Abstract
Alveolar bone destruction is a characteristic feature of periodontitis. Treponema denticola is known to be involved in periodontitis. To elucidate the role of T. denticola in alveolar bone destruction in periodontitis, the effects of lipooligosaccharide (LOS) from T. denticola on osteoclast formation and on expression of osteoclast differentiation factor (ODF) and osteoprotegerin (OPG) mRNAs were examined in a coculture system by using mouse calvaria and bone marrow cells. In addition, the effect of T. denticola LOS on expression of matrix metalloproteinases (MMPs), which are involved in bone resorption, was estimated in mouse calvaria-derived osteoblastic cells. When the mouse calvaria and bone marrow cells were challenged with LOS (0.1 to 10 μg/ml) for 4 days, the number of tartrate-resistant acid phosphatase-positive multinucleated cells increased in a dose-dependent manner. The expression of ODF mRNA increased, while OPG mRNA expression decreased. Polymyxin B changed the effect of LOS (10 μg/ml) on ODF and OPG mRNA expression to the control level. LOS (10 μg/ml) stimulated prostaglandin E2 (PGE2) production in the cocultures. Adding indomethacin, an inhibitor of prostaglandin synthesis, resulted in a reduction in the number of osteoclasts induced by LOS and eliminated the effect of T. denticola LOS on ODF and OPG mRNA expression. T. denticola LOS increased the levels of mRNAs encoding MMP-3, -8, -9, -10, -13, and -14. Expression of one of these mRNAs, MMP-9 mRNA, was significantly induced by T. denticola LOS. These findings suggest that LOS from T. denticola stimulates osteoclastogenesis and MMP expression. Up-regulation of ODF and down-regulation of OPG by a PGE2-dependent mechanism were involved in the osteoclastogenesis induced by T. denticola LOS.
Osteoclasts are multinucleated cells with bone-resorbing activity and play a crucial role in bone resorption. Osteoclast formation requires the presence of osteoblast or stromal cells (39). These cells express the osteoclast differentiation factor (ODF) (also known as a receptor activator of the nuclear factor-κB [RANK] ligand) that promotes osteoclastogenesis (21, 48). The osteoclast precursors that express RANK (the receptor for ODF) recognize ODF through a cell-to-cell interaction with osteoblasts and differentiate into osteoclasts. Osteoprotegerin (OPG), which is also secreted by osteoblast lineage cells, is a soluble decoy receptor that neutralizes the biological activity of ODF (34, 42, 47). Osteoclastogenesis is controlled by multiple factors, such as 1α,25-dihydroxyvitamin D3 [1α,25(OH)2D3], parathyroid hormone (PTH), prostaglandin E2 (PGE2), and interleukin-1 (IL-1) (26, 38). The regulation of ODF-OPG expression by these agents (1, 12-14, 25, 44, 47, 48) suggests that the effects of these factors on bone resorption may be mediated through control of ODF and/or OPG production and that osteoclast formation is determined principally by the ratio of ODF to OPG. Therefore, a shift to a higher ratio of ODF to OPG may be a major cause of bone loss in many metabolic disorders, including osteoporosis and periodontitis (15). Recently, it was reported that the ODF-RANK interaction is not the sole pathway that causes osteoclast progenitors to differentiate into osteoclasts (19). Tumor necrosis factor alpha (TNF-α), which is involved in bone resorption, can be substituted for the ODF to induce osteoclast differentiation.
The matrix metalloproteinases (MMPs) are a family of structurally and functionally related enzymes that are responsible for the proteolytic degradation of the extracellular matrix components. More than 20 different MMPs have been identified. These proteins can be classified into the following subgroups based on their substrate specificities and structural homologies: collagenase (MMP-1, -8, and -13), gelatinase (MMP-2 and -9), stromelysin (MMP-3, -10, and -11), membrane-type MMPs (MMP-14, -15, -16, -17, -23, -24, and -25), and other MMPs, including matrilysin (MMP-7) and metalloelastase (MMP-12) (11, 22, 36). Previous reports suggested that MMPs are involved in degrading the bone matrix. Various MMPs, including MMP-2, -3, -9, -11, -12, -13, and -14, are expressed in the osteoblasts. Bone resorption factors, such as PTH, 1α,25(OH)2D3, IL-1, IL-6, TNF-α, and PGE2, regulate the production of various MMPs in the osteoblasts (20, 40). Furthermore, a nonselective MMP inhibitor neutralized bone destruction stimulated by these bone resorption factors (20, 40, 41). This strongly suggests that the bone resorption factors function at least in part through MMP induction. Bone resorption first requires the osteoblasts to release collagenase to remove the nonmineralized organic matrix that covers the bone surfaces. Osteoclasts are then chemotactically attracted to the resorption site, where they settle onto the calcified matrix (2). Taken together, these reports suggest that the MMPs may be important for the bone resorption process.
Periodontitis is an inflammatory disease, and loss of alveolar bone is a hallmark of this disease (32). Treponema denticola is one of the bacteria which have been implicated in the etiology of periodontitis (6, 9, 35). This bacterium has multiple virulence factors that include adhesins, proteolytic and hydrolytic enzymes, cytopathic activity, and immunomodulation (3, 9). For the virulence factor associated with bone resorption, it was reported that the outer membrane of the bacterium increased Ca2+ release in an organ culture of fetal radii and ulnae (10). Although this suggested that the heat-stable material of T. denticola in the outer membrane might be involved in bone resorption, the components of this bacterium which stimulate bone resorption and the underlying mechanism have not been studied. Recently, we reported that whole-cell sonicates of Treponema lecithinolyticum associated with aggressive periodontitis induced osteoclast formation (5). In this case, heat-stable components were involved.
In this study, lipooligosaccharide (LOS) from T. denticola was isolated, and its effects on osteoclastogenesis and on expression of both ODF and OPG mRNAs were investigated in a coculture system consisting of mouse calvaria and bone marrow cells in order to determine the role of T. denticola in bone resorption. In addition, regulation of the expression of several MMPs by T. denticola LOS was estimated in mouse calvaria-derived osteoblastic cells. This is the first report showing that LOS from T. denticola induces osteoclast formation and that this process is dependent on up-regulation of ODF expression and down-regulation of OPG expression through PGE2 synthesis. We also found that T. denticola LOS increased the expression of the mRNAs of several MMPs in osteoblastic cells.
MATERIALS AND METHODS
Materials.
Mice (ICR strain) were obtained from Bio Korea Co. (Seoul, Korea). The α minimum essential medium (α-MEM), bovine serum albumin, and heat-inactivated fetal bovine serum (FBS) were purchased from GIBCO BRL (Grand Island, N.Y.). Indomethacin, polymyxin B, and a tartrate-resistant acid phosphatase (TRAP) (a marker of osteoclasts) staining kit were obtained from Sigma (St. Louis, Mo.).
Preparation of T. denticola sonicates.
