Abstract
The Arabidopsis disease resistance (R) genes RPW8.1 and RPW8.2 couple the recognition of powdery mildew pathogens of this plant with the subsequent induction of a localized necrosis, or hypersensitive response (HR). The HR restricts the spread of the infection and renders the plant resistant. One-third of Arabidopsis plants transformed with a genomic fragment containing RPW8.1 and RPW8.2 developed spontaneous HR-like lesions (SHL) in the absence of pathogens. We demonstrate that SHL occurs in transgenic lines that contain multiple copies of the transgene and express RPW8.1 and RPW8.2 at high levels. SHL is associated with salicylic acid (SA) accumulation, and at the site of the lesion, there is increased expression of RPW8.1, increased production of H2O2, and increased expression of pathogenesis-related genes. These lesions are physiologically similar to the pathogen-induced HR mediated by RPW8.1 and RPW8.2. Significantly, environmental conditions that suppress SHL suppress the transcription of RPW8.1 and RPW8.2 and also suppress resistance to powdery mildews, even in transgenic lines containing RPW8.1 and RPW8.2 that normally do not express SHL. Furthermore, treatment with SA increases the transcription of RPW8.1 and RPW8.2, induces SHL, and enhances resistance to powdery mildews. We conclude that HR requires the transcription of RPW8.1 and RPW8.2, which is regulated independently of the pathogen by SA-dependent feedback amplification.
INTRODUCTION
Plant disease resistance (R) gene products appear to interact with products of pathogen avirulence (Avr) genes and induce a localized necrosis, the hypersensitive response (HR), which restricts the spread of the pathogen (Staskawicz et al., 1995; Hammond-Kosack and Jones, 1997). The HR normally is associated with the production of reactive oxygen intermediates and the induction of pathogenesis-related (PR) genes, and may be a form of programmed cell death analogous to animal apoptosis (Morel and Dangl, 1997; Lam et al., 2001).
The predicted products of the R genes RPW8.1 and RPW8.2 (hereafter referred to as RPW8, unless indicated otherwise) of Arabidopsis are small, basic proteins with a putative N-terminal transmembrane domain and a coiled-coil domain (Xiao et al., 2001). They lack the nucleotide binding site and Leu-rich repeats that characterize the products of the other Arabidopsis R genes (Dangl and Jones, 2001). RPW8 confers resistance to all tested isolates of the four species of powdery mildew pathogens of Arabidopsis (Xiao et al., 2001). By contrast, most other R genes confer resistance to only one or a few isolates of a pathogen species carrying the corresponding Avr genes (Hammond-Kosack and Jones, 1997). Despite these differences, resistance mediated by RPW8 is characterized by an HR involving the formation of H2O2 and the induction of PR transcripts. The HR triggered by the RPW8 genes involves the defense signaling components salicylic acid (SA) and EDS1 (Xiao et al., 2001). Thus, disease resistance regulated by the RPW8 genes is similar to that regulated by the other Arabidopsis R genes.
Although the mechanisms by which R proteins induce HR are largely unknown, influx of calcium, protein phosphorylation and dephosphorylation, production of reactive oxygen intermediates and nitric oxide, and SA synthesis are associated with the onset of HR (Greenberg et al., 1994; Dangl et al., 1996; Lamb and Dixon, 1997; Grant et al., 2000; Glazebrook, 2001; Zhang and Klessig, 2001).
Most R genes induce HR only when expressed in the presence of the Avr product. However, a small number of R genes, including modified or mutant R genes, can induce HR-like lesions, and these provide an opportunity to study HR independently of the pathogen. Three genes resulting from recombination at the rp1 locus in maize, Rp1-D21, Rp1-MD19, and Rp1-NC3, induce lesions resembling those caused by the pathogen (Hulbert, 1997). A chimeric Mi gene, created by combining the N terminus of a nonfunctional Mi homolog with the Leu-rich repeat domain of the functional Mi gene, induces cell death in Nicotiana benthamiana when expressed transiently (Hwang et al., 2000). Similarly, transient expression of a constitutively active Pto gene, with mutations in the Pto kinase activation domain, induces cell death in N. benthamiana (Rathjen et al., 1999). Stable overexpression of Pto under the control of the 35S promoter of Cauliflower mosaic virus leads to spontaneous lesions, increased SA levels, and constitutive activation of plant defenses against a broad spectrum of pathogens in tomato (Tang et al., 1999). All of these studies highlight the potential of R genes and their products to activate cell death programs in the absence of pathogens (that ordinarily are induced only upon the recognition of an Avr gene product by the cognate R protein). Here, we show that the native R genes RPW8.1 and RPW8.2 activate cell death pathways independently of the pathogen.
A genomic fragment containing RPW8.1 and RPW8.2 from the powdery mildew–resistant ecotype Ms-0 was introduced into the powdery mildew–susceptible ecotype Col-0 by Agrobacterium tumefaciens–mediated transformation. Unexpectedly, one-third of the transformants developed cell death lesions in the absence of the pathogen, and of these, more than one-fifth did not survive to set seed. We show that these lesions resemble the HR induced by the powdery mildew pathogens. We used lines expressing the spontaneous HR-like lesions (SHL) to dissect RPW8.1- and RPW8.2-mediated cell death. We found that transcriptional activation of the RPW8.1 and RPW8.2 alleles of accession Ms-0 is required for SHL and HR development. We further show that this amplification depends on the accumulation of SA and is induced by exogenous application of SA. Hence, we conclude that activation of RPW8.1 and RPW8.2 transcription via a SA-dependent feedback amplification circuit stimulates a cell death program leading to SHL and HR. Interestingly, our results also demonstrate that RPW8-mediated SHL, HR, and mildew resistance involve environmentally sensitive mechanisms.
RESULTS
Transgenic Arabidopsis Plants with Multiple Copies of RPW8.1 and RPW8.2 Exhibit HR-Like Lesions in the Absence of the Pathogen
Arabidopsis accession Col-0 lacks RPW8.1 and RPW8.2 and is susceptible to powdery mildew pathogens. Genomic fragments containing both RPW8.1 and RPW8.2 and their promoters cloned from the mildew-resistant Arabidopsis accession Ms-0 (in constructs SE14 and EE6.2) were introduced into Col-0 by Agrobacterium-mediated transformation (Xiao et al., 2001). Surprisingly, ∼30% of transgenic T1 plants developed SHL (Table 1). Of these, approximately one-fifth died as a result of extensive cell death before setting seed. Genomic fragments of Ms-0 containing either RPW8.1 or RPW8.2 under the control of their native promoters (in constructs EP3.7 and XE3.8, respectively) also were introduced into Col-0 by Agrobacterium-mediated transformation. Examination of transgenic plants containing EP3.7 and XE3.8 indicated that SHL developed in ∼11 and 5% of T1 individuals, respectively. However, of the Col-0 plants containing RPW8.1 or RPW8.2 coding sequence under the control of the 35S promoter of Cauliflower mosaic virus, none developed necrotic lesions in the T1 or T2 generation (Table 1). As a control, we introduced the Ms-0 allele of SPK-2, a Ser/Thr protein kinase gene adjacent to RPW8.1, into Col-0 plants. None of these transgenic plants developed necrotic lesions (Table 1).
Table 1.
