Abstract
In searching the Staphylococcus aureus genome, we previously identified sarT, a homolog of sarA, which encodes a repressor for α-hemolysin synthesis. Adjacent but transcribed divergently to sarT is sarU, which encodes a 247-residue polypeptide, almost twice the length of SarA. Sequence alignment disclosed that SarU, like SarS, which is another SarA homolog, could be envisioned as a molecule with two halves, with each half being homologous to SarA. SarU, as a member of the SarA family proteins, disclosed conservation of basic residues within the helix-turn-helix motif and within the beta hairpin loop, two putative DNA binding domains within this protein family. The transcription of sarU is increased in a sarT mutant. Gel shift and transcriptional fusion studies revealed that SarT can bind to the sarU promoter region, probably acting as a repressor for sarU transcription. The expression of RNAII and RNAIII of agr is decreased in a sarU mutant. As RNAIII expression is up-regulated in a sarT mutant, we hypothesize that sarT may down regulate agr RNAIII expression by repressing sarU, a positive activator of agr expression. We propose that, in addition to the quorum sensing effect of the autoinducing peptide of agr, the sarT-sarU pathway may represent a secondary amplification loop whereby the expression of agr (e.g., those found in vivo) might repress sarT, leading to increased expression of sarU. Elevated sarU expression would result in additional amplification of the original agr signal.
Staphylococcus aureus is an important human pathogen in both the community and hospital settings (33). The spectrum of diseases caused by this organism is extremely broad, ranging from cutaneous to deep-seated infections such as pneumonia, endocarditis, and sepsis (33). Within its arsenal are virulence genes coding for proteins that facilitate tissue colonization, immune evasion, and tissue destruction (33). Superimposed upon these virulence genes is a network of regulatory genes that confer precise gene expression during different stages of infection (2, 6, 33). Expression of the regulatory elements, in turn, exerts transcriptional control of unlinked target genes. During growth in vitro, S. aureus expresses a number of cell wall-associated adhesins that are believed to promote host tissue colonization. In transition to the postexponential phase, the expression of cell wall proteins is repressed while the synthesis of exoproteins predominates, presumably to facilitate host cell lysis (33).
Postexponential protein expression in S. aureus is generally governed, in part, by global regulatory elements such as agr (20), sae (15), and sarA (10). The agr locus, a pleiotropic regulator of exoprotein synthesis in S. aureus, comprises two divergent transcripts, RNAII and RNAIII (20, 28, 29, 32), which encode agrDBCA and hld, respectively. AgrC and AgrA correspond to the sensor and activator of a two-component regulatory system (20). AgrD encodes a 46-residue peptide which, aided by AgrB, undergoes processing and cyclization to yield a quorum-sensing cyclic peptide (17). AgrC is the putative sensor for this cyclic peptide (23) and, upon phosphorylation, will lead to a second phosphorylation step of the activator AgrA. Phosphorylated AgrA is believed to bind to the agr promoter to activate transcription of RNAIII, which, as the regulatory molecule, up-modulates the transcription of exoprotein genes and down-modulates the expression of cell wall protein genes during the postexponential phase (32).
Contrary to agr, the sarA locus up-regulates the expression of many cell wall proteins (e.g., fibronectin binding protein A) and selected exoproteins (e.g., α [hla] and β hemolysins) (10) while repressing the transcription of the protein A gene (spa) (9). The sarA locus comprises a major 372-bp sarA open reading frame driven by three distinct promoters (4). DNA binding studies revealed that SarA, the major sarA effector molecule, binds to several target gene promoters (e.g., agr, hla, and spa) to modulate gene transcription (12), thus accounting for both agr-dependent and agr-independent pathways of regulation. As mutations in sarA or agr have been found to affect the transcription of over 100 genes (14), it is not surprising that other regulatory factors may be at work, in part to control SarA and agr expression and also to regulate genes downstream of the sarA-agr regulatory cascade. We and others have found several SarA homologs that are involved directly or indirectly in virulence gene regulation. SarR, being a SarA homolog with a molecular mass of 13.6 kDa (26), represses sarA (26), agr, hla, hlb, and spa expression during the postexponential phase (unpublished data), presumably by binding to the sarA or other target gene promoters. SarS (also called SarH1), a 250-residue SarA homolog (11, 37) repressible by agr, likely acts downstream of agr to up-regulate spa expression. An additional regulatory gene with partial homology to SarA, rot, is a repressor of hla and probably acts downstream of agr (27).
In searching the recently released S. aureus genomes (www.ncbi.nlm.nih.gov/genome/ and www.TIGR.org), we found additional SarA homologs. One of these homologs, SarT (a 118-residue protein), is a repressor of hla expression and is negatively controlled by sarA (36). Contiguous to sarT, but transcribed in opposite orientation, is sarU. The expression of sarU is negatively controlled by sarT. Phenotypic and transcriptional analysis revealed that sarU is a positive regulator of RNAIII and contributes to the expression of virulence genes controlled by agr.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
The bacterial strains and plasmids used in this study are listed in Table 1. CYGP, 0.3GL medium (30), and tryptic soy broth were used for the growth of S. aureus strains, while Luria-Bertani broth was used to cultivate Escherichia coli. Antibiotics were used at the following concentrations: erythromycin at 5 μg/ml, kanamycin at 50 μg/ml, tetracycline at 5 μg/ml, and chloramphenicol at 10 μg/ml for S. aureus; and ampicillin at 50 μg/ml, chloramphenicol at 30 μg/ml, and spectinomycin at 75 μg/ml for E. coli.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Reference or source | Comments |
|---|---|---|
| S. aureus | ||
| RN4220 | 31 | Mutant of 8325-4 that accepts foreign DNA |
| RN6390 | 31 | Laboratory strain that maintains its hemolytic pattern when propagated on sheep erythrocyte agar (parental strain) |
| RN6911 | 20 | agr mutant of RN6390 with an Δagr::tetM mutation |
| ALC1342 | Laboratory strain | A sarA mutant with deletion of open reading frame 3 and the sarA open reading frame and replaced with an ermC gene |
| ALC1713 | 26 | sarR mutant of RN6390 with ΔsarR::ermC |
| ALC1905 | 36 | sarT mutant of RN6390 with sarT::ermC |
| ALC2071 | 36 | sarT mutant ALC1905 carrying pSK236 with wild-type sarT gene |
| ALC1927 | 11 | sarS mutant of RN6390 with sarS::ermC |
| ALC2272 | This study | sarU mutant of RN6390 with a deletion of amino acids 1 to 153 of the N terminus of the sarU gene product and its replacement with an ermC gene |
| ALC2380 | This study | RN6390 with pALC2360 |
| ALC2381 | This study | ALC2272 with pALC2360 |
| ALC2601 | This study | RN6390 with pALC2591 |
| ALC2602 | This study | RN6390 with pALC2599 |
| ALC2604 | This study | RN6911 with pALC2591 |
