Abstract
Volatile sulfur compounds, including hydrogen sulfide (H2S), have been implicated in the development of periodontal disease. Glutathione is an important thiol source for H2S production in periodontal pockets. Our recent studies have delineated a pathway of glutathione metabolism in Treponema denticola that releases H2S. In this pathway, γ-glutamyltransferase (GGT) has been proposed to catalyze the first step of glutathione degradation. We have cloned the gene of GGT from T. denticola, which contains an open reading frame of 726 bp encoding a protein of 241 amino acids. Transformation of this gene into Escherichia coli led to the expression of a recombinant protein. After purification by chromatography, the recombinant protein showed enzymatic activity typical of GGT, catalyzing the degradation of Na-γ-glutamyl-4-nitroaniline (GNA) and the hydrolysis of glutathione, releasing glutamic acid or glutamine and cysteinylglycine. l-Cysteine is not a substrate of GGT. Importantly, GNA, when added to T. denticola, was able to compete with glutathione and inhibit the production of H2S, ammonia, and pyruvate. This was accompanied by the suppression of hemoxidative and hemolytic activities of the bacteria. Purified GGT was inactivated by TLCK (Nα-p-tosyl-l-lysine chloromethyl ketone) and proteinase K treatment. However, higher enzymatic activity was demonstrated in the presence of 2-mercaptoethanol and dithiothreitol. Our further experiments showed that the addition of recombinant GGT to Porphyromonas gingivalis, a bacterium without significant glutathione-metabolizing capacity, drastically increased the utilization of glutathione by the bacterium, producing H2S, ammonia, and pyruvate. This was again accompanied by enhanced bacterial hemoxidative and hemolytic activities. Together, the results suggest an important role for GGT in glutathione metabolism in oral bacteria.
Treponema denticola is a pathogenic member of the periodontopathic microbiota (16, 26). Among the many potential virulence factors produced by this microorganism is a family of volatile sulfur compounds, which is well exemplified by hydrogen sulfide (H2S) (13, 14, 17, 27, 34, 37). H2S has been found at levels of up to 2 mM in diseased periodontal pockets but at negligible levels in healthy sites (29). Highly toxic, H2S induces the injury or death of host cells, including gingival fibroblasts and epithelial cells (4, 31, 41).
Thiol sources for H2S production in vivo can be multiple, including serum proteins, l-cysteine, and glutathione (8, 9, 11, 30). Initial research focused mainly on l-cysteine (8, 11). However, the l-cysteine content in human plasma is very limited (25). On the other hand, a significantly higher amount of glutathione (γ-glutamylcysteinylglycine) is detected in mammalian cells, especially in the polymorphonuclear leukocytes (PMNs). In infected periodontal pockets and other inflamed gingival tissues, PMNs accumulate (5, 28), and significant amounts of glutathione may be released when PMNs in the pockets are damaged, thereby providing a large reservoir of glutathione for H2S formation by oral bacteria, including T. denticola.
A recent study examined glutathione metabolism in T. denticola (9). Based on these data, a pathway of three steps was proposed. Initially, glutathione is cleaved into glutamate or glutamine and the dipeptide cysteinylglycine (Cys-Gly). The last is then cleaved into glycine and l-cysteine. Finally, l-cysteine is degraded, releasing pyruvate, ammonia, and H2S. Key enzymes involved in glutathione metabolism may include γ-glutamyltransferase (GGT), cysteinylglycinase, and l-cysteine desulfhydrase (cystalysin) (9, 11, 20). In previous studies, the cystalysin from T. denticola has been cloned (8, 11). In the presence of l-cysteine, cystalysin catalyzes the production of H2S, expressing virulence in the forms of hemolysis and hemoxidation (10, 12, 21). In sharp contrast, cystalysin alone is incapable of digesting glutathione (9), suggesting an important role for upstream enzymes in glutathione metabolism.
A series of carefully conducted biochemical studies by Makinen and Makinen led to the purification of GGT from T. denticola (24). The enzyme was shown to hydrolyze Na-γ-glutamyl-4-nitroaniline (GNA) and glutathione. Moreover, an N-terminal sequence of 21 amino acids in this enzyme was determined (24). With this sequence, we have now cloned and expressed the entire gene for GGT from T. denticola. The expressed recombinant protein exhibited enzymatic activities typical of GGT. Importantly, in intact T. denticola cells, GNA competed with glutathione for GGT and inhibited the production of H2S, suggesting a critical role for GGT in glutathione metabolism. The role of GGT in glutathione metabolism is further supported by the observation that the addition of recombinant GGT to Porphyromonas gingivalis, a bacterium without significant glutathione-metabolizing capacity, drastically increased the utilization of glutathione and virulence expression by that bacterium.
MATERIALS AND METHODS
Materials.
Unless otherwise indicated, all chemicals and reagents were purchased from the Sigma Chemical Company, St. Louis, Mo.
Bacterial strains and culture conditions.
T. denticola strain ATCC 35405 (TD-5) was used as the source of genomic DNA for GGT gene cloning. The oral bacteria were cultured anaerobically in a Coy anaerobic chamber (5% CO2, 10% H2, 85% N2) in GM-1 medium (42). P. gingivalis W83 and Fusobacterium periodonticum were grown on the surface of enriched Trypticase soy agar plates with 5% (vol/vol) sheep blood (19) supplemented with 10% rabbit serum. pRsetA (Invitrogen, San Diego, Calif.) was used as a cloning vector. Escherichia coli TB-1 and DL21 were used as host strains for plasmid preparation and were routinely grown in Luria-Bertani broth or on Luria-Bertani agar plates supplemented with 50 μg of ampicillin/ml when appropriate.
Oligonucleotides and DNA amplification by PCR.
Based on the first N-terminal 16-amino-acid sequence (24), the entire genomic DNA sequence encoding GGT of T. denticola ATCC 35405 was identified from the Baylor T. denticola Database. The GGT coding region was PCR amplified from T. denticola TD-5 genomic DNA, using a forward primer (5′-CGGGATCCATGAAAAAACCGCCGCTTATAGG) and a reverse primer (5′-GGGGTACCATTAAACAGCCGGAATCAGC). Thermocycling was conducted with an initial denaturation step (5 min at 94°C) followed by 40 cycles of amplification (1 min at 94°C and 1 min at 48°C) and 3 min at 72°C in a PTC-100 thermocycler from MJ Research. The PCR products were gel resolved and purified for cloning into vectors (2).
