Abstract
The Salmonella enterica serovar Typhimurium type III secretion system (TTSS) encoded in Salmonella pathogenicity island 2 (SPI-2) promotes replication within host cells and systemic infection of mice. The SPI-2 TTSS is expressed following Salmonella internalization into host cells and translocates effectors across the membrane of the Salmonella-containing vacuole (SCV). Two effectors with similar amino-terminal domains, SseJ and SifB, localize to the SCV membrane in infected HEp-2 cells and subsequently traffic away from the SCV along Salmonella-induced-filaments (Sifs). Following infection of RAW cells, SseJ and SifB localize to the SCV as well as LAMP-1-positive, vesicular-appearing structures distant from the SCV. Trafficking of SseJ and SifB away from the SCV requires the SPI-2 effector SifA. Deletion of sseJ, but not sifB, leads to attenuation of Salmonella replication in mice following intraperitoneal inoculation. The contribution of SseJ to in vivo replication is identical in wild-type and sifA deletion backgrounds, suggesting that SseJ trafficking away from the SCV along Sifs is unnecessary for its virulence function.
Salmonellae are gram-negative enteric pathogens that cause diverse disease syndromes in a variety of animal hosts (15). The principal syndromes associated with Salmonella infections in humans are enteric (typhoid) fever and gastroenteritis. Enteric fever results from systemic infection with the exclusively human-adapted serovars Salmonella enterica serovar Typhi and S. enterica serovar Paratyphi. It is likely that these serovars rely on their ability to survive and replicate in the human macrophage to produce disseminated infection. In contrast, infection with the broad-host-range-adapted serovar S. enterica serovar Typhimurium usually produces a self-limited gastroenteritis in humans but causes a systemic infection resembling enteric fever in susceptible mice.
Many gram-negative pathogens, including the salmonellae, utilize type III secretion systems (TTSS) to subvert host cellular functions and promote host colonization (11). These complex protein machines translocate bacterial virulence proteins, termed effectors, directly from the bacterial cell into the host cell cytoplasm. Salmonella serovar Typhimurium possesses two virulence-associated TTSS, encoded in Salmonella pathogenicity islands 1 and 2 (SPI-1 and SPI-2, respectively) (9). The SPI-1 TTSS is expressed on contact with host cells and is required for invasion of intestinal epithelial cells and induction of intestinal inflammatory and secretory responses. In contrast, the SPI-2 TTSS is expressed within the Salmonella-containing vacuole (SCV) following Salmonella internalization into host cells and translocates effectors across the SCV membrane into the host cell cytoplasm (10). The SPI-2 TTSS is required for Salmonella replication within host cells and establishment of systemic infection in the murine typhoid model.
Recent work has identified a family of SPI-2 TTSS translocated effectors that share a conserved N-terminal domain (13). The SifA and SifB proteins are members of this protein family that are probably translocated by the SPI-2 TTSS, although this has not previously been shown for SifB. In addition to the conserved N-terminal domain, SifA and SifB also display sequence similarity in their C-terminal domains (26% identical, 43% similar). Salmonella serovar Typhimurium sifA deletion mutants demonstrate decreased intracellular replication and systemic mouse virulence (1, 3). In addition, sifA is required for the formation of Salmonella-induced filaments (Sifs) in infected tissue culture epithelial cells (20). Sifs are tubular-membranous structures that radiate away from the SCV at late time points following Salmonella invasion. These structures contain LAMP-1 and other markers characteristic of late endosomes. Although the role of Sifs in Salmonella pathogenesis remains unclear, they reflect the ability of Salmonella to modify endosomal compartments in infected cells and may promote intracellular replication.
The SseJ protein is an additional member of this family that also contains a domain with homology to several acyltransferases produced by Aeromonas and Vibrio species (6). These secreted toxins catalyze the transfer of an acyl group from glycerophospholipids to cholesterol at membrane-water interfaces. Ruiz-Albert et al. recently demonstrated that, following transient expression in HeLa cells, SseJ localizes to a LAMP-1-positive membranous compartment and induces formation of large membranous conglomerations that may represent aggregated endosomal compartments. (17). Expression of SseJ with a targeted mutation in the putative acyltransferase active site did not induce formation of these structures. The authors speculated that SseJ modifies the lipid composition of the SCV in a manner that alters its trafficking and maturation. The subcellular localization and function of SseJ following endogenous translocation across the SCV by intracellular bacteria remain unstudied.
This work examines the subcellular localization of the SseJ and SifB effector proteins following translocation by the SPI-2 TTSS in epithelial cells and macrophages and their contributions to Salmonella virulence.
MATERIALS AND METHODS
Bacterial strains, eukaryotic cell lines, and growth conditions.
Bacterial strains and plasmids used are listed in Table 1. Salmonella serovar Typhimurium was grown to stationary phase in Luria broth (LB) with aeration for infection of macrophages and mouse virulence assays and in LB with aeration to mid-log phase for infection of epithelial cells. RAW264.7 and HEp-2 cells were grown and maintained as previously described (14).
TABLE 1.
Strains and plasmids used in this study
| Designation | Description | Reference |
|---|---|---|
| Plasmids | ||
| pKAS32 | rpsL suicide vector for allelic exchange | 19 |
| pCAS61 | pWSK29 with HA epitope tag | This study |
| pJAF21 | ΔsifA in pKAS32 | This study |
| pJAF22 | ΔsifB in pKAS32 | This study |
| pJAF23 | ΔsseJ in pKAS32 | This study |
| pJAF111 | sseJ-HA in pCAS61 | This study |
| pJAF158 | sifB-HA in pCAS61 | This study |
| Salmonella serovar Typhimurium strains | ||
| CS401 | 14028s, phoN2 zxx::6251 Tn10d-Cm, Strr | 14 |
| CS600 | 14028s, phoN::Tn10d-Kan, Strr | This study |
| EM232 | CS401, ssaT::mTn5 Kmr | 14 |
| JAF57 | CS401, ΔsifA | This study |
| JAF41 | CS401, ΔsifB | This study |
| JAF43 | CS401, ΔsseJ | This study |
| JAF62 | CS401, ΔsifA ΔsifB | This study |
| JAF65 | CS401, ΔsifA ΔsseJ | This study |
| JAF89 | CS401, ΔsifB ΔsseJ | This study |
| JAF205 | CS401, ΔsifA phoN::Tn10d-Kan | This study |
Construction of deletion mutants.
