Abstract
Legionella pneumophila is the agent of Legionnaires' disease. It invades and replicates within eukaryotic cells, including aquatic protozoans, mammalian macrophages, and epithelial cells. The molecular mechanisms of the Legionella interaction with target cells are not fully defined. In an attempt to discover novel virulence factors of L. pneumophila, we searched for bacterial enzymes with transferase activity. Upon screening ultrasonic extracts of virulent legionellae, we identified a uridine diphospho (UDP)-glucosyltransferase activity, which was capable of modifying a 45-kDa substrate in host cells. An approximately 60-kDa UDP-glucosyltransferase was purified from L. pneumophila and subjected to microsequencing. An N-terminal amino acid sequence, as well as the sequence of an internal peptide, allowed us to identify the gene for the enzyme within the unfinished L. pneumophila genome database. The intact gene was cloned and expressed in Escherichia coli, and the recombinant protein was purified and confirmed to possess an enzymatic activity similar to that of the native UDP-glucosyltransferase. We designated this gene ugt (UDP-glucosyltransferase). The Legionella enzyme did not exhibit significant homology with any known protein, suggesting that it is novel in structure and, perhaps, in function. Based on PCR data, an enzyme assay, and an immunoblot analysis, the glucosyltransferase appeared to be conserved in L. pneumophila strains but was absent from the other Legionella species. This study represents the first identification of a UDP-glucosyltransferase in an intracellular parasite, and therefore modification of a eukaryotic target(s) by this enzyme may influence host cell function and promote L. pneumophila proliferation.
Legionella pneumophila is the principal agent of Legionnaires' disease, a pneumonic illness of humans. This bacterium is a facultative intracellular parasite that invades and proliferates in both professional and nonprofessional phagocytes (54). The host cells for L. pneumophila include freshwater protozoans and human alveolar macrophages and epithelial cells. Bacterial factors that have been identified as crucial for intracellular infection include, among others, the major outer membrane protein (25), Mip (13), Hsp60 (17, 18), PilD and type II protein secretion (4, 43), type IV pili (29, 51), flagella (41), the Dot/Icm type IV protein secretion system, and the products of the enh, lvh, mil, and pmi loci (2, 11, 14, 16, 35, 49). In addition to these virulence factors, a variety of enzymes have been identified in Legionella cultures, including a low-molecular-mass cytotoxin (30), a metalloprotease cytolysin (8, 9), phospholipases (3, 4, 6), lipases (3), phosphatases (5, 44) and phosphokinases (45). For many of these activities, the role in virulence is unclear and/or the eukaryotic substrate(s) is unknown. Thus, further studies aimed at identifying and characterizing novel L. pneumophila enzymes should be important for understanding both Legionnaires' disease and bacterial interactions with eukaryotic cells.
It is well known that certain types of transferases are major virulence factors in some bacteria and represent the enzymatic components of toxins. For example, whereas diphtheria toxin of Corynebacterium diphtheriae, cholera toxin of Vibrio cholerae, and exotoxin A of Pseudomonas aeruginosa are ADP-ribosyltransferases, toxins A and B of Clostridium difficile and the lethal toxin of Clostridium sordellii are uridine diphospho (UDP)-glucosyltransferases (48). Interestingly, in the gram-negative pathogens, these toxic transferases can be dependent upon type II protein secretion, a system that, as noted above, is also critical for L. pneumophila pathogenesis (4, 43, 56). Whereas ADP-ribosyltransferases have been found in both extra- and intracellular pathogens, UDP-glucosyltransferase toxins have been studied only in extracellular pathogens (1, 37, 39). For these various reasons, our research has been aimed at identification of similar types of enzymes in Legionella cultures. In the present study, we characterized a novel product of L. pneumophila that possesses UDP-glucosyltransferase activity and modifies a distinct component of eukaryotic host cells.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The following Legionella strains were used in this study: L. pneumophila Philadelphia I (= ATCC 33217) (serogroup 1), 130b (= ATCC BAA-74) (serogroup 1), ATCC 33823 (serogroup 7), and ATCC 35096 (serogroup 8), L. longbeachae ATCC 33462, L. gormanii ATCC 33297, and L. steigerwaltii ATCC 35302. Bacteria were grown on buffered charcoal yeast extract (BCYE) agar for 2 days at 37°C (15). Escherichia coli strain BL21(DE3) and the expression vector pET28b (Novagen, Madison, Wis.) were used for cloning and the production of recombinant protein. E. coli was maintained on Luria-Bertani agar supplemented with 50 μg of kanamycin per ml when necessary (46).
Protein purification and microsequencing.
