Abstract
Cell surface proteoglycans have been implicated in many aspects of plant growth and development, but genetic evidence supporting their function has been lacking. Here, we report that the Salt Overly Sensitive5 (SOS5) gene encodes a putative cell surface adhesion protein and is required for normal cell expansion. The sos5 mutant was isolated in a screen for Arabidopsis salt-hypersensitive mutants. Under salt stress, the root tips of sos5 mutant plants swell and root growth is arrested. The root-swelling phenotype is caused by abnormal expansion of epidermal, cortical, and endodermal cells. The SOS5 gene was isolated through map-based cloning. The predicted SOS5 protein contains an N-terminal signal sequence for plasma membrane localization, two arabinogalactan protein–like domains, two fasciclin-like domains, and a C-terminal glycosylphosphatidylinositol lipid anchor signal sequence. The presence of fasciclin-like domains, which typically are found in animal cell adhesion proteins, suggests a role for SOS5 in cell-to-cell adhesion in plants. The SOS5 protein was present at the outer surface of the plasma membrane. The cell walls are thinner in the sos5 mutant, and those between neighboring epidermal and cortical cells in sos5 roots appear less organized. SOS5 is expressed ubiquitously in all plant organs and tissues, including guard cells in the leaf.
INTRODUCTION
Proper regulation of cell expansion is essential for plant growth and development. The control of cell expansion is thought to depend on cell wall architecture, cytoskele-ton, wall–membrane interactions, and interactions between neighboring cells (Kohorn, 2000; Darley et al., 2001; Martin et al., 2001). The primary wall of plant cells is composed of a network of cellulose microfibrils, which is coated by cross-linking hemicelluloses and embedded in a pectin matrix and a network of structural proteins with varying amounts of linked carbohydrates (Cosgrove, 1999). These cell wall materials are assembled into a network that is mechanically strong but flexible enough to allow cell expansion. Cellulose is made at the outer surface of the plasma membrane and exists in the form of microfibrils. Mutational analysis has demonstrated a central role of cellulose in plant morphogenesis (Arioli et al., 1998; Favery et al., 2001). In many cell types, cellulose microfibrils have been shown to be oriented perpendicular to the primary direction of expansion (Giddings and Staehelin, 1991). The cytoskeleton, cross-linking glycans, pectic polysaccharides, and glycoproteins in the cell wall may play roles in the ordered assembly of cellulose microfibrils, providing the proper extensibility of the cell wall (Cosgrove, 2001). The importance of the association between cellulose microfibrils and cross-linking glycans was indicated by the finding that expansins involved in the disruption of noncovalent bonds between the two components cause the rapid induction of wall extension (McQueen-Mason and Cosgrove, 1994). Moreover, in the presence of xyloglucan, cellulose microfibrils become organized in a way that resembles the organization of plant cell walls to a remarkable extent, suggesting that the cross-linking glycans are important organizers of cellulose microfibrils (Whitney et al., 1995).
In addition to the cellulose cross-linking glycan network and pectin matrix, primary cell walls also contain small amounts of structural proteins. Cell wall structural proteins are thought to form an independent network that assists in the assembly or restructuring of the wall. Among these proteins, the arabinogalactan proteins (AGPs) represent a family of extensively glycosylated Hyp-rich proteoglycans. AGPs are found in intercellular spaces and cell walls, on plasma membranes, and in cytoplasmic vesicles, suggesting multiple roles for AGPs in various processes associated with cell growth and plant development (Majewska-Sawka and Nothnagel, 2000). AGPs have been implicated in fertilization, embryogenesis, cell–cell interaction, cell proliferation, and cell expansion (for review, see Showalter, 2001).
AGPs often contain >90% carbohydrates and a core protein backbone usually enriched in Hyp, Ala, Ser, and Thr. The recent identification of 15 genes encoding protein backbones of classic AGPs has improved our understanding of the protein moieties and the expression of the corresponding genes (Schultz et al., 2000). Typically, classic AGPs contain at least three distinct domains: an N-terminal signal sequence for secretion, a Pro/Hyp-rich domain, and a C-terminal hydrophobic transmembrane domain that is absent and replaced by a glycosylphosphatidylinositol (GPI) lipid anchor in the mature proteins. The processing of a GPI anchor has been identified in all known and putative classic AGPs and has been shown to be important in anchoring the proteins to the plasma membrane (Youl et al., 1998; Oxley and Bacic, 1999; Sherrier et al., 1999; Svetek et al., 1999, Zhao et al., 2002). A hydrophobic C-terminal domain for GPI modification is not present in any known nonclassic AGPs, which do not all share a common domain structure. Except for anchoring the proteins to the plasma membrane, the function of the GPI anchor is not fully understood. The recent isolation of the COBRA gene has shown the importance of a GPI anchor protein in cell expansion (Schindelman et al., 2001).
Numerous studies have used monoclonal antibodies that react with the carbohydrate epitopes of AGPs or the Yariv reagent to examine AGP expression and potential function (for review, see Knox, 1997). These studies have generated valuable information regarding the tissue- and cell-specific expression patterns of AGPs. The use of Yariv reagent to perturb AGPs in living cells and seedlings has suggested roles for AGPs in cell division, cell expansion, and programmed cell death (Majewska-Sawka and Nothnagel, 2000). However, because of the lack of plant mutants defective in AGPs, few of the proposed functions have been established. In Arabidopsis, the AGPs belong to a superfamily of proteins encompassing several subclasses (Schultz et al., 2002). Although some DNA insertion mutants have been identified for AGP core protein genes, finding phenotypes for AGP mutants remains a major challenge (Schultz et al., 2002). Some mutations were shown to cause decreased AGP contents (Takahashi et al., 1995; Ding and Zhu, 1997), but none of the mutants has been shown to have lesions in AGP or AGP-like genes.
Here, we report the identification and characterization of the Arabidopsis salt overly sensitive5 (sos5) mutant and the isolation of SOS5 as a gene encoding a putative cell surface adhesion protein with AGP-like domains and fasciclin-like domains. sos5 was isolated using the root-bending assay (Wu et al., 1996). In response to salt stress, the root tips of sos5 plants swell and root elongation is arrested. The root tips of sos5 plants have abnormal cell expansion, thinner walls, and reduced middle lamella. Our results suggest that SOS5 plays an important role in cell expansion in Arabidopsis.