Deletion of sseJ, sifB, and sifA was accomplished by using allelic-exchange plasmids. To construct the sseJ deletion plasmid, DNA flanking both the 5′ and 3′ ends of sseJ was amplified from Salmonella serovar Typhimurium chromosomal DNA by PCR with Pfu Turbo polymerase (Stratagene). The 5′ end was amplified with the primers 5′ ATATCTAGACAGGACGTAGTACCAGCCTC 3′ and 5′ AACGGTACCTGGCATAGTGTCCTCCTTAC 3′, and the 3′ end was amplified with the primers 5′ CATGGTACCACTGAATAAAGTTCCATCGG 3′ and 5′ AAGAATTCAGTGACGGTGCCTTTCATGT 3′. The flanking ends were sequentially cloned into the allelic-exchange vector pKAS32, yielding plasmid pJAF23. The sifB deletion plasmid was constructed in a similar manner, with the same parental plasmid and restriction enzymes. The 5′ end of sifB was amplified with the primers 5′ GCGTCTAGAGCAGCGGCGGATCACGGGCG 3′ and 5′ GCGGGTACCCATAATGTAGACCACAAGTG 3′, and the 3′ end was amplified with the primers 5′ GCGGGTACCGAAGAAAGTTCCTCTCATGG 3′ and 5′ GCGGAATTCCCGGTCATGATCACCAAACAC 3′. The resulting plasmid is pJAF22. To construct the sifA deletion plasmid, DNA flanking both the 5′ and 3′ ends of sifA was amplified from Salmonella serovar Typhimurium chromosomal DNA by PCR. The 5′ end was amplified with the primers 5′ GGTTATCTCAATGAATTCCTGCTGTGG 3′ and 5′ GCGGGTACCGTCCGCTTTTGCTTTGCCAG 3′, and the 3′ end was amplified with the primers 5′ GCGGGTACCCGCTCAGAACAACAAAGCGGC 3′ and 5′ GCGTCTAGACCAATAAAACGGTCGCCAGC 3′. The flanking ends were sequentially cloned into the allelic-exchange vector pKAS32, yielding plasmid pJAF21. Plasmids pJAF21, pJAF22, and pJAF23 were then introduced into strain CS401 by conjugation and selected by allelic exchange as previously described (16). The resulting deletion strains, JAF41 (ΔsifB), JAF43 (ΔsseJ), and JAF57 (ΔsifA), were verified by diagnostic PCR with Taq polymerase (Qiagen). Double-deletion mutants were generated by using the single-deletion mutants, JAF41, JAF43, and JAF57, as recipients of the allelic-exchange plasmids in place of CS401. The kanamycin-resistant sifA deletion strain was generated by transducing the Tn5 Kan from strain CS600 into strain JAF57, yielding strain JAF205.
Construction of HA-tagged SseJ.
Plasmid pCAS61 was constructed by annealing the primers 5′ TCGACTATCCTTATGATGTTCCTGATTATGCATAAC 3′ and 5′ TCGAGTTATGCATAATCAGGAACATCATAAGGATAG 3′, which encode the hemagglutinin (HA) epitope tag sequence and inserting this into plasmid pWSK29 digested with SalI and XhoI. The sseJ open reading frame and promoter region were amplified with the primers 5′ CGCGAATTCGTCAGATAATATGTACCAGGC 3′ and 5′ CGCGTCGACTTCAGTGGAATAATGATGAGC 3′ by PCR with Pfu Turbo. Plasmid pCAS61 and the PCR product were digested with EcoRI and SalI and ligated together, yielding plasmid pJAF111.
Construction of HA-tagged SifB.
The sifB open reading frame and promoter region were amplified with the primers 5′ CGATTTTAAAATATCCGGGCGATC 3′ and 5′ CGCGTCGACACTCTGGTGATGAGCCTCAT 3′ by PCR with Pfu Turbo. Plasmid pCAS61, a pWSK29-derived plasmid encoding a single HA epitope tag, and the PCR product were digested with EcoRI and SalI and ligated together, yielding plasmid pJAF158.
Immunofluorescent detection and microscopy.
Cultured mammalian cells were seeded onto 11-mm-diameter coverslips prior to infection. After infection, cells were washed three times with phosphate-buffered saline (PBS) and fixed with 10% formyl saline for 15 min before being washed and permeabilized with ice-cold acetone. After washing and 1 h of blocking with PBS plus 10% serum, samples were probed with various primary and secondary antibodies, mounted, and examined with a Zeiss Axiovert S100 TV microscope with a 100× or 63× lens. Images were acquired and deconvolved with the softWoRx program (Applied Precision). The HA epitope tag was detected with the monoclonal antibody HA.11 (Covance). LAMP-1 was detected with fluorescein isothiocyanate (FITC)-conjugated mouse monoclonal antibody H4A3 or 1D4B (Research Diagnostics). Salmonellae were detected with rabbit polyclonal antilipopolysaccharide (anti-LPS) antibodies (Difco). Secondary detection was performed with various anti-rabbit or anti-mouse antibodies conjugated to Cy5, FITC, or tetramethyl rhodamine isocyanate (Sigma and Chemicon). Triple staining for HA, LPS, and LAMP-1 was performed with tetramethyl rhodamine isocyanate-conjugated anti-HA (Covance) at 1:500, FITC-conjugated LAMP-1 (either anti-human or anti-mouse) at 1:250, and rabbit anti-LPS at 1:1,000, and secondary staining was performed with Cy5-conjugated anti-rabbit antibody at 1:1,000.