The L. pneumophila UDP-glucosyltransferase was purified from strain Philadelphia I. Bacterial cells from 100 BCYE agar plates were suspended in 20 ml of 20 mM Tris-HCl (pH 7.4)-150 mM NaCl (TBS), washed once by centrifugation (Sorvall, Newtown, Conn.) at 12,000 × g for 15 min (all centrifugation steps were performed at 4°C), and resuspended in 100 ml of TBS. The cell suspension was subjected to ultrasonication with five 1-min pulses with 2-min intervals between pulses at the high power setting (Techpan, Warsaw, Poland) on ice. Unbroken cells and cell wall fragments were pelleted by centrifugation at 18,000 × g for 60 min and discarded. The supernatant was treated with protamine sulfate (Sigma, St. Louis, Mo.) at a concentration of 2 mg/ml to sediment the majority of the bacterial DNA. After the sonicate was dialyzed overnight against 20 mM Tris-HCl (pH 7.4) (TB), it was loaded onto a Mono Q 10/10 anion-exchange column equilibrated with TB. The column was washed sequentially with TB and 0.05 M NaCl in TB, and fractions were tested for UDP-glucosyltransferase activity (see below). The enzymatically active preparation was then eluted with 20 ml of 0.1 M NaCl in TB. The eluate was dialyzed against 20 mM morpholineethanesulfonic acid (MES)-Na buffer (pH 6.25) (MB) and loaded onto a Mono S 5/5 cation-exchange column equilibrated with MB. The Mono S 5/5 column was then washed sequentially with MB, 0.05 M NaCl in MB, and 0.1 M NaCl in MB. The enzymatically active material was eluted with 3 ml of 0.15 M NaCl in MB. After the active samples were dialyzed against TB, they were loaded onto a Mono Q 5/5 anion-exchange column equilibrated with TB. Elution was performed with a linear 0.0 to 0.1 M NaCl gradient in TB. Active fractions, which eluted between 60 and 80 mM NaCl, were concentrated by ultrafiltration with a YM10 filter (Amicon, Danvers, Mass.) and subjected to gel chromatography on a TBS-equilibrated Superose 6 10/30 column, which was able to separate proteins having molecular masses ranging from 5 to 5,000 kDa. Fractions (0.4 ml) were collected, and the fractions with enzymatic activity, which eluted in a peak retention volume of 16.5 ml, were pooled and stored at −20°C as purified UDP-glucosyltransferase. All matrices used for liquid chromatography were obtained from Amersham Biosciences, Vienna, Austria. All chromatographic steps were done with the Amersham fast protein liquid chromatography system. Microsequencing of purified protein was performed at the Laboratory of Protein Microsequencing at the Institut Pasteur by standard procedures. In order to assay for UDP-glucosyltransferase activity in other strains of L. pneumophila or in other species of Legionella, crude ultrasonic extracts were prepared as described above for L. pneumophila Philadelphia I. Finally, in order to assay for a secreted UDP-glucosyltransferase activity, L. pneumophila Philadelphia I was grown in Proteose Peptone No. 3 (Difco, Detroit, Mich.) broth for 18 h at 37°C with constant shaking (200 rpm) (59). After centrifugation of the culture at 12, 000 × g for 15 min, the resultant supernatant was filter sterilized and concentrated 10-fold by using an Amicon YM10 filter.
PCR amplification and cloning of the UDP-glucosyltransferase gene.
Based on data from the L. pneumophila Philadelphia I genome project at Columbia University (http://genome3.cpmc.columbia.edu/∼legion/ngnp1033033), two DNA primers were designed for amplification of the putative UDP-glucosyltransferase gene (IDT, Coralville, Iowa). The sequence of the sense primer was 5′-ATGGGAGACGAGTATGAATTCAGCAAGAAG-3′ (an engineered EcoRI site is underlined), and the sequence of the antisense primer was 5′-TTCAAAATTTGTGTGTCGACATTAAGCTAC-3′ (an engineered SalI site is underlined). The PCR was performed under standard conditions (i.e., denaturation at 94°C, annealing at 55°C, and elongation at 72°C) with a Sprint thermocycler (Hybaid, Ashford, England). Legionella genomic DNA for the PCR was extracted by the phenolic method (46). For cloning of the UDP-glucosyltransferase gene, the PCR product that was amplified from L. pneumophila Philadelphia I DNA was digested with EcoRI and SalI and ligated into similarly digested pET28b. E. coli BL21(DE3) was transformed with the resulting plasmid by electroporation by using an Electroporator 2510 (Eppendorf, Hamburg, Germany).
For purification of a recombinant enzyme, the E. coli clone was grown in Luria-Bertani broth supplemented with kanamycin for 3 h on a shaker at 37°C. Hyperexpression of the cloned protein was then induced by supplementing the culture with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma) and incubating it for an additional 2 h. The bacterial cells from 100 ml of culture were harvested by centrifugation at 8,000 × g for 15 min, resuspended in 6 ml of TBS, and then lysed by ultrasonic treatment (three 20-s pulses with 1-min intervals between pulses at the medium power setting). Following clarification by centrifugation, the bacterial extract was subjected to chromatography on nickel-equilibrated chelating Sepharose Fast Flow (Amersham Biosciences) used according to the manufacturer's instructions. Purified recombinant UDP-glucosyltransferase was eluted with 0.5 M imidazole and desalted by using an FD10 column (Amersham Biosciences). Pure protein was stored at −20°C.
UDP-glucosyltransferase assay.
Protein samples derived from Legionella strains were tested for the ability to UDP-glucosylate substrates contained in HeLa, THP-1, CHO, and Vero cells, which were obtained from the Pasteur Institut and the Gamaleya Research Institute. The various cell lines were grown in 250-ml culture flasks (Becton Dickinson, Oxnard, Calif.) containing RPMI 1640 medium supplemented with 10% fetal calf serum (Gibco Laboratories, Grand Island, N.Y.). After 3 to 4 days of growth at 37°C in the presence of 5% CO2, cells were scrapped from the plastic surfaces, washed, and resuspended in 2 ml of ice-cold TBS per culture flask. Cell extracts were prepared by ultrasonication, which involved a 5- to 7-s pulse at medium power. UDP-glucosyltransferase reactions were carried out by using mixtures consisting of 5 μl of reaction buffer (150 mM triethanolamine [pH 7.5], 6 mM MgCl2, 0.9 mM GDP, 3 mM dithiothreitol), 2 μl of [14C]UDP-glucose (code CFB102-10μCi; Amersham Biosciences), 5 μl of bacterial preparation at a protein concentration of 0.05 to 1.5 mg/ml, and 5 μl of eukaryotic cell ultrasonic extract at a protein concentration of 1 mg/ml. Each mixture was incubated at 37°C for 1 h, after which the reaction was stopped by adding Laemmli sample buffer and heating the mixture at 100°C for 5 min. The samples were then subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) by using 7% polyacrylamide stacking and 12% polyacrylamide resolving gels (28). After this, each gel was washed in 7% acetic acid, dried, and exposed to Hyperfilm (Amersham Biosciences) for 3 days at −70°C.
Immunoblot analysis and protein determination.
