Abstract
Isoamylases are debranching enzymes that hydrolyze α-1,6 linkages in α-1,4/α-1,6–linked glucan polymers. In plants, they have been shown to be required for the normal synthesis of amylopectin, although the precise manner in which they influence starch synthesis is still debated. cDNA clones encoding three distinct isoamylase isoforms (Stisa1, Stisa2, and Stisa3) have been identified from potato. The expression patterns of the genes are consistent with the possibility that they all play roles in starch synthesis. Analysis of the predicted sequences of the proteins suggested that only Stisa1 and Stisa3 are likely to have hydrolytic activity and that there probably are differences in substrate specificity between these two isoforms. This was confirmed by the expression of each isoamylase in Escherichia coli and characterization of its activity. Partial purification of isoamylase activity from potato tubers showed that Stisa1 and Stisa2 are associated as a multimeric enzyme but that Stisa3 is not associated with this enzyme complex. Our data suggest that Stisa1 and Stisa2 act together to debranch soluble glucan during starch synthesis. The catalytic specificity of Stisa3 is distinct from that of the multimeric enzyme, indicating that it may play a different role in starch metabolism.
INTRODUCTION
Starch, the most common form of stored carbon in plants, is composed of two types of α-1,4–linked glucan polymer: essentially unbranched amylose and regularly branched amylopectin. Amylopectin is synthesized via three committed enzyme steps: ADP-Glc pyrophosphorylase, which synthesizes sugar nucleotide precursors; starch synthase, which extends the α-1,4–linked glucan chains using ADP-Glc; and starch-branching enzyme, which introduces α-1,6 branch points to form amylopectin. However, the activity of starch-debranching enzymes, which hydrolyze α-1,6 branches in glucans, is also important for amylopectin synthesis. Evidence for this has come from sweet corn varieties that carry mutations at the sugary1 (su1) locus of maize (Correns, 1901). These mutants synthesize reduced amounts of amylopectin but accumulate a soluble, homogeneously branched glucan, phytoglycogen, instead (Black et al., 1966). Phytoglycogen might accumulate in sweet corn as a result of a deficiency in starch-debranching enzyme activity, and Pan and Nelson (1984) demonstrated a close correlation between debranching enzyme activity and the dosage of wild-type Su1 alleles. The importance of debranching enzyme activity to amylopectin synthesis was demonstrated by James et al. (1995), who showed that the su1 locus of maize encodes a starch-debranching enzyme. A similar loss of activity is thought to be the cause of the sugary mutant phenotype in rice and the notch2 phenotype in barley, both of which show reduced starch synthesis and the accumulation of phytoglycogen in endosperm (Fujita et al., 1999; Kubo et al., 1999; Burton et al., 2002). In Chlamydomonas reinhardtii, sta7 mutants accumulate phytoglycogen and synthesize virtually no amylopectin. In these mutants, the activity of a debranching enzyme is lost (Mouille et al., 1996; Dauvillée et al., 2001a). Similarly, in Arabidopsis, phytoglycogen accumulation and reduced amylopectin synthesis have been shown to result from the deletion of a gene that encodes a debranching enzyme (DBE1) (Zeeman et al., 1998b).
Two models have been proposed to explain the role of debranching enzymes in starch biosynthesis. The glucan-trimming model suggests that debranching enzymes work on irregularly branched preamylopectin in the plastid stroma and at the edges of the starch granule (Ball et al., 1996; Myers et al., 2000). As the preamylopectin increases in size, debranching enzymes cleave the widely spaced branches and generate a regularly branched glucan structure that is “competent to crystallize.” However, at the outer edges of the starch granule, the glucan may not be crystalline, and the glucan chains of amylopectin there are branched by starch-branching enzymes. This extensive branching inhibits further crystallization. Debranching enzymes hydrolyze some of these α-1,6 branches, especially those that are widely spaced, to produce more regions of crystallization-competent glucan. This model proposes that the extensive branching of preamylopectin followed by trimming of the outer glucan chains to produce regions of glucan with the competence to crystallize may explain the regular distribution of α-1,6 branch clusters (the 9-nm repeat) in amylopectin. In debranching enzyme mutants, preamylopectin may become so branched at its outer edges that further extension is prevented, limiting amylopectin synthesis and the growth of starch granules. Preamylopectin in the stroma may become so branched that crystallization is prevented altogether and soluble phytoglycogen accumulates instead.
An alternative model, the water-soluble polysaccharide-clearing model, was suggested by Zeeman et al. (1998b). This model proposes that the principal substrate for starch-debranching enzymes during starch synthesis is branched, water-soluble glucan. This glucan is synthesized in the plastid stroma by starch synthases and starch-branching enzymes, and its accumulation inhibits starch synthesis because it is an alternative, competitive sink for ADP-Glc. In mutants with reduced debranching enzyme activity, branched water-soluble glucans are elaborated at the expense of amylopectin and phytoglycogen accumulates.