Col-0 T1 Plants Transgenic for RPW8 Develop SHL in the Absence of Pathogens
| Constructsa | T1 Plants with Lesions |
Total T1 Plants Surveyed |
Genes Contained |
|---|---|---|---|
| SE14 | 32 | 97 | RPW8.1, RPW8.2, SPK-2 |
| CC7 | 0 | 52 | SPK-2 |
| EE6.2 | 15 | 53 | RPW8.1, RPW8.2 |
| EP3.7 | 5 | 44 | RPW8.1 |
| XE3.8 | 3 | 57 | RPW8.2 |
| 35S-XE3.8b | 9 | 45 | RPW8.2 |
| 35S::RPW8.1 | 0 | 58 | RPW8.1 coding sequence |
| 35S::RPW8.2 | 0 | 56 | RPW8.2 coding sequence |
| 35S::SPK-2 | 0 | 65 | SPK-2 coding sequence |
T1 transgenic Col-gl plants were selected by Basta herbicide soon after seed germination, transplanted to 35- × 70-cm trays containing autoclaved soil, and grown in normal conditions for 6 weeks. Lesion formation was monitored by the naked eye.
These constructs were shown in Figure 1A and Note 11 of Xiao et al. (2001).
The same 3.8-kb fragment in XE3.8 was cloned into binary vector pSMB downstream of the 35S promoter.
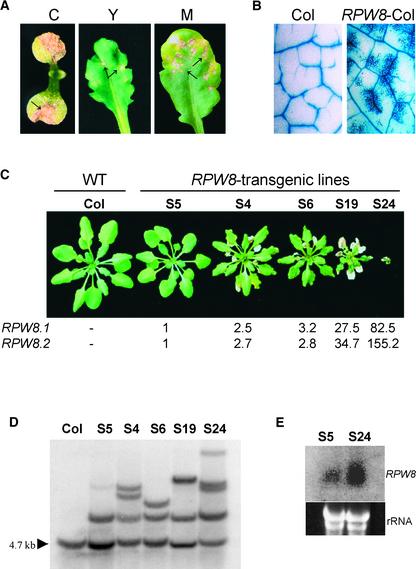
The age of the plant when SHL first appeared, and the size and density of the lesions, varied among the transgenic lines. Lesions occurred in cotyledons, young leaves, and mature leaves (Figure 1A). Microscopic examination of leaves stained with lactophenol–trypan blue, which stains fungal structures and dead plant cells (Koch and Slusarenko, 1990), confirmed that the development of SHL was not associated with pathogens: staining was localized to patches of dead cells that frequently occurred near veins (Figure 1B).
Figure 1.
High-Level Expression of RPW8.1 and RPW8.2 under the Control of Native Promoters Leads to SHL.
T2 progeny of RPW8 transgenic lines were first selected for the transgenes by spraying, soon after germination, herbicide containing 150 g/L glufosinate-ammonium (AgroEvo, Norfolk, UK) at a concentration of 0.02% (v/v). Plants were grown in soil under normal conditions for 4 weeks.
(A) SHL in leaves of T1 Col-0 transgenic plants carrying RPW8.1 and RPW8.2. Arrows indicate necrotic lesions. C, cotyledon; Y, young leaf; M, mature leaf.
(B) Lactophenol–trypan blue staining of leaves of Col-0 and a Col-0 transgenic line carrying RPW8.1 and RPW8.2 (RPW8-Col).
(C) Comparison of plant stature of 4-week-old wild-type (WT) Col-0 and five independent Col transgenic lines carrying construct SE14. Numbers below the plant photographs indicate relative mRNA levels of RPW8.1 and RPW8.2 determined by TaqMan chemistry (see Methods).
(D) DNA gel blot analysis for the copy number of the RPW8 transgenes. DNA extracted from bulked T2 plants of the five transgenic lines was digested with EcoRI, gel blotted, and probed with a DNA fragment in the insert-vector junction region of SE14. The arrowhead indicates the endogenous EcoRI fragment present in Col-0. The bands above the EcoRI band indicate the RPW8 transgenes.
(E) Gel blot analysis of RPW8 mRNA in S5 and S24. Total RNA (∼30 μg) extracted from bulked leaves of 3-week-old, soil-grown T2 transgenic seedlings of S5 and S24 was blotted and probed with a mixture of equal amounts of RPW8.1 and RPW8.2 genomic DNA.
To investigate the SHL phenotype in more detail, we selected transgenic lines with different severity of SHL. Col-0 plants were transformed with a 14-kb fragment of genomic DNA from Ms-0 (construct SE14), which contained RPW8.1 and RPW8.2 under the control of their native promoters. Five independent transgenic lines were selected, and their T2 and T3 progeny were examined. Line S5 had no lesions, lines S4, S6, and S19 had one to several lesions per leaf, and line S24 had numerous lesions on each leaf and was stunted (Figure 1C). DNA gel blot analysis indicated that S5 contained a single copy of the transgene, whereas S24 contained at least four copies (Figure 1D), indicating a correlation between the severity of SHL and the number of copies of the transgene.
RNA gel blot analysis indicated that RPW8.1 mRNA was more abundant in line S24 than in S5 (Figure 1E). Therefore, we used quantitative reverse transcriptase–mediated PCR to compare the abundance of RPW8.1 and RPW8.2 mRNA in S5, S4, S6, S19, and S24 (Figure 1C). Compared with S5, the RPW8.1 and RPW8.2 mRNAs were 2- to 3-fold higher in S4 and S6, 28- to 35-fold higher in S19, and 83- to 155-fold higher in S24. A T4 line containing 35S::RPW8.1 (coding sequence), but not displaying SHL, expressed RPW8.1 mRNA at a level 60-fold higher than S5. A T4 line containing 35S::RPW8.2 (coding sequence), but not displaying SHL, expressed RPW8.2 mRNA at a level ∼150-fold higher than S5. F1 plants from a cross between the 35S::RPW8.1 and 35S::RPW8.2 lines did not show SHL. However, when the 35S promoter was placed upstream of the native promoter of RPW8.2, the proportion of transgenic lines displaying SHL increased from ∼5% (XE3.8) to ∼20% (35S-XE3.8) (Table 1). These results indicate that SHL is associated with enhanced expression of RPW8.1 and/or RPW8.2 from their native promoters.
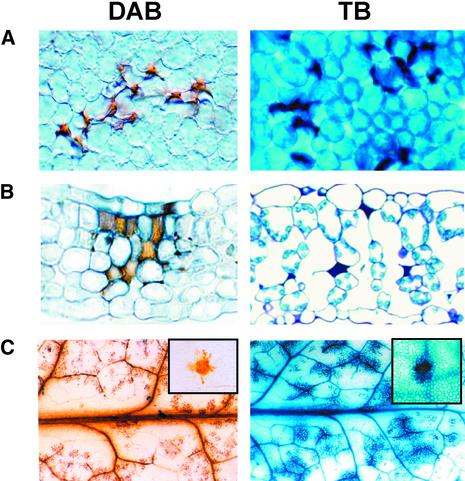
Spontaneous HR-Like Lesions in S24 Plants Develop in Mesophyll Cells and Are Associated with the Production of H2O2
Leaves of S24 plants were incubated with 3,3′-diaminobenzidine to detect H2O2 (Thordal-Christensen et al., 1997) or stained with trypan blue to reveal dead cells. Microscopic examination of young leaves revealed that H2O2 was produced in discrete clusters of mesophyll cells contacting epidermal cells and that these leaves also contained clusters of dead mesophyll cells (Figures 2A and 2B). Older leaves contained larger patches of H2O2-producing cells associated with the vascular bundles, and this matched the pattern of dead cells in these leaves (Figure 2C).
