Abstract
A two-sided model for DNA is employed to analyze fluctuations of the spatial distribution of condensed counterions and the effect of these fluctuations on transient bending. We analyze two classes of fluctuations. In the first, the number of condensed counterions on one side of the DNA remains at its average value, while on the other side, counterions are lost to bulk solution or gained from it. The second class of fluctuations is characterized by movement of some counterions from one side of the DNA to the other. The root-mean-square fluctuation for each class is calculated from counterion condensation theory. The amplitude of the root-mean-square fluctuation depends on the ionic strength as well as the length of the segment considered and is of the order 5–10%. Both classes of fluctuation result in transient bends toward the side of greater counterion density. The bending amplitudes are ∼15% of the total root-mean-square bends associated with the persistence length of DNA. We are thus led to suggest that asymmetric fluctuations of counterion density contribute modestly but significantly toward the aggregate of thermalized solvent fluctuations that cause bending deformations of DNA free in solution. The calculations support the idea that counterions may exert some modulating influence on the fine structure of DNA.
INTRODUCTION
A prominent feature of the double-helical sugar-phosphate skeleton of DNA is the negative unit charge on each phosphate group—DNA is a strong polyelectrolyte. There exists a body of experimental, computational, and theoretical evidence suggesting a significant contribution from phosphate electrostatics to the overall force balance that determines DNA structure. For example, DNA has been shown to bend away from a phantom protein modeled by a computer as merely a protein-shaped region of low dielectric constant (1). The bend is accomplished by large-scale opening of the minor groove as interstrand phosphate-phosphate repulsions seek lower energy by moving away from the low-dielectric protein to remain immersed in a high-dielectric environment. In another indication of structural electrostatic effects, DNA deposited on mica surfaces coated with positively charged poly-L-ornithine presents itself as a wormlike chain but with dramatically enhanced flexibility, its persistence length reduced by up to a factor of five from its familiar value of ∼150 basepairs in aqueous salt solution (2). Comparable fivefold reductions of the persistence length in single-molecule stretching experiments are induced by addition of the trivalent cation cobalt hexamine in stretch conditions preventing DNA condensation (3).
Much attention has focused on the consequences of laterally asymmetric reduction of DNA phosphate charge (4–7). Chemical substitution of neutral groups for phosphates on one side of the DNA results in a bend of the double-helical axis toward the neutralized side. Positively charged ions tethered near the phosphates on one side of DNA has the same effect; the DNA bends toward the side of electrostatic neutralization of the phosphate charge. Tethering negatively charged ions near phosphates on one side of DNA forces a bend toward the opposite side. These observations are in the laboratory and subject to conceivable if unlikely interpretations other than the most transparent one. A computer, however, can unequivocally turn off the ionic charge on selected phosphate groups, and when it does so on only one side of the DNA but not the other, finds a lowest energy conformation that bends toward the side with annihilated charge (8). The detailed structural changes in helicoidal parameters that are induced by the asymmetric phosphate neutralization, and that act in concert to produce a bend, have been catalogued (8).
A long lifetime molecular dynamics trajectory features the presence of counterions near sites of deformation of the double-helical axis of DNA (9). However, it was noted that a causal relationship remains to be established. No correlation of counterion proximity and minor groove width was detected in either this trajectory or another comparable one (10). Neither simulation appears to have addressed the question of asymmetric counterion fluctuations. What structural effects might occur if a transient imbalance in the counterion distribution around DNA creates greater net charge on one side of the DNA relative to the other? The question is significant, because asymmetric fluctuations of the solvent distribution around DNA, including counterions as a component of the solvent, are the cause of the random Brownian bending movements of DNA segments that determine DNA persistence length. In turn, the finite persistence length places limits on the rigidity of the DNA molecular architecture, which must change at least slightly to accommodate the bending motions.
This article is intended to provide some information toward clarifying possible structural effects of asymmetric counterion fluctuations. In this case, we cannot compare our results with experimental data or computer simulations, neither of which currently exist as far as we are aware. It is therefore important to have some confidence in the realism of the theoretical model employed. We use the same two-sided model that originally was able to predict significant bending of DNA toward the side where its phosphate charge is abolished (11,12). We recall the success of laboratory and computational efforts to test the conclusions from the model, as discussed above. The model correctly predicts not only the existence of the bend but also provides a realistic value of the bending angle. A modeled six-basepair DNA segment with no phosphate charge on one side was calculated to bend through 9°, whereas laboratory measurements and computer simulations have observed values ranging from a few degrees to ∼21°, depending on the methods used to neutralize the phosphate charge, as discussed and referenced above.