The principal difference between these two models is the nature of the glucan that is the primary target of debranching enzyme activity during starch synthesis (i.e., whether or not it goes on to form amylopectin) and consequently whether debranching enzymes play a direct or an indirect role in amylopectin synthesis.
Starch-debranching enzymes in plants are of two types. The pullulanases (which also have been referred to as R-enzymes or limit dextrinases) hydrolyze α-1,6 linkages in amylopectin and β-limit dextrin (glucans that are produced as a result of β-amylase activity during starch breakdown) but do not hydrolyze glycogen. These enzymes show greatest activity on the yeast glucan pullulan, which has regularly spaced α-1,6 linkages after every three α-1,4–linked Glc units. Isoamylase-type debranching enzymes are distinct from pullulanases in their substrate preferences. They are most active on amylopectin, but they also are active on glycogen and β-limit dextrin substrates. Isoamylases are inactive on pullulan. Although the two types of debranching enzyme have related primary amino acid sequences, pullulanases can be distinguished structurally from isoamylases by a number of distinct motifs (James et al., 1995; Beatty et al., 1999).
Although the primary lesion in both sugary1 in maize and sugary in rice is in a gene encoding an isoamylase-type debranching enzyme (James et al., 1995; Rahman et al., 1998; Fujita et al., 1999; Kubo et al., 1999), the activity of pullulanase also is decreased in these mutants, and its activity is inversely associated with the phytoglycogen content of the seed (Pan and Nelson, 1984; Nakamura et al., 1997; Kubo et al., 1999). Therefore, the relative contribution of each type of debranching enzyme to the synthesis of amylopectin (either quantitative or qualitative) in maize and rice is not known. However, mutations in the notch2 locus of barley, the DBE1 locus of Arabidopsis, and the STA7 locus of Chlamydomonas affect only isoamylase and not pullulanase activities (Mouille et al., 1996; Zeeman et al., 1998b; Dauvillée et al., 2000; Burton et al., 2002). These data support the view that it is the isoamylases that play the important role in starch synthesis.
Complicating this picture is the evidence for more than one isoform of isoamylase in higher plants. Doehlert and Knutson (1991) were able to separate two isoforms of isoamylase (distinct from pullulanase) from maize endosperm. There also is evidence that plant isoamylases operate in multimeric complexes, which may comprise more than one isoamylase isoform. Most reports of purified isoamylase activity from plant storage organs indicate a maximum mass for the purified native protein of ∼540 kD. Because isoamylase peptides are each ∼80 kD, the native protein probably is multimeric, consisting of up to six composite peptides. Ishizaki et al. (1983) first isolated isoamylase from potato as a native protein of 520 kD consisting of two distinct peptides of 95 and 83 kD in mass. A multimeric isoamylase also has been purified from rice (Fujita et al.,1999), but although two composite peptides were resolved by isoelectric focusing, N-terminal peptide sequencing indicated that the two peptides were identical and that the rice isoamylase was homomeric. In Chlamydomonas, the STA7 locus is thought to encode an isoamylase (Mouille et al., 1996), but mutants of the STA8 locus also lose some forms of the isoamylase detected on nondenaturing gels. Mutants at the STA8 locus accumulate phytoglycogen and show a modest decrease in isoamylase activity (Dauvillée et al., 2000, 2001a, 2001b), suggesting that STA8 encodes a second component of the isoamylase complex that regulates isoamylase activity.
We have focused on the role of isoamylase-debranching enzymes in starch synthesis in developing potato tubers. We have identified cDNA clones that encode three distinct isoamylase isoforms. All three genes are expressed in tubers that synthesize storage starch and in leaves that synthesize transitory starch. All three cDNAs encode sequences predicted to form chloroplast-targeting transit peptides at their N termini, and import assays confirm that all three isoforms can be imported into plastids. We have expressed each potato isoamylase isoform in Escherichia coli and shown that Stisa1 and Stisa3 have significant isoamylase activity alone, although their substrate specificities differ. Stisa2 has no discernible isoamylase activity when assayed on its own. However, mixing experiments show that Stisa1 and Stisa2 interact to give enhanced activity on some substrates. We have analyzed isoamylase activity from potato tubers. Two of the isoforms, Stisa1 and Stisa2, are associated as a multimeric enzyme. Stisa3 is not strongly associated with this multimer and may function as a monomer or in association with other proteins.
RESULTS
Molecular Characterization of the Isoamylase Isoforms from Potato
cDNA clones encoding isoamylases were identified by screening cDNA libraries prepared from mRNA from potato mini tubers grown in vitro on stem explants (Visser et al., 1989) and from developing tubers from greenhouse-grown plants. The probe used was an EST from Arabidopsis (At69012) that was selected as encoding an isoamylase by a Basic Local Alignment Search Tool (BLAST) search of Arabidopsis ESTs using the sequence of the su1 cDNA from maize (James et al., 1995) as the query sequence. Complete sequencing confirmed that the Arabidopsis EST encoded an isoamylase that was shown subsequently to be the product of the DBE1 locus (Zeeman et al., 1998b). The Arabidopsis EST was used because it provided a probe from a dicotyledonous plant, without the high G+C content of maize genes, for screening for structurally similar genes in potato. A total of 60,000 plaque-forming units from the unamplified tuber library and 60,000 plaque-forming units from the unamplified mini tuber library were screened using low-stringency washes (2 × SSC [1× SSC is 0.15 M NaCl and 0.015 M sodium citrate] and 0.5% SDS at 55°C), and 18 positive plaques (5 from the tuber library and 13 from the mini tuber library) were purified.
