Abstract
Transcriptional activation by CLOCK-CYCLE (CLK-CYC) heterodimers and repression by PERIOD-TIMELESS (PER-TIM) heterodimers are essential for circadian oscillator function in Drosophila. PER-TIM was previously found to interact with CLK-CYC to repress transcription, and here we show that this interaction inhibits binding of CLK-CYC to E-box regulatory elements in vivo. Coincident with the interaction between PER-TIM and CLK-CYC is the hyperphosphorylation of CLK. This hyperphosphorylation occurs in parallel with the PER-dependent entry of DOUBLE-TIME (DBT) kinase into a complex with CLK-CYC, where DBT destabilizes both CLK and PER. Once PER and CLK are degraded, a novel hypophosphorylated form of CLK accumulates in parallel with E-box binding and transcriptional activation. These studies suggest that PER-dependent rhythms in CLK phosphorylation control rhythms in E-box-dependent transcription and CLK stability, thus linking PER and CLK function during the circadian cycle and distinguishing the transcriptional feedback mechanism in flies from that in mammals.
Keywords: Circadian rhythms, transcriptional regulation, DNA binding, feedback regulation, protein phosphorylation, protein stability
The circadian timekeeping mechanism is composed of transcriptional feedback loops in organisms ranging from cyanobacteria to humans (Young and Kay 2001). A common feature of these feedback loops is that positive factors activate the transcription of negative factors, which then feedback to inhibit the positive factors. Once the negative factors are removed, the positive factors can start the next cycle of transcription. Among the best-characterized transcriptional feedback loop mechanisms is that in Drosophila, in which the basic helix–loop–helix PER-ARNT-SIM (PAS) transcription factors CLOCK (CLK) and CYCLE (CYC) form heterodimers that bind E-box elements to activate period (per) and timeless (tim) transcription, then PER accumulates in a delayed fashion as a heterodimer with TIM and feeds back to inhibit CLK-CYC activity (Hardin 2004). The delayed accumulation of PER-TIM heterodimers is necessary to maintain circadian period, and this delay is mediated by DOUBLE-TIME (DBT)-dependent and possibly casein kinase 2 (CK2)-dependent phosphorylation and degradation of PER (Kloss et al. 1998; Price et al. 1998; Lin et al. 2002; Akten et al. 2003). Continued phosphorylation-dependent degradation of PER by DBT ultimately eliminates PER, thereby promoting the next cycle of CLK-CYC transcription.
The mechanism by which PER-TIM, or PER alone (Rothenfluh et al. 2000b; Chang and Reppert 2003; Nawathean and Rosbash 2004), inhibits CLK-CYC transcription is not understood. Immunostaining and in vitro DNA binding experiments indicate that CLK (and thus CLK-CYC) is present at constant levels in oscillator cell nuclei throughout the daily cycle (Wang et al. 2001; Houl et al. 2006), which contrasts with the cycling levels of CLK on Westerns (Lee et al. 1998; Bae et al. 2000). PER-TIM or PER inhibit CLK-CYC binding to E-boxes in vitro (Lee et al. 1999), which suggests that rhythmic transcription is mediated by PER/PER-TIM-dependent rhythms in E-box binding by CLK-CYC in vivo. Such a mechanism would contrast with that defined in mammals, in which the CLK-CYC homologs, CLOCK-BMAL1, are bound to E-boxes whether or not they are bound by their feedback inhibitors CRYPTOCROME (mCRY) or mPER-mCRY (Lee et al. 2001). Rhythms in CLOCK-BMAL1-dependent transcription are regulated by mCRY/mPER-mCRY-dependent histone acetylation; CLOCK-BMAL1 promotes histone acetyltransferase binding to activate transcription and mCRY/mPER-mCRY inhibits his tone acetyltransferase binding and/or promotes histone deacetylase binding to inhibit transcription (Etchegaray et al. 2003; Curtis et al. 2004; Naruse et al. 2004). If rhythms in CLK-CYC binding to E-boxes control rhythmic transcription in flies, it would indicate that PER/PER-TIM function is mechanistically distinct from that of mCRY/mPER-mCRY.
Western analysis of CLK abundance in fly heads shows that CLK levels are high when per and tim transcription is low and CLK levels are low when per and tim transcription is high (Hardin et al. 1990; Sehgal et al. 1995; So and Rosbash 1997; Lee et al. 1998). This relationship is paradoxical given that CLK is required for per and tim transcription, but if small amounts of CLK (and thus CLK-CYC) that are free of PER/PER-TIM inhibition do maximally activate per and tim transcription, they would have to be extremely potent activators. Rhythms in CLK abundance occur in phase with the underlying rhythm in Clk mRNA (Bae et al. 1998; Lee et al. 1998) but are not dependent on Clk mRNA rhythms, indicating that CLK cycling is regulated at the post-transcriptional level (Kim et al. 2002). This rhythm in CLK abundance coincides with rhythms in PER and TIM abundance and phosphorylation (Edery et al. 1994; Hunter-Ensor et al. 1996; Myers et al. 1996; Zeng et al. 1996), thus the highest levels of CLK accumulate at times when CLK-CYC is bound to, and inhibited by, hyperphosphorylated PER/PER-TIM (Lee et al. 1998; Bae et al. 2000). Like PER and TIM, CLK is phosphorylated (Lee et al. 1998), thus providing a possible mode for post-transcriptional regulation of CLK levels. In mammals, the levels of CLK are relatively constant, but CLK phosphorylation cycles such that it is hyperphosphorylated when it is inhibited by mCRY/mPER-mCRY and hypophosphorylated when it transcribes target genes in the absence of mCRY/mPER-mCRY, thus correlating phosphorylation with transcriptional activity (Lee et al. 2001). However, the regulation and function of CLK/CLOCK phosphorylation is not well understood in either mammals or flies.
In this study, we show that CLK-CYC rhythmically binds to E-boxes in vivo, which implies that PER/PER-TIM inhibits transcription by preventing CLK-CYC binding to E-boxes. We also reveal that CLK phosphorylation cycles between hypophosphorylated and hyperphosphorylated states, just as CLK does in mammals. Hyperphosphorylated CLK accumulates as PER-dependent complexes containing DBT kinase and CLK are formed, then DBT destabilizes both CLK and PER, thereby coordinating their degradation. After hyperphosphorylated CLK and PER are removed, a novel hypophosphorylated form of CLK accumulates in parallel with the transcription of per, tim, and other CLK-CYC target genes. These results indicate that PER/PER-TIM and mCRY/mPER-mCRY inhibit transcription by different mechanisms, thus revealing a fundamental difference in feedback regulation between flies and mammals. However, PER/PER-TIM- and mCRY/mPER-mCRY-dependent transcriptional inhibition are associated with rhythms in CLK and CLOCK phosphorylation, respectively, which suggests that phosphorylation of this conserved transcriptional activator is critical for maintaining circadian oscillator function in both Drosophila and mammals.
