Abstract
Every eukaryotic chromosome requires a centromere for attachment to spindle microtubules for chromosome segregation. Although centromeric DNA sequences vary greatly among species, centromeres are universally marked by the presence of a centromeric histone variant, centromeric histone 3 (CenH3), which replaces canonical histone H3 in centromeric nucleosomes. Conventional chromatin is maintained in part by histone chaperone complexes, which deposit the S phase-limited (H3) and constitutive (H3.3) forms of histone 3. However, the mechanism that deposits CenH3 specifically at centromeres and faithfully maintains its chromosome location through mitosis and meiosis is unknown. To address this problem, we have biochemically purified a soluble assembly complex that targets tagged CenH3 to centromeres in Drosophila cells. Two different affinity procedures led to purification of the same complex, which consists of CenH3, histone H4, and a single protein chaperone, RbAp48, a highly abundant component of various chromatin assembly, remodeling, and modification complexes. The corresponding CenH3 assembly complex reconstituted in vitro is sufficient for chromatin assembly activity, without requiring additional components. The simple CenH3 assembly complex is in contrast to the multisubunit complexes previously described for H3 and H3.3, suggesting that centromeres are maintained by a passive mechanism that involves exclusion of the complexes that deposit canonical H3s during replication and transcription.
Keywords: chromosome segregation, Drosophila, histone variant, nucleosome assembly
The centromere is a defining feature of every eukaryotic chromosome, consisting of the unique site that is required for segregation at mitosis and meiosis. Large protein complexes assemble onto centromeres to mediate the attachment of microtubules that pull sister chromatids to opposite poles (1). During every cell cycle, and between meiotic generations, the location of the centromere on each chromosome remains invariant, and centromere positions are faithfully inherited over evolutionary time. This extraordinarily efficient mechanism of centromere maintenance does not seem to depend on specific DNA sequences found at centromeres. For example, although higher eukaryotic centromeres are embedded in multimegabase arrays of tandemly repetitive satellite sequences (2), in rare cases, functionally normal centromeres are found in regions that lack any common sequence features (3, 4).
Despite the sequence heterogeneity of centromeric DNA, centromeres are universally marked by the presence of a centromere-specific histone 3 variant [centromeric histone 3 (CenH3)] (5). Most of the DNA in a cell is packaged into nucleosomes in which ≈147 bp of DNA is wrapped around histones H3/H4/H2A/H2B. But at centromeres, CenH3 replaces canonical histone H3 (6, 7) and is essential for attachment to spindle microtubules. Most eukaryotic cells contain three distinct types of histone 3 variants (8), whose deposition leads to chromatin differentiation. Canonical histone H3 is synthesized during S phase and deposited by a chaperone complex containing chromatin assembly factor 1 (CAF-1), which has been shown to bind to the DNA replication clamp to mediate physical coupling of histone H3 deposition to DNA replication in vivo (9, 10). The constitutive histone variant H3.3 preferentially replaces and replenishes histones displaced by transcription and thus marks actively transcribed regions of the genome (11). A chaperone complex for H3.3 has been described for replication-independent deposition (9). Therefore, these two histone 3 variants are deposited via distinct chromatin assembly pathways that are coupled to replication (H3) and transcription (H3.3) and that may differentially mark active and inactive regions of the genome. However, little is known about how CenH3 is deposited and how its location on chromosomes is maintained. Like H3.3, CenH3 can be deposited throughout the cell cycle (12, 13); therefore, its assembly pathway is distinct from that of canonical H3 but similar to that of H3.3. However, overexpression of CenH3 results in low-level incorporation throughout euchromatic arms in addition to centromeres, whereas overexpressed H3.3 incorporates only throughout euchromatic arms, not centromeres. Taken together, these results indicate that three histone variants are assembled into chromatin at distinct domains of chromosomes by separate pathways.
Here, we describe the biochemical purification of the soluble complex that assembles Drosophila CenH3 [centromere identifier (CID)]. The simple composition of the purified CID nucleosome assembly complex and its in vitro activity suggest that CenH3 localization is maintained passively at centromeres, whereas H3 and H3.3 are assembled by complexes that restrict them to locations of polymerase-driven assembly processes.
Results
Purification of the CID Assembly Complex.
We used two different anti-CID antibodies (14) to detect soluble CID in Drosophila S2 cells and in early Drosophila embryo extracts; however, we were unable to reliably detect any non-chromatin-bound CID by Western blot analysis (Fig. 7, which is published as supporting information on the PNAS web site). Therefore, we overexpressed affinity-tagged versions of CID from a copper-inducible metallothionein promoter to obtain sufficient material for identification of its components by MS, analogous to the approach used for the identification of H3- and H3.3-specific assembly complexes from HeLa cells (9). We first used a tandem affinity purification (TAP) tag to determine the composition of pulled-down material. The TAP tag included a protein A domain, which binds IgG, and a calmodulin-binding sequence, separated by a tobacco etch virus (TEV) protease cleavage site (Fig. 1A) (15). After two sequential affinity purification steps, proteins that associate with CID were separated by SDS/PAGE, and Coomassie blue-stained bands not present in mock-purified material from nontransformed S2 cells were excised and analyzed by MS. Protein bands that appear in both mock and TAP-CID purification lanes were also analyzed by MS to ensure that there are no other genuine subunits of CID complex comigrating with contaminating proteins. Three proteins not present in mock-purified material were identified: RbAp48, CID, and histone H4 (Fig. 1B). The identities of RbAp48 and CID were confirmed by Western blot analysis (data not shown).