Nine of the purified clones were subcloned and sequenced. The sequences were predicted to encode three distinct isoamylase isoforms, which we named Stisa1 (two clones), Stisa2 (four clones), and Stisa3 (three clones). The lengths of the longest cDNA encoding each isoform were 2.7, 2.9, and 2.6 kb, respectively. Stisa1 encoded a 793–amino acid peptide that showed 82% similarity (70% identity) to the Su1 peptide of maize, Stisa2 encoded an 878–amino acid peptide that showed 57% similarity (35% identity) to Su1, and Stisa3 encoded a 766–amino acid peptide with 61% similarity (45% identity) to Su1. The predicted N-terminal amino acid sequences of Stisa1 and Stisa2 fitted the ChloroPVI.I criteria for plastid transit peptides well and were predicted to have 47 and 38 amino acid transit peptides, respectively. The prediction for Stisa3 was less clear, but a site fitting reasonably well with the prediction of Gavel and von Heijne (1990) was found (AFQPRLV↓AAAAKLQ) that would give a transit peptide of 69 amino acids. The predicted sizes for the mature Stisa1, Stisa2, and Stisa3 peptides were 746, 840, and 697 amino acids, respectively, giving predicted molecular masses of 84,206, 94,151, and 79,205 kD.
An alignment of the predicted amino acid sequences of Stisa1, Stisas2, and Stisa3 using CLUSTAL W was used to search the Homologous Structure Alignment Database (Mizuguchi et al., 1998) using FUGUE (Shi et al., 2001). The top hit was the catalytic domain of isoamylase from Pseudomonas amyloderamosa (residues 163 to 637) (Katsuya et al., 1998), with structural homology being certain (Z score = 53.7, with Z score ≥ 6.0 signifying ≥99% confidence). This result is consistent with the conformation of the potato proteins to the structural requirements of members of the α-amy-lase superfamily of (αβ)8 barrel proteins (Figure 1). All three isoforms contained eight regions of β-strand, each followed by eight regions of α-helix except for β-strand 5. Between β-strand 3 and β-strand 4 and between β-strand 6 and β-strand 7, there were additional regions of α-helix in each protein, as has been reported in other starch hydrolases of the α-amylase superfamily (Matsuura et al., 1984; Buisson et al., 1987; Jespersen et al., 1991, 1993; Klein et al., 1992).
Figure 1.
Alignment of the Predicted Protein Sequences of Stisa1, Stisa2, and Stisa3 with the Isoamylase from P. amyloderamosa.
Protein sequences were aligned using CLUSTAL W and FUGUE. Regions of β-strand in P. amyloderamosa isoamylase (Paisa) are indicated (b) below the alignment, regions of α-helix are indicated (a) below the alignment, and regions of 3/10 helix are indicated (3) below the alignment. For clarity, the regions of β-strand and the loops between these and the regions of α-helix in the (αβ)8 barrel structure are labeled in red and purple, respectively. The eight residues absolutely conserved in all active members of the α-amylase superfamily are indicated by green asterisks. The predicted first amino acids in the mature peptides of Stisa1, Stisa2, and Stisa3 are boxed in purple.
The active site of starch hydrolases, which includes sites for substrate binding and the catalytic amino acid side chains, is formed from the regions of β-strand (toward their C-terminal ends) and the loops between the regions of β-strand and α-helix within the barrel structure. Eight residues have been observed to be absolutely conserved in all members of the superfamily, and these belong to the active site. These residues are Asp-292, Val-294, His-297, Arg-373, Asp-375, Glu-435, His-509, and Asp-510 in P. amyloderamosa isoamylase. The three carboxylic acid groups Asp-375, Glu-435, and Asp-510 are essential for catalytic activity (MacGregor, 1993). All eight residues are conserved in Stisa1 and Stisa3 and lie within regions of strong similarity to the primary sequence of isoamylase from P. amyloderamosa (amino acids marked by green asterisks in Figure 1). However, in Stisa2, only two of the eight residues are conserved, Val-294 and His-297. Asp-292 is replaced by Glu, Arg-373 is replaced by Val, Asp-375 is replaced by Val, Glu-435 is replaced by Asp, His-509 is replaced by Asn, and Asp-510 is replaced by Ser (Figure 1). Although some of these changes represent conservative substitutions, the substitution of Arg-373 by Val, Asp-375 by Val, His-509 by Asn, and Asp-510 by Ser are likely to affect the catalysis of the enzyme profoundly. From these structural considerations, we predicted that Stisa2 was unlikely to have starch hydrolase activity. Interestingly, these substitutions of the active site residues in Stisa2 occur within regions that, overall, maintain their similarity to the sequences of other members of the α-amylase superfamily, suggesting that although Stisa2 may lack hydrolytic activity, it may retain the ability to bind glucans.