Results
A clock-dependent rhythm in CLK-CYC binding to E-boxes controls circadian transcription
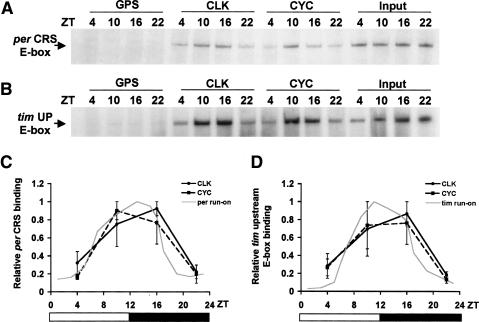
In Drosophila, in vitro DNA binding studies suggest that PER/PER-TIM regulates rhythmic transcription by inhibiting CLK-CYC binding to E-boxes (Lee et al. 1999), which contrasts with the situation in mammals, where mCRY/mPER-mCRY regulates rhythmic transcription by inhibiting CLOCK-BMAL1-dependent histone acetylation without a concomitant reduction in E-box binding (Etchegaray et al. 2003; Curtis et al. 2004; Naruse et al. 2004). To determine which of these regulatory mechanisms operate in flies, chromatin immunoprecipitation (ChIP) experiments were used to determine when CLK-CYC heterodimers were bound to E-boxes in vivo. Wild-type flies were collected every 6 h during a 12-h light/12-h dark (LD) cycle and subjected to ChIP using CLK, CYC, and control Guinea pig antisera. Fragments containing the circadian regulatory sequence (CRS) E-box from per that are necessary and sufficient to drive high-amplitude mRNA rhythms in vivo (Hao et al. 1997, 1999; Darlington et al. 2000; Lyons et al. 2000) were amplified from the immunoprecipitates and quantified. In CLK and CYC immunoprecipitates, the CRS E-box-containing fragments were three- to fivefold more abundant at ZT10 and ZT16 than at ZT2 and ZT22 (Fig. 1A,C), which is coincident with times when per mRNA is transcribed (Hardin et al. 1990; Sehgal et al. 1995; So and Rosbash 1997). Similar experiments were done using the upstream E-box from tim, which is necessary for high-amplitude mRNA cycling in vivo (McDonald et al. 2001; Wang et al. 2001). As with the per CRS, fragments containing the tim upstream E-box showed a robust cycle in CLK and CYC immunoprecipitates in line with tim transcription (Fig. 1B,D; Sehgal et al. 1995; So and Rosbash 1997). No rhythms in the abundance of an E-box from the constitutively expressed pigment dispersing factor (pdf) gene were observed in CLK and CYC immunoprecipitates (Park et al. 2000; data not shown). These results indicate that CLK and CYC rhythmically bind E-boxes upstream of per and tim under LD conditions.
Figure 1.
CLK-CYC heterodimers rhythmically bind E-boxes in vivo. (A) ChIP assays were performed on wild-type flies collected at the indicated times during LD cycles. Relative levels of the per CRS E-box present in each chromatin extract (Input) and immunoprecipitates from these chromatin extracts using CLK antibody (CLK), CYC antibody (CYC), and control Guinea pig serum (GPS) are seen as per CRS E-box containing PCR amplification products. (B) ChIP assays were performed as described in A, except that relative levels of the tim upstream E-box (tim UP E-box) are shown. (C) Quantification of per CRS ChIP assays. Error bars represent the range from two independent experiments. The gray line represents per transcription derived from nuclear run-on experiments (So and Rosbash 1997). (D) Quantification of tim UP E-box ChIP assays. Error bars represent the range from two independent experiments. The gray line represents tim transcription derived from nuclear run-on experiments (So and Rosbash 1997).
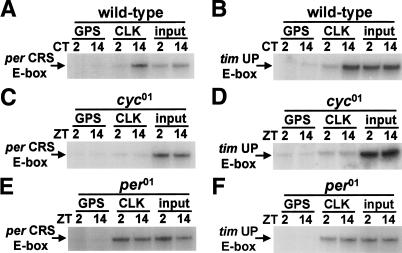
To verify that this rhythm in per and tim E-box immunoprecipitation by CLK and CYC is under circadian clock control, we performed ChIP assays on wild-type flies collected during constant darkness (DD) and arrhythmic cyc01 and per01 mutants collected during LD cycles. Rhythms in per and tim E-box immunoprecipitation by CLK antiserum persisted in wild-type flies during DD (Fig. 2A,B), but no binding was seen in cyc01 flies (Fig. 2C,D) and constant binding was seen in per01 flies (Fig. 2E,F). These results reflect the expression of per and tim mRNAs: rhythmic expression during DD, no expression in cyc01, and constant expression in per01 (Hardin et al. 1990; Sehgal et al. 1995; So and Rosbash 1997; Rutila et al. 1998). These data imply that CLK-CYC binds these per and tim E-boxes in a rhythmic manner, and that this rhythm is clock dependent. This in vivo rhythm in E-box binding supports a model in which PER or PER-TIM inhibits CLK-CYC binding to E-boxes. Such inhibition is likely to be direct since PER and TIM form complexes with CLK-CYC in vitro and in vivo (Lee et al. 1998, 1999; Bae et al. 2000).
Figure 2.
Rhythms in E-box binding by CLK-CYC are dependent on the circadian clock. (A) ChIP assays were performed on wild-type flies collected at the indicated circadian times (CTs) during DD. PCR amplification products representing levels of the per CRS E-box bound are presented as described in Figure 1A, except that CLK-CYC binding was assayed exclusively by CLK immunoprecipitation. (B) ChIP assays were performed as described for A, except that relative levels of the tim UP E-box are shown. (C) ChIP assays were performed on cyc01 flies collected at the indicated times. PCR amplification products representing levels of the per CRS E-box bound are presented as described in A. (D) ChIP assays were performed as described for C, except that relative levels of the tim UP E-box are shown. (E) ChIP assays were performed on per01 flies collected at the indicated times during LD cycles. PCR amplification products representing levels of the per CRS E-box bound are presented as described in A. (F) ChIP assays were performed as described for E, except that relative levels of the tim UP E-box are shown. All ChIP experiments described above were independently repeated with similar results.
CLK is rhythmically phosphorylated in a PER-dependent manner
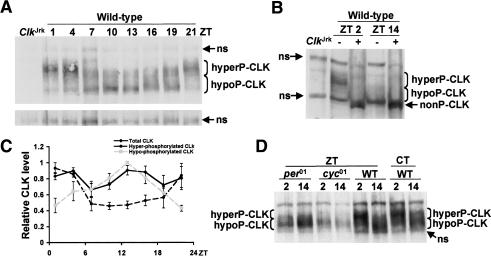
Contrary to expectation, CLK-CYC-dependent activation of per and tim transcription occurs at times when CLK levels are quite low on Western blots in fly head extracts (Lee et al. 1998). However, head extracts collected at different times of day show similar levels of CLK that are able to bind the tim E-box (Wang et al. 2001), and comparable levels of CLK immunostaining are detected in oscillator cells throughout the daily cycle (Houl et al. 2006). These in vitro DNA binding and immunostaining results suggest that CLK abundance does not cycle. The previous CLK Western results can be reconciled with CLK in vitro DNA binding and immunostaining results if CLK is not readily extracted from chromatin when CLK-CYC is bound to E-boxes. To explore this possibility, proteins were extracted from wild-type fly heads using a more stringent RBS procedure (see Materials and Methods). Western analysis using RBS extracts from wild-type flies shows two CLK bands: a slower migrating band that peaks in abundance during the late night and early morning (i.e., in phase with the previously characterized CLK band), and a novel faster migrating band that peaks during the late day and early night (Fig. 3A). Since CLK is known to be phosphorylated (Lee et al. 1998), RBS extracts were treated with phosphatase to determine whether phosphorylation accounted for the slower migrating CLK band. Phosphatase-treated head extracts produced a CLK band that migrated faster than either the slower or faster migrating CLK bands (Fig. 3B). Quantification of CLK levels in wild-type flies shows that CLK is hyperphosphorylated around dawn and hypophosphorylated around dusk, but overall levels of CLK do not cycle in abundance (Fig. 3C). The hypophosphorylated form of CLK was only detected when a more stringent extraction procedure was used that disrupts chromatin, which suggests that hypophosphorylated CLK is tightly bound to E-boxes. In support of this possibility, hypophosphorylated CLK accumulates in phase with E-box binding by CLK-CYC, whereas hyperphosphorylated CLK-CYC accumulates in phase with PER- or PER-TIM-dependent repression (Fig. 1A). Overall CLK abundance is relatively constant despite a peak in Clk mRNA levels around dawn, which suggests that CLK either is not synthesized efficiently or is unstable at this time.