Cloning and sequencing of the GGT gene from T. denticola.
The 0.7-kb PCR-amplified GGT gene fragment with BamHI-KpnI digestion sites was ligated into pUC 18 that had been predigested by BamHI-KpnI. The resultant construct was transformed into E. coli TB-1 by electroporation (2). After antibiotic selection, positive colonies containing the T. denticola GGT sequence were identified, and the insert was sequenced on both strands. The open reading frames of the sequence were determined and compared to the nonredundant combined protein database using the Baylor T. denticola Database supported by the Institute of Genomic Research, with the published N-terminal sequence of T. denticola GGT as a probe (24).
Expression and purification of recombinant GGT of T. denticola.
The coding region of the GGT gene was amplified by PCR and ligated, in frame, into the expression vector pRsetA. The recombinant plasmid was transformed into E. coli BL21 (DE3). The expression of T. denticola GGT was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to the cells containing the expression plasmid. E. coli cells in 1.5 liters of culture were then collected by 10 min of centrifugation at 6,500 × g and washed once with 20 mM phosphate (pH 7.4)-buffered normal saline (PBS). The cells were then suspended in 45 ml of 50 mM NaH2PO4-300 mM NaCl-20 mM imidazole (pH 8.0) with 2 mM protease inhibitor (phenylmethylsulfonyl fluoride [PMSF]) for sonication. The cells were broken by five sonication cycles (5 min at 40% intensity and a 40% duty cycle) with intermittent cooling on ice for 2 min, using a model 250 sonifier and a microtip probe from Branson Ultrasonics Corporation (Danbury, Conn.). The soluble cell fraction was obtained by centrifugation at 15,000 × g for 60 min and loaded onto a 2.5-ml Ni-nitrilotriacetic acid (NTA) gel column (Qiagen, Chatsworth, Calif.). The column was subsequently washed and eluted with a step gradient of imidazole (20 to 250 mM) to collect the recombinant GGT.
SDS-PAGE analysis.
The concentration of protein was determined using the bicinchoninic acid protein assay method described by Kennel and Holt (19). Proteins were resolved by the discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) system. The resolving gel consisted of 12% acrylamide in 0.125 M Tris(hydroxymethyl)aminomethane (pH 8.8), and the stacking gel contained 4% acrylamide in 0.125 M Tris(hydroxymethyl)aminomethane (pH 6.8). The gel was directly stained using 0.025% Coomassie brilliant blue R-250 to reveal the protein bands (22).
Enzymatic assay of GGT.
The activity of GGT was determined as described by Makinen (24) with minor modifications. Briefly, the reaction mixture contained 6 mM β-mercaptoethanol (2-ME) and 20 mM Tris buffer at pH 8.0 and 200 μM GNA. The reaction was started by the addition of 10 μl of enzyme samples to 1 ml of reaction mixture. Absorption at 410 nm in the reaction was monitored for 5 min in a cuvette at room temperature on a Beckman DU spectrophotometer. The enzyme activity of GGT was calculated based on the absorbance of standard GNA.
Mass spectrometric analysis of purified recombinant GGT.
Approximately 2 μg of purified recombinant protein was digested for 4 h at 37°C with 0.2 μg of trypsin in 50 μl of 20 mM ammonium bicarbonate, pH 8.0. Following digestion, 1 μl of the sample was applied to the matrix-assisted laser desorption ionization target. Matrix-assisted laser desorption ionization-time of flight mass spectra were acquired on an Applied Biosystems Voyager-DE STR operated in the reflectron mode. The monoisotopic masses of the tryptic peptides were compared with the hypothetical GGT tryptic fragments generated in silico by the program GPMAW (Lighthouse Data).
HPLC analysis of GGT-catalyzed glutathione degradation.
Purified recombinant GGT of T. denticola was combined at a concentration of 2 μg/ml with 5 mM glutathione in 20 mM Tris buffer, pH 8.0. After 10 h of incubation at room temperature, the mixture was subjected to ultrafiltration on a 10-kDa-cutoff Centricon concentrator. The resultant <10-kDa fraction was collected and concentrated using a SpeedVac concentrator for high-performance liquid chromatography (HPLC) analysis. HPLC analysis of amino acids was performed as described previously (9), using the 2690 Separations Module (Waters Co.).
Chemical analysis of H2S pyruvate and ammonia.
H2S was quantified by a method described previously (11, 12). Pyruvate was analyzed by a method modified from those of Chu et al. and Zheng et al. (9, 43). Ammonia contents were determined using the method of Bauer et al. (3). Modifications of these chemical methods in our studies have been described in detail in previous publications (9, 11). All analyses were carried out in triplicate unless otherwise indicated.
Assays of HeO-HeA activities.
Hemoxidative (HeO) activity was determined using sheep red blood cells. The method was modified from that of Leahy and Smith as described previously (7). Hemolytic (HeA) activity was determined as described in previous studies (6, 7).
Characterization of recombinant GGT of T. denticola.
Glutathione in a reduced form, Cys-Gly, and l-cysteine were used as substrates of the purified recombinant GGT and cystalysin from T. denticola. GGT was used at 2 μg/ml to hydrolyze GNA. The activity of untreated GGT was used as a control (100%).
Effects of thiol compounds.
Thiol compounds (2-ME, dithiothreitol, and l-cysteine) at 6 mM were incubated with 4 μg of GGT/ml at 37°C for 30 min. GNA at a concentration of 1 mM was added for another 60 min. GGT activity was indicated by GNA hydrolysis.
Effects of proteinase inhibitors and proteinases.
Protease inhibitors at 2 mM (TLCK [Nα-p-tosyl-l-lysine chloromethyl ketone], PMSF, and benzamidine) or proteases at 100 μg/ml (proteinase K and pronase) were added to 2-μg/ml GGT or cystalysin. After incubation at 37°C for 30 min, 0.2 mM GNA was added, and GGT enzyme activity was determined as described above.
Effects of selected treatments. (i) Heating.
GGT at a concentration of 2 μg/ml was heated at 56°C for 30 min in Tris buffer (pH 8.0) and then cooled on ice to 4°C. The enzyme was incubated with 0.2 mM GNA, and its activity was measured.
(ii) Freezing.
GGT at 2 μg/ml was stored overnight at −20°C. After being thawed at room temperature, the enzyme was incubated with GNA, and its enzyme activity was determined.
(iii) Sonication.