Determination of fold intracellular replication.
To determine intracellular replication within macrophages, RAW267.4 cells were seeded in 24-well dishes at 5 × 105 cells/well and infected with wild-type and mutant Salmonella serovar Typhimurium grown to stationary phase in LB at a multiplicity of infection (MOI) of 10 for 1 h. Extracellular bacteria were then removed by repeated washing and subsequent addition of RPMI medium supplemented with 15 mg of gentamicin per liter. At 2 and 16 h postinfection, the infected cells were washed repeatedly in PBS and lysed in 0.5% deoxycholate. Samples were then serially diluted and plated on selective media to enumerate the intracellular bacteria. Fold replication was then determined by dividing the intracellular load determined at 16 h by that determined at 2 h. To determine intracellular replication within epithelial cells, HEp-2 cells were seeded at 1.5 × 105 cells/well and infected with wild-type and mutant Salmonella serovar Typhimurium grown to mid-log phase in LB. Infections were carried out as described above, except that the HEp-2 cells were maintained in Dulbecco modified Eagle medium; the infected cells were lysed in 1% Triton X-100; and the amount of intracellular replication was determined at 6, 8, 10, 12, 14, 16, and 18 h postinfection. All infections were performed in triplicate.
Determination of mouse competitive indices.
Wild-type and mutant Salmonella serovar Typhimurium were grown to stationary phase in LB and used to infect BALB/c mice. Each mutant strain was marked with chloramphenicol resistance while wild type was marked with kanamycin resistance. Wild-type and mutant bacteria were diluted, and roughly equal numbers were mixed. An aliquot of the inoculum was then plated on selective media, and the numbers of wild-type and mutant bacteria were quantitated. A total of 105 bacteria in a volume of 0.2 ml were injected intraperitoneally into each mouse. In each experiment, four mice were infected with each mutant Salmonella serovar Typhimurium strain. Two days after infection, the mice were sacrificed and the infected spleens were removed and homogenized in sterile saline. Wild-type and mutant bacteria were then enumerated in each infected organ by serial dilution and plating on selective media. The competitive index was calculated by dividing the number of mutant bacteria isolated from infected animals by the number of wild-type bacteria recovered. This value was then corrected by the initial ratio of mutant to wild-type bacteria used to infect each animal.
RESULTS
SseJ localizes to the SCV upon translocation from intracellular bacteria.
To examine the subcellular localization of the SPI-2 effector SseJ following translocation by the SPI-2 TTSS, Salmonella serovar Typhimurium containing a carboxy-terminal HA epitope-tagged allele of sseJ was used to infect mammalian cells. Expression was driven by the native sseJ promoter to ensure that the amount of SseJ-HA expressed within mammalian cells was physiologic and appropriately regulated. SseJ-HA was shown to be produced upon growth of Salmonella serovar Typhimurium under in vitro SPI-2-inducing conditions and to be secreted into the culture supernatant in an SPI-2 TTSS-dependent manner (data not shown).
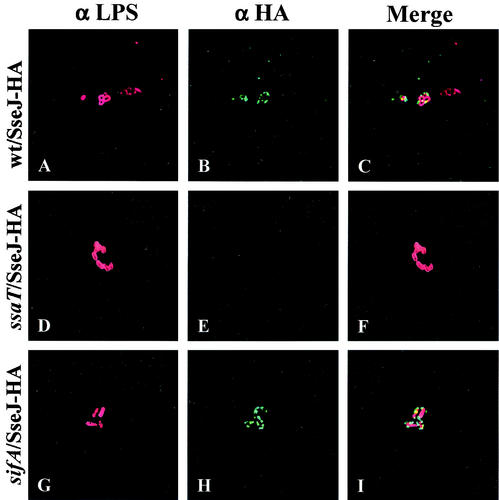
HEp-2 cells were infected with the wild-type strain CS401 and the ssaT mutant strain EM232, each harboring plasmid pJAF111, which encodes SseJ-HA. Four hours after infection, the HEp-2 cells were fixed, permeabilized, and stained for SseJ-HA (green) and bacteria (red). Intracellular bacteria were not permeabilized in this procedure, so that only SseJ-HA that was translocated or secreted from the bacteria was detected. As shown in Fig. 1, each infected HEp-2 cell contained only a few bacteria. SseJ-HA was detected immediately around the wild-type bacteria, apparently localizing to the SCV. In contrast, no SseJ-HA was detected in cells infected with the ssaT mutant strain. These results indicate that SseJ-HA is translocated into epithelial cells by the SPI-2 TTSS and suggest that SseJ-HA localizes to the SCV.
FIG. 1.
SseJ-HA is translocated by intracellular Salmonella serovar Typhimurium and localizes to the SCV in infected HEp-2 cells. HEp-2 cells were infected with wild-type and mutant Salmonella serovar Typhimurium expressing SseJ-HA for 1 h, followed by a 3-h incubation in medium containing gentamicin. Infected cells were then fixed, permeabilized, and stained for HA epitope (green) and Salmonella (red). (A to C) HEp-2 cells infected with the CS401 wild-type (wt) strain; (D to F) HEp-2 cells infected with the EM232 ssaT mutant strain; (G to I) HEp-2 cells infected with the JAF57 sifA deletion strain.
SseJ-HA colocalizes with LAMP-1 on Sifs upon bacterial intracellular growth.
