Abstract
Pneumococcal surface protein A (PspA), a virulence factor of Streptococcus pneumoniae, is exceptionally diverse, being classified into two major families which are over 50% divergent by sequence analysis. A family 1 PspA from strain WU2 was previously shown to impede the clearance of pneumococci from mouse blood and to interfere with complement deposition on the bacterial surface. To determine whether a family 2 PspA can perform the same role as family 1 PspA, the family 1 PspA (from strain WU2) was replaced with a family 2 PspA (from strain TIGR4) by molecular genetic methods to make an isogenic pair of strains expressing different PspA proteins. Surface binding of lactoferrin and interference with C3 deposition by the two types of PspA proteins were determined by flow cytometry, and virulence was assessed in a mouse bacteremia model. Although the family 2 PspA appeared to bind less human lactoferrin than did the family 1 PspA, both PspA proteins could interfere with complement deposition on the pneumococcal surface and could provide full virulence in the mouse infection model. A mutant form of the family 2 PspA with a deletion within the choline-binding region was also produced. Pneumococci with this mutant PspA failed to bind human lactoferrin even though the PspA was present on the pneumococcal surface. The mutant PspA only partially interfered with complement deposition and moderately attenuated virulence. These results suggest that family 1 and family 2 PspA proteins play similar roles in virulence and that surface accessibility of PspA is important for their function.
Streptococcus pneumoniae is a human pathogen that causes both mucosal and invasive diseases, including pneumonia, bacteremia, otitis media, and meningitis, throughout the world (12, 22). A number of surface structures have been identified that are expected to contribute to its pathogenicity. These include the polysaccharide capsule, teichoic acids, autolysin, pneumolysin, hyaluronidase, immunoglobulin A (IgA) protease, neuraminidase, and several members of a family of cell surface choline-binding proteins (6, 9, 17, 24, 26, 36).
Pneumococcal surface protein A (PspA) is a choline-binding protein tethered to the cell surface through its C-terminal choline-binding repeat region. Previous studies demonstrated that the PspA structure consists of five distinct domains. From the N terminus to the C terminus, these domains are as follows: a signal peptide, a highly charged alpha-helical domain, a proline-rich region, a choline-binding domain, and a C-terminal 17-amino-acid (17-aa) tail (33). The alpha-helical domain contains the protection-eliciting epitopes (3) and much of the variability in sequence among different PspA proteins. The approximately 100 C-terminal amino acids of the alpha-helical domain elicit most of the cross-protective immunity (20). Based on the sequence diversity of this region, PspA proteins have been classified into two major families that consist of five clades (3, 13). The choline-binding domain, which consists of 9 or 10 20-aa repeats, is required for the attachment of PspA to the pneumococcal cell surface (18, 34, 35). Deletion of the last five repeats and the 17-aa tail results in release into culture supernatant of family 1 PspA/Rx1 (34) (protein/strain name [described below]).
PspA is required for full virulence in mouse models of pneumococcal infection (21); PspA slows the clearance of pneumococci from mouse blood (4, 21). Our laboratory and others' have shown that family 1 PspA/WU2 leads to reduced complement activation in vivo and in vitro. Tu et al. demonstrated that mice infected with WU2 maintained higher levels of C3 in their blood than mice infected with an isogenic pspA-lacking strain (28). Tu et al. also showed that the pspA-lacking strain was cleared much faster from the blood of mice. The difference in virulence of the wild-type and pspA-lacking strains disappeared when C3 or factor B knockout mice were infected. Using enzyme-linked immunosorbent assay, Abeyta et al. showed that pspA-lacking strains bound more C3 than wild-type pspA-positive strains when pneumococci were incubated with mouse serum (1). Using the bystander complement fixation assay, Neeleman et al. also showed that PspA interfered with complement activation (23). All of these results indicate that PspA may interfere with complement's host defense functions. However, all the studies described above examined PspA proteins only from PspA family 1. Analysis of sequence data showed that family 1 and family 2 PspA proteins only share approximately 40% amino acid similarity over their N-terminal alpha-helical regions (13). Thus, it was important to determine whether or not a family 2 PspA could play the same role in host-pathogen interaction.
PspA also binds lactoferrin, a multifunctional glycoprotein (10, 11, 30, 31). The exact contribution of lactoferrin binding to virulence, if any, is not yet clear. Nevertheless, the fact that virtually all pneumococci bind to lactoferrin through PspA argues for the importance of lactoferrin binding in pneumococcal pathogenesis.
In this study, we have replaced the naturally occurring family 1 PspA/WU2 with family 2 PspA/TIGR4 in the WU2 genetic background. The WU2 wild-type strain and the WU2-derived strains expressing family 2 PspA were then compared in terms of virulence, complement activation, and binding of lactoferrin.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and genetic transformation.
S. pneumoniae strains used in this study are listed in Table 1. Strains were cultured as described previously (21). The following antibiotics were used at the indicated concentrations: erythromycin, 0.3 μg/ml for S. pneumoniae and 400 μg/ml for Escherichia coli; ampicillin, 100 μg/ml for E. coli.
TABLE 1.
Strains and plasmids used in this study
| S. pneumoniae strain or plasmid | Capsular serotype | Derivation and properties | PspA phenotype | PspA family | Reference |
|---|---|---|---|---|---|
| Strains | |||||
| WU2 | 3 | Encapsulated, parent, virulent | PspA+ | 1 | |
| JY1119 | 3 | WU2 derivative | PspA− | 34 | |
| JY1123 | 3 | WU2 derivative | PspAtra | 1 | 34 |
| BR93.1 | 3 | BR84 × WU2 | PspA+ | 2 | This study |
| BR92.1 | 3 | BR81 × WU2 | PspAi.d.b | 2 | This study |
| TIGR4 | 4 | Encapsulated, parent, virulent | PspA+ | 2 | |
| BR84 | 4 | pREN4 × TIGR4 | PspA+ | 2 | This study |
| BR81 | 4 | pREN4 × TIGR4 | PspAi.d. | 2 | This study |
| BR61.1 | 4 | pREN3 × TIGR4 | PspA− | This study | |
| Plasmids | |||||
| pJY4164 | ErmR, oriR E. coli | 34 | |||
| pREN3 | pJY4164::PCR fragment LSM12 and SKH79 | This study | |||
| pREN4 | pJY4164::PCR fragment SKH73 and LSM2 | This study |
tr, truncated at the C-terminal end. This particular PspA is constituted of only the α-helical region and is released into culture supernatant.
i.d., internal deletion. This PspA is missing six choline-binding repeats (121 amino acids) within the C-terminal end of the molecule.
