Abstract
We recently demonstrated that mutation of sarA in clinical isolates of Staphylococcus aureus results in a phenotype that is distinct by comparison to sarA mutants generated in the laboratory strain RN6390 (J. S. Blevins, K. E. Beenken, M. O. Elasri, B. K. Hurlburt, and M. S. Smeltzer, Infect. Immun. 70:470-480, 2002). This raises the possibility that studies demonstrating that RN6390 sarA mutants are attenuated do not accurately reflect the role of sarA in the pathogenesis of staphylococcal disease. To test this hypothesis, we used a murine model of musculoskeletal infection to assess the virulence of sarA and agr mutants generated in a clinical isolate of S. aureus (UAMS-1). By using this model, we confirmed that mutation of sarA and/or agr results in a reduced capacity to cause both septic arthritis and osteomyelitis.
The accessory gene regulator (agr) and the staphylococcal accessory regulator (sar) are the two best-characterized loci responsible for modulating the expression of Staphylococcus aureus virulence factors (27). Mutation of either locus has been shown to result in attenuation of S. aureus in several models of staphylococcal disease (1, 5, 9, 11, 19, 25). In most cases, characterization of these loci and evaluation of their role in pathogenesis were done with laboratory strain RN6390 and mounting evidence suggests that the regulatory events defined by using strain RN6390 are not representative of the events observed in clinical isolates of S. aureus (3, 10). Specifically, while RN6390 sarA mutants exhibit reduced hemolytic activity, mutation of sarA in other strains results in elevated hemolytic activity. Although sarA mutants generated in one such strain (DB) had reduced virulence in animal models of staphylococcal septic arthritis and endocarditis (11, 25), neither DB nor RN6390 encodes the collagen binding adhesin (cna) and therefore neither binds collagen (20). This is relevant because the ability to bind collagen has also been associated with virulence in septic arthritis, osteomyelitis, and endocarditis models (14, 21, 31), and we have demonstrated that mutation of sarA results in elevated transcription of cna and an enhanced capacity to bind collagen (3, 4, 20).
Recent reports have also identified several genotypic and phenotypic markers that appear to be characteristic of the most prominent S. aureus clinical isolates (6, 30). Included among these are the presence of cna and the absence of one of the two genes (fnbB) that encode fibronectin binding adhesins. These isolates also have a phenotype defined by a high binding capacity for host proteins and a relatively low level of exoprotein expression. Importantly, none of these characteristics have been observed in RN6390 (3, 20). In addition, RN6390 was recently shown to have a deletion in rsbU, which encodes a positive regulator of the stress response sigma factor SigB (16, 18, 23). This is relevant because mutation of sigB results in reduced sar transcription and a reduced capacity to produce SarA (2, 17), and the overall level of SarA has been shown to have an impact on the agr-dependent branch of the sarA regulatory pathway (12). Taken together, these factors make it important to assess the role of sarA in the pathogenesis of staphylococcal disease by using strains other than RN6390.
The specific strains included in this study are described in Table 1. UAMS-1 is a cna-positive osteomyelitis isolate that encodes fnbA but not fnbB (3). It also has a high binding capacity for host proteins and produces relatively low levels of most exoproteins (3). As noted above, all of these characteristics have been associated with prevalent clinical isolates of S. aureus (6, 30). Inocula were prepared and mice were infected as described by Elasri et al. (14). To determine viable counts and confirm the purity of each stock, the number of CFU was determined by dilution and plating on both nonselective and selective tryptic soy agar (TSA) by using 5 μg of tetracycline ml−1 for the agr-null mutation (28), 50 μg of kanamycin ml−1 and 50 μg of neomycin ml−1 for the sarA::kan insertion (7), and 10 μg of chloramphenicol ml−1 for pSARA (4).
TABLE 1.
Strains used in this study
| Strain | Relevant genotypea | Comment or parent strain | Reference |
|---|---|---|---|
| RN6390 | cna | 8325-4 | 28 |
| UAMS-957 | cna sarA | RN6390 | 3 |
| UAMS-1 | fnbB | Osteomyelitis isolate | 19 |
| UAMS-155 | fnbB agr | UAMS-1 | 3 |
| UAMS-929 | fnbB sarA | UAMS-1 | 3 |
| UAMS-930 | fnbB agr sarA | UAMS-1 | 3 |
| UAMS-969 | fnbB sarA | UAMS-929 containing pSARA | 3 |
Genotype descriptions are limited to chromosomal genes associated with a negative phenotype due to either mutation (agr or sarA) or the absence of the gene (cna or fnbB).