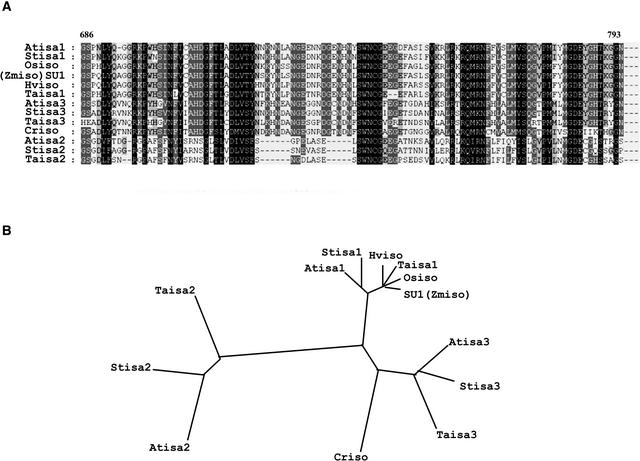
The predicted peptide sequences for Stisa1, Stisa2, and Stisa3 were used to screen the sequence databases for homologous sequences from other plants. The complete Arabidopsis genome sequence contains three genes that have homology with isoamylases in their predicted protein products. Atisa1 is most similar to Stisa1 and the Su1 gene product from maize. It resides on chromosome 2 in Arabidopsis. Atisa2 is most similar to Stisa2 and is encoded by the DBE1 locus (Zeeman et al., 1998b). It lies on chromosome 1 in Arabidopsis. Atisa3 is most similar to Stisa3 and is encoded by a gene on chromosome 4. Among the other plant EST sequences available in the public databases are partial sequences for different isoamylase genes from wheat. Alignment of these partial sequences against the equivalent peptide sequences of the isoforms from potato and Arabidopsis revealed that wheat also has three distinct genes that encode different isoamylase isoforms (Figure 2A). These align with the isoform types from Arabidopsis and potato such that we can conclude that three isoforms are present in both monocots and dicots and that they fall into structurally distinct isoform classes (Figure 2B).
Figure 2.
Phylogenetic Relationships between Isoamylases from Different Species.
(A) Alignment of fragments of peptide sequences of isoamylases from ESTs available in the public databases by CLUSTAL W. Highly conserved amino acid residues found in all sequences are boxed in black, those with related side groups present in 75% or more of the peptides are boxed in dark gray, and those with related side groups present in 50% or more of the peptides are boxed in light gray. The numbers above the alignment refer to the amino acid positions denoted in Figure 1.
(B) Dendrogram showing the relatedness of the isoamylase peptide sequences from different species. Three distinct subgroups of isoamylase, typified by Stisa1, Stisa2, and Stisa3, are apparent. Atisa1 is At2g39930, Atisa2 is At1g03310, and Atisa3 is At4g09020 from Arabidopsis. Taisa1 is AF438328, Taisa2 is BG262546, and Taisa3 is BE492683 from wheat. Hviso is an isoamylase identified from barley (AF142589; Sun et al., 1999), SU1 (Zmiso) is the product of the sugary1 gene (U18908) from maize (James et al., 1995), and Osiso is the isoamylase from rice (AB015615; Fujita et al., 1999). Criso is deduced from EST AV630278 from Chlamydomonas.
Perhaps the most interesting observation from this comparison was that the primary structure of the Atisa2 protein is very similar to that of Stisa2, including the substitution of six of the eight conserved amino acids of the active site. The substitutions for four of the six “absolutely conserved residues” are the same in Stisa2 and Atisa2. This finding suggests that despite the unlikelihood of Stisa2 encoding an active isoamylase, the amino acid substitutions that have replaced the “essential amino acids” have been conserved over wide evolutionary distances. These structural features suggested that Stisa2/Atisa2 play conserved, noncatalytic roles in determining isoamylase activity.
Other isoamylases that have been characterized are the isoamylase from rice and one from barley (Fujita et al., 1999; Sun et al., 1999). These align most closely with Su1, Stisa1, and Atisa1, showing that all of these proteins belong to the isa1 subgroup of isoamylases in plants, which fits well with the similarities in the mutant phenotypes associated with the loss of function of these genes (Pan and Nelson, 1984; Nakamura et al., 1996; Burton et al., 2002). A search of EST sequences from Chlamydomonas revealed a series of overlapping sequences that could be combined to form a contiguous fragment of cDNA encoding a fragment of an isoamylase peptide. This peptide is most similar to the isa3 isoforms from Arabidopsis, potato, and wheat. A second Chlamydomonas EST that encodes sequences homologous with isoamylases does not overlap with the EST sequences more 3′ in the cDNA. Therefore, this sequence could come from a second gene that encodes isoamylase or from the 5′ end of the same cDNA that gave rise to the other clones. The predicted peptide sequence of this EST is most similar to that of Stisa3. Within the Chlamydomonas EST sequences currently available, we found no evidence of genes that encode isoamylase isoforms similar to Stisa1 or Stisa2.