GGT at 2 μg/ml was sonicated for 5 min on ice (as mentioned above) and then incubated with GNA.
Effects of T. denticola GGT on glutathione metabolism in P. gingivalis.
P. gingivalis W83 was harvested from enriched Trypticase soy agar-blood plates and resuspended in 20 ml of PBS. The cells were then washed twice in PBS and diluted to a final concentration of 0.5 mg of protein/ml. The cells were mixed with 2 μg of recombinant T. denticola GGT/ml in the presence of glutathione, Cys-Gly, or l-cysteine. The mixtures were incubated at 37°C for 1 h and centrifuged at 13,000 × g for 1 min to collect the supernatant to determine the production of H2S and ammonia.
Effects of GGT on HeO-HeA activities of P. gingivalis in the presence of thiol compounds.
P. gingivalis cells were mixed with 2 μg of recombinant GGT/ml and 2 mM glutathione, Cys-Gly, or l-cysteine. The mixture was incubated at 37°C for 10 h in glass tubes with gentle shaking. A mixture without GGT was used as a control. Samples were then taken for measurement of HeO-HeA activities using the methods described above.
RESULTS
Cloning and sequencing of GGT of T. denticola.
We initially searched the Baylor T. denticola Database supported by the Institute of Genomic Research, using the published N-terminal sequence of T. denticola GGT as a probe (24). The database search led to the identification of the entire GGT gene of T. denticola. The open reading frame of the sequence was predicted to encode a protein of 241 amino acids (Fig. 1, positions 40 to 281). We subsequently designed primers based on the ends of the coding sequence to PCR amplify the genomic DNA of T. denticola. The amplification produced a major fragment ∼740 bp long, which was consistent with the predicted GGT coding sequence. The fragment was purified and ligated into the vector pUC18 and then transformed into E. coli TB-1. Positive colonies were selected, and the insert in the plasmid was sequenced to confirm the presence of the T. denticola GGT gene. The sequence consisting of 726 bp agreed completely with that of T. denticola GGT reported in the database. The deduced amino acid sequence of T. denticola GGT is shown in Fig. 1. The protein is expected to have a molecular weight of 26849.8 and an isoelectric point of 5.9. It contains 33 basic amino acids (17 Lys, 8 Arg, and 8 His) and 28 acidic amino acids (17 Glu and 11 Gln). Of interest, four cysteine residues have been identified in this protein that may have an important role in its enzymatic activity (24).
FIG. 1.
Amino acid sequence of GGT deduced from its gene in T. denticola ATCC 35405. The peptides detected by mass spectrometry (Table 2) are shown in boldface letters. The sequence of GGT of T. denticola covered 241 amino acids (positions 40 to 280).
Expression and purification of T. denticola GGT.
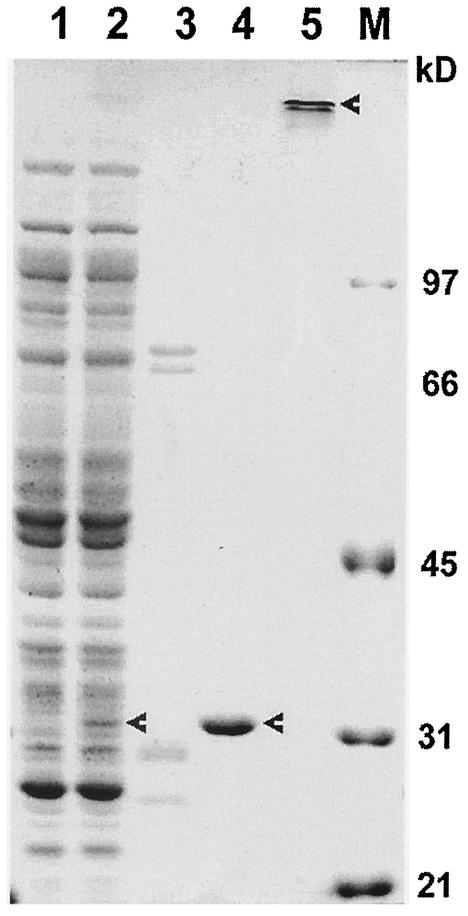
In order to investigate the properties of T. denticola GGT, we expressed and purified a recombinant protein of the enzyme. The pRsetA-ggt construct was transformed into E. coli BL21(DE3) cells. A positive colony that expressed a recombinant protein of ∼31 kDa was identified (Fig. 2, lane 2). This colony expressed dramatically high GGT enzymatic activities when analyzed in reactions containing the chromogenic substrate GNA (not shown). To purify the recombinant protein, E. coli cells of the colony were subjected to sonication to collect soluble and insoluble fractions. As shown in Table 1, >85% of the GGT enzymatic activity was recovered in the soluble fraction. We subsequently loaded the soluble fraction on an Ni-NTA gel column. After flowthrough, the column was first washed with 50 to 110 mM imidazole; this wash did not a contain significant amount of recombinant protein or GGT enzymatic activity (Fig. 2, lane 3). The majority of the recombinant protein was eluted from the column with 150 to 210 mM imidazole (Fig. 2, lane 4). The whole procedure of purification led to a 121.8-fold enrichment of the protein and the recovery of 68.7% of the total GGT enzymatic activity (Table 1). The purified recombinant GGT protein exhibited an apparent molecular mass of ∼31 kDa, which was expected to include 39 amino acid residues at the N terminus in addition to the T. denticola GGT peptide. The first two peptides and a portion of the third peptide, WGSELS, were derived from the vector sequence of 39 amino acids (Table 2). Under nondenaturing conditions, the recombinant protein aggregated to a complex of >200 kDa (Fig. 2, lane 5). This property has been demonstrated for the natural GGT protein isolated from T. denticola (24).
FIG. 2.
SDS-PAGE analysis of purification of recombinant T. denticola GGT expressed in E. coli. Wild-type and transformed E. coli were subjected to sonication to break up the cells. The soluble fractions were then collected and analyzed or loaded onto Ni-NTA columns for sequential elution by 50 to 110 and 150 to 210 mM imidazole. Lane 1, soluble cell fraction of wild-type E. coli; lane 2, soluble cell fraction of pRsetA-ggt-transformed E. coli; lane 3, elution by 50 to 110 mM imidazole; lanes 4 and 5, elution by 150 to 210 mM imidazole; lane M, low-molecular-mass standards. Prior to SDS-PAGE, the samples (except that in lane 5) were boiled for 5 min. The arrowheads indicate the bands of interest.