T. denticola ATCC 33521 was cultured anaerobically in OMIZ-PAT broth for 3 to 5 days, as described previously (46). The bacterial cells were harvested by centrifugation at 5,000 × g for 10 min at 4°C. The cells were then washed three times with phosphate-buffered saline. The bacterial cells were disrupted for 5 min with an ultrasonic processor (Sonic Dismembrator; Fisher Scientific, Pittsburgh, Pa.) by using an output power of 8 W with 20-s intervals. The cell debris was removed after centrifugation at 15,000 × g for 5 min at 4°C, and the supernatant was collected. The protein concentrations were determined by using a Coomassie brilliant blue protein assay reagent (Pierce, Rockford, Ill.).
Isolation of LOS.
T. denticola LOS was isolated by the method described by Walker et al. (45). The bacteria were cultured and harvested as described above. The cell pellets were repeatedly frozen and thawed for 40 cycles and then centrifuged at 6,000 × g for 10 min to remove the cellular debris. The supernatant was centrifuged at 36,000 × g for 30 min at 4°C, and the resulting pellet containing the membrane fraction was washed twice in 60 ml of 0.05 M Tris-HCl (pH 7.2) and suspended in Tris-HCl. The detergent-soluble outer membrane fraction was obtained by extracting the membrane fraction with 1% Zwittergent 3.14 (Calbiochem Co., La Jolla, Calif.). To obtain the LOS, the detergent-soluble fraction was digested with proteinase K (50 μg/ml) at 37°C overnight. Two volumes of 0.375 M MgCl2 in 95% ethanol (−20°C) was added, and the preparation was kept at −20°C for 40 min and then centrifuged at 15,000 × g for 20 min. The pellet was suspended in a solution containing 2% sodium dodecyl sulfate, 0.1 M EDTA, and 10 mM Tris-HCl (pH 8.0). The proteinase K digestion-ethanol precipitation procedure was repeated twice. The LOS was purified by centrifugation at 48,000 × g for 2 h at 4°C and suspended in a small volume of distilled water. After the LOS was heated at 90°C for 30 min, it was quantified by lyophilization, and the weight was measured. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (15% polyacrylamide gel) of the LOS was performed, and the gels were silver stained.
Preparation of primary calvaria and bone marrow cells.
The osteoblastic cells were isolated from the calvariae of 1- to 2-day-old ICR mice as previously described (5). The calvariae were digested in 10 ml of α-MEM containing 0.2% collagenase (Wako Pure Chemicals, Osaka, Japan) and 0.1% dispase (GIBCO BRL) for 20 min at 37°C with vigorous shaking and then centrifuged at 1,500 × g for 5 min. The first supernatant was discarded, another 10 ml of the collagenase-dispase enzyme solution was added, and the preparation was incubated for 20 min. The digestion procedure was repeated four times, and the cells isolated by the last three digestions were combined as an osteoblastic cell population. They were cultured in α-MEM containing 10% FBS and an antibiotic solution (100 U of penicillin per ml, 100 μg of streptomycin per ml, 25 μg of amphotericin B per ml) and used for the coculture system. The bone marrow cells were collected from 5- to 8-week-old mice. The ends of the tibiae and femurs were removed, and each marrow cavity was flushed by slowly injecting medium at one end with a 25-gauge needle. The marrow cells were washed and used for the coculture.
Osteoclast formation assay.
The isolated calvaria cells were seeded at a concentration of 106 cells per 10-cm culture dish and grown to confluence. The cells were then detached from the culture dishes with trypsin-EDTA (GIBCO BRL). Subsequently, the cells (1 × 104 cells/well) were cocultured with the bone marrow cells (1 × 105 cells/well) in α-MEM containing 10% FBS in 48-well plates (Corning Inc., Corning, N.Y.). The culture volume was adjusted to 400 μl per well with α-MEM containing 10% FBS. Either a bacterial sonicate or LOS was added to each coculture with or without polymyxin B or indomethacin after the medium was exchanged on day 3. The coculture was then maintained for an additional 4 days. Osteoclast differentiation was monitored by using a TRAP staining kit according to the manufacturer's instructions. TRAP-positive multinucleated cells having more than three nuclei per well were counted as osteoclasts. ODF and OPG mRNA expression in the cocultures was determined after the mRNA was isolated with TRIzol reagent (Life Technologies, Inc., Grand Island, N.Y.).
Osteoblastic cell cultures.
To analyze expression of mRNAs of MMPs and tissue inhibitors of metalloproteinases (TIMPs), osteoblastic cells isolated from mouse calvariae were seeded into 24-well dishes at a density of 8 × 104 cells/well in 800 μl of α-MEM containing 10% FBS. When the cells had grown to 80% confluence, the medium was changed to α-MEM containing 1 mg of bovine serum albumin per ml. After incubation for 12 h, the cells were exposed to T. denticola LOS alone or in combination with polymyxin B for 48 h. The mRNA was isolated from the cultured osteoblastic cells by using TRIzol reagent according to the manufacturer's protocol (Life Technologies, Inc.).
RT-PCR.
Expression of ODF, OPG, MMP, and TIMP mRNAs was determined by reverse transcription (RT)-PCR. Total RNA (1 μg) from nontreated or treated cells was used as a template for cDNA synthesis in a 20-μl reaction mixture performed with an RT kit (CLONTECH, Palo Alto, Calif.) used according to the manufacturer's instructions. The RNA (1 μg) and oligo(dT)18 primers (1 mM) were denatured at 70°C for 5 min and incubated for 1 to 2 min on ice. The denatured RNA and oligo(dT)18 primers were added to the reaction mixture (1 U of Moloney murine leukemia virus reverse transcriptase per μl, 1× reaction buffer, 500 μM dATP, 500 μM dCTP, 500 μM dGTP, 500 μM dTTP, 20 U of recombinant RNase inhibitor) and incubated at 42°C for 60 min, followed by 94°C for 5 min.
The cDNA (4 μl) was amplified by PCR in a 50-μl reaction mixture containing 1× PCR buffer, each deoxynucleoside triphosphate at a concentration of 200 μM, 200 pM forward primer, 200 pM reverse primer, and 0.5 U of Taq DNA polymerase (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, United Kingdom) in a DNA thermal cycler (Biometra, Göttingen, Germany). The amplification reaction was performed for 35 cycles, and the primer sequences and annealing temperatures used are shown in Table 1. The PCR products were separated by electrophoresis on a 1.5% agarose gel and visualized by ethidium bromide staining. The relative intensities of the gel bands were measured by using an image-analyzing program (TINA 2.0e; Neuro-Image Analysis Centre, Oxford, United Kingdom). In order to exclude contaminating DNA from the isolated RNA, the RNA was subjected to PCR without cDNA synthesis. In all preparations, no band was detected after PCR.
TABLE 1.