Pneumococci were transformed with plasmids extracted from E. coli for the insertion-duplication mutagenesis or with S. pneumoniae cell lysates containing chromosomal DNA by procedures described previously (5). The transformants were selected on blood agar plates supplemented with erythromycin. When strains were constructed by transformation with chromosomal DNA from a donor strain, the initial transformants were backcrossed three times into the parental background. For each backcross, cell lysates were prepared from the previous passage and used as a donor for transformation with a competent parental strain. The final constructs all had a smooth morphology similar to that of the wild-type parent strains, suggesting that the strains within each background had similar levels of capsular polysaccharide.
Cloning and plasmid construction.
The 3′ region of the TIGR4 pspA gene was amplified by PCR using primer pair SKH73 (5′GGAAACAAGAAAACGGTATGTG3′) and LSM2 (5′GCGCGTCGACGGCTTAAACCCATTCACCATTGG3′). The amplified PCR product was first cloned into pTOPO (Invitrogen, Carlsbad, Calif.), and then the insert was released from pTOPO by digestion with EcoRI and subcloned into the vector pJY4164 (34) encoding the erythromycin resistance gene. This formed plasmid pREN4. Similarly, the 5′ region of the TIGR4 pspA gene was amplified by PCR using primer pair LSM12 (5′CCGGATCCAGCGTCGCTATCTTAGGGGCTGGTT3′) and SKH79 (5′CCTTTAGAGTTGCATAATCATAT3′). A 500-bp fragment near the start codon of pspA of TIGR4 was produced, cloned into pTOPO, and then subcloned into pJY4164. The constructed plasmid was designated pREN3. To determine the pspA sequences from constructed strains BR92.1 and BR93.1, the pspA genes were amplified with primer pair LSM12 and LSM2 and cloned into pTOPO for sequencing.
Insertion-duplication mutagenesis.
The plasmids pREN4 and pREN3 were used to transform S. pneumoniae TIGR4. Erythromycin-resistant transformants were obtained after plasmid integration via homologous recombination into chromosomes at the site of the cloned fragment. Plasmid pREN4 contains the 3′ end of the pspA gene and so does not interrupt PspA expression upon insertion. This was used to link the selectable marker, erythromycin resistance, to the pspA/TIGR4 gene for later transfer of the gene to WU2. To serve as a negative control for family 2 PspA in antibody binding and lactoferrin binding experiments, a TIGR4 pspA-lacking strain was also constructed using pREN3. This plasmid had an internal portion of pspA/TIGR4 allowing the pspA gene to be inactivated by the insertion-duplication process (32). The resultant strain was designated BR61.1.
Western blot analysis.
To identify each PspA, S. pneumoniae cultures were grown to exponential phase and centrifuged. The bacterial cell pellets were incubated with lysis buffer (0.01% sodium dodecyl sulfate, 0.1% sodium deoxycholate, 0.15 M sodium citrate) at 37°C for 10 min. Lysates from different strains were boiled in sodium dodecyl sulfate and electrophoresed on 10% polyacrylamide gels. After their separation, the proteins were transferred to nitrocellulose membranes and probed with rabbit antisera specific to family 1 or family 2 PspA proteins (29). The secondary antibody, goat anti-rabbit IgG (heavy and light chain)-biotin, was used in conjunction with streptavidin-alkaline phosphatase (Southern Biotechnology Associates, Inc., Birmingham, Ala.).
To examine differences in association of PspA with the bacterial surface, the presence of PspA in different culture fractions was detected by Western blot analysis as described previously (34) with some modification. Frozen bacterial stock was thawed, diluted with Todd-Hewitt broth containing 0.5% yeast extract (THY), and grown to exponential phase. To minimize contamination by PspA released from dead bacteria from the frozen stock, the exponential cultures were diluted to an optical density at 600 nm (OD600) of <0.1 with THY and then regrown to an OD600 of 0.5. One milliliter of the bacterial cell culture was centrifuged, and the supernatant was removed, passed through a 0.2-μm-pore-size filter, and collected. The bacterial cell pellet was washed with phosphate-buffered saline (PBS) (pH 7.0) and lysed with lysis buffer, and water was added to adjust to the original volume. Another 1-ml culture volume was withdrawn, and the bacterial cells were washed with PBS and then washed with 1 ml of 2% choline chloride in PBS. After centrifugation, the supernatant was collected, passed through a 0.2-μm-pore-size filter, and saved. The pellet was washed, incubated with lysis buffer, and adjusted to a 1-ml total volume with water. Equivalent amounts of all four fractions, representing 40 μl of unconcentrated cultures, were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting, as described above.
Detection of surface PspA.
Cultures were grown in THY medium to an OD600 of 0.5, and approximately 107 CFU were withdrawn. Bacterial cells were enumerated by plating on blood agar plates. After washing with PBS, pneumococci were incubated with 100 μl of rabbit anti-PspA family-specific serum (29) (1:100 dilution with PBS) for 30 min at 37°C. After washing with PBS, the bacteria were incubated with 100 μl of fluorescein isothiocyanate-conjugated goat anti-rabbit IgG (heavy and light chain) (Southern Biotechnology Associates, Inc.) for 30 min on ice. After the final wash with PBS, bacteria were suspended in 300 μl of 2% paraformaldehyde and analyzed by flow cytometer (FACScan; Becton Dickinson, Franklin Lakes, N.J.). The results are expressed as median fluorescence intensity.
Binding of lactoferrin.
Purified human iron-saturated lactoferrin (Sigma, St. Louis, Mo.) was biotinylated using the biotin-labeling kit according to the manufacturer's instructions (Boehringer Mannheim GmbH, Mannheim, Germany). Cultures were grown to OD600 = 0.5, and 107 CFU bacteria were incubated with 100 μl of biotinylated lactoferrin at various concentrations (5, 10, and 50 μg/ml in PBS) for 30 min at 37°C. Bacteria were washed with PBS and incubated with 100 μl of Alexa Fluor 488-conjugated streptavidin (10 μg/ml in PBS; Molecular Probes, Eugene, Oreg.) for 30 min on ice. The binding of lactoferrin to different pneumococcal strains is expressed as median fluorescence intensity.
Measurement of C3 deposition.
To quantitate the deposition of C3 on the pneumococcal surface, the same numbers of CFU of different strains (107 CFU of culture at an OD of 0.5) were washed with PBS and incubated with 100 μl of 10% normal human serum (NHS) (Quidel, San Diego, Calif.) diluted in gelatin Veronal buffer (GVB) (Sigma) for 30 min at 37°C. Goat anti-human C3 serum (Quidel) was biotinylated as described above. After washing with PBS, the bacteria were incubated with biotinylated goat anti-human C3 antibody (1:100 dilution in PBS) for 30 min at 37°C. The bacterial suspensions were then mixed with Alexa Fluor 488-conjugated streptavidin, and finally the samples were analyzed by flow cytometry. The percentage of Alexa Fluor 488-positive bacteria (fluorescence intensity greater than 10) was used as a measure of the amount of C3 deposition.