The article is organized in the following way. We begin with specification of the model and a qualitative overview of the analysis and results. The formal part of the article is launched with a calculation of the electrostatic free energy and its minimization to find the equilibrium state of the counterions. Next, formulas are derived for the root-mean-square of the amplitude of asymmetric fluctuations in the counterion distribution. Finally, we give formulas for the transient bending angles generated by these asymmetries. Numerical results are presented alongside the formulas for the transients of counterion distribution and bending. A concluding section summarizes the results and also touches on the atomistic origin of Brownian movement, including the random bending motions that determine the persistence length of a polymer. There it is pointed out that an approach to the problem by molecular dynamics simulations would have to identify the infrequent coordinated sequences of asymmetric solvent fluctuations responsible for an observable elementary Brownian step or bend.
DESCRIPTION OF THE MODEL AND OVERVIEW OF THE ANALYSIS AND RESULTS
We employ a minimal model that, as discussed in the Introduction, has been successful in the treatment of problems related to the one of interest here (11). DNA is idealized as a pair of parallel lines, each equipped with uniformly spaced sites of unit negative charges representing the phosphate groups. The spacing between consecutive charge sites on each line is designated as b, while the perpendicular distance between the lines is a. There are P charge sites on each line, signifying a DNA of P basepairs. The spacing b has the value 0.34 nm, the step height between basepairs of B-form DNA. The distance a between the two parallel lines equals 2 nm, close to the value of the diameter of the B-form double helix. In this way, with only two structural parameters fixed by the actual structure of DNA, the model captures the reality that the electrostatic charge on one lateral face of DNA can be modulated independently of the charge on a diametrically opposite face.
Counterions condense on each of the two parallel lines. Each charge site on each line has charge −q, with q the charge on a proton, representing the unit negative valence of the DNA phosphate group. However, this charge is reduced by the factor (1 – θi) to account for reduction of the net charge by condensed counterions. The quantity θi is the number of condensed counterions per phosphate group, and the subscript i enumerates the two sides, i = 1, 2, so that there are Pθi counterions condensed on side i. The notation θ for the number of condensed counterions may not be optimal in an article that will stress bending angles, but we retain it for consistency with previous work.
The mathematical analysis of the model proceeds through counterion condensation theory. A recent discussion of the theory is available for three models, a single line of charge, a single helix, and a double helix (13). See also an earlier reference for the single line of charge (14). The two helical models are intractable for the problem at hand. The single line of charge does not contain enough information for the present problem, but a reader who wishes to follow in detail the calculations below for the model of two parallel charged lines may find the indicated references useful. Here, we point out only some qualitative features applicable to all the models.
The first part of the analysis is to find the free energy minimum, that is, the equilibrium state. There are two quantities that must be determined, the number of condensed counterions, and the internal free energy, or partition function, of the condensed layer of counterions. The former is determined mathematically by removing a divergence in the free energy. The physical meaning of removal of the divergence is that the favorable entropy of dissociation of counterions from the polyion is balanced by the unfavorable energy of dissociation. The entropy of dissociation, as in any thermodynamic system, is logarithmic in concentration. The energy of dissociation, unlike most systems, is also logarithmic in concentration due to the logarithmic potentials of the essentially cylindrical polyion models. For B-form DNA the number of undissociated, or condensed, counterions equals 0.76 times the number of phosphates (14), regardless of the model used (13).
The predicted number of condensed counterions has been verified repeatedly, most recently by both Monte Carlo and molecular dynamics simulations (9,15). An outer inflection point in a plot of number of counterions as a function of distance from the simulated DNA has been observed (15). The physical meaning of the inflection point is that there is a spatial separation, a gap, between the condensed layer of counterions and the more diffuse counterion cloud that lies further out. As the gap is crossed by a variable point that moves out from the polyion, counterions do not accumulate, hence the inflection. The physical distinction between condensed and diffuse counterion layers is visually obvious in computer graphics (9,15). In the molecular dynamics simulations, the number of counterions lying inside the inflection point (dubbed “the Manning radius” by the authors) is observed numerically to equal 0.76 times the number of phosphates (9,15).