RESULTS
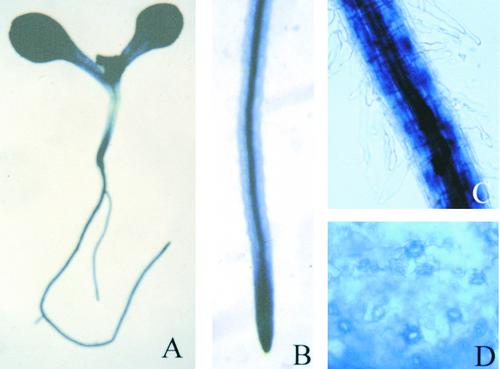
Isolation of the sos5 Mutant
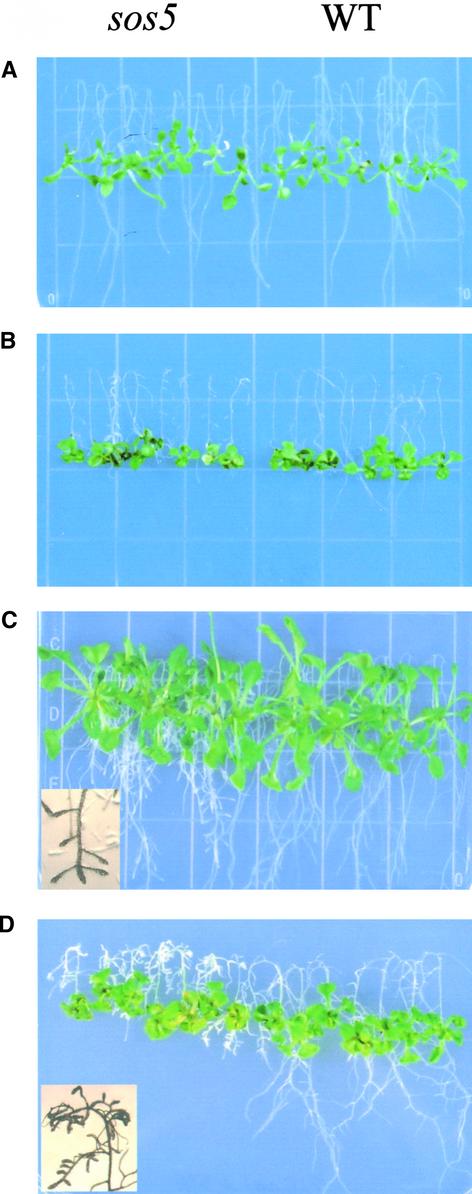
sos5 was identified from an ethyl methanesulfonate–mutagenized M2 population in a screen for sos mutants by the root-bending assay on 100 mM NaCl–containing agar medium (Shi et al., 2002). Figure 1 shows the phenotype of sos5 mutant seedlings. When grown on Murashige and Skoog (1962) (MS) nutrient medium for 1 week, sos5 seedlings were indistinguishable from wild-type seedlings (Figure 1A). Under NaCl stress, root growth in sos5 was more inhibited than that in the wild type. The root tips of the NaCl-treated sos5 seedlings were visibly swollen (Figure 1B). The shoot part of sos5 seedlings did not appear to be significantly more sensitive to NaCl stress than the wild-type shoot (Figures 1B and 1D). With prolonged growth on MS nutrient medium without NaCl supplementation, sos5 mutant roots appeared fatter and the root tips became swollen, although the overall growth of sos5 plants was similar to that of wild-type plants (Figure 1C).
Figure 1.
Root-Bending Assay of Wild-Type and sos5 Mutant Seedlings.
Four-day-old seedlings grown on MS agar medium were transferred to MS agar medium without NaCl (A) and (C) or with 100 mM NaCl (B) and (D). WT, wild type.
(A) and (B) One week after transfer.
(C) and (D) Two weeks after transfer. Insets show the enlarged root tip region displaying swollen root tips.
The sos5 mutant was crossed to other sos mutants (sos1, sos2, sos3, and sos4) identified in our laboratory. All of the resulting F1 plants showed a wild-type phenotype under NaCl stress (data not shown), suggesting that sos5 defines a new locus. An ∼3:1 segregation of wild-type to sos5 mutant phenotypes was observed in the F2 progeny resulting from a backcross (data not shown), which indicates that sos5 is a recessive mutation in a single nuclear gene.
Root Growth of sos5 Plants Is Hypersensitive to Salt Stress
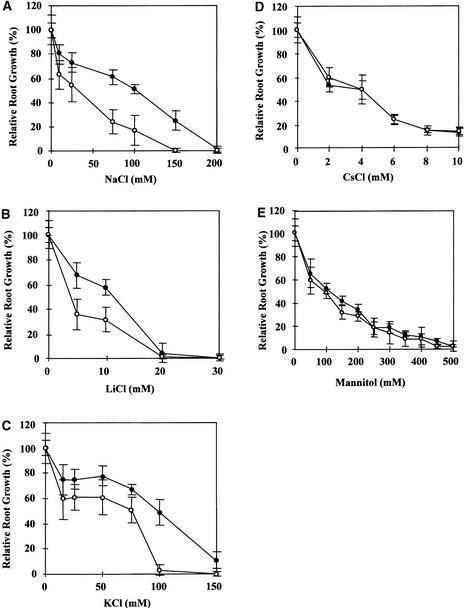
As a convenient indicator of root growth, root elongation was measured to determine the effects of various salts on sos5. Figure 2 shows that root elongation in sos5 was hypersensitive to NaCl, LiCl, and KCl but not to CsCl. This finding is distinct from the results with sos1, sos2, and sos3, which are specifically hypersensitive to sodium and lithium salts but not to potassium salts (Wu et al., 1996; Liu and Zhu, 1997; Zhu et al., 1998). When grown on MS medium supplemented with 100 mM KCl, the root tips of sos5 seedlings also swelled (data not shown), suggesting that high concentrations of NaCl and KCl have similar effects on sos5 roots. Although the hypersensitivity of sos5 root growth to both NaCl and KCl suggests a potential general osmotic effect, measurement of root elongation in response to different concentrations of mannitol indicated otherwise. Figure 2E shows that sos5 root growth was not hypersensitive to osmotic stress caused by mannitol. The root tips of sos5 seedlings also did not show a swollen phenotype in response to high concentrations of mannitol (data not shown).
Figure 2.
Sensitivity of sos5 Root Elongation to Various Salts and Osmotic Stress.
(A) Sensitivity to NaCl.
(B) Sensitivity to LiCl.
(C) Sensitivity to KCl.
(D) Sensitivity to CsCl.
(E) Sensitivity to mannitol.
Closed circles, wild type; open circles, sos5. Values shown are averages ± se (n = 15).
Abnormal Cell Expansion in sos5 Plants
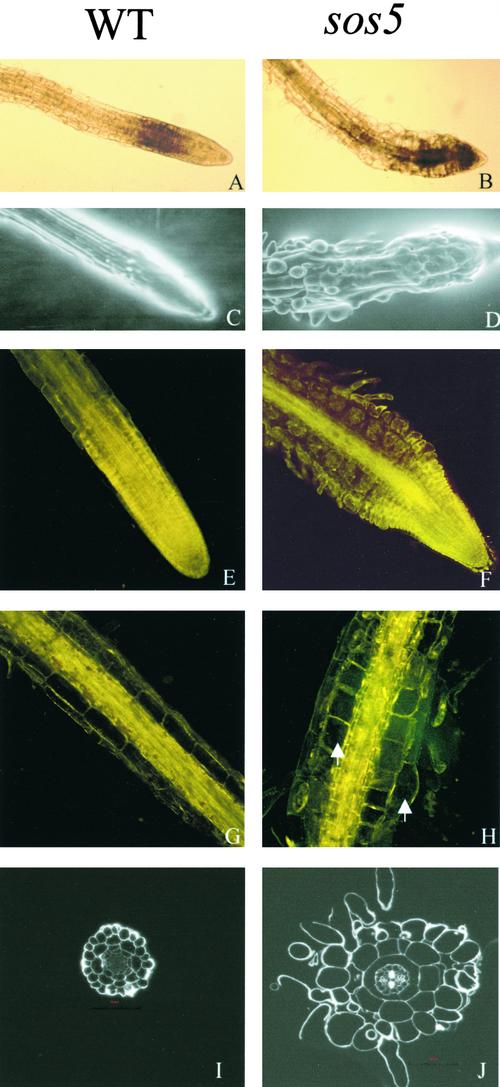
Cell expansion is tightly controlled during plant growth and development. Even under NaCl stress, the roots of wild-type seedlings exhibited highly organized cell files and defined cell shapes (Figure 3A). However, the root tips of sos5 mutant seedlings displayed abnormal cell expansion under NaCl stress (Figure 3B). The tissue organization and epidermal cell files in sos5 mutant roots became disrupted, whereas the integrity of wild-type root tips remained intact after 100 mM NaCl treatment for 1 week (Figures 3C and 3D). Because the severe inhibition of root growth and the disruption of root tip structure in salt-stressed sos5 plants make it difficult to determine which cell layer(s) undergoes abnormal cell expansion, we used the feature that without salt stress, sos5 roots also show a swollen phenotype when cultured for long periods of time. After growth on MS nutrient medium for 2 to 3 weeks or longer, sos5 plants exhibited fatter and abnormally shaped root tips compared with wild-type plants (Figures 3E and 3F). Microscopic observations revealed that both epidermal cells and cortical cells in the root tips of sos5 expanded irregularly (Figure 3F). Importantly, increased extracellular spaces between cortical cells, between cortical and epidermal cells, and between epidermal cells were observed in sos5 (Figure 3F). This observation indicates that the sos5 mutation may affect cell-to-cell adhesion. sos5 mutant plants also had thicker (in diameter) mature roots than wild-type plants under normal growth conditions. This finding is attributable to the greater radial expansion of cortical cells and some epidermal cells (Figures 3G and 3H).