TABLE 1.
Purification of recombinant GGT of T. denticolaa
| Preparation | Total protein (mg) | Enzymatic activity (mmol/mg/min) | Purification (n-fold original GGT activity) | Yield of activity (%) |
|---|---|---|---|---|
| Whole cell sonicated | 425.5 | 1.3 | 1.0 | 100.0 |
| Soluble cell fraction | 250.2 | 1.9 | 1.5 | 85.9 |
| Elution from affinity gel | 2.4 | 158.4 | 121.8 | 68.7 |
GNA was used as a substrate for the GGT enzymatic assay. A majority of the GGT activity (68.7%) was recovered in the final purified protein.
TABLE 2.
Peptides in recombinant GGT identified by mass spectrometry
| Residues | [M+H]+a
|
Delta | Sequence | |
|---|---|---|---|---|
| Experimental | Calculated | |||
| 1-33 | 3,879.90 | 3,879.59 | 0.31 | MRGSHHHHHHGMASMTGGQQMGRDLYDDDDKDR |
| 3-23 | 2,277.99 | 2,277.97 | 0.02 | GSHHHHHHGMASMTGGQQMGR |
| 34-41 | 979.47 | 979.46 | 0.01 | WGSELEMK |
| 66-97 | 3,464.68 | 3,464.71 | −0.03 | MYTNADYVNSVLAAGGVPLMLPIIDDEDAIQR |
| 132-141 | 1,235.72 | 1,235.70 | 0.02 | RDVYELSLIK |
| 156-176 | 2,267.16 | 2,267.21 | −0.05 | GMQILNVAFGGSLYQDLSLIK |
| 156-177 | 2,423.23 | 2,423.31 | −0.08 | GMQILNVAFGGSLYQDLSLIKR |
| 178-186 | 1,108.63 | 1,108.61 | 0.02 | DIQIQHVQK |
| 187-192 | 756.41 | 756.41 | 0.0 | ARPQER |
| 193-197 | 585.32 | 585.34 | −0.02 | THSIK |
| 198-206 | 978.48 | 978.49 | −0.01 | TEAASIMQK |
| 211-223 | 1,560.70 | 1,560.69 | 0.01 | EDMVNSYHHMAVK |
| 267-273 | 793.42 | 793.45 | −0.03 | ASLDLFK |
| 274-278 | 678.35 | 678.36 | −0.01 | EFINR |
Oxidation of Met observed in the analysis. Mass spectrometry was employed to determine the amino acid sequences of the peptides after trypsin digestion of purified recombinant GGT. The 14 peptides identified by mass spectrometry shown in this table represent >50% coverage of the deduced protein sequence (Fig. 1). Note that the first 39 amino acids (residues 1 to 39) of the recombinant protein are derived from the peptide sequence of the cloning vector, pRsetA.
Analysis of recombinant T. denticola GGT by mass spectrometry.
To further confirm the identity of the recombinant protein, we analyzed it by mass spectrometry. Upon trypsin digestion, 14 peptides of the recombinant protein were identified (Table 2). Other peptides were all identified in the sequence of T. denticola GGT, covering >50% (Fig. 1) of the entire sequence. On mass spectrometry, the recombinant protein showed a molecular mass of 31,675 Da, which included T. denticola GGT and the peptide from the pRsetA vector.
Characterization of enzymatic activity of GGT.
We subsequently characterized the enzymatic activity of recombinant GGT, using GNA as a substrate (24). The results are shown in Fig. 3. Treatment of the recombinant protein with the protease inhibitor TLCK, but not PMSF or benzamidine, resulted in a significant loss (>90%) of GGT activity. Moreover, recombinant GGT could be inactivated by proteinase K and pronase treatments (Table 3). On the other hand, thiol compounds were able to activate the recombinant GGT, with a 1.5- to 2.5-fold increase in enzymatic activity in the presence of 2-ME, dithiothreitol, or l-cysteine. GGT, unlike cystalysin, was not affected by being frozen at −20°C but was inactivated by being heated at 56°C. Also of interest, 600 mM KCl was shown to increase the enzymatic activity of GGT (Table 3), while the addition of NaCl was without effect. These results are similar to those presented previously (24).
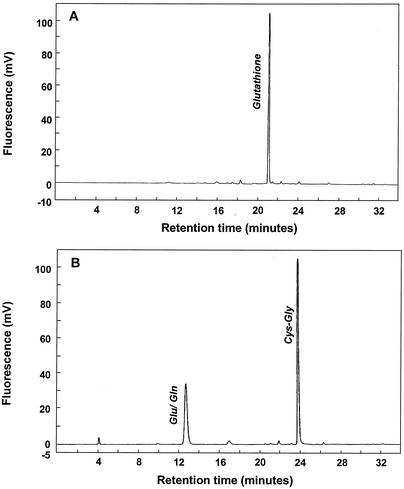
FIG. 3.
HPLC analysis of glutathione degradation catalyzed by recombinant T. denticola GGT. Glutathione at 5 mM was incubated for 8 h at room temperature alone (A) or with 2 μg of recombinant GGT/ml (B). Glutathione and its metabolites were analyzed by HPLC. In the presence of GGT, glutathione was completely degraded into glutamic acid-glutamine and Cys-Gly.
TABLE 3.
Characterization of T. denticola GGTa
| Treatment | Conditions | Enzyme activity (% of control)
|
|
|---|---|---|---|
| GGT | Cystalysin | ||
| Control | Buffer only | 100.0 ± 5.6 | 100.0 ± 5.6 |
| Proteinase inhibitor | 100 μg/ml | ||
| TLCK | 2 mM | 5.8 ± 0.5 | 96.8 ± 10.2 |
| PMSF | 2 mM | 95.8 ± 5.8 | 98.4 ± 8.8 |
| Benzamidine | 2 mM | 98.5 ± 8.9 | 95.5 ± 6.7 |
| Thiol compound | |||
| 2-ME | 6 mM | 255.1 ± 16.8 | 189.5 ± 12.6 |
| l-Cysteine | 6 mM | 155.9 ± 13.7 | ND |
| Dithiothreitol | 6 mM | 219.8 ± 18.9 | 168.9 ± 8.6 |
| Proteinase | |||
| Proteinase K | 100 μg/ml | 4.9 ± 0.6 | 91.8 ± 7.6 |
| Pronase | 100 μg/ml | 15.4 ± 1.8 | 95.4 ± 8.9 |
| Salt | |||
| NaCL | 600 mM | 105.8 ± 9.6 | 101.5 ± 12.4 |
| KCL | 600 mM | 155.6 ± 8.9 | 98.4 ± 10.2 |
| Heating | 56°C | 20.5 ± 2.1 | 92.5 ± 8.9 |
| Freezing | −20°C | 98.5 ± 6.8 | 100.5 ± 11.9 |
Purified recombinant GGT and cystalysin of T. denticola were compared with regard to their sensitivities to various treatments. Significant differences between the two enzymes in their sensitivities to TLCK, proteinase, heating, and KCl treatment are shown. ND, not determined.