It might be thought that the calculation of the internal free energy of the condensed layer would present difficulties. However, it is obtained in a most straightforward way, once the logarithmic divergence is removed, from the obvious minimization condition that the derivative of total free energy with respect to number of condensed counterions vanish. No new parameters are introduced. The free energy of the condensed layer depends on the model used. In a simple model, the condensed counterions can be portrayed as freely translating within a cylindrical shell surrounding a DNA cylinder. The thickness of the shell predicted by the theory, with the introduction of no new parameters, then equals 7 Å. The most recent of the molecular dynamics computations discussed above observes the inflection point that marks the boundary of the condensed layer to be located at 9 Å from the surface of the simulated all-atom DNA (9).
The result for the equilibrium state of the two-sided model required in this article is as expected. The total number of condensed counterions is 76% of the total number of phosphate groups, and the condensed counterions are equally distributed on each of the two identical sides of the model DNA. The internal free energy of the condensed counterions is not presented, as it is not needed for the problem at hand.
In a next stage of the calculation, thermal fluctuations of the condensed counterion distribution away from uniformity are considered. For example, due to a fluctuation, the number of condensed counterions on one side of the DNA may transiently deviate from 76% of the number of phosphate groups on that side, while the number of counterions condensed on the other side remains at its equilibrium value. The calculation is an application of textbook statistical mechanics. No new parameters are introduced into the model at this stage. We conclude that root-mean-square asymmetric fluctuations away from the equilibrium number 0.76 of condensed counterions are ∼5–10%. For example, the number of counterions condensed on one face of the DNA might be 72% of the number of phosphates on that face, while the number on the other side might be 80% of the number of phosphates.
In the equilibrium state the electrostatic forces on each side of the DNA are in balance, since the net charge on each side (phosphate charge minus charge of condensed counterions) is the same. But if the net charge on the two sides is transiently different, the balance of forces is upset, and there will be a tendency of the DNA to tilt, or bend, toward the side with the smaller net charge. We are able to calculate the force imbalance (more precisely, the electrostatic bending torque) as an application of counterion condensation theory without the introduction of new assumptions or parameters.
The tendency to bend produced by the counterion imbalance is resisted by the elastic stiffness of DNA against bending. If the stiffness is sufficiently great, the bend will be insignificant. For an estimate of the bending amplitudes using standard elasticity theory, we need the numerical value of the Hooke's Law bending constant B for DNA. There is a direct correlation between B and the persistence length of DNA through a well-known formula of polymer theory. We use the measured value of the persistence length. Our calculations are in 0.1 M aqueous NaCl, and measurements of persistence length at this ionic strength from different laboratories converge on a consensus value of ∼150 DNA basepairs, or 50 nm. Finally, we obtain our main result, that transient electrostatic force imbalances created by asymmetric fluctuations of the distribution of counterions can generate modest but structurally significant transient bending deformations of a few degrees angular amplitude.
ELECTROSTATIC FREE ENERGY AND THE EQUILIBRIUM STATE
Let Gα be a generic free energy component, and reduce it to dimensionless form gα = Gα/kBT, where kBT is Boltzmann's constant times temperature. The first two of these components (13,14) are the free energies involved in transferring Pθi counterions from bulk solution, where the counterion concentration is c, to the condensed counterion layer on side i, i = 1, 2,
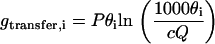 |
(1) |
Here, Q is an internal partition function for the condensed counterions, which we take to be the same for both sides. The assumption that short-range interactions among condensed counterions can be neglected in the present analysis allows us to take Q as independent of the condensed fractions θi. Its units are chosen as cm3, and a simple physical interpretation identifies it as the local volume of the condensed layer per charge site (14). The factor 1000 converts to liters, since c is in units of molarity.
The next two free energy components (13,14) are the charge-charge repulsions within each side i, i = 1, 2,
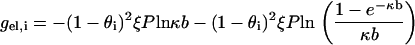 |
(2) |
A quantity ξ has been introduced in this formula. It is a measure of charge density on each side (identical for both sides), and it is dimensionless. Its definition is
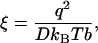 |
(3) |
where D is the dielectric constant of bulk solvent. Notice that ξ is the ratio of the Bjerrum length q2/DkBT to the charge spacing b of each side. The Bjerrum length is the distance at which the electrostatic interaction energy of two point unit charges equals thermal energy kBT. It equals ∼0.71 nm in water at room temperature. For B-form DNA, ξ is approximately equal to 2.1. Note again that ξ is a reduced charge density for each side; the familiar value 4.2 for the single-line electrostatic model of DNA (14) is given by 2ξ in this article.
