (data not shown). We determined the 2.5 Å resolution structure of this form of Osh4 in its apo conformation (Fig. 3c, d).
Figure 3. Conformational changes in Osh4.
a. Tryptic digestion of Osh4. The arrows indicate the N-terminal amino acid sequences of the bands. b. The ribbon model is colored by the average B-factor of the residue to show their relative mobility. Trypsin cleavage sites are indicated by arrows. c. Apo (blue) and cholesterol complex (red for lid region and yellow elsewhere) structures were superimposed. The sulfate ions bound to the apo structure in positions that membrane phospholipid phosphates are shown. d. Superposition of apo and cholesterol complex structures, with the location of the membrane indicated by yellow shading.
The apo structure undergoes a dramatic conformational change (2.9 Å r.m.s. for all common Cα positions) with respect to the complexes. The change is triggered when the ligand-lid interactions are lost. The movement of the lid away from the rest of the protein unlocks α7 and basic-residue containing loops from their complexed conformations. In the apo structure the tunnel is open and there is no longer any barrier to ligand exchange. Helix α7 pivots about its N-terminus such that its C-terminus moves 15 Å. The β1 strand and several loop regions near the tunnel opening move by up to 7 Å. The Cα of the conserved basic residue Lys109 moves 6 Å in this conformational change. These shifts result in the reorganization of the entire conserved basic cluster at the tunnel entrance (Fig. 3c). The apo conformation presents putative phosphate binding sites and a flattened, unobstructed surface surrounding the tunnel opening, none of which are present in the bound conformation (Fig. 3d).
We tested whether the conserved basic residues at the tunnel entrance, as well as structural core, lid, and sterol-binding residues were important for biological function by assessing their ability to complement a yeast strain in which all seven ORPs have either been deleted or are present as a temperature-sensitive allele 3. Lys 109, the His 143- His 144 pair, and Lys 336 are essential for function13, while two basic residues farther from the tunnel entrance, Lys 168 and Arg 344, are not (Fig. 2b, Supplementary Fig. 6). None of these residues directly contacts cholesterol, and their mutations at most modestly impairs cholesterol binding (Fig. 2c). In contrast, some mutations within the tunnel such as Y97F, and structure core mutations near the tunnel such as L111D and E117A, abrogate both cholesterol binding and biological function (Fig. 2a,c). K109A, K168A, and K336A all show sharp reductions in basal cholesterol transport (Raychaudhuri et al, in preparation), while HHAA shows a smaller reduction, hence the major biochemical consequence of the basic cluster mutations is a consistent defect in transport.
In the complexes, the ɛ-amino groups of Lys 109 and Lys 336 are ~3.6 Å away from each other in an unfavorable semi-buried environment (Fig. 1f). In the apo structure they move 8.7 Å apart. Two ordered sulfate ions are bound in the apo structure, one between the new positions of Lys 109 and Lys 336, and the other near the conserved His pair (Fig. 3c). We believe these sulfate ions mimic the roles of phospholipid phosphate groups because they bind in the position expected for the plane of the membrane surface during sterol exchange. The unfavorable geometry of the Lys 109-Lys 336 pair in the complex suggests that the closed structure is in a destabilized “tense” state. We propose that the open state is selectively promoted by the availability of phosphate groups at the membrane surface, which bind to these Lys only in the open state. Thus, these residues appear to function as an electrostatic spring-loaded lock that controls access to the tunnel. Based on these observations, we propose a sterol transfer and signaling cycle (Fig. 4).
Figure 4. Mechanism of sterol transfer.
The proposed cholesterol transfer cycle is depicted. Oxysterols are shown binding via the cytosol rather than the membrane because of their higher solubility. Cholesterol and oxysterol-dependent signals are shown as occurring via the bound conformation in the cytosol as found for OSBP5.
OSBP is a cholesterol sensor that regulates phosphatase complexes5. 25-HC antagonizes cholesterol activation. We find that 25-HC and cholesterol bind to the same site and induce similar gross structural changes. However, 25-HC is unique among oxysterols in that it forms a water-mediated hydrogen bond with the critical Lys 109. Differences in packing between the different sterol side-chains and the surrounding side-chains of Ile 17, Leu 24, and Leu 27 lead to shifts in the lid main-chain position by up 1.5 Å as compared to cholesterol (Supplementary Fig. 7). Of the oxysterol complexes, the 25-HC structure shows the largest differences from the cholesterol complex. The possible role these subtle structural changes in explaining 25-HC/cholesterol antagonism warrants further study.