Following SDS-PAGE (28), an immunoblot analysis was performed by using standard procedures (57), a 1/5,000 dilution of primary rabbit serum, and a 1/4,000 dilution of anti-rabbit horseradish conjugate (Bio-Rad, Moscow, Russia). For production of primary antiserum, 2- to 3-kg rabbits, obtained from the animal facility at the Gamaleya Research Institute, were immunized three times with 100 μg of native, purified UDP-glucosyltransferase in TBS at 2-week intervals. Ten days after the last injection, the rabbits were exsanguinated by sublethal cardiac puncture, and the resultant serum was stored. Protein concentrations were determined by a Coomassie brilliant blue R250 assay (Serva, Heidelberg, Germany) by using bovine serum albumin as a standard (10).
RESULTS
Identification of a novel UDP-glucosyltransferase activity.
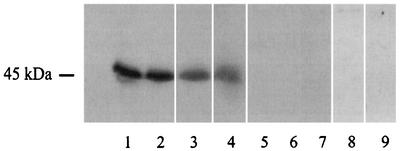
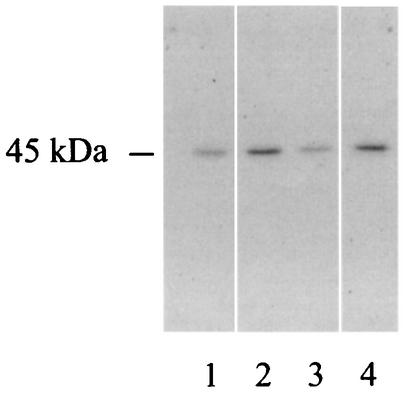
As part of our work aimed at identifying virulence factors of the legionellae, we sought a UDP-glucosyltransferase in cultures of L. pneumophila. To do this, culture supernatant from the L. pneumophila Philadelphia I strain and ultrasonic lysates of cells from several strains were tested in reaction mixtures consisting of HeLa cells extracts and [14C]UDP-glucose. The HeLa cell line is one of a number of human epithelial cell lines that are susceptible to L. pneumophila invasion and intracellular replication (18). When sonicates were used, but not when supernatants were used, we observed incorporation of radioactive label into a ca. 45-kDa protein band (Fig. 1 and data not shown). The activity was observed for all four L. pneumophila strains tested, including representatives of serogroups 1, 7, and 8. There was no protein labeling if the bacterial lysate or HeLa cell extract was omitted from the mixture (Fig. 1), indicating that the reaction resulted from the interaction of prokaryotic and eukaryotic factors. Interestingly, no reaction was observed when sonicates from other Legionella species were used (Fig. 1). To determine whether the labeling reaction required factors peculiar to HeLa or epithelial cells, the experiment was repeated with extracts from human monocytic THP-1 cells as well as from Vero and CHO cells. Like HeLa cells, THP-1, Vero, and CHO cells have been shown to support L. pneumophila intracellular growth (27, 33, 38). In all cases, similar UDP-glucosylation reactions were observed (Fig. 2). Based on these observations, as well as the fact that other bacterial enzymes (i.e., clostridial toxins) can transfer UDP-glucose to mammalian cell substrates (23, 24, 40), we hypothesized that L. pneumophila possesses a UDP-glucosyltransferase that is active against a conserved 45-kDa component of eukaryotic cells.
FIG. 1.
Identification of a new UDP-glucosyltransferase activity. Ultrasonic extracts from different Legionella species were incubated in the presence of HeLa cell extract and [14C]UDP-glucose. After this, the samples were subjected to SDS-PAGE, dried, and exposed to X-ray film. Lane 1, L. pneumophila Philadelphia I; lane 2, L. pneumophila 130b; lane 3, L. pneumophila ATCC 33823; lane 4, L. pneumophila ATCC 35096; lane 5, L. longbeachae ATCC 33462; lane 6, L. gormanii ATCC 33297; lane 7, L. steigerwaltii ATCC 35302; lane 8, L. pneumophila Philadelphia I without eukaryotic substrate; lane 9, HeLa cell extract without bacterial extract. The data represent data obtained in several experiments.
FIG. 2.
Identification of a substrate for L. pneumophila UDP-glucosyltransferase in different eukaryotic cell extracts. Ultrasonic extracts from the L. pneumophila Philadelphia I strain were incubated in the presence of HeLa (lane 1), Vero (lane 2), CHO (lane 3), and THP-1 (lane 4) cell preparations and [14C]UDP-glucose. After this, the samples were subjected to SDS-PAGE, dried, and exposed to X-ray film.
Purification and partial sequence of an L. pneumophila UDP-glucosyltransferase.
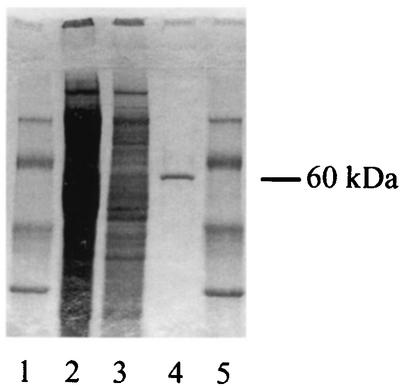
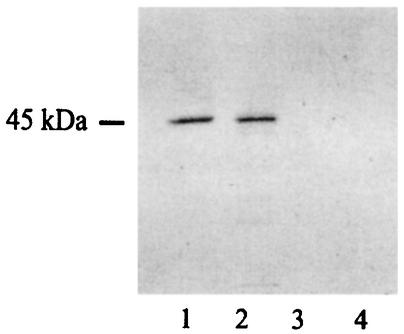
To investigate the existence of a Legionella UDP-glucosyltransferase, we tested various fractions from ultrasonic extracts of L. pneumophila Philadelphia I for the ability to generate, when coupled with HeLa cell extracts, the radiolabeled, 45-kDa protein band. The biochemical procedure developed consisted of an initial combination of anion- and cation-exchange chromatography and a final gel chromatography step (see Materials and Methods). As shown by SDS-PAGE, the preparation from the last step appeared to consist of a single major protein with a molecular mass of approximately 60 kDa (Fig. 3). The purified protein yielded the same result as the sonicate; i.e., it facilitated radiolabeling of a 45-kDa protein band (Fig. 4). The yield obtained with this preparative scheme was 0.7 mg of pure product per 150 mg of starting crude protein. Since the molecular mass of the reactive material purified from the bacteria was 60 kDa instead of 45 kDa, we concluded that L. pneumophila encodes the UDP-glucosyltransferase and not the substrate for a eukaryotic enzyme. As a first step toward understanding the molecular nature of the Legionella UDP-glucosyltransferase, we subjected the purified protein to a microsequencing analysis. The N-terminal peptide sequence was found to be DQQLSXLRMRFFSAL, while an internal peptide sequence was (F/Q)NISLIDIDSVR. BLASTP searches (cost to open gap, 11; expect value, 10) of the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) failed to detect any relationship between these peptide sequences and known proteins, indicating that the Legionella UDP-glucosyltransferase is a previously unrecognized protein.