Expression of Stisa1, Stisa2, and Stisa3
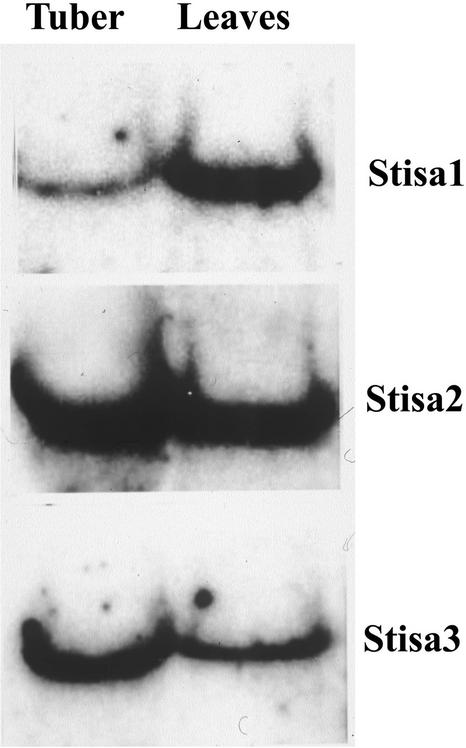
To determine whether the genes that encode the different isoforms of potato isoamylase operate in different starch-synthesizing tissues, we examined the expression of each in RNA from developing tubers and from leaves of plants harvested during the day. RNA gel blots revealed each gene to be expressed in both tubers and leaves (Figure 3). The blots were washed at high stringency to avoid cross-hybridization between the probes and the transcripts of the other Stisa isoforms. RNA gel blots showed all three genes to be expressed in both storage starch– and transitory starch–synthesizing tissues.
Figure 3.
Expression of mRNA of Isoamylase Isoforms in Leaves and Tubers of Potato.
Total RNA (20 μg per sample) was separated on denaturing agarose gels and blotted onto nitrocellulose. Duplicate blots were probed with 32P-labeled cDNA encoding Stisa1, Stisa2, or Stisa3.
Subcellular Localization of Isoamylase Isoforms
One explanation for the presence of multiple isoforms of isoamylase in starch-synthesizing tissues of potato could be that the enzymes have different subcellular localizations. The isoamylase activity of maize endosperm is plastidial (Doehlert and Knutson, 1991; Yu et al., 1998), and there is some evidence that the isoamylase activity in Arabidopsis leaves is plastidial (Zeeman et al., 1998a, 1998b). In developing pea embryos, isoamylase activity is largely or entirely confined to the amyloplasts, whereas pullulanase activity is present both inside and outside the plastids (Zhu et al., 1998). To test the localization of the isoamylase isoforms of potato, chloroplast import assays were performed using isolated pea chloroplasts and in vitro–translated proteins synthesized from the cDNA clones encoding Stisa1, Stisa2, and Stisa3; the results of this analysis are shown in the supplemental data online. These experiments demonstrated that all three isoforms carry plastid-targeting transit peptides at their N termini and confirmed the plastidial localization of all of the isoforms. After import, all three isoamylases were localized in the plastid stroma.
Debranching Enzyme Activity of the Isoamylase Isoforms
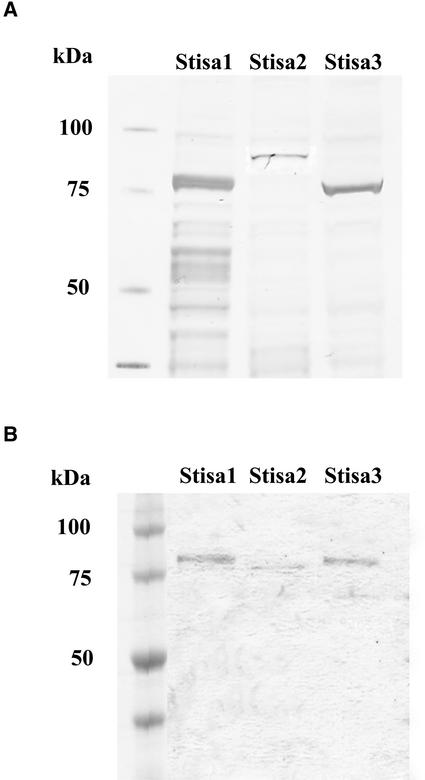
To examine the activity of the different isoamylase isoforms, each was expressed in E. coli. The cDNA sequences that encode the predicted mature proteins were cloned into the expression vector pSTAG (Edwards et al., 1999) such that each was fused, in frame, behind the 15–amino acid S-TAG peptide from RNaseS. The synthesis of each isoform of potato isoamylase in E. coli was confirmed using SDS-PAGE followed by Coomassie blue staining of proteins and by blotting the proteins onto nitrocellulose and developing with biotinylated S-protein to detect the tagged proteins (Figure 4A). All three isoforms were produced in the soluble phase when the bacteria were grown under the appropriate conditions. All three proteins were of the appropriate size, as predicted from the molecular mass of the mature isoamylase plus the size of the S-TAG peptide.