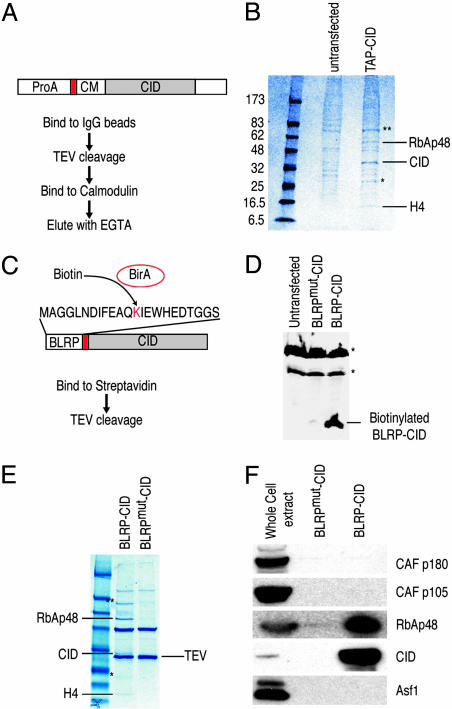
Fig. 1.
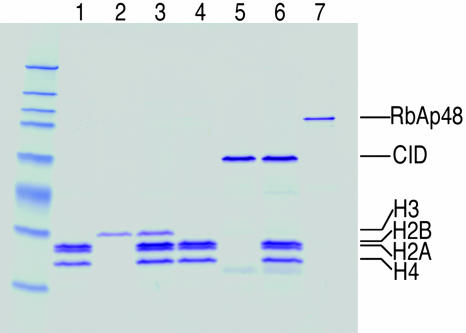
Purification of the CID assembly complex from S2 cells. (A) Purification scheme using TAP-tagged CID. ProA, protein A domain; red box, TEV cleavage site; CM, calmodulin-binding peptide. (B) TAP-CID-associated complex was purified and separated on a gradient gel. Mock purification was performed with extracts from untransfected cells. After Coomassie blue staining, protein bands that are only present in the TAP-CID lane were excised and identified by MS. (C) Biotin-mediated purification scheme. Coexpressed E. coli biotin ligase (BirA) transfers biotin to the lysine residue of BLRP tag. Red box, TEV cleavage site. A point mutation in BLRP (BLRPmut-CID) that changes K to R, rendering it nonbiotinylatable, was also made (not shown). (D) Western blot analysis with streptavidin-horseradish peroxidase on untransfected, BLRPmut-CID, and BLRP-CID cells. Asterisks indicate endogenously biotinylated proteins in the cell. Western blots with anti-CID antibody detect both BLRP-CID and BLRPmut-CID equally (data not shown). (E) The BLRP-CID-associated complex contains the same proteins found in the TAP-purified complex. In B and E, ∗ indicates partially degraded CID, and ∗∗ indicates a protein enriched but present in the mock-purified material. (F) Western blot analysis was performed on samples shown in the Coomassie blue-stained gel (E), with various antibodies confirming the identities of RbAp48 and CID. There are no detectable levels of other subunits of CAF-1 complex (p180 and p105) or Asf1 in the purified material.
We were surprised by the simplicity of the soluble complex identified by TAP-tag purification and considered the possibility that subunits were lost during the sequential steps. Therefore, we used a simpler, single-step technique that nevertheless had been shown to yield highly purified multicomponent complexes (16). We fused the 23-aa biotin ligase recognition peptide (BLRP) to the N terminus of CID, which, when biotinylated by Escherichia coli BirA in vivo, can be pulled down by using streptavidin (Fig. 1C) (17). The exceptionally high affinity and specificity of streptavidin for biotin allowed purification to be performed with only a single affinity step under physiological conditions. Coexpression of the BLRP-CID construct with a construct encoding E. coli BirA produced biotinylated CID in vivo (Fig. 1D). As a control, we used a construct in which the biotinylation target lysine residue was substituted with arginine (BLRPmut-CID), rendering it nonbiotinylatable (Fig. 1D). Biotin affinity purification identified three proteins not present in BLRPmut-CID control purifications (Fig. 1E). MS analysis revealed that these proteins are the same three identified by TAP-tag affinity purification: RbAp48, CID, and H4. We confirmed the presence of RbAp48 and CID in the purified fraction (BLRP-CID) and their absence in the BLRPmut-CID control by Western blot analysis (Fig. 1F). The purification of exactly the same complex under physiological conditions using very different affinity purification procedures strongly suggests that CID/H4-RbAp48 is the complete CID assembly complex. The presence of H4 in the complex is consistent with results from numerous studies that indicate that CenH3 replaces canonical histone H3 in the centromeric nucleosomes (6, 7) and that H4 is an obligatory partner of all histone 3 variants (8).
