Abstract
Lipopolysaccharide (LPS), as the major surface molecule of gram-negative bacteria, interacts with the host in complex ways, both inducing and protecting against aspects of inflammatory and adaptive immunity. The membrane-distal repeated carbohydrate structure of LPS, the O antigen, can prevent antibody functions and may vary as a mechanism of immune evasion. Genes of the wbm locus are required for the assembly of O antigen on the animal pathogen Bordetella bronchiseptica and the human pathogen B. parapertussis. However, the important human pathogen B. pertussis lacks these genes and a number of in vitro and in vivo characteristics associated with O antigen in other organisms. To determine the specific functions of O antigen in these closely related Bordetella subspecies, we compared wbm deletion (Δwbm) mutants of B. bronchiseptica and B. parapertussis in a variety of assays relevant to natural respiratory tract infection. Complement was not activated or depleted by wild-type bordetellae expressing O antigen, but both Δwbm mutants activated complement and were highly sensitive to complement-mediated killing in vitro. Although the O-antigen structures appear to be substantially similar, the two mutants differed strikingly in their defects within the respiratory tract. The B. parapertussis Δwbm mutant was severely defective in colonization of the tracheas and lungs of mice, while the B. bronchiseptica Δwbm mutant showed almost no defect. While in vitro characteristics such as serum resistance may be attributable to O antigen directly, the role of O antigen during infection appears to be more complex, possibly involving factors differing among the closely related bordetellae or different interactions between each one and its host.
The genus Bordetella currently contains eight species, of which the three referred to as the classical bordetellae are exceptionally closely related respiratory tract pathogens (17, 35). Bordetella pertussis and B. parapertussis are human pathogens, causing whooping cough or pertussis, and are endemic in both vaccinated and unvaccinated populations worldwide (11, 12, 20, 30). B. bronchiseptica is a very common cause of respiratory tract infections in many animals, causing atrophic rhinitis in pigs, snuffles in rabbits, and kennel cough in dogs, but rarely infects humans (7, 29). These three organisms have recently been reclassified as subspecies (34) and express substantially similar sets of virulence factors regulated by the BvgAS two-component system (13, 14). Interestingly, differences between their abilities to infect, persist in, and cause disease in different hosts correlates with differences in their lipopolysaccharide (LPS) structures. Furthermore, we have previously shown that mutants of these three bordetellae expressing truncated LPS have very different defects during in vivo infection, suggesting that their LPS structures play roles in infection that are specific to each organism (19). B. pertussis LPS comprises a lipid A domain and a branched-chain core oligosaccharide that together form the structure called band B. This may be further modified by the addition of a complex trisaccharide (band B plus trisaccharide) and called band A (9, 23). B. bronchiseptica expresses band B and band A LPS but also expresses an O antigen that is a homopolymer of 2,3-dideoxoy-2,3-di-N-acetylgalactosaminuronic acid and is attached somewhere in the core-trisaccharide region. While B. parapertussis expresses the same O antigen as B. bronchiseptica, it expresses a band B LPS that is electrophoretically distinguishable from that of B. pertussis and B. bronchiseptica, indicating that the B. parapertussis core is smaller than that of the other two species. B. parapertussis also does not express band A LPS, possibly because of a mutation in the wlbH gene involved in band A trisaccharide biosynthesis (4). A comparison of their LPS compositions is shown in Table 1.
TABLE 1.
LPS composition of the Bordetella strains used in this studya
| Strain | Band A
|
O antigen | ||
|---|---|---|---|---|
| Band B
|
Trisaccharide | |||
| Lipid A | Core | |||
| B. bronchiseptica RB50 | + | + | + | + |
| B. bronchiseptica RB50Δwbm | + | + | + | − |
| B. bronchiseptica RB50Δwlb | + | + | − | − |
| B. parapertussis CN2591 | + | + | +b | + |
| B. parapertussis CN2591Δwbm | + | + | +c | − |
| B. parapertussis CN2591Δwlb | + | + | − | − |
| B. pertussis Bp536 | + | + | + | − |
| B. pertussis Bp536Δwlb | + | + | − | − |
A plus sign denotes the presence, and a minus sign denotes the absence, of the indicated LPS structure.
There is no apparent band A component visible on gels of this species because it appears to be substituted with O antigen (4).
This species does make a sugar group, but it may only be a disaccharide or a mixture of di-and trisaccharides, possibly because of an inefficient wlb H-encoded transferase (25).
A B. pertussis strain with a deletion in the wlb locus (Δwlb) expresses only band B LPS, indicating that these genes are required for the addition of the trisaccharide of band A. In B. bronchiseptica, this mutation results in the loss of both the band A trisaccharide and O antigen (presumably because of loss of the core acceptor site), leaving a lipid A-inner core molecule that was electrophoretically indistinguishable from that of the B. pertussis Δwlb mutant (Table 1). B. parapertussis Δwlb mutants express an LPS molecule that lacks both band A trisaccharide and O antigen but is more electrophoretically mobile than that of the Δwlb mutants of the other two subspecies because of the apparently smaller lipid A-inner core structures (4, 26). Although the LPS structures of Δwlb mutants of B. pertussis and B. bronchiseptica were similar and the LPS structures of both B. parapertussis and B. bronchiseptica Δwlb mutants lack expression of the band A trisaccharide and O antigen, there were substantial differences among the phenotypes of the three mutants in a mouse infection model. While the B. pertussis Δwlb mutant was severely defective in colonization of the nasal cavities of BALB/c mice, neither of the other mutants showed any defect in the colonization of or persistence at this site. The B. parapertussis Δwlb mutant was severely defective in colonizing the lungs, being recovered in numbers 1/10,000 of those of the wild-type strain by day 3 postinoculation. However, the B. bronchiseptica mutant was recovered from the lungs in numbers similar to those of the wild-type strain until day 7 postinoculation (19). The variety of defects of these three mutants was interpreted to reflect the diversity of functions of LPS in infection. It is interesting that resistance to serum complement, which has been attributed to O-antigen expression in other bacteria, was observed with wild-type B. bronchiseptica and B. parapertussis, which express O antigen, but not with wild-type B. pertussis or the wlb mutants, which do not (19).
Adjacent to the wlb locus in both B. bronchiseptica and B. parapertussis is the wbm locus, which is responsible for the expression of the 2,3-dideoxy-2,3-di-N-acetylgalactosaminuronic acid homopolymeric O antigen. Deletion of wbmB through the N-terminal portion of wbmE (Δwbm) in B. bronchiseptica and B. parapertussis leads to loss of O-antigen expression in these bacteria (25, 26). Since B. pertussis naturally lacks O antigen, it is worth noting that deletion of the wbm locus removes the major structural difference between the LPSs of B. bronchiseptica and B. parapertussis and that of B. pertussis (Table 1) (25, 26).