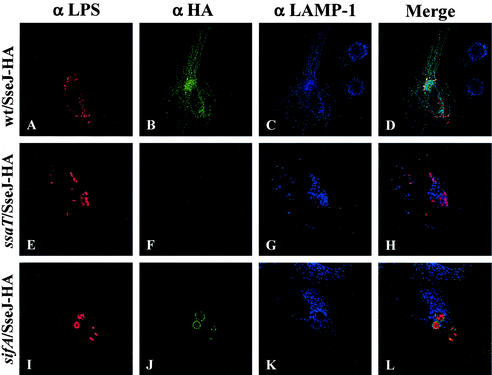
In order to determine if the subcellular localization of translocated SseJ-HA changed upon intracellular growth of Salmonella serovar Typhimurium, HEp-2 cells were infected with wild-type and ssaT mutant bacteria carrying pJAF111 for 20 h and analyzed by indirect immunofluorescence. As shown in Fig. 2, no translocation of SseJ-HA was observed from the ssaT SPI-2 TTSS mutant bacteria, as was seen at the earlier time point. In HEp-2 cells infected with wild-type bacteria, the subcellular localization of SseJ-HA was different at 20 h postinfection. As seen in Fig. 2, at 20 h postinfection, SseJ-HA was still detected around some intracellular bacteria. The majority of the SseJ-HA, however, was not necessarily associated with the bacteria but appeared to localize as discrete dots or upon thin filaments. Thus, it appeared that SseJ-HA was localizing to vesicles which may or may not be associated with the bacteria.
FIG. 2.
SseJ-HA colocalizes with LAMP-1 on Sifs in infected HEp-2 cells. Subcellular localization of SseJ-HA was examined 20 h after the infection of HEp-2 cells with wild-type and mutant Salmonella serovar Typhimurium. Infected cells were fixed and stained for HA epitope (green), Salmonella (red), and LAMP-1 (blue). (A to D) HEp-2 cells infected with the CS401 wild-type (wt) strain; (E to H) HEp-2 cells infected with the EM232 ssaT mutant strain; (I to L) HEp-2 cells infected with the JAF57 sifA deletion strain.
The filamentous localization pattern seen for SseJ-HA at 20 h after infection is very reminiscent of Sifs, aggregations of LAMP-1-containing vesicles seen in epithelial cells infected with SPI-2 TTSS-proficient Salmonella serovar Typhimurium (7, 8). LAMP-1 has also been observed to localize to the SCV. To determine if SseJ-HA is localizing to these LAMP-1-containing filaments, HEp-2 cells infected with bacteria expressing SseJ-HA for 20 h were fixed and stained for bacteria (red), SseJ-HA (green), and LAMP-1 (blue). As shown in Fig. 2, the SseJ-HA filamentous staining was coincident with LAMP-1 staining, indicating that SseJ-HA is localizing to Sifs. In contrast, in HEp-2 cells infected with ssaT mutant bacteria, no LAMP-1 filaments were observed, consistent with previous analyses demonstrating that SPI-2 TTSS is required for Sif formation (8).
Sif formation has previously been shown to require the SPI-2 effector protein SifA (20). To determine if SseJ-HA localization was dependent on SifA, plasmid pJAF111 was introduced into the sifA deletion strain JAF57. HEp-2 cells were then infected with this strain, and the subcellular localization of SseJ-HA was determined at both 4 and 20 h postinfection. As shown in Fig. 1, SseJ-HA localizes to the SCV of cells infected with the sifA deletion strain at 4 h postinfection. At 20 h, however, SseJ-HA was still localized only to the SCV. At this late time point, many SCVs had grown in size and had become roughly spherical, containing many bacteria. SseJ-HA appeared to localize to the periphery of these spheres, resulting in a ring-like localization pattern. At 20 h, the localization of LAMP-1 was also determined. LAMP-1 was shown to localize to this enlarged SCV as well, although no Sifs were observed. Many sifA mutant bacteria were not associated with LAMP-1 staining and were presumably residing in the cytoplasm (60% ± 10% bacteria without associated LAMP-1 [data not shown]). This is consistent with prior reports demonstrating that sifA contributes to maintaining the integrity of the SCV (1). In addition, SseJ staining was not observed around bacteria in the absence of associated LAMP-1 staining.
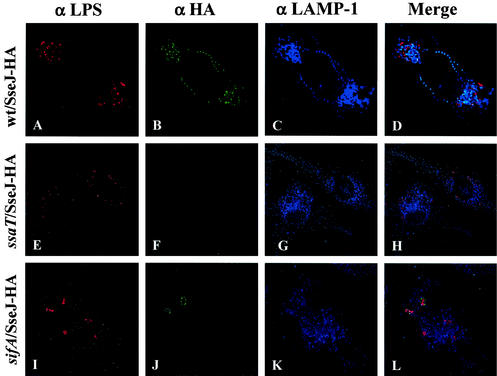
In infected macrophages, SseJ-HA localizes both to the SCV and to LAMP-1-positive structures distant from the SCV. The subcellular localization of SseJ-HA upon translocation into infected RAW264.7 cells was also analyzed. RAW264.7 cells were infected with the wild-type strain CS401 and the sifA deletion strain JAF57, each carrying pJAF111. Sixteen hours postinfection, the infected cells were fixed and stained for SseJ-HA, LAMP-1, and Salmonella. As shown in Fig. 3, SseJ-HA localized to the SCV of RAW264.7 cells infected with both wild-type and sifA mutant bacteria. In cells infected with wild-type bacteria, SseJ-HA was also observed away from intracellular bacteria in association with LAMP-1-positive, vesicular-appearing structures. Although these structures occasionally appeared to line up within the cell, there were no associated filamentous structures, and collection of images in multiple Z sections with deconvolution failed to identify connecting LAMP-1-positive filamentous structures in any plane. Although LAMP-1-positive filaments were not observed, SseJ-HA clearly traffics away from the SCV of infected macrophages into discrete LAMP-1-positive vesicular structures.
