Abstract
We have cloned and characterized the kex1 gene of Pneumocystis jiroveci. Unlike the case for Pneumocystis carinii, in which the homologous PRT-1 genes are multicopy, kex1 is a single-copy gene encoding a protein homologous to fungal serine endoproteases, which localize to the Golgi apparatus. Thus, substantial biological differences can be seen among Pneumocystis species.
Pneumocystis jiroveci (see reference 20 for the new nomenclature for Pneumocystis species) is an opportunistic fungus that causes pneumonia in immunocompromised patients, particularly in those infected with human immunodeficiency virus (16). Pneumocystis fungi infecting different mammalian hosts are known to be genetically divergent (15, 21). Two multicopy gene families have been identified from rat-derived Pneumocystis carinii. One encodes the major surface glycoprotein (MSG), which is the most abundant cell surface protein of Pneumocystis (5, 8), and the other encodes the kexin homologue PRT-1, which is also a surface protein (12, 13, 17). To date the only antigens of P. jiroveci, the organism that causes disease in humans, that have been cloned and expressed are members of the MSG gene family, although a small fragment (366 bp) of a putative PRT-1 gene has been reported elsewhere (13). In an attempt to identify genes encoding additional antigens of P. jiroveci, we undertook to clone and characterize members of the PRT-1 family of genes from P. jiroveci.
Using primers based on the previously reported fragment (13); genomic DNA extracted from a P. jiroveci-infected human lung; and a combination of techniques including a two-step PCR with magnetic beads (Dynal Biotech Inc.) (19), inverse PCR (14), and genomic library screening (10), we obtained the complete genomic sequence (∼2.9 kb) of kex1.
To obtain the cDNA sequence, reverse transcription-PCR was performed with RNA extracted by using RNAzol B (Tel-Test Inc.) from P. jiroveci-infected human lung tissue and primers designed from the kex1 genomic sequence. Three sets of overlapping primers spanning nucleotides 536 to 3068 of the genomic sequence gave amplification products. To obtain the 3′ end, 3′ rapid amplification of cDNA ends (RACE) (3′-RACE kit; Life Technologies) was performed with a primer corresponding to nucleotides 3044 to 3063. Our attempts to determine the 5′ end of the cDNA sequence by 5′ RACE were unsuccessful and were limited by our ability to obtain only small amounts of undegraded RNA from autopsy lung samples. However, based on a comparison with the genomic sequence, the cDNA sequence appears to include the complete coding sequence. There are two potential initiation codons, at nucleotide positions 536 and 539 of the genomic sequence. Although the second one appears to be the more favorable translation start site (9), we are unable to determine which codon is in fact utilized. The stop codon TAA is at position 3245. Comparison of the cDNA with the genomic sequence identified eight introns whereas, in comparison, in P. carinii this gene is reported elsewhere to have only seven introns (12, 17). The cDNA encodes a protein containing 779 amino acids.
Comparison of the deduced amino acid sequence with those of other fungal serine endoproteases (Fig. 1) allowed identification of characteristic domains, including a signal peptide, a prodomain, a subtilisin-like catalytic domain, a P domain, a serine/threonine-rich region, and a transmembrane domain (4, 23). A hydrophobic transmembrane domain followed by a hydrophilic intracytoplasmic region, as predicted by the transmembrane hidden Markov model (18), is present at the carboxy terminus and is characteristic of Golgi apparatus-associated yeast kexins (7). This is different from most PRT-1 clones of rat-derived P. carinii but is similar to clone 71 (17) as well as Kex1 from mouse-derived Pneumocystis muris (11). The prodomain that can be removed by autocatalytic cleavage has a potential cleavage site (KR) at amino acid positions 113 and 114 (6). The deduced amino acid sequence of Kex1 from P. jiroveci showed 39% identity to P. carinii surface protease (SPRT) clone 12; 37% identity to clone 71; and 34% identity to Kex1 of P. muris, Kex2 of Saccharomyces cerevisiae, and Krp of Schizosaccharomyces pombe. However, the catalytic and P domains were more highly conserved. Amino acid residues D190, H228, and S400, the catalytic triad, are conserved in all kexins (23). There is an unusually proline-rich domain present in kexins from P. carinii and P. muris that is absent in Kex1 of P. jiroveci as well as other fungal kexins (2, 7, 12, 17).