Figure 3.
The state of CLK phosphorylation cycles in a clock-dependent manner. (A) Western containing RBS extracts from wild-type and ClkJrk fly heads. Wild-type flies were collected at the indicated times, and ClkJrk was collected at ZT2. Hyperphosphorylated CLK (hyperP-CLK) and hypophosphorylated CLK (hypoP-CLK) run as broad bands at ∼150 kDa and 120 kDa, respectively. A nonspecific (ns) band, shown in a longer exposure below, indicates the relative amount of protein loaded in each lane. (B) RBS extracts from the heads of wild-type flies collected at the indicated times were either treated (+) or not treated (−) with phosphatase. In addition to the hyperP-CLK and hypoP-CLK bands, a nonphosphorylated CLK (nonP-CLK) band is shown. Extracts from ClkJrk flies serve as a negative control for CLK. Two nonspecific (ns) bands are detected in these extracts. (C) Quantification of hypophosphorylated (dashed gray line), hyperphosphorylated (dashed black line), and total (solid black line) CLK abundance during an LD cycle. In each case, the peak level within a time course was normalized to 1.0. Results are based on three independent time courses. Error bars represent SEM. (D) RBS extracts from the heads of per01, cyc01, and wild-type flies collected at the indicated times were used to determine CLK levels and phosphorylation state.
Arrhythmic clock mutants might be expected to eliminate the rhythm in CLK phosphorylation. Indeed, the rhythm in CLK phosphorylation is abolished in per01 and cyc01 mutants (Fig. 3D). The CLK produced in per01 and cyc01 mutants appears to run more slowly than hypophosphorylated CLK in wild-type flies but remains primarily hypophosphorylated at both time points. For example, 55% ± 6% of CLK runs within the hypophosphorylated range at ZT2 and 54% ± 8% of CLK runs within the hypophosphorylated range at ZT14 in per01 flies, whereas 31% ± 6% of CLK runs within the hypophosphorylated range at ZT2 and 65% ± 7% of CLK runs within the hypophosphorylated range at ZT14 in wild-type flies. Since neither of these mutants expresses PER, this result suggests that CLK hyperphosphorylation is dependent on PER. Unexpectedly, overall CLK levels are 32% ± 10% higher at ZT14 than at ZT2 in per01 flies (Fig. 3D). This phenomenon is also seen in per01;ARK flies (see below), but the reason for this difference in CLK abundance is not known. The rhythm in CLK phosphorylation persists under DD conditions (Fig. 3D), thus demonstrating that it is a true circadian rhythm. As a whole, these results demonstrate that rhythms in CLK phosphorylation are controlled by the Drosophila circadian clock.
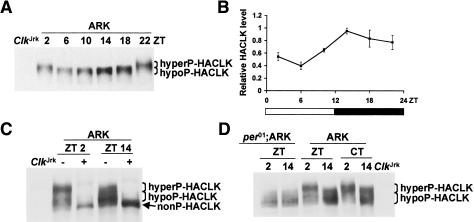
CLK stability correlates with CLK phosphorylation state
To independently verify rhythms in CLK phosphorylation, we employed transgenic ARK flies that express a per promoter-driven hemagglutinin (HA)-tagged version of CLK (Kim et al. 2002). Western blots prepared using RBS extracts from ARK fly heads were probed with an antibody against HA to reveal rhythms in HA-CLK mobility in phase with CLK phosphorylation cycles in wild-type flies (Fig. 4A). As in wild-type flies, phosphatase-treated head extracts from ARK flies produced an HA-CLK band that migrated faster than either the slower or faster migrating HA-CLK bands (Fig. 4C), thus demonstrating that HA-CLK also cycles between hyperphosphorylated and hypophosphorylated forms. This rhythm in HA-CLK phosphorylation persists under DD conditions but is abolished when the ARK insert is placed in a per01 background, where HA-CLK remains in a hypophosphorylated state (Fig. 4D). These results further demonstrate that the rhythm in CLK phosphorylation is clock dependent.
Figure 4.
CLK abundance and phosphorylation cycle in ARK flies. (A) Western containing RBS extracts from the heads of ARK flies collected at the indicated times and ClkJrk flies were probed with HA antibody. Hyperphosphorylated HACLK (hyperP-HACLK) and hypophosphorylated HACLK (hypoP-HACLK) bands are denoted. Note that this gel was not run or exposed as long as those in C and D, thus separation between CLK forms is not as great and weak bands are not as apparent. (B) Quantification of total HA-CLK levels in ARK flies. In each case, the peak level within a time course was normalized to 1.0. Results are based on three independent time courses. Error bars represent SEM. (C) RBS extracts from the heads of ARK flies collected at the indicated times were either treated (+) or not treated (−) with phosphatase. In addition to the hyperP-HACLK and hypoP-HACLK bands, a nonphosphorylated HACLK (nonP-HACLK) band is shown. Extracts from ClkJrk flies serve as a negative control for HACLK. (D) RBS extracts from the heads of per01;ARK and ARK flies collected at the indicated times were used to determine HACLK levels and phosphorylation state. Extracts from ClkJrk flies serve as a negative control for HACLK. Hyperphosphorylated HACLK (hyperP-HACLK) and hypophosphorylated HACLK (hypoP-HACLK) bands are denoted.
In contrast to the constant levels of total CLK protein in wild-type flies, the overall levels of HA-CLK cycle in ARK flies with a peak near dusk (Fig. 4A,B). This rhythm in HA-CLK abundance is in phase with that of per promoter-driven HA-Clk mRNA and, consequently, cycles in the opposite phase as endogenous Clk mRNA (Kim et al. 2002). Accumulation of HA-CLK in phase with HA-Clk mRNA at dusk contrasts with the lack of CLK accumulation when Clk mRNA reaches high levels near dawn in wild-type flies (Bae et al. 1998; Darlington et al. 1998). Since CLK is hyperphosphorylated at dawn and hypophosphorylated at dusk, these results suggest that high levels of phosphorylation destabilize CLK to limit its accumulation when Clk mRNA levels are high, and low levels of phosphorylation stabilize CLK so that it can accumulate from declining levels of Clk mRNA at dusk in wild-type flies.
PER is required for the DBT-dependent destabilization of CLK
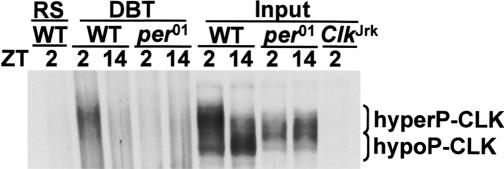
Phosphorylated forms of PER accumulate in the nucleus coincident with the accumulation of hyperphosphorylated CLK (Figs. 3, 4; Edery et al. 1994; Lee et al. 1998; Kim et al. 2002). Lack of hyperphosphorylated CLK in per01 flies suggests that PER directly or indirectly activates a kinase that phosphorylates CLK (Figs. 3D, 4D). PER is directly bound by DBT kinase, which phosphorylates and destabilizes PER (Kloss et al. 1998; Price et al. 1998). DBT is translocated along with PER into the nucleus (Kloss et al. 2001), where PER-DBT and/or PER-TIM-DBT complexes bind to and inhibit CLK-CYC (Lee et al. 1998, 1999; Bae et al. 2000). These data predict that DBT is present in the same complex as CLK, and that this DBT- and CLK-containing complex would be PER dependent. We immunoprecipitated EB3-S head extracts from wild-type and per01 flies at ZT2 and ZT14 using DBT antisera and determined whether these immunoprecipitates contained CLK. In wild-type flies, CLK was part of a complex with DBT at ZT2 but not ZT14 (Fig. 5), consistent with the formation of complexes between PER-DBT/PER-TIM-DBT and CLK-CYC. CLK is not part of a complex with DBT in per01 flies (Fig. 5), indicating that the formation of such complexes is indeed dependent on PER.