Figure 3.
Root Tip Swelling in the sos5 Mutant.
(A), (C), (E), (G), and (I) Wild-type (WT) roots as controls.
(B), (D), (F), (H), and (J) sos5 mutant roots.
(A) and (B) Light microscopy images of root tips 3 days after transfer from MS agar medium to MS agar medium supplemented with 100 mM NaCl.
(C) and (D) Scanning electron microscopy images of root tips treated for 5 days with 100 mM NaCl.
(E) to (H) Confocal images of root tips and mature primary roots grown on the surface of MS agar (0.8%) medium for 3 weeks. Arrows point to cells with abnormal expansion.
(I) and (J) Thin cross-sections at 0.5 cm away from the apex of roots grown on the surface of MS agar (0.8%) medium for 3 weeks.
Each wild-type and corresponding sos5 image has the same magnification.
To determine whether structural alterations occurred and to localize more precisely the cell layers of abnormal cell expansion in sos5 roots, root cross-sections were examined. The organization of tissue layers and cell files in sos5 roots was similar to that of wild-type roots (Figures 3I and 3J). Abnormal cell expansion was observed in the cells of epidermis, cortex, and endodermis (Figure 3J).
We noticed that when plants were grown in MS nutrient medium without salt stress, the root-swelling phenotype of sos5 was position dependent. Roots grown on an agar surface displayed the most severe swelling in both the root tip and the mature root. Swelling also was observed when sos5 roots grew at the interface between the agar medium and the plastic Petri dish, although the swelling was not as strong as when the roots grew on the agar surface and were exposed to air. Interestingly, root swelling was not observed when sos5 roots grew inside the agar. These observations suggest that the abnormal cell expansion in sos5 roots might be caused by a weakening of the cell wall, because external physical constraints could prevent or limit root swelling.
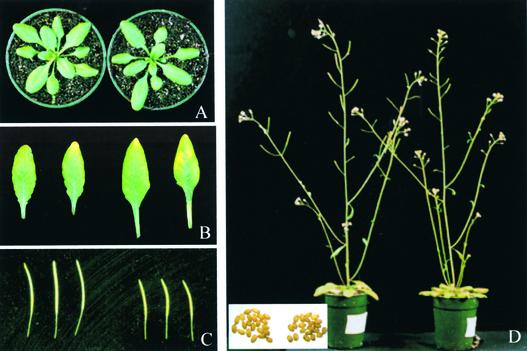
sos5 mutant plants can grow, develop, and set seeds as wild-type plants do (Figure 4). Under normal growth conditions, the rate of growth and development of sos5 plants was similar to that of wild-type plants. During vegetative growth in soil, sos5 rosette-stage plants had slightly larger leaves with longer petioles than wild-type plants (Figures 4A and 4B). Although the reproductive development of sos5 plants generally was similar to that of wild-type plants (Figure 4D), sos5 plants produced shorter siliques (Figure 4C). No significant difference in seed size was observed between sos5 and the wild type (Figure 4D), although the sos5 mutant produced fewer seeds per silique (data not shown).
Figure 4.
Morphological Phenotypes of sos5 Aerial Parts.
(A) Rosette-stage plants.
(B) First and second true leaves.
(C) Siliques.
(D) Plants with inflorescences. The inset shows seeds.
Left, wild type. Right, sos5 mutant.
Positional Cloning of SOS5
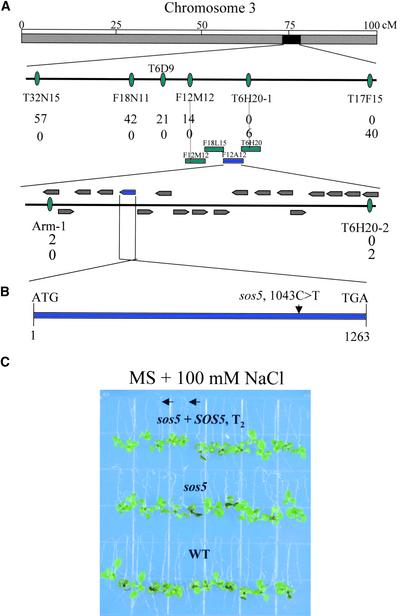
To understand the mechanism of SOS5 function in cell expansion, a map-based cloning approach was used to isolate the SOS5 gene. A segregating F2 family was generated from a cross between sos5 mutants in the Columbia background and wild-type plants in the Landsberg background. A total of 807 sos5 mutant plants were selected from the F2 population, and DNA was extracted from each plant for genetic mapping. SOS5 was mapped initially to chromosome 3 between the simple sequence length polymorphism (SSLP) markers MSJ3 and F28P10. Further mapping localized the SOS5 locus to the low arm of chromosome 3 between markers T32N15 and T17F15 (Figure 5A). Fine mapping with SSLP markers within this chromosomal section narrowed the SOS5 locus to an ∼70-kb region within BAC clone F12A12 (Figure 5A). Within this region, 18 putative genes were predicted from the TAIR database. To find the mutation in sos5, all 18 open reading frames in this region were amplified and sequenced. A single base pair mutation from C to T was found in the sos5 mutant in the hypothetical gene At3g46550 that was annotated by the Arabidopsis Genome Initiative (http://www.arabidopsis.org/info/agi.html) as encoding an endosperm specific protein-like protein (Figure 5B). No other mutation was detected in any of the open reading frames in this region of the sos5 mutant.
Figure 5.
Positional Cloning of the SOS5 Gene.
(A) SOS5 is located on the low arm of chromosome 3. The SSLP markers for fine genetic mapping and the number of recombinant genes identified from 1614 chromosomes for the respective loci are indicated. The contigs and putative genes were assembled based on information in the Arabidopsis database (http://www.arabidopsis.org).
(B) SOS5 is an intronless gene. The location of the sos5 mutation is indicated.