Figure 2.
SHL in S24 Occurs in Clusters of Mesophyll Cells and Is Preceded by H2O2 Accumulation.
Four-week-old homozygous S24 plants were transplanted from MS-agar to perlite. H2O2 production and cell death were revealed by 3,3′-diaminobenzidine (DAB) staining and trypan blue (TB) staining, respectively. H2O2 production (indicated by reddish-brown stain) and cell death (indicated by dark blue stain) were detected at ∼4 days and 5 to 6 days after transplanting, respectively, in mesophyll cells of mature leaves of S24, as shown by a top surface view (A) and by a transverse section (B) at ×500 magnification. The amplification and spread of H2O2 production (at day 8) and cell death (at day 10) in the whole leaf is shown in a large section of a leaf (C) at ×5 magnification. The insets in (C) show localized spots at ×100 magnification.
Spontaneous HR-Like Lesions Are Suppressed in S24 Plants Growing on Agar Medium, in Low Light, in High Humidity, or at High Temperature
Plant cell death caused by lesion-mimic mutations, misregulation of R genes, and HR is influenced by environmental conditions, including light (intensity or duration) (Dietrich et al., 1994; Greenberg et al., 1994; Tang et al., 1999; Stone et al., 2000), humidity (Hosford, 1978; Jambunathan et al., 2001; Yoshioka et al., 2001), and temperature. For example, high temperature suppresses cell death mediated by a chimeric Mi gene (Hwang et al., 2000), the HR controlled by the N gene in tobacco (Whitham et al., 1994), the resistance of wheat to Puccinia graminis tritici (Harder et al., 1979), and the resistance of soybean to Phytophthora sojae (Gijzen et al., 1996).
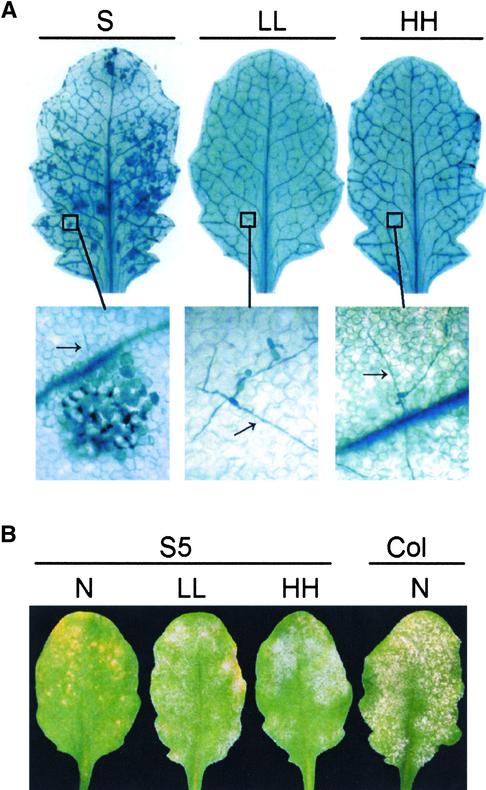
T2 progeny of the transgenic line S24 developed SHL when grown in soil but not when grown on Murashige and Skoog (1962) (MS)-agar medium (Figure 3A). Therefore, we examined the effect of environmental conditions on SHL in the T7 progeny of S24, which are homozygous for the transgenes, and in the T4 progeny of S5, which also are homozygous for the transgenes. For these experiments, seedlings were germinated and grown for 4 weeks on MS-agar in standard conditions (see Methods). SHL was not detected in any of these plants. Four-week-old plants were transferred subsequently under aseptic conditions to Magenta jars containing either 125 mL of perlite, an inert soil substitute, and 50 mL of half-strength MS, or MS-agar, and grown under the standard conditions. SHL developed on the S24 plants at 8 days after transfer to perlite but not during a 30-day period on MS-agar. We examined whether changes in light, humidity, or temperature could suppress SHL after transfer to perlite.
Figure 3.
Formation of SHL in S24 Is Environmentally Sensitive and Is Associated with Enhanced Expression of RPW8.1, RPW8.2, and PR.
(A) Phenotypes of S24 under different conditions. For the test of growth medium, S24 seeds were sown directly in soil or on MS-agar without transplanting. Photographs were taken at 3 weeks after seed germination. For the other three paired tests, 4-week-old S24 plants grown on MS-agar plates were transplanted to perlite and shifted to standard conditions (see Methods) but in continuous high light (∼85 μmol·m−2·s−1) or continuous low light (∼14 μmol·m−2·s−1), at 22 or 30°C, and at RH of 80 to 85% or ≥96%. Photographs were taken at 4 days after transplanting for the light treatment and at 14 days after transplanting for the temperature and humidity treatments.
(B) Relative mRNA levels of RPW8.1, RPW8.2, and PR1 in S24. For the medium test, 4-week-old S24 plants grown on MS-agar were transplanted to perlite or fresh MS-agar. For the other three paired tests, S24 plants were treated as described in (A). Leaf samples for mRNA analysis were collected at 8 days after transplanting, when SHL was about to be visible, except for the treatment with light, in which samples were collected at 24 h after transplanting. The mRNA levels of the three genes in S24 plants under SHL-suppressive conditions were calculated relative to those under the paired SHL-permissive conditions. These experiments were repeated three times with similar results, and the mRNA data from one experiment are presented.
(C) PR1 and PR2 expression reported by GUS. Three-week-old F1 plants of S24 × PR1::GUS and S24 × BGL2 (PR2)::GUS were transplanted from MS-agar plates to fresh MS-agar or perlite. Mature leaves were assayed for GUS activity at 2 weeks after transplanting.
To examine the effect of light intensity on the development of SHL, S24 plants were transplanted to perlite and grown in standard conditions except that they received continuous “high” light (∼85 μmol·m−2·s−1) or “low” light (∼14 μmol·m−2·s−1). The S24 plants held in high light started to develop visible chlorotic spots at ∼36 h after transplanting, and these covered the entire leaf in 4 days. SHL developed on younger leaves in 3 to 4 days (Figure 3A). The S24 plants held in low light did not develop chlorotic spots until 8 days after transplanting to perlite, and these were less severe than those on plants in the higher light intensity. No chlorosis or SHL developed in leaves of S5 or Col-0 plants.
To examine the effect of humidity on the development of SHL, plants were transplanted to perlite and kept at RH of either 80 to 85% or ≥96%. The S24 plants held at RH of 80 to 85% started to develop SHL at 8 days after transplanting, but the S24 plants held at RH of ≥96% started to develop SHL later, at 11 to 13 days after transplanting (Figure 3A). No SHL developed in S5 or Col-0 plants.
To examine the effect of temperature on the development of SHL, plants transplanted to sterile perlite were kept at either 22 or 30°C. The S24 plants held at 22°C started to develop SHL at 8 days after transplanting, and the S24 plants held at 30°C did not develop any SHL during the 30-day period of observation (Figure 3A).