| ALC2605 | This study | RN6911 with pALC2599 |
| ALC2607 | This study | ALC1905 with pALC2591 |
| ALC2608 | This study | ALC1905 with pALC2599 |
| ALC2609 | This study | ALC2272 with pALC2591 |
| ALC2610 | This study | ALC2272 with pALC2599 |
| ALC2714 | This study | RN6390 with pALC2707 |
| ALC2716 | This study | ALC1905 with pALC2707 |
| ALC2717 | This study | ALC2272 with pALC2707 |
| E. coli | ||
| XL-1 Blue | 25 | Host strain for cloning |
| Topo InvαF′ | Invitrogen | Host strain for the TA cloning vector |
| BL21(DE3)pLysS | Novagen | Host strain for the pET expression system |
| Plasmids | ||
| pCL52.2 | 34 | Temperature-sensitive E. coli-S. aureus shuttle vector |
| pCR2.1 | Invitrogen | E. coli cloning vector for direct cloning of PCR products |
| pET14b | Novagen | Expression vector in E. coli |
| pSK236 | 19 | Shuttle vector containing pUC19 cloned into the HindIII site of pC194 |
| pUC19 | 25 | E. coli cloning vector |
| pALC1484 | 18 | Modified pSK236 shuttle vector with a promoterless gfpuvr reporter gene preceded by an S. aureus ribosome binding site |
| pALC1740 | This study | pALC1484 with the hla promoter |
| pALC1742 | 36 | pALC1484 with the agr P2 promoter |
| pALC1743 | 18 | pALC1484 with the agr P3 promoter |
| pALC1904 | This study | pET14b containing the 345-bp sarT gene at the XhoI/BamHI sites |
| pALC2208 | This study | pCR2.1 containing a 2.3-kb PCR sarU-sarT fragment with flanking upstream and downstream sequences |
| pALC2229 | This study | pUC19 containing a 2.3-kb EcoRV-KpnI fragment from pALC2208 at the HincII and KpnI sites |
| pALC2240 | This study | pCL52.2 containing a 2.9-kb DNA fragment, which has a deletion of a 547-bp ClaI-HincII fragment that includes residue 1 to 153 of the N terminus of the sarU gene product and an insertion of the 1.2-kb ermC gene at the EcoRI site |
| pALC2360 | This study | pSK236 with a 1.5-kb DNA fragment containing the sarU gene with a 392-bp upstream sequence at the EcoRI site |
| pALC2591 | This study | pALC1484 with a 310-bp promoter fragment of the sarU gene fused with the gfpuvr reporter gene at the EcoRI and XbaI sites |
| pALC2599 | This study | pALC1484 with a 147-bp promoter fragment of the sarU gene fused with the gfpuvr reporter gene at the EcoRI and XbaI sites |
| pALC2707 | This study | pALC1484 with a 310-bp promoter fragment of sarT fused with the gfpuvr reporter gene at the EcoRI site |
Genetic manipulations in E. coli and S. aureus.
Based on homology with SarA, the sarU gene product was identified in the S. aureus genome database (www.TIGR.org/). To construct a sarU mutant, the sarU gene together with flanking sequences on both sides was amplified by PCR with the primers 5′-TGACGATTTCGGCTGAACTTC-3′ and 5′-TGGAACACGAAATGGTGAAC-3′, with chromosomal DNA from strain RN6390 being used as the template. The 2.3-kb PCR fragment was cloned into cloning vector pCR2.1 (Invitrogen, San Diego, Calif.) in E. coli. The KpnI-EcoRV DNA fragment containing the 2.3-kb fragment was then cloned into the KpnI and HincII sites of pUC19. A 547-bp fragment comprising the N-terminal 153 amino acids of SarU was deleted by restricting with HincII and ClaI and then replaced with an ∼1.2-kb ermC fragment. The fragment containing an ermC insertion into the partially deleted sarU gene was cloned into the temperature-sensitive shuttle vector pCL52.2 (22). The recombinant pCL52.2 was transformed into RN4220 by electroporation (35). Plasmid isolated from RN4220 was restriction digested for authenticity and introduced into RN6390 by electroporation. Transformants were selected at 30°C on erythromycin- and tetracycline-containing agar plates. S. aureus RN6390, harboring the recombinant pCL52.2 construct, was grown overnight at 30°C in liquid medium in the presence of erythromycin, diluted 1:1,000 in fresh medium, and propagated at 42°C, a nonpermissive temperature for the replication of pCL52.2. This cycle was repeated four times, and the cells were plated onto 03GL plates containing erythromycin and erythromycin-tetracycline to select for tetracycline-sensitive but erythromycin-resistant colonies, representing mutants with double crossovers (a frequency of 10−2 after the fourth passage). The mutations were confirmed by PCR and Southern hybridization with sarU and ermC probes as described previously (25). One clone, designated ALC2272, was selected for further study.
To complement the sarU mutation, a 1.5-kb fragment encompassing the sarU gene and a 392-bp sequence upstream of the sarU transcription start site (see below) was cloned into shuttle plasmid pSK236. The recombinant plasmid was electroporated into RN4220, selecting for chloramphenicol-resistant colonies. The transformant was verified by plasmid restriction analysis. The plasmid from RN4220 was then electroporated into parental strain RN6390 and the sarU mutant (ALC2272).
Immunoblot analysis.
To assess SarA expression in different S. aureus strains, cellular proteins were extracted from RN6390, the sarU mutant (ALC2272), and the complemented strains (ALC2380 and ALC2381). In brief, after being pelleted, the cells were suspended in 1 ml of TEG buffer (25 mM Tris-Cl, 5 mM EGTA; pH 8) and cell extracts were prepared from lysostaphin-treated cells as described previously (13). Cell extracts were immunoblotted onto nitrocellulose membranes. For the detection of SarA, monoclonal antibody 1D1 (1:6,000 dilution) was added to the blot and incubated for 3 h, followed by another hour of incubation with a 1:10,000 dilution of goat anti-mouse alkaline phosphatase conjugate (Jackson Immuno Research, West Grove, Pa.). Reactive bands were detected with developing substrates as described previously (5).
Isolation of RNA and Northern blot hybridization.
Overnight cultures of S. aureus were diluted 1:50 in CYGP or tryptic soy broth and grown to mid-log (optical density at 650 nm [OD650] = 0.7), late log (OD650=1.1), and early postexponential (OD650=1.7) phases. The cells were pelleted and processed with a Trizol isolation kit (Gibco BRL, Gaithersburg, Md.) in combination with 0.1-mm-diameter sirconia-silica beads in a Biospec reciprocating shaker, as described previously (8). Ten or 15 μg of each sample was electrophoresed through a 1.5% agarose-0.66 M formaldehyde gel in morpholinepropanesulfonic acid (MOPS) running buffer (20 mM MOPS, 10 mM sodium acetate, 2 mM EDTA, pH 7.0). Blotting of RNA onto Hybond N+ membranes (Amersham, Arlington Heights, Ill.) was performed with a Turbo blotter alkaline transfer system (Schleicher & Schuell, Keene, N.H.). For detection of specific transcripts (agr, sarA, sarU, sarT, spa, coa, agr, and hla), gel-purified DNA probes were radiolabeled with [α-32P]dCTP with a random-primed DNA labeling kit (Roche Diagnostics GmbH) and hybridized under aqueous-phase conditions at 65°C (7). The blots were subsequently washed, autoradiographed, and developed with Kodak BioMax film or Kodak Blue film.
Primer extension analysis.
Mapping of the 5′ end of the sarU transcript was performed with the primer 5′-GTTGCTTTAACTCTTGAGTGAG-3′, which is complementary to the coding strand for sarU and is located at nucleotide positions 85 to 64 downstream from the putative initiation codon GTG. Primer extension was carried out as described previously (3, 4).
Transcriptional fusion studies of RNAII, RNAIII, hla, and sarU and sarT promoters linked to the gfpuvr reporter gene.