Figure 5.
DBT forms PER-dependent complexes with CLK. EB3-S extracts were prepared from the heads of wild-type (WT), per01, and ClkJrk flies collected at the indicated times. Extracts were immunoprecipitated with DBT antisera (DBT) or rat serum (RS) or were used directly (input) to prepare Western blots. These Westerns were probed with CLK antiserum to detect hyperphosphorylated CLK (hyperP-CLK) and hypophosphorylated CLK (hypoP-CLK).
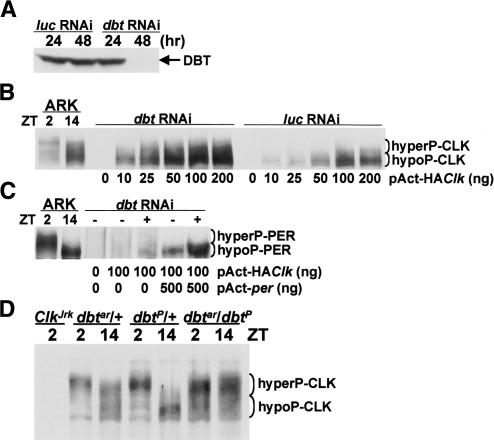
Since DBT is present in the same complex with CLK, DBT may act to phosphorylate and destabilize CLK as it does for PER. This possibility was initially tested in cultured Drosophila Schneider 2 (S2) cells, which were used previously to show that reducing DBT expression increases PER abundance and increasing DBT expression stimulates PER phosphorylation (Ko et al. 2002; Nawathean and Rosbash 2004). S2 cells were treated with either dbt RNA interference (RNAi) to reduce DBT levels or control luciferase (luc) RNAi and then transfected with different amounts of pAct-HAClk. CLK phosphorylation and abundance were measured 48 h after transfection since DBT was virtually eliminated by dbt RNAi at this time (Fig. 6A). Cells treated with luc RNAi and transfected with pAct-HAClk had low levels of CLK compared with cells treated with dbt RNAi and transfected with pAct-HAClk (Fig. 6B). This difference was most apparent at the lowest concentration of transfected pAct-HAClk, presumably because higher levels of pAct-HAClk produced saturating levels of CLK even in the luc RNAi control. This DBT-dependent reduction in CLK levels suggests that DBT destabilizes CLK. Since PER is necessary for DBT to form complexes with CLK in the nucleus, PER is predicted to be present in cells containing DBT. Although we do not detect appreciable amounts of PER in pAct-HAClk-transfected S2 cells, PER does accumulate when DBT levels are reduced in these cells via dbt RNAi (Fig. 6C). This result argues that low levels of PER are present in pAct-HAClk-transfected S2 cells, but can only be detected when PER is stabilized by reducing DBT. PER is apparently generated in pAct-HAClk-transfected S2 cells via HA-CLK-CYC-dependent activation of endogenous per.
Figure 6.
DBT destabilizes CLK. (A) S2 cells were treated with luciferase RNAi (luc RNAi) or dbt RNAi and incubated for the number of hours indicated. Western blots were prepared using RBS extracts from these cells and probed with DBT antisera. (B) S2 cells treated with luc RNAi or dbt RNAi were transfected with the indicated amounts of pAct-HAClk. Western blots prepared using RBS extracts from these cells and from the heads of ARK flies collected at ZT2 and ZT14 were probed with HA antiserum to detect hyperphosphorylated HACLK (hyperP-HACLK) and hypophosphorylated HACLK (hypoP-HACLK). (C) Untreated (−) and dbt RNAi-treated (+) S2 cells were transfected with the indicated amounts of pAct-HAClk and pAct-per. Western blots were prepared as described in B and probed with PER antiserum to reveal hyperphosphorylated PER (hyperP-PER) and hypophosphorylated PER (hypoP-PER). (D) RBS extracts were prepared from dbtar/TM3,Sb (dbtar/+), dbtP/TM3,Sb (dbtP/+), and dbtP/dbtar flies collected at the indicated times. dbtar/+ and dbtP/+ serve as controls for dbtP/dbtar flies, and ClkJrk serves as a negative control for CLK. The hyperphosphorylated CLK (hyperP-CLK) and hypophosphorylated CLK (hypoP-CLK) bands are denoted.
The increased CLK levels in S2 cells treated with dbt RNAi predicts that mutants with reduced DBT activity will also have higher levels of CLK. Among the most severe dbt mutants are dbtP, which has little or no functional DBT and is a homozygous pupal lethal (Kloss et al. 1998; Price et al. 1998), and dbtar, which is severely hypomorphic and behaviorally arrhythmic (Rothenfluh et al. 2000a). To determine whether CLK levels increase when DBT function is compromised, CLK levels were measured in RBS extracts from arrhythmic dbtP/dbtar flies (Rothenfluh et al. 2000a), which are more hardy than dbtar homozygotes in our hands. Compared with dbtar/+ and dbtP/+ controls (Rothenfluh et al. 2000a), the levels of CLK were higher by two- to fivefold (three independent experiments) in dbtP/dbtar flies (Fig. 6D). Importantly, CLK appears to be predominantly hyperphosphorylated in dbtP/dbtar flies (54% ± 3% at ZT2 and 59% ± 3% at ZT14), indicating that DBT is not necessary for the majority of CLK phosphorylation. These dbt mutant results, along with the dbt RNAi results in S2 cells, indicate that DBT destabilizes CLK.
Discussion
Rhythmic transcription via PER-dependent bindingof CLK-CYC to E-boxes
Our ChIP studies demonstrate that CLK-CYC is only bound to E-boxes when target genes are being actively transcribed (Figs. 1, 2). Since PER-DBT/PER-TIM-DBT complexes interact with CLK-CYC to inhibit transcription (Darlington et al. 1998; Lee et al. 1998), these data imply that these PER-containing complexes inhibit transcription by removing CLK-CYC from E-boxes. It is also possible that binding of these PER-containing complexes to CLK-CYC effectively blocks CLK and CYC antibody access, in which case PER complexes would inhibit transcription while CLK-CYC is bound to E-boxes. Given that the polyclonal CLK and CYC antibodies used here were raised against full-length proteins and have been used to immunoprecipitate PER-containing complexes (Lee et al. 1998; Bae et al. 2000), it is highly unlikely that all CLK and CYC epitopes are fully blocked by PER complex binding. Thus, we conclude that CLK-CYC rhythmically binds E-boxes in concert with target gene activation.
PER complex binding could remove CLK-CYC from E-boxes by directly altering their conformation or by promoting CLK phosphorylation. The region of PER that inhibits CLK-CYC transcription, called the CLK-CYC inhibitory domain or CCID, is near the C terminus (Chang and Reppert 2003). The CCID can act independently of the N terminus of PER (Chang and Reppert 2003), where the DBT-binding domain resides (Kloss et al. 1998). This observation argues that PER does not inhibit CLK-CYC binding to E-boxes by promoting DBT-dependent CLK phosphorylation. DBT- and CK2-dependent phosphorylation nevertheless enhances transcriptional repression in S2 cells by potentiating PER inhibition (Nawathean and Rosbash 2004) or by inhibiting CLK activity directly. Unfortunately, these disparate results from S2 cells do not allow us to distinguish between the different effects of PER complex binding to inhibit transcription outlined above.