Five- to 8-week-old male, outbred NIH-Swiss mice (Harlan, Indianapolis, Ind.) were infected via tail vein injection with 108 CFU. After 2 weeks, mice were euthanized and the left hind limb was removed and prepared for histological analysis as described by Skinner et al. (32). For pathological analysis, three 5-μm sections were stained with hematoxylin and eosin (H&E). For visualization of bacteria, Gram stains of two additional sections were prepared. For bacteriological analysis of the knee joint, the right leg was dissected aseptically and bacteria were collected from the joint with calcium alginate swabs and then plated on nonselective TSA. Viable counts greater than 10 were considered positive following confirmation by plating on selective/differential CHROMagar plates (Hardy Diagnostics, Santa Maria, Calif.). A joint yielding fewer than 10 colonies was considered positive only after verification of the identity of isolated colonies by plating on S. aureus CHROMagar and confirmation of the presence of cna, the absence of fnbB, and the appropriate sarA and/or agr mutation.
The presence of inflammation in the joint was used to determine the incidence of arthritis, while the degree of inflammation was taken as an indication of severity. The degree of inflammation was scored from 0 to 3 (0, no infiltration of polymorphonuclear leukocytes; 1, mild acute inflammation; 2, moderate acute inflammation; 3, severe acute inflammation). Joints were also scored for abscess formation and erosion of articular cartilage and/or cortical bone (Fig. 1). To assess the overall extent of disease, we calculated a composite score ranging from 0 to 6 based on a combination of all parameters (inflammation score plus one point for each of the other positive parameters). The composite comparison was included because the degree of inflammation was not always directly correlated with the overall effect on the joint (i.e., some mice with a low inflammation score showed signs of erosion of joint tissues).
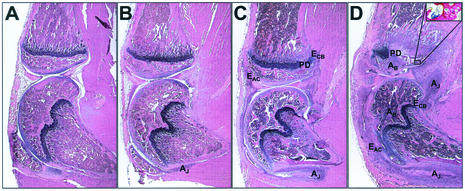
FIG. 1.
Histological evidence of musculoskeletal infection. Photomicrographs of H&E-stained sections 14 days after infection with S. aureus. (A) Section scored as unaffected; inflammation score = 0. (B) Section showing a joint with mild synovitis; 1+ inflammation with a small joint abscess (AJ) and no demonstrable bone involvement. (C) Section with moderate synovitis and mild osteomyelitis; 2+ inflammation in joint and 1+ inflammation in bone. The severity of synovial inflammation has led to erosion of articular cartilage (EAC) and cortical bone (ECB) and secondary osteomyelitis with physis destruction (PD). (D) Section showing a hind limb with severe synovitis and osteomyelitis; 3+ inflammation scores in both the bone and synovium. In addition to the aforementioned parameters, two bone abscesses (AB) are also present, one in the tibia (upper bone) and the other in the femur (lower bone). The inset shows a Gram-stained serial section with gram-positive cocci present in the subchondral bone of the tibia. Magnifications: panels A to D, ×16; panel D inset, ×200.
A similar scheme was used to assess evidence of osteomyelitis. The incidence of osteomyelitis was determined by the presence of inflammation in either the tibia or the femur. The degree of inflammation was also scored from 0 to 3. Sections were further scored for the presence of abscesses and destruction of the physis (Fig. 1D). As with arthritis, the composite osteomyelitis score takes into account all disease parameters by combining the score for inflammation with a score of one added for each positive parameter. Some mice developed signs of osteomyelitis in the absence of synovitis (Fig. 2). However, in most cases, mice that developed osteomyelitis also had clear signs of synovitis. In these cases, the severity of the arthritis sometimes made it difficult to determine whether there was a primary focus of infection in the bone prior to the development of erosive synovitis. For this reason, we did not attempt to distinguish between primary (bone directly seeded via the hematogenous route) and secondary (bone infected via spread from the infection in the adjacent synovium) osteomyelitis.