FIG. 3.
Purification of a UDP-glucosyltransferase from L. pneumophila. Samples were subjected to SDS-PAGE and stained with Coomassie brilliant blue R250. Lanes 1 and 5, molecular mass markers; (from top to bottom, 94, 67, 43, and 30 kDa; Amersham Biosciences); lanes 2 and 3, ultrasonic lysate of L. pneumophila Philadelphia I containing 30 and 15 μg of protein, respectively; lane 4, purified, 60-kDa UDP-glucosyltransferase (ca. 0.5 μg).
FIG. 4.
Enzymatic activity of UDP-glucosyltransferase purified from L. pneumophila and recombinant E. coli. Approximately 0.5 μg of purified native protein (lanes 1 and 3) or recombinant protein (lanes 2 and 4) was incubated in the presence of [14C]UDP-glucose with (lanes 1 and 2) or without (lanes 3 and 4) HeLa cell extract. After this, the samples were subjected to SDS-PAGE, dried, and exposed to X-ray film.
Cloning and PCR detection of the L. pneumophila UDP-glucosyltransferase gene.
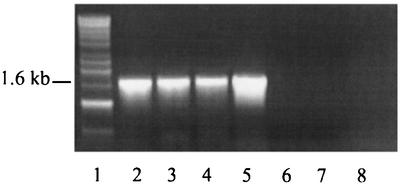
In an attempt to identify the gene encoding the Legionella UDP-glucosyltransferase, we used our protein sequence data to search the incomplete genomic database of L. pneumophila Philadelphia I (http://genome3.cpmc.columbia.edu/∼legion/ngnp1033033). An open reading frame whose translated sequences matched the sequences of isolated peptides was found on contig CTG.BC.5E37D10.01.44.07280. The 1.57-kb gene, which had not yet been assigned a possible function, was predicted to encode a protein product with a molecular mass of 57.8 kDa, a size that was compatible with the size of the purified UDP-glucosyltransferase (i.e., ca. 60 kDa). There was an apparent discrepancy between the N-terminal sequences of the purified protein (DQQLSXLRMRFFSAL) and the translated gene product (MKARRDQQLSKLRMRFFSAL). The absence of the amino acid string MKARR in the sequenced protein might have resulted from posttranslational modification of the enzyme or from proteolytic degradation during storage or purification. BLASTX searches (cost to open gap, 11; expect value, 10) with the entire open reading frame sequence only retrieved hits possessing scores of less than 36 bits and E values of more than 1.1. These results demonstrated that there was a lack of overall homology between the Legionella protein and known proteins in the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). In order to determine whether the gene which we examined in fact encodes the L. pneumophila UDP-glucosyltransferase, we sought to clone it and test its protein product for activity in our HeLa cell extract assay. To do this, we utilized the pET28 expression system, which allows cloning of recombinant proteins in frame with an upstream polyhistidine tag and thereby permits protein purification by nickel affinity chromatography. Using PCR and primers based on sequences in the Legionella database, we amplified a 1.6-kb fragment from Philadelphia I genomic DNA (Fig. 5, lane 2). The PCR fragment was expected to contain the entire UDP-glucosyltransferase open reading frame plus 16 bp upstream and 29 bp downstream. By incorporating convenient restriction sites into the primers, the DNA fragment was easily cloned into pET28b, resulting in plasmid p28b-13. E. coli BL21(p28b-13) was confirmed by SDS-PAGE to express a considerable amount of the cloned gene product (Fig. 6). When tested in the UDP-glucosylation assay, the recombinant protein modified a substrate in a HeLa cell extract with the same molecular mass (45 kDa) as the substrate modified by the native enzyme (Fig. 4). These results confirm that the L. pneumophila gene encodes UDP-glucosyltransferase activity. We designated this gene ugt (UDP-glucosyltransferase). The ugt gene mapped between a putative heat shock protein gene and a predicted deaminase gene in the unfinished L. pneumophila genome project sequence. The previously described lspFGHIJK genes, which are associated with type II protein secretion (19, 43), were approximately 4 kb downstream of ugt. PCR analysis with the primers described above revealed that all L. pneumophila strains tested possessed a 1.6-kb ugt-containing fragment (Fig. 5). In contrast, representatives of L. longbeachae, L. gormanii, and L. steigerwaltii were negative in the PCR (Fig. 5).
FIG. 5.
PCR detection of the L. pneumophila UDP-glucosyltransferase gene. Based on nucleotide sequence data for L. pneumophila Philadelphia I, specific primers were used to PCR amplify DNA fragments from purified genomic preparations. Following amplification, the DNA mixtures were subjected to agarose gel electrophoresis and stained with ethidium bromide. Lane 1, molecular mass markers (from top to bottom, 10, 8, 6, 5, 4, 3, 2.5, 2, 1.5, 1 [threefold intensity], and 0.5 kb; Amersham Biosciences); lane 2, L. pneumophila Philadelphia I; lane 3, L. pneumophila 130b; lane 4, L. pneumophila ATCC 33823; lane 5, L. pneumophila ATCC 35096; lane 6, L. longbeachae ATCC 33462, lane 7, L. gormanii ATCC 33297; lane 8, L. steigerwaltii ATCC 35302.