One possible explanation for the simplicity of the CID complex is that overexpression of tagged CID led to formation of nonfunctional complexes consisting of only the most abundant components. If this explanation were true, then the majority of tagged CID protein would be found outside centromeres. However, we find that BLRP-CID is incorporated into chromatin (data not shown). Moreover, cytological detection of biotinylated BLRP-CID revealed a characteristic centromere staining pattern, indicating that BLRP-CID, which is complexed with H4 and RbAp48, is properly targeted (Fig. 2).
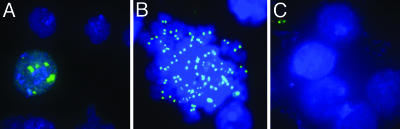
Fig. 2.
BLRP-CID localizes specifically to centromeres. (A and B) Cytological detection of BLRP-CID by streptavidin-Alexa Fluor 488 (green) on interphase (A) or mitotic (B) chromosomes of S2 cells (DAPI shown in blue) shows a characteristic centromere-staining pattern with only a faint noncentromeric signal. (C) No specific staining of BLRPmut-CID cells is seen. Immunological methods are described in Supporting Materials and Methods.
CID/H4-RbAp48 Is Distinct from Known Nucleosome Assembly Complexes.
RbAp48 is a common subunit of H3- and H3.3-specific nucleosome assembly complexes isolated from HeLa cells (9); therefore, it is the only protein that is found in nucleosome assembly complexes for all histone 3 variants identified so far. RbAp48 was initially identified as a protein that binds retinoblastoma protein (Rb) (18) and functions in recruitment of corepressor complexes to Rb target genes. RbAp48 and its homologs in other species have been found in many other chromatin-associated complexes. For this reason, it is known by other names [NURFp55 or p55 in Drosophila (19, 20), Mis16 in Schizosaccharomyces pombe (21), Msi1 in Arabidopsis (22), and Msi1 in Saccharomyces cerevisiae (23)]. To avoid confusion, we will refer to it as RbAp48, the name most commonly given to this protein.
RbAp48 is a WD40 protein that binds histones in vitro (24, 25). WD40 proteins have multiple binding surfaces that are often involved in multiple protein–protein interactions (26). RbAp48 of fission yeast has been implicated in CenH3 deposition, because loss of RbAp48 function interferes with centromere localization of CenH3s (21). In Drosophila, RbAp48 is highly abundant [220,000 molecules per diploid nucleus (20)] and is in large excess over the soluble pool of CID. If we assume that ≈200 kb of DNA is packaged into CenH3 nucleosomes (6) on four chromosomes per haploid genome and an average nucleosome repeat length of 180 bp, we estimate that there are ≈18,000 CID molecules in a normal diploid cell (200 kb × 4 chromosomes × 2 CID molecules per nucleosome × 2 genomes per diploid ÷ 180 bp). Therefore, the fraction of RbAp48 associated with CID would be only ≈8% even if 100% of the CID in the cell were associated with RbAp48. However, essentially all CID in S2 cells is found in insoluble chromatin (Fig. 8, which is published as supporting information on the PNAS web site), so that at most a trace fraction of soluble RbAp48 can be CID-associated at any time.
The much higher abundance of RbAp48 than CID and the potential of RbAp48 for multiple protein–protein interactions led us to consider the possibility that overexpressed CID is fortuitously associating with the abundant pools of RbAp48. Because RbAp48 is complexed with CAF-1 for replication-coupled nucleosome assembly and with HIRA (the yeast homolog of Hir1p) for transcription-coupled assembly, we would expect that a fortuitous association between CID and RbAp48 would carry along some substoichiometric amounts of subunits of these complexes. Therefore, we performed Western blot analysis of purified CID/H4-RbAp48 by using antibodies against other components of CAF-1 subunits (p180 and p105) and a common chaperone (Asf1) found in both H3.1 and H3.3 nucleosome assembly complexes. Fig. 1F shows that our purified preparation of CID/H4-RbAp48 does not contain detectable amounts of these proteins. Western blot analysis of TAP-purified material yielded similar results (data not shown). Therefore, the copurification of RbAp48 with CID is unlikely to be the result of promiscuous association of RbAp48-containing complexes.
Identification of a histone chaperone suggests that we have isolated a complex that is responsible for the deposition of this centromeric histone. Copurification of histones H2A and H2B would be expected if the chromatin-bound form of CID were being isolated; however, we do not detect any protein bands corresponding to H2A/H2B on gels stained with Coomassie blue (Fig. 1 B and E) or H2A/H2B peptides from MS analysis of purified material (data not shown). Therefore, the presence of a common histone chaperone and the absence of H2A/H2B provide further evidence that we have purified a nonnucleosomal complex that is responsible for the CID incorporation into centromeric chromatin. Although a previous study detected no interaction between fission yeast RbAp48 (Mis16) and CenH3 (Cnp1) by coimmunoprecipitation assays, given the abundance of RbAp48, there may not have been sufficient Cnp1 present in the soluble fraction to be detected, as the researchers suggested (21). This lack of detection of soluble fission yeast Cnp1 is consistent with our inability to detect a soluble pool of CID in either S2 cells or early Drosophila embryos (Fig. 7) and might reflect the regulation of CenH3 levels seen in budding yeast (27).
Drosophila RbAp48 Interacts with CID and H4 in Vitro.