The lid and the basic cluster at the tunnel opening are conserved in all ORPs, while parts of the cholesterol binding site are not well conserved, suggesting that many ORPs might transport non-sterol ligands. The conservation of the lid and basic cluster lead us to believe that the transport cycle will be maintained throughout this protein family. Similarly, cholesterol-dependent signaling will require the same apparatus as transfer in order to take up cholesterol at membranes.
Methods
Protein expression and purification
DNA encoding Osh4 residues 2–434 was amplified by PCR and subcloned into the BamHI and XhoI sites of a pGEX-4T vector modified to contain a TEV protease recognition site. The fusion protein was expressed in E. coli BL21(DE3) overnight at 30 °C. The lysate was applied to glutathione sepharose. The GST was removed by digestion with TEV protease in the column and Osh4 was eluted with 500 mM NaCl and 50 mM NaH2PO4 (pH7.5). The protein was concentrated and then purified on a Superdex 200 column (Pharmacia) in 20 mM Tris-HCl (pH 8.0), 100 mM NaCl.
Crystallization
25-hydroxycholesterol was prepared in ethanol and added to 15 mg/ml protein at a final concentration of 1 mM. Crystals were grown by vapor-diffusion at 25 °C over a reservoir of 100 mM MES-NaOH (pH 6.5), 12% PEG 20000 over one week. Ergosterol, cholesterol, 7-hydroxy cholesterol, and 20-hydroxy cholesterol complex crystals were grown similarly. Crystals were cryoprotected in reservoir solution supplemented with 20% (v/v) glycerol and flash frozen under N2 gas at 95 K. Apo Osh4 crystals were grown in 100 mM MES-NaOH (pH 6.0) and 1.6 M ammonium sulfate using micro- and macro-seeding. The apo crystals were cryoprotected in reservoir solution containing 32% (v/v) glycerol. Structure determination is described in the supplementary methods.
Tryptic digestion and N-terminal amino acid sequencing
25-HC bound and apo-Osh4 were digested with increasing trypsin concentrations at 37°C for 2 hour. 25-HC bound Osh4 was prepared by adding 25-HC to the protein solution (0.3 mM) to a final concentration of 1 mM. 30 ug of each 25-HC bound Osh4 or apo-Osh4 was digested with increasing trypsin concentrations at 37°C for 2 hrs. From left to right samples were digested with 2.6, 0.45, 0.26, 0.052 μg/ml of trypsin. Fragments of the Osh4 protein prepared by the limited proteolysis using trypsin were separated by SDS-PAGE, transferred to PVDF membrane, and stained with Coomassie Blue R-250. Bands were cut out and the N-terminal amino acids were sequenced using a 492cLC Protein Sequencer (Applied Biosystems).
Ligand binding assay
For analysis of mutant Osh4 binding at a single concentration (Fig. 2a), [3H]-cholesterol and unlabeled cholesterol were mixed at a ratio of 1:9 to a final concentration of 10 μM in ethanol. Each reaction was carried out at room temperature in a final volume of 100 μl of 20 mM Tris pH 8.5, 30 mM NaCl, and 100 pmol Osh4. 15 pmol of cholesterol was added to the protein solution and incubated for 1 hr. Then 20 μl of Hightrap Q resin (70% (w/v)) (Amershan Biosciences) pre-equilibrated with Buffer A (20 mM Tris-HCl pH 8.5) was added and incubated for 5 min. The tubes were centrifuged at 13,000g for 1 min and washed with 1 ml of buffer A four times. The protein was eluted from the resin with 700 μl of buffer A containing 1 M NaCl. After centrifuging at 13,000g for 10 min, 200 μl of supernatant was taken and the radioactivity was measured in a liquid scintillation counter. In the low-stringency washing protocol (Fig. 2b), radiolabeled cholesterol (40 pmol) was dried under nitrogen and resuspended in 100 μL of 20 mM Tris, pH 8.5, 30 mM NaCl, 0.05% Triton-X-100. Purified Osh4p was added to a final concentration of 200nM and incubated at 30°C. After incubation for one hour, 15 μL of Hi-Trap Q resin (Amersham Pharmacia) was added and the samples were votexed and left at room temperature for five minutes. The resin was pelleted by spinning in a microcentrifuge at top speed for five minutes. After three washes with 20 mM Tris (pH 8.5), the protein was eluted from the resin with 500 μL of 20 mM Tris, pH 8.5, 1 M NaCl. After centrifugation, the radiolabeled cholesterol in the supernatant was determined by scintillation counting. To determine nonspecific binding, 500 μM unlabelled cholesterol was included in the incubation.