A 229-bp fragment in forward (nt 1528 to 1756) and reverse (nt 1756 to 1528) positions (20), representing RNAII and RNAIII promoter fragments, respectively, was amplified by PCR with genomic DNA of S. aureus strain RN6390 as the template and cloned into the TA cloning vector pCR2.1. Similarly, a 440-bp hla promoter fragment, representing sequence upstream of the ribosomal binding sites of hla, was cloned into pCR2.1. Various lengths of the sarU promoter region (310 and 148 bp) extending from the sequence upstream of the ribosomal binding sites were amplified by PCR by using chromosomal DNA of S. aureus strain RN6390 and primers with flanking EcoRI or XbaI sites. EcoRI and XbaI fragments containing RNAII, RNAIII, hla, or various lengths of sarU promoter fragments were cloned into shuttle plasmid pALC1484 (18), generating transcriptional fusions to the gfpuvr reporter gene as pALC1743, pALC1742, pALC1740, pALC2591, and pALC2599. As sarT is transcribed contiguously but divergently from sarU, we also cloned the 310-bp sarU promoter fragment in reverse orientation to the gfpuvr gene to yield a sarT promoter fragment driving the reporter gene (pALC2707). The construction of plasmid pALC1484 and modification of gfpuv (Clontech, Palo Alto, Calif.) to gfpuvr with a S65T mutation to yield a red shift (excitation maxima from 395 to 488 nm) were described earlier (18). The orientation and authenticity of the promoter fragments were confirmed by restriction analysis and DNA sequencing. The recombinant plasmids containing the respective RNAIII, RNAII, and hla and sarU promoters driving the gfpuvr reporter were first introduced into S. aureus strain RN4220 by electroporation (35). Plasmids containing RNAII, RNAIII, and hla promoters were electroporated into RN6390 and its isogenic sarU and sarT mutants, while the sarU and sarT promoter constructs were introduced into RN6390, the isogenic agr, sarT, or sarU mutant.
After overnight culture, S. aureus strains harboring the recombinant plasmids were diluted 1:100 and grown at 37°C with shaking in tryptic soy broth containing chloramphenicol (10 μg/ml). Aliquots (100 μl) were transferred hourly to microtiter plates to assay for cell density (OD650) and fluorescence for 10 h in a FL600 fluorescence reader (BioTek Instrument, Winooski, Vt.). Promoter activation was plotted as mean fluorescence per OD unit over time, using the average values from triplicate readings.
Overexpression and purification of SarT in a pET vector.
The 420-bp DNA fragment containing the full-length sarT gene was amplified by PCR by using chromosomal DNA from S. aureus RN6390 as the template and primers containing flanking restriction sites (XhoI and BamHI for sarT) to facilitate cloning into the expression vector. The purified PCR product was digested with XhoI and BamHI, gel purified, ligated into the expression vector pET14b (Novagen, Madison, Wis.), and transformed into E. coli XL-1 Blue. The recombinant plasmid containing a full-length sarT coding region was confirmed by restriction digestion and DNA sequencing. The recombinant plasmids were then transformed to E. coli BL21(DE3) pLys.S. The resulting plasmid (pALC1904; see Table 1) contained the entire sarT coding region in frame with an N-terminal His tag. The expression of recombinant protein was induced by adding isopropyl-1-thio-β-d-galactopyranoside (ITPG; final concentration, 1 mM) to a growing culture (37°C) at an OD600 of 0.5. After a 4-h induction, the cells were harvested, resuspended in binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl, pH 7.9), frozen by dipping in liquid N2, and thawed overnight in ice. Cellular debris were removed by centrifugation (TL-100 tabletop ultracentrifuge; Beckman, Palo Alto, Calif.) at 45,000 rpm for 60 min, and the clarified supernatant was applied to a nickel affinity column (Novagen) in accordance with the manufacturer's instruction. The protein was eluted with the elution buffer (1 M imidazole, 0.5 M NaCl, 20 mM Tris-HCl, pH 7.9), followed by dialysis in a buffer containing 20 mM Tris-Cl (pH 8.0), 50 mM NaCl, 1 mM EDTA, 10% glycerol, and 1 mM DTT. The authenticity of the purified SarT protein was confirmed by N-terminal sequencing, and the size and purity of the recombinant protein was verified by sodium dodecyl sulfate gels stained with Coomassie brilliant blue R-250.
Gel shift assays.
To determine whether the recombinant SarT protein binds to the sarU promoter, a 150-bp fragment representing nucleotide positions 13 to 160 bp upstream of the sarU start codon was end labeled with [γ-32P]ATP by using the T4 polynucleotide kinase. Labeled fragment (0.1 ng) was incubated at room temperature for 20 min with various amounts of purified SarT protein in 25 μl of binding buffer (25 mM Tris-Cl, pH 7.5, 0.1 mM EDTA, 75 mM NaCl, 1 mM dithiothreitol, and 10% glycerol) containing 0.5 μg of calf thymus DNA. The reaction mixtures were analyzed in an 8.0% nondenaturing polyacrylamide gel. The band shifts were detected by exposing dried gels to X-ray film.
RESULTS
Identification of the sarU gene.
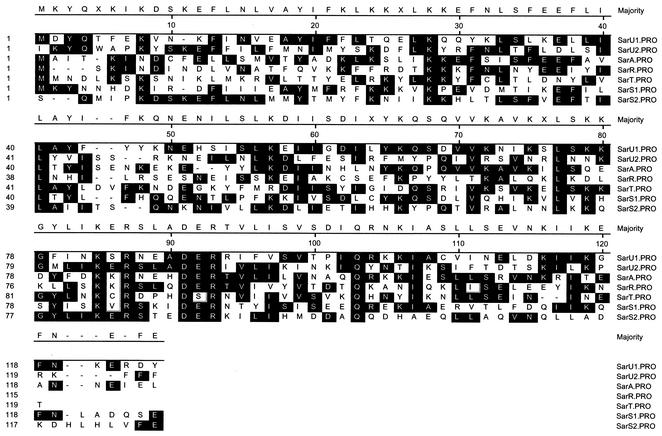
In searching for SarA homologs in the S. aureus N315 genome (www.ncbi.nlm.nih.gov/genome/staphylococcus), we found at least 12 proteins homologous to SarA (protein GI 13700508) with a default setting of 30 as the cutoff in the BLAST search (21). Some of these homologs, including SarR (protein GI 13702095, with an e value of 9e−12), SarS (also called SarH1; protein GI 13700028, with an e value of 1e−16), SarT (also called SarH3; protein GI 13702448, with an e value of 9e−17), and Rot (protein GI 13701558, with an e value of 0.003 and a 50% homology) (27) have been previously studied. The remaining homologs, many of which we are characterizing, are SarX (protein GI 13700559, with an e value of 1e−04 and a 46% homology), SarV (protein GI 13702067, with an e value of 5e−04 and a 48% homology), SarZ (protein GI 13702335, with an e value of 0.002 and a 53% homology), and three proteins of unknown function (proteins GI 13701144, GI 13700062, and GI 13702441, with e values of 0.003, 0.030, and 0.066 and homologies of 48, 48, and 45%, respectively). SarR is a 113-residue protein that binds to the sarA promoter to down-modulate SarA protein expression (26). SarS, a 250-residue protein whose expression is normally repressed by sarA and agr (11, 37), acts as an activator of protein A synthesis. SarT, identified initially by its homology to SarA, is a 118-residue protein that functions as a repressor for hla transcription (36). Adjacent to sarT but transcribed in the opposite direction is a gene, designated sarU, that encodes another SarA homolog (protein GI 13702449 with an e value of 2e−15). SarU is the third protein that came up in our BLAST search for SarA homologs, with SarT, SarS, and SarR being the first, second, and fourth hits, respectively. SarU, a 247-residue polypeptide (29,276 Da), is almost twice the size of the smaller SarA homologs (e.g., SarA, SarR, and SarT). Similar to SarS, SarU, which has a 64% homology to SarA, can be considered to be a molecule with two halves (namely, SarU1, consisting of residues 1 to 124, and SarU2, consisting of residues 125 to 247), with each half sharing sequence similarity to the smaller SarA homologs (Fig. 1). SarU1 has a 25% sequence identity to SarU2 and shares 56, 59, and 51% homology to SarA, SarT, and SarR, respectively. Likewise, SarU2 has 56, 53, and 57% homology to these respective proteins. Interestingly, when SarU was analyzed with BLAST against the entire microbial data bank, most of the hits with significant e values (e−36 to 0.05) were proteins from S. aureus strains as well as from Staphylococcus epidermidis. Only the MarR protein from Bacillus subtilis yielded a significant e value of 0.045 (see Discussion, below).