Copurification of RbAp48 with CID/H4 suggests that RbAp48 directly binds either CID or H4 or both. In vitro binding assays with recombinant GST fusion proteins and35S-labeled proteins translated in vitro show an association between CID and RbAp48 (Fig. 3). Mammalian RbAp48 binds directly to the first α-helix of histone H4 (24, 25), and we also observed an interaction between RbAp48 and H4 by using recombinant GST-RbAp48 and H4 translated in vitro (Fig. 3B). The binding of RbAp48 to histone H4 and the presence of RbAp48 in H3.1-, H3.3-, and CenH3-assembly complexes suggests that the interface between RbAp48 and H4 is similar in these three complexes. The direct association between RbAp48 and CID but not between RbAp48 and H3 (24, 25) further indicates that RbAp48 makes unique contacts with CID for assembly of centromeric chromatin. In contrast to the simple composition of the CID complex, which contains only RbAp48 and H4, both H3.1 and H3.3 complexes contain other proteins in addition to RbAp48. The physical interactions among all three proteins in CID/H4-RbAp48 might serve to stabilize the trimolecular chaperone complex and help to prevent aggregation in the dense protein environment in living cells.
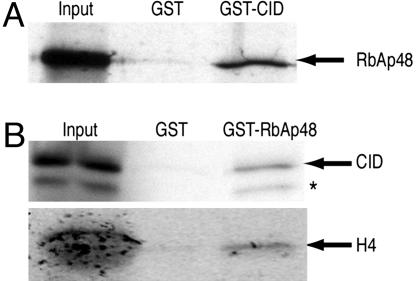
Fig. 3.
RbAp48 and CID directly interact in vitro. (A) A GST-CID fusion protein binds 35S-labeled RbAp48 translated in vitro. (B) A GST-RbAp48 fusion protein binds 35S-labeled CID and H4 translated in vitro. Some CID is either degraded or prematurely terminated (∗). An abundant protein present in the in vitro translation reaction (Promega) comigrates with the in vitro translated H4 in the input lane, causing a broad diffuse appearance. Input corresponds to 20% of the material used for binding assays. GST pull-down is described in Supporting Materials and Methods.
CID/H4-RbAp48 Assembles Nucleosomes in Vitro.
To measure CID chromatin assembly in vitro, we used a plasmid supercoiling assay in which the wrapping of DNA around the histone core particle induces supercoiling in relaxed, closed, circular DNA. Initial attempts using pUC19 plasmid DNA did not appreciably assemble chromatin in our system (Fig. 4E). However, the human CenH3 (CENP-A) has been successfully assembled into nucleosomes in vitro onto a tandem array of alpha-satellite DNA (28). Therefore, we cloned in tandem eight copies of a pericentric Drosophila satellite repeat unit into a pCR4 vector and used this construct (pCR4–360 × 8) in our assembly reactions. After an assembly reaction on relaxed pCR4–360 × 8, DNA was deproteinized, and plasmid topoisomers were resolved by agarose gel electrophoresis. Incubation of purified CID/H4-RbAp48 with pCR4–360 × 8 resulted in the introduction of several supercoils compared with the control complex (Fig. 4A). However, routinely performing in vitro chromatin assembly assays with CID/H4-RbAp48 purified from S2 cells was impractical because of its limited amount.
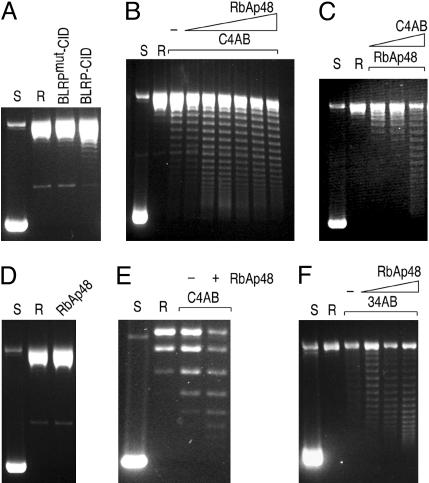
Fig. 4.
Analysis of nucleosome assembly by plasmid supercoiling assays. Supercoiled pCR4–360 × 8 plasmid was purified from E. coli (S) and relaxed by addition of topoisomerase I (R). (A) Chromatin assembly was performed by incubating the relaxed plasmid with the indicated protein complexes purified from S2 cells. (B) CID chromatin assembly reactions were performed by incubating the relaxed plasmid with CID/H4/H2A/H2B (recombinant CID and HPLC-purified H4/H2A/H2B) and increasing amounts of recombinant RbAp48. −, no RbAp48 added. (C) Same as in B except that the amount of RbAp48 was fixed, and increasing amounts of C4AB were added to the reaction. (D) A control assembly reaction with only RbAp48 does not yield any supercoils. (E) A plasmid supercoiling assay on pUC19 is markedly less efficient compared with pCR4–360 × 8. (F) Same as in B except that H3 was substituted for CID.