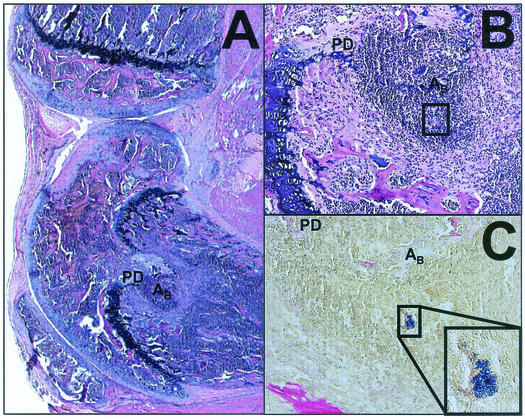
FIG. 2.
Histological evidence of primary osteomyelitis. Photomicrographs of H&E- and Gram-stained sections 14 days after infection with S. aureus. (A) Section scored as negative for synovitis with 3+ inflammation in the bone, abscess formation (AB), and physis destruction (PD). To maintain orientation, the AB and PD labels are in the same relative positions in each panel. Magnification, ×20. (B) Higher magnification, ×200, of the section shown in panel A. (C) Gram staining of a serial section demonstrating the presence of bacteria in the abscess; magnification, ×200. The framed region in panels B and C coincides with the inset; magnification ×400.
Investigators responsible for scoring H&E-stained sections were blinded with respect to the strain used to infect each mouse. Statistical analysis was done by using SigmaStat (SSPS Science, Chicago, Ill.). The chi-square test with Yates' correction was used to evaluate differences in the incidence of disease, pathology, or colonization. A pairwise multiple comparison of the inflammation and composite scores was carried out by using the Kruskal-Wallis one-way analysis of variance. For statistical analysis of colony counts from infected mice, the Mann-Whitney rank sum test was applied. In all cases, P < 0.05 was considered significant.
Two independent trials with at least eight mice per strain were carried out for UAMS-1 and each of its regulatory mutants. With respect to the ability to cause arthritis, the UAMS-1 sarA and sarA agr mutants were significantly less virulent than the parent strain (Table 2). Although mutation of agr in UAMS-1 appeared to result in reduced virulence, the difference was not statistically significant. However, the difference was reproducible in that it was apparent when the agr mutant was compared to the parent strain and when the sarA agr mutant was compared to the sarA mutant (Table 2). Taken together, these results support the hypothesis that mutation of agr in UAMS-1 also results in a decreased capacity to cause septic arthritis as defined by this model.
TABLE 2.
Incidence and severity of arthritis in mice infected with UAMS-1 and RN6390 regulatory mutantsa
| Strain (no. of mice) | Genotype | Incidence (%) of:
|
Inflammation score | Composite score | |||
|---|---|---|---|---|---|---|---|
| Arthritis | Abscess | Erosion of articular cartilage | Erosion of cortical bone | ||||
| UAMS-1 (28) | Wild type | 96.3 ± 3.7 | 96.3 ± 3.7 | 92.6 ± 7.4 | 89.6 ± 6.4 | 2.39 ± 0.16 | 5.18 ± 0.27 |
| UAMS-929 (30) | sarA | 63.5 ± 11.8b | 53.8 ± 17.4b | 49.7 ± 10.1b | 37.7 ± 16.4b | 1.40 ± 0.23b | 2.80 ± 0.47b |
| UAMS-155 (21) | agr | 66.8 ± 3.2b | 66.8 ± 3.2b | 66.8 ± 3.2b | 66.7 ± 3.2 | 1.81 ± 0.31 | 3.81 ± 0.61 |
| UAMS-930 (21) | sarA/agr | 33.2 ± 3.2b | 24.1 ± 5.9b | 23.7 ± 3.7b | 18.7 ± 8.7b | 0.57 ± 0.20b | 1.24 ± 0.43b |
| UAMS-969 (10) | sarA (pSARA) | 100.0 | 70.0 | 100.0 | 100.0 | 2.40 ± 0.27 | 5.10 ± 0.38 |
| RN6390 (8) | Wild type | 75.0 | 75.0 | 62.5 | 75.0 | 1.88 ± 0.48 | 4.00 ± 0.95 |
| UAMS-957 (9) | sarA | 33.3 | 33.3 | 22.2 | 22.2 | 0.56 ± 0.29b | 1.33 ± 0.69b |
Results for UAMS-969, RN6390, and UAMS-957 are based on one experiment. Results for other groups are reported as the mean ± the standard error of the mean.
Difference between the parent and the mutant that is statistically significant (P < 0.05).