Sequences of primers for ODF, OPG, MMPs, TIMPs, and β-actin
| Molecule | Direction | Primer sequence | Annealing temp (°C) | Product size (bp) |
|---|---|---|---|---|
| ODF | Forward | 5′-ATCAGAAGACAGCACTCACT-3′ | 45.3 | 750 |
| Reverse | 5′-ATCTAGGACATCCATGCTAATGTTC-3′ | |||
| OPG | Forward | 5′-TGAGTGTGAGGAAGGGCGTTAC-3′ | 45.5 | 636 |
| Reverse | 5′-TTCCTCGTTCTCTCAATCTC-3′ | |||
| MMP-3 | Forward | 5′-GTACAGAGCTGTGGGAAGTCAATG-3′ | 60 | 287 |
| Reverse | 5′-ATCAGCTCCATAGTGTTGGAGTCC-3′ | |||
| MMP-7 | Forward | 5′-TGTTGATGGCAGCTATGCAGCTCA-3′ | 60 | 387 |
| Reverse | 5′-CTAAGTTCACTGGGATCTGCATAC-3′ | |||
| MMP-8 | Forward | 5′-TGACTCTGGTGATTTCTTGCTAA-3′ | 60 | 164 |
| Reverse | 5′-GTGAAGGTCAGGGGCGATGC-3′ | |||
| MMP-9 | Forward | 5′-CTGTCCAGACCAAGGGTACAGCCT-3′ | 60 | 263 |
| Reverse | 5′-GTGGTATAGTGGGACACATAGTGG-3′ | |||
| MMP-10 | Forward | 5′-GATGTACCCAGTCTACAGGTTCTC-3′ | 60 | 409 |
| Reverse | 5′-GTAGCCTGCTTGGACTTCATTTCC-3′ | |||
| MMP-11 | Forward | 5′-GGAGAAGACAGACCTCACCTATAG-3′ | 60 | 376 |
| Reverse | 5′-CTTAGCTGCTGTGGTGTGTTGTAG-3′ | |||
| MMP-12 | Forward | 5′-CCAGGAAATGCAGCAGTTCTTTGG-3′ | 60 | 252 |
| Reverse | 5′-CTTAGAGGAGTCACATCACTCCAG-3′ | |||
| MMP-13 | Forward | 5′-CATTCAGCTATCCTGGCCACCTTC-3′ | 60 | 250 |
| Reverse | 5′-CAAGTTTGCCAGTCACCTCTAAGC-3′ | |||
| MMP-14 | Forward | 5′-GAGATCAAGGCCAATGTTCGGAGG-3′ | 60 | 382 |
| Reverse | 5′-TTAGATCCTCATTTTGGACAGTCC-3′ | |||
| TIMP-1 | Forward | 5′-CCTTATACCAGCCGTTATAAGATCAAGAT-3′ | 60 | 346 |
| Reverse | 5′-GTCCACAAACAGTGAGTGTCACTC-3′ | |||
| TIMP-2 | Forward | 5′-GCAATGCAGACGTAGTGATCAGAG-3′ | 60 | 371 |
| Reverse | 5′-GATCATGGGACAGCGAGTGATCTT-3′ | |||
| β-Actin | Forward | 5′-GGACTCCTATGGTGGGTGACGAGG-3′ | 58 | 366 |
| Reverse | 5′-GGGAGAGCATAGCCCTCGTAGAT-3′ |
PGE2 production assay.
Osteoblastic cells (1 × 104 cells/well) were cocultured with bone marrow cells (1 × 105 cells/well) in 200 μl of α-MEM containing 10% FBS in 48-well plates (Corning Inc.). After the cells had grown to confluence, the cocultures were treated with LOS (10 μg/ml) in either the presence or the absence of polymyxin B and incubated for an additional 24 h. PGE2 production in the cocultures was determined by using a PGE2 enzyme immunoassay kit according to the manufacturer's instructions (Amersham Pharmacia Biotech).
Statistical analyses.
The statistical significance of differences was determined by the Mann-Whitney U test. A P value of <0.05 was considered significant.
RESULTS
Effect of T. denticola LOS on osteoclast formation in a coculture system.
The purified lipopolysaccharide (LPS) preparation from T. denticola did not show a typical ladder-like band pattern in sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels after staining with silver nitrate, as previously described (28, 33). It migrated to a position between 7 and 14 kDa (based on the protein marker) and was not stained with Coomassie brilliant blue (data not shown). These data mean that the LPS preparation from T. denticola is LOS.
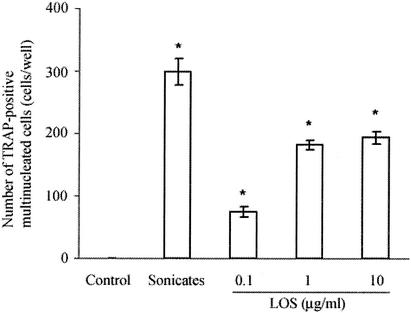
In the coculture treated with either the T. denticola sonicate (10 μg/ml) or LOS (0.1 to 10 μg/ml) for 4 days, a number of TRAP-positive multinucleated cells were formed, while the nontreated cultures did not contain TRAP-positive multinucleated cells. LOS increased the number of osteoclasts in a dose-dependent manner (Fig. 1).
FIG. 1.
Formation of TRAP-positive multinucleated cells in the coculture system treated with whole-cell sonicates and LOS from T. denticola. Mouse bone marrow and calvaria cells were cocultured to confluence and treated with T. denticola sonicates (10 μg/ml) or LOS (0.1 to 10 μg/ml) for an additional 4 days. The cells were then stained for TRAP. The TRAP-positive multinucleated cells containing more than three nuclei were counted as osteoclasts. The data are the means ± standard errors for four cultures. An asterisk indicates that the P value is <0.05 for a comparison with the results for the nontreated cultures.
Effect of T. denticola LOS on expression of ODF and OPG mRNAs.