Here we used Δwbm mutants of B. bronchiseptica and B. parapertussis to determine the contribution of O antigen to the in vivo and in vitro defects observed in wlb mutants that lack both O antigen and outer core trisaccharide. Both Δwbm mutants lack the in vitro complement resistance of the respective wild-type strains. The B. parapertussis mutant was severely defective in colonization of BALB/c mice, being recovered from the trachea and lungs in numbers 1/1,000 of those of the wild-type strain. The B. bronchiseptica mutant, however, showed almost no defect in these mice. Similar results were observed in the colonization of SCID-beige mice, indicating that the B. parapertussis mutant was defective in the absence of adaptive immunity. Only the wild-type strains are lethal in SCID-beige mice, indicating that O antigen is required for the virulence of both subspecies in this model. These results reveal characteristics that are shared by the two Δwbm mutants and may be directly attributable to O antigen, and others that differ, suggesting more complex and subspecies-specific roles for O antigen.
MATERIALS AND METHODS
Bacterial strains and growth.
B. bronchiseptica RB50 (wild type) was obtained after a single passage from the original rabbit isolate (13). B. parapertussis wild-type strain CN2591 and the Δwbm mutant forms of both B. bronchiseptica and B. parapertussis wild-type strains have been previously described (3, 25). Mating with Escherichia coli SM10λpir containing the allelic exchange vector pEH10 was used to generate gentamicin-resistant Δwbm mutants of both bordetellae. In order to generate the plasmid pEH10, first the KpnI-SphI fragment of pBA61 (2), containing the 3′ end and terminator region of the flaA gene, was inserted into KpnI-SphI-cut pUC19. The resulting plasmid was then cut with SmaI and SnaBI, and the SmaI-SnaBI fragment of pMTL24Gm (10), containing a gentamicin resistance cassette, was inserted to make a construct with this antibiotic marker inserted between the flaA gene and its terminator. This locale would not be likely to interfere with transcription, translation, or virulence, as the flaA gene is not expressed during Bvg+-phase growth of bordetellae (1). The flaA gene gentamicin resistance fragment was excised by first cutting with KpnI and then blunt-end formation with the Klenow fragment of DNA polymerase (5), followed by HindIII restriction. The fragment was inserted into HindIII-EcoRV-cut pEG7 (15) to generate pEH8. The sacBR gene cartridge from pUM24 (27) was inserted as a BamHI fragment into the BglII site of pEH8 to generate pEH10. All enzymes were obtained from Promega (Madison, Wis.). Bacteria were maintained on Bordet-Gengou (BG) agar (Difco) supplemented with 7.5% defibrinated sheep blood (Remel or Hema Resources), 20 μg of streptomycin per ml, and, when indicated, 20 μg of gentamicin per ml. Bacteria were grown in Stainer-Scholte broth with supplements to mid-log phase (optical densities at 600 nm of ≤0.3) at 37°C on a roller drum for experiments.
Animal experiments.
Four- to six-week-old, male and female BALB/c mice were obtained from Jackson Laboratories, and breeding colonies were maintained at The Pennsylvania State University. SCID-beige mice were obtained from Charles River Laboratories (C.B-17/Icr/Crl-scid/bgBR background). Mice were lightly sedated with isoflurane (Abbott Laboratories, Chicago, Ill.) and inoculated by pipetting onto the tip of the external nares. For the time course experiments (see Fig. 2 and 3), groups of four animals were inoculated with 1,000 CFU in 50 μl of phosphate-buffered saline (PBS) and sacrificed on various days postinoculation. For the SCID-beige virulence experiment, a low-dose regimen consisted of 500 CFU of bacteria in 5 μl of PBS pipetted onto the external nares while the high-dose regimen consisted of 5 × 105 CFU of bacteria in 50 μl of PBS pipetted onto the external nares. After the progression of disease became clear, moribund animals were euthanized to prevent unnecessary suffering. For the colonization experiment (see Fig. 5A and B), SCID-beige mice were sacrificed 14 days postinfection with the high-dose regimen. Colonization levels were determined by homogenizing the indicated organ in PBS. The nasal cavity, trachea, spleen, kidneys, large and small intestines, and heart were homogenized in 500 μl of PBS, whereas the lungs and liver were homogenized in 1 ml of PBS. The homogenates and necessary dilutions were plated in 50-μl volumes onto BG agar with streptomycin. Colonies were counted after 2 (B. bronchiseptica) or 3 (B. parapertussis) days of incubation at 37°C. Animals were handled in accordance with institutional guidelines. The statistical significance of differences in the data was determined with Student's unpaired t test.
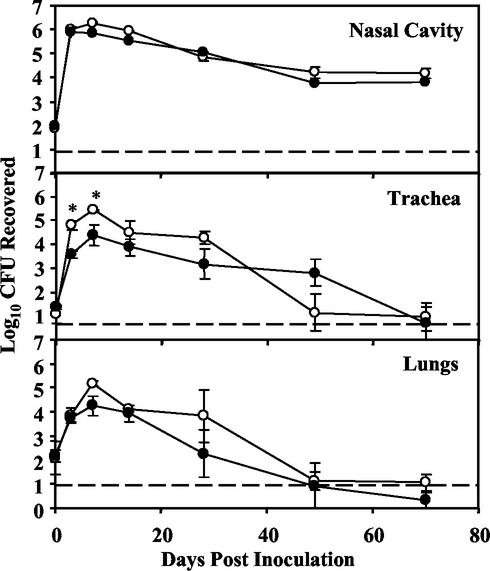
FIG. 2.
Colonization of wild-type B. bronchiseptica (open circles) and the corresponding Δwbm mutant (closed circles) in the respiratory tracts of BALB/c mice. Groups of four 4- to 6-week-old mice were inoculated intranasally with 1,000 CFU of either the wild-type or Δwbm mutant strains in 50 μl of PBS. Data are presented as the average number (log10) of CFU recovered at each time point with the standard error. Statistically significant differences are indicated by asterisks (P < 0.05). The lower limit of detection by the assay is indicated by a dashed line.
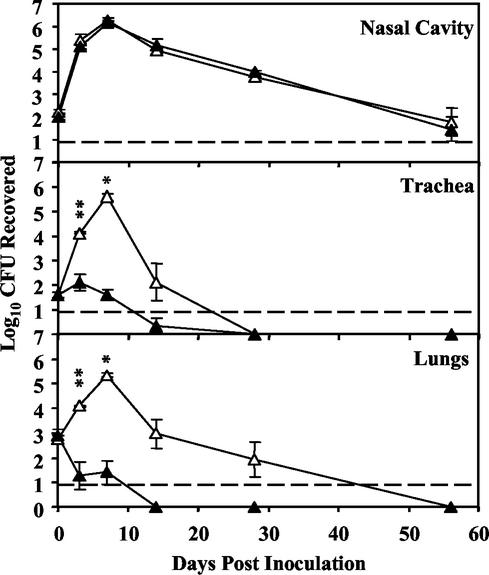
FIG. 3.