Although there was no statistically significant difference between the agr and sarA mutants, the sarA mutant was at least as attenuated as the agr mutant, if not more so, and this was somewhat surprising given the prominent role of agr in other animal models (1, 9, 19). It is feasible that the impact of agr is more modest in UAMS-1 because this strain already produces a limited amount of the agr-encoded regulatory RNA (RNAIII) and a limited amount of exoproteins (3). On the other hand, sarA has functions that are both agr dependent and agr independent (4, 7, 8). Indeed, we did observe a statistically significant reduction in the virulence of the sarA agr double mutant even by comparison with the corresponding agr mutant.
The same trend was observed in our assessment of osteomyelitis in mice infected with UAMS-1 and the corresponding regulatory mutants. Specifically, mutation of sarA resulted in a statistically significant reduction in the incidence and severity of osteomyelitis (Table 3). Similarly, the differences between UAMS-1 and its agr mutant were statistically significant in every aspect of osteomyelitis pathology. These results agree with our earlier findings obtained with a rabbit model of acute posttraumatic osteomyelitis, which showed attenuation of a UAMS-1 agr mutant (18). Once again, the UAMS-1 sarA mutant appeared to be less virulent than the UAMS-1 agr mutant but the difference was not statistically significant. Similarly, the degree of attenuation in the UAMS-1 sarA agr mutant appeared to be greater than that of either of the single mutants but the difference was not statistically significant.
TABLE 3.
Incidence and severity of osteomyelitis in mice infected with UAMS-1 and RN6390 regulatory mutantsa
| Strain (no. of mice) | Genotype | Incidence (%) of:
|
Inflammation score | Composite score | ||
|---|---|---|---|---|---|---|
| Osteomyelitis | Abscess | Physis destruction | ||||
| UAMS-1 (28) | Wild type | 96.3 ± 3.7 | 86.5 ± 6.8 | 89.6 ± 6.4 | 2.43 ± 0.15 | 4.18 ± 0.25 |
| UAMS-929 (30) | sarA | 37.0 ± 13.8b | 26.9 ± 13.7b | 31.0 ± 13.9b | 0.80 ± 0.21b | 1.36 ± 0.35b |
| UAMS-155 (21) | agr | 61.8 ± 1.8b | 47.3 ± 7.3b | 24.1 ± 5.9b | 1.14 ± 0.21b | 1.86 ± 0.37b |
| UAMS-930 (21) | sarA agr | 18.7 ± 8.7b | 9.6 ± 0.5b | 14.1 ± 4.1b | 0.29 ± 0.14b | 0.52 ± 0.27b |
| UAMS-969 (10) | sarA (pSARA) | 90.0 | 70.0 | 70.0 | 1.20 ± 0.25 | 2.60 ± 0.43 |
| RN6390 (8) | Wild type | 100.0 | 75.0 | 50.0 | 2.13 ± 0.30 | 3.38 ± 0.53 |
| UAMS-957 (9) | sarA | 44.4b | 11.1b | 0.0 | 0.56 ± 0.24b | 0.67 ± 0.33b |
Results for UAMS-969, RN6390, and UAMS-957 are based on one experiment. Results for other groups are reported as the mean ± the standard error of the mean.
Difference between the parent and the mutant that is statistically significant (P < 0.05).
To evaluate our results in the context of previous studies, one additional trial was carried out with RN6390 and its sarA mutant form. With respect to arthritis, mutation of sarA had an effect similar to that observed with UAMS-1 although statistical significance could be established only with respect to the degree of inflammation and overall severity of disease (Table 2). It is important to emphasize, however, that the focus of our experiments was UAMS-1. For that reason, the RN6390 experiments were not repeated, and it is certainly possible that this accounts for the failure to establish statistical significance. Indeed, with respect to osteomyelitis, the differences between RN6390 and its sarA mutant were significant in every case except physis destruction (Table 3).
To confirm that the reduced virulence of UAMS-929 was due to interruption of the sarA locus, the mutation was complemented by using a plasmid containing a fragment of the sarA locus coding for the sarA transcript (pSARA) (3, 4). Three groups of at least 10 mice were infected with UAMS-1, UAMS-929, or UAMS-969. A comparison of the severity of arthritis observed in mice infected with UAMS-1 with that observed in mice infected with UAMS-969 (Table 2) showed that the virulence of the pSARA-complemented sarA mutant was restored to wild-type levels with regard to every parameter, with the exception of abscess formation. However, this was not the case when these same mice were assessed for evidence of osteomyelitis (Table 3). Specifically, while the number of infected mice was essentially the same in the two groups of mice, the overall severity of infection was reduced in the complemented strain. The reason for differential complementation with respect to arthritis and osteomyelitis is unknown. It is possible that, since arthritis is primarily an immunopathological disease (15), fewer wild-type bacteria are required to induce synovitis. It is also possible that, while the complemented strain can cause septic arthritis, it may still have an attenuated ability to induce secondary osteomyelitis.