(C) Complementation of sos5 by the wild-type SOS5 gene. Four-day-old seedlings of the wild type, sos5, and sos5 transformed with the wild-type SOS5 gene (T2) grown on MS agar medium were transferred to MS agar medium containing 100 mM NaCl in a root-bending assay. The photograph was taken 1 week after the transfer. Arrows indicate sos5 mutants that were segregated from the T2 population.
To demonstrate that the mutation in At3g46550 is responsible for the sos5 mutant phenotype, we transformed sos5 mutant plants with an ∼2.8-kb genomic fragment that includes the entire At3g46550 gene. Twenty independent T1 transgenic lines were selected. An examination of T2 plants produced from these transformants showed that the T2 progeny segregated for wild-type and sos5 mutant phenotypes on MS medium containing 100 mM NaCl (Figure 5C). Analysis of the transgene by PCR confirmed the cosegregation of the transgene with the wild-type phenotype. This result confirms the notion that At3g46550 is the SOS5 gene.
SOS5 Encodes a Putative Adhesion Protein
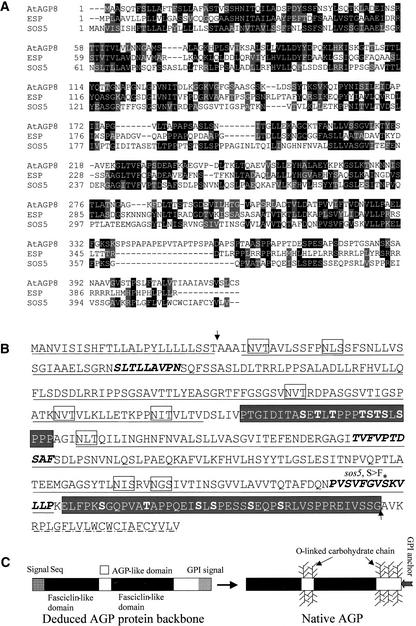
SOS5 cDNA was cloned by reverse transcriptase–PCR. Alignment of the cDNA with the corresponding genomic sequence revealed that SOS5 is an intronless gene (Figure 5B), which is consistent with the computer-based annotation by the Arabidopsis Genome Initiative. SOS5 is predicted to encode a polypeptide of 420 amino acid residues that is abundant in Ser (13.8%), Leu (13.8%), Val (9.8%), Pro (8.1%), Thr (8.1%), and Ala (7.6%). BLASTP (Basic Local Alignment Search Tool) searches revealed that SOS5 shares 32% identity and 48% similarity over a stretch of 260 amino acid residues with AtAGP8, an AGP recently identified biochemically (Figure 6A) (Schultz et al., 2000). SOS5 also shares 26% identity and 42% similarity with an endosperm-specific protein from maize over a stretch of 331 amino acid residues (Figure 6A) (Carlson and Chourey, 1997). Furthermore, SOS5 shows substantial similarities with several cell-to-cell adhesion proteins from nonplant organisms. For instance, over a stretch of 357 amino acid residues, SOS5 shares 21% identity and 36% similarity with an osteoblast-specific factor 2–related protein from Deinococcus radiodurans (data not shown). From amino acid residues 207 to 331 of SOS5, there is 23% identity and 48% similarity with an RGD-CAP protein from Gallus gallus. SOS5 also shows 28% identity and 48% similarity with a putative cell adhesion protein, Sym32, from Anthopleura elegantissima.
Figure 6.
SOS5 Encodes a Putative Adhesion Protein Containing AGP-Like Domains and Fasciclin-Like Domains.
(A) Alignment of the deduced amino acid sequence of SOS5 with Arabidopsis AtAGP8 (Schultz et al., 2000) and maize endosperm-specific protein (ESP) (Carlson and Chourey, 1997). Amino acids identical in at least two proteins are highlighted in black, and conservative substitutions are highlighted in gray. The sequences were aligned using the program CLUSTAL W (http://searchlauncher.bcm.tmc.edu/multi-align/Options/clustalw.html).
(B) Predicted primary structure and post-translational modifications. The fasciclin-like domains are underlined with solid lines. The AGP-like domains are shown in white letters on a gray background. The O-linked glycosylation of Ser and Thr residues was predicted by the program NetOGlyc 2.0 (http://www.cbs.dtu.dk/services/NetOGlyc/) and is shown in boldface within the AGP-like domains. Motifs within the fasciclin-like domains that are highly conserved are shown in boldface italics. Potential sites for N-linked glycosylation are boxed. The N-terminal signal sequence is underlined with dots. The C-terminal hydrophobic GPI anchor signal sequence was predicted by both the big-PI Predictor (http://mendel.imp.univie.ac.at/sat/gpi/gpi_server.html) and DGPI (http://129.194.186.123/GPI-anchor/DGPI_demo_en.html) programs and is underlined with alternatively arranged lines and dots. Arrows indicate predicted cleavage sites for signal peptides. The asterisk indicates the single amino acid substitution in the sos5 mutant.
(C) Scheme of SOS5 structure before and after processing and post-translational modification. After the N-terminal signal sequence is removed, the C-terminal GPI anchor signal is recognized, cleaved, and replaced by a GPI anchor. Pro residues are hydroxylated to Hyp. The O-linked carbohydrate chains are added to the AGP-like domains.
SOS5 contains two alternatively organized fasciclin-like domains and two putative AGP-like domains (Figure 6B). The AGP-like domains are rich in Hyp residues for the addition of O-linked arabinogalactan chains. Putative O-linked Ser and Thr glycosylation sites also are predicted to be localized predominantly in the AGP-like domains (Figure 6B). Like classic AGPs, SOS5 is predicted to have a N-terminal signal peptide for plasma membrane localization and a C-terminal signal for the addition of the GPI anchor (Figure 6B). Therefore, the deduced SOS5 protein backbone contains an N-terminal signal sequence, two AGP-like domains, two fasciclin-like domains, and a GPI anchor signal sequence (Figure 6C). After post-translational processing, the N-terminal signal peptide and the C-terminal GPI anchor signal would be cleaved from the protein, and the mature SOS5 protein would contain two fasciclin-like domains, two AGP-like domains with attached carbohydrate chains, and a GPI anchor to the C terminus (Figure 6C).
The C-to-T nucleotide substitution in the sos5 mutant is predicted to change the polar Ser-348 to the hydrophobic Phe in the junction region between the second fasciclin-like domain and the second AGP-like domain. This region is highly conserved among AGP-like proteins (Figure 6A) and has been proposed to be a subdomain within the fasciclin-like domain (Figure 6B) (Schultz et al., 2000). The sos5 mutant phenotypes clearly indicate a critical role for Ser-348 in the proper function of SOS5.