FIG. 3.
SseJ-HA traffics from the SCV in infected RAW264.7 cells. RAW264.7 cells were infected with wild-type and mutant Salmonella serovar Typhimurium expressing SseJ-HA. Sixteen hours postinfection, the cells were fixed and stained for HA epitope (green), LAMP-1 (blue), and Salmonella (red). (A to D) RAW264.7 cells infected with the CS401 wild-type (wt) strain; (E to H) RAW264.7 cells infected with the EM232 ssaT mutant strain; (I to L) RAW264.7 cells infected with the JAF57 sifA deletion strain.
The localization pattern of SseJ-HA in RAW264.7 cells infected with the sifA deletion strain was also significantly different from that observed in HEp-2 cells. Although SseJ-HA was observed in association with LAMP-1 around some bacteria, presumably on the SCV, these SCV contained only one or a few bacteria, instead of the large, spherical vacuoles seen in epithelial cells that contained many bacteria. In contrast to cells infected with wild-type bacteria, SseJ-HA was not observed away from the SCV in cells infected with sifA deletion mutants. In addition, infected RAW264.7 cells also contained many enlarged bacteria that were not associated with LAMP-1 or SseJ-HA (80% ± 10% bacteria without associated LAMP-1 staining). These bacteria presumably represent sifA mutants which have escaped from the SCV and are residing within the macrophage cytoplasm.
SifB has similar localization as SseJ in infected HEp-2 and RAW264.7 cells.
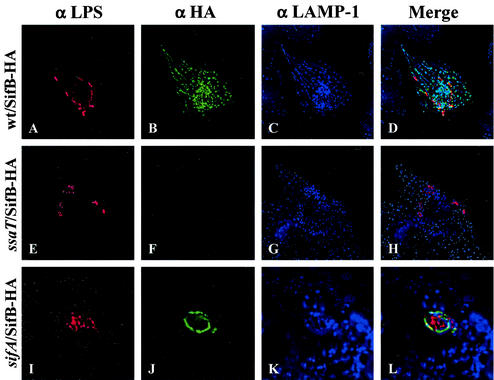
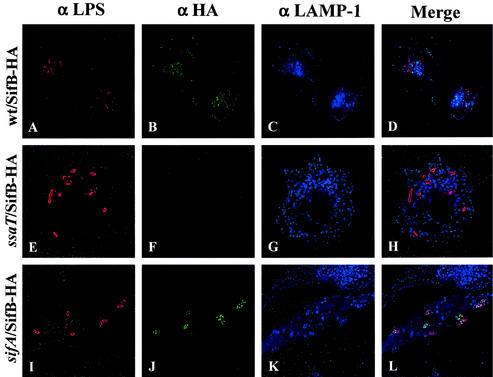
SifB shares a similar N terminus as and transcriptional regulation with SifA and SseJ and is therefore also likely to be an SPI-2 TTSS translocated effector. To examine this possibility, SifB fused to a C-terminal HA epitope tag was expressed from a low-copy-number plasmid under the regulation of its native promoter. Western blotting confirmed that this fusion was expressed under SPI-2-inducing conditions. Translocation of SifB-HA into infected HEp-2 cells was investigated by immunofluorescence microscopy in an identical manner as described for SseJ above. It was not possible to detect SifB at 4 h following infection. However, as shown in Fig. 4, at 20 h after infection, SifB localized to the SCV and Sifs in association with LAMP-1. In cells lacking Sifs, SifB showed staining on the SCV. In a sifA deletion background, SifB localized to the SCV of large, single collections of bacteria. SifB was not detected in a ssaT mutant, confirming that SifB is translocated by the SPI-2 TTSS. Infection of HEp-2 cells with sifB deletion mutants led to formation of Sifs, as visualized with LAMP-1 staining, at similar rates as infection with wild-type bacteria (data not shown), indicating that sifB is not required for Sif formation. In addition, infection of RAW264.7 macrophages for 16 h revealed SifB-HA staining similar to that observed for SseJ-HA in wild-type and sifA mutant backgrounds, respectively (Fig. 5).
FIG. 4.
SifB is translocated and localizes to Sifs in infected HEp-2 cells. HEp-2 cells were infected with wild-type and mutant Salmonella serovar Typhimurium expressing SifB-HA. Twenty hours postinfection, cells were fixed and stained for HA epitope (green), LAMP-1 (blue), and Salmonella (red). (A to D) HEp-2 cells infected with the CS401 wild-type (wt) strain; (E to H) HEp-2 cells infected with the EM232 ssaT mutant strain; (I to L) HEp-2 cells infected with the JAF57 sifA deletion strain.
FIG. 5.
SifB is translocated into infected RAW264.7 cells and traffics away from the SCV. RAW264.7 cells were infected with wild-type and mutant Salmonella serovar Typhimurium expressing SifB-HA in an identical manner as described for Fig. 3. Sixteen hours postinfection, the cells were fixed and stained for HA epitope (green), LAMP-1 (blue), and Salmonella (red). (A to D) RAW264.7 cells infected with the CS401 wild-type (wt) strain; (E to H) RAW264.7 cells infected with the EM232 ssaT mutant strain; (I to L) RAW264.7 cells infected with the JAF57 sifA deletion strain.
Contribution of SPI-2 effectors to Salmonella serovar Typhimurium replication within mammalian cells and mouse virulence.