Glutathione degradation catalyzed by recombinant GGT.
Our recent studies have suggested a stepwise pathway for glutathione metabolism in T. denticola involving three enzymes: GGT, cysteinylglycinase, and cystalysin. Thus, after characterization of the recombinant GGT, we went on to examine whether GGT catalyzes glutathione metabolism and to describe the mechanism. To this end, recombinant GGT at 2 μg/ml was incubated with 5 mM glutathione. After incubation, the reaction mixture was analyzed by HPLC. The results are shown in Fig. 3. Clearly, glutamic acid and the dipeptide Cys-Gly were generated (Fig. 3B). Of note, these two products were detected at molarities similar to the starting amount of glutathione and glutathione was undetectable after the incubation, indicating a complete digestion of this tripeptide (Fig. 3). The data suggest that GGT may catalyze the first step of glutathione degradation in vivo, releasing glutamic acid and the dipeptide Cys-Gly.
Inhibition of glutathione metabolism in T. denticola by GNA.
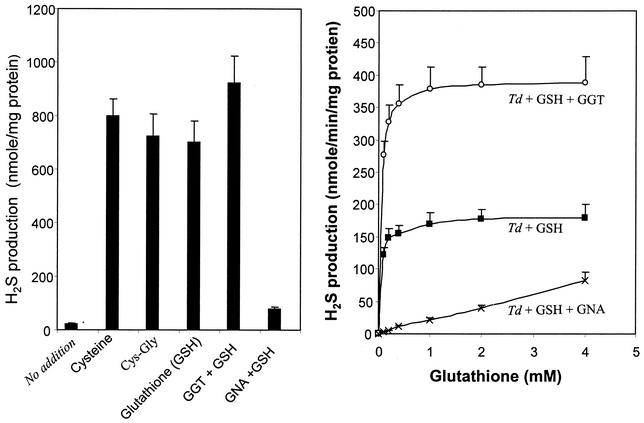
GNA was initially utilized in our experiments as a chromogenic substrate in the GGT enzymatic assay (Tables 1 and 3). As an exogenous substrate, it was reasoned that GNA might compete with glutathione and inhibit GGT activity toward the tripeptide. Under this assumption, we examined the effects of GNA on glutathione metabolism in T. denticola. Consistent with previous observations (9), T. denticola was able to metabolize l-cysteine and Cys-Gly, as well as glutathione, releasing H2S, ammonia, and pyruvate (shown in Fig. 4, left, only for H2S). The addition of recombinant GGT to T. denticola did not enhance l-cysteine or Cys-Gly metabolism (not shown) but increased H2S production from glutathione (Fig. 4, left). Importantly, GNA inhibited glutathione metabolism in T. denticola (Fig. 4, left).
FIG. 4.
Effects of GGT and GNA on H2S production from glutathione (GSH) by T. denticola. (Left) H2S formation from thiol compounds in T. denticola and effects of recombinant GGT and GNA. Significant amounts of H2S were produced in the presence of 2 mM l-cysteine, Cys-Gly, or glutathione. (Right) Glutathione dose-dependent H2S production in T. denticola (Td) and effects of recombinant GGT and GNA. The production of H2S was enhanced by recombinant GGT but was suppressed by GNA. The bars and data points represent means, and the error bars indicate standard deviations; n = 3.
To further pursue these observations, we examined H2S production by T. denticola in the presence of various amounts of glutathione. As shown in Fig. 4, right, the relationship between the rate of H2S formation and glutathione concentrations approximated the hyperbolic Michaelis-Menten kinetics curve (γ = 0.996). The maximum initial rate of H2S formation was calculated at 182 nmol/min/mg of cellular protein. The half-maximal rate (K[infi]m) was obtained at 15.5 ± 1.2 μM glutathione. The addition of 2 μg of GGT/ml to T. denticola increased the maximum initial rate of H2S formation to 385 ± 44 nmol/min/mg of cellular protein. Significantly, GNA suppressed H2S production from glutathione in an impressive manner (Fig. 4, right).
Effects of GNA on virulence expression in T. denticola in the presence of glutathione.
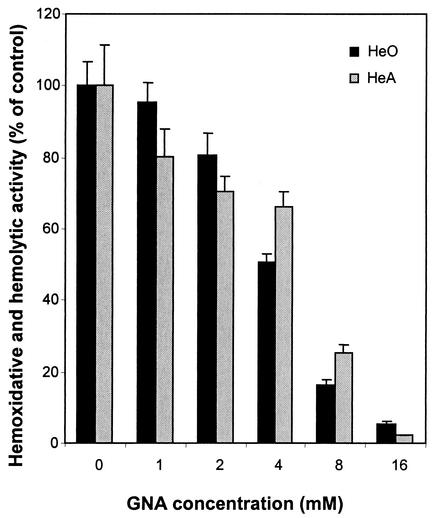
As reported recently, glutathione metabolism significantly increases HeO-HeA activities in T. denticola (9). Since GNA was able to inhibit H2S production from glutathione in T. denticola (Fig. 4), we went on to examine whether this inhibition was accompanied by the suppression of bacterial virulence expression. The results are shown in Fig. 5. Clearly, GNA inhibited the HeO-HeA activities of T. denticola in a dose-dependent manner, achieving maximal inhibition at 16 mM.
FIG. 5.
GNA suppression of HeO-HeA activities of T. denticola in the presence of glutathione. T. denticola was cultured in the presence of 2 mM glutathione and 0 to 16 mM GNA. HeO-HeA activities were analyzed as described in Materials and Methods. The bars represent means plus standard deviations; n = 3.
Recombinant GGT facilitates glutathione metabolism in P. gingivalis.