Figure 4.
Expression of Stisa Proteins in E. coli.
Stisa proteins were expressed as S-tagged fusion proteins in E. coli.
(A) Separation of soluble extracts of E. coli expressing Stisa1, Stisa2, or Stisa3 by SDS-PAGE. Proteins were blotted onto nitrocellulose and visualized by developing the blots with biotinylated S-protein (Invitrogen, Carlsbad, CA). Molecular mass is indicated in kD.
(B) Stisa1, Stisa2, and Stisa3 were purified by binding to S-agarose. The bound proteins were released by thrombin cleavage, separated by SDS-PAGE, and stained with Coomassie blue. The Stisa proteins appear pure by this method. Stisa2 was cleaved by thrombin within the isoamylase peptide as well as at the point of fusion to the S-TAG, so it was reduced in size compared with the mature protein.
It was necessary to express the isoamylases as tagged fusion proteins because it was not possible to measure isoamylase activity in crude extracts of E. coli as a result of interfering activities from amylases and other glucanases. Consequently, a one-step purification procedure was used to remove interfering activities. Crude extracts containing S-tagged proteins were incubated with S-agarose beads. The beads were washed, and then the isoamylase activity was detected on the S-agarose support. The one-step purification gave a considerable enrichment of each isoamylase (Figure 4B) and reduced the background activity in the isoamylase assays effectively to zero.
We tested whether the S-TAG or agarose support affected the activity of the isoamylases by measuring the activity of each on S-agarose beads and then cleaving the fusion protein with biotinylated thrombin to remove the tag. For Stisa1 and Stisa3, we recovered 84 and 88% of the activity, respectively, after thrombin cleavage. Thrombin cleaved the mature Stisa2 protein (detected by SDS-PAGE) as well as removing the S-TAG (Figure 4), so we were unable to test whether or not the S-TAG affected Stisa2 activity. Overall, our results indicated that the S-TAG had little or no effect on activity when fused N terminally to the isoamylases.
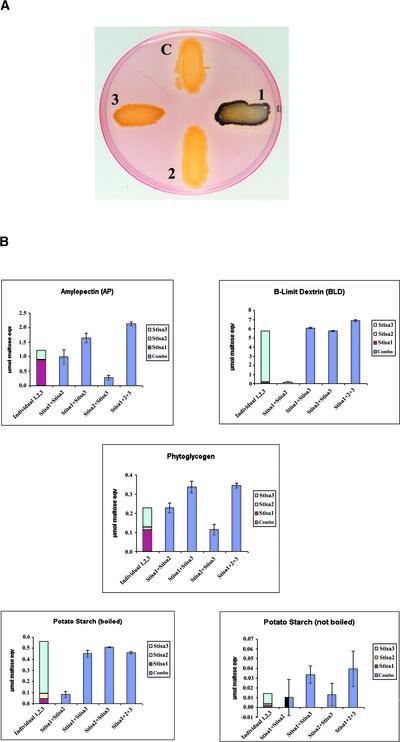
As an initial screen for activity, E. coli strains expressing each isoamylase isoform were stained with iodine vapor to determine which, if any, could affect the structure of glycogen synthesized by E. coli. Controls stained weakly the red/brown color normal for glycogen from E. coli, but lines expressing Stisa1 or Stisa3 stained bluer with iodine, suggesting that the activity of these isoforms had an effect on glycogen structure. There was no observable effect of Stisa2 on glycogen in E. coli (Figure 5A).
Figure 5.
Activity of Stisa Proteins in Debranching Glucan.
(A) E. coli expressing Stisa1 (1), Stisa2 (2), and Stisa3 (3) grown to the stationary phase and stained with iodine vapor. C indicates a control strain carrying empty vector. Colonies expressing Stisa1 and Stisa3 stained blue, whereas the control and Stisa2 colonies stained very light red/brown.
(B) Activities of mixtures of Stisa proteins on amylopectin, β-limit dextrin, phytoglycogen, boiled potato starch, and nonboiled starch granules. The equivalent average activity for each isoamylase alone is indicated by a colored bar in the first column: red for Stisa1, yellow for Stisa2, and light blue for Stisa3. These activities of each isoform are represented by the size of the bar of the appropriate color only but are shown one atop the other for ease of comparison and to identify additive interactions in the mixtures. The dark blue columns indicate the activities of the mixtures of isoforms as labeled below each column, with the standard deviations calculated from three different assays with two different preparations of proteins from E. coli. Combo refers to the activity of Stisa1, Stisa2, and Stisa3 mixed together.