FIG. 6.
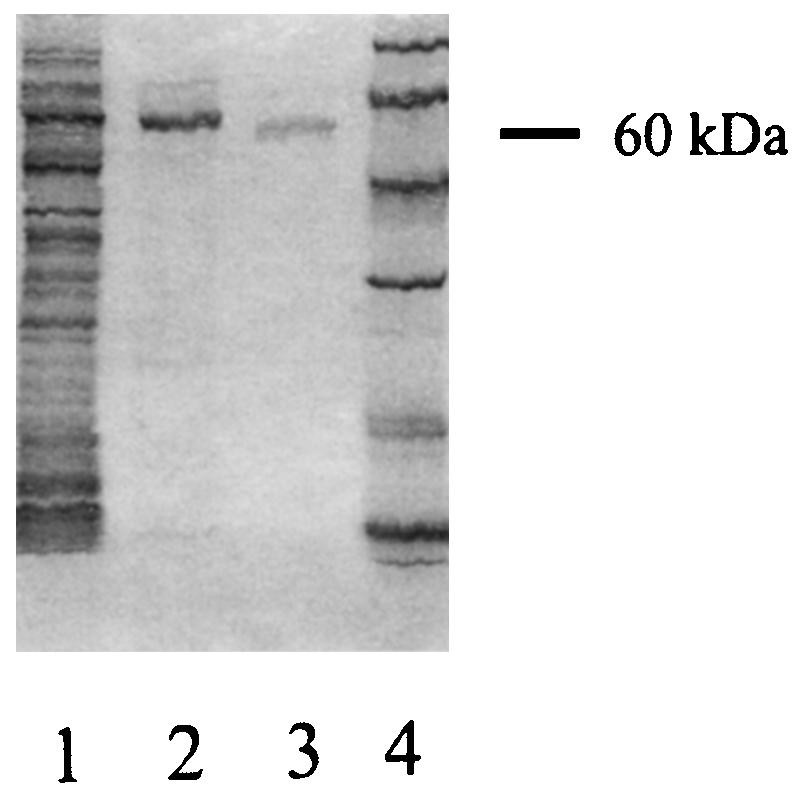
SDS-PAGE analysis of recombinant L. pneumophila UDP-glucosyltransferase. Lane 1, crude lysate of E. coli BL21(p28b-13); lane 2, purified recombinant UDP-glucosyltransferase; lane 3, UDP-glucosyltransferase purified from L. pneumophila; lane 4, molecular mass markers (94, 67, 43, 30, 20, and 14 kDa). Because of its N-terminal polyhistidine tag, the recombinant enzyme migrated more slowly than the native enzyme.
Immunoblot assay of the L. pneumophila UDP-glucosyltransferase.
In the next stage of our investigation, polyclonal anti-UDP-glucosyltransferase antibodies were generated, and Western blotting was used to test a panel of bacterial extracts for the presence of the UDP-glucosyltransferase. All L. pneumophila strains tested contained a major ca. 60-kDa protein that reacted with the antiserum, while strains representing other Legionella species were negative in the immunoblot analysis (Fig. 7). Thus, our results demonstrated that there was a good correlation among UDP-glucosyltransferase activity (Fig. 1), PCR amplification (Fig. 5), and Ugt cross-reactive proteins. Furthermore, they indicated that the UDP-glucosyltransferase may be peculiar to L. pneumophila.
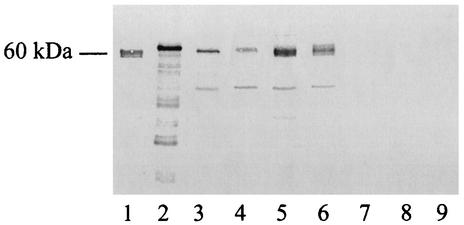
FIG. 7.
Immunoblot analysis of different Legionella strains. Ultrasonic extracts were obtained from various legionellae grown on BCYE agar, and then samples containing approximately 5 μg of protein were subjected to SDS-PAGE, transferred onto a nitrocellulose filter, and probed with an antiserum raised against purified L. pneumophila UDP-glucosyltransferase. Lane 1, UDP-glucosyltransferase purified from L. pneumophila; lane 2, purified recombinant UDP-glucosyltransferase; lane 3, L. pneumophila Philadelphia I; lane 4, L. pneumophila 130b; lane 5, L. pneumophila ATCC 33823; lane 6, L. pneumophila ATCC 35096; lane 7, L. longbeachae ATCC 33462; lane 8, L. gormanii ATCC 33297; lane 9, L. steigerwaltii ATCC 35302. Minor bands observed in recombinant protein and some L. pneumophila preparations probably resulted from proteolytic degradation of a major 60-kDa enzyme.
DISCUSSION
Based on the behavior of purified native and recombinant proteins, we discovered a new L. pneumophila enzyme, a UDP-glucosyltransferase. Enzymes capable of transferring the UDP-glucose moiety have been described previously in viruses, bacteria, plants, and animals (12, 36, 42, 53, 60). Importantly, though, the L. pneumophila Ugt enzyme is the first example of a glucosyltransferase that is produced by a facultative intracellular parasite. As a group, the UDP-glucosyltransferases have been linked with a variety of processes, including, in bacteria, biosynthesis of antibiotics and modification of substrates having different chemical structures (34, 36). Based on the overall lack of homology between the L. pneumophila enzyme and known proteins and the fact that the legionellae are intracellular parasites of protozoan and mammalian cells, Ugt is likely a new type of UDP-glucosyltransferase and may catalyze a novel reaction. Indeed, the enzyme transferred UDP-glucose to an unidentified 45-kDa protein within eukaryotic cells. Since Ugt-containing Legionella extracts failed to undergo autoglucosylation, there do not appear to be additional substrates in the bacterial cell. Thus, the L. pneumophila enzyme may have evolved to interact specifically with a host cell target.