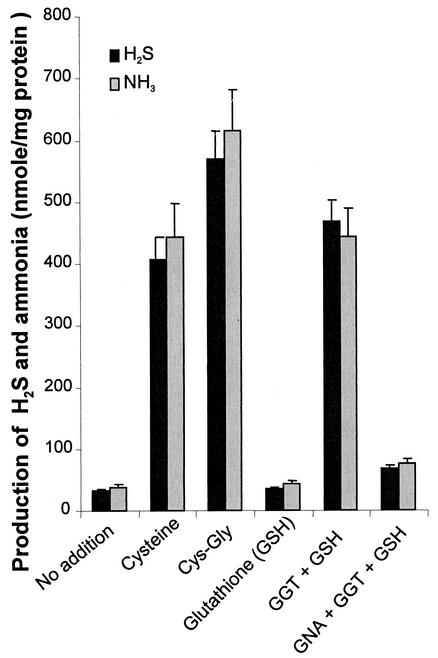
The ability to metabolize thiol compounds varies significantly among oral bacteria (30). While T. denticola exhibits a high capacity for glutathione metabolism, another important oral pathogen, P. gingivalis, did not show significant H2S or ammonia production in the presence of glutathione (Fig. 6, Glutathione). On the other hand, P. gingivalis was able to metabolize l-cysteine and Cys-Gly (Fig. 6, Cysteine and Cys-Gly). More importantly, in the presence of recombinant GGT, P. gingivalis became capable of utilizing glutathione, producing H2S and ammonia (Fig. 6, GGT + GSH). As expected, GNA was shown to inhibit glutathione metabolism by P. gingivalis under these conditions (Fig. 6, GNA + GGT + GSH)
FIG. 6.
Effects of recombinant GGT and its inhibitor GNA on glutathione utilization in P. gingivalis. P. gingivalis W83 was incubated without (No addition) or with 2 mM l-cysteine, Cys-Gly, or glutathione. GGT was added at 2 μg/ml (GGT + GSH), and GNA was added at 16 mM (GNA + GGT + GSH). P. gingivalis was able to metabolize l-cysteine and Cys-Gly, but not glutathione, to produce significant amounts of H2S and ammonia. The addition of GGT enhanced the bacterial capacity for metabolizing glutathione. The effect of GGT was ameliorated by the presence of GNA. The bars represent means plus standard deviations; n = 3.
Effects of GGT on virulence expression in P. gingivalis in the presence of glutathione.
Based on the effects of GGT on glutathione metabolism in P. gingivalis, we reasoned that the virulence expression of this bacterium in the presence of glutathione would be significantly enhanced by recombinant GGT. Such assumptions were validated by our experiments measuring the HeO-HeA activities of P. gingivalis (data not shown). In the absence of thiol compounds, P. gingivalis expressed low HeO-HeA activities. The addition of Cys-Gly or l-cysteine to the bacterium led to a four- to fivefold increase in HeO-HeA activities, while glutathione was not effective. However, significantly higher HeO-HeA activities were observed in P. gingivalis when recombinant GGT was added along with glutathione. The increase in HeO-HeA activities was again suppressed by the addition of GNA. The results suggest an important role for GGT in an enzyme pathway to produce H2S with HeO-HeA activities from glutathione and in potential virulence expression in oral bacteria.
DISCUSSION
In the present study, we cloned the gene for GGT from T. denticola. Expression of this gene in E. coli led to the production of a recombinant protein, which was subsequently purified to homogeneity on SDS-PAGE. The identity of the recombinant protein as T. denticola GGT was confirmed by mass spectrometry. The recombinant protein exhibited enzymatic activity typical of GGT, degrading GNA and glutathione. Importantly, further experiments showed that GNA as a substrate and competitive inhibitor of GGT was able to suppress glutathione metabolism in T. denticola. Moreover, recombinant GGT greatly facilitated the metabolism of glutathione in P. gingivalis, a bacterium otherwise having very limited glutathione-metabolizing capacity. Together, these results suggest a critical role for GGT in glutathione metabolism in vivo.
GGT of T. denticola was first purified and characterized in 1997 by P. L. Makinen and K. K. Makinen (24). In a series of carefully performed experiments, they identified a 26-kDa protein which exhibited significant hydrolytic activity toward GNA and glutathione. The same study also determined the N-terminal sequence of 21 amino acids. These results were essential for the cloning strategy used to isolate the gene. In this study, we utilized the reported sequence information to clone and express the GGT gene. The expressed recombinant protein showed several characteristic features of GGT isolated from T. denticola. First, the recombinant protein demonstrated significant hydrolytic activity toward GNA and glutathione. Second, the enzymatic activity of the protein, similar to that of homologous GGT, was dependent on the presence of free thiols. Finally, without being boiled prior to SDS-PAGE, the recombinant protein existed as high-molecular-weight aggregates, as did the GGT protein isolated from T. denticola (24). These observations suggest that the recombinant protein might be a useful model for the characterization of GGT and the investigation of its metabolic role.
Several microorganisms, including E. coli, exhibit GGT activity (1, 15, 24, 39). At ∼27 kDa, T. denticola GGT is significantly smaller than the 39-kDa E. coli GGT. In addition, the enzymatic activity of E. coli GGT is much weaker than that of T. denticola GGT. The low enzymatic activity of E. coli GGT might be caused by a single amino acid change in its sequence from l-cysteine to serine (39). As suggested by Makinen and Makinen, T. denticola GGT closely resembles mammalian GGT, with cysteine residues at locations that are critical in maintaining high enzymatic activity (23, 24, 36, 38) (Fig. 1).
The results from this study have demonstrated an important role for GGT in glutathione metabolism in vivo. Previous studies have shown that GGT isolated from T. denticola is capable of degrading glutathione (24). The present study confirmed those observations, clearly showing the breakdown of glutathione into glutamic acid or glutamine and Cys-Gly in the presence of recombinant GGT. Since no further degradation was detected for Cys-Gly during incubation, the results also suggest that GGT catalyzes only the first step of glutathione metabolism. The role of GGT in glutathione metabolism in vivo is further supported by our inhibition experiments using GNA. GNA was initially utilized as an exogenous chromogenic substrate in a GGT enzymatic assay. We reasoned that GNA may compete with the endogenous substrate glutathione for GGT and inhibit GGT-mediated glutathione hydrolysis. This assumption was validated by subsequent experiments, in which GNA indeed suppressed H2S and ammonia production from glutathione in T. denticola. This observation is very important, because it supports an essential role for GGT in glutathione metabolism in T. denticola. Of note, GNA did not significantly affect the degradation of l-cysteine or Cys-Gly in T. denticola (data not shown). The significance of GGT in glutathione hydrolysis is also suggested by the evidence from our experiments on P. gingivalis. P. gingivalis was able to metabolize l-cysteine and Cys-Gly, but its capacity for metabolizing glutathione was very limited. However, glutathione utilization in the bacterium was drastically increased when the recombinant GGT was added to the incubation medium. Together, these results suggest a pivotal role for GGT in the first step of glutathione degradation.