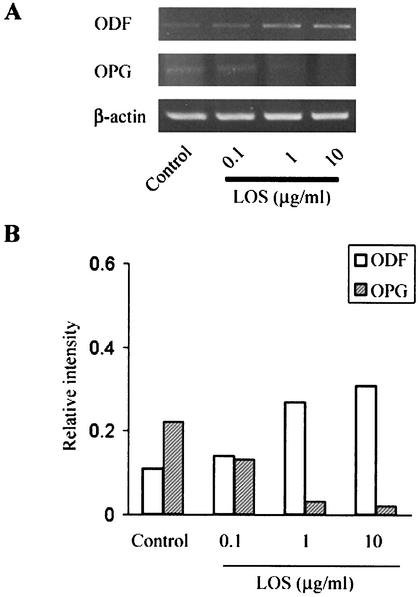
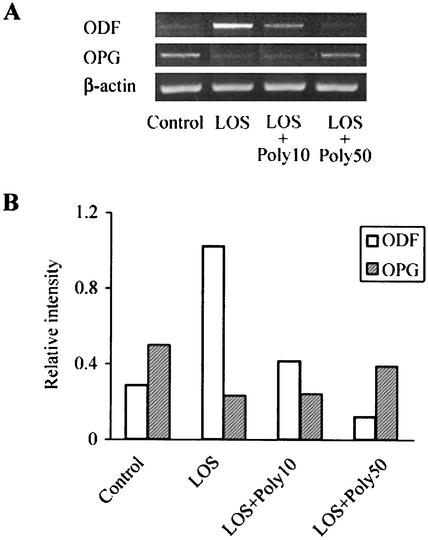
Expression of ODF and OPG mRNAs was investigated with the cocultures treated with LOS for 4 days by RT-PCR (Fig. 2). The nontreated cells exhibited steady-state levels of ODF and OPG mRNA expression, as described in other reports (16, 25). ODF mRNA expression increased and OPG mRNA expression decreased after stimulation with T. denticola LOS (0.1 to 10 μg/ml). To confirm the effect of LOS on ODF and OPG mRNA expression, an inhibition assay was performed with polymyxin B, which is known to form a stable complex with the lipid A of LPS and to neutralize LPS activity (24). Polymyxin B (50 μg/ml) eliminated the effect of LOS (10 μg/ml) on ODF and OPG mRNA expression (Fig. 3).
FIG. 2.
ODF and OPG mRNA expression in cocultures treated with T. denticola LOS. After the cocultures were treated with the LOS from T. denticola (0.1 to 10 μg/ml), as described in the text, the ODF, OPG, and β-actin mRNA levels were examined by RT-PCR (A). Signals in the RT-PCR were quantified and normalized to β-actin mRNA expression by using an image analyzer (B). The experiments were repeated three times, and similar results were obtained in all experiments.
FIG. 3.
Effect of polymyxin B on the ODF and OPG mRNAs induced by T. denticola LOS. After cocultures were treated with LOS (10 μg/ml) in the presence or absence of 10 μg of polymyxin B per ml (Poly10) or 50 μg of polymyxin B per ml (Poly50) for 4 days, the ODF, OPG, and β-actin mRNA levels were examined by RT-PCR (A). The signals in the RT-PCR were quantified and normalized to β-actin mRNA expression by using an image analyzer (B). The experiments were repeated three times, and similar results were obtained in all experiments.
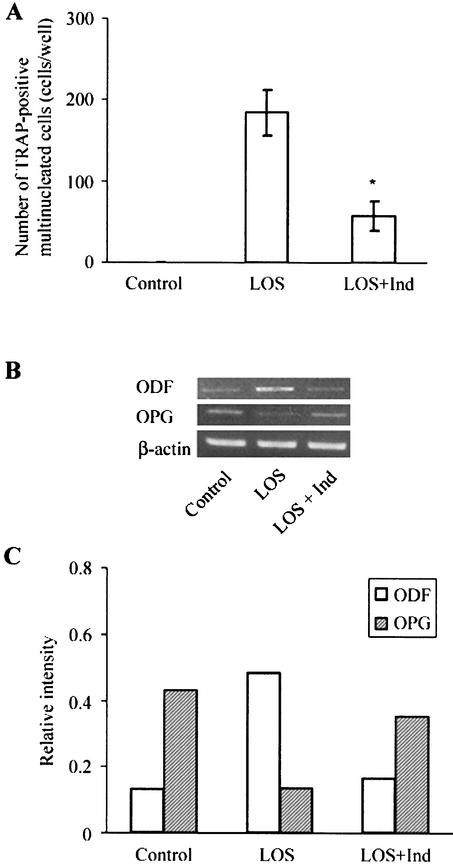
Effect of indomethacin on osteoclast formation and expression of ODF and OPG mRNAs regulated by T. denticola LOS.
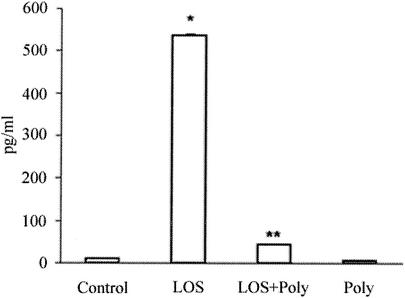
In order to determine whether PGE2 was involved in osteoclastogenesis induced by T. denticola LOS, PGE2 production in the coculture treated with LOS (10 μg/ml) in the presence or absence of polymyxin B was determined. A low level of PGE2 was detected in the untreated cultures, and the PGE2 concentration was higher in the cultures treated with LOS. Polymyxin B (50 μg/ml) decreased the PGE2 production stimulated by LOS (Fig. 4). To confirm the involvement of PGE2 in osteoclast formation and the expression of ODF and OPG mRNAs, cocultures were treated with LOS in the presence or absence of indomethacin, which is a prostaglandin inhibitor. In the coculture treated simultaneously with indomethacin (1 μM) and LOS (10 μg/ml) for 4 days, the number of TRAP-positive multinucleated cells was significantly less than the numbers of such cells in the cultures treated with LOS alone (Fig. 5A). The ODF mRNA was down-regulated and the OPG mRNA was up-regulated by indomethacin (Fig. 5B and C). Indomethacin alone did not affect osteoclast formation and the expression of ODF and OPG mRNAs (data not shown).
FIG. 4.
Effect of LOS on PGE2 production in cocultures. The cocultures were treated with LOS (10 μg/ml) with or without polymyxin B (Poly) (50 μg/ml) for 24 h. The PGE2 concentration was determined by using a PGE2 enzyme immunoassay kit. The data are the means ± standard errors for three cultures. One asterisk indicates that the P value is <0.05 for a comparison with the results for the nontreated cultures. Two asterisks indicate that the P value is <0.05 for a comparison with the results for the LOS-treated cultures.
FIG. 5.
Effect of indomethacin on osteoclastogenesis and the expression of ODF and OPG mRNAs modulated by T. denticola LOS. Cocultures were simultaneously treated with LOS (10 μg/ml) with or without indomethacin (Ind) (1 μM) for 4 days. The cells were then stained for TRAP to count the number of osteoclasts. The data are means ± standard errors for three cultures (A). The RNA was isolated from the cultured cells, and the ODF, OPG, and β-actin mRNA levels were analyzed by RT-PCR (B). The RT-PCR signals shown were quantified and normalized to the β-actin mRNA expression by using an image analyzer (C). Representative results of three experiments that yielded similar results are shown. An asterisk indicates that the P value is <0.05 for a comparison with the results for the LOS-treated cultures.