In summary, SHL developed in S24 plants transferred to perlite, and this effect was delayed by low light or high humidity and was suppressed by high temperature. No SHL was detected on S5 or Col-0 plants under any of the conditions tested.
Development of Spontaneous HR-Like Lesions in S24 Plants Correlates with the Expression of RPW8.1, RPW8.2, and PR Genes
We measured the relative levels of RPW8.1, RPW8.2, and PR1 mRNAs from S24 plants at the time when SHL first became visible and compared these to the levels of the mRNAs from S24 plants of the same age but grown under SHL-suppressive conditions. Figure 3B shows that the mRNA levels of RPW8.1 and RPW8.2 in S24 plants in SHL-permissive conditions (i.e., perlite and standard growth conditions) were reduced when plants were grown in conditions that suppressed SHL (i.e., on agar medium, in high humidity, or at high temperature). However, transcript levels in S24 plants grown in SHL-permissive high light were lower than those in S24 plants grown in SHL-suppressive low light (Figure 3B). This effect may be caused by the death of a large proportion of leaf cells with increased expression of RPW8.1 and RPW8.2 under continuous high-light conditions before sample collection. Alternatively, a light-derived signal coincident with or downstream of RPW8 may be required to drive the SA-amplification loop that would be registered by an increase in PR1. Significantly, all plants under SHL-permissive conditions had much higher levels of PR1 transcript (49- to 264-fold) compared with those grown under SHL-suppressive conditions (Figure 3B).
The correlation between SHL development and PR gene expression was further confirmed using β-glucuronidase (GUS) reporter lines. The F1 progeny of S24 plants crossed to plants homozygous for GUS reporters for the PR1 and PR2 promoters displayed induced GUS activity localized to necrotic lesions that developed on these plants grown on perlite. GUS activity was not detected in the F1 plants grown on SHL-suppressive MS-agar medium (Figure 3C).
Conditions That Suppress SHL in Line S24 Also Suppress Resistance to Powdery Mildews Mediated by RPW8.1 and RPW8.2
We examined the effects of conditions that suppress SHL on the development of disease in susceptible Col-0 plants. Col-0 plants were transplanted to Magenta jars containing agar medium or perlite, inoculated with Erysiphe cichoracearum UCSC1, and maintained in continuous low light (14 μmol·m−2·s−1), in high humidity (RH ≥ 96%), or at high temperature (30°C). Disease did not develop on Col-0 plants grown in MS-agar medium or in perlite at 30°C. However, disease did develop on Col-0 plants grown in perlite at RH ≥ 96% and on plants grown in perlite in continuous low light.
S24 plants were raised aseptically on agar medium, transplanted to Magenta jars containing perlite and MS, inoculated with E. cichoracearum UCSC1, and maintained under standard conditions, standard conditions with high humidity (RH ≥ 96%), or standard conditions with continuous low light. Two days after inoculation, leaves were harvested and stained with trypan blue. On plants grown under standard conditions, >95% of germinated conidia induced HR. On plants grown in low light or at high RH, <25% of the germinated fungal conidia induced HR; rather, the fungus developed as it did on susceptible Col-0 plants in the first 3 days (Figure 4A).
Figure 4.
HR and Mildew Resistance Mediated by RPW8 Exhibit Similar Environmental Sensitivity as SHL in S24.
(A) Four-week-old S24 plants grown on MS-agar plates were inoculated with E. cichoracearum UCSC1 and transplanted immediately to perlite in Magenta jars under standard SHL-permissive conditions (S), in low light (∼14 μmol·m−2·s−1) (LL), or in high humidity (RH ≥ 96%) (HH). Inoculated leaves were collected at 48 h after inoculation and stained with trypan blue to visualize cell death and fungal structure. The micrographs at bottom were taken with a light microscope at ×400 magnification. Arrows indicate fungal hyphae.
(B) Four-week-old S5 and Col-0 plants were inoculated with E. cichoracearum UCSC1. After inoculation, plants were shifted to normal conditions (see Methods) (N), low light (∼14 μmol·m−2·s−1) (LL), or high humidity (RH ≥ 85%) (HH). Photographs were taken at 8 days after inoculation.
We also examined whether high humidity and low light, which suppress SHL in S24, also suppress resistance controlled by RPW8.1 and RPW8.2 in plants grown under more natural conditions. Col-0 and S5 plants grown in soil were inoculated with E. cichoracearum UCSC1 and maintained under normal conditions (see Methods), normal conditions with reduced light (∼14 μmol·m−2·s−1), or normal conditions with high humidity (RH ≥ 85%). Under normal conditions, S5 plants were resistant, but in low light or high humidity, they were more susceptible to infection (Figure 4B). These observations indicate that the environmental conditions that suppress SHL also suppress RPW8.1- and RPW8.2-mediated resistance to powdery mildews.
SA Is a Positive Regulator of a Feedback Amplification Circuit Stimulated by RPW8.1 and RPW8.2
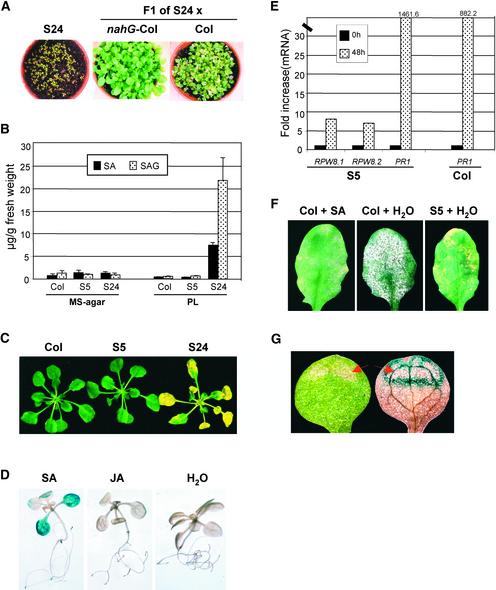
SA is required for RPW8.1- and RPW8.2-mediated HR and resistance (Xiao et al., 2001). Therefore, we examined the contribution of SA to SHL in S24. S24 plants were crossed to Col-0 plants homozygous for the nahG transgene, which encodes a salicylate hydroxylase that converts free SA to catechol, and to Col-0 plants as a control. F1 seeds were germinated on soil under standard conditions. As shown in Figure 5A, S24 seedlings developed SHL on cotyledons soon after germination, and most died within 3 weeks. F1 progeny of S24 × Col-0 developed SHL at 7 days after germination, whereas F1 progeny of S24 × Col-0/nahG did not develop SHL during 30 days of observation. These results indicate that SA also mediates RPW8-dependent SHL. The gene EDS1 is placed before SA in defense responses (Falk et al., 1999) and is required for RPW8-mediated resistance to powdery mildew pathogens (Xiao et al., 2001). Crosses of S24 to the wild type gave F2 progeny, of which 2 of 32 showed no SHL, indicating that two or more loci contribute to SHL in S24. From a cross of S24 to the eds1-2 mutant, F2 progeny containing the transgene were selected, and of these, 7 of 27 had no apparent SHL. This finding is consistent with the suppression of SHL in plants homozygous for eds1-2 (P > 0.05).