The three isoamylases were extracted and, after optimization for pH, temperature, and substrate concentration, assayed on different substrates: amylopectin, β-limit dextrin, pullulan, phytoglycogen, and potato starch granules (Table 1). Stisa2 showed no activity on any of these substrates. Stisa1 was most active on amylopectin but showed some activity on phytoglycogen. It had relatively low activity on the β-limit dextrin of amylopectin. These data for the substrate specificity and specific activity of Stisa1 agree strongly with the data of Rahman et al. (1998) for the maize Su1 protein, supporting the view that the Stisa1 isoform of potato is functionally equivalent to the Su1 isoform of maize. By contrast, Stisa3 had relatively high activity on β-limit dextrin. Its activity on amylopectin and phytoglycogen was much less (15-fold less). Neither Stisa1 nor Stisa3 had activity on pullulan, confirming that these enzymes are isoamylase-type debranching enzymes.
Table 1.
Activity of Isoamylase Isoforms from Potato on Different Glucan Substrates
| Glucan
|
||||||
|---|---|---|---|---|---|---|
| Isoamylase | Amylopectin | β-Limit Dextrin | Pullulan | Phytoglycogen | Potato Starch (Boiled) | Potato Starch (Not Boiled) |
| Stisa1 | 5.871 ± 0.461 | 1.072 ± 0.081 | 0.222 ± 0.102 | 0.747 ± 0.078 | 0.268 ± 0.163 | 0.013 ± 0.078 |
| Stisa2 | 0.069 ± 0.047 | 0.020 ± 0.207 | 0.251 ± 0.144 | 0.092 ± 0.148 | 0.346 ± 0.245 | 0.010 ± 0.029 |
| Stisa3 | 4.328 ± 1.090 | 74.135 ± 1.590 | 0.310 ± 0.235 | 1.345 ± 0.344 | 6.257 ± 0.479 | 0.143 ± 0.063 |
Mean activity (μmol·h−1·mg−1 protein) and standard deviations were calculated from triplicate assays of proteins bound to S-agarose from each of two independent preparations.
Despite Stisa1 being most active on amylopectin and Stisa3 preferring β-limit dextrin to amylopectin, neither of these glucans is likely to represent the type of glucan most readily available to debranching enzymes in tuber cells that synthesize starch. Amylopectin rapidly crystallizes at the granule surface and so becomes unavailable to debranching enzymes, whereas β-limit dextrins may be produced during starch degradation but probably are not present at high levels in cells that undertake net synthesis of starch. Therefore, we tested the activity of the isoamylases on two other substrates, which are, arguably, more similar to the glucans available in starch-synthesizing cells: phytoglycogen and starch granules.
Stisa2 showed essentially no activity on either substrate. Stisa1 showed no activity on starch granules but significant activity on phytoglycogen. Stisa3 had the highest specific activity of the three isoamylases on both substrates, although its activity on these substrates was between 12- and 500-fold lower than its activity on β-limit dextrin.
We also tested for potential interactions between the different isoforms on different substrates. Extracts containing pair-wise combinations of each isoamylase and all three together were mixed before the addition of the S-agarose beads, and debranching enzyme activity was measured on the beads. We reasoned that coexpression experiments would not provide relevant data because each Stisa protein has to be imported into the plastid before they can interact. The data from the mixing experiments are shown in Figure 5B. A slight interaction between Stisa1 and Stisa3 isoforms was observed using amylopectin as a substrate (34% extra activity). This effect also was seen when all three isoforms were combined (75.8% extra activity). This increase in activity was not observed when β-limit dextrin was used as a substrate. On phytoglycogen, there was a synergistic increase in activity when Stisa1 and Stisa2 were mixed (81% extra activity). Mixing Stisa1 and Stisa3 gave 58.2% extra activity on phytoglycogen. No additional increase was observed when all three isoforms were mixed. On starch granules (either native or solubilized), there was no activity of Stisa1 or Stisa2 and no evidence for interaction between these isoforms. The activity of Stisa3 on solubilized starch granules (boiled) was not enhanced by interaction with either Stisa1 or Stisa2. The activity of Stisa3 on intact starch granules was very low compared with its activity on other substrates.
The interpretation of these interactions is complex because the substrates themselves are complex. The enhanced activity of Stisa1 and Stisa3 together on amylopectin and phytoglycogen may result from the activity of one isoform making more substrate available to the second isoform, as was suggested recently for starch-branching enzyme isoforms (Seo et al., 2002). For example, removal of short glucan chains by Stisa3 may make longer branches accessible to Stisa1 and vice versa. However, our data also show that Stisa1 and Stisa2 interact synergistically to enhance their debranching activity on the soluble glucan phytoglycogen, an interaction that cannot be explained by the complex nature of the substrate, because Stisa2 has no activity on its own. It is possible that this interactive activity also works for other similar soluble, branched glucans that might be produced transiently in plastids that synthesize starch.
Nature of the Isoamylase Activity in Potato Tubers
To increase our understanding of the relationship between the isoforms of isoamylase, we analyzed isoamylase activity from tuber extracts after separation from other starch-hydrolyzing activities.