Infection of mice.
Female CBA/CaHN-XID/J (CBA/N) mice, 6 to 12 weeks old, were obtained from The Jackson Laboratories (Bar Harbor, Maine). Frozen infection stocks containing a known concentration of viable bacteria were diluted in lactated Ringer's solution to achieve the desired concentrations of bacteria. The numbers of bacteria injected were confirmed by plating on blood agar plates. For the median-time-to-death studies, CBA/N mice were challenged intravenously (i.v.) with 200 CFU of pneumococci per mouse in 200 μl of Ringer's solution. To assess the pneumococcal net growth and clearance in mouse blood in the early phase of infection, mice were challenged i.v. with 2 × 105 CFU per mouse. Blood samples were collected from the retro-orbital plexus at indicated time points. Samples were serially diluted and plated on the blood agar with, or without, erythromycin to determine the number of viable S. pneumoniae cells in the blood. When mice were coinfected with WU2 and its isogenic mutant (BR93.1 or BR92.1), 105 CFU of WU2 and 105 CFU of the mutant were mixed and injected into mice i.v. in a 0.2-ml volume. In this case, half of each blood sample was plated on blood agar, and the other half was plated on blood agar containing erythromycin. The numbers of BR93.1 and BR92.1 were determined by colony counts on the erythromycin-containing plates, and the numbers of WU2 were calculated by subtracting the numbers of CFU growing on the erythromycin plates from those growing on blood agar plates.
Sequencing and DNA analysis.
DNA sequencing was completed using automated DNA sequencing (model ABI 377; Applied Biosystems, Inc., Foster City, Calif.). Sequence analyses were performed using MacVector 6.5 (Oxford Molecular, San Diego, Calif.) and Sequencer 3.0 (GeneCodes, Ann Arbor, Mich.). PspA/TIGR4 sequence data (27) were obtained from The Institute for Genomic Research website at http://www.tigr.org.
RESULTS
Construction of WU2 derivatives expressing family 2 PspA from strain TIGR4.
WU2 is a capsule type 3 strain that has a family 1 pspA gene; it is in this background that the majority of data regarding PspA's effects on virulence and complement activation have been previously obtained (23, 28; M. Abeyta, G. G. Hardy, and J. Yother, submitted for publication). Because capsule and several non-PspA bacterial proteins are also expected to affect complement deposition, it was important to compare different PspA proteins in the same strain or genetic background. TIGR4 has a family 2 PspA, and it is only 32% identical to PspA/WU2 in the N-terminal alpha-helical region. To determine if this family 2 PspA could function similarly to the family 1 PspA, the pspA gene in WU2 was replaced with the pspA gene from TIGR4 as follows: (i) the C-terminal portion of PspA/TIGR4 was amplified and cloned into the vector pJY4164 to make pREN4; (ii) pREN4 was inserted into the chromosome of TIGR4 by the insertion-duplication procedure (32) to form strains BR81 and BR84, which carried an erythromycin resistance marker downstream of pspA; (iii) chromosomal DNA from strain BR81 or BR84 was used to transform strain WU2, and erythromycin-resistant transformants (BR92.1 and BR93.1, respectively) were selected; (iv) the transfer of pspA/TIGR4 was confirmed based on the size and specificity of PCR amplicons produced using family-specific primers (29); (v) three additional backcrosses were performed to remove unlinked genes that resulted from the original transformation with chromosomal DNA from the TIGR4 genetic background.
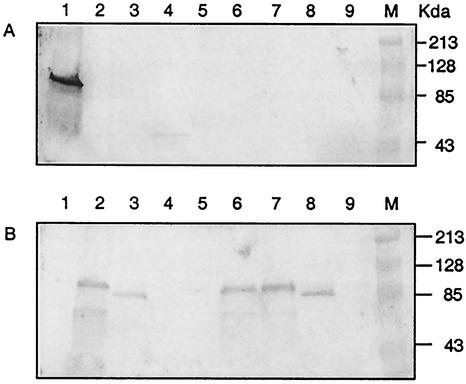
The replacement of PspA was verified by Western blot analysis using rabbit antisera specific for family 1 and 2 PspA proteins (Fig. 1). PspA proteins from WU2 and JY1123 were recognized by anti-family 1 PspA serum, but not by anti-family 2 PspA serum. The band for cell-associated PspA/JY1123 was very faint, since most of the PspA is secreted into the culture supernatant (described below). PspA proteins from TIGR4, BR81, BR84, BR93.1, and BR92.1 were recognized by anti-family 2 PspA, but not by anti-family 1 PspA serum. PspA-negative mutants JY1119 (WU2 background) and BR61.1 (TIGR4 background) were included as controls; neither of these reacted with either anti-PspA family 1 serum or anti-PspA family 2 serum (Fig. 1).
FIG. 1.
Western blots reacted with rabbit antisera specific for family 1 or 2 PspA. Bacterial cell lysates were separated on duplicate SDS-PAGE gels and transferred onto nitrocellulose membranes. Blots were reacted with anti-family 1 PspA serum (A) or anti-family 2 PspA serum (B). Lanes: 1, WU2; 2, BR93.1; 3, BR92.1; 4, JY1123; 5, JY11119; 6, TIGR4; 7, BR84; 8, BR81; 9, BR61.1.
Characterization of a mutant family 2 PspA lacking six choline-binding repeats.
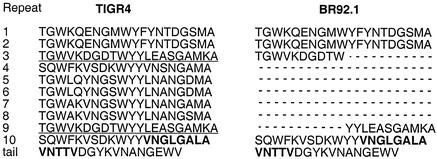
Taken together, the results above demonstrated that the family 1 PspA of WU2 was replaced by the family 2 PspA/TIGR4 in both BR93.1 and BR92.1. However, although PspA/BR93.1 and PspA/BR84 had the same apparent molecular size as wild-type PspA/TIGR4, PspA/BR92.1 and PspA/BR81 were smaller. Since BR92.1 was constructed by transforming WU2 with BR81, and BR81 was constructed by transforming TIGR4 with pREN4, it was speculated that a deletion occurred when the plasmid incorporated into the TIGR4 chromosome to form BR81. By using various primer pairs to amplify different regions of pspA, we confirmed the presence of a deletion and demonstrated that the deletion occurred in the 3′ region of the gene (data not shown). To further define the mutation, the pspA genes of BR93.1 and BR92.1 were amplified with primer pair LSM12 and LSM2 and sequenced. The sequence of pspA/BR93.1 matched the sequence of pspA/TIGR4, but pspA/BR92.1 had an internal deletion (Fig. 2). As a result, PspA/BR92.1 contained only four choline-binding repeats in the C-terminal region instead of ten choline-binding repeats as in the wild-type PspA/TIGR4. The six internal repeats had been deleted, including all of the fourth to eighth repeats and complementary portions of the third and ninth repeats. The third and the ninth choline-binding repeats of pspA/TIGR4 are identical, making it likely that the deletion occurred through recombination between the third repeat on the TIGR4 chromosome and the ninth repeat on pREN4 when pREN4 was inserted into the chromosome in TIGR4.