Given the different lifestyles of L. pneumophila, Ugt could serve any number of roles, including as a virulence determinant. In one scenario, the enzyme may promote intracellular infection of host cells. Such a hypothesis is supported by the fact the 45-kDa substrate was conserved within monocytes and epithelial cells, two types of cells that support L. pneumophila intracellular growth. In another scenario, Ugt may facilitate extracellular survival in the mammalian host and/or in natural aquatic environments. Two observations suggest that the Legionella UDP-glucosyltransferase may promote pathogenesis, participating in intracellular infection and/or extracellular survival. First, enzymatic activity and cross-reactive proteins appeared to be specific to L. pneumophila, the most pathogenic of the Legionella species. Second, UDP-glucosyltransferases can play critical roles in bacterial virulence. In the genus Clostridium, UDP-glucosyltransferases represent the enzymatic components of the so-called large clostridial toxins, which include toxins A and B of C. difficile, as well as the lethal and hemorrhagic toxins of C. sordellii (48). Another large clostridial toxin, the alpha-toxin of Clostridium novyi, uses UDP-N-acetylglucosamine instead of UDP-glucose as its cofactor (50). The 250- to 300-kDa clostridial toxins modify low-molecular-mass G proteins of the Rho and Ras subfamilies by attaching a sugar moiety to a threonine residue in the effector domain of the GTPase (1, 40, 50). The diseases caused by these clostridia have different clinical and epidemiological manifestations; C. difficile is an agent of antibiotic-associated diarrhea and pseudomembranous colitis in humans, C. sordellii produces gas gangrene, diarrhea, and enterotoxemia in humans and animals, and C. novyi causes gas gangrene in humans and animals (21, 31). However, a common feature of these diseases is that they all involve bacterial extracellular proliferation in the face of an inflammatory response (7). It has been speculated that inhibition of phagocytosis may result from the toxins' ability to target Ras-related proteins, which are important regulators of actin cytoskeleton rearrangements (1, 20, 32, 47). Indeed, the clostridial glucosyltransferases result in suppression of actin polymerization and rapid disorganization of the cytoskeleton network (1). Since the molecular masses of the Rho and Ras proteins are only 21 to 25 kDa (20), it is unlikely that the Legionella and Clostridium enzymes catalyze the same reaction. However, this does not preclude a role for Ugt in the cytoskeleton rearrangements or organelle trafficking alterations that are associated with L. pneumophila intracellular infection (55); e.g., there are multiple molecular mechanisms by which bacterial toxins influence the host cell cytoskeleton (40, 52). Thus, the L. pneumophila UDP-glucosyltransferase may promote pathogenesis by facilitating an intimate interaction with cells (e.g., macrophages) that is favorable for intracellular growth or by minimizing, as the clostridial toxins do, association with cells (e.g., polymorphonucleocytes) that are nonpermissive.
Clearly, future studies should include the generation of L. pneumophila mutants specifically defective for the glucosyltransferase and then an examination of their behavior in protozoan and mammalian models of intracellular infection, as well as in animal models of legionellosis. Any attenuation would indicate that the transferase enzyme has a role in virulence and definitely justify further identification of the eukaryotic substrates for the L. pneumophila transferase. Such investigations not only should shed light on the function of this enzyme in Legionella biology but also may increase our understanding of eukaryotic cell physiology. If the UDP-glucosyltransferase were implicated in infection, then we would also need to further investigate its locations in and/or processing by the bacterial cell. Although our data indicate that the enzyme is inside the Legionella cell, bacterial toxins can be cell associated and exhibit their pathogenic activity only after cell lysis (22, 26). Alternatively, it is conceivable that Ugt either is subject to type II secretion but only under certain growth conditions or is delivered into target cells by another specialized secretion apparatus, such as the Legionella type IV secretion system (19, 43, 49, 58). Another interesting question for possible future investigation is where and how Ugt is transported into host cells. Since the Legionella enzyme did not exhibit homology with the C-terminal, receptor-binding domain of the clostridial toxins, it may interact with host cell plasma membranes in a unique way. Alternately, it may be delivered into the host cell cytosol only after the bacteria have entered the replicative phagosome.
In summary, in this paper we describe the identification and cloning of a novel UDP-glucosyltransferase enzyme from L. pneumophila. Based on its apparently unique structure and the existence of a eukaryotic substrate, this enzyme may represent an important virulence factor of L. pneumophila. Additional study of this enzyme and its gene should yield new insights into the biochemistry of UDP-glucosyltransferases, the biology of a unique facultative intracellular parasite, and the pathogenesis of Legionnaires' disease.
Acknowledgments
We thank J. D'Alayer for protein microsequencing, members of M. R. Popoff's laboratory for assistance, A. Flieger for helpful suggestions, and S. Starkenburg for Legionella culturing.
This work was supported in part by NIH grant AI43987 awarded to N.P.C. and by a Regional Public Russian Medicine Support Fund grant awarded to I.B.