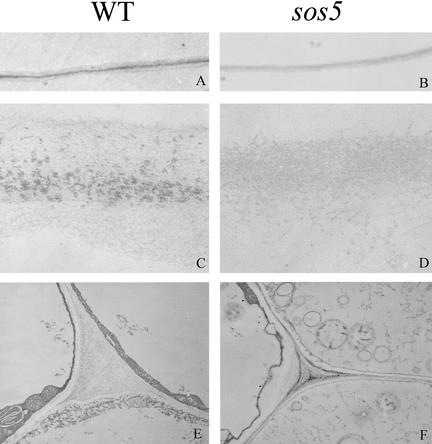
The sos5 Mutation Alters Cell Wall Structure
AGPs have been proposed to play multiple roles in plant development, including roles in cell expansion and cell-to-cell adhesion (Majewska-Sawka and Nothnagel, 2000; Showalter, 2001). Because SOS5 has some sequence similarity to AtAGP8 and some cell-to-cell adhesion proteins from nonplant organisms and sos5 mutant roots show abnormal cell expansion, we investigated whether SOS5 is important for the ultrastructure of the cell wall. Under low magnification, cell walls of neighboring epidermal cells and cortical cells in the wild type showed three distinct layers: two layers with low electron opacity separated by a middle lamella with high electron opacity (Figure 7A). However, these wall layers could not be distinguished in the sos5 mutant (Figure 7B). Under high magnification, the middle lamella in the wild type appeared as particulate materials between two fibrous primary cell walls (Figure 7C). The high-electron-opacity middle lamella presumably consists of polysaccharides of interstitial adhesive matrix. In the sos5 mutant, the middle lamella was missing and the primary cell walls appeared less organized than those of the wild type (Figure 7D). In addition, the cell walls in sos5 were thinner than those in the wild type (Figures 7C to 7F). In the tips of roots treated with 100 mM NaCl, the cell membrane appeared less tightly attached to the wall in sos5 than in the wild type (Figures 7E and 7F).
Figure 7.
Changes in Cell Wall Structure in the sos5 Mutant.
All pictures are transmission electron microscope images from ultrathin sections at 0.5 to 1.0 cm away from the root apex. The cell walls shown in (A) to (D) are those bordering epidermal and cortical cells. (A), (C), and (E) show the wild type (WT), and (B), (D), and (F) show the sos5 mutant. Each wild-type and corresponding sos5 image has the same magnification.
(A) and (B) Low-magnification images of cell walls.
(C) and (D) High-magnification images of cell walls.
(E) and (F) Membrane-wall interfaces and cell corners.
Expression Pattern of SOS5
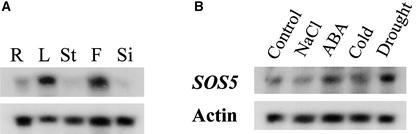
To determine the expression pattern and regulation of the SOS5 gene, we performed RNA gel blot analysis. The SOS5 transcript was detected in roots, leaves, stems, flowers, and siliques, with relatively higher abundances in leaves and flowers (Figure 8A). Because of our general interest in abiotic stress responses, we tested the influence of ABA and several stresses on SOS5 expression. The level of SOS5 transcript in young seedlings was upregulated slightly by ABA, cold, and drought treatments but remained unchanged after NaCl treatment (Figure 8B). To examine the tissue and cell distribution of SOS5 gene expression, we performed SOS5 promoter–β-glucuronidase (GUS) analysis. An ∼1.3-kb promoter region upstream of the ATG start codon of SOS5 was fused to the GUS reporter gene, and the resulting construct was introduced into wild-type Arabidopsis plants. Strong GUS expression was detected throughout the transgenic seedlings (Figure 9A). The strong GUS staining at the root tip (Figure 9B) is consistent with the root tip swelling phenotype of the sos5 mutant. In mature roots, strong GUS staining was detected in cortical cells and vascular tissues, whereas relatively weaker GUS expression was seen in epidermal cells and root hairs (Figure 9C). Interestingly, a high level of GUS expression was seen in guard cells in the leaf (Figure 9D). Although the ubiquitous GUS expression pattern is consistent with our RNA gel blot results, we note that mRNA in situ hybridization was not performed to confirm the GUS reporter results.
Figure 8.
RNA Gel Blot Analysis of SOS5 Expression.
A DNA fragment corresponding to the second fasciclin-like domain of SOS5 was used as a probe.
(A) Expression of SOS5 in different plant parts. R, root; L, leaf; St, stem; F, flower; Si, silique.
(B) SOS5 expression under different stress conditions.
Treatments are as follows: control, no treatment; NaCl, 300 mM NaCl for 5 h; ABA, 100 μM ABA for 3 h; cold, 0°C for 24 h; and drought, dehydration for 30 min.
Figure 9.
SOS5 Promoter–GUS Analysis in Arabidopsis Transgenic Seedlings.
(A) A young seedling showing strong GUS staining throughout the plant.
(B) A young root showing strong GUS expression at the root tip.
(C) Mature root region.
(D) A leaf showing strong expression of GUS preferentially in guard cells.
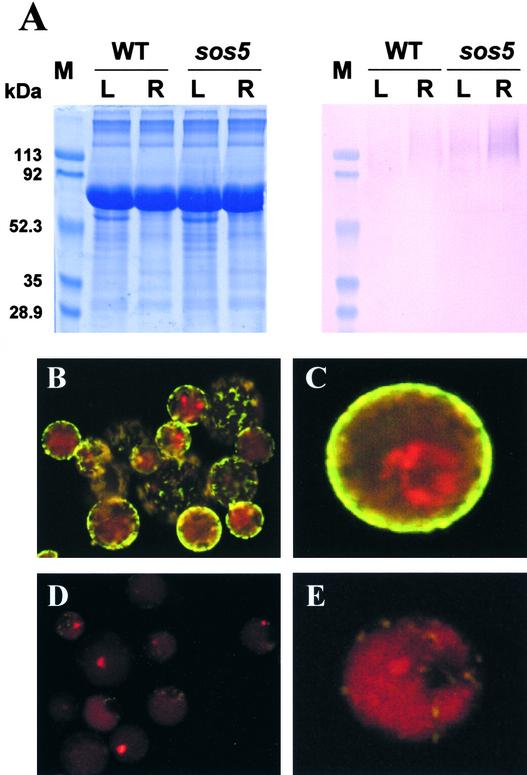
Immunological Characterization of SOS5
Polyclonal antisera against SOS5 were produced by injecting rabbits with synthetic peptides (5′-DFLSDSDLRRIP-PSGS-3′) corresponding to the first fasciclin-like domain of SOS5. The polyclonal antiserum decorated a diffuse, slow-moving band in SDS-PAGE of Arabidopsis leaf and root extracts (Figure 10A), whereas preimmune serum did not detect any signal (data not shown). Broad bands on SDS–poly-acrylamide gels are very typical of proteoglycans, especially of plasma membrane AGPs (Pennell et al., 1989; Komalavilas et al., 1991). The smear signal was stronger in the root extract than in the leaf extract. The higher level of SOS5 in the root suggests possible post-transcriptional regulation, because the SOS5 mRNA was less abundant in the root than in the leaves (Figure 8). In sos5 mutant seedlings, the SOS5 signal still was present and at higher levels than in the corresponding wild-type samples (Figure 10A). The diffuse and slow-moving signal suggests that SOS5 might be heavily glycosylated. These results also suggest that the sos5 mutation impairs the function of SOS5 without reducing the protein level.
Figure 10.
Immunological Detection of the SOS5 Protein.
(A) Immunoblot analysis of leaf and root extracts with anti-SOS5 antiserum. Left, Coomassie blue–stained gel; right, immunoblot probed with anti-SOS5 antiserum. L, leaf; M, molecular mass markers; R, root; WT, wild type.
(B) and (C) Immunofluorescence labeling of intact Arabidopsis protoplasts with anti-SOS5 antiserum.
(D) and (E) Immunofluorescence labeling with preimmune serum as a control.
For (B) to (E), the green fluorescence indicates the presence of SOS5, and the red fluorescence shows the presence of chloroplasts.