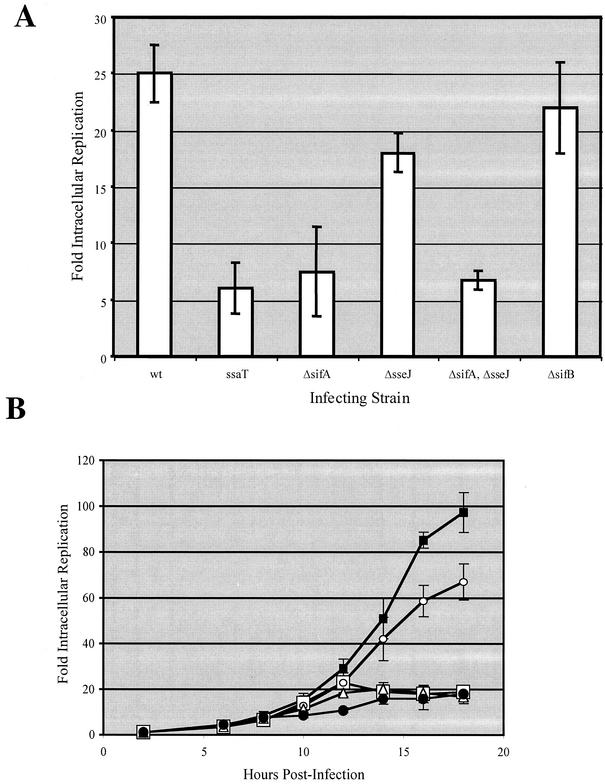
The sifA-dependent localization of SseJ and SifB suggests that the large virulence defect of a sifA mutant may be due in part to altered targeting of multiple other SPI-2 effector proteins. If this is the case, then deletion of sifA may have a dominant effect on mutations in other effectors in in vivo virulence assays. In order to test this hypothesis, nonpolar in-frame deletions were constructed in the sifA, sifB, and sseJ genes, either alone or in combination. These deletion mutant strains were tested for the ability to survive and replicate within a monocyte-derived cell line by a gentamicin protection assay. RAW264.7 cells were infected with wild-type and mutant Salmonella serovar Typhimurium, and intracellular bacteria were enumerated at 2 and 16 h postinfection. Fold replication, as shown in Fig. 6A, was then determined by dividing the number of bacteria recovered at 16 h by that recovered at 2 h. The wild-type strain CS401 replicated roughly 25-fold within RAW264.7 cells. In contrast, the ssaT mutant strain EM232 was defective in intracellular replication, replicating only about fivefold. The sifA deletion strain JAF57 was nearly as defective in replication, while the sifB deletion strain JAF41 was not defective at all. The sseJ deletion strain JAF43 displayed a minor replication defect, replicating at approximately 66% of the level of the wild-type strain CS401. The sseJ sifA double mutant showed similar replication as the sifA mutant.
FIG. 6.
Intracellular replication of wild-type and mutant Salmonella serovar Typhimurium. (A) RAW264.7 cells were infected with wild-type and mutant Salmonella serovar Typhimurium grown to stationary phase in LB at an MOI of 10 for 1 h. Infected macrophages were subsequently incubated in medium containing gentamicin and then lysed at 2 and 16 h postinfection. Intracellular bacteria were then enumerated, and fold replication was determined by dividing the number of bacteria recovered at 16 h by the number recovered at 2 h. Values reported are the means of experiments performed in triplicate, and error bars represent 1 standard deviation above and below the mean. (B) HEp-2 cells were infected with wild-type and mutant Salmonella serovar Typhimurium grown to mid-log phase in LB at an MOI of 10. Fold replication was then determined at 6, 8, 10, 12, 14, 16, and 18 h postinfection. Values represent the means from experiments performed in triplicate, and error bars represent 1 standard deviation above and below the mean. Closed black squares represent the wild-type strain CS401, open circles represent the sseJ deletion strain JAF43, open squares represent the sifA deletion strain JAF57, closed circles represent the ssaT mutant strain EM232, and open triangles represent the sifA sseJ double deletion strain JAF65.
The ability of these mutants to replicate within HEp-2 cells, an epithelium-like cell line, was also measured. Previous studies have reported that sifA deletion mutants display increased replication within epithelial cells at intermediate time points (5, 20). This increase in replication has been postulated elsewhere to result from an ability of the sifA mutant bacteria to escape from the SCV (1). In order to take this possibility into account, an intracellular replication time course assay was performed. HEp-2 cells were infected, and intracellular bacteria were enumerated at 2, 6, 8, 10, 12, 14, 16, and 18 h postinfection. Fold replication was determined for each time point. As shown in Fig. 6B, the wild-type strain CS401 replicates rather slowly for the initial 10 h of infection, after which the bacteria begin to replicate exponentially, ultimately reaching a fold replication of nearly 100 by 18 h postinfection. The ssaT mutant, on the other hand, did not display this exponential replication phase but did replicate as well as the wild type did for the first 8 h. The sifA deletion strain JAF57 did not display an elevated replication at any time point analyzed. Instead, the sifA deletion strain replicated as well as the wild type did for about 12 h, at which time replication ceased for the remainder of the experiment. Interestingly, although at the last time points equal numbers of ssaT and sifA mutant strains were recovered, at intermediate time points, the sifA deletion strain replicated twice as well as the ssaT mutant strain. The sseJ deletion strain JAF43 replicated as well as the wild type did for the initial 10 h, after which this strain, too, began to replicate exponentially. The growth rate for the sseJ deletion mutant, however, was reduced compared to that of the wild type, resulting in a fold replication roughly 66% of the level of wild type, just as seen within the infected macrophages. The sseJ sifA double mutant again showed similar replication as the sifA double mutant.