Strains and complementation analysis
Complementation analysis of was performed by introducing plasmids expressing mutant Osh4p into either CBY9264 or NDY63 (MATa sec14-3 osh4::kanMX4 ura3-1 his3-11,-15 leu2-5,-112). CBY926 contains a temperature-sensitive allele of OSH4 (osh4-1) and deletions of the other six OSH genes. NDY93 is a sec14-1 Δ osh4 strain that cannot grow at 37°C if it contains a plasmid encoding functional Osh4p; complementing plasmids prevent NDY93 from growing at 37°C13. To assess complementation, strains containing plasmids were plated on synthetic complete dextrose medium minus uracil and grown at 37°C.
Materials
Correspondence and requests for materials should be sent to JHH. Coordinates have been deposited with the Protein Data Bank with accession numbers 1ZHT, 1ZHB, 1ZHW, 1ZHX, 1ZHY, 1ZHZ, and 1ZI7.
Acknowledgments
We thank T. Levine, V. Bankaitis, and M. Brown for discussions and sharing unpublished data, N. DeAngelis for excellent technical assistance, R. Craigie for assistance with protein sequencing, J. Kim for advice on the early stages of this project, G. Miller and H. Shi for collecting synchrotron data, C. Beh and R. Scheckman for yeast strains, F. Dyda for maintaining the home x-ray facility, and the staff of beamline X25, NSLS and of SER-CAT, APS, ANL for assistance with data collection. Y. J. I. thanks S. H. Eom for mentoring and support. Y. J. I. was partly supported by the Korea Science and Engineering Foundation. Research carried out at the National Synchrotron Light Source, Brookhaven National Laboratory, is supported by the U.S. Department of Energy, Division of Materials Sciences and Division of Chemical Sciences under Contract No. DE-AC02-98CH10886. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract No.W-31-109-Eng-38.
References
- 1.Dawson PA, Ridgway ND, Slaughter CA, Brown MS, Goldstein JL. cDNA Cloning and Expression of Oxysterol-Binding Protein, an Oligomer with a Potential Leucine Zipper. J Biol Chem. 1989;264:16798–16803. [PubMed] [Google Scholar]
- 2.Olkkonen VM, Levine TP. Oxysterol binding proteins: in more than one place at one time? Biochem Cell Biol. 2004;82:87–98. doi: 10.1139/o03-088. [DOI] [PubMed] [Google Scholar]
- 3.Beh CT, Cool L, Phillips J, Rine J. Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics. 2001;157:1117–1140. doi: 10.1093/genetics/157.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beh CT, Rine J. A role for yeast oxysterol-binding protein homologs in endocytosis and in the maintenance of intracellular sterol-lipid distribution. J Cell Sci. 2004;117:2983–2996. doi: 10.1242/jcs.01157. [DOI] [PubMed] [Google Scholar]
- 5.Wang P, Weng J, Anderson RGW. OSBP is a cholesterol-regulated scaffolding protein in control of ERK1/2 activation. Science. 2005;307:1472–1476. doi: 10.1126/science.1107710. [DOI] [PubMed] [Google Scholar]
- 6.Kandutsch AA, Chen HW, Heiniger HJ. Biological-Activity of Some Oxygenated Sterols. Science. 1978;201:498–501. doi: 10.1126/science.663671. [DOI] [PubMed] [Google Scholar]
- 7.Taylor FR, Saucier SE, Shown EP, Parish EJ, Kandutsch AA. Correlation between Oxysterol Binding to a Cytosolic Binding-Protein and Potency in the Repression of Hydroxymethylglutaryl Coenzyme-A Reductase. J Biol Chem. 1984;259:12382–12387. [PubMed] [Google Scholar]
- 8.Schroepfer GJ. Oxysterols: Modulators of cholesterol metabolism and other processes. Physiol Rev. 2000;80:361–554. doi: 10.1152/physrev.2000.80.1.361. [DOI] [PubMed] [Google Scholar]
- 9.Levine TP, Munro S. The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr Biol. 1998;8:729–739. doi: 10.1016/s0960-9822(98)70296-9. [DOI] [PubMed] [Google Scholar]
- 10.Loewen CJR, Roy A, Levine TP. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ridgway ND, Dawson PA, Ho YK, Brown MS, Goldstein JL. Translocation of Oxysterol Binding-Protein to Golgi-Apparatus Triggered by Ligand-Binding. J Cell Biol. 1992;116:307–319. doi: 10.1083/jcb.116.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu YQ, Liu YL, Ridgway ND, McMaster CR. Novel members of the human oxysterol-binding protein family bind phospholipids and regulate vesicle transport. J Biol Chem. 2001;276:18407–18414. doi: 10.1074/jbc.M101204200. [DOI] [PubMed] [Google Scholar]