SOS5 was detected readily by immunofluorescence labeling of intact Arabidopsis protoplasts with the SOS5 antiserum but not with the preimmune serum (Figures 10B to 10E). The presence of SOS5 on the outer surface of the plasma membrane is consistent with its role in cell wall structure and cell expansion and with the sequence-based prediction that it is a GPI-anchored protein (Figure 6).
DISCUSSION
The root-bending assay has been very useful for the isolation of salt-hypersensitive mutants of Arabidopsis, which has led to the elucidation of a signaling pathway for ion homeostasis and salt tolerance (Zhu, 2000). Here, we report a novel salt-hypersensitive mutant, sos5, identified from a screen for sos mutants by paying special attention to the alteration of root morphology after salt treatment. The root growth of sos5 seedlings is arrested under high salt stress because of abnormal cell expansion at the root tip (Figure 1). Even without salt stress, prolonged culture of sos5 seedlings on agar medium results in abnormal cell expansion and defective cell-to-cell adhesion (Figure 3). SOS5 was cloned and found to encode a putative GPI-anchored protein with similarities to AtAGP8 and cell-to-cell adhesion proteins (Figure 6). Immunological characterization suggests that SOS5 probably is highly glycosylated and located on the outer surface of the plasma membrane, although it also may be present in other cellular compartments (Figure 10). Microscopic characterization suggests that SOS5 is required for the formation of proper cell wall structure and for cell expansion (Figure 7).
Another interesting phenotype of sos5 mutant plants is their shorter siliques. The siliques in sos5 contain fewer seeds than those of wild-type plants, but the size of sos5 seeds is similar to that of wild-type seeds (Figure 4). This phenotype implies that SOS5 also is important in plant reproduction. SOS5 has high sequence similarity to an endosperm-specific protein from maize (Figure 6A) and to some pollen surface proteins (data not shown), suggesting a possible role for SOS5 in pollination and/or embryogenesis. Some AGPs also have been shown to play important roles in plant reproduction (Wu et al., 2001).
SOS5 encodes a protein that has an overall primary structure similar to that of AtAGP8 but is distinct from other classic and nonclassic AGPs (Du et al., 1996; Schultz et al., 2000). SOS5 has an N-terminal signal sequence, two AGP-like domains rich in Pro, Ala, Ser, and Thr, two fasciclin-like domains, and a C-terminal GPI anchor signal (Figures 6B and 6C). The AGP-like domains in SOS5 are rich in Pro, Ala, Ser, and Thr but are divergent in sequences from those in AtAGP8. The predicted O-linked glycosylation sites for Ser and Thr are present mainly in the two AGP-like domains (Figure 6B). These glycosylation sites, together with the putative sugar attachments to Hyp residues in the two AGP-like domains, suggest that SOS5 is a highly glycosylated protein. Indeed, the electrophoretic feature on SDS–polyacrylamide gels is consistent with SOS5 being a proteoglycan.
Importantly, the two fasciclin-like domains strongly suggest a role for SOS5 in cell adhesion. Fasciclin domains are present in proteins that are known to function as adhesion molecules in animals, insects, algae, and microbes (Kawamoto et al., 1998). One motif within the second fasciclin domain, TVFVPTDSAF, not only is highly conserved in the three plant AGPs (Figure 6A) but also is highly similar to the conserved repeat domain H1 found in fasciclin-like proteins, which has the consensus sequence 5′-TV(L)F(L)A(V)PT-(S)N(D)-AF(W)-3′ (Kawamoto et al., 1998). The presence of fasciclin-like domains in SOS5 suggests that mature SOS5 proteins may aggregate together or interact with other cell wall proteins to form an independent network. This network would contain large amounts of attached polysaccharides, which may interact with the cellulose microfibril network and/or the pectin network to make up a finely structured cell wall. TTS, an AGP located predominantly in the intercellular matrix of the stylar transmitting track, has been reported to oligomerize in vitro and suggested to be highly adhesive (Cheung et al., 1995). Constitutive expression of the SOS5 coding region in Escherichia coli caused severe inhibition of bacterial colony formation (data not shown), conceivably as a result of a disruption of cell adhesion that is essential for the colonization of E. coli cells.
Severe swelling in sos5 plants often was observed in the elongation zone at the root tip. It is known that Ca2+ concentrations are kept quite low in meristems and elongating cell walls (Carpita and McCann, 2000). The low concentration of calcium possibly means less Ca2+-mediated cross-linking of pectin chains and consequently high cell wall extensibility. The abnormal cell expansion caused by salt stress possibly is caused by a further weakening of the Ca2+-mediated cross-linking of pectins by salts. Soil-grown sos5 plants have longer leaf petioles (Figure 4B) and fatter inflorescent stems (data not shown), suggesting that SOS5 not only is important in root cells but also is functional in leaf and stem cells. This conclusion is consistent with the expression pattern of SOS5 revealed by both RNA gel blot and promoter-GUS analyses. The SOS5 gene is expressed ubiquitously in all organs and tissues tested (Figures 8 and 9). Because the cell wall of guard cells is subjected to strong forces during frequent and rapid swelling and contraction for stomatal opening and closing, the preferentially strong expression of the SOS5 gene in this highly specialized cell type suggests that AGPs may contribute to the special physical properties of guard cell walls.
METHODS
Isolation of the sos5 Mutant and Genetic Analysis
The isolation of the sos5 mutant of Arabidopsis thaliana was performed as described previously (Wu et al., 1996; Shi et al., 2002). Briefly, ethyl methanesulfonate–mutagenized M2 seeds were surface-sterilized and sown in rows onto medium containing Murashige and Skoog (1962) (MS) salts, 3% Suc, and 1.2% agar, pH 5.7. To identify mutants, 4-day-old seedlings were transferred to the same medium supplemented with 100 mM NaCl for root-bending assays (Wu et al., 1996). The sos5 mutant was identified as a plant arrested in root elongation and altered in root cell expansion after NaCl treatment. Mutant seedlings were transferred to pot medium (Metro Mix 350, Scotts-Sierra Horticultural Products, Marysville, OH) and grown to maturity. Mutant plants were backcrossed at least four times to wild-type plants to remove other mutations. sos5 mutants also were crossed to other sos mutants for allelic tests.
Growth Measurements
Four-day-old wild-type and sos5 mutant seedlings grown on vertical MS agar plates were transferred to MS agar plates supplemented with various salts or mannitol. The plates were placed vertically, and the root tip of each seedling was marked. Root elongation was measured 1 week after the transfer. Relative root growth was calculated as a percentage relative to that on MS medium.