Measurement of intracellular replication by gentamicin protection assay is a relatively insensitive measure of virulence compared to in vivo competition experiments. Therefore, the chloramphenicol-resistant deletion mutants were analyzed for virulence for mice by competition with the isogenic kanamycin-resistant wild-type strain CS600. Colonization of the spleen of intraperitoneally infected animals was determined 48 h after the inoculation of a total of 105 bacteria. As a control for the method, the parental chloramphenicol-resistant wild-type strain CS401 was competed against CS600. As shown in Table 2, when mice were inoculated with equal numbers of the two wild-type strains, equal numbers were recovered (log competitive index of 0). As another control, the ssaT SPI-2 secretion mutant strain EM232 was competed against CS600. Disruption of the SPI-2 TTSS has previously been shown to result in a reduction of Salmonella serovar Typhimurium virulence for mice (1, 18). Strain EM232 was severely attenuated in this competition assay, displaying a log competitive index of −2.32. The three effector deletion mutants displayed various phenotypes, as shown in Table 2. While deletion of sifB had little effect on virulence, the sifA deletion strain JAF57 was significantly outcompeted by the wild-type strain. The sseJ deletion strain JAF43, on the other hand, displayed a small but significant virulence defect.
TABLE 2.
Virulence of SPI-2 mutant Salmonella serovar Typhimurium for mice
| Strain 1 genotype | Strain 2 genotype | Log10 competitive indexa (strain 1/strain 2) |
|---|---|---|
| Wild typeb | Wild typec | 0.00 ± 0.09 |
| ssaT | Wild typec | −2.32 ± 0.18 |
| ΔsifA | Wild typec | −1.11 ± 0.14 |
| ΔsseJ | Wild typec | −0.32 ± 0.15 |
| ΔsifB | Wild typec | −0.08 ± 0.06 |
| ΔsifA ΔsifB | Wild typec | −1.00 ± 0.12 |
| ΔsifA ΔsseJ | Wild typec | −1.44 ± 0.17 |
| ΔsseJ ΔsifB | Wild typec | −0.31 ± 0.11 |
| ssaT | ΔsifA | −1.39 ± 0.19 |
| ΔsifA ΔsseJ | ΔsifA | −0.36 ± 0.09 |
Ratio of the numbers of strain 1 to strain 2 bacteria recovered from infected spleens removed 2 days after the intraperitoneal injection of 105 total bacteria into mice. The mean competitive index from the infection of at least four mice is reported with the corresponding standard deviation.
Wild-type chloramphenicol-resistant strain CS401, parent strain for all mutant strains.
Wild-type kanamycin-resistant strain CS600, isogenic to CS401.
In a previous analysis, mutation of sifA was shown to result in severe attenuation as determined by a similar competition assay (1). In that report, a sifA mutant was as attenuated as an SPI-2 secretion mutant. In this analysis, however, the sifA deletion mutant, although defective in virulence, appeared to be significantly more virulent than the ssaT mutant. To confirm this result, the sifA deletion strain JAF57 was competed directly against the ssaT mutant strain EM232. As shown in Table 2, strain JAF57 outcompeted strain EM232, indicating that deletion of sifA does not result in a virulence phenotype equivalent to that resulting from loss of the SPI-2 TTSS.
As shown in Table 2, the addition of a sifB deletion to either the sifA or the sseJ deletion did not further augment the virulence defect of these single mutants. The sifA sseJ double deletion strain JAF65, however, was more attenuated than either of the single mutants. In fact, the double mutant appeared to display a virulence defect equal to the sum of those of the individual mutants. To further examine this, the sifA sseJ double-deletion strain JAF65 was competed against the kanamycin-resistant sifA deletion strain JAF205. As shown in Table 2, the sifA mutant outcompeted the double-deletion mutant to the same degree as the wild-type strain outcompeted the sseJ deletion strain. Therefore, deletion of sifA and sseJ resulted in an additive virulence defect, and these results do not indicate that the defect conferred by deletion of sifA is a result of a dominant effect resulting from altered localization of other effectors.
DISCUSSION
This work describes the subcellular localization of the SPI-2 TTSS effector proteins SseJ and SifB following translocation across the SCV membrane in epithelial cells and macrophages and quantifies the contribution of these effectors to in vivo replication in tissue culture cells and a mouse systemic infection model. SseJ localizes to the SCV at early time points (4 h) following invasion and subsequently moves away from intracellular bacteria along forming Sifs in epithelial cells. SifB also localizes to Sifs in infected HEp-2 cells, and these images represent the first direct demonstration of SifB translocation by the SPI-2 TTSS. Both SseJ and SifB localize to the SCV and LAMP-1-positive structures distant from the SCV in infected macrophages. Deletion of sseJ, but not sifB, leads to minor but reproducible attenuation in Salmonella virulence. Interestingly, deletion of sseJ produces an identical decrease in virulence in both wild-type and sifA deletion backgrounds, suggesting that the role of SseJ in virulence is independent of its localization to Sifs.
SseJ and SifB share a similar subcellular localization with the previously described SPI-2 effectors, SifA and PipB, which also concentrate on the SCV and along Sifs (2, 12). Three of these effectors (SifA, SifB, and SseJ) share homologous N termini, suggesting that this domain may contain a conserved localization motif. The absence of this conserved sequence in PipB indicates that there may be inapparent structural conservation or that multiple mechanisms may direct the localization of different SPI-2 effectors.
Several mechanisms may explain the redistribution of SseJ from its initial localization on the SCV to its subsequent distribution along the membranes of forming Sifs. Brumell et al. have hypothesized that Sifs result from recruitment and aggregation of late-endosomal membranous compartments along microtubules, leading to elongation of the SCV (4). As Sifs form, SseJ may diffuse away from the elongating SCV along continuous membranes. A more intriguing possibility is that SseJ may traffic away from the SCV in vesicles budding from the SCV membrane or become associated with additional membranous compartments through transient “kiss-and-run” interactions between these compartments and the SCV. This is supported by our observation of occasional SseJ-positive, LAMP-1-positive vesicular structures distant from the SCV in infected RAW cells and from the SCV and Sif network in HEp-2 cells.