While complementation with respect to osteomyelitis was not complete, the scores obtained with the pSARA-complemented sarA mutant were still higher than those of the sarA mutant without the plasmid. In contrast, in vitro phenotypic characterization of the pSARA-complemented sarA mutant has shown that the phenotype of this strain is comparable to that of the wild type in every respect (3). One possible explanation for partial complementation in vivo is loss of the plasmid during the course of infection. To test this hypothesis, six mice were infected with 108 CFU of UAMS-969. At 7 and 14 days postinfection (p.i.), three mice were euthanized and a bacteriological assessment was performed on the knee, kidneys, and spleen. The spleen and kidneys were removed aseptically and homogenized on ice. All tissue samples were processed individually. Tenfold dilutions of the homogenates were plated on TSA. The number of CFU per organ was determined following overnight incubation at 37°C. Colonies were then picked to TSA containing either chloramphenicol (for the pSARA plasmid) or kanamycin-neomycin (for the sarA mutation) and to CHROMagar to verify that all colonies were S. aureus. When the number of CFU obtained from the tissue was <30, all colonies were picked to selective TSA and CHROMagar. When the number was >30, at least 30 colonies were examined. Joint swabs were plated on TSA and then processed as described above.
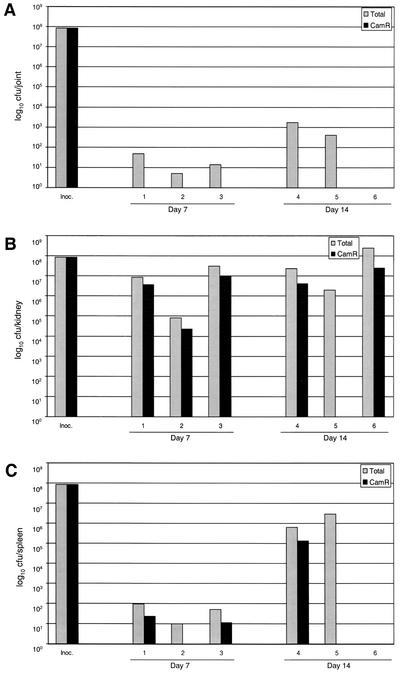
At 7 and 14 days p.i., the knee joints of five of six mice were positive for S. aureus (Fig. 3A). More importantly, in all five mice, the only isolates obtained were kanamycin-neomycin resistant (Kanr) and chloramphenicol sensitive (Cams) (P = 0.033). This indicates loss of the pSARA plasmid. When counts from the kidneys were considered (Fig. 3B), the difference was not as dramatic; however, in every case in which we isolated bacteria, the number of Kanr Cams colonies was greater than the number of Kanr Camr colonies. Specifically, at 7 days p.i., the proportion of Camr colonies was 34.7% ± 4.9% of the total number of CFU recovered. This difference was even more pronounced at 14 days p.i., when the average proportion of Camr colonies was only 9.3% ± 5.1% of the total number of isolates. A similar reduction was seen in the spleen, with the proportion of Camr colonies recovered being 16% ± 8.0% and 10% ± 10.0% at days 7 and 14, respectively (Fig. 3C). While it is difficult to say what specific effect a reduction of this magnitude would have on virulence, it is possible that the partial complementation observed in the osteomyelitis pathology was due to loss of pSARA. In fact, the loss of the plasmid from UAMS-969 was most pronounced in the tissue in which we are the most interested (i.e., the joint). This suggests that the reduced virulence of the complemented mutant was due to loss of pSARA during the course of infection.
FIG. 3.
Plasmid stability in vivo. Mice were infected with the pSARA-complemented UAMS-1 sarA mutant (UAMS-969). Samples were obtained from a joint (A), a kidney (B), and the spleen (C) at days 7 and 14 p.i. Results are reported as the total number of CFU recovered and the number that were chloramphenicol resistant (Camr). Inoc., inoculum.