Colonization of wild-type B. parapertussis (open triangles) and the corresponding Δwbm mutant (closed triangles) in the respiratory tracts of BALB/c mice. Groups of four 4- to 6-week-old mice were inoculated intranasally with 1,000 CFU of either the wild-type or the Δwbm mutant B. parapertussis strain in 50 μl of PBS. Each point represents the average number (log10) of CFU recovered at each time point with the standard error. Statistically significant differences are indicated by asterisks (*, P < 0.05; **, P < 0.001). The lower limit of detection by the assay is indicated by a dashed line.
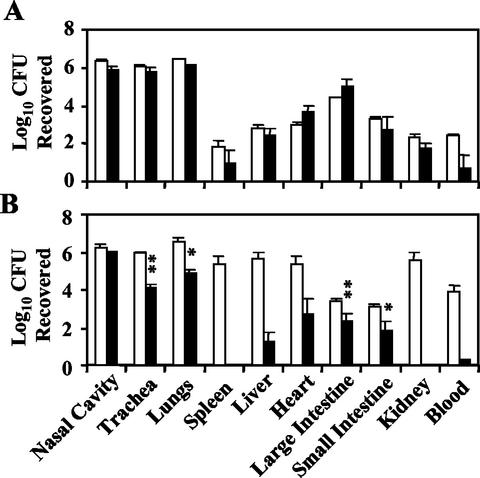
FIG. 5.
Levels of organ colonization in SCID-beige mice by wild-type B. bronchiseptica (open bars) and the corresponding Δwbm mutant (closed bars) (A) and wild-type B. parapertussis (open bars) and the corresponding Δwbm mutant (closed bars) (B). Groups of four SCID-beige mice were intranasally inoculated with 5 × 105 CFU in 50 μl of PBS and sacrificed 14 days postinoculation. Data are presented as the average number (log10) of CFU recovered with the standard error. Statistically significant differences are indicated by asterisks (*, P < 0.05; **, P < 0.001).
Serum assays and antibody responses.
Serum assays were performed with Bordetella-free rabbit serum purchased from Charles River Laboratories or Covance Research Products, Inc. (see Fig. 1, 6, and 7), or human immune serum collected at The Pennsylvania State University (normal human serum contains vaccine-induced antibodies). Bacteria were grown for 8 to 10 h in Stainer-Scholte broth to mid-log phase before their absorbance at 600 nm was measured. Optical density was used to calculate bacterial concentration, and the bacteria were diluted accordingly. For serum-killing assays, bacteria were diluted to 100 or 200 CFU/μl in PBS, depending on the experiment. Various concentrations of serum diluted in PBS were added to 5 μl of bacteria in a final volume of 50 or 100 μl, again depending on the specific experiment. Following 1 h of incubation at 37°C, 50 μl was plated on BG-blood agar with streptomycin (20 μg/ml). To test complement depletion, a novel assay was developed by initial incubation of serum with bacteria that may potentially activate and deplete it (depletion bacteria), followed by incubation with serum-sensitive target bacteria. The gentamicin-resistant target bacteria were diluted to a concentration of 100 CFU/μl in PBS. Depletion bacteria were collected by centrifugation, washed, and then resuspended in PBS to appropriate concentrations of 106 to 1010 CFU/ml. Depletion bacteria were incubated with serum diluted in PBS to a final concentration of 10% at 37°C for 10 min in a total volume of 50 μl. After 10 min, 5 μl of target bacteria was added (500 CFU total) and the reaction mixture was incubated at 37°C for an additional hour. Samples were either diluted in PBS and spread on BG-blood agar with streptomycin (20 μg/ml) for determination of survival of the depletion bacteria or spread on BG-blood agar with streptomycin (20 μg/ml) and gentamicin (20 μg/ml) to determine the survival of the target bacteria. Colonies were enumerated after 2 (B. bronchiseptica) or 3 (B. parapertussis) days of incubation at 37°C. Indirect enzyme-linked immunosorbent assays (ELISA) were performed as previously described (13).
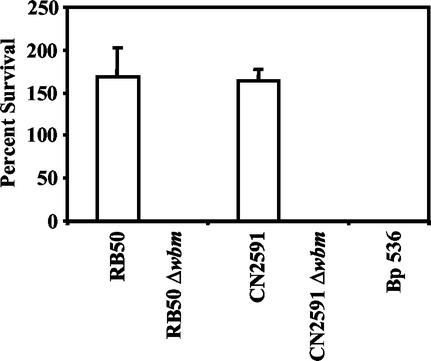
FIG. 1.
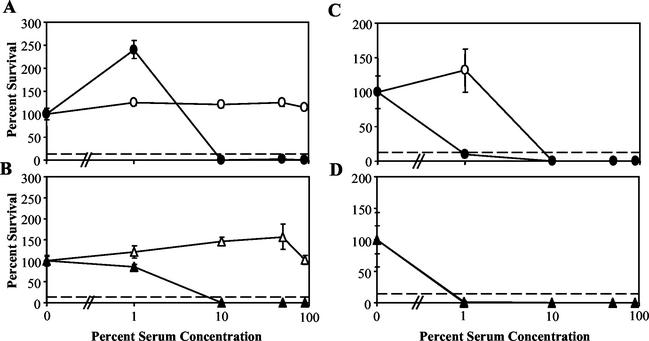
Survival of B. bronchiseptica (RB50), B. parapertussis (CN2591), and B. pertussis (Bp 536) wild-type strains and the B. bronchiseptica and B. parapertussis Δwbm mutants (RB50 Δwbm and CN2591 Δwbm, respectively) in naive-rabbit serum. Bacteria were grown to mid-log phase (optical density at 600 nm, ≤0.3) in Stainer-Scholte broth and diluted in PBS. A total of 500 CFU of bacteria were incubated at 37°C for 1 h in 50 μl of 90% naive-rabbit serum. Serum resistance is presented as percent survival relative to that of a PBS control with standard-error bars (n = 3).
FIG. 6.
Serum resistance of the B. bronchiseptica wild type (open circles) and Δwbm mutant (closed circles) and the B. parapertussis wild type (open triangles) and Δwbm mutant (closed triangles). Bacteria were grown to mid-log phase (optical density at 600 nm, ≤0.3) in Stainer-Scholte broth and diluted in PBS. In a 100-μl total volume, 200 CFU of bacteria were diluted in PBS containing various concentrations of naive (Bordetella antibody-free)-rabbit serum (A and B) or immune human serum (normal human serum) (C and D) and incubated for 1 h at 37°C. Serum resistance is presented as percent survival relative to that of a PBS control with the standard error (n = 3). The lower limit of detection by the assay is indicated by a dashed line.
FIG. 7.