FIG. 1.
Comparison of the deduced amino acid sequence of Kex1 from human-derived P. jiroveci with those of other fungal kexin-like proteases. Sequences of Kex1 of P. jiroveci (HPC Kex1), Kex1 of P. muris (MPC Kex1), SPRTs (clone 71 and clone 12) of P. carinii (RPC SPRT #71 and RPC SPRT #12, respectively), Kex2 of S. cerevisiae (SC Kex2), and Krp of S. pombe (SP Krp) (GenBank accession numbers AY127566, AF093132, U82999, U62910, M24201, and X82435, respectively) were analyzed by using Clustal W. The C-terminal end was aligned manually. Identical amino acid residues are boxed. An asterisk denotes the conserved catalytic triad of amino acid residues.
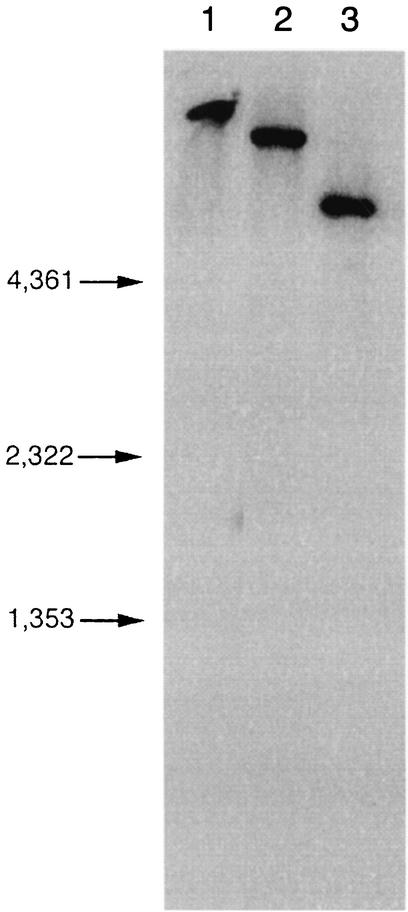
Northern blot analysis showed that kex1 mRNA is ∼2.4 kb in size. To determine the copy number of kex1 from P. jiroveci, Southern blot analysis (10) was done with genomic DNA isolated from P. jiroveci-infected human lung tissue digested with restriction enzyme EcoRI, PstI, or HindIII (Fig. 2). A PCR product corresponding to nucleotides 967 to 1928 of kex1 from P. jiroveci was used as the probe. Southern blotting showed a single band, demonstrating that kex1 is present as a single-copy gene in P. jiroveci. This is distinctly different from P. carinii, in which a family of related genes encode the homologous PRT-1, most of which have a carboxy terminus consistent with a glycosylphosphatidylinositol-type anchor and a surface location (12, 13, 17). Thus, P. carinii apparently duplicated the single-copy, presumably Golgi apparatus form of PRT-1 (with a simple frameshift to change to a glycosylphosphatidylinositol-type anchor) (17) at some point after the divergence of these two strains. In fact, the duplication must have occurred after the divergence of the otherwise very closely related species, P. carinii and P. muris, since the latter also has been reported elsewhere to have only a single copy of this gene (11). This is very different from the MSG gene family, since the genomes of all Pneumocystis species examined to date contain multiple copies of MSG genes, whose duplication must have occurred in a very early ancestor of most or all of the present-day Pneumocystis species (5, 8, 24).
FIG. 2.
Southern blot analysis of DNA from P. jiroveci. Genomic DNA was digested with EcoRI (lane 1), PstI (lane 2), or HindIII (lane 3) and probed with a 961-bp kex1 PCR product. Molecular weight markers are shown on the left.