To overcome this limitation, we reconstituted the CID complex from purified components, supplemented with equimolar amounts of histone H2A and H2B (Fig. 5). Both CID and RbAp48 were purified under denaturing conditions as His-tagged fusion proteins expressed in E. coli. H4, H2A, and H2B were purified by salt extraction from S2 cells, followed by reverse-phase HPLC (Fig. 8). Because of the large differences in the hydrophobicity of H3/H3.3 and other histones, HPLC fractionation provides an effective method to separate H2A/H2B/H4 from H3/H3.3. CID prepared from bacteria was mixed in equimolar amounts with HPLC-purified H2A/H2B/H4 and dialyzed to allow refolding (Fig. 5). To determine whether RbAp48 acts as a chaperone in the formation of CID chromatin, we held the amount of purified histones constant (C4AB for CID/H4/H2A/H2B) and varied the amount of RbAp48 in the plasmid supercoiling assay (Fig. 4B). The assembly reaction resulted in the introduction of supercoils, indicative of CID chromatin assembly in vitro. RbAp48 increased the efficiency of assembly in a dose-dependent manner up to an amount equimolar to C4AB, as judged from Coomassie blue-stained polyacrylamide gels (data not shown). A similar dose dependence was found for C4AB (Fig. 4C). As expected, RbAp48 alone failed to introduce supercoils in the plasmid DNA (Fig. 4D), thus confirming that the induction of supercoils depends on deposition of the histones. Assembly onto pUC19 plasmid was inefficient, resulting in only one or two induced supercoils (Fig. 4E), which raised the possibility that CID chromatin assembly in vitro is specific to some features in the repeated array of the 360-bp satellite. However, assembly does not appear to be sequence-specific, because randomly chosen 3-kb Drosophila genomic fragments (the same size as the array of eight copies of 360-bp satellite) were capable of being assembled into CID chromatin (data not shown). Therefore, the inability to assemble CID chromatin onto pUC19 plasmid is likely to be the result of originating in bacteria where it never encountered nucleosomes.
Fig. 5.
Reconstitution of the CID assembly complex from purified components. In vitro reconstituted CID assembly complex components: lanes 1 and 4, HPLC-purified histone H4/H2A/H2B; lane 2, HPLC-purified H3; lane 3, refolded H3/H4/H2A/H2B; lane 5, recombinant CID; lane 6, refolded CID/H4/H2A/H2B; lane 7, recombinant RbAp48.
We also found that RbAp48 is sufficient for assembly of H3-containing nucleosomes from HPLC-purified H3 and H4/H2A/H2B (34AB) under the same conditions used for C4AB (Fig. 4F), although our qualitative assay might not have detected assembly rate differences between CID and H3. RbAp48-dependent assembly of 34AB is consistent with numerous studies that show that chromatin assembly from histones and DNA can be accomplished by simple chaperones, such as NAP1 (29), polyglutamic acid (30), and high salt concentration (31). Because assembly complexes for H3, H3.3, and CID all contain RbAp48, the absence of other protein subunits in the CID/H4-RbAp48 complex is its distinguishing feature, rather than the presence of RbAp48. The relatively large H3-specific complex with at least seven nonhistone subunits (9) is incapable of chromatin assembly even in the presence of crude extracts unless accompanied by DNA replication. In contrast, the simplicity and sufficiency of CID/H4-RbAp48 suggests that CID localization is achieved by passive accumulation at centromeres, whereas H3 and H3.3 are assembled by complexes that restrict them to locations of polymerase-driven assembly processes.
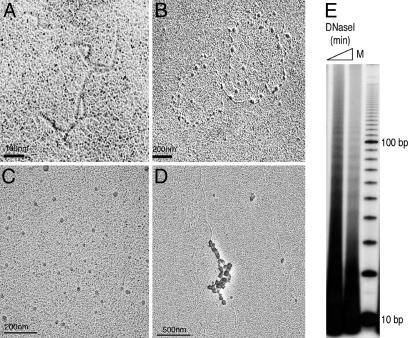
In vitro chromatin assembly onto the pCR4–360 × 8 plasmid was confirmed by transmission EM. This simple assembly reaction resulted in characteristic “beads on a string” structures resembling chromatin (Fig. 6A–D), suggesting that our reconstituted complex is capable of depositing histones onto DNA successfully. We further confirmed the product of assembly by assaying for the periodic exposure of minor grooves on the outside of the nucleosome structure. DNaseI digestion of assembled CID chromatin generated a characteristic ladder with ≈10-bp periodicity, as expected from DNA being wrapped around histone core particles (Fig. 6E). Thus, CID/H4-RbAp48 that we have purified from Drosophila cells is found to assemble CID chromatin, as shown by using three independent assays: plasmid supercoiling, EM, and DNaseI protection.
Fig. 6.
Analysis of in vitro assembled CID nucleosomes. (A–D) EM images of naked DNA before assembly into chromatin (A), circular DNA assembled into CID chromatin (B), and linear DNA assembled into CID chromatin and diluted into physiological salt (C) or assembly buffer (D) before imaging. Sample grids were rotary-shadowed. Images are representative of ≈100 molecules counted during three independent assembly reactions. EM methods are described in Supporting Materials and Methods. (E) DNase I digestion of in vitro assembled CID chromatin. A ladder of ≈10-bp periodicity indicates that DNA had been wrapped around histones. M, 10-bp ladder marker.
Discussion
We have biochemically purified a protein complex associated with the non-chromatin-bound pool of Drosophila centromeric histone CID by two independent affinity purification methods. The complex contains a single chaperone RbAp48, which binds both CID and histone H4 in vitro. Surprisingly, CID/H4-RbAp48 is capable of assembling centromeric nucleosomes in vitro in the absence of any other cellular processes.