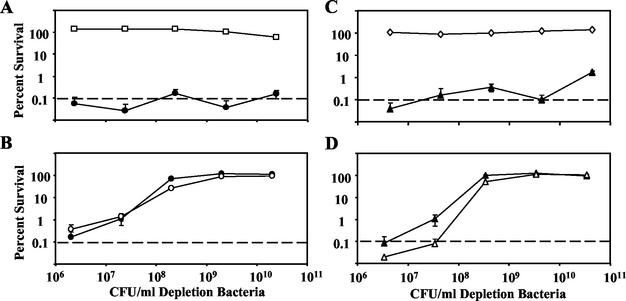
Activation and depletion of complement by the B. bronchiseptica wild type (A, RB50, open squares) or Δwbm mutant (B, RB50Δwbm, open circles) with the B. bronchiseptica Δwbm mutant (RB50 Δwbm Gmr, closed circles) as a sensitive target with which to detect remaining complement activity. Activation of complement by the B. parapertussis wild type (C, CN2591, open diamonds) or Δwbm mutant (D, CN259Δwbm, open triangles) with the B. parapertussis Δwbm mutant (CN2591 Δwbm Gmr; closed triangles) as a sensitive target with which to detect remaining complement activity. All bacteria were grown to mid-log phase in Stainer-Scholte broth and diluted in 1× PBS. Depletion bacteria (streptomycin resistant) were incubated in 10% naive-rabbit serum at various concentrations between 106 and 1011 CFU/ml for 10 min at 37°C. Target bacteria (5 μl of 100 CFU of streptomycin- and gentamicin-resistant bacteria per ml) were then added to the depletion reaction mixture, which was incubated at 37°C for an additional hour. Percent survival of both the depletion and target bacteria is compared to that of a PBS control with the standard error and plotted against the number of CFU of depletion bacteria per milliliter of reaction mixture (n = 4). The lower limit of detection by the assay is indicated by a dashed line.
RESULTS
O antigen is required for resistance to serum complement.
As previously shown, both B. bronchiseptica and B. parapertussis wild-type bacteria are resistant to the effects of 90% rabbit serum free of Bordetella-specific antibodies (naive serum) (Fig. 1) (19). However, wild-type B. pertussis, unable to synthesize O antigen because of the absence of the wbm locus (Table 1), is killed by naive serum (Fig. 1). The Δwlb mutants of B. bronchiseptica, B. parapertussis, and B. pertussis, lacking the sugars comprising the distal core trisaccharide, as well as the O-antigen repeats of LPS (Table 1), were also defective in naive-serum survival (19). To determine the contribution of O antigen to serum sensitivity, B. bronchiseptica and B. parapertussis Δwbm mutants, which do not express O antigen but do express the lipid A-core-trisaccharide, were examined (Fig. 1). In each assay, 500 CFU of the B. bronchiseptica or B. parapertussis Δwbm mutant was incubated in 90% serum for 1 h at 37°C in a total volume of 50 μl. Less than 1% of these bacteria survived, indicating that these mutants, like their Δwlb counterparts, are also sensitive to the effects of naive serum. The corresponding parental strains survived exposure to the same concentration of naive serum. These data indicate that O antigen protects B. bronchiseptica and B. parapertussis from the antimicrobial activities of serum in the absence of an antibody response.
B. bronchiseptica does not require wbm genes for respiratory tract colonization in BALB/c mice.
It has been previously shown that the B. bronchiseptica Δwlb mutant is defective in colonization of the tracheas of BALB/c, C57/BL6, and C5−/− mice from day 3 postinoculation onward. A defect in colonization of the lungs of BALB/c mice was observed only after 1 week postinoculation, and no defect in colonization was seen in the lungs of SCID-beige mice (BALB/c genetic background), suggesting that impaired interaction with adaptive immunity is responsible for this reduction in colonization of the lungs (19). To determine what contribution O antigen makes to these phenotypes, we inoculated groups of BALB/c mice with wild-type and Δwbm mutant strains of B. bronchiseptica (1,000 CFU in a total volume of 50 μl of PBS delivered intranasally). Colonization levels in the nasal cavities, tracheas, and lungs were determined at various time points postinoculation (Fig. 2). In the nasal cavity, both the wild-type and Δwbm mutant B. bronchiseptica strains increased in numbers 10,000-fold by day 3 postinfection and by 1 month they had stabilized at approximately 104 to 105 CFU, levels previously observed to be stable for the life of the animal (19). In the trachea and lungs, wild-type bacterial numbers increased 10,000-fold by day 7 before decreasing to nearly undetectable numbers by day 70. Although the Δwbm mutant was indistinguishable from wild-type B. bronchiseptica in the nasal cavity and lungs, it was recovered at approximately 1/10 of wild-type levels on days 3 and 7 in the trachea. These findings indicate that B. bronchiseptica O antigen, while not necessary for colonization of the nasal cavity or lungs, may contribute to efficient colonization of the trachea.
Since the outer surface of the outer membrane of gram-negative bacteria is made up primarily of LPS, in bacteria that present O antigen on their LPS, such as B. bronchiseptica, O antigen is one of the most prominent surface antigens present. It was therefore important to determine if a lack of O antigen affects the antibody response to B. bronchiseptica. To do this, serum and tissue samples were collected at various time points during the course of infection and ELISA with immobilized wild-type or Δwbm mutant bacteria were used to quantitate the antibody titers. Anti-Bordetella antibody titers of total immunoglobulin G (IgG), IgG1, IgG2a, and IgA were detectable by day 14 in serum and peaked by day 28 postinfection. The wild-type and mutant B. bronchiseptica strains induced similar antibody titers in the blood on day 28 postinoculation (wild type, 7,000; Δwbm mutant, 6,000), suggesting that B. bronchiseptica O antigen has little or no effect on the antibody responses. This may mean that the colonization defects previously observed in the B. bronchiseptica Δwlb mutant were due to the absence of the outer core trisaccharide of the LPS rather than to the absence of O antigen.
B. parapertussis requires wbm genes for tracheal and lung colonization in BALB/c mice.
Previous experiments with a B. parapertussis Δwlb mutant lacking both outer core and O-antigen LPS structures showed that there were dramatic defects in the mutant's ability to colonize the tracheas and lungs of BALB/c mice (19). To determine the role of O antigen in reducing this colonization, BALB/c mice were inoculated as described above with wild-type or Δwbm mutant B. parapertussis and sacrificed at various time points postinoculation (Fig. 3). In the nasal cavity, the numbers of wild-type and Δwbm mutant bacteria were indistinguishable, increasing approximately 10,000-fold by day 7 and decreasing thereafter to approximately the original colonization levels by day 56. In the trachea, the wild-type strain increased in numbers 100-fold by day 7 but was eventually cleared by day 28. In the lungs, wild-type bacterial numbers increased approximately 1,000-fold by day 7 postinfection and were cleared by day 56. However, as previously observed with the Δwlb mutant, the Δwbm mutant did not increase in numbers in the trachea and lungs and both organs were cleared by day 14. These data suggest that B. parapertussis requires O antigen for efficient colonization of both the trachea and lungs.