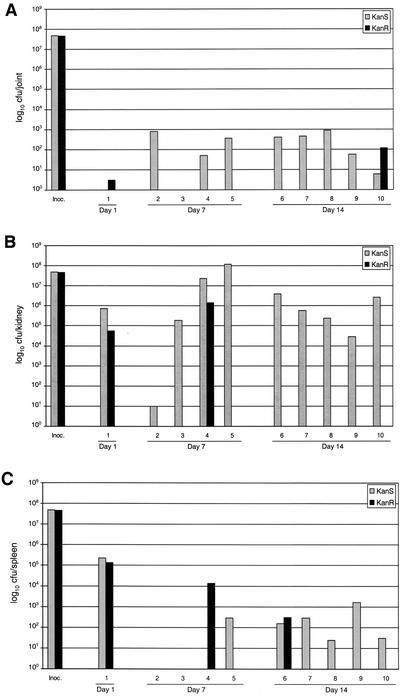
Since complementation of the sarA mutation in UAMS-1 was not complete, we carried out a third trial in which 10 mice were infected with a mixed culture containing 5 × 107 CFU of both UAMS-1 and UAMS-929. With the exception of one mouse that died on day 1, mice were euthanized at 7 and 14 days p.i. Bacteria isolated from all of the animals were first recovered on nonselective medium and then tested for the marker that defines the sarA mutation (Kanr). Although the number of bacteria recovered was very low, the mouse that died at 1 day p.i. yielded only Kanr colonies from the joint (Fig. 4A). Roughly equal numbers of Kans and Kanr colonies were obtained from kidney and spleen samples from this mouse (Fig. 4B). With the exception of one mouse, no Kanr colonies were recovered from the joints at day 7 or 14 (P = 0.045) (Fig. 4A). The mean number of CFU from the joint samples of all mice was 302 ± 106 Kans (UAMS-1) versus 12 ± 11 Kanr (UAMS-929) (P = 0.009). A similar difference was observed in the kidney (Fig. 4B). Specifically, in all but two mice only the wild-type strain was isolated (P = 0.033). The spleen results were less dramatic in that five of the eight samples contained only UAMS-1 (Fig. 4C). Taken together, these results provide further support for the hypothesis that mutation of sarA attenuates the virulence of UAMS-1 in the murine model of musculoskeletal infection.
FIG. 4.
Coinfection studies. Mice were coinfected with equal numbers of cells of UAMS-1 and its isogenic sarA mutant (UAMS-929). One mouse died on day 1 p.i. All other samples from a joint (A), a kidney (B), and the spleen (C) were taken at 7 and 14 days p.i. Results are reported as the number of colonies that were sensitive to kanamycin-neomycin (Kans) and the number that were resistant (Kanr). Inoc., inoculum.
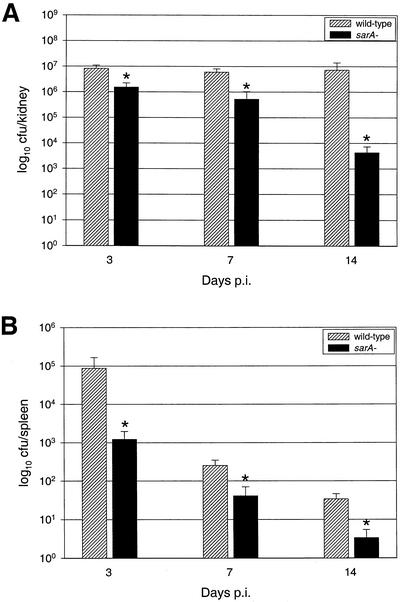
In an attempt to determine whether the UAMS-1 sarA mutant was more susceptible to clearance by host defenses or had a specific defect in the ability to colonize musculoskeletal tissues, we infected 15 mice with UAMS-1 and 15 with UAMS-929. At 3, 7, and 14 days p.i., five mice infected with each strain were euthanized and evaluated as described above. There was no statistically significant difference in the isolation rate of each strain from the joint at any time point. However, we did recover bacteria from the joints of 100% of the mice infected with UAMS-1 and 60% of the mice infected with the sarA mutant at 14 days p.i. (data not shown). While this may reflect a defect in the ability of UAMS-929 to colonize bone and joint tissues at later stages of infection, it could also be due to a reduced capacity of the sarA mutant to persist in the host. In agreement with the latter suggestion are the results obtained with the kidney (Fig. 5A), which is classically taken as an indicator of persistence (25). Specifically, fewer CFU were obtained from the kidneys of mice infected with UAMS-929 at all time points: 18.5, 8.7, and 0.06% of the number obtained from the kidneys of mice infected with UAMS-1 at days 3, 7, and 14, respectively (P < 0.05 for all). The same trend was seen in the counts obtained from the spleen (Fig. 5B). Although it is difficult to definitively distinguish between a reduced capacity to persist in the host and a reduced capacity to colonize the joint, these results suggest that UAMS-929 retains the capacity to colonize the joint but is more susceptible to host defenses and clearance.