The Debye screening parameter κ has also been introduced in Eq. 2. Its reciprocal is the Debye screening length of the univalent/univalent salt solution (like NaCl) in which the DNA is immersed. The numerical value of 1/κ is ∼0.96 nm in aqueous 0.1 M NaCl at room temperature. Values at other salt concentrations may be calculated from the fact that κ is proportional to the square-root of salt concentration c. Notice that in assumed conditions of excess salt over DNA concentration, the salt concentration and the counterion concentration (which appears in Eq. 1) are both equal to c. The general formula for κ is readily available in textbooks covering Debye-Hückel theory.
Eq. 2 itself is obtained in the following way (13,14). The repulsive screened Coulomb interaction energy in units of kBT between a pair of charge sites on side i separated by a distance that is an integral multiple nb of the spacing b is (1 – θi)2ξ exp( – κnb)/n, since each site bears net absolute charge (after counterion condensation) (1 – θi)q. Equation 2 reflects summation over all pairs from nearest neighbors on to infinite separation. Recalling that κ ∼ c1/2, we recognize that the series is logarithmically divergent as counterion concentration c → 0. The divergence is separated out as the first term on the right-hand side of Eq. 2, leaving as the second term a small concentration correction that converges to zero in the limit c → 0.
The next and final free energy component describes the electrostatic coupling between the two sides (16),
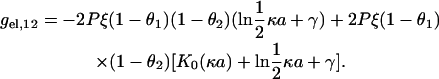 |
(4) |
This equation is obtained by summing screened Coulomb potentials between all charge pairs, one member of the pair from side 1 with net absolute charge (1 – θ1)q, the other member of the pair from the side 2 with net absolute charge (1 – θ2)q. The distance between sides is a (for DNA, it is the diameter of the double helix), and the modified Bessel function of the second kind K0(κa) appears when the sum is replaced by an integral. The Bessel function is logarithmically divergent as c → 0,
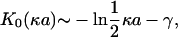 |
(5) |
where γ is Euler's constant, γ = 0.5772…, and the divergence is separated out as the first term on the right-hand side of Eq. 4, while the second term is a concentration correction that converges to zero as c → 0.
The total reduced free energy g is the sum of the components,
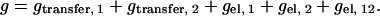 |
(6) |
Each of the five components of g contributes to a divergent overall ln c term,
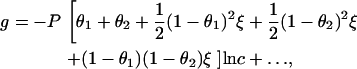 |
(7) |
where we have again recalled that κ ∼ c1/2, and where the omitted terms converge as c → 0.
Following the standard theory of counterion condensation (13,14), we take a next step of differentiating with respect to θ1,
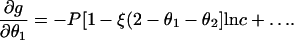 |
(8) |
To have an equilibrium state in dilute solution, we remove the logarithmic singularity by setting the bracketed factor to zero,
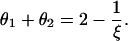 |
(9) |
Working with θ2 instead of θ1 gives exactly the same equation, so thus far, we have only a single equation for the two unknowns, the equilibrium values of θ1 and θ2. There is, however, an obvious second condition for the equilibrium (average) values, namely, that they must be equal, since the two sides are identical. With 〈θ〉 designating the equal average values of θ1 and θ2, Eq. 9 is solved as
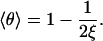 |
(10) |
Counterion condensation emerges from the competition of long-range energy and entropy (14). Eq. 10 is therefore an expected result, as it states that the average condensation fraction for two parallel lines at a fixed distance, each of charge density ξ, is the same as for a single line of charge sites with twice the charge density.
For a complete equilibrium state, it is necessary that the omitted convergent expression in Eq. 8 also equal zero, as well as the corresponding expression in ∂g/∂θ2. This requirement places a self-consistency condition on the internal partition function, which we do not pursue here, since there is no explicit need for it in subsequent development.
ASYMMETRIC COUNTERION FLUCTUATIONS
Class I counterion fluctuations
We define Class I fluctuations as those for which θ1 fluctuates while θ2 remains fixed at its average value. The physical event is that some of the counterions condensed on side 1 are transiently lost to bulk, or additional counterions from bulk condense on side 1.
We need the second derivative of g with respect to θ1 with θ2 fixed,
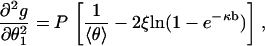 |
(11) |
where it is understood that both θ1 and θ2 have been evaluated at their common average value 〈θ〉, Eq. 10.