FIG. 2.
Comparison of sequences of PspA C-terminal regions from TIGR4 and BR92.1. The missing six choline-binding repeats in PspA/BR92.1 are indicated as dashed lines. The sequences of the third and ninth repeats in PspA/TIGR4 are identical and are underlined. The C-terminal hydrophobic stretch is indicated in boldface type.
Surface expression of parental PspA proteins and the mutant PspA lacking six choline-binding repeats.
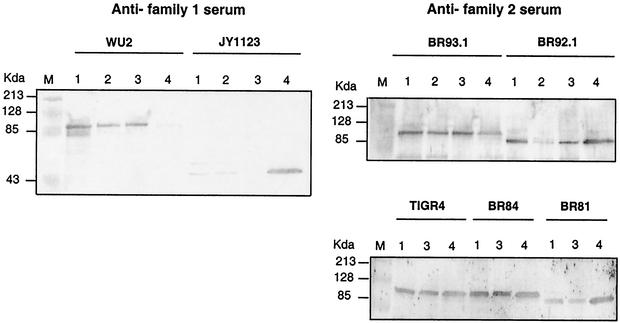
The choline-binding repeats of PspA bind to phosphocholine residues of the cell membrane lipoteichoic acid and attach PspA to the pneumococcal cell surface (34). Accordingly, the pneumococcal surface attached PspA can be released by washing with 2% choline chloride (35). To determine whether the family 2 wild-type and mutant PspA proteins were as strongly attached to the WU2 surface as family 1 PspA, we examined the PspA content of the intact bacteria, culture supernatant, and 2% choline chloride wash of the bacteria (Fig. 3). As previously described (7), 46-kDa truncated PspA of JY1123 only containing the N-terminal alpha-helical region was almost completely secreted into culture supernatant. A residual amount of PspA/JY1123 was present, however, in cell pellet of grown cells and the pellet of the same cells washed with 2% choline chloride. In the case of WU2, a trace amount of PspA was observed in supernatant, and the majority of PspA was located in the pellet of grown WU2 pneumococci. About half of the cell-associated PspA was released from the cell surface when WU2 was washed with 2% choline chloride.
FIG. 3.
Western blot analysis of the release of PspA proteins from the pneumococcal surface. Cultures taken at mid-exponential phase were centrifuged, and the culture supernatants were filtered. The cell pellets were incubated with 2% choline chloride, the mixture was centrifuged, and the supernatants were filtered. The pellets were lysed by incubation with lysis buffer. Equivalent amounts of all fractions, based on original culture volume, were analyzed by Western blotting. Lanes: 1, whole-cell pellets; 2, cell pellets after 2% choline wash; 3, supernatant fluid of 2% choline wash; 4, culture supernatant fluid.
Compared to the results with WU2, more PspA appeared in the supernatant fractions of strains TIGR4, BR84, and BR93.1. These results suggested that the family 2 PspA/TIGR4 was not attached on the cell surface as tightly as the family 1 PspA/WU2 and that the attachment of the two PspA proteins was not affected by bacterial surface structure since the poorer attachment of PspA/TIGR4 occurred with both the capsular serotype 3 strain, BR93.1, and the serotype 4 strain, TIGR4.
When strain BR92.1 was examined, an even larger fraction of PspA appeared in the supernatant and choline wash than that observed for BR93.1. Earlier reports showed that truncated PspA/Rx1 containing five choline-binding repeats, but missing the 17-aa tail, was totally secreted into culture supernatant. In contrast we observed that there was a significant amount of the mutant family 2 PspA/BR92.1 associated with the cell surface, even though this PspA contained only 4 of the normal 10 choline-binding repeats.
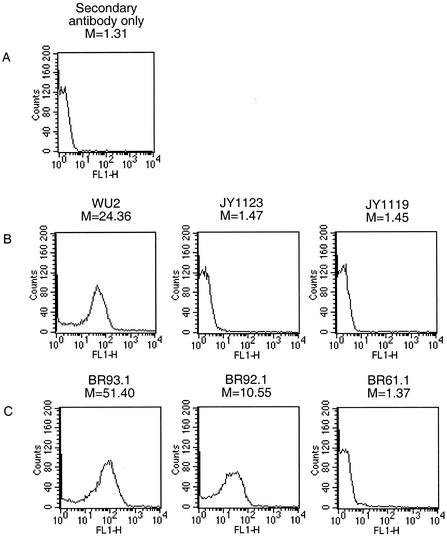
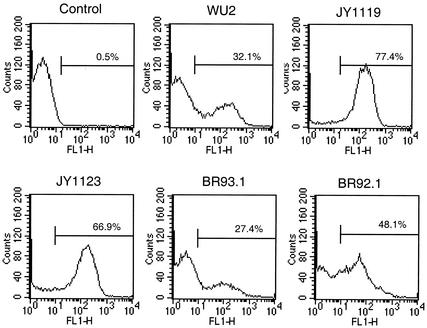
To further verify the presence of PspA on the bacterial cell surface in the newly constructed strains, flow cytometry was performed (Fig. 4). The median fluorescence intensities of JY1119, JY1123, and BR61.1 were comparable to those of the control sample incubated with Alexa Fluor 488-conjugated streptavidin alone. This procedure verified a lack of PspA on the surface of these strains. The median intensities of BR93.1 and BR92.1 were 51.40 and 10.55, respectively. The binding intensity of BR92.1 was only 21% of that of BR93.1 but was much greater than that of the TIGR4 PspA-negative strain, BR61.1. These results support the conclusion based on the results in Fig. 3 showing that PspA/BR92.1 is bound on the bacterial surface even though its attachment is not as robust as that of full-length family 2 PspA.
FIG. 4.
Binding of anti-PspA antibodies to the surface. Bacteria were incubated with rabbit anti-family 1 (B) or 2 (C) PspA serum and then washed with PBS. Bacterium-bound anti-PspA antibodies were measured by flow cytometry using fluorescein isothiocyanate-labeled goat anti-rabbit IgG as the probe. The intensity of the fluorescence associated with the different strains was registered. The value (M) represents the median intensity of fluorescence for each sample. In each case, the data shown are representative of at lease three experiments.
Binding of human lactoferrin to WU2 wild-type and mutant strains.