In mammals, mCRY complexes bind to CLOCK-BMAL1 and repress transcription without removing CLOCK-BMAL1 from E-boxes (Lee et al. 2001; Etchegaray et al. 2003). This contrasts with the situation in flies, where PER complexes inhibit transcription by inhibiting CLK-CYC E-box binding, and suggests that these PER and mCRY complexes repress transcription via different mechanisms. Although mCRY complexes do not remove CLOCK-BMAL1 from E-boxes, they repress transcription by inhibiting the CLOCK-BMAL1-induced acetylation of histones by blocking p300 histone acetyl transferase function or introducing a histone deacetylase (Etchegaray et al. 2003; Curtis et al. 2004; Naruse et al. 2004). Even though PER complexes repress transcription by inhibiting CLK-CYC binding to E-boxes, this does not exclude the possibility that rhythms in histone acetylation are also involved in regulating rhythmic transcription in flies. Since chromatin remodeling is generally accepted as a prerequisite for transcription initiation (Orphanides and Reinberg 2002), it would be surprising if rhythms in transcription were not accompanied by rhythms in histone acetylation or some other form of chromatin remodeling.
PER-dependent rhythms in CLK phosphorylation
We have defined a rhythm in CLK phosphorylation in which hyperphosphorylated CLK predominates during times of transcriptional repression and hypophosphorylated CLK predominates during times of transcriptional activation (Figs. 3, 4). This rhythm occurs in parallel to the rhythm in PER phosphorylation (Edery et al. 1994); hyperphosphorylated PER and CLK accumulate in nuclei during the late night and early morning, then these forms are degraded and hypophosphorylated forms of PER and CLK accumulate in the cytoplasm and nucleus, respectively, during the late day and early evening. The rhythm in CLK and PER phosphorylation are not merely coincidental; the accumulation of hyperphosphorylated CLK is PER dependent (Figs. 3, 4). Although PER is not itself a kinase, it is bound by DBT kinase (Kloss et al. 1998, 2001). PER brings DBT into the nucleus (Kloss et al. 2001), where PER-DBT or PER-TIM-DBT complexes bind CLK-CYC to inhibit transcription (Lee et al. 1998, 1999).
Since DBT enters a complex containing CLK-CYC at times when CLK becomes hyperphosphorylated, DBT may also act to phosphorylate CLK. However, an in vitro assay for DBT phosphorylation is not available, thus we do not know whether DBT directly phosphorylates CLK. DBT acts to reduce CLK levels in S2 cells even though PER levels are very low (Fig. 6B,C). It is therefore possible that DBT can act to destabilize CLK in a PER-independent manner, although we believe this is unlikely to be the case since CLK hyperphosphorylation and complex formation with DBT are both PER dependent (Figs. 3D, 4, 5).
CLK is phosphorylated to some extent in the absence of PER and is hyperphosphorylated in the absence of functional DBT (Figs. 3D, 4D, 6D), indicating that other kinases act to phosphorylate CLK. The accumulation of hyperphosphorylated CLK in dbtAR/dbtP flies suggests that DBT triggers CLK degradation subsequent to CLK hyperphosphorylation. A similar situation is seen for PER, where hyperphosphorylated PER accumulates in the absence of functional DBT (Rothenfluh et al. 2000a), and phosphorylation by CK2 precedes DBT-dependent phosphorylation and PER destabilization (Lin et al. 2002; Akten et al. 2003). In addition, rhythmically expressed phosphatases may also contribute to CLK phosphorylation rhythms (Sathyanarayanan et al. 2004).
Rhythms in CLK phosphorylation may function to modulate CLK stability, subcellular localization, and/or activity. CLK levels do not change appreciably throughout the daily cycle despite approximately fivefold higher levels of Clk mRNA at dawn than at dusk (Bae et al. 1998; Darlington et al. 1998). If a less stable hyperphosphorylated form of CLK accumulates when Clk mRNA is high and a more stable hypophosphorylated form of CLK accumulates when Clk mRNA is low, they would tend to equalize total CLK levels over the daily cycle. This possibility is supported by results in ARK flies, which express Clk mRNA in the opposite circadian phase (i.e., Clk mRNA peak at dusk rather than dawn). The overall level of CLK cycles in ARK flies with a peak in (hypophosphorylated) CLK around dusk (Fig. 4B), consistent with hypophosphorylated CLK being more stable than hyperphosphorylated CLK. This possibility is also supported by DBT-dependent destabilization of CLK in S2 cells since DBT associates with CLK as hyperphosphorylated CLK accumulates in wild-type flies. If hypophosphorylated CLK is relatively stable, higher levels of CLK might be expected to accumulate in per01 flies. However, constant low levels of Clk mRNA likely limit CLK accumulation in per01 flies (Glossop et al. 1999). CLOCK phosphorylation is coupled to its degradation in cultured mammalian cells (Kondratov et al. 2003), yet degradation of phosphorylated CLOCK does not lead to a rhythm in CLOCK abundance even though Clock mRNA levels are constant (Glossop and Hardin 2002).
Studies in cultured mammalian cells also demonstrate that CLOCK phosphorylation promotes CLOCK-BMAL1 nuclear localization (Kondratov et al. 2003), although the significance of this nuclear localization is not clear given that CLOCK-BMAL1 binding to E-boxes is either constant or more robust during transcriptional repression in vivo (Lee et al. 2001; Etchegaray et al. 2003). In contrast, CLK is nuclear throughout the daily cycle in flies (Houl et al. 2006), indicating that phosphorylation state does not alter CLK subcellular localization.
The coincidence between CLOCK phosphorylation and transcriptional repression in mice supports the possibility that phosphorylation inhibits CLOCK-BMAL1 activity, perhaps by promoting HDAC binding or inhibiting HAT binding (Etchegaray et al. 2003; Curtis et al. 2004; Naruse et al. 2004). Likewise, hypophosphorylated and hyperphosphorylated CLK accumulate in parallel with target gene activation and repression, respectively, in flies. This relationship suggests that the state of CLK phosphorylation may alter its ability to activate target genes. Given that target gene activation occurs when CLK-CYC is bound to E-boxes and that E-box binding coincides with the accumulation of hypophosphorylated CLK (Figs. 1, 2, 3, 4), it is possible that CLK hyperphosphorylation compromises CLK-CYC binding to E-boxes and, consequently, target gene transcription is repressed. Precedent for such a regulatory mechanism is seen in the Neurospora clock, where limiting levels of FREQUENCY (FRQ) promote phosphorylation of WHITE COLLAR 1 (WC1) and WHITE COLLAR 2 (WC2), thereby inhibiting WC1-WC2 binding to C-box regulatory elements and repressing transcription (Schafmeier et al. 2005). In contrast to FRQ in Neurospora, PER is considerably more abundant than CLK in Drosophila and forms stable complexes with CLK-CYC (Lee et al. 1998, 1999; Bae et al. 2000). In addition, PER/PER-TIM can release CLK-CYC from E-boxes in vitro (Lee et al. 1999), thus demonstrating that PER/PER-TIM binding is sufficient to release CLK-CYC from E-boxes independent of CLK phosphorylation. Taken together with the in vitro E-box binding results, the high levels of PER relative to CLK and the formation of stable PER-TIM-CLK-CYC complexes in flies argue that PER/PER-TIM binding may also be sufficient to inhibit E-box binding by CLK-CYC in vivo, although they do not rule out a role for CLK hyperphosphorylation in inhibiting CLK-CYC E-box binding. For instance, PER/PER-TIM binding could function to initially remove CLK-CYC from E-boxes, and subsequent CLK phosphorylation could maintain CLK-CYC in a form that is incapable of binding E-boxes.
The constant levels and rhythmic phosphorylation of CLK defined here are similar to those previously characterized for mammalian CLOCK (Lee et al. 2001). This similarity extends beyond metazoans to fungi, where positive elements of the Neurospora circadian feedback loop; i.e., WC1 and WC2, are also rhythmically phosphorylated (He and Liu 2005; Schafmeier et al. 2005). In each of these organisms, phosphorylation of positive factors increases when they interact with their respective negative feedback regulators, and decreases when they activate target gene transcription in the absence of these feedback inhibitors (Figs. 3, 4; Lee et al. 2001; He and Liu 2005; Schafmeier et al. 2005). This remarkable similarity suggests that phosphorylation controls one or more critical aspects of positive element function, and consequently, the rhythm in the positive element phosphorylation has become a conserved feature of circadian feedback loops in eukaryotes.