Editor: D. L. Burns
REFERENCES
- 1.Aktories, K. 1997. Bacterial toxins that target Rho proteins. J. Clin. Investig. 99:827-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, H. L., J. P. Vogel, and R. R. Isberg. 1998. Identification of linked Legionella pneumophila genes essential for growth and evasion of the endocytic pathway. Infect. Immun. 66:950-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aragon, V., O. Rossier, and N. P. Cianciotto. 2002. Legionella pneumophila genes that encode lipase and phospholipase C activities. Microbiology 148:2223-2231. [DOI] [PubMed] [Google Scholar]
- 4.Aragon, V., S. Kurtz, A. Flieger, B. Neumeister, and N. P. Cianciotto. 2000. Secreted enzymatic activities of wild type and pilD-deficient Legionella pneumophila. Infect. Immun. 68:1855-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aragon, V., S. Kurtz, and N. P. Cianciotto. 2001. Legionella pneumophila major acid phosphatase and its role in intracellular infection. Infect. Immun. 69:177-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baine, W. B. 1985. Cytolytic and phospholipase C activity in Legionella species. J. Gen. Microbiol. 131:1383-1391. [DOI] [PubMed] [Google Scholar]
- 7.Belyi, I. F. 2002. Actin machinery of phagocytic cells: universal target for bacterial attack. Microsc. Res. Tech. 57:432-440. [DOI] [PubMed] [Google Scholar]
- 8.Belyi, Y. F., I. S. Tartakovskii, Y. V. Vertiev, Y. V. Ezepchuk, and S. V. Prosorovskii. 1988. Characterization of cytolysin produced by Legionella pneumophila. Zh. Mikrobiol. Epidemiol. Immunobiol. 2:4-7. [PubMed] [Google Scholar]
- 9.Black, W. J., F. D. Quinn, and L. S. Tompkins. 1990. Legionella pneumophila zinc metalloprotease is structurally and functionally homologous to Pseudomonas aeruginosa elastase. J. Bacteriol. 172:2608-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 11.Brand, B. C., A. B. Sadowky, and H. A. Shuman. 1994. The Legionella pneumophila icm locus: a set of genes required for intracellular multiplication in human macrophages. Mol. Microbiol. 14:797-808. [DOI] [PubMed] [Google Scholar]
- 12.Bustamante, J., G. Bersier, R. A. Badin, C. Cymeryng, A. Parodi, and A. Boveris. 2002. Sequential NO production by mitochondria and endoplasmic reticulum during induced apoptosis. Nitric Oxide 6:333-341. [DOI] [PubMed] [Google Scholar]
- 13.Cianciotto, N. P., B. I. Eisenstein, C. H. Mody, and N. C. Engleberg. 1990. A mutation in the mip gene results in an attenuation of Legionella pneumophila virulence. J. Infect. Dis. 162:121-126. [DOI] [PubMed] [Google Scholar]
- 14.Cirillo, S. L., L. E. Bermudez, S. H. El-Etr, G. E. Duhamel, and J. D. Cirillo. 2001. Legionella pneumophila entry gene rtxA is involved in virulence. Infect. Immun. 69:508-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edelstein, P. H. 1981. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J. Clin. Microbiol. 14:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, L.-Y., B. J. Stone, J. K. Brieland, and Y. Abu Kwaik. 1998. Different fates of Legionella pneumophila pmi and mil mutants within macrophages and alveolar epithelial cells. Microb. Pathog. 25:291-306. [DOI] [PubMed] [Google Scholar]
- 17.Garduno, R. A., E. Garduno, and P. S. Hoffman. 1998. Surface-associated Hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infect. Immun. 66:4602-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garduno, R. A., G. Faulkner, M. A. Trevors, N. Vats, and P. S. Hoffman. 1998. Immunolocalization of Hsp60 in Legionella pneumophila. J. Bacteriol. 180:505-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hales, L. M., and H. A. Shuman. 1999. Legionella pneumophila contains a type II general secretion pathway required for growth in amoebae as well as for secretion of the Msp protease. Infect. Immun. 67:3662-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science 279:509-514. [DOI] [PubMed] [Google Scholar]
- 21.Hatheway, C. L. 1990. Toxigenic clostridia. Clin. Microbiol. Rev. 3:66-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirst, T. R., L. L. Randall, and S. J. S. Hardy. 1984. Cellular location of heat-labile enterotoxin in Escherichia coli. J. Bacteriol. 157:637-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Just, I., J. Selzer, F. Hofmann, G. A. Green, and K. Aktories. 1996. Inactivation of Ras by Clostridium sordellii lethal toxin-catalyzed glucosylation. J. Biol. Chem. 271:10149-10153. [DOI] [PubMed] [Google Scholar]
- 24.Just, I., J. Selzer, M. Wilm, C. von Eichel-Streiber, M. Mann, and K. Aktories. 1995. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature 375:500-503. [DOI] [PubMed] [Google Scholar]
- 25.Krinos, C., A. S. High, and F. G. Rodgers. 1999. Role of the 25 kDa major outer membrane protein of Legionella pneumophila in attachment to U-937 cells and its potential as a virulence factor for chick embryos. J. Appl. Microbiol. 86:237-244. [DOI] [PubMed] [Google Scholar]
- 26.Kume, K., T. Nakai, Y. Samejima, and C. Sugimoto. 1986. Properties of dermonecrotic toxin prepared from sonic extracts of Bordetella bronchiseptica. Infect. Immun. 52:370-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunishima, H., H. Takemura, H. Yamamoto, K. Kanemitsu, and J. Shimada. 2000. Evaluation of the activity of antimicrobial agents against Legionella pneumophila multiplying in a human monocytic cell line, THP-1, and an alveolar epithelial cell line, A549. J. Infect. Chemother. 6:206-210. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Liles, M. R., V. K. Viswanathan, and N. P. Cianciotto. 1998. Identification and temperature regulation of Legionella pneumophila genes involved in type IV pilus biogenesis and type II protein secretion. Infect. Immun. 66:1776-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lochner, J. E., R. H. Bigley, and B. H. Iglewski. 1985. Defective triggering of polymorphonuclear leukocyte oxidative metabolism by Legionella pneumophila toxin. J. Infect. Dis. 151:42-46. [DOI] [PubMed] [Google Scholar]