In S. cerevisiae, Kex2 is involved in the processing of α mating factor and killer toxin (3). Recently it has been shown that Kex2 mutants of Candida glabrata are more sensitive than the wild type to antifungal drugs and chemicals targeting the cell membrane (1). Kex2 may be involved in the processing of proteins that maintain the cell surface integrity. Therefore, it has been proposed elsewhere that Kex2 inhibitors along with other antifungal agents could be useful for the treatment of fungal infections (1). The role of Kex1 of P. jiroveci in processing proteins that maintain cell surface integrity remains to be investigated. Kex1 may also be involved in the proteolytic processing of MSG, which is important in the pathogenicity of this organism, by removing the invariant upstream conserved sequence whose expression on the surface of the organism would counteract the antigenic diversity provided by the variant MSGs (22). Thus, inhibition of Kex1 may provide a new therapeutic approach to the management of P. carinii pneumonia.
Nucleotide sequence accession number.
The genomic sequence of kex1 was deposited in GenBank under accession number AY130996. The cDNA sequence was deposited in GenBank under accession number AY127566.
Editor: T. R. Kozel
REFERENCES
- 1.Bader, O., M. Schaller, S. Klein, J. Kukula, K. Haack, F. Muhlschlegel, H. C. Korting, W. Schafer, and B. Hube. 2001. The KEX2 gene of Candida glabrata is required for cell surface integrity. Mol. Microbiol. 41:1431-1444. [DOI] [PubMed] [Google Scholar]
- 2.Davey, J., K. Davis, Y. Imai, M. Yamamoto, and G. Matthews. 1994. Isolation and characterization of krp, a dibasic endopeptidase required for cell viability in the fission yeast Schizosaccharomyces pombe. EMBO J. 13:5910-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuller, R. S., A. Brake, and J. Thorner. 1989. Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc. Natl. Acad. Sci. USA 86:1434-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuller, R. S., A. J. Brake, and J. Thorner. 1989. Intracellular targeting and structural conservation of a prohormone-processing endoprotease. Science 246:482-486. [DOI] [PubMed] [Google Scholar]
- 5.Garbe, T. R., and J. R. Stringer. 1994. Molecular characterization of clustered variants of genes encoding major surface antigens of human Pneumocystis carinii. Infect. Immun. 62:3092-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Germain, D., F. Dumas, T. Vernet, Y. Bourbonnais, D. Y. Thomas, and G. Boileau. 1992. The pro-region of the Kex2 endoprotease of Saccharomyces cerevisiae is removed by self-processing. FEBS Lett. 299:283-286. [DOI] [PubMed] [Google Scholar]
- 7.Julius, D., A. Brake, L. Blair, R. Kunisawa, and J. Thorner. 1984. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-alpha-factor. Cell 37:1075-1089. [DOI] [PubMed] [Google Scholar]
- 8.Kovacs, J. A., F. Powell, J. C. Edman, B. Lundgren, A. Martinez, B. Drew, and C. W. Angus. 1993. Multiple genes encode the major surface glycoprotein of Pneumocystis carinii. J. Biol. Chem. 268:6034-6040. [PubMed] [Google Scholar]
- 9.Kozak, M. 1995. Adherence to the first-AUG rule when a second AUG codon follows closely upon the first. Proc. Natl. Acad. Sci. USA 92:2662-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kutty, G., L. Ma, and J. A. Kovacs. 2001. Characterization of the expression site of the major surface glycoprotein of human-derived Pneumocystis carinii. Mol. Microbiol. 42:183-193. [DOI] [PubMed] [Google Scholar]