FIG. 5.
Impact of sarA mutation on colonization of and survival in mouse tissues. Mice infected intravenously with UAMS-1 or its isogenic sarA mutant (UAMS-929) were euthanized at days 3, 7, and 14, and bacteria were isolated from a kidney (A) and the spleen (B). Results are reported as the mean number of CFU ± the standard error of the mean, and each mean represents a group of five mice. The asterisk indicates a statistically significant difference (P < 0.05).
Overall, our results demonstrate that mutation of sarA results in a reduced capacity of both UAMS-1 and RN6390 to cause disease. This is somewhat surprising because mutation of sarA results in a distinct phenotype in these two strains (3). It should be emphasized that our inclusion of RN6390 in these experiments was only done as a control, with the more important question being whether the enhanced hemolytic activity and enhanced capacity to bind collagen observed in the UAMS-1 sarA mutant translate to an enhanced capacity to cause disease. This is a relevant question in that both of these factors have been shown to contribute to the pathogenesis of septic arthritis (14, 22, 24, 26, 29). The fact that the UAMS-1 sarA mutant was attenuated implies that UAMS-1 encodes additional virulence factors that are relevant to the pathogenesis of staphylococcal musculoskeletal infection and that at least some of these are under the regulatory control of sarA. In that regard, it is interesting that microarray-based transcriptional profiling recently confirmed that mutation of sarA has a more global impact than previously recognized (13). We are currently pursuing a similar comparison by using UAMS-1 and its sarA mutant with the hope of ultimately identifying the sarA-regulated factor(s) involved in the pathogenesis of staphylococcal musculoskeletal disease.
Acknowledgments
We thank Don Blevins for technical assistance.
This work was supported by a grant (AI43356) to M.S.S. from the National Institute of Allergy and Infectious Diseases.
Editor: A. D. O'Brien
REFERENCES
- 1.Abdelnour, A., S. Arvidson, T. Bremell, C. Ryden, and A. Tarkowski. 1993. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect. Immun. 61:3879-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bischoff, M., J. M. Entenza, and P. Giachino. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 183:5171-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blevins, J. S., K. E. Beenken, M. O. Elasri, B. K. Hurlburt, and M. S. Smeltzer. 2001. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blevins, J. S., A. F. Gillaspy, T. M. Rechtin, B. K. Hurlburt, and M. S. Smeltzer. 1999. The staphylococcal accessory regulator (sar) represses transcription of the Staphylococcus aureus collagen adhesin gene (cna) in an agr-independent manner. Mol. Microbiol. 33:317-326. [DOI] [PubMed] [Google Scholar]
- 5.Booth, M. C., R. V. Atkuri, S. K. Nanda, J. J. Iandolo, and M. S. Gilmore. 1995. Accessory gene regulator controls Staphylococcus aureus virulence in endophthalmitis. Investig. Ophthalmol. Vis. Sci. 36:1828-1836. [PubMed] [Google Scholar]
- 6.Booth, M. C., L. M. Pence, P. Mahasreshti, M. C. Callegan, and M. S. Gilmore. 2001. Clonal associations among Staphylococcus aureus isolates from various sites of infection. Infect. Immun. 69:345-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, P. F., and S. J. Foster. 1998. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J. Bacteriol. 180:6232-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, A. L., M. G. Bayer, and J. H. Heinrichs. 1997. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J. Bacteriol. 179:3963-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung, A. L., K. J. Eberhardt, E. Chung, M. R. Yeaman, P. M. Sullam, M. Ramos, and A. S. Bayer. 1994. Diminished virulence of a sar−/agr− mutant of Staphylococcus aureus in the rabbit model of endocarditis. J. Clin. Investig. 94:1815-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung, A. L., J. M. Koomey, C. A. Butler, S. J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 89:6462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung, A. L., M. R. Yeaman, P. M. Sullam, M. D. Witt, and A. S. Bayer. 1994. Role of the sar locus of Staphylococcus aureus in induction of endocarditis in rabbits. Infect. Immun. 62:1719-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chien, Y., A. C. Manna, and A. L. Cheung. 1998. SarA level is a determinant of agr activation in Staphylococcus aureus. Mol. Microbiol. 30:991-1001. [DOI] [PubMed] [Google Scholar]