- 31.Lyerly, D. M., H. C. Krivan, and T. D. Wilkins. 1988. Clostridium difficile disease and toxins. Clin. Microbiol. Rev. 1:1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackay, D., and A. Hall. 1998. Rho GTPases. J. Biol. Chem. 273:20685-20688. [DOI] [PubMed] [Google Scholar]
- 33.McCusker, K. T., B. A. Braaten, M. W. Cho, and D. A. Low. 1991. Legionella pneumophila inhibits protein synthesis in Chinese hamster ovary cells. Infect. Immun. 59:240-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mulichak, A. M., H. C. Losey, C. T. Walsh, and R. M. Garavito. 2001. Structure of the UDP-glucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics. Structure (Cambridge) 9:547-557. [DOI] [PubMed] [Google Scholar]
- 35.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295:679-682. [DOI] [PubMed] [Google Scholar]
- 36.Nakajima, N., K. Ishihara, H. Hamada, S. Yamane, K. Nakamura, and T. Furuya. 1999. Multi-enzymatic glucosylation using eucalyptus UDP-glucosyltransferase coupled UDP-glucose fermentation by bakers' yeast. Biosci. Biotechnol. Biochem. 63:934-936. [DOI] [PubMed] [Google Scholar]
- 37.Narayanan, J., P. A. Hartman, and D. J. Graves. 1989. Assay of heat-labile enterotoxins by their ADP-ribosyltransferase activities. J. Clin. Microbiol. 27:2414-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogawa, M., A. Takade, H. Miyamoto, H. Taniguchi, and S. Yoshida. 2001. Morphological variety of intracellular microcolonies of Legionella species in Vero cells. Microbiol. Immunol. 45:557-562. [DOI] [PubMed] [Google Scholar]
- 39.Otto, H., D. Tezcan-Merdol, R. Girisch, F. Haag, M. Rhen, and F. Koch-Nolte. 2000. The spvB gene-product of the Salmonella enterica virulence plasmid is a mono(ADP-ribosyl)transferase. Mol. Microbiol. 37:1106-1115. [DOI] [PubMed] [Google Scholar]
- 40.Popoff, M. R., E. Chaves-Olarte, E. Lemichez, C. von Eichel-Streiber, M. Thelestam, P. Chardin, D. Cussac, B. Antonny, P. Chavrier, G. Flatau, M. Giry, J. de Gunzburg, and P. Boquet. 1996. Ras, Rap, and Rac small GTP-binding proteins are targets for Clostridium sordellii lethal toxin glucosylation. J. Biol. Chem. 271:10217-10224. [DOI] [PubMed] [Google Scholar]
- 41.Pruckler, J. M., R. F. Benson, M. Moyenuddin, W. T. Martin, and B. S. Fields. 1995. Association of flagellum expression and intracellular growth of Legionella pneumophila. Infect. Immun. 63:4928-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rausell, C., J. Llorca, and M. D. Real. 1997. Separation by FPLC chromatofocusing of UDP-glucosyltransferases from three developmental stages of Drosophila melanogaster. Arch. Insect Biochem. Physiol. 34:347-358. [DOI] [PubMed] [Google Scholar]
- 43.Rossier, O., and N. P. Cianciotto. 2001. Type II protein secretion is a subset of the PilD-dependent processes that facilitate intracellular infection by Legionella pneumophila. Infect. Immun. 69:2092-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saha, A. K., J. N. Dowling, K. L. LaMarco, S. Das, A. T. Remaley, N. Olomu, M. T. Pope, and R. H. Glew. 1985. Properties of an acid phosphatase from Legionella micdadei which blocks superoxide anion production by human neutrophils. Arch. Biochem. Biophys. 243:150-160. [DOI] [PubMed] [Google Scholar]
- 45.Saha, A. K., J. N. Dowling, N. K. Mukhopadhyay, and R. H. Glew. 1988. Demonstration of two protein kinases in extracts of Legionella micdadei. J. Gen. Microbiol. 134:1275-1281. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 47.Satoh, T., M. Nakafuku, and Y. Kaziro. 1992. Function of Ras as a molecular switch in signal transduction. J. Biol. Chem. 267:24149-24152. [PubMed] [Google Scholar]
- 48.Schmitt, C. K., K. C. Meysick, and A. D. O'Brien. 1999. Bacterial toxins: friends or foes? Emerg. Infect. Dis. 5:224-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Segal, G., J. J. Russo, and H. A. Shuman. 1999. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol. Microbiol. 34:799-809. [DOI] [PubMed] [Google Scholar]
- 50.Selzer, J., F. Hofmann, G. Rex, M. Wilm, M. Mann, I. Just, and K. Aktories. 1996. Clostridium novyi α-toxin-catalyzed incorporation of GlcNAc into Rho subfamily proteins. J. Biol. Chem. 41:25173-25177. [DOI] [PubMed] [Google Scholar]
- 51.Stone, B. J., and Y. Abu Kwaik. 1998. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect. Immun. 66:1768-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stubbs, S., M. Rupnik, M. Gibert, J. Brazier, B. Duerden, and M. Popoff. 2000. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol. Lett. 186:307-312. [DOI] [PubMed] [Google Scholar]
- 53.Sullivan, T. A., J. L. Jakobek, and P. B. Lindgren. 2001. Cloning and characterization of a bean UDP-glucosyltransferase cDNA expressed during plant-bacterial interactions. Mol. Plant Microbe Interact. 14:90-92. [DOI] [PubMed] [Google Scholar]
- 54.Swanson, M. S., and B. K. Hammer. 2000. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54:567-613. [DOI] [PubMed] [Google Scholar]
- 55.Swanson, M. S., and R. R. Isberg. 1995. Formation of the Legionella pneumophila replicative phagosome. Infect. Agents Dis. 2:269-271. [PubMed] [Google Scholar]
- 56.Tauschek, M., R. J. Gorrell, R. A. Strugnell, and R. M. Robins-Browne. 2002. Identification of a protein secretory pathway for the secretion of heat-labile enterotoxin by an enterotoxigenic strain of Escherichia coli. Proc. Natl. Acad. Sci. USA 99:7066-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 59.Warren, W. J., and R. D. Miller. 1979. Growth of Legionnaires disease bacterium (Legionella pneumophila) in chemically defined medium. J. Clin. Microbiol. 10:50-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wormleaton, S. L., and D. Winstanley. 2001. Phylogenetic analysis of conserved genes within the ecdysteroid UDP-glucosyltransferase gene region of the slow-killing Adoxophyes orana granulovirus. J. Gen. Virol. 82:2295-2305. [DOI] [PubMed] [Google Scholar]