The antibody responses to B. parapertussis wild-type and Δwbm strains were determined by ELISA and were at maximum levels from day 28 onward. Wild-type B. parapertussis induced antibody titers that were higher on plates coated with wild-type bacteria (titer, 7,400) than on plates coated with mutant bacteria (titer, 1,900), suggesting that O antigen is a prominent antigen. In contrast, the anti-B. parapertussis Δwbm serum antibody titers measured on plates coated with the Δwbm mutant were low (titer, 700) and when measured on plates coated with wild-type bacteria, the titers generated by the mutant were even lower (titer, 270), suggesting that O antigen substantially blocks many of the antigens recognized by this serum. The lower antibody titers induced by Δwbm infection may be the result of the decreased colonization levels of this mutant (Fig. 3).
wbm genes are required for virulence of B. bronchiseptica and B. parapertussis in SCID-beige mice.
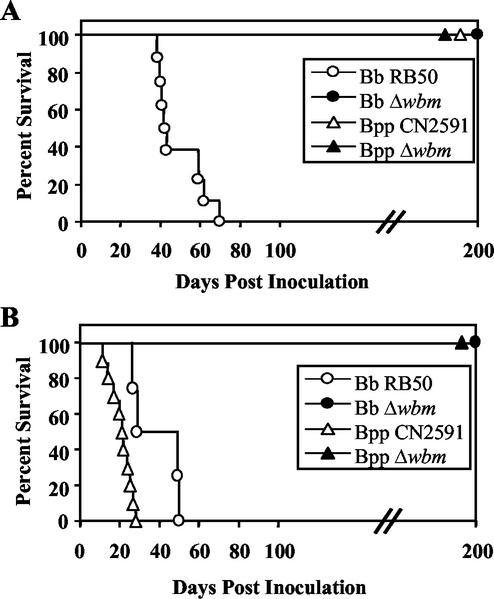
We have previously used SCID-beige mice (BALB/c genetic background, lacking B, T, and NK cells) to examine interactions between bacterial factors and innate immune functions, showing that both adenylate cyclase toxin and the wlb locus are required for virulence in this model but the type III secretion system is not (18, 19, 40). In this model, virulence is associated with systemic spread of bacteria, which might require bordetellae to have some resistance to serum microbicidal effects. Since the wbm mutants are highly susceptible to serum complement, we hypothesized that they would be less virulent in SCID-beige mice. To determine the role of O antigen in virulence in the SCID-beige model, these mice were intranasally inoculated with a low dose (500 CFU/μl in 5 μl of PBS; Fig. 4A), which only delivers bacteria to the nasal cavity. Only wild-type B. bronchiseptica proved to be lethal with this low-dose regimen, killing all of the mice by day 70 (Fig. 4A). Mice infected with a low dose of Δwbm mutant B. bronchiseptica or with either wild-type or Δwbm mutant B. parapertussis survived for the entire time course of the experiment (more than 200 days; Fig. 4A). With a high-dose inoculation regimen (5 × 105 CFU in 50 μl of PBS; Fig. 4B), which deposits bacteria throughout the respiratory tract, wild-type B. parapertussis and wild-type B. bronchiseptica were lethal by days 30 and 50, respectively (Fig. 4B). Neither the Δwbm mutant of B. bronchiseptica nor that of B. parapertussis was lethal in the high-dose regimen. Since neither Δwbm mutant was lethal regardless of the inoculation regimen used, it is clear that O antigen is required for virulence in the SCID-beige mouse model.
FIG. 4.
Survival of SCID-beige mice following inoculation with wild-type B. bronchiseptica (Bb; open circles) and B. parapertussis (Bpp; open triangles) and the corresponding Δwbm mutants (closed symbols). Groups of four 4- to 6-week-old female mice were inoculated intranasally by a low-dose, low-volume regimen consisting of 500 CFU in 5 μl of PBS (A) or a high-dose, high-volume regimen consisting of 5 × 105 CFU in 50 μl of PBS (B). Data are presented as percent survival over time.
B. parapertussis requires wbm genes for systemic infection of SCID-beige mice, but B. bronchiseptica does not.
To determine the role of Bordetella O antigen in bacterial survival during systemic infection, SCID-beige mice were infected with 5 × 105 CFU via the high-dose regimen described above. At 14 days postinfection, mice were euthanized and their nasal cavities, tracheas, lungs, spleens, livers, hearts, large and small intestines, kidneys, and whole-blood samples were collected for determination of bacterial colonization levels (Fig. 5A and B). Approximately 106 CFU of both wild-type and Δwbm mutant B. bronchiseptica were recovered from the nasal cavities, tracheas, and lungs of these animals, indicating that the wbm locus is not required for respiratory tract colonization, as was observed in BALB/c mice. Both wild-type and mutant bacteria were also recovered in similar numbers from various solid organs, indicating that the wbm locus is not required for systemic infection, even though the mutant does not kill these animals. Together, these data indicate that O antigen is required for virulence in SCID-beige mice but is not necessary for B. bronchiseptica to efficiently infect the respiratory tract and spread systemically in these animals.
In contrast to the B. bronchiseptica mutant, the B. parapertussis Δwbm mutant was severely defective in both colonization of the respiratory tract and the ability to spread systemically. The Δwbm mutant was recovered from the trachea, lungs, liver, heart, large intestine, small intestine, and blood at levels that varied between 1/10 and 1/100,000 of wild-type levels (Fig. 5B). No Δwbm mutant bacteria were recovered from the spleen or kidneys. These data indicate that B. parapertussis O antigen is necessary for efficient colonization of the lower respiratory tract, as observed in BALB/c mice and for full systemic colonization in SCID-beige mice.
wbm genes are required to protect Bordetella bacteria from antibody-independent complement activation.
Concentrations of complement in respiratory secretions are lower than that of serum (24, 37). To determine if O antigen affects complement-mediated killing at lower concentrations of complement, wild-type and Δwbm mutants of B. bronchiseptica and B. parapertussis were incubated for 1 h at 37°C in various dilutions of naive serum (free of antibodies to Bordetella bacteria) (Fig. 6A and B). Naive-rabbit serum was used because of an inability to collect naive-human serum. At serum concentrations as high as 90%, wild-type strains of both bordetellae survived or expanded in numbers (Fig. 6A and B). In contrast, the Δwbm mutants of B. bronchiseptica and B. parapertussis were sensitive to antibody-independent complement-mediated lysis and more than 99% of these bacteria were killed by naive-serum concentrations as low as 10% (Fig. 6A and B). Serum killing was inhibited by heat treatment or chelation of Mg2+ by EDTA (data not shown). These data indicate that O antigen is required by both B. bronchiseptica and B. parapertussis for resistance to antibody-independent complement-mediated lysis.