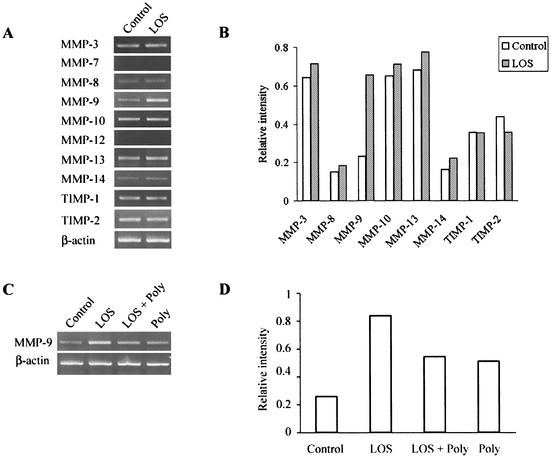
Expression of mRNAs of MMPs and TIMPs of osteoblastic cells treated with T. denticola LOS.
MMPs, including MMP-3, -7, -8, -9, -10, -12, -13, and -14, and TIMPs, including TIMP-1 and -2, were examined by RT-PCR to determine expression of their mRNAs in the osteoblastic cells treated with T. denticola LOS (10 μg/ml) for 48 h (Fig. 6A and B). The MMP-3, -8, -9, -10, -13, and -14 and TIMP-1and -2 mRNAs were expressed in the untreated osteoblastic cells, whereas MMP-7 and -12 mRNA expression could not be detected in treated or untreated cells. T. denticola LOS (10 μg/ml) increased the MMP-3, -8, -9, -10, -13, and -14 mRNA levels. Of these, MMP-9 mRNA expression was markedly enhanced by T. denticola LOS. T. denticola LOS slightly decreased TIMP-2 mRNA expression, whereas it did not influence TIMP-1 mRNA expression. To confirm the effect of T. denticola LOS on MMP expression, the levels of expression of MMP-9, whose mRNA exhibited a marked change in expression, were compared for cells treated with LOS (10 μg/ml), with polymyxin B (50 μg/ml), and with LOS plus polymyxin B (Fig. 6C and D). Polymyxin B (50 μg/ml) increased MMP-9 expression to some extent compared to the expression in the untreated cells. However, polymyxin B (50 μg/ml) decreased the MMP-9 expression stimulated by T. denticola LOS to the same level observed for the cells treated with polymyxin B alone.
FIG. 6.
Expression of MMP and TIMP mRNAs in mouse calvaria-derived osteoblastic cells treated with T. denticola LOS. After the osteoblastic cells were treated with T. denticola LOS (10 μg/ml) in the absence (A) or in the presence (C) of polymyxin B (Poly) (50 μg/ml) for 48 h, the RNA was isolated from the cells, and the MMP and TIMP mRNA levels were analyzed by RT-PCR. Representative results of three experiments that yielded similar results are shown. The RT-PCR signals shown in panels A and C were quantified and normalized to β-actin mRNA expression by using an image analyzer (B and D).
DISCUSSION
To determine the role of T. denticola in bone resorption, the effects of T. denticola sonicates and LOS on osteoclast formation and MMP expression were examined. Osteoblasts or stromal cells play an essential role in osteoclastogenesis through ODF expression (48). Therefore, to investigate the effect of T. denticola on osteoclastogenesis, a coculture system consisting of mouse calvaria cells which contained primary osteoblasts and bone marrow cells which included osteoclast precursors was used. In the preliminary study, T. denticola sonicates stimulated osteoclastogenesis in the coculture. This result prompted us to search for a component of T. denticola involved in osteoclastogenesis.
Yotis et al. (49) reported that T. denticola possesses an LPS-like molecule (8 to 14 kDa) that exhibits Limulus amoebocyte lysate clotting activity. Other research groups (28, 33) purified an approximately 14- to 21-kDa LOS from T. denticola. This LOS stimulated nitric oxide and TNF-α production in mouse macrophages, and the induction was inhibited by polymyxin B (28). In addition, Schultz et al. (31) reported that T. denticola LOS is quite different from the LPS of other gram-negative bacteria. Therefore, it was of interest to investigate the ability of T. denticola LOS to stimulate osteoclastogenesis. Gopalsami et al. (10) reported that the outer membrane of T. denticola increased Ca2+ release in an organ culture of radii and ulnae, and heat treating the outer membrane did not alter the effect on Ca2+ release. These authors concluded that a heat-stable LPS-like material is present in the outer membrane of T. denticola, which might be responsible for bone resorption. Taken together, these findings imply that the LOS from T. denticola may be involved in osteoclastogenesis. In the present study, T. denticola LOS was shown to stimulate osteoclast differentiation in a dose-dependent manner by using a coculture of osteoblast and osteoclast precursors. This means that T. denticola LOS induces osteoclastogenesis. However, T. denticola sonicates induced more TRAP-positive cells than LOS induced. In addition, when T. denticola sonicates were heat treated, the osteoclast formation activity of sonicates was not completely inhibited (data not shown). Therefore, the possibility that some heat-labile components of T. denticola are involved in osteoclastogenesis cannot be ruled out.
It was reported previously that the osteoblast or stromal cell lines that support osteoclastogenesis showed a much higher ODF mRNA level, whereas the level of OPG mRNA was drastically reduced in the presence of either PGE2 or 1α,25(OH)2D3 (25). Another group of workers also reported that 1α,25(OH)2D3, PTH, or IL-11 prompted an increase in the ratio of ODF to OPG (16). These findings indicate that the effects of these bone resorptive factors are mediated through regulation of the production of ODF and its endogenous receptor antagonist, OPG, and that the ODF/OPG ratio appears to be an essential factor that determines the ability of osteoblastic cells to induce osteoclast formation. Since the discovery of ODF, it has been believed that ODF is the sole factor responsible for inducing osteoclast differentiation. However, it has been shown that TNF-α stimulates osteoclast formation via an ODF-independent mechanism (19). Our results suggest that the stimulatory effect of T. denticola LOS on osteoclast formation is mediated through ODF up-regulation and OPG down-regulation.
PGE2 has been shown to play a role in osteoclastogenesis (17). Two studies on the involvement of PGE2 in ODF and OPG mRNA expression in osteoblastic cells by LPS from Escherichia coli had different results. Kikuchi et al. (18) reported that LPS increased the ODF mRNA level and that an inhibitor of PGE2 synthesis failed to block the effect of LPS after 2 h of exposure, while OPG gene expression remained constant after LPS stimulation. These authors suggested that LPS induced ODF mRNA in osteoblasts directly, not via PGE2, and did not affect OPG expression. In contrast, Sakuma et al. (30) observed the effect of LPS for a longer period (24 h). They showed that LPS exposure induced ODF mRNA expression in a time-dependent manner. Induction for 4 h was not inhibited by indomethacin. However, induction for 24 h was partially inhibited by indomethacin. Furthermore, LPS exposure increased OPG mRNA expression, but the OPG levels that were induced by LPS exposure were not affected by indomethacin. This report suggested that ODF is induced by LPS in a PGE2-dependent manner and in a PGE2-independent manner and that OPG expression by LPS is increased independent of PGE2. To determine if PGE2 is involved in the effect that T. denticola LOS has on ODF and OPG mRNA expression, we estimated the numbers of osteoclasts and the levels of ODF and OPG mRNA expression in cocultures treated with LOS in the presence and in the absence of indomethacin for 4 days. T. denticola LOS stimulated PGE2 production and osteoclast formation by LOS was reduced by indomethacin in coculture, which suggests the possibility that PGE2 is involved in LOS-regulated ODF and OPG expression. Like LPS, T. denticola LOS increased ODF mRNA expression, and this stimulating activity was inhibited by indomethacin. However, unlike regulation by LPS, OPG mRNA was down-regulated by T. denticola LOS, and the OPG mRNA level recovered to the control level after treatment with indomethacin. Taken together, these findings indicate that PGE2 is involved in the regulation of ODF and OPG gene expression by T. denticola LOS. The down-regulation of OPG gene expression by T. denticola LOS is somewhat different from the results for LPS reported previously (18, 30). The discrepancy may result from the different culture conditions used (i.e., the cell type and the stimulation time) or from the structural differences between LPS and LOS.