Let Δg be the deviation g(θ1) – g(〈θ〉) of the free energy from its average, and expand Δg out to quadratic order with vanishing first derivative,
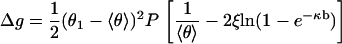 |
(12) |
Fluctuation of θ1 at fixed θ2 constitutes a single degree of freedom, and the thermal average value of Δg equals 1/2. Averaging both sides of Eq. 12 then produces the mean-square fluctuation of θ1 as
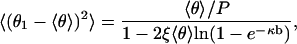 |
(13) |
where 〈θ〉 is given by Eq. 10.
Table 1 gives some calculated values of the root-mean-square fluctuation of θ1 as a function of segment length N in DNA basepairs (N = P, the number of charges on each side of our model DNA). For example, at 0.1 M salt concentration, a 10-basepair segment has 76 ± 12% of the phosphate charge on one side compensated by condensed counterions, whereas, if the statistical count is performed on a longer 30-basepair segment, the average charge compensation on one side with fluctuation limits is 76 ± 7%.
TABLE 1.
Class I counterion fluctuations at 0.1 M NaCl
| N, bp | δθ |
|---|---|
| 10 | 0.125 |
| 30 | 0.072 |
| 50 | 0.056 |
Class II counterion fluctuations
In this family of fluctuations, some of the counterions condensed on one side of the DNA diffuse over to the other side. The total number of condensed counterions Pθ1 + Pθ2 remains constant at its average value 2P〈θ〉. In the reduced free energy g(θ1, θ2), we may therefore make the substitution θ2 = 2〈θ〉 – θ1, calculate the second derivative with respect to θ1, and proceed as before. The result for the mean-square fluctuation,
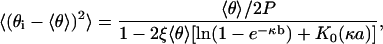 |
(14) |
may be applied to either side i, but it must be remembered that the fluctuations on the two sides are coupled; if counterions have moved from side 2 to side 1, then θ1 – 〈θ〉 = 〈θ〉 – θ2.
Numerical evaluation of this coupled fluctuation as the root-mean-square (square-root of right-hand side of Eq. 14) is illustrated by the entries in Table 2. As examples, charge compensation by condensed counterions on one side of a 10-basepair segment can increase in a root-mean-square fluctuation from 76% to 85%, while on the other side charge compensation has fallen to 67% (i.e., δθ = ±0.09). For a 30-basepair segment, the corresponding fluctuation is 81% on one side and 71% on the other.
TABLE 2.
Class II counterion fluctuations at 0.1 M NaCl
| N, bp | δθ |
|---|---|
| 10 | 0.092 |
| 30 | 0.053 |
| 50 | 0.041 |
BENDING FLUCTUATIONS
In the previous section, we calculated root-mean-square fluctuations in the condensed counterion distribution that result in momentary asymmetry of the charge density in our two-sided DNA model. Here we estimate the amplitudes of the bending fluctuations caused by the counterion asymmetries (or, more precisely, by a succession of concerted asymmetries (17)).
The starting point is a calculation of the stretching force on each of the two DNA sides due to phosphate-phosphate repulsion (11). Let G be the total unreduced polyelectrolyte free energy from the previous section (G = kBTg, g from Eq. 6). The electrostatic force that stretches side i, i = 1, 2, is
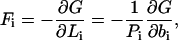 |
(15) |
where in the derivatives the length Lj and the charge spacing bj of the other side, j ≠ i, are held fixed. In the formula for G in the previous section, the spacings b1 and b2 must be made explicit before setting each of them to their common value b (but the number of charges P1 and P2 on each side have their common value P throughout the stretching process, even if we make them explicit for the sake of physical transparency). Thus, in Eq. 2, for the intrastrand repulsive free energy we replace the factor ξP by ξiPi, i = 1, 2, and we symmetrize the coupling electrostatic free energy of Eq. 4 with replacement of ξP by (1/2)(P1ξ2 + P2ξ1). We then find that
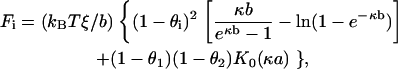 |
(16) |
where the values ξ and b common to the two sides have been resubstituted after differentiation.