Figure 5.
SA Is a Component of a Feedback Amplification Circuit of RPW8-Mediated Cell Death and Resistance.
(A) Plants were grown in soil under normal conditions for 3 weeks.
(B) Four-week-old seedlings were transplanted from MS-agar to fresh MS-agar or perlite (PL). Leaves were collected from these plants at 8 days after transplanting and were assayed for SA and SAG levels. Data represent the average of two duplicate experiments ± sd.
(C) Three-week-old seedlings grown on MS-agar plates were transplanted to MS-agar containing 100 μm SA for 3 days.
(D) Col-0 seedlings transgenic for RPW8.1::GUS (T3 progeny homozygous for the transgene) were germinated on MS-agar. Ten-day-old seedlings were transferred to fresh MS-agar containing 100 μM SA, 50 μM jasmonic acid (JA), or water for 2 days and then assayed for GUS activity.
(E) Three-week-old seedlings grown in soil under normal conditions were sprayed with 500 μM SA twice with a 24-h interval. Total RNA was prepared from leaves of these seedlings before the first spray and 24 h after the second spray, and cDNA was synthesized from 1 μg of total RNA and used for transcript analysis by TaqMan chemistry.
(F) Four-week-old, soil-grown Col-0 and S5 plants were sprayed with 500 μM SA or water twice (once every 24 h) and inoculated with E. cichoracearum UCSC1 on day 3. Photographs were taken at 8 days after inoculation.
(G) F1 individuals of Col-0 homozygous for RPW8.1::GUS crossed to S24 were grown in soil for 2 weeks and then assayed for GUS activity. Cotyledons of seedlings displaying SHL were examined before (left) and after (right) GUS staining. The arrow indicates SHL.
Further evidence for the role of SA in RPW8-dependent SHL came from measurements of free SA and SA-Glc conjugate (SAG) in Col-0, S5, and S24 plants grown in standard conditions, either on MS-agar medium (SHL suppressive) or on perlite (SHL permissive). SA and SAG levels were low in Col-0 and S5 in all growth conditions (Figure 5B). SA and SAG levels were similarly low in S24 plants grown under SHL-suppressive conditions, but they were ∼6- and 24-fold higher, respectively, in plants grown in SHL-permissive conditions for 8 days (Figure 5B), at which time SHL first appeared. These results indicate that SA accumulation is associated with enhanced expression of RPW8.1 and RPW8.2 (Figure 3B) and SHL.
To determine whether SHL in S24 causes or results from SA accumulation, SA was applied exogenously to plants maintained on MS-agar, whereby both SHL and SA accumulation were suppressed. Three-week-old seedlings of Col-0, S5, and S24 were transferred from MS-agar to MS-agar containing 100 μM SA. Leaves of S24 plants developed chlorotic lesions within 2 days, and whole leaves were chlorotic within 3 days (Figure 5C). The leaves that formed subsequently were small and distorted. By contrast, Col-0 and S5 developed no lesions (Figure 5C). These findings indicate that SA alone is sufficient to induce SHL in S24 plants and that growth on MS-agar probably suppresses SHL by blocking SA accumulation.
Having identified SA as the causative signal in SHL, we examined whether the localized SHL could be accounted for by localized, SA-dependent expression of RPW8.1 and RPW8.2. To test this possibility, we fused the putative promoter region 1 kb upstream from the translational start codon of RPW8.1 to the GUS gene. This construct was introduced into Col-0 plants by Agrobacterium-mediated transformation, and transgenic lines homozygous for this transgene were selected. Ten-day-old seedlings of three independent transgenic lines were transferred from MS-agar to MS-agar containing 100 μM SA, 50 μM jasmonic acid, or water. GUS activity in these seedlings was monitored at 2 days after transplanting. We found that SA induced GUS expression, whereas jasmonic acid had no effect (Figure 5D).
To confirm that SA activates the transcription of RPW8.1 and RPW8.2, we treated plants with SA and measured the levels of RPW8.1 and RPW8.2 mRNA. S5 and Col-0 seedlings grown in soil under normal conditions were sprayed with SA. Leaves were harvested 48 h later, and mRNA levels of RPW8.1, RPW8.2, and PR1 were determined. In S5, the SA treatment induced an ∼8-fold increase in RPW8.1 and RPW8.2 mRNA and a 1460-fold increase in PR1 mRNA. In Col-0, the same treatment caused an 882-fold increase in PR1 mRNA (Figure 5E). However, the leaves of Col-0 sprayed with SA before inoculation displayed enhanced resistance to E. cichoracearum UCSC1 compared with leaves of mock-treated Col-0 plants (Figure 5F), consistent with the role of SA as an inducer of systemic acquired resistance (Dempsey et al., 1999).
Finally, to examine whether the development of SHL is associated with the localized expression of RPW8.1, we crossed a transgenic line carrying the GUS reporter for the putative RPW8.1 promoter to S24. F1 plants were grown in soil for 14 days until SHL began to develop, and then GUS activity was detected histochemically. GUS activity was localized at the margins of visible lesions (Figure 5G) and to small spots that may represent incipient lesions.
Together, these data indicate that SA is a positive regulator of a feedback amplification circuit stimulated by RPW8.1 and RPW8.2 that leads to SHL and probably to HR and disease resistance.
DISCUSSION
Arabidopsis plants with multiple copies of genomic fragments containing RPW8.1 and RPW8.2, transcriptionally regulated by their native promoters, developed apparently SHL that were associated with enhanced expression of RPW8.1, RPW8.2, and PR genes. In the transgenic line S24, SHL appeared as isolated necrotic spots that enlarged to form necroses that resembled the HR induced by powdery mildew pathogens on plants containing RPW8.1 and RPW8.2, such as resistant accession Ms-0 and Col-0 transgenic line S5. SHL developed in S24 and other lines in which the transgenes RPW8.1 and RPW8.2 were under the control of their native promoters but not in lines in which these transgenes were under the control of the 35S promoter (Table 1). However, the level of the transcripts for RPW8.1 and RPW8.2 was not greatly different between these different lines. To investigate this discrepancy, we used a reporter for the RPW8.1 promoter and observed that in progeny of line S24, this reporter was activated only in cells at the margin of spreading lesions, which represented <10% of the leaf tissue. Apparently, there would have been at least 10 times more transcripts of RPW8.1 and RPW8.2 in these S24 cells destined to become necrotic than in cells of lines in which the genes were expressed from the 35S promoter. These results indicate that the native promoters of RPW8.1 and RPW8.2 probably are required for SHL. It is possible that any lines with similarly high expression of the transgenes under the control of the 35S promoter would not survive. By contrast, overexpression of the R gene Pto controlled by the 35S promoter in tomato results in the nonpropagative spontaneous death of isolated cells (Tang et al., 1999). However, these lesions were barely visible to the naked eye and much smaller than the HR lesions induced by avirulent strains of the pathogen.