FIG. 1.
Sequence alignment of SarU, SarA, SarR, SarT, and SarS. SarU is a 247-residue polypeptide. Based on regional homology, SarU can be divided into two domains, U1 and U2, of 124 and 123 residues, respectively. The homology of SarU with a two-domain protein like SarS is higher, with 39% identity and 64% homology. The conserved residues, seen in the black boxes, are designed as the majority at each position when at least half of the residues have the same amino acid. The alignment was done by the Clustal program of DNASTAR.
Translational and transcriptional start site and promoter structure of sarU.
To verify that the start codon of the sarU gene is indeed GTG, a valine (V) residue, we resequenced our cloned sarU fragment (pALC2208) by using primer 5′-GTTGCTTTAACTCTTGAGTGAG-3′ (nucleotide positions 85 to 64 downstream from the putative start codon GTG). Repeated sequencing confirmed GTG to be the start codon, with a strong ribosome-binding site AGGAGA that is located 7 bp upstream. To determine the transcriptional start site and the promoter sequences, primer extension was performed with an identical primer, and total RNA was isolated from the wild-type strain RN6390 and the isogenic sarT mutant (data not shown). The transcriptional start site of sarU was mapped, based on data from both strains, to an A residue located 90 bp upstream of the initiation codon. Based upon the transcriptional start site, the predicted promoter boxes are TATAAA (−10)-N16-TTTATA (−35), which has close resemblance to the −10 and −35 consensus sequences of σA-dependent promoters (16). The organization of the sarU-sarT intergenic region is complex, as the two genes are divergently transcribed and separated by 323 bp, and the −10 region of the sarU and sarT genes are located 97 and 233 bp upstream of their corresponding start sites, respectively. The putative TATAAT (−10)-N24-AAGACA (−35) promoter region for sarT is predicated upon transcript size, and sequence prediction and precise mapping have not been done (36).
Expression of sarU in RN6390 and its isogenic agr, sarA, sarR, sarS, and sarT mutants.
To assess the role of sarU within the sarA-agr regulatory cascade, we constructed a sarU mutant by allelic replacement, essentially replacing the sarU gene with an ermC cassette (see Materials and Methods). A 547-bp region, containing the N-terminal 153 amino acids of SarU and an 88-bp sequence upstream of the translation start, was deleted and replaced with an ∼1.2-kb ermC fragment. PCR amplification with primers outside the ermC insertion yielded a fragment that was 1.2 kb larger than the wild type, consistent with an ermC insertion into the sarU gene. This result also corroborated Southern hybridization data with selected ermC and sarU probes (data not shown).
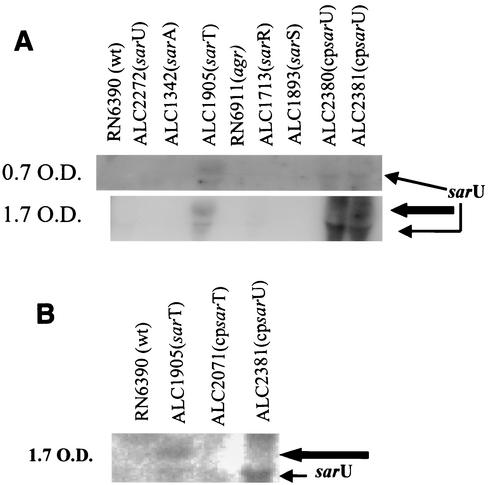
A Northern blot with a sarU probe (820 bp) that encompassed the coding region disclosed that the sarU gene was poorly transcribed in the parental strain. Interestingly, the sarU transcript, consisting of ∼1,200 nt, was enhanced in the sarT mutant but not in the sarA, sarR, sarS, or agr mutants (Fig. 2). Interestingly, the level of the sarU transcript was higher during the postexponential phase (OD, 1.7) than in the exponential phase (OD, 0.7). The size of the transcript (650 nm) also hinted at the possible monocistronic nature of the sarU transcript. In addition to the 1,200-nt transcript, we also observed an ∼1,400-nt transcript that hybridized with the sarU probe in the sarT mutant. However, upon introducing the complemented plasmid into the sarU mutant or the parental strain, we were not able to restore the larger transcript (strains ALC2381 and ALC2380) in repeated attempts. A representative blot is shown in Fig. 2A. This finding suggests that the larger transcript might arise from neighboring genes or a cross-hybridizing band (see Discussion, below). Nevertheless, the possibility of a cryptic promoter in this region cannot be entirely ruled out. Both transcripts were abolished in the complemented sarT mutant (ALC2071 in Fig. 2B). The sarU transcript was undetectable by Northern blotting in strain SH1000, an rsbU+ strain, as well as in RN6390 (data not shown).
FIG. 2.
(A) Northern analysis of the sarU transcript in sarA, agr, sarT, sarR, and sarS mutants and in complemented strains at exponential (OD650, ∼0.7) (upper panel) and postexponential (OD650, ∼1.7) (lower panel) phases of growth. Expression of the sarU transcript appeared to be higher during the postexponential phase than during the exponential phase in the sarT mutant, a conclusion drawn from multiple repeated experiments. A total 15 μg of cellular RNA was loaded onto each lane. The blot was probed with an 820-bp sarU probe containing the entire open reading frame of the sarU gene. The thin arrow points to the ∼1,200-nt sarU transcript and the thick arrow points to a 1,400-nt transcript (see Discussion). (B) Northern hybridization with the same fragment to the wild-type RN6390, sarT mutant (ALC1905), and to ALC1905 complemented with a wild copy of the sarT gene (ALC2071) and complemented sarU mutant (ALC2381).
Gel shift assays of the SarT protein with sarU promoter fragments.