It has been reported that PGE2 and IL-1 are involved in osteoclast formation by Actinobacillus actinomycetemcomitans LPS and that Porphyromonas gingivalis LPS promotes bone resorption, which is mediated by IL-1, IL-6, TNF-α, and PGE2 (4, 23, 43, 50). These previous studies suggest that cytokines, such as IL-1, TNF-α, and IL-6, are also involved in LPS-induced osteoclastogenesis. We are currently studying the involvement of proinflammatory cytokines in osteoclastogenesis by T. denticola LOS.
The bone matrix consists of various proteins, including collagen, proteoglycans, and glycoproteins (27, 29). Recent observations show that MMPs may play a role not only in dissolving the bone matrix but also in initiating bone resorption, which determines where and when bone resorption occurs. Removing nonmineralized matrix from the bone surface is essential for initiating osteoclastic bone resorption, because osteoclasts cannot attach to nonmineralized osteoid (2). In order to determine the involvement of MMPs in bone resorption by T. denticola LOS, we analyzed the expression of mRNAs of various MMPs. The most apparent increase was observed for MMP-9. MMP-9 has broad substrate specificity, including proteoglycans, glycoproteins, and gelatin, a denatured form of collagen (22). In addition to gelatinase activity, MMP-9 exerts chemotactic activity on osteoclasts. MMP-9 releases extracellular matrix-bound vascular endothelial growth factor, which acts as a chemoattractant for osteoclasts (8). TNF-α is a bone resorption-inducing factor, and it has been reported that the proteolytic processing of this factor from its precursor to an active form is carried out by MMP-9 (22). These reports imply that MMP-9 may play an important role in the bone resorption process by dissolving the bone matrix, releasing the chemotactic factor on osteoclasts, and activating the cytokines involved in bone resorption. Although the expression of mRNAs of MMP-3, -8, -10, -13, and -14 is increased less than that of mRNA of MMP-9 by T. denticola LOS in osteoblastic cells, increased expression of the mRNAs of these MMPs was observed in all three experiments. Therefore, in the future, the significance of these MMPs in the bone resorption induced by T. denticola LOS should be confirmed.
TIMPs are the major endogenous inhibitors that down-regulate MMP activity (7). Four TIMPs have been identified. Although each TIMP inhibits most of the MMPs, a certain degree of specificity has been observed. TIMP-1 and -2 are inhibitors of MMP-9. TIMP-1 preferentially binds MMP-9 and inhibits its activity (7, 37). The balance between the activities of the MMPs and the TIMPs is believed to determine the rate of matrix degradation. T. denticola LOS did not affect TIMP-1 mRNA expression and slightly decreased TIMP-2 mRNA expression in osteoblastic cells. These observations imply that TIMPs have a minor effect on MMP-9 activity.
In conclusion, here we provide evidence that the LOS from T. denticola stimulates osteoclastogenesis and the expression of several MMPs, including MMP-9, in osteoblastic cells. ODF up-regulation and OPG down-regulation via PGE2 are involved in osteoclastogenesis. T. denticola is known to be one of the major putative pathogens of periodontitis, a polymicrobial infection. Our results show that the pathogenesis induced by T. denticola may be one of the mechanisms of bone destruction in periodontitis.
Acknowledgments
This work was supported by the research fund of Yonsei University College of Dentistry for 2001.
Editor: J. D. Clements
REFERENCES
- 1.Brändström, H., T. Bjorkman, and Ö. Ljunggren. 2001. Regulation of osteoprotegerin secretion from primary cultures of human bone marrow stromal cells. Biochem. Biophys. Res. Commun. 280:831-835. [DOI] [PubMed] [Google Scholar]
- 2.Chambers, T. J., J. A. Darby, and K. Fuller. 1985. Mammalian collagenase predisposes bone surfaces to osteoclastic resorption. Cell Tissue Res. 241:671-675. [DOI] [PubMed] [Google Scholar]
- 3.Chan, E. C. S., and R. McLaughlin. 2000. Taxonomy and virulence of oral spirochetes. Oral Microbiol. Immunol. 15:1-9. [DOI] [PubMed] [Google Scholar]
- 4.Chiang, C. Y., G. Kyritsis, D. T. Graves, and S. Amar. 1999. Interleukin-1 and tumor necrosis factor activities partially account for calvarial bone resorption induced by local injection of lipopolysaccharide. Infect. Immun. 67:4231-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, B. K., S. H. Ohk, H. J. Lee, J. H. Kang, G. J. Jeong, and Y. J. Yoo. 2001. Effects of whole cell sonicates of Treponema lecithinolyticum on osteoclast differentiation. J. Periodontol. 72:1172-1177. [DOI] [PubMed] [Google Scholar]
- 6.Dewhirst, F. E., M. A. Tamer, R. E. Ericson, C. N. Lau, V. A. Levanos, S. K. Boches, J. L. Galvin, and B. J. Paster. 2000. The diversity of periodontal spirochetes by 16S rRNA analysis. Oral Microbiol. Immunol. 15:196-202. [DOI] [PubMed] [Google Scholar]
- 7.Dong, J. C., H. Dong, A. Campana, and P. Bischof. 2002. Matrix metalloproteinases and their specific tissue inhibitors in menstruation. Reproduction 123:621-631. [DOI] [PubMed] [Google Scholar]
- 8.Engsig, M. T., Q.-J. Chen, T. H. Vu, A.-C. Pedersen, B. Therkidsen, L. R. Lund, K. Henriksen, T. Lenhard, N. T. Foged, Z. Werb, and J.-M. Delaissé. 2000. Matrix metalloproteinase 9 and vascular endothelial growth factor are essential for osteoclast recruitment into developing long bones. J. Cell Biol. 151:879-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenno, J. C., and B. C. McBride. 1998. Virulence factors of oral treponemes. Anaerobe 4:1-17. [DOI] [PubMed] [Google Scholar]