Genetic Mapping
sos5 mutant plants (Columbia ecotype) were crossed to Landsberg wild-type plants. A total of 807 sos5 mutant plants were selected from the selfed F2 population by the root-bending assay (Wu et al., 1996) combined with the identification of the root-swelling phenotype. DNA was isolated from individual mutant plants and analyzed for recombination events with simple sequence length polymorphism (SSLP) markers. The following primer pairs for SSLP markers that are polymorphic between Columbia and Landsberg were developed. MSJ3: forward, 5′-GAAACGCAAGAGATTCGACG-3′; reverse, 5′-CCATCGGAAATTGTGAATGC-3′. K11J14: forward, 5′-AGAGGAAGA-AGAAAGCTTGC-3′; reverse, 5′-GTGGGCCCACTTATATCACC-3′. T32N15: forward, 5′-TGAAAATCCTTGCGTGAGTG-3′; reverse, 5′-GACGAATAGTGAAAGGAGAG-3′. F18N11: forward, 5′-TGATTCGCA-GGTTTCTCGTC-3′; reverse, 5′-ACCCGACATGCTTGTGAAAC-3′. T6D9: forward, 5′-CTTTTCAGGTCAAAACCTCTG-3′; reverse, 5′-CCAAAGACCAAAGAGACACG-3′. F12M12: forward, 5′-CACCGA-TTGAAATACGACTG-3′; reverse, 5′-TATCGTAAGGTACATGCCAC-3′. Arm: forward, 5′-GCGTTCGTATGTCTATACATAG-3′; reverse, 5′-GTTCACTTCAACCTCAGCTTC-3′. T6H20-2: forward, 5′-GTGGAGCAACGAAGTTGATG-3′; reverse, 5′-ACATGTGAAGAAGGAGAC-CC-3′. T6H20-1: forward, 5′-TGCATTGGTTTCTCTGCTTG-3′; reverse, 5′-CCATGAATATCACCTGCAAC-3′. T17F15: forward, 5′-ATCCCACATTCGTTCTTCGC-3′; reverse, 5′-CATACTATATGGATGCATTTGG-3′. F18B3: forward, 5′-CTTGCGTGTGTGTAACTAAAG-3′; reverse, 5′-ACGGTCAGATTGAGTGATTC-3′. F28P10: forward, 5′-CGATCCAAGTGGAACCAACC-3′; reverse, 5′-TTAGAGGACATT-CCGGTTCG-3′.
Molecular Techniques
To identify the sos5 mutation within the ∼70-kb region between SSLP markers Arm-1 and T6H20-2, the coding regions of all 18 predicted putative genes in this region were amplified and sequenced. Both strands of overlapping fragments for each gene were sequenced. Mutation was further confirmed by sequencing 16 individual sos5 mutant plants and the cDNAs from both mutant and wild-type plants.
cDNA containing the entire SOS5 open reading frame was amplified by reverse transcriptase–PCR. The amplified fragments were cloned into pCR-Blunt vector (Invitrogen, Carlsbad, CA), and six independent plasmids were sequenced. The cDNA sequence was aligned with genomic sequence to determine the gene organization.
For complementation testing, an ∼2.8-kb genomic fragment including the SOS5 promoter region, the coding region, and the 3′ untranslated region was amplified from wild-type genomic DNA using the primer pair 5′-GCAGAGATTTGCGTGTGAAGC-3′ and 5′-GAA-ACTGGGAATAACCTTCG-3′. The fragment was cloned into pCR-Blunt vector and sequenced. The fragment then was subcloned into the plant binary vector pCAMBIA 1200. The resulting construct was introduced into Agrobacterium tumefaciens strain GV3101 and transferred into sos5 mutant plants using the vacuum infiltration method (Bechtold et al., 1993). Twenty T2 transgenic lines were examined for complementation by salt sensitivity and root-swelling phenotypes.
An ∼1.3-kb promoter region upstream of the start codon was amplified by PCR using the primer pair 5′-CAGAGATTTGCGTGTGAAGC-3′ and 5′-GAGATTGGTAAAGAGAAGAACAC-3′ to introduce a HindIII site at the 5′ end and a BamHI site at the 3′ end to facilitate cloning. The fragment was cloned into the binary vector pCAMBIA 1391Z, resulting in a transcriptional fusion of the SOS5 promoter with the β-glucuronidase (GUS) coding region. The SOS5 promoter–GUS fusion construct then was introduced into Agrobacterium and transferred into plants as described above. Twenty T2 transgenic lines were subjected to GUS assays. GUS staining was performed as described previously (Shi et al., 2002). RNA isolation and RNA gel blot analysis were performed essentially as described (Shi et al., 2002).
Immunoblot Analysis
Rabbit anti-peptide serum against the peptide sequence (5′-DFL-SDSDLRRIPPSGS-3′) within the first fasciclin-like domain of SOS5 was generated by Covance Research Products (Richmond, CA). For immunoblot analysis, wild-type and sos5 plants were grown vertically on MS agar medium for 4 weeks. Total proteins were extracted from the leaf and root parts, respectively. Protein amount was determined by the Bradford (1976) assay. Sixty micrograms of total protein from each sample was separated by 7.5% SDS-PAGE. The proteins were transferred onto nitrocellulose membranes by electroblotting at 4°C. The membranes were incubated with a blocking solution containing 3% BSA in 1× Tris-buffered saline for 3 h at room temperature before reacting for 1 h at room temperature with a preimmune serum diluted to 1:1000 or with anti-SOS5 serum diluted to 1:1000. After washing three times with 1× TTBS (50 mM Tris-HCl, pH 7.5, 0.9% NaCl, 7.1% Tween) for 20 min, an alkaline phosphatase–conjugated secondary antibody in 0.5% BSA in 1× Tris-buffered saline was added. After final washing with 1× TTBS, the membrane was developed with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate reagents.
Immunofluorescence Labeling
Protoplasts were isolated from wild-type Arabidopsis (Columbia ecotype) suspension cell culture with an enzyme solution consisting of 1% Cellulysin Cellulase (Calbiochem, La Jolla, CA), 0.2% Macerase Pectinase (Calbiochem), 0.4 M mannitol, and 20 mM Mes, pH 5.7, in MS liquid medium. Protoplasts were washed twice with W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM Mes, pH 5.7) and resuspended in MMg solution (0.4 M mannitol, 15 mM MgCl2, and 4 mM Mes, pH 5.7). Protoplasts then were incubated with the rabbit anti-SOS5 antiserum or the rabbit preimmune serum for 1 h at a dilution of 1:50. After washing three times with MMg solution, protoplasts were incubated with fluorescein-conjugated goat anti-rabbit IgG (Calbiochem) for 30 min at a dilution of 1:10. The protoplasts then were washed three times with MMg and viewed with a Bio-Rad 1024 confocal laser scanning microscope.
Microscopy
Light microscopic observations were made using a Leica (Wetzlar, Germany) microscope. Three-week-old roots of wild-type and sos5 mutant seedlings grown on the surface of MS agar medium were fixed as described (Zhu et al., 1993). For cell wall staining and confocal imaging, fixed roots were washed with PBS and stained with 0.01% fluorescent brightener 28 (Sigma) in PBS for 1 min. Stained materials were washed with PBS, placed onto a glass microscope slide under a cover slip, and visualized using a Bio-Rad 1024 laser scanning confocal microscope attached to a Nikon Optiphot 2 microscope (Tokyo, Japan). For scanning electron microscopy, roots were fixed in situ on the agar plates using 2.5% glutaraldehyde in MS solution and postfixed with 1% osmium tetroxide in water. After dehydration and critical point drying, the samples were attached to stubs, coated with gold, and examined by scanning electron microscopy. For cross-sectional studies, the materials were infiltrated with LR White resin after fixation and dehydration as mentioned above. Thin sections (1.0 μm thick) and ultra-thin sections (0.1 μm thick) were collected and examined using light and electron microscopes, respectively.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We are indebted to Jiping Liu and David Bentley of the biotechnology division of Arizona Research Labs for excellent assistance. This work was supported by U.S. Department of Agriculture National Research Initiative Competitive Grants Program Grant 2001-35100-10687 and National Institutes of Health Grant R01GM59138 to J.-K.Z.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.007872.