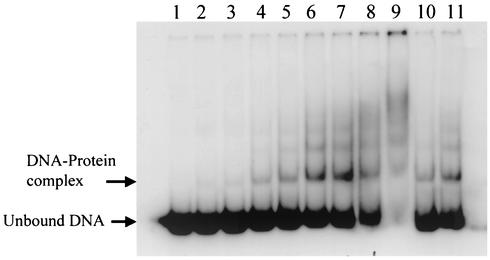
Since the transcription of sarU, a gene adjacent to but divergently transcribed from sarT, is increased in a sarT mutant (Fig. 2) but not vice versa (data not shown), we speculated that SarT might bind to the sarU promoter to modulate sarU expression. Taking advantage of the fact that the coding regions of sarU and sarT are only 323 bp apart, we amplified by PCR a 148-bp sarU promoter fragment directly upstream of the sarU ribosomal binding site. The fragment was end labeled with [γ-32P]ATP and used in gel shift assays with increasing concentrations of purified SarT protein (Fig. 3). Retarded protein-DNA complexes can be detected with as little as 0.2 μg of protein (6.8 pM). As the concentrations of SarT increased, multiple retarded DNA-protein complexes were discerned, suggesting that multiple proteins may bind to the sarU promoter region. Alternatively, SarT may bind to multiple sites on the sarU promoter region. Based on the data, we estimated the dissociation constant (Kd) to be ∼102.5 pM.
FIG. 3.
Autoradiogram of an 8.0% nondenaturing polyacrylamide gel showing a gel shift assay for purified SarT protein with a sarU promoter fragment. Purified SarT protein was allowed to bind a 148-bp radiolabeled sarU promoter fragment. Lanes 1 to 9, mobility of the 148-bp DNA fragment with 0, 0.2, 0.3, 0.5, 0.75, 1.0, 2.0, 2.5, and 3.0 μg of purified SarT protein, respectively; lanes 10 and 11, mobility of the same fragment in the presence of 1.0 μg of the purified protein and a 40-fold excess (molar ratio) of unlabeled 148-bp sarU fragment and a 273-bp nonspecific DNA fragment, respectively.
sarU and sarT promoter-gfpuvr fusion studies in isogenic sarT and sarU strains.
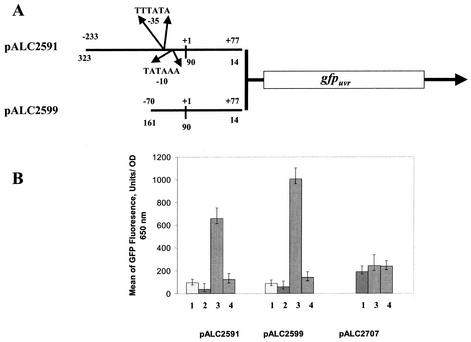
To confirm the involvement of sarT in sarU expression, we constructed transcriptional fusions of two sarU promoter fragments, representing 148 and 310 bp upstream of the sarU ribosomal binding site, to the gfpuvr reporter gene. The constructs, pALC2591 and pALC2599, containing 310- and 148-bp fragments, respectively, were introduced into the wild-type strain RN6390, the sarT mutant (ALC1905), the agr mutant RN6911, and the sarU mutant (ALC2272) and assayed for green fluorescence emitted from GFPuvr. The results, presented in Fig. 4B, disclosed that there are ∼7- to 11-fold increases in mean fluorescences attributable to sarU promoter activity in the sarT mutant (1,007 ± 95 and 657 ± 65 fluorescence units plus or minus the indicated standard deviations for pALC2599 and pALC2591, respectively) compared to those in the parental strain (89 ± 20 and 95 ± 25 units for pALC2599 and pALC2591, respectively). The levels of sarU promoter activity in the sarU mutant (140 ± 30 and 124 ± 50 units for pALC2599 and pALC2591, respectively) were not significantly higher than those in the parental strain, consistent with a lack of significant autoregulatory effect. Although the mean fluorescence values in the parental strain are not very high, the level of sarU promoter activity in an isogenic agr mutant (58 ± 20 and 38 ± 25 units versus 89 ± 20 and 95 ± 25 in the parental strain for pALC2599 and pALC2591, respectively) was even lower. These results were consistent in several repeated experiments. Collectively, these data confirmed the role of sarT in repressing sarU expression and probably in slightly lowering sarU promoter activity in the agr mutant relative to the parental strain.
FIG. 4.
Promoter activation of the sarU promoter fused to a gfpuvr reporter gene, as evaluated with a fluorescence spectrophotometer (FL600; BioTek Instruments). (A) Graphical representation of the 310- and 147-bp DNA fragments of sarU promoter region fused to a promoterless gfpuvr gene with an S. aureus ribosome-binding site. The transcriptional start site, labeled as +1, was identified by primer extension, and the putative promoter −10 and −35 boxed sequences are also indicated. The numbers at the lines (both top and bottom) are marked according to the transcriptional start site and to the start codon of the sarU gene, respectively. (B) Recombinant shuttle constructs pALC2591, pALC2599, and pALCALC2707 containing 310-bp, 148-bp, and reversed-orientation 310-bp sarU promoter fragments, respectively, were introduced into the wild type and its isogenic mutant strains, namely, wild-type RN6390 (lanes 1), agr mutant RN6911 (lanes 2), sarT mutant ALC1905 (lanes 3), and sarU mutant ALC2272 (lanes 4). To minimize variations in fluorescence attributable to cell density, the data are presented as the averages of reported fluorescence per OD650 unit in triplicate samples obtained during the postexponential phase, when sarU promoter activity is expected to be higher (see Fig. 2). The experiment was repeated several times, and one of the typical experiments is shown. A negative control with the shuttle vector pALC1484 alone lacking any promoter fragment was performed and showed no significant background (less than 50 mean fluorescence units) (data not shown).
We also attempted to verify whether sarT regulates its own expression. For this experiment, we introduced pALC2707, a gfpuvr construct carrying a 310-bp sarU promoter fragment with an orientation opposite to that found in pALC2591, thus yielding a sarT promoter-gfpuvr construct, into various strains and then subjected them to fluorescence spectrometry. There were only slight differences in GFP expression among the wild-type RN6390 (190 ± 30 units), sarU mutant ALC2272 (236 ± 50 units), and sarT mutant ALC1905 (243 ± 45 units), thus confirming an absence of a major autoregulatory effect by sarT. In addition, sarT regulates sarU but not vice versa.
GFP fusion assays for RNAII, RNAIII, and hla promoters in a sarU mutant.
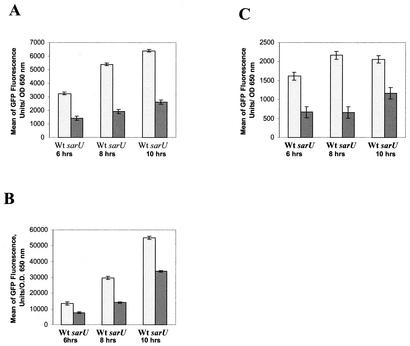
In addition to the regulation of sarU by sarT, we also found that sarU plays a regulatory role in target gene expression. In previous studies, we have found that sarT can repress the expression of RNAII and RNAIII (36). Cognizant of the SarT-mediated repression of sarU (delineated above), we hypothesized that sarT may repress RNAII and RNAIII promoter activities by down-regulating sarU. If this hypothesis were correct, one would expect RNAII and RNAIII expression to be decreased in a sarU mutant. To verify this possibility, we introduced plasmids containing an RNAII (pALC1742) or RNAIII (pALC1743) promoter linked to gfpuvr into wild-type RN6390 and the isogenic sarU mutant (ALC2272). Upon monitoring bacterial growth and serial GFP expression over a 10-h period, we found that the growth rates were comparable between these two constructs, with early stationary phase appearing after ∼6 h of growth. However, the expression of RNAII and RNAIII was significantly lower in the sarU mutant than in the parental strain (Fig. 5A and B). Recognizing that RNAIII activates hla transcription, we also explored the effect of the sarU mutation on hla promoter activity (Fig. 5C). In parallel with the RNAII and RNAIII data, the GFP-mediated fluorescence attributable to the hla promoter activity was lower in the sarU mutant than in the parental strain. Northern blotting with respective probes also confirmed the reduction in RNAII, RNAIII, and hla expression in the sarU mutant relative to that of the wild-type strain (data not shown).