Previously, Hammerschmidt et al. reported that 88% of the clinical S. pneumoniae isolates bound to lactoferrin and that PspA was one of the lactoferrin ligands on the bacterial surface (11). Later, Hakansson et al. observed that human lactoferrin specifically bound to PspA and the primary binding site resides in the C-terminal part of the alpha-helical domain of PspA (10).
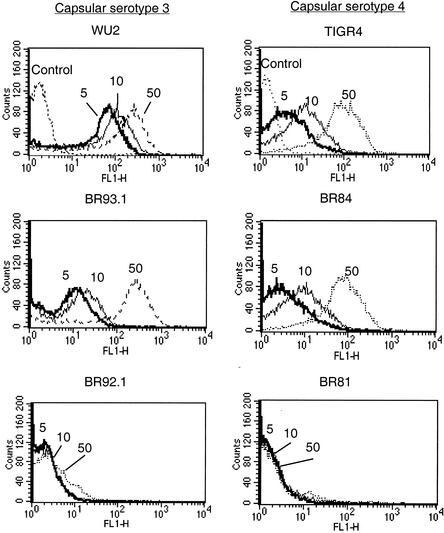
We compared the lactoferrin binding of the family 1 (PspA/WU2) and family 2 (PspA/TIRG4) PspA proteins at three different concentrations of lactoferrin (5, 10, and 50 μg/ml) (Fig. 5). PspA-negative strains JY1119 and BR61.1 did not bind lactoferrin at any of the three concentrations (data not shown). At lactoferrin concentrations of 5 and 10 μg/ml, the lactoferrin binding of WU2 was stronger than that of TIGR4. BR93.1 showed the same result as TIGR4. The median fluorescence intensity of WU2 at a concentration of 5 μg/ml was 36.9, almost nine times as much as the medians of BR93.1 and TIGR4 (4.2 and 3.8, respectively).
FIG. 5.
Binding of lactoferrin by full-length PspA proteins and internal deleted PspA in two genetic backgrounds. Bacteria were incubated with biotin-labeled lactoferrin at concentrations of 5, 10, and 50 μg/ml (indicated as 5, 10, and 50). Samples treated with PBS instead of lactoferrin were used as controls. With lactoferrin concentrations at 5, 10, and 50 μg/ml, the medians of fluorescence intensities of each strain were as follows: for WU2, 36.9, 54.7, and 125.2; for BR93.1, 4.2, 6.9, and 121.9; for BR92.1, 1.6, 1.7, and 2.29; for TIGR4, 3.8, 10.5, and 83.5; for BR84, 2.9, 7.8, and 73.0; for BR81, 1.6, 1.4, and 1.4. The medians of control samples of WU2 and TIGR4 were 1.5 and 1.3, respectively.
However, when the concentration of lactoferrin increased to 50 μg/ml, the binding of lactoferrin to BR93.1 and TIGR4 was similar to that of WU2. BR84 showed the same result as TIGR4. The difference in lactoferrin binding between WU2 and BR93.1 (or TIGR4) at low but not high concentrations of lactoferrin suggested that family 1 PspA probably has higher binding affinity to lactoferrin than family 2 PspA.
We were surprised to observe that the mutant PspA/BR92.1, missing six choline-binding repeats, did not bind lactoferrin even at a concentration of 50 μg/ml. The failure of this shorter surface-anchored PspA to bind lactoferrin was not a property of the genetic background of the strain, since both BR92.1 and BR81 showed the same result. Lactoferrin's inability to bind to BR92.1 and BR81 could not be fully explained by a lack of PspA on the cell surface, since the anti-PspA antibody detected at least 20% as much of the mutant PspA on the surface compared with the full-length family 2 PspA (Fig. 4).
C3 deposition on WU2 wild-type and mutant strains.
After incubation with NHS, the amount of C3 deposition was monitored by measuring the percentage of fluorescence-positive bacteria by flow cytometry (Fig. 6). As expected, the amount of C3 that could be detected on PspA-negative strains JY1119 and JY1123 was substantially greater than the amount of C3 detected on PspA-positive strain WU2. The same amount of C3 was detected on BR93.1 as on WU2, indicating that both family 1 and family 2 PspA proteins can decrease complement activation on the WU2 surface. Deposition of C3 on BR92.1 was higher than those on BR93.1 and WU2, but less than those on JY1119 and JY1123. This observation was consistent with the relative amount of surface-associated proteins as detected by anti-PspA antibodies above (Fig. 4).
FIG. 6.
Comparison of complement deposition. Live bacteria were incubated with GVB (Control) or 10% NHS (all other panels) in GVB, washed with PBS and then incubated with biotin-labeled goat anti-human C3 serum. The relative amount of C3 bound on cell surface was detected by flow cytometry after staining with Alexa Fluor 488-conjugated streptavidin. Bacteria were then fixed prior to flow cytometry. The percentage of bacteria with fluorescence intensity higher than 10 is shown for each strain. Data representative of at least three different experiments are shown.
Evaluation of the virulence effects of family 1 and family 2 PspA proteins on the WU2 genetic background.
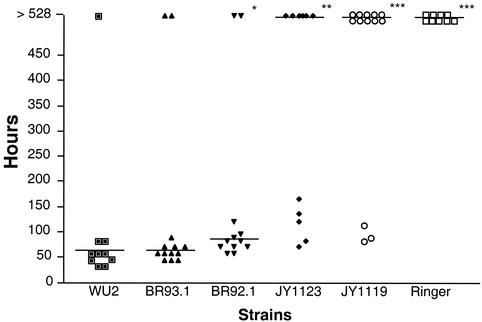
When CBA/N mice were challenged i.v. with 200 CFU of bacteria, it was observed that wild-type WU2 and its derivative expressing family 2 PspA, BR93.1, exhibited the same high levels of virulence. The median time to death was 58 h in both cases (Fig. 7). In contrast, the PspA-negative mutant, JY1119, and the mutant JY1123 expressing secreted PspA, showed significant reductions in virulence as has been previously shown (1, 4). More than half of the mice infected with JY1119 and JY1123 were still alive at 528 h postinoculation when the experiment was terminated. The family 2 PspA mutant, BR92.1, showed slightly lower virulence than WU2 or BR93.1 and resulted in a median time to death of 82 h. The result with BR92.1 was different from those with WU2 and BR93.1, at P = 0.03 and 0.08, respectively (Fig. 7).
FIG. 7.
Median time to death of CBA/N mice infected with WU2 and its derivatives. Ten to thirteen CBA/N mice for each group were infected i.v. with 200 CFU of bacteria. Median time to death is indicated. Significance of the result with WU2 is indicated as follows: *, P < 0.05; **, P < 0.01; ***. P < 0.001. Significance was assessed using the Mann-Whitney test.