The characteristics of GGT and cystalysin suggest that these enzymes exhibit hydrolytic activities toward different specific substrates. As detailed in previous studies, cystalysin catalyzes the degradation of l-cysteine, releasing H2S, ammonia, and pyruvate; the enzyme has no activity toward either glutathione or Cys-Gly (9, 11, 12). On the other hand, the results from the present study have demonstrated an enzymatic activity of GGT for glutathione hydrolysis, while it is not effective in catalyzing Cys-Gly or l-cysteine degradation. Apparently, the substrate specificities of these two enzymes are defined by their unique molecular structures and biochemical characteristics. In our experiments, it was shown that the enzymes displayed quite different sensitivities to treatment with TLCK, heating, and proteases. Nevertheless, they cooperate in vivo to degrade glutathione in a stepwise manner.
The observation that recombinant GGT was able to facilitate glutathione utilization in P. gingivalis may have significant implications for bacterial interaction at the metabolic level. In infected periodontal pockets, a number of oral bacteria have been identified (16, 18, 32, 33, 35, 40). These include not only T. denticola, but also P. gingivalis, Peptostreptococcus, Fusobacterium, Prevotella, Eubacterium, Selenomonas, Bacteroides, and other genera and species. Although little has been learned about their interactions, these microorganisms may work together, expressing virulence to damage host cells and culminating in periodontal-tissue pathology. With the observation of GGT facilitation of glutathione utilization in P. gingivalis, it would not be a far-fetched scenario that T. denticola may assist P. gingivalis in vivo with glutathione metabolism. GGT-catalyzed glutathione degradation into Cys-Gly by T. denticola is thought to greatly enhance the use of this important thiol source by oral bacteria, including P. gingivalis, which otherwise has a very limited capacity for glutathione metabolism. The interaction between oral bacteria at the metabolic level may play a significant role in the progression of periodontal disease.
Acknowledgments
We are grateful to Susan Weintraub, Chris Carroll, and Steven L. Mouton for scientific input and technical assistance in mass spectrometric and HPLC analyses. We also thank Stanley C. Holt and David Kolodrubetz for scientific discussions on this project.
This work was supported by grant DE-13819 from NIDCR to Lianrui Chu.
Editor: J. D. Clements
REFERENCES
- 1.Allison, R. D. 1985. γ-Glutamyl transpeptidase: kinetics and mechanism. Methods Enzymol. 113:419-437. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M. (ed.). 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Bauer, J. D., P. G. Ackermann, and P. Toro. 1974. Clinical laboratory methods, p. 399-401. The C. V. Company, Saint Louis, Mo.
- 4.Beauchamp, R. O., Jr., J. S. Bus, J. A. Popp, C. J. Boreiko, and D. A. Andjelkovich. 1984. A critical review of the literature on hydrogen sulfide toxicity. Crit. Rev. Toxicol. 13:25-97. [DOI] [PubMed] [Google Scholar]
- 5.Beilzer, M., and B. H. Lauterburg. 1991. Glutathione metabolism in activated human neutrophils: stimulation of glutathione synthesis and consumption of glutathione by reactive oxygen species. Eur. J. Clin. Investig. 21:316-322. [DOI] [PubMed] [Google Scholar]
- 6.Chu, L., and S. C. Holt. 1994. Purification and characterization of a 45 kDa hemolysin from Treponema denticola ATCC 35404. Microb. Pathog. 16:197-212. [DOI] [PubMed] [Google Scholar]
- 7.Chu, L., W. Kennell, and S. C. Holt. 1994. Characterization of hemolysis and hemoxidation activities by Treponema denticola. Microb. Pathog. 16:183-195. [DOI] [PubMed] [Google Scholar]
- 8.Chu, L., A. Burgum, D. Kolodrubetz, and S. C. Holt. 1995. The 46-kilodalton-hemolysin gene from Treponema denticola encodes a novel hemolysin homologous to aminotransferases. Infect. Immun. 63:4448-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu, L., Z. Dong, X. Xu, D. L. Cochran, and J. L. Ebersole. 2002. Role of glutathione metabolism of Treponema denticola in bacterial growth and virulence expression. Infect. Immun. 70:1113-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu, L., J. L. Ebersole, and S. C. Holt. 1999. Hemoxidation and binding of the 46-kDa cystalysin of Treponema denticola leads to a cysteine-dependent hemolysis of human erythrocytes. Oral Microbiol. Immunol. 14:293-303. [DOI] [PubMed] [Google Scholar]
- 11.Chu, L., J. L. Ebersole, G. P. Kurzben, and S. C. Holt. 1997. Cystalysin, a 46-kilodalton cysteine desulfhydrase from Treponema denticola, with hemolytic and hemoxidative activities. Infect. Immun. 65:3231-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu, L., J. L. Ebersole, G. P. Kurzben, and S. C. Holt. 1999. Cystalysin, a 46-kDa l-cysteine desulfhydrase from Treponema denticola: biochemical and biophysical characterization. Clin. Infect. Dis. 28:442-450. [DOI] [PubMed] [Google Scholar]
- 13.Deng, Q. D., Y. Han, X. Xia, and H. Kuramitsu. 2001. Effects of the oral spirochete Treponema denticola on interleukin-8 expression from epithelial cells. Oral Microbiol. Immunol. 16:185-187. [DOI] [PubMed] [Google Scholar]
- 14.Fenno, J. C., and B. C. McBride. 1998. Virulence factors of oral treponemes. Anaerobe 4:1-17. [DOI] [PubMed] [Google Scholar]
- 15.Hasegawa, M., and I. Matsubara. 1978. γ-Glutamylpeptide formative activity of Corynebacterium glutamicum by the reverse reaction of the γ-glutamylpeptide hydrolytic enzyme. Agric. Biol. Chem. 42:371-381. [Google Scholar]