FIG. 5.
GFP expression was driven by agr RNAII (A), agr RNAIII (B), and hla (C) promoters in the wild type (Wt) and in an isogenic sarU strain of S. aureus. Bar diagrams represent various time points of the growth phase after inoculation. Samples obtained at 6 h of growth represent early postexponential phase, a time point at which RNAII, RNAIII and hla promoter activities can easily be detected.
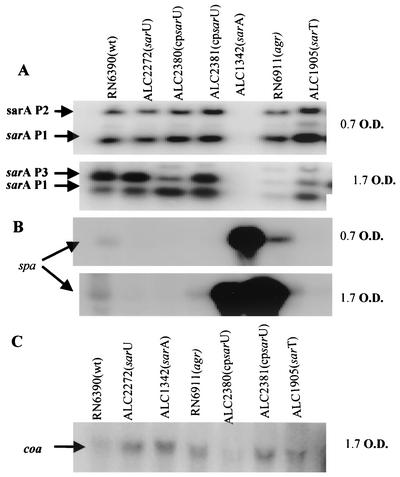
Expression of sarA, spa, and coa (coagulase) in the sarU mutant. The above data indicated that sarU positively regulates agr RNAII and RNAIII expression. To determine whether sarU also controls sarA, we assayed for sarA transcripts (sarA P2, sarA P3, and sarA P1 transcripts originating from P2, P3, and P1 promoters of the sarA locus, respectively) in the sarU mutant. Our data indicated that sarA P2 and sarA P1 transcripts expressed during the log phase were not significantly altered in the sarU mutant compared with the parental strain (Fig. 6A). However, the sarA P3 transcript appeared to be slightly increased in the sarU mutant during the early stationary phase, whereas sarA P1 and P3 transcripts were increased and decreased, respectively, in the wild type carrying sarU in a shuttle plasmid (ALC2380), and both sarA P1 and P3 transcripts were enhanced in the complemented mutant strain (ALC2381). As differential activation of the three sarA promoters leads to varying SarA protein levels, we also determined the overall sarU mutation effect on the expression of SarA, the regulatory molecule of the sarA locus. Cell extracts from parental, mutant, and complemented strains during the stationary phase were obtained, and various amounts of extracts (5, 10, and 20 μg) were analyzed by Western blotting with anti-SarA monoclonal antibody 1D1. Significant alterations in SarA expression were not observed among the wild type (RN6390), sarU mutant (ALC2272), and complemented strains (ALC2380 and ALC2381). These data suggest that, despite the mild effect of sarU on individual sarA promoters, sarU does not have a significant effect on the expression of SarA.
FIG. 6.
Northern blots of sarA, spa, and coa transcripts in the wild type, sarU mutant, trans-complemented strains, sarA, agr, and sarT mutants. A total of 10 μg of RNA was applied to each lane. RNAs extracted from the wild type (RN6390), various mutants, and complemented strains are from the exponential (OD650, 0.7) and postexponential (OD650, 1.7) phases. (A) The blot was probed with a 400-bp fragment containing the sarA open reading frame. The transcripts are the larger sarA P2 (1.2 kb), sarA P3 (0.8 kb), and the smaller sarA P1 (0.5 kb) transcripts, respectively (4). (B and C) The blot was hybridized with a 1.4-kb DNA fragment containing the spa gene (B) and a labeled 4.5-kb coa fragment of S. aureus as a probe (C).
The effect of the sarU mutation on the expression of protein A was more variable, with a slight decrease in spa transcription in the mutant (Fig. 6B), thus indicating that sarU, besides its effect on agr, may have a direct effect on the target gene as well. One effect of agr is the repression of cell wall-associated protein genes, such as that for coagulase. To ascertain the effect of sarU on coa, Northern blot analyses were conducted, revealing that the coa transcript level was higher in the sarU mutant than in the parental strain (Fig. 6C). As a control, we confirmed that coa expression was higher in the agr mutant than in the parental strain. Upon providing the sarU in trans in the parental strain (ALC2380), the transcript remained low. Curiously, complementation of the sarU mutant in trans (ALC2381) did not lead to a reduction in coa transcription as anticipated. Conceivably, this may be due to a higher gene dosage or to possible action of SarU in conjunction with other factor(s) that cannot be readily restored with the sarU fragment.
DISCUSSION
In prior studies, we have observed that SarT, a SarA homolog, represses hla expression in part by down-modulating agr RNAIII expression (36). In deciphering the S. aureus genomes (www.TIGR.org and www.ncbi.nlm.nih.gov), we found a divergently transcribed gene, sarU, adjacent to sarT. Remarkably, the transcription of sarU, not readily detected in the parental strain, was enhanced in a sarT mutant but not vice versa, indicating that sarT may repress sarU transcription. The gene product of sarU is 247 residues in length, with a molecular mass of 29,276 Da. As with many of the SarA homologs, SarU is a basic protein with a pI of 9.81. Similar to SarS (sarH1), which is a 250-residue SarA homolog, SarU can be envisioned as a molecule with two halves, with each half sharing sequence similarity to the smaller homologs such as SarA, SarR, and SarT (Fig. 1). SarU has good homology with the MarR protein of B. subtilis. The MarR proteins have been implicated in the negative regulation of multiple antibiotic resistances in gram-negative species (1). In particular, the expression of marAB, the genes encoding the antibiotic efflux pump itself, is controlled by the MarR repressor. A large number of compounds induce the transcription of the marAB operon, presumably by binding to MarR and thus preventing the binding of MarR to the marAB promoter (1).
Within the S. aureus genome, a family of proteins homologous to SarA can now be identified. We called this group of proteins the SarA protein family. Included in this family are SarA, -R, -S, -T, and -U; we are in the midst of characterizing other mutants within this protein family, including SarV (protein GI 13702067), SarX (protein GI 13700559), SarZ (protein GI 13702335), and SarY (protein GI 13702097)(unpublished data). To understand the mechanism of gene regulation in this protein family, we recently determined the crystal structure of SarR, a member of the SarA protein family (24). SarR is a dimeric structure comprising five α-helices, three β-strands, and several flexible loops (α1α2-β1α3α4-β2β3-α5). The SarR structure can also be envisioned as a three-domain structure, with a central helical core and two winged helix motifs. A short turn between α3 and α4 constitutes a typical helix-turn-helix motif (HTH) within each of the winged helix motifs. Deletion analysis indicated that the HTH motif is essential to the function of SarA, the prototypic member of the SarA protein family (7). In examining the amino acid sequence alignment (Fig. 1), the HTH region corresponds to residues 53 to 76 in the first half and to residues 178 to 201 in the second half of the SarU molecule. In the SarR dimeric structure, this region has been modeled to be the DNA binding domain that maintains contact with the major groove of DNA (24). Interestingly, many basic residues (Lys54, Lys63, and Arg69 in SarU1 and Lys179 and Arg194 in SarU2) within this region are conserved within the SarA protein family. Besides the HTH motif, the SarR structure also predicts the beta hairpin loop (β2-β3) corresponding to residues 81 to 97 in the first half and residues 206 to 212 in the second half to be another DNA-binding domain. In particular, the basic residues Lys82 (residue 207), Arg84 (residue 209), Arg90 (residue 215), and acidic residues Asp88 (residue 213) and Glu89 (residue 214) of SarU within the putative beta hairpin loop are absolutely conserved. These data are consistent with the notion that the DNA-binding domains might be highly conserved within this protein family. The structure of MarR in E. coli has recently been described; interestingly, MarR is also a dimeric structure with two winged helix motifs (1).