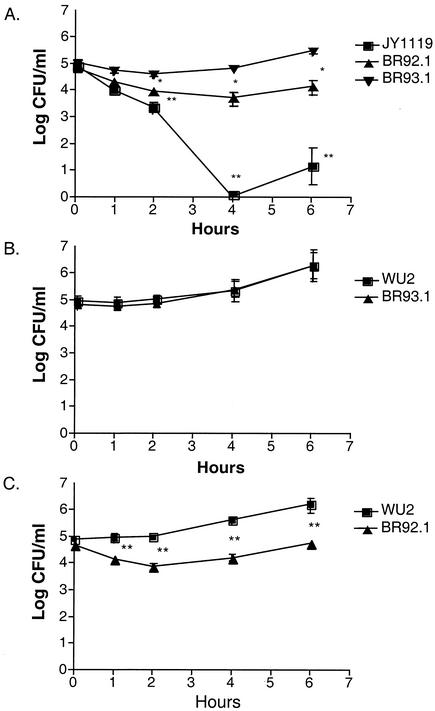
To obtain a better comparison of the difference in virulence between BR92.1 and BR93.1, we challenged the CBA/N mice i.v. separately with these two strains (2 × 105 CFU/mouse) as well as JY1119 and assessed the changes in CFU per milliliter of blood of the infected mice (Fig. 8A). During the first 4 h after infection, the number of BR92.1 CFU continued to decrease and dropped to the lowest point at 4 h postinoculation. In contrast, the number of BR93.1 CFU only decreased slightly in the first 2 h and continued to increase after that. There was a 10-fold difference between BR93.1 and BR92.1 at 4 h postinoculation. As expected, the PspA-negative strain JY1119 showed a marked attenuation of virulence, and was almost cleared from mouse blood within four hours postinoculation. An increase of the number of bacteria in the blood after first few hours is commonly seen in mouse infection with pneumococci and probably reflects some adaptation of the bacteria to the host (21, 28).
FIG. 8.
Growth of WU2 and/or its derivatives in mouse blood. CBA/N mice (five mice per group) were infected i.v. with 2 × 105 CFU and bled retro-orbitally at various time points postinoculation (A). In coinfection experiments, CBA/N mice were challenged i.v. with WU2 (1 × 105 CFU) plus the same number of BR93.1 (B) or BR92.1 (C) CFU. The number of CFU per milliliter was determined by plating the blood sample on blood agar. Error bars indicate standard errors. All experiments were repeated three times with equivalent results. Significance of the result compared to BR93.1 (A) or WU2 (B and C) is indicated as follows: *, P < 0.05; **, P < 0.01 (Mann-Whitney test).
To provide confirmation of the difference in bacteremia caused by BR93.1 and BR92.1 and to compare these strains directly with WU2, we preformed coinfection of CBA/N mice with WU2 plus BR93.1 or BR92.1 (Fig. 8B and C). The CFU of BR93.1 and BR92.1 could be differentiated from those of WU2 due to antibiotic resistance marker in the former two strains. A mixture of WU2 (105 CFU) and BR93.1 or BR92.1 (105 CFU) was injected into mice. The growth of BR93.1 in mouse blood was comparable to that of WU2; the numbers of CFU of both strains rose steadily, and the ratio of the number of BR93.1 CFU to that of WU2 remained near 1 over the period of observation. In contrast, the ratio of the number of CFU of BR92.1 to that of WU2 decreased from 1 to 0.1 over the period of observation. These results made it clear that both family 1 and family 2 PspA proteins had the same effect on virulence of WU2, but the mutant family 2 PspA which lacked six choline-binding repeats caused a partial attenuation of virulence in capsular serotype 3 strain (WU2).
DISCUSSION
It has been previously shown that PspA binds to the pneumococcal surface through the choline-binding domain on the C terminus (35). From a molecular model of PspA, it has been predicted that an antiparallel coiled-coil alpha-helical domain extends into and through the capsular layer and is tethered by the proline-rich region to the surface-attached choline-binding domain (3, 14). In the present study, more family 2 PspA/TIGR4 was observed in culture supernatant than family 1 PspA/WU2. The surface availability of family 2 PspA was the same on both capsular serotype 3 strain, WU2, and capsular serotype 4 strain, TIGR4, backgrounds. Sequence analysis of the C-terminal region of PspA (including the choline-binding repeats and the tail domain) from several strains has revealed considerable conservation among different families of PspA proteins (3, 18). Even so, we observed differences in the binding and/or release of PspA/WU2 and PspA/TIGR4, even when both were expressed on the same genetic background. We assume that this observation was due to small differences in the structure of the choline-binding or proline-rich domains of the proteins.
The original studies about the attachment mechanism of PspA demonstrated that the anchoring of PspA to the bacterial cell surface requires choline-mediated interactions between the membrane-associated lipoteichoic acid and the C-terminal repeat region (35). However, there is a short and weakly hydrophobic stretch containing 13 aa crossing the last half-repeat and extending into the C-terminal 17-aa tail (Fig. 2). This sequence is highly conserved (18). It appeared to be too short to function as a membrane-spanning anchor, and Yother et al. previously reported that the elimination of the last 20 aa, which included more than half of this hydrophobic region, did not result in loss of attachment of PspA in strain Rx1 (35). It was also shown that PspA lacking the last five choline-binding repeats plus the C-terminal tail was no longer bound to the cell surface of Rx1 or WU2 (1, 34), indicating that the five remaining repeats were not sufficient for surface attachment. These results had suggested that more than five choline-binding repeats, but not the 17-aa tail, were required for surface attachment of PspA.
Our results showed that PspA/BR92.1, with four repeats, was still anchored on the cell surface of WU2 or TIGR4. However, because the deletion occurred in the internal repeat region instead of at the C-terminal end, the mutant PspA/BR92.1 still contains the last repeat and the 17-aa C-terminal tail with its hydrophobic stretch intact. Therefore, our data considered in the light of the previous studies (1, 34, 35) suggest that the C-terminal 17-aa tail can function together with the repeat units to help anchor PspA to pneumococcal surface.
We also observed that family 1 and 2 PspA proteins showed different binding activities for lactoferrin. These differences were inherited in the PspA proteins themselves, since changing genetic background did not affect the lactoferrin binding activity of PspA/TIGR4. At low concentration (5 or 10 μg/ml), family 1 PspA/WU2 binds more lactoferrin than family 2 PspA/TIGR4. It is unlikely that the different binding is due to the release of the PspA from cell surface, since both PspA proteins bound similar amounts of lactoferrin at high concentration (50 μg/ml).