There was a general positive correlation between SHL development and level of RPW8.1 and RPW8.2 transcripts (Figure 3B). This result, together with our previous finding that RPW8.1 and RPW8.2 were induced by E. cichoracearum pathogens (Xiao et al., 2001), led us to speculate that RPW8-mediated HR and SHL involve a self-amplification mechanism. By introducing a GUS reporter for the putative RPW8.1 promoter into a background containing multiple copies of RPW8.1 and RPW8.2, we clearly demonstrated that transcription of RPW8.1 was further enhanced in the localized area where SHL developed, most likely as a consequence of the expression of RPW8.1 and RPW8.2 above a threshold level that has yet to be determined. Thus, the promoters of RPW8.1 and probably RPW8.2 seem to be critical for the transcriptional self-amplification. Analysis of the 1-kb sequence upstream of the RPW8.1 and RPW8.2 translational starts revealed three W-box elements in the promoter region of RPW8.1 (TTGACC at −162 bp, TTGACT at −282 bp, and AGTCAA at −135 bp in front of the translational start) and two in the promoter region of RPW8.2 (TTGACT at −281 and −526 bp). W boxes are cis-acting elements often found in promoters of many SA- and pathogen-responsive genes, such as NPR1 and PR1 (Lebel et al., 1998; Yu et al., 2001). They are binding sites for the WRKY family of transcription factors for the transcriptional regulation of defense-related genes (Rushton and Somssich, 1998; Eulgem et al., 2000). Therefore, we anticipated that RPW8.1 and RPW8.2 would be induced by SA.
SA is required for HR and for the expression of disease resistance in many plants (Malamy and Klessig, 1992; Shirasu et al., 1997; Dempsey et al., 1999; Alvarez, 2000). In Arabidopsis, SA also may function in a feedback amplification circuit involving the defense signaling genes EDS1, PAD4, and EDS5 that amplifies the resistance response (Falk et al., 1999; Jirage et al., 1999; Feys et al., 2001; Rusterucci et al., 2001). These responses have similarity to the cell death in the lsd5 mutant, which is SA dependent and SA induced (Weymann et al., 1995).
SA also is essential to RPW8.1- and RPW8.2-dependent HR, and plants containing the nahG transgene did not express the HR (Xiao et al., 2001). Here, we show that SA is critical for RPW8-mediated SHL, because the nahG transgene suppressed SHL in line S24 (Figure 5A). Moreover, S24 plants in which SHL was initiated also had high levels of endogenous SA, and this finding correlated positively with the enhanced expression of RPW8.1 and RPW8.2 (Figures 3B and 5B). Significantly, environmental conditions that suppressed SHL also suppressed both the accumulation of SA and the enhanced transcription of RPW8.1 and RPW8.2. These data suggest that the expression of RPW8.1 and RPW8.2 above a threshold level leads to SA accumulation, because SA did not accumulate, nor did SHL develop, in the single-copy line S5. Paradoxically, perhaps, we have shown by SA application to S24 plants that SA regulates RPW8.1 and RPW8.2 expression directly, enhancing SHL in otherwise suppressive conditions. Significantly, Col-0 plants containing the GUS reporter for the putative RPW8.1 promoter did not develop lesions when treated with SA, but GUS activity increased after SA application (Figure 5D).
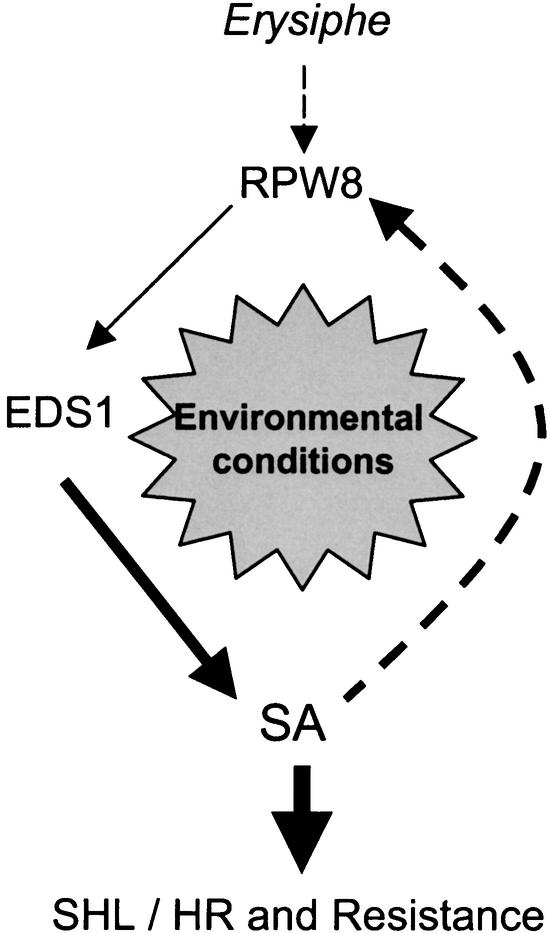
Our studies support a model for the RPW8-dependent SHL, HR, and resistance in which SA mediates the transcriptional self-amplification of RPW8 (Figure 6). This feedback circuit amplifies the defense response stimulated by RPW8 independently of the pathogen. We further propose that unknown local stimuli trigger this amplification circuit in line S24, leading to local necroses. Our RPW8.1 promoter–GUS reporter experiments provide compelling independent evidence that SA enhances RPW8.1 transcription through the promoter element, even in lines lacking the RPW8 genes. We provide further evidence for this model by showing that the RPW8.1 promoter is activated only in incipient lesions and at the leading edges of developing lesions (Figure 5G). This finding indicates that RPW8.1 expression, and we presume RPW8.2 expression as well, is programmed to occur just ahead of the advancing lesion but not throughout the leaf. A corollary to this is that local signaling molecules, which include SA, must define the advance of lesions into tissue in which the transcription of RPW8 genes is activated. This model also could explain why SHL did not develop in 35S::RPW8 lines: this promoter is not activated by SA.
Figure 6.
Model for RPW8-Mediated SHL, HR, and Resistance.
In this model, the recognition of Erysiphe pathogens by RPW8 triggers defense responses via a SA-dependent pathway (Xiao et al., 2001), leading to SA accumulation. Increased SA further enhances the expression of RPW8.1 and RPW8.2 via a feedback amplification circuit, leading to HR and resistance or SHL. Environmental conditions that suppress this amplification circuit suppress HR and disease resistance or SHL.
We conclude that SA-dependent transcription of RPW8.1 and RPW8.2 forms part of an amplification circuit that leads to the accumulation of SA and SHL or HR. Presumably, therefore, SA formed during the expression of resistance to different pathogens also would induce the accumulation of transcripts of RPW8.1 and RPW8.2. This raises the possibility that RPW8.1 and RPW8.2, and possibly other members of this gene family as well (Xiao et al., 2001), contribute to the expression of HR against other pathogens. However, RPW8.1 and RPW8.2 couple the specific recognition of powdery mildew pathogens to the induction of this defense response, and a future challenge is to understand the basis of this specificity.
METHODS
Plant Materials and Cultivation
The constructs used for RPW8 expression analysis were reported previously (Xiao et al., 2001), except for 35S-XE3.8, which was generated by cloning the 3.8-kb fragment of Ms-0 containing RPW8.2 and its native promoter into the XhoI and EcoRI sites of pSMB (Mylne and Botella, 1998) downstream of the 35S promoter of Cauliflower mosaic virus. The RPW8 promoter–β-glucuronidase (GUS) fusion construct was generated by cloning a fragment of 1 kb upstream of the RPW8.1 translational start in front of the GUS gene in pBI101 into the HindIII and XbaI sites. Transgenic Arabidopsis thaliana plants were generated as described (Xiao et al., 2001).