We have shown that RbAp48 is sufficient for centromeric chromatin assembly in vitro, but is it necessary for this process in vivo? RbAp48 is found in various chromatin-associated protein complexes, where it is thought to play a common role in mediating their interactions with histones (32). Although no mutations have been reported to eliminate Drosophila RbAp48 (NURFp55), mutations in other components of RbAp48-associated complexes are lethal [Nurf-38 (33), E(z) (34), sin3 (35), and many others]; therefore, we would expect that removal of RbAp48 would have pleiotropic effects. Indeed, knock-down of RbAp48 by RNAi in Drosophila S2 cells resulted in S phase arrest (36) and derepression of various Rb/E2F target genes (37). These pleiotropic effects caused by reduction in RbAp48 levels would mask any centromere defect, and, in any case, such a defect would not be expected to occur immediately, because disruption of fission yeast RbAp48 did not affect chromosome segregation until the second round of mitosis (21).
The single chaperone that we have purified by using tagged CID contrasts with the multiple subunits found in purified chaperone complexes using tagged H3.1 and H3.3. The H3.1-specific replication-coupled assembly complex contains more than seven nonhistone subunits, and the H3.3-specific replication-independent complex contains at least five (9). Furthermore, H3.1- and H3.3-specific assembly reactions were performed in the presence of crude lysates, suggesting requirements for additional components that might restrict deposition to polymerase-driven processes. In contrast, both purified and reconstituted CID/H4-RbAp48 are sufficient for chromatin assembly in the absence of any other processes.
The formation of chromatin from histones and DNA is a thermodynamically favorable reaction, and it is thought that histone chaperones are needed to prevent nonproductive aggregation between highly positively charged histones and highly negatively charged DNA in a dense protein environment. Both replication-coupled assembly of H3.1/H4 (38) and transcription-coupled assembly of H3.3/H4 (39, 40) take place in the highly dynamic context of multisubunit polymerase transit, and assembly in both cases might require a large number of subunits to facilitate tethering of assembly complexes for rapid histone deposition (9). However, the basic assembly reaction appears to have minimal requirements, and conventional nucleosomes can be assembled in the presence of the NAP1 protein chaperone (29), polyglutamate (30), or high concentration of salt (31). We suggest that the simplicity of CID/H4-RbAp48 reflects a simple in vivo situation in which assembly occurs in the absence of rapidly transiting polymerases and associated factors. Although both H3.1- and H3.3-specific complexes also contain RbAp48 (9) and RbAp48 alone can assemble H3 nucleosomes (Fig. 4F), other components in these complexes might prevent spontaneous deposition at gaps in chromatin due to steric hindrance, whereas the much simpler CID/H4-RbAp48 would gain access to these chromatin gaps without impediment. In other words, H3- and H3.3-specific chromatin assembly complexes may have evolved to strictly couple their activities to replication and transcription, respectively, to increase the efficiency of these cellular processes, and to delineate assembly pathways of different histone 3 variants. There is precedence for such a variant-dependent exclusion mechanism: H3 appears to be prevented from assembling by replication-independent deposition anywhere in the genome, whereas H3.3 appears to deposit anywhere except at centromeres (41). When overproduced, CID deposits in a euchromatic pattern that is similar to that seen for H3.3, suggesting that CenH3s have fewer constraints than either H3 or H3.3 and that other chaperones in these complexes are the best candidates for mediating differential exclusion. Any CenH3 that incorporates in euchromatin at transient gaps created by transcription would be continuously replaced by transcription-coupled assembly of H3.3 (11); in this way, CenH3 would be passively retained at centromeres but actively removed from transcriptionally active regions.
Exclusion of H3 and H3.3 but not CenH3 from centromeric chromatin, such as by steric hindrance or RNA-mediated targeting (42), might help account for the deposition of CenH3s at a wide variety of sequences within a genome, including human neocentromeres (43), nematode holocentromeres (44), and gene-rich rice centromeres (4). Furthermore, budding yeast CenH3 (Cse4p) can localize properly to human centromeres and rescue a CENP-A depletion phenotype (45). Because of the high degree of divergence between Cse4p and CENP-A relative to the near invariance of H3, it is unlikely that a protein complex that normally recognizes CENP-A can associate with Cse4p and deposit it only at the centromeres. Rather, assembly of CenH3-H4 into centromeric chromatin in other organisms might be achieved by a simple H4-binding chaperone, such as RbAp48. Perhaps what distinguishes a CenH3 from a canonical H3 is that it is not accepted by H3- or H3.3-specific chaperone complexes.
The efficient propagation of centromeric chromatin domains during every cell cycle requires the correct localization of CenH3s. The robustness and precision of this process is extraordinary; for example, the location of centromeres have not changed in our lineage for 30 million years. It has been proposed that the compact structure of the CENP-A/H4 protein tetramer leads to the perpetuation of correct CENP-A localization (46), but it is not clear how compactness by itself can facilitate the faithful recruitment of additional CENP-A/H4 protein tetramers during every cell division. The apparent simplicity of CenH3 assembly can provide a mechanism to delineate this assembly pathway from that of H3 and H3.3. Torsional stress induced at centromeres at anaphase may be an efficient mechanism to clear H3 or H3.3 from centromeres and to create gaps for CenH3 deposition (47). Thus, the assembly of centromeric nucleosomes at gaps, which are created by the very process that requires CenH3, would provide a robust self-enforcing mechanism to maintain centromeres indefinitely.