Using recombinant fragments of family 1 and family 2 PspA proteins, Hankansson et al. showed that lactoferrin binds to the C-terminal end of the PspA alpha-helical region (10), the same region that elicits cross-protective immune responses (19). Alignment of the whole alpha-helical regions of family 1 PspA/WU2 and family 2 PspA/TIGR4 revealed that this family 2 PspA (418 aa) is much longer than the family 1 PspA (278 aa), and they share 32% identity overall. At the C-terminal end of the alpha-helical region where lactoferrin is thought to bind, there is only 23% identity between PspA/WU2 and PspA/TIGR4 (10, 13, 27). Thus, the disparate sequences in this region probably contribute to the different binding activity to lactoferrin. We suspect that a lactoferrin concentration of 50 μg/ml may be saturating, which therefore explains our inability to detect differential binding of family 1 and 2 PspA proteins.
It was surprising to observe that no lactoferrin binding was detected on the surfaces of BR92.1 and BR81, as the mutant family 2 PspA is missing only six choline-binding repeats and the alpha-helical region maintains intact. When the whole-cell lysate of BR92.1 was analyzed by SDS-PAGE and transferred onto nitrocellulose, the immobilized mutant PspA was found to bind lactoferrin (data not shown). Thus, the lack of binding by lactoferrin is probably not because of any change in the linear sequence of lactoferrin-binding site. Our results suggest that when PspA/BR92.1 is expressed on the cell surface, the reduction in length caused by the 121-aa deletion in the repeat region may prevent the binding site from being as accessible as it is on wild-type full-length PspA.
By incubating S. pneumoniae with normal mouse serum in vitro, both Tu et al. and Abeyta et al. observed, with Western blot or enzyme-linked immunosorbent assay methodology, that PspA-negative pneumococci bind more C3 than their isogenic PspA-positive wild-type strains (1, 28; M. Abeyta, G. G. Hardy, and J. Yother, submitted). Here, we have shown that PspA has the same inhibiting effect on deposition of human complement as was previously shown for mouse complement. In our studies, complement deposition was quantified using FACS analysis and intact living pneumococci, thus further increasing our confidence in the relevance of these findings.
The surface attachment of PspA appears to be essential for PspA to interfere with complement deposition onto pneumococci. Although PspA/JY1123 expresses the entire N-terminal functional region, the PspA is secreted and does not remain on the bacterial surface. The amount of C3 fixed on JY1123 was comparable to that of PspA-negative strain, JY1119. The amount of C3 on BR92.1 fell between the amount of C3 on BR93.1 and that on JY1119, and the difference in C3 deposition between BR92.1 and BR93.1 correlated with the results of anti-PspA antibody staining on the surface of these two strains and with the differences in virulence of these two strains. The latter observation provides the best evidence so far that the ability of PspA to interfere with complement activation plays a role in virulence.
Although the high diversity of PspA sequence would be consistent with a hypothesis that different PspA families may not have the same function, our results failed to support this idea. Family 1 and family 2 PspA proteins only share approximately 40% similarity overall, and the alpha-helical region of family 2 PspA is much longer than that of family 1 PspA. In spite of these differences, both exhibit the same virulence in a mouse bacteremia model. These similarities of our in vivo virulence results were in agreement with the virtually identical levels of in vitro complement deposition on BR93.1 and WU2.
These results suggest that these two PspA proteins, representing the two major PspA families, exert the same negative effects on complement deposition. Because of the sequence variation in PspA, it is likely that the ability of PspA proteins to interfere with complement deposition may depend upon a shared area of conformations of family 1 and 2 PspA proteins rather than upon the specific conservation of a linear amino acid sequence. Our present findings are consistent with a hypothesis that the major effect of different PspA proteins on virulence is mediated by their common ability to inhibit complement deposition on pneumococci.
Lactoferrin is a multifunctional protein which plays an important role in innate immunity. It is cytotoxic for bacteria and inhibits bacterial adherence and colonization on mucosal surfaces (2, 8). Importantly, it was also reported that lactoferrin affects the activation of the classical complement pathway (15, 16, 25). In our studies, family 1 and family 2 PspA proteins did not show the same binding activity to lactoferrin at low concentrations (5 or 10 μg/ml), which are near the lactoferrin concentrations in NHS. However, they showed the same effects on complement activation. Moreover, the truncated PspA/BR92.1 did not bind to lactoferrin but still maintained the anticomplementary function in human serum and was still partially virulent in mice. These results may indicate that the function of inhibiting human complement deposition probably does not depend on the PspA binding with serum lactoferrin. The results suggest, but do not prove, that lactoferrin may not play an important role in the pneumococcal bacteremia or sepsis model. It must be remembered that our lactoferrin binding studies were done with human lactoferrin sharing only 69% sequence identity with mouse lactoferrin. Thus, based on these data, we cannot completely exclude an important role of the PspA-human lactoferrin binding in bacteremia or sepsis in humans. The PspA-lactoferrin interaction may play an important role in nasopharyngeal colonization, which is important for both pneumococcal acquisition and spread to other humans.
Acknowledgments
We thank John F. Kearney, Flavius Martin, and Moon H. Nahm for their assistance with the flow cytometric analysis and thank Janet Yother, Anders Hakansson, Gunnar Lindahl, James Watt, and Alexis Brooks-Walter for helpful discussions.
This work was supported by NIH grant AI42183 (A.J.S.) and by NIH grant AI21548 and the Carsten Cole Buckley Pediatric Meningitis Research Fund (D.E.B.).
Editor: D. L. Burns
REFERENCES
- 1.Abeyta, M. 1999. Ph.D. thesis. University of Alabama at Birmingham, Birmingham.