- 13.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellerman, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elasri, M. O., J. R. Thomas, R. A. Skinner, J. S. Blevins, K. E. Beenken, C. L. Nelson, and M. S. Smeltzer. 2002. Staphylococcus aureus collagen adhesin contributes to the pathogenesis of osteomyelitis. Bone 30:275-280. [DOI] [PubMed] [Google Scholar]
- 15.Gentry, L. O. 1997. Osteomyelitis and other infections of bones and joints, p. 455-473. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 16.Gertz, S., S. Engelmann, R. Schmid, K. Ohlsen, J. Hacker, and M. Hecker. 1999. Regulation of σB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol. Gen. Genet. 261:558-566. [DOI] [PubMed] [Google Scholar]
- 17.Gertz, S., S. Engelmann, R. Schmid, A. K. Ziebandt, K. Tischer, C. Scharf, J. Hacker, and M. Hecker. 2000. Characterization of the σB regulon in Staphylococcus aureus. J. Bacteriol. 182:6983-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giachino, P., S. Engelmann, and M. Bischoff. 2001. σB activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillaspy, A. F., S. G. Hickmon, R. A. Skinner, J. R. Thomas, C. L. Nelson, and M. S. Smeltzer. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63:3373-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillaspy, A. F., C. Y. Lee, S. Sau, A. L. Cheung, and M. S. Smeltzer. 1998. Factors affecting the collagen binding capacity of Staphylococcus aureus. Infect. Immun. 66:3170-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hienz, S. A., T. Schennings, A. Heimdahl, and J. I. Flock. 1996. Collagen binding of Staphylococcus aureus is a virulence factor in experimental endocarditis. J. Infect. Dis. 174:83-88. [DOI] [PubMed] [Google Scholar]
- 22.Kielian, T., A. Cheung, and W. F. Hickey. 2001. Diminished virulence of an alpha-toxin mutant of Staphylococcus aureus in experimental brain abscesses. Infect. Immun. 69:6902-6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menzies, B. E., and D. S. Kernodle. 1994. Site-directed mutagenesis of the alpha-toxin gene of Staphylococcus aureus: role of histidines in toxin activity in vitro and in a murine model. Infect. Immun. 62:1843-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsson, I. M., T. Bremell, C. Ryden, A. L. Cheung, and A. Tarkowski. 1996. Role of the staphylococcal accessory gene regulator (sar) in septic arthritis. Infect. Immun. 64:4438-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nilsson, I. M., O. Hartford, T. Foster, and A. Tarkowski. 1999. Alpha-toxin and gamma-toxin jointly promote Staphylococcus aureus virulence in murine septic arthritis. Infect. Immun. 67:1045-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novick, R. P. 2001. Pathogenicity factors and their regulation, p. 392-407. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 28.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Callaghan, R. J., M. C. Callegan, J. M. Moreau, L. C. Green, T. J. Foster, O. M. Hartford, L. S. Engel, and J. M. Hill. 1997. Specific roles of alpha-toxin and beta-toxin during Staphylococcus aureus corneal infection. Infect. Immun. 65:1571-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papakyriacou, H., D. Vaz, A. Simor, M. Louie, and M. J. McGavin. 2000. Molecular analysis of the accessory gene regulator (agr) locus and balance of virulence factor expression in epidemic methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 181:990-1000. [DOI] [PubMed] [Google Scholar]
- 31.Patti, J. M., T. Bremell, D. Krajewska-Pietrasik, A. Abdelnour, A. Tarkowski, C. Ryden, and M. Hook. 1994. The Staphylococcus aureus collagen adhesin is a virulence determinant in experimental septic arthritis. Infect. Immun. 62:152-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skinner, R. A., S. G. Hickmon, C. L. Nelson, and R. A. Germer. 1992. Modified stain for identification of Staphylococcus aureus in osteomyelitis. J. Histotechnol. 15:303-306. [Google Scholar]