The additive contributions of SseJ and SifA to Salmonella virulence indicate that these effectors probably function independently. Although deletion of sifA prevents movement of SseJ away from the SCV along Sifs, it has no impact on the contribution of SseJ to virulence. Therefore, the virulence function of SseJ is probably unrelated to its trafficking away from the SCV, although it may still require localization to the SCV membrane directly surrounding bacteria. One may speculate that the putative acyltransferase activity of SseJ modifies the lipid composition of the SCV, thus altering its trafficking and maturation. In addition, previous studies have reported that sifA deletion mutants escape from the SCV and reside in the cytoplasm of infected cells and that these mutants display increased intracellular replication in certain cell types (1, 20). Although we frequently observe sifA deletion mutants without associated LAMP-1 staining in HEp-2 and RAW264.7 cells, and therefore presumably residing in the cytoplasm, we do not observe increased intracellular replication of these mutants in HEp-2 cells at any time point.
Deletion of sifB, either alone or in combination with sifA or sseJ, does not impact splenic colonization following intraperitoneal injection. SifB may be important for other stages of infection, for example, in intestinal epithelia or Peyer's patches, or may share a redundant function with other effectors. Such redundancy exists within the SPI-1 TTSS effector set, where it is necessary to delete three effectors (SopE, SopE2, and SopB) to obtain an impact on host cell invasion (21). It will be necessary to determine the molecular functions of individual SPI-2 effectors to determine if such redundancy exists.
Acknowledgments
This work was supported by RO1 AI48683 from the National Institutes of Health. M. Ohl was supported by a Postdoctoral Research Fellowship for Physicians from the Howard Hughes Medical Institute.
We thank the laboratory of Stanley Fields for use of the Deltavision microscope.
J.A.F. and M.E.O. contributed equally to this work.
Editor: B. B. Finlay
REFERENCES
- 1.Beuzon, C. R., S. Meresse, K. E. Unsworth, J. Ruiz-Albert, S. Garvis, S. R. Waterman, T. A. Ryder, E. Boucrot, and D. W. Holden. 2000. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 19:3235-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brumell, J. H., D. L. Goosney, and B. B. Finlay. 2002. SifA, a type III secreted effector of Salmonella typhimurium, directs Salmonella-induced filament (Sif) formation along microtubules. Traffic 3:407-415. [DOI] [PubMed] [Google Scholar]
- 3.Brumell, J. H., C. M. Rosenberger, G. T. Gotto, S. L. Marcus, and B. B. Finlay. 2001. SifA permits survival and replication of Salmonella typhimurium in murine macrophages. Cell. Microbiol. 3:75-84. [DOI] [PubMed] [Google Scholar]
- 4.Brumell, J. H., P. Tang, S. D. Mills, and B. B. Finlay. 2001. Characterization of Salmonella-induced filaments (Sifs) reveals a delayed interaction between Salmonella-containing vacuoles and late endocytic compartments. Traffic 2:643-653. [DOI] [PubMed] [Google Scholar]
- 5.Brumell, J. H., P. Tang, M. L. Zaharik, and B. B. Finlay. 2002. Disruption of the salmonella-containing vacuole leads to increased replication of Salmonella enterica serovar Typhimurium in the cytosol of epithelial cells. Infect. Immun. 70:3264-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brumlik, M. J., and J. T. Buckley. 1996. Identification of the catalytic triad of the lipase/acyltransferase from Aeromonas hydrophila. J. Bacteriol. 178:2060-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-del Portillo, F., M. B. Zwick, K. Y. Leung, and B. B. Finlay. 1993. Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proc. Natl. Acad. Sci. USA 90:10544-10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guy, R. L., L. A. Gonias, and M. A. Stein. 2000. Aggregation of host endosomes by Salmonella requires SPI2 translocation of SseFG and involves SpvR and the fms-aroE intragenic region. Mol. Microbiol. 37:1417-1435. [DOI] [PubMed] [Google Scholar]
- 9.Hansen-Wester, I., and M. Hensel. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3:549-559. [DOI] [PubMed] [Google Scholar]
- 10.Hensel, M. 2000. Salmonella pathogenicity island 2. Mol. Microbiol. 36:1015-1023. [DOI] [PubMed] [Google Scholar]
- 11.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knodler, L. A., J. Celli, W. D. Hardt, B. A. Vallance, C. Yip, and B. B. Finlay. 2002. Salmonella effectors within a single pathogenicity island are differentially expressed and translocated by separate type III secretion systems. Mol. Microbiol. 43:1089-1103. [DOI] [PubMed] [Google Scholar]
- 13.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 97:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miao, E. A., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Baumler, and S. I. Miller. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850-864. [DOI] [PubMed] [Google Scholar]
- 15.Ohl, M. E., and S. I. Miller. 2001. Salmonella: a model for bacterial pathogenesis. Annu. Rev. Med. 52:259-274. [DOI] [PubMed] [Google Scholar]
- 16.Rakeman, J. L., H. R. Bonifield, and S. I. Miller. 1999. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J. Bacteriol. 181:3096-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz-Albert, J., X. J. Yu, C. R. Beuzon, A. N. Blakey, E. E. Galyov, and D. W. Holden. 2002. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol. Microbiol. 44:645-661. [DOI] [PubMed] [Google Scholar]
- 18.Shea, J. E., C. R. Beuzon, C. Gleeson, R. Mundy, and D. W. Holden. 1999. Influence of the Salmonella typhimurium pathogenicity island 2 type III secretion system on bacterial growth in the mouse. Infect. Immun. 67:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 20.Stein, M. A., K. Y. Leung, M. Zwick, F. Garcia-del Portillo, and B. B. Finlay. 1996. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol. Microbiol. 20:151-164. [DOI] [PubMed] [Google Scholar]
- 21.Zhou, D., L. M. Chen, L. Hernandez, S. B. Shears, and J. E. Galan. 2001. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol. Microbiol. 39:248-259. [DOI] [PubMed] [Google Scholar]