Despite sequence similarity among members of the SarA family, each member is also unique in its own function. This view is supported by the lesser degree of conservation within the activation domain among family members, thus accounting for diverse functions of the family members (24). For example, SarS activates the synthesis of protein A, while SarT represses the expression of α-hemolysin (11, 36). Northern and transcriptional analyses revealed that sarU likely activates agr, as reflected by lower RNAII and RNAIII expression in a sarU mutant than in the parental strain. Additionally, the expression of sarU was also enhanced during the postexponential phase in the sarT mutant, corresponding to a period when agr activation is expected to be highest. This effect on agr is independent of sarA, since the expression of SarA, the sarA regulatory molecule, was not significantly altered in the sarU mutant compared to that in the parental strain. We also confirmed that the transcription of hla, a target gene of agr, was diminished in a sarU mutant, in concordance with the decreased agr transcription in the sarU mutant relative to that in the parental strain. Moreover, the expression of one of the cell wall-associated proteins (i.e., coagulase) controlled by agr was also increased in the sarU mutant relative to the parental strain. However, the effect of the sarU mutation on protein A expression is more variable, with slightly lower expression in the sarU mutant than in the parental strain, contrary to what one would expect from reduced agr expression in a sarU mutant. This finding suggests that besides its effect on agr and the ensuing target genes, sarU may also regulate target genes via an agr-independent mechanism, possibly like SarA-dependent regulation of virulence genes (12).
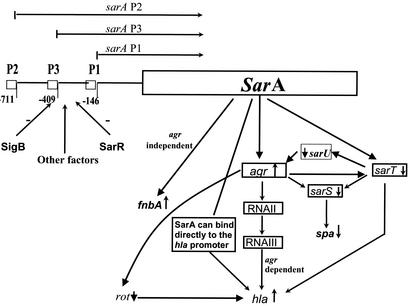
In prior studies, we have made the observation that a mutation in sarT has led to an increase in agr RNAIII expression compared to that in the parental strain. Paradoxically, sarT expression is also elevated in an agr mutant (36). Thus, the identification of sarU, a gene repressed by sarT, has provided the plausible link for a putative pathway whereby a feedback loop on agr amplification can be discerned (Fig. 7). In this scheme, activation of agr would repress sarT, leading to a moderate increase in sarU transcription, presumably occurring as a result of decreased SarT binding to the sarU promoter. Secondary activation of sarU would promote transcription of RNAII and RNAIII promoters of agr, leading to a further increase in agr transcription. This model also predicts sarU promoter activity to be lower in the agr mutant than in the parent (Fig. 7). Given the constraint that the sarU promoter was not highly active in the parent strain, we have consistently found that the fluorescence attributable to the sarU promoter was lower in the agr mutant than in the parent (Fig. 4B). Viewed in this context, activation of agr via the sarT-sarU pathway may represent an alternative mechanism to the primary quorum-sensing mechanism mediated by the cyclic autoinducing peptide (AIP). Although the RNAIII promoter cannot be activated in an agr mutant in vitro, Xiong et al. have found in a recent study that the RNAIII promoter can be activated at a significant level in an agr mutant in a rabbit endocarditis model (38). Whether the sarT-sarU activation mechanism is deployed in vivo in response to host signals in the absence of agr is not known. Nevertheless, the identification of the sarT-sarU pathway has provided a plausible mechanism to explain agr activation in vivo.
FIG. 7.
A model of the agr-sarT-sarU pathway for regulating the expression of virulence genes in S. aureus. SarA is synthesized during the exponential phase based on differential activation of three sarA promoters (sarA P2, P3, and P1). SarR and SigB can down-regulate SarA expression, and other factors may up-regulate it. The basal level of RNAII expression, in part in response to SarA, would lead to synthesis of the AIP. Upon bacterial accumulation, the AIP would activate RNAIII transcription via a quorum-sensing mechanism. Besides the quorum-sensing pathway, we also propose that RNAIII activation would lead to repression of SarT. SarT repression would result in elevated sarU and decreased sarS expression; Rot protein acts as a repressor for hla expression (27). As SarU is an activator of RNAIII synthesis, this would provide a secondary amplification of the original agr signal. Accordingly, this model predicts a reduction in RNAIII promoter activity in a sarU mutant as well as decreased sarU promoter activity in an agr mutant compared to activities in the parental strain.
The sarU and sarT genes are divergently transcribed and are separated by a 323-bp intergenic sequence between the two coding regions. Transcriptional analysis indicated that sarU expression is repressed by the sarT gene product and not vice versa. Mapping of the transcriptional start site indicated that the start site is located 90 bp upstream of the initiation codon of the sarU gene, with typical σA-dependent −10 and −35 promoter boxes. In gel shift studies, we found that purified SarT can bind to the sarU promoter region with fairly high affinity (Kd, ∼102.5 pM). As multiple protein-DNA complexes are discerned, we speculate that multiple SarT dimers may interact with each other to bind to the sarU promoter region. This is based on our modeling data showing that three SarA dimers may bind to a single 135-bp agr promoter fragment (unpublished data). Nevertheless, we cannot exclude the possibility that a single dimer may bind to multiple sites on a 148-bp sarU promoter fragment.
Given that SarT is a repressor of sarU promoter activity, we speculated that sarU is primarily transcribed in the absence of SarT. Whether other factors are involved in augmenting sarU transcription in a sarT mutant is not defined in this study. It is curious that two hybridizing bands reacted with a sarU probe that encompassed only the coding region in the sarT mutant. Complementation of the sarU mutant with a 1.5-kb fragment encompassing both the sarU gene and an ∼392-bp upstream sequence restored only the smaller hybridizing band of ∼1,200 nt, presumably representing the monocistronic form of the sarU RNA message. Complementation with a 2.3-kb fragment containing 392-bp upstream and 1,158-bp downstream sequences of sarU plus the sarU coding region did not restore the larger, ∼1,400-nt transcript (data not shown). Thus, the origin of the larger hybridizing band remains unclear. The gene immediately downstream of sarU encodes a 288-residue polypeptide, representing UTP-glucose-1-phosphate uridyltransferase (gtaB). The gtaB gene is transcribed divergently from the cDNA strand, with the stop codons of both genes being separated by 210 bp. Northern hybridization with a 2.15-kb DNA probe containing open reading frames of sarU and gtaB yielded an additional ∼1,000-nt transcript (data not shown) distinct from the ∼1.2-kb sarU RNA message. These data suggested that the gtaB gene is likely to be controlled from its own promoter and that the larger, ∼1,400-nt transcript that hybridized with the sarU probe is not part of the gtaB RNA message. Upstream of sarT-sarU is a gene that encodes a hypothetical protein (SA2285) homologous to these proteins with the serine-aspartic acid repeats (e.g., clumping factors A and B), and this gene is transcribed in the same orientation as sarT. Given the size of the transcript (∼1,400 nt), this gene is unlikely to be part of that message. Collectively, these data suggested that the larger, ∼1,400-nt transcript might be a cross-hybridizing band present in a sarT mutant. However, we did not entirely rule out the possibility that a cryptic promoter may originate upstream of sarU and that it is transcribed in opposite orientation to sarT or other genes. Clearly, the molecular architecture of the sarT-sarU loci needs to be further defined before we can clearly assign the origin of the larger transcript.