The stretching forces Fi(θ1, θ2) in Eq. 16 pertain to arbitrary binding fractions θi. The forces needed for the bending calculation are the excess forces ΔFi = Fi(θ1, θ2) – Fi(〈θ〉, 〈θ〉) relative to the equilibrium values from Eq. 10,
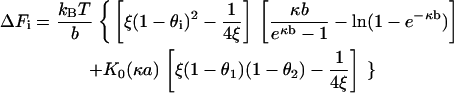 |
(17) |
Class IA bending fluctuations
Bending fluctuations of Class IA are defined to be those caused by a certain subfamily of Class I counterion fluctuations. In Class I counterion fluctuations, the bulk solution is a source or sink of counterions for side 1 of the DNA model, while the number of counterions on side 2 remains fixed at its equilibrium value. Class IA bending fluctuations are caused by transient increase of counterions condensed on side 1 (bulk solution is a source of condensed counterions for side 1), thus causing excess neutralization of the phosphate charge on side 1. The electrostatic forces stretching both sides are therefore diminished, but asymmetrically, with the weakening of the stretch being much more substantial on side 1 than on side 2. The net result is equivalent to the effect produced by asymmetrically placed compressive loads on the DNA that tend to bend it toward side 1.
Let w1 be the compression load on side 1. As a positive quantity, it equals the absolute value of ΔF1 from Eq. 17 with i = 1, θ1 greater than 〈θ〉, and θ2 = 〈θ〉 = 1 – (1/2ξ). In writing the following expression, we simplify by dropping the subscript on θ1,
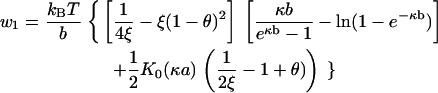 |
(18) |
For the smaller compression load w2 on side 2, we have the absolute value of ΔF2, again from Eq. 17 but with i = 2, and again with θ1 greater than 〈θ〉 and θ2 = 〈θ〉 = 1 – (1/2ξ). The subscript is dropped from θ1,
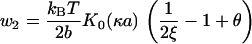 |
(19) |
A rod subjected to longitudinal compressive loads on both sides of its central axis clearly must bend toward the side subject to the heavier load. The rod assumes the shape of a portion of a sine wave with maximum curvature halfway along its length. The mechanical problem has been analyzed previously, resulting in a formula for the radius R of maximum curvature (11),
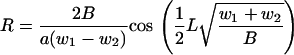 |
(20) |
The factor 2 appears here, because the eccentricity, that is, the offset distance of the loads from the central axis, is equal to half the diameter a of the rod. The loads w1 and w2 are given by Eqs. 18 and 19, respectively, which in turn are functions of the nonequilibrium number θ of counterions condensed on the inner side of the bend. The numerical value of the Hooke's Law bending modulus B, which causes elastic resistance to the bend, is obtained from a well-known formula (18) as the product kBT times the experimentally determined persistence length of DNA, ∼150 bp, or 50 nm. We evaluate R when θ exceeds its equilibrium value by a root-mean-square fluctuation, given by Eq. 13.
It is easier to grasp the geometrical meaning of an angle than of a radius of curvature, so we also compute the total bending angle α = L/R (angle between the directions at the two ends of the rod segment) under the assumption that the radius of curvature R given by Eq. 20 is uniform along the rod length. This approximation to the angle through which the deformed rod segment is bent is an overestimate. Table 3 lists some values of α as a function of the length of the segment for DNA parameters with length L converted to number of basepairs N through L = (N – 1)b. Table 3 also lists the angle per basepair, which is a direct measure of the curvature of the segment. The curvature decreases slightly as the number of basepairs in the segment increases.
TABLE 3.
Class IA bending fluctuations at 0.1 M NaCl
| N | α, deg | α/N, deg/bp |
|---|---|---|
| 10 | 1.7 | 0.17 |
| 30 | 3.9 | 0.13 |
| 50 | 6.1 | 0.12 |
Class IB bending fluctuations
In class IA fluctuations of the condensed counterion distribution, bulk solution is a source of additional counterion condensation on side 1, while the number of counterions condensed on side 2 remains invariant. Class IB fluctuations are characterized by bulk solution acting as a sink for side 1. Thus, side 1 relinquishes counterions to bulk, so that θ1 becomes less than 〈θ〉, while θ2 remains equal to the average value 1 – (1/2ξ). There is a deficit of counterions on side 1, so the phosphate-phosphate stretching force on side 1, and to a lesser extent on side 2 (which interacts with side 1), increases over its average value. The loads w1 and w2 are now tensile (stretching). They are equal to the absolute values of ΔF1 and ΔF2, respectively; and hence, with the Class IB conditions on θi, to the negatives of the right-hand sides of Eqs. 18 and 19.