- 16.Holt, S. C., and T. E. Bramanti. 1991. Factors in virulence expression and their role in periodontal disease pathogenesis. Crit. Rev. Oral Biol. Med. 2:177-281. [DOI] [PubMed] [Google Scholar]
- 17.Horowitz, A., and L. E. Folke. 1973. Hydrogen sulfide production in the periodontal environment. J. Periodontol. 44:390-395. [DOI] [PubMed] [Google Scholar]
- 18.Kasuga Y., K. Ishihara, and K. Okuda. 2000. Significance of detection of Porphyromonas gingivalis, Bacteroides forsythus and Treponema denticola in periodontal pockets. Bull. Tokyo Dent. Coll. 41:109-117. [DOI] [PubMed] [Google Scholar]
- 19.Kennell, W., and S. C. Holt. 1990. Comparative studies of the outer membranes of Bacteroides gingivalis, strains ATCC 33277, W50, W83, 381. Oral Microbiol. Immunol. 5:121-130. [DOI] [PubMed] [Google Scholar]
- 20.Krupka, H. I., R. Huber, S. C. Holt, and T. Clausen. 2000. Crystal structure of cystalysin from Treponema denticola: a pyridoxal-dependent protein acting as haemolytic enzyme. EMBO J. 19:3168-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurzben, G. P., L. Chu, J. L. Ebersole, and S. C. Holt. 1999. Sulfhemoglobin formation in human erythrocytes by cystalysin, an l-cysteine desulfhydrase from Treponema denticola. Oral Microbiol. Immunol. 14:153-164. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Laperche, Y., F. Bulle, T. Aissani, M. N. Chobert, M. Aggerbeck, J. Hanoune, and G. Guellaen. 1986. Molecular cloning of rat kidney γ-glutamyl transpeptidase cDNA. Proc. Natl. Acad. Sci. USA 83:937-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makinen, P.-L., and K. K. Makinen. 1997. γ-Glutamyltransferase from the outer cell envelope of Treponema denticola ATCC 35405. Infect. Immun. 65:685-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mills, B. J., J. P. Richie, and C. A. Lang. 1990. Sample processing alters glutathione and cysteine values in blood. Anal. Biochem. 184:263-267. [DOI] [PubMed] [Google Scholar]
- 26.Moore, W. E. C. 1987. Microbiology of periodontal disease. J. Periodont. Dis. 22:335-341. [DOI] [PubMed] [Google Scholar]
- 27.Morita, M., and H.-L. Wang. 2001. Relationship of sulcular sulfide level to severity of periodontal disease and BANA Test. J. Periodontol. 72:74-78. [DOI] [PubMed] [Google Scholar]
- 28.Page, R. C., and H. E. Schroeder. 1982. Periodontitis in man and other animals: a comparative review. Karger, Basel, Switzerland.
- 29.Persson, S. 1992. Hydrogen sulfide and methyl mercaptan in periodontal pockets. Oral Microbiol. Immunol. 7:378-379. [DOI] [PubMed] [Google Scholar]
- 30.Persson, S., M.-B. Edlund, R. Claesson, and J. Carlsson. 1990. The formation of hydrogen sulfide and methyl mercaptan by oral bacteria. Oral Microbiol. Immunol. 5:195-201. [DOI] [PubMed] [Google Scholar]
- 31.Reiffenstein, R. J., W. C. Hulbert, and S. H. Roth. 1992. Toxicology of hydrogen sulfide. Annu. Rev. Pharmacol. Toxicol. 32:109-134. [DOI] [PubMed] [Google Scholar]
- 32.Riviere, G. R., K. S. Smith, N. Carranza, Jr., E. Tzagaronlaki, S. L. Kay, and M. Dock. 1995. Subgingival distribution of Treponema denticola, Treponema socranskii, and pathogen-related oral spirochetes: prevalence and relationship to periodontal status of sampled sites. J. Periodontol. 66:829-837. [DOI] [PubMed] [Google Scholar]
- 33.Rocas, I. N., J. F. Siqueira, Jr., K. R. Santos, and A. M. Coelho. 2001. “Red complex” (Bacteroides forsythus, Porphyromonas gingivalis, and Treponema denticola) in endodontic infections: a molecular approach. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 91:468-471. [DOI] [PubMed] [Google Scholar]
- 34.Rosen, G., R. Naor, E. Rahamin, R. Yishai, and M. N. Sela. 1995. Protease of Treponema denticola outer sheath and extracellular vesicles. Infect. Immun. 63:3973-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simonson, L., K. T. McMahon, D. W. Childers, and H. E. Morton. 1992. Bacterial synergy of Treponema denticola and Porphyromonas gingivalis in a multinational population. Oral Microbiol. Immunol. 7:111-112. [DOI] [PubMed] [Google Scholar]
- 36.Smith, T. K., and A. Meister. 1994. Active deglycosylated mammalian γ-glutamyl transpeptidase. FASEB J. 8:661-664. [DOI] [PubMed] [Google Scholar]
- 37.Solis, M. C., K. N. Rustogi, and A. Gaffar. 1980. Hydrogen sulfide production from gingival crevicular fluid. J. Periodontol. 51:603-606. [DOI] [PubMed] [Google Scholar]
- 38.Song, L. J., M. Q. Ye, M. Troyanovskaya, E. Wilk, S. Wilk, and D. P. Healy. 1994. Rat kidney glutamyl aminopeptidase (aminopeptidase A): molecular identity and cellular localization. Am. J. Physiol. 36:F546-F557. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki, H., H. Kumagai, T. Echigo, and T. Tochikura. 1989. DNA sequence of the Escherichia coli K-12 γ-glutamyltranspeptidase gene, ggt. J. Bacteriol. 171:5169-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeuchi, Y., M. Umeda, M. Sakamoto, Y. Benno, Y. Huang, and I. Ishikawa. 2001. Treponema socranskii, Treponema denticola, and Porphyromonas gingivalis are associated with severity of periodontal tissue destruction. J. Periodontol. 72:1354-1363. [DOI] [PubMed] [Google Scholar]
- 41.U.S. National Research Council. 1979. Hydrogen sulfide, p. 1-183. University Park Press, Baltimore, Md.
- 42.Weinberg, A., and S. C. Holt. 1990. Interaction of Treponema denticola TD-4, GM-1, and MS25 with human gingival fibroblasts. Infect. Immun. 58:1720-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng, L., R. H. White, V. L. Cash, R. F. Jack, and D. R. Dean. 1993. Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc. Natl. Acad. Sci. USA. 90:2754-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]