In sum, the sarU gene product participates in the activation of agr and some of its target genes (e.g., hla and coa). Additionally, sarU may also regulate target genes via an agr-independent pathway. To further define the exact function of the sarU gene product, we are in the process of constructing a double knockout of sarU and sarT; the combination of a sarU knockout together with mutations in sarA, sarR, sarS, agr, and other regulatory genes may provide a clearer understanding of the mechanism of virulence gene activation by SarU and other members of the SarA protein family.
Acknowledgments
The contribution of the S. aureus genome database at TIGR, NIH, and the University of Oklahoma to this work is gratefully acknowledged. We thank Brain Bateman and MaryBeth Maloney for their technical help. We also thank Simon Foster for strain SH1000, Katherine Schmidt for sharing results, and Willem Van Wamel for constructing some GFP fusions.
This work was supported in part by NIH grants AI37142 and AI50678 to A.L.C.
Editor: A. D. O'Brien
REFERENCES
- 1.Alekshun, M. N., S. B. Levy, T. R. Mealy, B. A. Seaton, and J. F. Head. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance at 2.3A resolution. Nat. Struct. Biol. 8:710-714. [DOI] [PubMed] [Google Scholar]
- 2.Arvidson, S., and K. Tegmark. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291:159-170. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. Wiley, New York, N.Y.
- 4.Bayer, M. G., J. H. Heinrichs, and A. L. Cheung. 1996. The molecular architecture of the sar locus in Staphylococcus aureus. J. Bacteriol. 178:4563-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blake, M. S., K. H. Johnston, G. J. Russell-Jones, and E. C. Gotschlich. 1984. A rapid sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal. Biochem. 136:175-179. [DOI] [PubMed] [Google Scholar]
- 6.Cheung, A. L. 2001. Global regulation of virulence determinants in Staphylococcus aureus, p. 295-322. In A. L. Honeyman, H. Friedman, and M. Bendinelli (ed.), Staphylococcus aureus infection and disease. Kluwer Academic Plenum Publishers, New York, N.Y.
- 7.Cheung, A. L., M. G. Bayer, and J. H. Heinrichs. 1997. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J. Bacteriol. 179:3963-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, A. L., K. Eberhardt, and V. A. Fischetti. 1994. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal. Biochem. 222:511-514. [DOI] [PubMed] [Google Scholar]
- 9.Cheung, A. L., K. Eberhardt, and J. H. Heinrichs. 1997. Regulation of protein A synthesis by the sar and agr loci of Staphylococcus aureus. Infect. Immun. 65:2243-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung, A. L., J. M. Koomey, C. A. Butler, S. J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 89:6462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung, A. L., K. A. Schmidt, B. Bateman, and A. C. Manna. 2001. SarS, a SarA homolog repressible by agr, is an activator of protein A synthesis in Staphylococcus aureus. Infect. Immun. 69:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chien, Y.-T., A. C. Manna, S. J. Projan, and A. L. Cheung. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar dependent gene regulation. J. Biol. Chem. 274:37169-37176. [DOI] [PubMed] [Google Scholar]
- 13.Chien, Y-T., and A. L. Cheung. 1998. Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J. Biol. Chem. 273:2645-2652. [DOI] [PubMed] [Google Scholar]
- 14.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcriptional profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giraudo, A. T., A. L. Cheung, and R. Nagel. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 168:53-58. [DOI] [PubMed] [Google Scholar]
- 16.Haldenwang, W. G. 1995. The sigma factors of Bacillus subtilis. Microbiol. Rev. 59:506-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji, G., R. C. Beavis, and R. P. Novick. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 92:12055-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kahl, B., M. Goulian, W. Van Wamel, M. Herrmann, S. Simon, G. Kaplan, G. Peters, and A. L. Cheung. 2000. Staphylococcus aureus RN6390 replicates and induces apoptosis in a pulmonary epithelial cell line derived from a cystic fibrosis patient. Infect. Immun. 68:5385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan, S. A., M. E. Gaskill, R. Mahmood, H. C. Wong, M. B. Johns, Jr., and J. M. Zock. 1990. Studies on the staphylococcal enterotoxin B gene, p. 289-299. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, New York, N.Y.
- 20.Kornblum, J., B. Kreiswirth, S. J. Projan, H. Ross, and R. P. Novick. 1990. Agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus, p. 373-402. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, New York, N.Y.
- 21.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, I. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kalto, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 22.Lin, W. S., T. Cunneen, and C. Y. Lee. 1994. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J. Bacteriol. 176:7005-7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lina, G., S. Jarraud, G. Ji, T. Greenland, A. Pedraza, J. Etienne, R. P. Novick, and F. Vandenesch. 1998. Transmembrane topology and histine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol. Microbiol. 28:655-662. [DOI] [PubMed] [Google Scholar]
- 24.Liu, Y., A. Manna, R. Li, W. E. Martin, R. C. Murphy, A. L. Cheung, and G. Zhang. 2001. Crystal structure of the SarR protein from Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 98:6877-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Manna, A., and A. L. Cheung. 2001. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect. Immun. 69:885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNamara, P. J., K. C. Milligan-Monroe, S. Khalili, and R. A. Proctor. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 182:3197-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morfeldt, E., K. Tegmark, and S. Arvidson. 1996. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol. Microbiol. 21:1227-1237. [DOI] [PubMed] [Google Scholar]
- 29.Novick, R. P., S. J. Projan, J. Kornblum, H. F. Ross, G. Ji, B. Kreiswirth, F. Vandenesch, and S. Moghazeh. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248:446-458. [DOI] [PubMed] [Google Scholar]
- 30.Novick, R. P. 1990. The staphylococcus as a molecular genetic system, p. 1-40. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, New York, N.Y.
- 31.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 32.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Projan, S. J., and R. P. Novick. 1997. The molecular basis of pathogenicity, p. 55-81. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human diseases. Churchill Livingstone, New York, N.Y.
- 34.Sau, S., L. Sun, and C. Y. Lee. 1997. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J. Bacteriol. 179:1614-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 94:133-138. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt, K. A., A. C. Manna, S. Gill, and A. L. Cheung. 2001. SarT: a repressor of α-hemolysin synthesis in Staphylococcus aureus. Infect. Immun. 69:4749-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tegmark, K., A. Karlsson, and S. Arvidson. 2000. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 37:398-409. [DOI] [PubMed] [Google Scholar]
- 38.Xiong, Y.-Q., W. Van Wamel, C. C. Nast, M. R. Yeaman, A. L. Cheung, and A. S. Bayer. 2002. Activation and transcription interaction between agr RNAII and RNAIII in Staphylococcus aureus in vitro and in an experimental endocarditis model. J. Infect. Dis. 186:668-677. [DOI] [PubMed] [Google Scholar]