- 11.Lee, L. H., F. Gigliotti, T. W. Wright, P. J. Simpson-Haidaris, G. A. Weinberg, and C. G. Haidaris. 2000. Molecular characterization of KEX1, a kexin-like protease in mouse Pneumocystis carinii. Gene 242:141-150. [DOI] [PubMed] [Google Scholar]
- 12.Lugli, E. B., A. G. Allen, and A. E. Wakefield. 1997. A Pneumocystis carinii multi-gene family with homology to subtilisin-like serine proteases. Microbiology 143:2223-2236. [DOI] [PubMed] [Google Scholar]
- 13.Lugli, E. B., E. T. Bampton, D. J. Ferguson, and A. E. Wakefield. 1999. Cell surface protease PRT1 identified in the fungal pathogen Pneumocystis carinii. Mol. Microbiol. 31:1723-1733. [DOI] [PubMed] [Google Scholar]
- 14.Ma, L., L. Borio, H. Masur, and J. A. Kovacs. 1999. Pneumocystis carinii dihydropteroate synthase but not dihydrofolate reductase gene mutations correlate with prior trimethoprim-sulfamethoxazole or dapsone use. J. Infect. Dis. 180:1969-1978. [DOI] [PubMed] [Google Scholar]
- 15.Ma, L., H. Imamichi, A. Sukura, and J. A. Kovacs. 2001. Genetic divergence of the dihydrofolate reductase and dihydropteroate synthase genes in Pneumocystis carinii from 7 different host species. J. Infect. Dis. 184:1358-1362. [DOI] [PubMed] [Google Scholar]
- 16.Masur, H., M. A. Michelis, J. B. Greene, I. Onorato, R. A. Stouwe, R. S. Holzman, G. Wormser, L. Brettman, M. Lange, H. W. Murray, and S. Cunningham-Rundles. 1981. An outbreak of community-acquired Pneumocystis carinii pneumonia: initial manifestation of cellular immune dysfunction. N. Engl. J. Med. 305:1431-1438. [DOI] [PubMed] [Google Scholar]
- 17.Russian, D. A., V. Andrawis-Sorial, M. P. Goheen, J. C. Edman, P. Vogel, R. E. Turner, D. L. Klivington, C. W. Angus, and J. A. Kovacs. 1999. Characterization of a multicopy family of genes encoding a surface-expressed serine endoprotease in rat Pneumocystis carinii. Proc. Assoc. Am. Physicians 111:347-356. [DOI] [PubMed] [Google Scholar]
- 18.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences, p. 175-182. In J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff, and C. Sensen (ed.), Proceedings of Sixth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif. [PubMed]
- 19.Sorensen, A. B., M. Duch, P. Jorgensen, and F. S. Pedersen. 1993. Amplification and sequence analysis of DNA flanking integrated proviruses by a simple two-step polymerase chain reaction method. J. Virol. 67:7118-7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stringer, J. R., C. B. Beard, R. F. Miller, and A. E. Wakefield. 2002. A new name (Pneumocystis jiroveci) for Pneumocystis from humans. Emerg. Infect. Dis. 8:891-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stringer, J. R., S. L. Stringer, J. Zhang, R. Baughman, A. G. Smulian, and M. T. Cushion. 1993. Molecular genetic distinction of Pneumocystis carinii from rats and humans. J. Eukaryot. Microbiol. 40:733-741. [DOI] [PubMed] [Google Scholar]
- 22.Sunkin, S. M., M. J. Linke, F. X. McCormack, P. D. Walzer, and J. R. Stringer. 1998. Identification of a putative precursor to the major surface glycoprotein of Pneumocystis carinii. Infect. Immun. 66:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van de Ven, W. J., A. J. Roebroek, and H. L. Van Duijnhoven. 1993. Structure and function of eukaryotic proprotein processing enzymes of the subtilisin family of serine proteases. Crit. Rev. Oncog. 4:115-136. [PubMed] [Google Scholar]
- 24.Wright, T. W., T. Y. Bissoondial, C. G. Haidaris, F. Gigliotti, and P. J. Haidaris. 1995. Isoform diversity and tandem duplication of the glycoprotein A gene in ferret Pneumocystis carinii. DNA Res. 2:77-88. [DOI] [PubMed] [Google Scholar]