T7 seeds of line S24, which is homozygous for the multiple copies of the transgene containing RPW8.1 and RPW8.2, were obtained by growing ∼20 individuals for each generation in Murashige and Skoog (1962) (MS)-agar medium under short-day conditions (8 h of light and 16 h of dark) for 4 weeks and subsequently transferring them to perlite wetted with half-strength MS liquid medium under long-day conditions (16 h of light and 8 h of dark). Seeds from a single plant displaying the most extensive spontaneous HR-like lesions (SHL) were chosen to propagate, and seeds for the T7 generation were collected. All of the T7 progeny of S24 displayed homogeneous SHL in soil or perlite.
For comparative analysis with S24, S5, and Col-0, the experiments were standardized as follows: 4-week-old seedlings were raised in MS-agar medium supplemented with 1.5% Suc with one change of fresh medium. The seedlings were transferred individually to Magenta jars (77 × 77 mm; Sigma) containing ∼125 mL of perlite wetted with 50 mL of half-strength MS liquid medium without Suc. The lids of these boxes were fitted with a 0.22-μm membrane vent, and plants were kept in aseptic conditions at all times. Unless indicated otherwise, the standard growth conditions were short days (8 h of light [∼85 μmol·m−2·s−1] and 16 h of dark), 22°C, and RH of 80 to 85%.
For experiments with soil-grown plants, seeds were sown in autoclaved soil for germination, and seedlings were grown under short days for 2 weeks and then transplanted and maintained in a growth room under normal conditions (16 h of light [∼85 μmol·m−2·s−1] and 8 h of dark, 22°C, and RH of 40 to 50%), unless indicated otherwise.
Pathogen Strains and Inoculation
Erysiphe cichoracearum UCSC1 was used for the analysis of RPW8-dependent hypersensitive response and resistance. Methods of inoculation and disease scoring were the same as those described previously (Xiao et al., 1997).
Histochemical Detection of H2O2 and Cell Death
The protocol for the detection of H2O2 accumulation in leaf tissues by 3,3′-diaminobenzidine (DAB) staining was modified from Thordal-Christensen et al. (1997). Inoculated leaves were excised at the base of the petiole, placed in 1 mg/mL DAB (Sigma), and incubated for 8 h at 22°C with illumination. Leaves were fixed, cleared, and stained according to Reuber et al. (1998). H2O2 reacts with DAB to form a reddish-brown stain, whereas fungal structures are stained blue by trypan blue, both of which were visualized with a light microscope. Cell death was detected by lactophenol–trypan blue staining as described by Koch and Slusarenko (1990). Leaf material for sectioning was fixed in 3.7% formaldehyde, 5% acetic acid, and 50% ethanol, dehydrated, and embedded in paraffin. Embedded leaves were sectioned on a microtome at a thickness of 6 to 10 μm. Leaf sections were mounted on microscope slides, dewaxed, and rehydrated.
Transcript Analysis
Total RNA was extracted using RNeasy plant mini kits (Qiagen, Valencia, CA). For quantitative reverse transcriptase–mediated PCR analysis, 1 μg of total RNA was used for cDNA synthesis using Superscript II reverse transcriptase (Gibco BRL) and random hexamers (Pharmacia). PCR was performed on an ABI 7700 Sequence Detection System (TaqMan; Perkin-Elmer) according to the manufacturer's instructions. All reactions were performed in triplicate.
The Arabidopsis ACT2 gene was chosen as a normalization standard for TaqMan analysis of RPW8.1 and RPW8.2. The sequences of the primers and the cDNA-specific probe for ACT2 were those described previously by Feys et al. (2001). Probes for RPW8.1 and RPW8.2 also were made cDNA specific by designing them across the intron. The ribosomal 18S gene was used as a normalization standard for the analysis of PR1 and the transgenes of 35S::RPW8.1 and 35S::RPW8.2, because all them are intronless. Primers and probes were designed using Primer Express software (Perkin-Elmer), and the sequences were as follows: RPW8.1-F, 5′-GTTTTTGTTGAGGCTTATCCGAAA-3′; RPW8.1-R, 5′-TTATGCTTCTTAATGCAAGTT-CTATCGT-3′; RPW8.1-probe, 5′-TCGATTGCTTTGATGTACCTGTACTTCCTGAGTACTT-3′; RPW8.2-F, 5′-CGGAGCTGAGACGCA-GAAA-3′; RPW8.2-R, 5′-ATCTACCACCCATCGTAATTTAGCTT-3′; RPW8.2-probe, 5′-AACTCTTTGATATCTCTCATGTACCTGAACTTCTTGCGTA-3′; 18S-F, 5′-CGTCCTAGTCTCAACCATAAACGAT-3′; 18S-R, 5′-GGTGCCAGCGGAGTCCTAT-3′; 18S-probe, 5′-AACATC-CGCTGATCCCTGGTCGG-3′; PR1-F, 5′-AGAGGCAACTGCAGACTC-ATACAC-3′; PR1-R, 5′-AGCCTTCTCGCTAACCCACAT-3′; and PR1- probe, 5′-TTCACGGCGGAGACGCCAGACAAGTC-3′.
For RNA gel blot analysis, 30 μg of total RNA, separated on a 1.2% formaldehyde agarose gel, was blotted to Hybond NX (Amersham) according to the manufacturer's instructions. The DNA mixture, containing equal amounts of RPW8.1 and RPW8.2 genomic DNA, was 32P radiolabeled with the Megaprime DNA labeling system (Amersham) and used as a probe for hybridization.
Measurement of Salicylic Acid
Salicylic acid and its conjugates were measured by a method reported by Yalpani et al. (1991).
Other Analyses
DNA gel blot analysis was performed as described previously (Xiao et al., 1997). Histochemical staining for β-glucuronidase activity was according to Xie et al. (1998).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank Bart Feys (The Sainsbury Laboratory) for providing advice on TaqMan analysis, Allan Shapiro (University of Delaware) for seeds of Col-0 transgenic for PR-1 promoter::GUS, and Xinnian Dong (Duke University, Durham, NC) for seeds of Col-0 transgenic for BGL2::GUS and seeds of Col-0 transgenic for 35S::nahG. We thank David Alden and Richard Evan-Gowing for technical support and Alexi Balmuth for critical reading of the manuscript. This work was supported by Biotechnology and Biological Science Research Council Grant P15697.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006940.