To determine the effect of O antigen on the sensitivity of bordetellae to antibody-dependent complement-mediated lysis (ADCML), wild-type and Δwbm mutant strains were incubated, as described above, in normal human serum (Fig. 6C and D). This serum contains antibodies to Bordetella primarily due to vaccination. Wild-type B. bronchiseptica was killed by immune serum at concentrations of 10% or greater, whereas even 1% serum killed the Δwbm mutant derivative, suggesting that the loss of O antigen increases the bacterium's sensitivity to ADCML (Fig. 6C). Similar results were obtained with immune (convalescent-phase) rabbit serum (data not shown). An additional experiment showed that the B. bronchiseptica Δwbm mutant was sensitive to serum concentrations as low as 0.5%; however, the wild type was resistant to concentrations as high as 5% (data not shown). Both wild-type B. parapertussis and the Δwbm mutant derivative were killed by 1% immune serum, indicating that they have similar levels of sensitivity to ADCML (Fig. 6D). These data indicate that O antigen is not sufficient to prevent antibody-mediated lysis, even when complement is diluted 100-fold, suggesting that the complement cascade proceeds efficiently after antibody-mediated activation. O antigen may therefore protect bacteria by preventing complement activation in the absence of antibody (naive serum) rather than inhibiting a downstream step in the pathway.
Complement activation requires a preliminary step that, once initiated, generally results in the rapid expansion of an enzymatic cascade that consumes the available complement components (36). If the role of O antigen is to protect from complement activation, then complement activity should remain after incubation with wild-type, but not O-antigen-deficient, bacteria. Alternatively, if complement is activated but a downstream step such as membrane attack complex formation is inhibited by O antigen, then complement components would be consumed by both wild-type and O-antigen-deficient bacteria. To determine which of these two possibilities occurs in Bordetella bacteria, we developed a novel immunological assay designed to measure complement activation by monitoring the consumption of complement components, as suggested by an assay done by Smith et al. (31). We used two types of bacteria in this assay. The first type of bacteria, the depletion bacteria, was preincubated at a high concentration with serum in the reaction mixture for a short time to potentially deplete complement components. The second type, a highly serum-sensitive target, was then added, and the mixture was incubated for 1 h to determine the amount of complement activity remaining in the reaction mixture. We found that even at levels as high as 2 × 1010 CFU/ml, neither the B. bronchiseptica nor the B. parapertussis wild-type strain depleted the complement in 10% naive serum (Fig. 7A and C). In contrast, the Δwbm mutants depleted the complement at concentrations as low as 2 × 107 CFU/ml and full depletion occurred at 2 × 109 CFU/ml (Fig. 7B and D). This suggests that O antigen protects wild-type B. bronchiseptica and B. parapertussis from complement by preventing its activation, as opposed to protecting the bacteria from a downstream effect of activated complement, as has been suggested (8).
DISCUSSION
The O antigen is the membrane-distal region of the LPS molecule that is in intimate contact with the bacterial environment and is likely to directly participate in host-bacterial interactions that dictate the outcome of infection. Being at the outermost layer of the bacterium, O antigen also forms part of the first line of defense against host immune responses. Much of our knowledge of the role of O antigen in infection has come from the study of Enterobacteriaceae, in which O antigen is required for resistance to both serum components and aspects of the innate immune response, including antimicrobial peptides (22, 32). If these were the primary roles of O antigen, expression of O antigen by pathogens would be expected to be high in the host. However, for some pathogens, like Pseudomonas aeruginosa and Yersinia enterocolitica, this does not appear to be the case. For example, P. aeruginosa strains cultured in vitro express LPS containing O antigens made up of long-chain repeats while isolates from the lungs of cystic fibrosis patients express little or no O antigen, possibly because of the accumulation of mutations within the wbp O-antigen biosynthetic locus that occur during replication in the host lung (28). Activation of migA by the respiratory mucus of cystic fibrosis patients may also contribute to this reduction of O antigen by P. aeruginosa in the host (39). In Y. enterocolitica, O-antigen expression is repressed at 37°C compared to its expression between 22 and 25°C. Repression occurs at the level of transcription of the O-antigen biosynthesis genes and involves both the Wzz O-antigen chain length determinant and the RosAB efflux pump/potassium antiporter, possibly affecting signaling by the CpxAR two-component system (6). In both P. aeruginosa and Y. enterocolitica, conditions that resemble the host environment yield a reduction in the overall amount of O antigen produced.
B. bronchiseptica and B. parapertussis LPS structures also vary in response to environmental conditions, involving regulation by the Bvg virulence regulator locus (33). In the Bvg+ phase, lipid A is modified by PagP-mediated palmitoylation (A. Preston, E. Maxim, E. Toland, E. J. Pishko, E. T. Harvill, M. Caroff, and D. J. Maskell, submitted for publication). In addition, a switch from the Bvg− phase to the Bvg+ phase results in either a decrease in O-antigen substitution or a decrease in O-antigen chain length (33), ultimately resulting in a decrease in the amount of O antigen expressed. A lacZ reporter fused to the wbmD gene was expressed at higher levels in the Bvg− phase, suggesting that regulation is at the transcription level (Liu et al., submitted). Since the Bvg+ phase has been shown to be necessary and sufficient for respiratory tract infection (1, 13, 21), the decreased levels of O antigen appear to be sufficient for efficient and persistent colonization. It may be that decreasing the proportion of LPS molecules containing long O-antigen repeats increases the accessibility to host tissues of other Bvg+-phase factors, such as adhesins, or facilitates secretion of toxins or immunomodulatory proteins while allowing the bacteria to express a basal level of O antigen for protection against host defenses such as complement-mediated killing.
When we tested the ability of serum complement to kill bordetellae, we found that wild-type B. bronchiseptica and B. parapertussis were unaffected by high concentrations of naïve-rabbit, -rat, or -mouse serum while the Δwbm mutants were sensitive to very low concentrations of serum. Serum killing was inhibited by heat treatment or chelation of Mg2+ by EDTA, suggesting that O antigen provides protection from the bactericidal activities of serum complement. The fact that these mutants are not killed by serum from mice genetically deficient in the C3 component of complement (data not shown) also supports this view. It appears that O antigen is sufficient to protect the bacteria from antibody-independent complement killing. This may explain why both B. bronchiseptica and B. parapertussis Δwbm mutants appear to be recovered at reduced levels in the blood of SCID/beige mice.
If O antigen protects bordetellae from complement by preventing complement activation, then complement activity should remain after serum is incubated with wild-type, but not Δwbm, bacteria. Conversely, if O antigen prevents some downstream effect of complement, then components involved in the first steps of the cascade will be depleted by bacterium-induced complement activation. Since very high concentrations of wild-type bacteria (more than 1010 CFU/ml) were not sufficient to significantly deplete complement, whereas both mutants lacking O antigen substantially consumed complement components at concentrations as low as 107 CFU/ml, it appears that O antigen protects bordetellae from complement by inhibiting complement activation. These observations, as well as interstrain differences, may explain apparently conflicting reports of different abilities of bordetellae to survive complement-mediated killing. Assays involving 107 CFU of B. pertussis (lacking O antigen) per ml in 10% serum (8) are likely to result in limiting concentrations of complement components, especially when the serum has undergone treatments that are likely to reduce the concentration of highly labile complement components. Under these conditions, small differences in the bacterial numbers used in an assay may result in large differences in the number of surviving bacteria identified, in a manner that is independent of any complement resistance mechanism. This would introduce substantial complexity into the interpretation of serum sensitivity or resistance reported in such experiments.