- 13.Li XM, et al. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J Cell Biol. 2002;157:63–77. doi: 10.1083/jcb.200201037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchanan SK. beta-Barrel proteins from bacterial outer membranes: structure, function and refolding. Curr Opin Struct Biol. 1999;9:455–461. doi: 10.1016/S0959-440X(99)80064-5. [DOI] [PubMed] [Google Scholar]
- 15.Holm L, Sander C. Dali - a Network Tool for Protein-Structure Comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 16.Tsujishita Y, Hurley JH. Structure and lipid transport mechanism of a StAR-related domain. Nat Struct Biol. 2000;7:408–414. doi: 10.1038/75192. [DOI] [PubMed] [Google Scholar]
- 17.Romanowski MJ, Soccio RE, Breslow JL, Burley SK. Crystal structure of the Mus musculus cholesterol-regulated START protein 4 (StarD4) containing a StAR-related lipid transfer domain. Proc Natl Acad Sci USA. 2002;99:6949–6954. doi: 10.1073/pnas.052140699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roderick SL, et al. Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nat Struct Biol. 2002;9:507–511. doi: 10.1038/nsb812. [DOI] [PubMed] [Google Scholar]
- 19.Tilley SJ, et al. Structure-function analysis of phosphatidylinositol transfer protein alpha bound to human phosphatidylinositol. Structure. 2004;12:317–326. doi: 10.1016/j.str.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Schouten A, et al. Structure of apophosphatidylinositol transfer protein alpha provides insight into membrane association. EMBO J. 2002;21:2117–2121. doi: 10.1093/emboj/21.9.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoder MD, et al. Structure of a multifunctional protein - Mammalian phosphatidylinositol transfer protein complexed with phosphatidylcholine. J Biol Chem. 2001;276:9246–9252. doi: 10.1074/jbc.M010131200. [DOI] [PubMed] [Google Scholar]
- 22.Sha BD, Phillips SE, Bankaitis VA, Luo M. Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein. Nature. 1998;391:506–510. doi: 10.1038/35179. [DOI] [PubMed] [Google Scholar]
- 23.Meier R, Tomizaki T, Schulze-Briese C, Baumann U, Stocker A. The molecular basis of vitamin E retention: Structure of human alphatocopherol transfer protein. J Mol Biol. 2003;331:725–734. doi: 10.1016/s0022-2836(03)00724-1. [DOI] [PubMed] [Google Scholar]
- 24.Min KC, Kovall RA, Hendrickson WA. Crystal structure, of human alphatocopherol transfer protein bound to its ligand: Implications for ataxia with vitamin E deficiency. Proc Natl Acad Sci USA. 2003;100:14713–14718. doi: 10.1073/pnas.2136684100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright CS, Li SC, Rastinejad F. Crystal structure of human GM2-activator protein with a novel beta-cup topology. J Mol Biol. 2000;304:411–422. doi: 10.1006/jmbi.2000.4225. [DOI] [PubMed] [Google Scholar]
- 26.Friedland N, Liou HL, Lobel P, Stock AM. Structure of a cholesterol-binding protein deficient in Niemann-Pick type C2 disease. Proc Natl Acad Sci USA. 2003;100:2512–2517. doi: 10.1073/pnas.0437840100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choinowski T, Hauser H, Piontek K. Structure of Sterol Carrier Protein 2 at 1.8 A Resolution Reveals a Hydrophobic Tunnel Suitable for Lipid Binding. Biochemistry. 2000;39:1897–1902. doi: 10.1021/bi992742e. [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Garcia F, et al. NMR structure of the sterol carrier protein-2: implications for the biological role. J Mol Biol. 2000;295:595–603. doi: 10.1006/jmbi.1999.3355. [DOI] [PubMed] [Google Scholar]
- 29.Malinina L, Malakhova ML, Teplov A, Brown RE, Patel DJ. Structural basis for glycosphingolipid transfer specificity. Nature. 2004;430:1048–1053. doi: 10.1038/nature02856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams S, et al. X-ray crystal structure of the liver X receptor beta ligand binding domain - Regulation by a histidine-tryptophan switch. J Biol Chem. 2003;278:27138–27143. doi: 10.1074/jbc.M302260200. [DOI] [PubMed] [Google Scholar]