Model for the regulation of rhythmic transcription in Drosophila
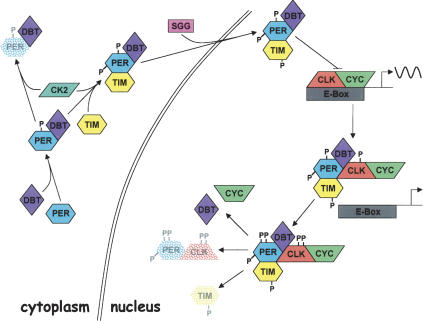
PER-dependent regulation of CLK-CYC binding to E-boxes, PER-dependent formation of PER-DBT and/or PER-DBT-TIM complexes with CLK-CYC, and PER-dependent rhythms in CLK phosphorylation suggest a model for the regulation of rhythmic transcription (Fig. 7). During the late day and early evening, hypophosphorylated CLK-CYC binds E-boxes to activate transcription of per, tim, and other genes within and downstream of the transcriptional feedback loop. Accumulating levels of per mRNA peak during the early evening (Hardin et al. 1990), but PER accumulation is delayed due to DBT-dependent (and possibly CK2-dependent) phosphorylation (Kloss et al. 1998; Price et al. 1998; Lin et al. 2002; Akten et al. 2003; Nawathean and Rosbash 2004), which destabilizes PER. PER is subsequently stabilized via TIM binding, which inhibits further phosphorylation of PER by DBT (Kloss et al. 1998; Price et al. 1998; Ko et al. 2002; Nawathean and Rosbash 2004). Phosphorylation of TIM by SGG then promotes the translocation of TIM-PER-DBT complexes into the nucleus (Martinek et al. 2001), where they bind (hypophosphorylated) CLK-CYC (Lee et al. 1999) and repress transcription by inhibiting E-box binding and promoting CLK hyperphosphorylation and degradation. These transcriptional repression mechanisms are not mutually exclusive; CLK hyperphosphorylation may inhibit E-box binding as well as promote CLK degradation. DBT is able to enter the nucleus in per01 flies (Kloss et al. 2001), but does not associate with CLK in the absence of PER. This suggests that PER is required to either bring DBT into a complex with CLK-CYC, enable phosphorylation of CLK after DBT enters the complex, or both. Once the TIM-PER-DBT-CLK-CYC complex has formed, hyperphosphorylated PER and CLK levels decline in a coordinated fashion by mid-day. TIM is eliminated prior to hyperphosphorylated PER and CLK via separate light-dependent and light-independent mechanisms (Hunter-Ensor et al. 1996; Lee et al. 1996; Myers et al. 1996; Zeng et al. 1996; Naidoo et al. 1999). As hyperphosphorylated CLK and Clk mRNA decline during the day, hypophosphorylated CLK accumulates. This hypophosphorylated CLK forms complexes with CYC and binds E-boxes in the absence of nuclear TIM-PER-DBT complexes, thus initiating the next cycle of transcription.
Figure 7.
Model for the regulation of circadian transcription in Drosophila. PER (blue hexagon) is generated during the early evening, but DBT (purple diamond) binding and phosphorylation (P) and possibly CK2 (green parallelogram) phosphorylation lead to PER degradation (stippled blue hexagon). TIM (yellow hexagon) binds to and stabilizes PER, and phosphorylation of PER by CK2 and TIM by SGG (pink rectangle) promotes nuclear localization of the PER-TIM-DBT complex. Nuclear PER-TIM-DBT complexes then bind to and remove hypophosphorylated CLK (red polygon)-CYC (green polygon) heterodimers from E-boxes (gray rectangle) to inhibit transcription. Coincident with transcriptional inhibition and PER-TIM-DBT-CLK-CYC complex formation, DBT phosphorylates PER and CLK to produce unstable hyperphosphorylated forms of PER and CLK. Degradation of hyperphosphorylated PER (stippled blue hexagon) and hyperphosphorylated CLK (stippled red polygon) and tyrosine phosphorylated TIM (stippled yellow hexagon) release DBT and CYC, and enable the accumulation of hypophosphorylated CLK to begin the next round of E-box binding and transcriptional activation. (Arrows) Subsequent steps in the process; (bar) inhibitory interaction; (wavy line) transcriptional activity; (double line) nuclear envelope.
Materials and methods
ChIP
ChIP assays were performed as described (Orlando et al. 1997), with modifications to accommodate fly head samples. Frozen fly heads (∼300 μL per sample) were gently homogenized in ∼3 volumes of NEB buffer (10 mM HEPES at pH 8.0, 10 mM NaCl, 0.1 mM EGTA, 0.5 mM EDTA, 1 mM DTT, 0.5% Tergitol NP-10). The homogenate was filtered with 100-μm nylon mesh to remove cuticle and then centrifuged at 6000g for 10 min. The pellet was resuspended in NEB and layered to a 0.8 M and 1.6 M sucrose cushion followed by 19,900g centrifugation for 20 min. The resulting intact nuclei were suspended in NEB and cross-linked in 1% formaldehyde for 10 min at room temperature. The reaction was stopped by the addition of glycine to a final concentration of 0.125 M, and nuclei were washed three times in NEB. Cross-linked nuclei were sonicated six times for 15 sec in sonication buffer (20 mM HEPES at pH 7.5, 2 mM EDTA, 1% SDS, 0.2% Triton X-100) using Artek Sonic Dismembrator (Model 150) set at 0.4. The average size of sheared DNA fragment was ∼500 base pairs (bp) on agarose gel. After centrifugation at 15,000g for 10 min, 1/20 volume of supernatants were set aside as input, and the remainder was diluted fivefold in IP buffer (20 mM HEPES at pH 7.5, 60 mM NaCl, 2 mM EDTA, 0.2% Triton X-100). This diluted fraction was precleared overnight with 70 μL of a 50% protein A-Sepharose beads (Amersham) slurry. These beads were prepared by incubating in 35 μL of normal Guinea pig serum (GPS) (Sigma) containing 500 μL of PBS and three washings in PBS. The precleared supernatant was subjected to immunoprecipitation for overnight by the addition of 3 μL of GPS, anti-CYC (Edery GP-122), or anti-CLK (Edery GP-90) antibodies. Immune complexes were recovered by 2-h incubation with 30 μL of poly dI-dC (3 μg/μLbeads) blocked protein A-Sepharose beads. Precipitates were sequentially washed twice with wash buffer I (20 mM HEPES at pH 7.5, 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), wash buffer II (20 mM HEPES at pH 7.5, 500 mM NaCl, 2 mM EDTA, 1% Triton X-100), wash buffer III (10 mM HEPES at pH 7.5, 250 mM LiCl, 1 mM EDTA, 0.5% NP-40, 0.5% sodium deoxycholate), and 1× TE (pH 8.0) and then eluted. All buffers above contained 10 μg/mL aprotinin, 10 μg/mL leupeptin, 2 μg/mL pepstatin A, 0.5 mM PMSF, 0.5 mM spermidine, and 0.15 mM spermine. All procedures above were performed on ice or at 4°C except where otherwise indicated. Immune complexes and input were then treated with RNase A and proteinase K, and the cross-linking was reversed as described (Orlando et al. 1997). DNA was purified by phenol/chloroform/isoamyl alcohol extraction and ethanol precipitation. Purified DNA was resuspended in 50 μL of 1× TE. This suspension was subjected to hot PCR after being diluted at 1:20 for GPS, CLK, and CYC immunoprecipitated DNA and at 1:100 for input DNA. For each 20 μL of PCR, 1 μL of diluted DNA was used as template.