- 10.Gopalsami, C., W. Yotis, K. Corrigan, S. Schade, J. Keene, and L. Simonson. 1993. Effect of outer membrane of Treponema denticola on bone resorption. Oral Microbiol. Immunol. 8:121-124. [DOI] [PubMed] [Google Scholar]
- 11.Hoekstra, R., F. A. L. M. Eskens, and J. Verweij. 2001. Matrix metalloproteinase inhibitors: current developments and future perspectives. Oncologist 6:415-427. [DOI] [PubMed] [Google Scholar]
- 12.Hofbauer, L. C., C. R. Dunstan, T. C. Spelsberg, B. L. Riggs, and S. Khosla. 1998. Osteoprotegerin production by human osteoblast lineage cells is stimulated by vitamin D, bone morphogenetic protein-2, and cytokines. Biochem. Biophys. Res. Commun. 250:776-781. [DOI] [PubMed] [Google Scholar]
- 13.Hofbauer, L. C., F. Gori, B. L. Riggs, D. L. Lacey, C. R. Dunstan, T. C. Spelsberg, and S. Khosla. 1999. Stimulation of osteoprotegerin ligand and inhibition of osteoprotegerin production by glucocorticoids in human osteoblastic lineage cells: potential paracrine mechanisms of glucocorticoid-induced osteoporosis. Endocrinology 140:4382-4389. [DOI] [PubMed] [Google Scholar]
- 14.Hofbauer, L. C., D. L. Lacey, C. R. Dunstan, T. C. Spelsberg, B. L. Riggs, and S. Khosla. 1999. Interleukin-1β and tumor necrosis factor-α, but not interleukin-6, stimulate osteoprotegerin ligand gene expression in human osteoblastic cells. Bone 25:255-259. [DOI] [PubMed] [Google Scholar]
- 15.Hofbauer, L. C., S. Khosla, C. R. Dunstan, D. L. Lacey, W. J. Boyle, and B. L. Riggs. 2000. The roles of osteoprotegerin and osteoprotegerin ligand in the paracrine regulation of bone resorption. J. Bone Miner. Res. 15:2-12. [DOI] [PubMed] [Google Scholar]
- 16.Horwood, N. J., J. Elliott, T. J. Martin, and M. T. Gillespie. 1998. Osteotropic agents regulate the expression of osteoclast differentiation factor and osteoprotegerin in osteoblastic stromal cells. Endocrinology 139:4743-4746. [DOI] [PubMed] [Google Scholar]
- 17.Kaji, H., T. Sugimoto, M. Kanatani, M. Fukase, M. Kumegawa, and K. Chihara. 1996. Prostaglandin E2 stimulates osteoclast-like cell formation and bone-resorbing activity via osteoblasts: role of cAMP-dependent protein kinase. J. Bone Miner. Res. 11:62-71. [DOI] [PubMed] [Google Scholar]
- 18.Kikuchi, T., T. Matsuguchi, N. Tsuboi, A. Mitani, S. Tanaka, M. Matsuoka, G. Yamamoto, T. Hishikawa, T. Noguchi, and Y. Yoshikai. 2001. Gene expression of osteoclast differentiation factor is induced by lipopolysaccharide in mouse osteoblasts via Toll-like receptors. J. Immunol. 166:3574-3579. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi, K., N. Takahashi, E. Jimi, N. Udagawa, M. Takami, S. Kotake, N. Nakagawa, M. Kinosaki, K. Yamaguchi, N. Shima, H. Yasuda, T. Morinaga, K. Higashio, T. J. Martin, and T. Suda. 2000. Tumor necrosis factor α stimulates osteoclast differentiation by a mechanism independent of the ODF/RANKL-RANK interaction. J. Exp. Med. 191:275-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusano, K., C. Miyaura, M. Inada, T. Tamura, A. Ito, H. Nagase, K. Kamoi, and T. Suda. 1998. Regulation of matrix metalloproteinases (MMP-2, -3, -9, and -13) by interleukin-1 and interleukin-6 in mouse calvaria: association of MMP induction with bone resorption. Endocrinology 139:1338-1345. [DOI] [PubMed] [Google Scholar]
- 21.Lacey, D. L., E. Timms, H.-L. Tan, M. J. Kelley, C. R. Dunstan, T. Burgess, R. Elliott, A. Colombero, G. Elliott, S. Scully, H. Hsu, J. Sullivan, N. Hawkins, E. Davy, C. Capparelli, A. Eli, Y.-X. Qian, S. Kaufman, I. Sarosi, V. Shalhoub, G. Senaldi, J. Guo, J. Delaney, and W. J. Boyle. 1998. Osteoprotegerin (OPG) ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 93:165-176. [DOI] [PubMed] [Google Scholar]
- 22.McCawley, L. J., and L. M. Matrisian. 2001. Matrix metalloproteinases: they're not just for matrix anymore! Curr. Opin. Cell Biol. 13:534-540. [DOI] [PubMed] [Google Scholar]
- 23.Miyata, Y., H. Takeda, S. Kitano, and S. Hanazawa. 1997. Porphyromonas gingivalis lipopolysaccharide-stimulated bone resorption via CD14 is inhibited by broad-spectrum antibiotics. Infect. Immun. 65:3513-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrison, D. C., and D. M. Jacobs. 1976. Binding of polymyxin B to the lipid A portion of bacterial lipopolysaccharides. Immunochemistry 13:813-818. [DOI] [PubMed] [Google Scholar]
- 25.Nagai, M., and N. Sato. 1999. Reciprocal gene expression of osteoclastogenesis inhibitory factor and osteoclast differentiation factor regulates osteoclast formation. Biochem. Biophys. Res. Commun. 257:719-723. [DOI] [PubMed] [Google Scholar]
- 26.Reddy, S. V., and G. D. Roodman. 1998. Control of osteoclast differentiation. Crit. Rev. Eukaryot. Gene Expr. 8:1-17. [DOI] [PubMed] [Google Scholar]
- 27.Robey, P. G. 1986. Bone matrix proteoglycans and glycoproteins, p. 155-165. In J. P. Bilezikian, L. G. Raisz, and G. A. Rodan (ed.), Principles of bone biology, 1st ed. Academic Press, San Diego, Calif.
- 28.Rosen, G., M. N. Sela, R. Naor, A. Halabi, V. Barak, and L. Shapira. 1999. Activation of murine macrophages by lipoprotein and lipooligosaccharide of Treponema denticola. Infect. Immun. 67:1180-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossert, J., and B. D. Crombrugghe. 1986. Type I collagen: structure, synthesis, and regulation, p. 127-142. In J. P. Bilezikian, L. G. Raisz, and G. A. Rodan (ed.), Principles of bone biology, 1st ed. Academic Press, San Diego, Calif.