The rod is subjected to longitudinal tensile loads w1 and w2 eccentrically placed, respectively, on either side of the central axis. The resulting deformation is the same as would be produced by a tension w1 + w2 along the central axis and a torque w1 – w2 on the arm a/2. The centered tension does not contribute to bending, and we neglect its effect (the rod is taken as inextensible). The torque generates a uniform bend with radius of curvature R into side 2 (where the extent of phosphate neutralization is greater),
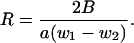 |
(21) |
Table 4 shows the corresponding bending angle α, as well as the bending angle per basepair (directly correlated with curvature) for DNA segments of varying lengths under root-mean-square counterion fluctuations from Eq. 13. The table also lists values of the ratio of the bending angle α to the root-mean-square bending angle αp that is dictated by the experimentally determined persistence length of DNA (150 bp) according to a standard formula (18). In other words, we are interested in comparing α to the overall bending fluctuation αp that is caused by the totality of equilibrated interactions with solvent, including both counterions (condensed and diffuse atmospheres) and water molecules. We did not make this comparison in Table 3 for Model IA, because the bending in Model IA is actually a buckling mode, in the sense that the direction of the segment at its two ends does not change.
TABLE 4.
Class IB bending fluctuations at 0.1 M NaCl
| N | α, deg | α/N, deg/bp | 100 × (α/αp), % |
|---|---|---|---|
| 10 | 2.9 | 0.29 | 14 |
| 30 | 4.9 | 0.16 | 14 |
| 50 | 6.2 | 0.12 | 13 |
Class II bending fluctuations
In Class II counterion fluctuations some of the counterions condensed on side 2 diffuse over to side 1, producing a counterion deficit on side 2 and an excess on side 1. Therefore an excess compression w1 emerges on side 1, and an excess tension w2 on side 2, where as usual these quantities are defined to be positive. The compression w1 on side 1 is given by the negative of the right-hand side of Eq. 17 with i = 1. The tension on side 2 is given by the right-hand side of Eq. 17 with i = 2. For the condensed counterion fractions θ1 and θ2, we use their root-mean-square values; for i = 1, 2,
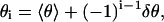 |
(22) |
where δθ equals the positive square-root of the right-hand side of Eq. 14.
The longitudinal stresses w1 and w2 combine to produce a tension w2 – w1 along the central axis. Indeed, we have verified numerically (not shown) that the tension w2 is greater than the compression w1, so that the net force along the center is a stretch. As in Class IB, it does not contribute to bending, hence is not further considered. Additionally, a torque w2 + w1 acts on the arm a/2 to generate a pure bend with uniform radius of curvature R into side 1,
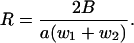 |
(23) |
Table 5 shows the corresponding bending angle α for DNA segments of varying lengths. The bend per basepair is included as a direct measure of curvature, as well as comparison with the overall bending angle calculated from the persistence length.
TABLE 5.
Class II bending fluctuations at 0.1 M NaCl
| N | α, deg | α/N, deg/bp | 100 × (α/αp), % |
|---|---|---|---|
| 10 | 3.4 | 0.34 | 17 |
| 30 | 6.2 | 0.21 | 17 |
| 50 | 8.1 | 0.16 | 18 |
DISCUSSION
We set out in this article to determine whether fluctuations in the distribution of condensed counterions could cause a force imbalance sufficient to “tip over” an otherwise stiff DNA segment and hence to bend it. We have concluded that asymmetric counterion fluctuations, for example, fluctuations whereby some counterions move from one side of the DNA to the other, can indeed cause significant bending. Specifically, Tables 3–5 indicate that the three models of fluctuations cause bending of a few degrees.
We have also compared the bending angles caused by counterion fluctuations to the overall root-mean-square bending amplitude corresponding to the experimentally known persistence length of DNA. Tables 4 and 5 show in the framework of our models that counterion fluctuations can contribute ∼15% of the total statistical bending amplitude that determines the persistence length. Of course, we have not tried to determine the relative frequencies of counterion and water fluctuations, so the effect of the former may be an overestimate. On the other hand, we have calculated only the effect of condensed counterions, and have not considered fluctuations in the residual diffuse atmosphere, so our estimate of 15% may be less than the total counterion contribution. Perhaps the main point is that counterions do seem to exert forces on the DNA structure sufficient to affect it significantly, at least insofar as the structural features that determine bending.