Semiquantitative hot PCR
Primers were labeled using γ-32P-ATP and T4 polynucleotide kinase. PCR was performed for 10 cycles using 0.2 μM of cold primers, then hot primers were added at a final concentration of 0.5 μM for another 10 cycles of amplification. PCR products were purified using QIAquick columns and separated on 6% TBE denaturing polyacrylamide gel. Bands were quantified by densitometry. Background signals from GPS were subtracted in each set from CLK and CYC. CLK or CYC signals were then normalized to inputs. For 6-h time courses, the input-normalized intensity was further normalized to peak level to get relative intensity as shown in Figure 1.
Primers for hot PCR were as follows: for CRS E box, 5′-CCAGTGCCAGTGCGAGTTC-3′ and 5′-GATGCCAAGTGT CAATCCAAGC-3′; for tim upstream E-box, 5′-GCGGCACGT TGTGATTACAC-3′ and 5′-ACACTGACCGAAACACCCAC TC-3′.
Western blotting and immunoprecipitation
For preparing RBS extract, ∼100 μL of frozen fly heads were homogenized in ∼3 volumes of RBS buffer (10 mM HEPES at pH 7.5, 5 mM Tris at pH 7.5, 50 mM KCl, 10% glycerol, 2 mM EDTA, 1 mM DTT, 1% Triton X-100, 0.4% NP-40, 10 μg/mL aprotinin, 10 μg/mL leupeptin, 2 μg/mL pepstatin A, 0.5 mM PMSF, 1 mM Na3VO4, 10 mM r-nitrophenyl phosphate). Na3VO4 and r-nitrophenyl phosphate were not contained in RBS extract for λ phosphatase treatment. This homogenate was sonicated as described above for ChIP assays, except sonication was repeated five times for 10 sec and then centrifuged at 15,000g for 10 min. The supernatant was taken as RBS extract. EB3-S extract was prepared the same as EB3 (Kim et al. 2002), except that the extract was sonicated as described for ChIP assays following homogenization. Immunoprecipitations were performed as described (Kim et al. 2002) using EB3-S extract and anti-DBT antibody (Preuss et al. 2004). For SDS-PAGE, 120 μg of RBS, 80 μg of EB3-S extracts, or eluted immunoprecipitates were run on 5% Criterion Tris-HCl gels (Bio-Rad) and transferred to nitrocellulose membrane. Immunoblots were probed with anti-CLK (GP-47; incubated at room temperature for 2 h [Figs. 3A, 5] or overnight [Fig. 3B,D] with dilution of 1:2000 in presence of 100 μg/mL of ClkJrk cytoplasm) or anti-HA (Roche, Clone 3F10) at a final concentration of 1:500 and visualized with ECL (Amersham). Hyperphosphorylated and hypophosphorylated CLK was quantified by densitometry. For each lane, the CLK signal above and below a line drawn equidistant from the hypophosphorylated and hyperphosphorylated bands was quantified. Background was removed by subtracting signal from the corresponding area from ClkJrk samples. The signal from each lane was normalized to the same nonspecific band, and relative CLK levels were calculated by dividing the signal in each lane by the peak value for each time course.
Cell culture and transfection
HA-Clk fragment digested from pGEM-HA-Clk (Kim et al. 2002) was subcloned into pAc5.1/V5-His B (Invitrogen) vector to generate pAct-HA-CLK. Double-strand RNA (dsRNA) was in vitro synthesized for RNAi treatment in cultured S2 cells essentially as described elsewhere (Kao and Megraw 2004). Briefly, PCR was performed using dbt cDNA clone or pGL3-Basic as templates and primers of the target gene-specific sequences: ds/dbt-N, 16–686; ds/dbt-c, 540–1168; ds/luc, 171–864. Each primer contained a 5′ T7 RNA polymerase-binding site (TTAA TACGACTCACTATAGGGAGA). The MEGA T7 kit (Ambion) was used to synthesize RNA from the purified PCR products according to manufacturer’s instructions. RNA was annealed and then checked on 1% agarose gel before use. S2 cells were maintained as described (Tanoue et al. 2004). One day before RNAi treatment, cells were seeded (1 × 106 cells per well) in six-well plates. After washing, the dsRNA was added directly into the serum-free media to a final concentration of 40 nM. The cells were incubated for 1 h at room temperature followed by the addition of 2 mL of serum-containing medium. Cells were allowed to recover for 48 h at 24°C before transfections were performed using Cellfectin (Invitrogen) according to manufacturer’s instructions. After further 48 h of incubation, cells were collected in RBS buffer and total proteins were extracted using RBS procedure for Western analysis as described above for fly heads.
Acknowledgments
We are grateful to Jeff Hall for providing anti-PER antibody, Jeff Price for providing DBT antibody, and Isaac Edery for providing anti-CLK and anti-CYC antibodies. We are indebted to members of the Hardin laboratory for helpful discussions of results and to Dr. Nick Glossop for comments on the manuscript. This work was supported by NIH grants MH61423 and NS051280 to P.E.H.
Footnotes
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1404406
References
- Akten B., Jauch E., Genova G.K., Kim E.Y., Edery I., Raabe T., Jackson F.R. A role for CK2 in the Drosophila circadian oscillator. Nat. Neurosci. 2003;6:251–257. doi: 10.1038/nn1007. [DOI] [PubMed] [Google Scholar]
- Bae K., Lee C., Sidote D., Chuang K.Y., Edery I. Circadian regulation of a Drosophila homolog of the mammalian Clock gene: PER and TIM function as positive regulators. Mol. Cell. Biol. 1998;18:6142–6151. doi: 10.1128/mcb.18.10.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K., Lee C., Hardin P.E., Edery I. dCLOCK is present in limiting amounts and likely mediates daily interactions between the dCLOCK-CYC transcription factor and the PER-TIM complex. J. Neurosci. 2000;20:1746–1753. doi: 10.1523/JNEUROSCI.20-05-01746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D.C., Reppert S.M. A novel C-terminal domain of Drosophila PERIOD inhibits dCLOCK:CYCLE-mediated transcription. Curr. Biol. 2003;13:758–762. doi: 10.1016/s0960-9822(03)00286-0. [DOI] [PubMed] [Google Scholar]
- Curtis A.M., Seo S.B., Westgate E.J., Rudic R.D., Smyth E.M., Chakravarti D., FitzGerald G.A., McNamara P. Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J. Biol. Chem. 2004;279:7091–7097. doi: 10.1074/jbc.M311973200. [DOI] [PubMed] [Google Scholar]
- Darlington T.K., Wager-Smith K., Ceriani M.F., Staknis D., Gekakis N., Steeves T.D., Weitz C.J., Takahashi J.S., Kay S.A. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- Darlington T.K., Lyons L.C., Hardin P.E., Kay S.A. The period E-box is sufficient to drive circadian oscillation of transcription in vivo. J. Biol. Rhythms. 2000;15:462–471. doi: 10.1177/074873040001500603. [DOI] [PubMed] [Google Scholar]
- Edery I., Zwiebel L.J., Dembinska M.E., Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc. Natl. Acad. Sci. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray J.P., Lee C., Wade P.A., Reppert S.M. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- Glossop N.R., Hardin P.E. Central and peripheral circadian oscillator mechanisms in flies and mammals. J. Cell Sci. 2002;115:3369–3377. doi: 10.1242/jcs.115.17.3369. [DOI] [PubMed] [Google Scholar]
- Glossop N.R., Lyons L.C., Hardin P.E. Interlocked feedback loops within the Drosophila circadian oscillator. Science. 1999;286:766–768. doi: 10.1126/science.286.5440.766. [DOI] [PubMed] [Google Scholar]
- Hao H., Allen D.L., Hardin P.E. A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol. Cell. Biol. 1997;17:3687–3693. doi: 10.1128/mcb.17.7.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H., Glossop N.R., Lyons L., Qiu J., Morrish B., Cheng Y., Helfrich-Forster C., Hardin P. The 69 bp circadian regulatory sequence (CRS) mediates per-like developmental, spatial, and circadian expression and behavioral rescue in Drosophila. J. Neurosci. 1999;19:987–994. doi: 10.1523/JNEUROSCI.19-03-00987.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin P.E. Transcription regulation within the circadian clock: The E-box and beyond. J. Biol. Rhythms. 2004;19:348–360. doi: 10.1177/0748730404268052. [DOI] [PubMed] [Google Scholar]