Although O antigen is likely to be the most prevalent antigenic structure on the bacterial surface, it does not appear to affect the generation of anti-B. bronchiseptica antibodies since the wild-type and Δwbm mutant strains of B. bronchiseptica induced similar antibody titers. Although the Δwbm mutant strain of B. parapertussis generated lower antibody titers than did the wild-type parent strain, this is probably related to substantially lower bacterial numbers and more transient infection of the lower respiratory tract by the mutant. In addition, serum from convalescent-phase animals (immune serum) very efficiently killed both wild-type strains in vitro, suggesting that O antigen does not result in the generation of blocking antibodies (19). Although the B. bronchiseptica Δwbm mutant showed a small defect in colonization of the trachea after 1 week postinoculation, the striking defect of the B. parapertussis mutant was observed by day 3, too early for antibodies to be present in substantial concentrations. Defective colonization by this mutant was also seen in SCID-beige mice. Together, these data suggest that the antigenic nature of O antigen has little to do with its role in infection.
Since the B. parapertussis Δwbm mutant was defective in tracheal and lung colonization early in infection, prior to the generation of adaptive immunity, we suspected that O antigen might be involved in interactions with innate immunity. To focus on the innate response, we used the SCID-beige mouse model of infection, where no adaptive immune response is present. We previously showed that both B. bronchiseptica and B. parapertussis are virulent and cause lethal infections in these animals but B. pertussis does not. The Δwlb mutants of B. bronchiseptica and B. parapertussis also do not cause lethal infections in SCID-beige mice, indicating that membrane-distal LPS structures are required for virulence in this model (19). Since these structures in B. bronchiseptica and B. pertussis are similar except for the expression of O antigen, we examined the role of O antigen in virulence in SCID-beige mice (26). Although only the B. parapertussis Δwbm mutant was defective in BALB/c mice, both Δwbm mutants failed to cause lethal infections in SCID-beige mice, indicating that O antigen is required for virulence in this model. Colonization of various organs in these mice revealed that the B. bronchiseptica Δwbm mutant was capable of colonizing the respiratory tract and systemic organs at levels comparable to those of the wild-type strain. The B. parapertussis Δwbm mutant, however, showed significant defects in the colonization of most organs, indicating a requirement for O antigen for full respiratory and systemic colonization in the SCID-beige mouse model.
The two Δwbm mutants have phenotypes in common, such as loss of resistance to alternative-pathway complement activation and loss of virulence in SCID-beige mice, that we propose may be directly attributable to the effects of O antigen. Interestingly, B. pertussis, which is naturally devoid of O antigen, is similar to the O-antigen mutants in these assays. Although B. pertussis expresses BrkA, a BvgAS-regulated protein implicated in complement resistance (16, 38), and appears to have developed an additional mechanism by which to acquire resistance to complement during respiratory tract colonization (D. J. Betting, E. J. Pishko, and E. T. Harvill, unpublished data), neither of these mechanisms is sufficient to allow B. pertussis to cause lethal sepsis in SCID-beige animals (19). With respect to phenotypic differences between the two Δwbm mutants, such as the marked difference in respiratory tract colonization, the specific role of O antigen is more difficult to interpret. Infection is a complex process that is likely to involve many bacterial and host factors, any number of which could be affected by a large change in the outer bacterial surface, such as the loss of O antigen. This makes it somewhat surprising that the Δwbm mutation does not cause a significant change in the course of B. bronchiseptica infection in mice. Since B. bronchiseptica does not require O antigen to colonize mice and B. pertussis naturally lacks O antigen, the requirement of O antigen for respiratory tract colonization is specific to B. parapertussis and may reflect the effect of O antigen on host-bacterial interactions that are unique to this organism or may indicate that underlying structures interact with the host in a different way in the absence of O antigen.
Acknowledgments
This work was funded by a grant from Neose Corp., Pennsylvania Department of Agriculture grant ME440678, U.S. Department of Agriculture grant 2002-35204-11684 to E.T. H., and Wellcome Trust Programme grant 054588 to D.J.M.
We thank Sheila Plock for excellent technical assistance.
Editor: D. L. Burns
REFERENCES
- 1.Akerley, B. J., P. A. Cotter, and J. F. Miller. 1995. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80:611-620. [DOI] [PubMed] [Google Scholar]
- 2.Akerley, B. J., and J. F. Miller. 1993. Flagellin gene transcription in Bordetella bronchiseptica is regulated by the BvgAS virulence control system. J. Bacteriol. 175:3468-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, A., and D. Maskell. 1996. The identification, cloning and mutagenesis of a genetic locus required for lipopolysaccharide biosynthesis in Bordetella pertussis. Mol. Microbiol 19:37-52. [DOI] [PubMed] [Google Scholar]
- 4.Allen, A. G., R. M. Thomas, J. T. Cadisch, and D. J. Maskell. 1998. Molecular and functional analysis of the lipopolysaccharide biosynthesis locus wlb from Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Mol. Microbiol. 29:27-38. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, and K. Struhl (ed.). 1988. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 6.Bengoechea, J. A., L. Zhang, P. Toivanen, and M. Skurnik. 2002. Regulatory network of lipopolysaccharide O-antigen biosynthesis in Yersinia enterocolitica includes cell envelope-dependent signals. Mol. Microbiol. 44:1045-1062. [DOI] [PubMed] [Google Scholar]
- 7.Burns, E. H., Jr., J. M. Norman, M. D. Hatcher, and D. A. Bemis. 1993. Fimbriae and determination of host species specificity of Bordetella bronchiseptica. J. Clin. Microbiol. 31:1838-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrd, D. W., R. M. Roop, H. P. Veit, and G. G. Schurig. 1991. Serum sensitivity and lipopolysaccharide characteristics in Bordetella bronchiseptica, B. pertussis and B. parapertussis. J. Med. Microbiol. 34:159-165. [DOI] [PubMed] [Google Scholar]
- 9.Caroff, M., R. Chaby, D. Karibian, J. Perry, C. Deprun, and L. Szabo. 1990. Variations in the carbohydrate regions of Bordetella pertussis lipopolysaccharides: electrophoretic, serological, and structural features. J. Bacteriol. 172:1121-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chambers, S. P., S. E. Prior, D. A. Barstow, and N. P. Minton. 1988. The pMTL nic- cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene 68:139-149. [DOI] [PubMed] [Google Scholar]