References
- Alvarez, M.E. (2000). Salicylic acid in the machinery of hypersensitive cell death and disease resistance. Plant Mol. Biol. 44, 429–442. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., Dietrich, R.A., and Richberg, M.H. (1996). Death don't have no mercy: Cell death programs in plant-microbe interactions. Plant Cell 8, 1793–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Dempsey, D.A., Shah, J., and Klessig, D.F. (1999). Salicylic acid and disease resistance in plants. Crit. Rev. Plant Sci. 18, 547–575. [Google Scholar]
- Dietrich, R.A., Delaney, T.P., Uknes, S.J., Ward, E.R., Ryals, J.A., and Dangl, J.L. (1994). Arabidopsis mutants simulating disease resistance response. Cell 77, 565–577. [DOI] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P.J., Robatzek, S., and Somssich, I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Falk, A., Feys, B.J., Frost, L.N., Jones, J.D.G., Daniels, M.J., and Parker, J.E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96, 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J., Moisan, L.J., Newman, M.A., and Parker, J.E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20, 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijzen, M., MacGregor, T., Bhattacharyya, M., and Buzzell, R. (1996). Temperature induced susceptibility to Phytophthora sojae in soybean isolines carrying different Rps genes. Physiol. Mol. Plant Pathol. 48, 209–215. [Google Scholar]
- Glazebrook, J. (2001). Genes controlling expression of defense responses in Arabidopsis: 2001 status. Curr. Opin. Plant Biol. 4, 301–308. [DOI] [PubMed] [Google Scholar]
- Grant, M., Brown, I., Adams, S., Knight, M., Ainslie, A., and Mansfield, J. (2000). The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J. 23, 441–450. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., Guo, A.L., Klessig, D.F., and Ausubel, F.M. (1994). Programmed cell-death in plants: A pathogen-triggered response activated coordinately with multiple defense functions. Cell 77, 551–563. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D.G. (1997). Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 575–607. [DOI] [PubMed] [Google Scholar]
- Harder, D.E., Samborski, D.J., Rohringer, R., Rimmer, S., Kim, W., and Chong, J. (1979). Electron microscopy of susceptible and resistant near-isogenic (sr6/sr6) lines of wheat infected by Puccinia graminis tritici. III. Ultrastructure of incompatible reactions. Can. J. Bot. 57, 2626–2634. [Google Scholar]
- Hosford, R.M., Jr. (1978). Effects of wetting period on resistance to leaf spotting of wheat, barley, and rye by Leptosphaeria herpotrichoides. Phytopathology 68, 591–594. [Google Scholar]
- Hulbert, S.H. (1997). Structure and evolution of the rp1 complex conferring rust resistance in maize. Annu. Rev. Phytopathol. 35, 293–310. [DOI] [PubMed] [Google Scholar]
- Hwang, C.F., Bhakta, A.V., Truesdell, G.M., Pudlo, W.M., and Williamson, V.M. (2000). Evidence for a role of the N terminus and leucine-rich repeat region of the Mi gene product in regulation of localized cell death. Plant Cell 12, 1319–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambunathan, N., Siani, J.M., and McNellis, T.W. (2001). A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance. Plant Cell 13, 2225–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage, D., Tootle, T.L., Reuber, T.L., Frost, L.N., Feys, B.J., Parker, J.E., Ausubel, F.M., and Glazebrook, J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96, 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, E., and Slusarenko, A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, E., Kato, N., and Lawton, M. (2001). Programmed cell death, mitochondria and the plant hypersensitive response. Nature 411, 848–853. [DOI] [PubMed] [Google Scholar]
- Lamb, C., and Dixon, R.A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 251–275. [DOI] [PubMed] [Google Scholar]
- Lebel, E., Heifetz, P., Thorne, L., Uknes, S., Ryals, J., and Ward, E. (1998). Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 16, 223–233. [DOI] [PubMed] [Google Scholar]
- Malamy, J., and Klessig, D.F. (1992). Salicylic acid and plant disease resistance. Plant J. 2, 643–654. [Google Scholar]
- Morel, J.B., and Dangl, J.L. (1997). The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 4, 671–683. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Mylne, J., and Botella, J.R. (1998). Binary vectors for sense and antisense expression of Arabidopsis ESTs. Plant Mol. Biol. Rep. 16, 257–262. [Google Scholar]
- Rathjen, J.P., Chang, J.H., Staskawicz, B.J., and Michelmore, R.W. (1999). Constitutively active Pto induces a Prf-dependent hypersensitive response in the absence of avrPto. EMBO J. 18, 3232–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber, T.L., Plotnikova, J.M., Dewdney, J., Rogers, E.E., Wood, W., and Ausubel, F.M. (1998). Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 16, 473–485. [DOI] [PubMed] [Google Scholar]
- Rushton, P.J., and Somssich, I.E. (1998). Transcriptional control of plant genes responsive to pathogens. Curr. Opin. Plant Biol. 1, 311–315. [DOI] [PubMed] [Google Scholar]
- Rusterucci, C., Aviv, D.H., Holt, B.F., Dangl, J.L., and Parker, J.E. (2001). The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13, 2211–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu, K., Nakajima, H., Rajasekhar, V.K., Dixon, R.A., and Lamb, C. (1997). Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell 9, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz, B.J., Ausubel, F.M., Baker, B.J., Ellis, J.G., and Jones, J.D.G. (1995). Molecular genetics of plant disease resistance. Science 268, 661–667. [DOI] [PubMed] [Google Scholar]
- Stone, J.M., Heard, J.E., Asai, T., and Ausubel, F.M. (2000). Simulation of fungal-mediated cell death by fumonisin B1 and selection of fumonisin B1-resistant (fbr) Arabidopsis mutants. Plant Cell 12, 1811–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X.Y., Xie, M.T., Kim, Y.J., Zhou, J.M., Klessig, D.F., and Martin, G.B. (1999). Overexpression of Pto activates defense responses and confers broad resistance. Plant Cell 11, 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen, H., Zhang, Z.G., Wei, Y.D., and Collinge, D.B. (1997). Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. [Google Scholar]
- Weymann, K., Hunt, M., Uknes, S., Neuenschwander, U., Lawton, K., Steiner, H.Y., and Ryals, J. (1995). Suppression and restoration of lesion formation in Arabidopsis lsd mutants. Plant Cell 7, 2013–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham, S., Dinesh-Kumar, S.P., Choi, D., Hehl, R., Corr, C., and Baker, B. (1994). The product of the tobacco mosaic virus resistance gene N: Similarity to toll and the interleukin-1 receptor. Cell 78, 1101–1115. [DOI] [PubMed] [Google Scholar]
- Xiao, S., Ellwood, S., Findlay, K., Oliver, R.P., and Turner, J.G. (1997). Characterization of three loci controlling resistance of Arabidopsis thaliana accession Ms-0 to two powdery mildew diseases. Plant J. 12, 757–768. [DOI] [PubMed] [Google Scholar]
- Xiao, S.Y., Ellwood, S., Calis, O., Patrick, E., Li, T.X., Coleman, M., and Turner, J.G. (2001). Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291, 118–120. [DOI] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Yalpani, N., Silverman, P., Wilson, T.M., Kleier, D.A., and Raskin, I. (1991). Salicylic acid is a systemic signal and an inducer of pathogenesis-related proteins in virus-infected tobacco. Plant Cell 3, 809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka, K., Kachroo, P., Tsui, F., Sharma, S.B., Shah, J., and Klessig, D.F. (2001). Environmentally sensitive, SA-dependent defense responses in the cpr22 mutant of Arabidopsis. Plant J. 26, 447–459. [DOI] [PubMed] [Google Scholar]
- Yu, D., Chen, C., and Chen, Z. (2001). Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13, 1527–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S.Q., and Klessig, D.F. (2001). MAPK cascades in plant defense signaling. Trends Plant Sci. 6, 520–527. [DOI] [PubMed] [Google Scholar]