References
- Arioli, T., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316, 1194–1199. [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Carlson, S.J., and Chourey, P.S. (1997). Cloning of a novel maize endosperm-specific protein with partial sequence homology to a pollen surface protein. Biochim. Biophys. Acta 1341, 101–104. [DOI] [PubMed] [Google Scholar]
- Carpita, N., and McCann, M. (2000). The cell wall. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 52–108.
- Cheung, A.Y., Wang, H., and Wu, H.M. (1995). A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82, 383–393. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (1999). Enzymes and other agents that enhance cell wall extensibility. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 391–417. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2001). Wall structure and wall loosening: A look backwards and forwards. Plant Physiol. 125, 131–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darley, C.P., Forrester, A.M., and McQueen-Mason, S.J. (2001). The molecular basis of plant cell wall extension. Plant Mol. Biol. 47, 179–195. [PubMed] [Google Scholar]
- Ding, L., and Zhu, J.K. (1997). A role for arabinogalactan-proteins in root epidermal cell expansion. Planta 203, 289–294. [DOI] [PubMed] [Google Scholar]
- Du, H., Clarke, A.E., and Bacic, A. (1996). Arabinogalactan-proteins: A class of extracellular matrix proteoglycans involved in plant growth and development. Trends Cell Biol. 6, 411–414. [DOI] [PubMed] [Google Scholar]
- Favery, B., Ryan, E., Foreman, J., Linstead, P., Boudonck, K., Steer, M., Shaw, P., and Dolan, L. (2001). KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev. 15, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings, T.H.J., and Staehelin, L.A. (1991). Microtubule-mediated control of microfibril deposition: A re-examination of the hypothesis. In The Cytoskeletal Basis of Plant Growth and Form, C.W. Lloyd, ed (London: Academic Press), pp. 85–99.
- Kawamoto, T., Noshiro, M., Shen, M., Nakamasu, K., Hashimoto, K., Kawashima-Ohya, Y., Gotoh, O., and Kato, Y. (1998). Structural and phylogenetic analyses of RGD-CAP/beta ig-h3, a fascic-lin-like adhesion protein expressed in chick chondrocytes. Biochim. Biophys. Acta 1395, 288–292. [DOI] [PubMed] [Google Scholar]
- Knox, J.P. (1997). The use of antibodies to study the architecture and developmental regulation of plant cell walls. Int. Rev. Cytol. 171, 79–120. [DOI] [PubMed] [Google Scholar]
- Kohorn, B.D. (2000). Plasma membrane-cell wall contacts. Plant Physiol. 124, 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komalavilas, P., Zhu, J.-K., and Nothnagel, E.A. (1991). Arabinogalactan-proteins from the suspension culture medium and plasma membrane of rose cells. J. Biol. Chem. 266, 15956–15965. [PubMed] [Google Scholar]
- Liu, J., and Zhu, J.-K. (1997). An Arabidopsis mutant that requires increased calcium for potassium nutrition and salt tolerance. Proc. Natl. Acad. Sci. USA 94, 14960–14964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska-Sawka, A., and Nothnagel, E.A. (2000). The multiple roles of arabinogalactan proteins in plant development. Plant Physiol. 122, 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, C., Bhatt, K., and Baumann, K. (2001). Shaping in plant cells. Curr. Opin. Plant Biol. 4, 540–549. [DOI] [PubMed] [Google Scholar]
- McQueen-Mason, S., and Cosgrove, D.J. (1994). Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc. Natl. Acad. Sci. USA 91, 6574–6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Oxley, D., and Bacic, A. (1999). Structure of the glycosylphosphatidylinositol anchor of an arabinogalactan protein from Pyrus communis suspension-cultured cells. Proc. Natl. Acad. Sci. USA 96, 14246–14251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell, R.I., Knox, J.P., Scofield, G.N., Selvendran, R.R., and Roberts, K. (1989). A family of abundant plasma membrane-associated glycoproteins related to the arabinogalactan proteins is unique to flowering plants. J. Cell Biol. 108, 1967–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelman, G., Morikami, A., Jung, J., Baskin, T.I., Carpita, N.C., Derbyshire, P., McCann, M.C., and Benfey, P.N. (2001). COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 15, 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, C.J., Johnson, K.L., Currie, G., and Bacic, A. (2000). The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell 12, 1751–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, C.J., Rumsewicz, M.P., Johnson, K.L., Jones, B.J., Gaspar, Y.M., and Bacic, A. (2002). Using genomic resources to guide research directions: The arabinogalactan protein gene family as a test case. Plant Physiol. 129, 1448–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrier, D.J., Prime, T.A., and Dupree, P. (1999). Glycosylphosphatidylinositol-anchored cell-surface proteins from Arabidopsis. Electrophoresis 20, 2027–2035. [DOI] [PubMed] [Google Scholar]
- Shi, H., Xiong, L., Stevenson, B., Lu, T., and Zhu, J.-K. (2002). The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell 14, 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter, A.M. (2001). Arabinogalactan-proteins: Structure, expression and function. Cell. Mol. Life Sci. 58, 1399–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetek, J., Yadav, M.P., and Nothnagel, E.A. (1999). Presence of a glycosylphosphatidylinositol lipid anchor on rose arabinogalactan proteins. J. Biol. Chem. 274, 14724–14733. [DOI] [PubMed] [Google Scholar]
- Takahashi, T., Gasch, A., Nishizawa, N., and Chua, N.-H. (1995). The DIMINUTO gene of Arabidopsis is involved in regulating cell elongation. Genes Dev. 9, 97–107. [DOI] [PubMed] [Google Scholar]
- Whitney, S.E.C., Brigham, J.E., Darke, A.H., Reid, J.S.G., and Gidley, M.J. (1995). In vitro assembly of cellulose/xyloglucan networks: Ultrastructural and molecular aspects. Plant J. 8, 491–504. [Google Scholar]
- Wu, H., de Graaf, B., Mariani, C., and Cheung, A.Y. (2001). Hydroxyproline-rich glycoproteins in plant reproductive tissues: Structure, functions and regulation. Cell. Mol. Life Sci. 58, 1418–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S.-J., Lei, D., and Zhu, J.-K. (1996). SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8, 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youl, J.J., Bacic, A., and Oxley, D. (1998). Arabinogalactan-proteins from Nicotiana alata and Pyrus communis contain glycosylphosphatidylinositol membrane anchors. Proc. Natl. Acad. Sci. USA 95, 7921–7926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z.D., Showalter, A.M., Lamport, D.T.A., and Kieliszewski, M.J. (2002). Tomato LeAGP-1 arabinogalactan-protein purified from transgenic tobacco corroborates the Hyp contiguity hypothesis. Plant J. 31, 431–444. [DOI] [PubMed] [Google Scholar]
- Zhu, J.-K. (2000). Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol. 124, 941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.-K., Bressan, R.A., and Hasegawa, P.M. (1993). Loss of arabinogalactan-proteins from the plasma membrane of NaCl-adapted tobacco cells. Planta 190, 221–226. [Google Scholar]
- Zhu, J.-K., Liu, J., and Xiong, L. (1998). Genetic analysis of salt tolerance in Arabidopsis: Evidence for a critical role of potassium nutrition. Plant Cell 10, 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]