- 30.Sakuma, Y., K. Tanaka, M. Suda, Y. Komatsu, A. Yasoda, M. Miura, A. Ozasa, S. Narumiya, Y. Sugimoto, A. Ichikawa, F. Ushikubi, and K. Nakao. 2000. Impaired bone resorption by lipopolysaccharide in vivo in mice deficient in the prostaglandin E receptor EP4 subtype. Infect. Immun. 68:6819-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schultz, C. P., V. Wolf, R. Lange, E. Mertens, J. Wecke, D. Naumann, and U. Zähringer. 1998. Evidnece for a new type of outer membrane lipid in oral spirochete Treponema denticola. J. Biol. Chem. 273:15661-15666. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz, Z., J. Goultschin, D. D. Dean, and B. D. Boyan. 1997. Mechanisms of alveolar bone destruction in periodontitis. Periodontol. 2000 14:158-172. [DOI] [PubMed] [Google Scholar]
- 33.Sela, M. N., A. Bolotin, R. Naor, A. Weinberg, and G. Rosen. 1997. Lipoproteins of Treponema denticola: their effect on human polymorphonuclear neutrophils. J. Periodontal Res. 32:455-466. [DOI] [PubMed] [Google Scholar]
- 34.Simonet, W. S., D. L. Lacey, C. R. Dunstan, M. Kelley, M.-S. Chang, R. Lüthy, H. Q. Nguyen, S. Wooden, L. Bennett, T. Boone, G. Shimamoto, M. DeRose, R. Elliott, A. Colombero, H.-L. Tan, G. Trail, J. Sullivan, E. Davy, N. Bucay, L. Renshaw-Gegg, T. M. Hughes, D. Hill, W. Pattison, P. Campbell, S. Sander, G. Van, J. Tarpley, P. Derby, R. Lee, and W. J. Boyle. 1997. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell 89:309-319. [DOI] [PubMed] [Google Scholar]
- 35.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 36.Sternlicht, M. D., and Z. Werb. 2001. How matrix metalloproteinases regulate cell behavior. Annu. Rev. Cell Dev. Biol. 17:463-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strongin, A. Y., I. Collier, G. Bannikov, B. L. Marmer, G. A. Grant, and G. I. Goldberg. 1995. Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloproteinase. J. Biol. Chem. 270:5331-5338. [DOI] [PubMed] [Google Scholar]
- 38.Suda, T., I. Nakamura, E. Jimi, and N. Takahashi. 1997. Regulation of osteoclast function. J. Bone Miner. Res. 12:869-879. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi, N., T. Akatsu, N. Udagawa, T. Sasaki, A. Yamaguchi, J. M. Moseley, T. J. Martin, and T. Suda. 1988. Osteoblastic cells are involved in osteoclast formation. Endocrinology 123:2600-2602. [DOI] [PubMed] [Google Scholar]
- 40.Uchida, M., M. Shima, T. Shimoaka, A. Fujieda, K. Obara, H. Suzuki, Y. Nagai, T. Ikeda, H. Yamato, and H. Kawaguchi. 2000. Regulation of matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinase (TIMPs) by bone resorptive factors in osteoblastic cells. J. Cell. Physiol. 185:207-214. [DOI] [PubMed] [Google Scholar]
- 41.Uchida, M., M. Shima, D. Chikazu, A. Fujieda, K. Obara, H. Suzuki, Y. Nagai, H. Yamato, and H. Kawaguchi. 2001. Transcriptional induction of matrix metalloproteinase-13 (collagnase-3) by 1α,25-dihydroxyvitamin D3 in mouse osteoblastic MC3T3-E1 cells. J. Bone Miner. Res. 16:221-230. [DOI] [PubMed] [Google Scholar]
- 42.Udagawa, N., N. Takahashi, H. Yasuda, A. Mizuno, K. Itoh, Y. Ueno, T. Shinki, M. T. Gillespie, T. J. Martin, K. Higashio, and T. Suda. 2000. Osteoprotegerin produced by osteoblasts is an important regulator in osteoclast development and function. Endocrinology 141:3478-3484. [DOI] [PubMed] [Google Scholar]
- 43.Ueda, N., M. Koide, M. Ohguchi, Y. Ishihara, T. Noguchi, N. Okahashi, and T. Nishihara. 1998. Involvement of prostagladin E2 and interleukin-1α in the differentiation and survival of osteoclasts induced by lipopolysaccharide from Actinobacillus actinomycetemcomitansY4. J. Periodontal Res. 33:509-516. [DOI] [PubMed] [Google Scholar]
- 44.Vidal, O. N. A., K. Sjögren, B. I. Eriksson, Ö. Ljunggren, and C. Ohlsson. 1998. Osteoprotegerin mRNA is increased by interleukin-1α in the human osteosarcoma cell line MG-63 and in the human osteoblast-like cells. Biochem. Biophys. Res. Commun. 248:696-700. [DOI] [PubMed] [Google Scholar]
- 45.Walker, S. G., J. L. Ebersole, and S. C. Holt. 1997. Identification, isolation and characterization of the 42-kilodalton major outer membrane protein (MompA) from Treponema pectinovorum ATCC 33768. J. Bacteriol. 179:6441-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wyss, C., B. K. Choi, P. Schupbach, B. Guggenheim, and U. B. Gobel. 1996. Treponema maltophilum sp. nov., a small oral spirochete isolated from human periodontal lesions. Int. J. Syst. Bacteriol. 46:745-752. [DOI] [PubMed] [Google Scholar]
- 47.Yasuda, H., N. Shima, N. Nakagawa, S.-I. Mochizuki, K. Yano, N. Fujise, Y. Sato, M. Goto, K. Yamaguchi, M. Kuriyama, T. Kanno, A. Murakami, E. Tsuda, T. Morinaga, and K. Higashio. 1998. Identity of osteoclastogenesis inhibitory factor (OCIF) and osteoprotegerin (OPG): a mechanism by which OPG/OCIF inhibits osteoclastogenesis in vitro. Endocrinology 139:1329-1337. [DOI] [PubMed] [Google Scholar]
- 48.Yasuda, H., N. Shima, N. Nakagawa, K. Yamaguchi, M. Kinosaki, S.-I. Mochizuki, A. Tomoyasu, K. Yano, M. Goto, A. Murakami, E. Tsuda, T. Morinaga, K. Higashio, N. Udagawa, N. Takahashi, and T. Suda. 1998. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc. Natl. Acad. Sci. USA 95:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yotis, W. W., F. Macaluso, and C. Gopalsami. 1995. Immunochemical features of a macromolecule of Treponema denticola. J. Basic Microbiol. 35:255-268. [DOI] [PubMed] [Google Scholar]
- 50.Zubery, Y., C. R. Dunstan, B. M. Story, L. Kesavalu, J. L. Ebersole, S. C. Holt, and B. F. Boyce. 1998. Bone resorption caused by three periodontal pathogens in vivo in mice is mediated in part by prostaglandin. Infect. Immun. 66:4158-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]