- 11.Cherry, J. D. 1992. Pertussis: the trials and tribulations of old and new pertussis vaccines. Vaccine 10:1033-1038. [DOI] [PubMed] [Google Scholar]
- 12.Cherry, J. D. 1997. The role of Bordetella pertussis infections in adults in the epidemiology of pertussis. Dev. Biol. Stand. 89:181-186. [PubMed] [Google Scholar]
- 13.Cotter, P. A., and J. F. Miller. 1994. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect. Immun. 62:3381-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotter, P. A., and J. F. Miller. 1998. In vivo and ex vivo regulation of bacterial virulence gene expression. Curr. Opin. Microbiol. 1:17-26. [DOI] [PubMed] [Google Scholar]
- 15.Cotter, P. A., and J. F. Miller. 1997. A mutation in the Bordetella bronchiseptica bvgS gene results in reduced virulence and increased resistance to starvation, and identifies a new class of Bvg-regulated antigens. Mol. Microbiol. 24:671-685. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez, R. C., and A. A. Weiss. 1994. Cloning and sequencing of a Bordetella pertussis serum resistance locus. Infect. Immun. 62:4727-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerlach, G., F. von Wintzingerode, B. Middendorf, and R. Gross. 2001. Evolutionary trends in the genus Bordetella. Microbes Infect. 3:61-72. [DOI] [PubMed] [Google Scholar]
- 18.Harvill, E. T., P. A. Cotter, M. H. Yuk, and J. F. Miller. 1999. Probing the function of Bordetella bronchiseptica adenylate cyclase toxin by manipulating host immunity. Infect. Immun. 67:1493-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harvill, E. T., A. Preston, P. A. Cotter, A. G. Allen, D. J. Maskell, and J. F. Miller. 2000. Multiple roles for Bordetella lipopolysaccharide molecules during respiratory tract infection. Infect. Immun. 68:6720-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heininger, U., J. D. Cherry, P. D. Christenson, T. Eckhardt, U. Goering, P. Jakob, W. Kasper, D. Schweingel, S. Laussucq, J. G. Hackell, et al. 1994. Comparative study of Lederle/Takeda acellular and Lederle whole-cell pertussis-component diphtheria-tetanus-pertussis vaccines in infants in Germany. Vaccine 12:81-86. [DOI] [PubMed] [Google Scholar]
- 21.Martinez de Tejada, G., P. A. Cotter, U. Heininger, A. Camilli, B. J. Akerley, J. J. Mekalanos, and J. F. Miller. 1998. Neither the Bvg− phase nor the vrg6 locus of Bordetella pertussis is required for respiratory infection in mice. Infect. Immun. 66:2762-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicolas, P., and A. Mor. 1995. Peptides as weapons against microorganisms in the chemical defense system of vertebrates. Annu. Rev. Microbiol. 49:277-304. [DOI] [PubMed] [Google Scholar]
- 23.Peppler, M. S. 1984. Two physically and serologically distinct lipopolysaccharide profiles in strains of Bordetella pertussis and their phenotype variants. Infect. Immun. 43:224-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Persson, C. G., I. Erjefalt, U. Alkner, C. Baumgarten, L. Greiff, B. Gustafsson, A. Luts, U. Pipkorn, F. Sundler, C. Svensson, et al. 1991. Plasma exudation as a first line respiratory mucosal defense. Clin. Exp. Allergy 21:17-24. [DOI] [PubMed] [Google Scholar]
- 25.Preston, A., A. G. Allen, J. Cadisch, R. Thomas, K. Stevens, C. M. Churcher, K. L. Badcock, J. Parkhill, B. Barrell, and D. J. Maskell. 1999. Genetic basis for lipopolysaccharide O-antigen biosynthesis in bordetellae. Infect. Immun. 67:3763-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Preston, A., and D. Maskell. 2001. The molecular genetics and role in infection of lipopolysaccharide biosynthesis in the bordetellae. J. Endotoxin Res. 7:251-261. [PubMed] [Google Scholar]
- 27.Ried, J. L., and A. Collmer. 1987. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 57:239-246. [DOI] [PubMed] [Google Scholar]
- 28.Rocchetta, H. L., L. L. Burrows, and J. S. Lam. 1999. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 63:523-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rutter, J. M. 1981. Quantitative observations on Bordetella bronchiseptica infection in atrophic rhinitis of pigs. Vet. Rec. 108:451-454. [DOI] [PubMed] [Google Scholar]
- 30.Senzilet, L. D., S. A. Halperin, J. S. Spika, M. Alagaratnam, A. Morris, and B. Smith. 2001. Pertussis is a frequent cause of prolonged cough illness in adults and adolescents. Clin. Infect. Dis. 32:1691-1697. [DOI] [PubMed] [Google Scholar]
- 31.Smith, R. I., M. J. Coloma, and S. L. Morrison. 1995. Addition of a mu-tailpiece to IgG results in polymeric antibodies with enhanced effector functions including complement-mediated cytolysis by IgG4. J. Immunol. 154:2226-2236. [PubMed] [Google Scholar]
- 32.Taylor, P. W. 1983. Bactericidal and bacteriolytic activity of serum against gram-negative bacteria. Microbiol. Rev. 47:46-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van den Akker, W. M. 1998. Lipopolysaccharide expression within the genus Bordetella: influence of temperature and phase variation. Microbiology 144:1527-1535. [DOI] [PubMed] [Google Scholar]
- 34.van der Zee, A., F. Mooi, J. Van Embden, and J. Musser. 1997. Molecular evolution and host adaptation of Bordetella spp.: phylogenetic analysis using multilocus enzyme electrophoresis and typing with three insertion sequences. J. Bacteriol. 179:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Wintzingerode, F., A. Schattke, R. A. Siddiqui, U. Rosick, U. B. Gobel, and R. Gross. 2001. Bordetella petrii sp. nov., isolated from an anaerobic bioreactor, and emended description of the genus Bordetella. Int. J. Syst. E vol. Microbiol. 51:1257-1265. [DOI] [PubMed] [Google Scholar]
- 36.Walport, M. J. 2001. Complement. First of two parts. N. Engl. J. Med. 344:1058-1066. [DOI] [PubMed] [Google Scholar]
- 37.Watford, W. T., A. J. Ghio, and J. R. Wright. 2000. Complement-mediated host defense in the lung. Am. J. Physiol. Lung Cell Mol. Physiol. 279:L790-L798. [DOI] [PubMed] [Google Scholar]
- 38.Weiss, A. A., P. S. Mobberley, R. C. Fernandez, and C. M. Mink. 1999. Characterization of human bactericidal antibodies to Bordetella pertussis. Infect. Immun. 67:1424-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang, H., M. Matewish, I. Loubens, D. G. Storey, J. S. Lam, and S. Jin. 2000. migA, a quorum-responsive gene of Pseudomonas aeruginosa, is highly expressed in the cystic fibrosis lung environment and modifies low-molecular-mass lipopolysaccharide. Microbiology 146:2509-2519. [DOI] [PubMed] [Google Scholar]
- 40.Yuk, M. H., E. T. Harvill, and J. F. Miller. 1998. The BvgAS virulence control system regulates type III secretion in Bordetella bronchiseptica. Mol. Microbiol. 28:945-959. [DOI] [PubMed] [Google Scholar]