Finally, we comment on the possibility of more detailed atomistic approaches through molecular dynamics simulations. The bending considered here results from concerted movements of a few counterions. For example, the value δθ = 0.092 in Table 2 for a 10-bp segment means that two counterions move from one side of the segment to the other. Depending on the comparative timescales of the lifetime of the counterion fluctuation and the bending movement, several such counterion fluctuations in succession may be necessary before a perceptible bend occurs (see the discussion of Brownian motion in (17)). The infrequency of these relatively rare events should be kept in mind.
References
- 1.Elcock, A. H., and J. A. McCammon. 1996. The low dielectric interior of proteins is sufficient to cause major structural changes in DNA on association. J. Am. Chem. Soc. 118:3787–3788. [Google Scholar]
- 2.Podestà, A., M. Indrieri, D. Brogioli, G. S. Manning, P. Milani, R. Guerra, L. Finzi, and D. Dunlap. 2005. Positively charged surfaces increase the flexibility of DNA. Biophys. J. 89:2558–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumann, C. G., V. A. Bloomfield, S. B. Smith, C. Bustamante, M. D. Wang, and S. M. Block. 2000. Stretching of single collapsed DNA molecules. Biophys. J. 78:1965–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strauss, J. K., and L. J. Maher. 1994. DNA bending by asymmetric phosphate neutralization. Science. 266:1829–1834. [DOI] [PubMed] [Google Scholar]
- 5.Maher, L. J. 1998. Mechanisms of DNA bending. Curr. Opin. Chem. Biol. 2:688–694. [DOI] [PubMed] [Google Scholar]
- 6.Williams, L. D., and L. J. Maher. 2000. Electrostatic mechanism of DNA deformation. Annu. Rev. Biophys. Biomol. Struct. 29:497–521. [DOI] [PubMed] [Google Scholar]
- 7.Hardwidge, P. R., J. Wu, S. L. Williams, K. M. Parkhurst, L. J. Parkhurst, and L. J. Maher. 2002. Biochemistry. 41:7732–7742. [DOI] [PubMed] [Google Scholar]
- 8.Kosikov, K. M., A. A. Gorin, X. J. Lu, W. K. Olson, and G. S. Manning. 2002. Bending of DNA by asymmetric charge neutralization: all-atom energy simulations. J. Am. Chem. Soc. 124:4838–4847. [DOI] [PubMed] [Google Scholar]
- 9.Ponomarev, S. Y., K. M. Thayer, and D. L. Beveridge. 2004. Ion motions in molecular dynamics simulations on DNA. Proc. Natl. Acad. Sci. USA. 101:14771–14775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Várnai, P., and K. Zakrzewska. 2004. DNA and its counterions: a molecular dynamics study. Nucleic Acids Res. 32:4269–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manning, G. S., K. K. Ebralidse, A. D. Mirzabekov, and A. Rich. 1989. An estimate of the extent of folding of nucleosomal DNA by laterally asymmetric neutralization of phosphate groups. J. Biomol. Struct. Dyn. 6:877–889. [DOI] [PubMed] [Google Scholar]
- 12.Manning, G. S. 1995. The theory of DNA bending. Science. 268:188. [DOI] [PubMed] [Google Scholar]
- 13.Manning, G. S. 2002. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Biophys. Chem. 101–102:461–473. [DOI] [PubMed] [Google Scholar]
- 14.Manning, G. S. 1978. Electrostatic free energy of the DNA double helix in counterion condensation theory. Quart. Rev. Biophys. 11:179–246. [DOI] [PubMed] [Google Scholar]
- 15.Young, M. A., B. Jayaram, and D. L. Beveridge. 1997. Intrusion of counterions into the spine of hydration in the minor groove of B-DNA: fractional occupancy of electronegative pockets. J. Am. Chem. Soc. 119:59–69. [Google Scholar]
- 16.Ray, J., and G. S. Manning. 2000. Formation of loose clusters in polyelectrolyte solutions. Macromolecules. 33:2901–2908. [Google Scholar]
- 17.Nelson, P. Biological Physics. W. H. Freeman, New York, 2004.
- 18.Grosberg, A. Y., and A. R. Khokhlov. 1994. Statistical Physics of Macromolecules. AIP Press, New York.