Materials and Methods
Plasmid Transfection and Cell Culture.
TAP-CID, BLRP-CID, and BirA were all cloned into the pRMHA3 vector, which drives the expression of these genes from a copper-inducible metallothionein promoter (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site). S2 cells were cultured in serum-free media (SFM, Invitrogen) before transfection (Supporting Materials and Methods). TAP-CID cell lines were generated by double transfection with pRMHA3-TAP-CID and pCoPuro (48) in 19:1 mass ratio. BLRP-CID and BLRPmut-CID cell lines were produced by triple cotransfection with pRMHA3-BLRP-CID (or BLRPmut-CID), pRMHA3-BirA, and pCoPuro in 10:10:1 mass ratio.
Assembly Complex Purifications.
TAP-tag purification from S2 cell lysates was performed essentially as described in ref. 15. For BLRP-CID purification, extract was mixed with streptavidin beads (Amersham Pharmacia) for 30 min at 4°C. The extract/beads slurry was packed into a disposable column and washed with 40 column volumes of PBS350 (PBS supplemented with NaCl to a final concentration of 350 mM) with 0.1% Tween 20 and then washed with 10 column volumes of TEV buffer (PBS/0.1% Tween 20/1 mM DTT). The packed beads were resuspended with an equal volume of TEV buffer, and TEV (200 units) was added, digesting for 1 h at room temperature. Released complex was recovered by repacking the slurry into a column and flushing with PBS350 with 0.1% Tween 20 by gravity flow. MS analysis of excised protein bands identified the following peptides (amino acid sequences in parentheses are part of the TAP tag). For RbAp48, SDNAAESFDDAVEER, TPSSDVLVFDYTK, LMIWDTR, TVALWDLR, and LHVWDLSK; for CID, (GELK), (RRWK), (NFIAVSAANR), (KISSSGALGGGS)MPR, SEPEDGTDYGLEFTTSQLTLQDNNRR, QPAAR, RRK, EIRR, and LADSYMLTK; and for H4, DNIQGITKPAIR, ISGLIYEETR, VFLENVIR, TVTAMDVVYALK, and TLYGFGG. The entire process was also performed in PBS (instead of in PBS350) with identical results.
In Vitro Reconstitution and Chromatin Assembly Assays.
CID and RbAp48 were cloned into pET16b (Novagen) to produce N-terminal His-tagged proteins. Bulk histones were prepared from S2 cells by salt extraction (49) and fractionated by HPLC (Supporting Materials and Methods). Supercoiling, DNaseI, and EM assays for chromatin assembly are described in Supporting Materials and Methods.
Supplementary Material
Acknowledgments
We thank P. Gafken for identification of peptides by MS; B. Milless for separation of histones by HPLC; J. Tyler (University of Colorado Health Sciences Center, Aurora) for the gift of NURFp55, CAF-1 p180, p105, and Asf1 antibodies; T. Iwaki (University of Notre Dame, Notre Dame, IN) for kindly providing the pCoPuro plasmid; and S. Furuyama, S. Biggins, and P. J. Harte for comments on the manuscript. T.F. was supported by National Institutes of Health Fellowship 1F32GM71230-01.
Abbreviations
- CenH3
centromeric histone 3
- BLRP
biotin ligase recognition peptide
- TAP
tandem affinity purification
- CID
centromere identifier
- CAF-1
chromatin assembly factor 1
- TEV
tobacco etch virus
- CENP-A
human CenH3
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Choo K. H. The Centromere. London: Oxford Univ. Press; 1997. [Google Scholar]
- 2.Malik H. S., Henikoff S. Curr. Opin. Genet. Dev. 2002;12:711–718. doi: 10.1016/s0959-437x(02)00351-9. [DOI] [PubMed] [Google Scholar]
- 3.Amor D. J., Choo K. H. Am. J. Hum. Genet. 2002;71:695–714. doi: 10.1086/342730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagaki K., Cheng Z., Ouyang S., Talbert P. B., Kim M., Jones K. M., Henikoff S., Buell C. R., Jiang J. Nat. Genet. 2004;36:138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- 5.Henikoff S., Ahmad K., Malik H. S. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 6.Blower M. D., Sullivan B. A., Karpen G. H. Dev. Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meluh P. B., Yang P., Glowczewski L., Koshland D., Smith M. M. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 8.Malik H. S., Henikoff S. Nat. Struct. Biol. 2003;10:882–891. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- 9.Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z., Shibahara K., Stillman B. Nature. 2000;408:221–225. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- 11.Henikoff S., Furuyama T., Ahmad K. Trends Genet. 2004;20:320–326. doi: 10.1016/j.tig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Ahmad K., Henikoff S. J. Cell Biol. 2001;153:101–110. doi: 10.1083/jcb.153.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shelby R. D., Monier K., Sullivan K. F. J. Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henikoff S., Ahmad K., Platero J. S., van Steensel B. Proc. Natl. Acad. Sci. USA. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Seraphin B. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 16.de Boer E., Rodriguez P., Bonte E., Krijgsveld J., Katsantoni E., Heck A., Grosveld F., Strouboulis J. Proc. Natl. Acad. Sci. USA. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beckett D., Kovaleva E., Schatz P. J. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian Y. W., Wang Y. C., Hollingsworth R. E., Jr., Jones D., Ling N., Lee E. Y. Nature. 1993;364:648–652. doi: 10.1038/364648a0. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Balbas M. A., Tsukiyama T., Gdula D., Wu C. Proc. Natl. Acad. Sci. USA. 1998;95:132–137. doi: 10.1073/pnas.95.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyler J. K., Bulger M., Kamakaka R. T., Kobayashi R., Kadonaga J. T. Mol. Cell. Biol. 1996;16:6149–6159. doi: 10.1128/mcb.16.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., Yanagida M. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Hennig L., Taranto P., Walser M., Schonrock N., Gruissem W. Development (Cambridge, U.K.) 2003;130:2555–2565. doi: 10.1242/dev.00470. [DOI] [PubMed] [Google Scholar]
- 23.Ruggieri R., Tanaka K., Nakafuku M., Kaziro Y., Toh-e A., Matsumoto K. Proc. Natl. Acad. Sci. USA. 1989;86:8778–8782. doi: 10.1073/pnas.86.22.8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vermaak D., Wade P. A., Jones P. L., Shi Y. B., Wolffe A. P. Mol. Cell. Biol. 1999;19:5847–5860. doi: 10.1128/mcb.19.9.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verreault A., Kaufman P. D., Kobayashi R., Stillman B. Curr. Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 26.Neer E. J., Schmidt C. J., Nambudripad R., Smith T. F. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 27.Collins K. A., Furuyama S., Biggins S. Curr. Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 28.Yoda K., Ando S., Morishita S., Houmura K., Hashimoto K., Takeyasu K., Okazaki T. Proc. Natl. Acad. Sci. USA. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fyodorov D. V., Kadonaga J. T. Methods Enzymol. 2003;371:499–515. doi: 10.1016/S0076-6879(03)71037-4. [DOI] [PubMed] [Google Scholar]
- 30.Stein A. Methods Enzymol. 1989;170:585–603. doi: 10.1016/0076-6879(89)70066-5. [DOI] [PubMed] [Google Scholar]
- 31.Dyer P. N., Edayathumangalam R. S., White C. L., Bao Y., Chakravarthy S., Muthurajan U. M., Luger K. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 32.Roth S. Y., Allis C. D. Cell. 1996;87:5–8. doi: 10.1016/s0092-8674(00)81316-1. [DOI] [PubMed] [Google Scholar]
- 33.Spradling A. C., Stern D., Beaton A., Rhem E. J., Laverty T., Mozden N., Misra S., Rubin G. M. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips M. D., Shearn A. Genetics. 1990;125:91–101. doi: 10.1093/genetics/125.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pennetta G., Pauli D. Dev. Genes Evol. 1998;208:531–536. doi: 10.1007/s004270050212. [DOI] [PubMed] [Google Scholar]
- 36.Pile L. A., Schlag E. M., Wassarman D. A. Mol. Cell. Biol. 2002;22:4965–4976. doi: 10.1128/MCB.22.14.4965-4976.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor-Harding B., Binne U. K., Korenjak M., Brehm A., Dyson N. J. Mol. Cell. Biol. 2004;24:9124–9136. doi: 10.1128/MCB.24.20.9124-9136.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verreault A., Kaufman P. D., Kobayashi R., Stillman B. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 39.Janicki S. M., Tsukamoto T., Salghetti S. E., Tansey W. P., Sachidanandam R., Prasanth K. V., Ried T., Shav-Tal Y., Bertrand E., Singer R. H., Spector D. L. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwartz B. E., Ahmad K. Genes Dev. 2005;19:804–814. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahmad K., Henikoff S. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 42.Topp C. N., Zhong C. X., Dawe R. K. Proc. Natl. Acad. Sci. USA. 2004;101:15986–15991. doi: 10.1073/pnas.0407154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lo A. W., Magliano D. J., Sibson M. C., Kalitsis P., Craig J. M., Choo K. H. Genome Res. 2001;11:448–457. doi: 10.1101/gr.167601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore L. L., Morrison M., Roth M. B. J. Cell Biol. 1999;147:471–480. doi: 10.1083/jcb.147.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wieland G., Orthaus S., Ohndorf S., Diekmann S., Hemmerich P. Mol. Cell. Biol. 2004;24:6620–6630. doi: 10.1128/MCB.24.15.6620-6630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Black B. E., Foltz D. R., Chakravarthy S., Luger K., Woods V. L., Jr., Cleveland D. W. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 47.Henikoff S., Dalal Y. Curr. Opin. Genet. Dev. 2005;15:177–184. doi: 10.1016/j.gde.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 48.Iwaki T., Figuera M., Ploplis V. A., Castellino F. J. BioTechniques. 2003;35:482–484, 486. doi: 10.2144/03353bm08. [DOI] [PubMed] [Google Scholar]
- 49.Stein A., Mitchell M. J. Mol. Biol. 1988;203:1029–1043. doi: 10.1016/0022-2836(88)90127-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.