- 2.Arnold, R. R., M. F. Cole, and J. R. McGhee. 1977. A bactericidal effect for human lactoferrin. Science 197:263-265. [DOI] [PubMed] [Google Scholar]
- 3.Briles, D. E., S. K. Hollingshead, E. Swiatlo, A. Brooks-Walter, A. Szalai, A. Virolainen, L. S. McDaniel, K. A. Benton, P. White, K. Prellner, A. Hermansson, P. C. Aerts, H. Van Dijk, and M. J. Crain. 1997. PspA and PspC: their potential for use as pneumococcal vaccines. Microb. Drug Resist. 3:401-408. [DOI] [PubMed] [Google Scholar]
- 4.Briles, D. E., J. Yother, and L. S. McDaniel. 1988. Role of pneumococcal surface protein A in the virulence of Streptococcus pneumoniae. Rev. Infect. Dis. 10(Suppl. 2):S372-S374. [DOI] [PubMed] [Google Scholar]
- 5.Brooks-Walter, A., D. E. Briles, and S. K. Hollingshead. 1999. The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect. Immun. 67:6533-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, Q., D. Finkel, and M. K. Hostetter. 2000. Novel purification scheme and functions for a C3-binding protein from Streptococcus pneumoniae. Biochemistry 39:5450-5457. [DOI] [PubMed] [Google Scholar]
- 7.Crain, M. J., W. D. Waltman, J. S. Turner, J. Yother, D. F. Talkington, L. S. McDaniel, B. M. Gray, and D. E. Briles. 1990. Pneumococcal surface protein A (PspA) is serologically highly variable and is expressed by all clinically important capsular serotypes of Streptococcus pneumoniae. Infect. Immun. 58:3293-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deriy, L. V., J. Chor, and L. L. Thomas. 2000. Surface expression of lactoferrin by resting neutrophils. Biochem. Biophys. Res. Commun. 275:241-246. [DOI] [PubMed] [Google Scholar]
- 9.Gosink, K. K., E. R. Mann, C. Guglielmo, E. I. Tuomanen, and H. R. Masure. 2000. Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect. Immun. 68:5690-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hakansson, A., H. Roche, S. Mirza, L. S. McDaniel, A. Brooks-Walter, and D. E. Briles. 2001. Characterization of binding of human lactoferrin to pneumococcal surface protein A. Infect. Immun. 69:3372-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammerschmidt, S., G. Bethe, P. H. Remane, and G. S. Chhatwal. 1999. Identification of pneumococcal surface protein A as a lactoferrin-binding protein of Streptococcus pneumoniae. Infect. Immun. 67:1683-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henriques Normark, B., R. Novak, A. Ortqvist, G. Kallenius, E. Tuomanen, and S. Normark. 2001. Clinical isolates of Streptococcus pneumoniae that exhibit tolerance of vancomycin. Clin. Infect. Dis. 32:552-558. [DOI] [PubMed] [Google Scholar]
- 13.Hollingshead, S. K., R. Becker, and D. E. Briles. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun. 68:5889-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jedrzejas, M. J., E. Lamani, and R. S. Becker. 2001. Characterization of selected strains of pneumococcal surface protein A. J. Biol. Chem. 276:33121-33128. [DOI] [PubMed] [Google Scholar]
- 15.Kievits, F., and A. Kijlstra. 1985. Inhibition of C3 deposition on solid-phase bound immune complexes by lactoferrin. Immunology 54:449-456. [PMC free article] [PubMed] [Google Scholar]
- 16.Kijlstra, A., and S. H. Jeurissen. 1982. Modulation of classical C3 convertase of complement by tear lactoferrin. Immunology 47:263-270. [PMC free article] [PubMed] [Google Scholar]
- 17.Kostyukova, N. N., M. O. Volkova, V. V. Ivanova, and A. S. Kvetnaya. 1995. A study of pathogenic factors of Streptococcus pneumoniae strains causing meningitis. FEMS Immunol. Med. Microbiol. 10:133-137. [DOI] [PubMed] [Google Scholar]
- 18.McDaniel, L. S., D. O. McDaniel, S. K. Hollingshead, and D. E. Briles. 1998. Comparison of the PspA sequence from Streptococcus pneumoniae EF5668 to the previously identified PspA sequence from strain Rx1 and ability of PspA from EF5668 to elicit protection against pneumococci of different capsular types. Infect. Immun. 66:4748-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDaniel, L. S., B. A. Ralph, D. O. McDaniel, and D. E. Briles. 1994. Localization of protection-eliciting epitopes on PspA of Streptococcus pneumoniae between amino acid residues 192 and 260. Microb. Pathog. 17:323-337. [DOI] [PubMed] [Google Scholar]
- 20.McDaniel, L. S., J. S. Sheffield, P. Delucchi, and D. E. Briles. 1991. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular type. Infect. Immun. 59:222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDaniel, L. S., J. Yother, M. Vijayakumar, L. McGarry, W. R. Guild, and D. E. Briles. 1987. Use of insertional inactivation to facilitate studies of biological properties of pneumococcal surface protein A (PspA). J. Exp. Med. 165:381-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Musher, D. M. 1992. Infections caused by Streptococcus pneumoniae: clinical spectrum, pathogenesis, immunity, and treatment. Clin. Infect. Dis. 14:801-807. [DOI] [PubMed] [Google Scholar]
- 23.Neeleman, C., S. P. Geelen, P. C. Aerts, M. R. Daha, T. E. Mollnes, J. J. Roord, G. Posthuma, H. van Dijk, and A. Fleer. 1999. Resistance to both complement activation and phagocytosis in type 3 pneumococci is mediated by the binding of complement regulatory protein factor H. Infect. Immun. 67:4517-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paton, J. C., and J. K. Morona. 2000. Streptococcus pneumoniae capsular polysaccharide, p. 201-213. In V. A. Fishchetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 25.Rainard, P. 1993. Activation of the classical pathway of complement by binding of bovine lactoferrin to unencapsulated Streptococcus agalactiae. Immunology 79:648-652. [PMC free article] [PubMed] [Google Scholar]
- 26.Smith, B. L., and M. K. Hostetter. 2000. C3 as substrate for adhesion of Streptococcus pneumoniae. J. Infect. Dis. 182:497-508. [DOI] [PubMed] [Google Scholar]
- 27.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 28.Tu, A. H., R. L. Fulgham, M. A. McCrory, D. E. Briles, and A. J. Szalai. 1999. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect. Immun. 67:4720-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vela Coral, M. C., N. Fonseca, E. Castaneda, J. L. Di Fabio, S. K. Hollingshead, and D. E. Briles. 2001. Pneumococcal surface protein A of invasive Streptococcus pneumoniae isolates from Colombian children. Emerg. Infect. Dis. 7:832-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vorland, L. H. 1999. Lactoferrin: a multifunctional glycoprotein. APMIS 107:971-981. [DOI] [PubMed] [Google Scholar]
- 31.Vorland, L. H., H. Ulvatne, J. Andersen, H. H. Haukland, O. Rekdal, J. S. Svendsen, and T. J. Gutteberg. 1999. Antibacterial effects of lactoferricin B. Scand. J. Infect. Dis. 31:179-184. [DOI] [PubMed] [Google Scholar]
- 32.Yother, J. 2000. Genetics of Streptococcus pneumoniae, p. 232-243. In V. A. Fishchetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 33.Yother, J., and D. E. Briles. 1992. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J. Bacteriol. 174:601-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yother, J., G. L. Handsome, and D. E. Briles. 1992. Truncated forms of PspA that are secreted from Streptococcus pneumoniae and their use in functional studies and cloning of the pspA gene. J. Bacteriol. 174:610-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yother, J., and J. M. White. 1994. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J. Bacteriol. 176:2976-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zysk, G., B. K. Schneider-Wald, J. H. Hwang, L. Bejo, K. S. Kim, T. J. Mitchell, R. Hakenbeck, and H. P. Heinz. 2001. Pneumolysin is the main inducer of cytotoxicity to brain microvascular endothelial cells caused by Streptococcus pneumoniae. Infect. Immun. 69:845-852. [DOI] [PMC free article] [PubMed] [Google Scholar]