- Hardin P.E., Hall J.C., Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- He Q., Liu Y. Molecular mechanism of light responses in Neurospora: From light-induced transcription to photoadaptation. Genes & Dev. 2005;19:2888–2899. doi: 10.1101/gad.1369605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houl J.H., Yu W., Dudek S.M., Hardin P.E. Drosophila CLOCK is constitutively expressed in circadian oscillator and non-oscillator cells. J. Biol. Rhythms. 2006 doi: 10.1177/0748730405283697. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter-Ensor M., Ousley A., Sehgal A. Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell. 1996;84:677–685. doi: 10.1016/s0092-8674(00)81046-6. [DOI] [PubMed] [Google Scholar]
- Kao L.R., Megraw T.L. RNAi in cultured Drosophila cells. Methods Mol. Biol. 2004;247:443–457. doi: 10.1385/1-59259-665-7:443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.Y., Bae K., Ng F.S., Glossop N.R., Hardin P.E., Edery I. Drosophila CLOCK protein is under posttranscriptional control and influences light-induced activity. Neuron. 2002;34:69–81. doi: 10.1016/s0896-6273(02)00639-6. [DOI] [PubMed] [Google Scholar]
- Kloss B., Price J.L., Saez L., Blau J., Rothenfluh A., Wesley C.S., Young M.W. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iε. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- Kloss B., Rothenfluh A., Young M.W., Saez L. Phosphorylation of period is influenced by cycling physical associations of double-time, period, and timeless in the Drosophila clock. Neuron. 2001;30:699–706. doi: 10.1016/s0896-6273(01)00320-8. [DOI] [PubMed] [Google Scholar]
- Ko H.W., Jiang J., Edery I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- Kondratov R.V., Chernov M.V., Kondratova A.A., Gorbacheva V.Y., Gudkov A.V., Antoch M.P. BMAL1-dependent circadian oscillation of nuclear CLOCK: Posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes & Dev. 2003;17:1921–1932. doi: 10.1101/gad.1099503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Parikh V., Itsukaichi T., Bae K., Edery I. Resetting the Drosophila clock by photic regulation of PER and a PER-TIM complex. Science. 1996;271:1740–1744. doi: 10.1126/science.271.5256.1740. [DOI] [PubMed] [Google Scholar]
- Lee C., Bae K., Edery I. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron. 1998;21:857–867. doi: 10.1016/s0896-6273(00)80601-7. [DOI] [PubMed] [Google Scholar]
- Lee C., Bae K., Edery I. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: A basis for circadian transcription. Mol. Cell. Biol. 1999;19:5316–5325. doi: 10.1128/mcb.19.8.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Etchegaray J.P., Cagampang F.R., Loudon A.S., Reppert S.M. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Lin J.M., Kilman V.L., Keegan K., Paddock B., Emery-Le M., Rosbash M., Allada R. A role for casein kinase 2α in the Drosophila circadian clock. Nature. 2002;420:816–820. doi: 10.1038/nature01235. [DOI] [PubMed] [Google Scholar]
- Lyons L.C., Darlington T.K., Hao H., Houl J., Kay S.A., Hardin P.E. Specific sequences outside the E-box are required for proper per expression and behavioral rescue. J. Biol. Rhythms. 2000;15:472–482. doi: 10.1177/074873040001500604. [DOI] [PubMed] [Google Scholar]
- Martinek S., Inonog S., Manoukian A.S., Young M.W. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell. 2001;105:769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- McDonald M.J., Rosbash M., Emery P. Wild-type circadian rhythmicity is dependent on closely spaced E boxes in the Drosophila timeless promoter. Mol. Cell. Biol. 2001;21:1207–1217. doi: 10.1128/MCB.21.4.1207-1217.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers M.P., Wager-Smith K., Rothenfluh-Hilfiker A., Young M.W. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- Naidoo N., Song W., Hunter-Ensor M., Sehgal A. A role for the proteasome in the light response of the timeless clock protein. Science. 1999;285:1737–1741. doi: 10.1126/science.285.5434.1737. [DOI] [PubMed] [Google Scholar]
- Naruse Y., Oh-hashi K., Iijima N., Naruse M., Yoshioka H., Tanaka M. Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol. Cell. Biol. 2004;24:6278–6287. doi: 10.1128/MCB.24.14.6278-6287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawathean P., Rosbash M. The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol. Cell. 2004;13:213–223. doi: 10.1016/s1097-2765(03)00503-3. [DOI] [PubMed] [Google Scholar]
- Orlando V., Strutt H., Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Park J.H., Helfrich-Forster C., Lee G., Liu L., Rosbash M., Hall J.C. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc. Natl. Acad. Sci. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss F., Fan J.Y., Kalive M., Bao S., Schuenemann E., Bjes E.S., Price J.L. Drosophila doubletime mutations which either shorten or lengthen the period of circadian rhythms decrease the protein kinase activity of casein kinase I. Mol. Cell. Biol. 2004;24:886–898. doi: 10.1128/MCB.24.2.886-898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J.L., Blau J., Rothenfluh A., Abodeely M., Kloss B., Young M.W. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- Rothenfluh A., Abodeely M., Young M.W. Short-period mutations of per affect a double-time-dependent step in the Drosophila circadian clock. Curr. Biol. 2000a;10:1399–1402. doi: 10.1016/s0960-9822(00)00786-7. [DOI] [PubMed] [Google Scholar]
- Rothenfluh A., Young M.W., Saez L. A TIMELESS-independent function for PERIOD proteins in the Drosophila clock. Neuron. 2000b;26:505–514. doi: 10.1016/s0896-6273(00)81182-4. [DOI] [PubMed] [Google Scholar]
- Rutila J.E., Suri V., Le M., So W.V., Rosbash M., Hall J.C. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- Sathyanarayanan S., Zheng X., Xiao R., Sehgal A. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell. 2004;116:603–615. doi: 10.1016/s0092-8674(04)00128-x. [DOI] [PubMed] [Google Scholar]
- Schafmeier T., Haase A., Kaldi K., Scholz J., Fuchs M., Brunner M. Transcriptional feedback of Neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell. 2005;122:235–246. doi: 10.1016/j.cell.2005.05.032. [DOI] [PubMed] [Google Scholar]
- Sehgal A., Rothenfluh-Hilfiker A., Hunter-Ensor M., Chen Y., Myers M.P., Young M.W. Rhythmic expression of timeless: A basis for promoting circadian cycles in period gene autoregulation. Science. 1995;270:808–810. doi: 10.1126/science.270.5237.808. [DOI] [PubMed] [Google Scholar]
- So W.V., Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 1997;16:7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue S., Krishnan P., Krishnan B., Dryer S.E., Hardin P.E. Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in. Drosophila. Curr. Biol. 2004;14:638–649. doi: 10.1016/j.cub.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Wang G.K., Ousley A., Darlington T.K., Chen D., Chen Y., Fu W., Hickman L.J., Kay S.A., Sehgal A. Regulation of the cycling of timeless (tim) RNA. J. Neurobiol. 2001;47:161–175. doi: 10.1002/neu.1024. [DOI] [PubMed] [Google Scholar]
- Young M.W., Kay S.A. Time zones: A comparative genetics of circadian clocks. Nat. Rev. Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- Zeng H., Qian Z., Myers M.P., Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]