The quantification of isoamylase activity in extracts of higher plant organs generally is impossible because of interference in assays from other starch-degrading enzymes. A specific assay developed for the enzyme in Chlamydomonas extracts (Dauvillée et al., 2001a) was not specific for isoamylases in extracts of potato as a result of the multiplicity of starch-hydrolyzing enzymes in potato tubers. Instead, we visualized isoamylase activity on native gels containing glucan substrate (amylopectin or β-limit dextrin). After the incubation of gels at an appropriate pH, followed by staining with iodine solution, regions containing activities of starch-degrading enzymes appeared as clear or colored bands against a dark background. Isoamylase activity was observed as a slow-migrating blue band. The only other glucan-degrading enzyme to give a blue band (indicative of linear chains) was the pullulanase-type debranching enzyme. Pullulanase can be distinguished from isoamylase by transferring proteins from the native gel to a gel containing pullulan linked to a colored dye. Pullulanase hydrolyzes pullulan, leaving a clear band on the pullulan gel, whereas isoamylase does not. Based on these criteria, we identified isoamylase as a major blue-staining band on native amylopectin gels of crude extracts of tubers.
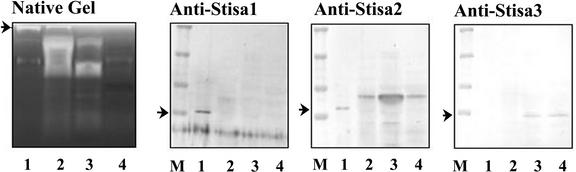
To discover which of the three forms of isoamylase expressed in the potato tuber was responsible for this activity, we first prepared antisera to peptides specific to each of the three proteins. The specificity of the antisera was verified by testing against extracts of E. coli expressing the recombinant isoforms. As expected, each antiserum recognized only the isoform containing the peptide to which it was raised (see supplemental data online).
The antisera were used to discover which of the three isoforms was present in the isoamylase separable from partially purified tuber extracts on native gels. In a series of ammonium sulfate precipitations of crude, soluble extracts of tubers, most of the isoamylase activity detectable as the blue band on native gels was present in material precipitating at between 0 and 20% saturation with ammonium sulfate (Figure 6). Immunoblot analysis revealed that proteins recognized by antisera to Stisa1 and Stisa2 also precipitated at between 0 and 20% saturation with ammonium sulfate. However, the protein recognized by the Stisa3 antiserum precipitated at higher ammonium sulfate concentrations than most of the activity recognized on the native gels (Figure 6). Thus, the major isoamylase activity detectable as a separable blue band on native, amylopectin-containing gels is a function of Stisa1 and/or Stisa2 but is not attributable to Stisa3.
Figure 6.
Detection of Stisa Proteins and Isoamylase Activity in Ammonium Sulfate Fractions of Crude Soluble Extracts of Tubers.
Proteins precipitating at ammonium sulfate concentrations of 0 to 20% (lane 1), 20 to 30% (lane 2), 30 to 40% (lane 3), and 40 to 50% (lane 4) saturation were redissolved and subjected to electrophoresis either on a native, amylopectin-containing gel that was stained subsequently with iodine solution (left gel) or on SDS–polyacrylamide gels that were blotted subsequently onto nitrocellulose. Blots were developed with antisera raised to synthetic peptides unique to Stisa1, Stisa2, or Stisa3 (at dilutions of 1:10,000 [Stisa1 and Stisa3] or 1:20,000 [Stisa2]). Lanes 1 to 4 contain protein from the four ammonium sulfate precipitates. Each lane contains the same fraction of the precipitated protein. M indicates molecular mass markers. The arrow on the native gel indicates the position of the blue band of isoamylase activity. Arrows on the protein gel blots indicate the positions of the Stisa1, Stisa2, and Stisa3 peptides detected by the antisera.
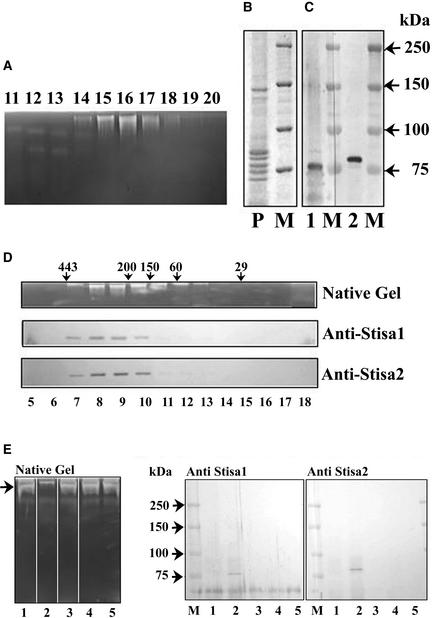
To investigate the relationship between Stisa1, Stisa2, and the isoamylase activity detected on native gels, activity was purified further from tubers. The initial step was either ammonium sulfate precipitation or precipitation at pH 5. The results discussed below were the same regardless of the initial step: precipitation at pH 5 gave a purer final preparation but a lower yield than ammonium sulfate precipitation. The precipitation step was followed by ion-exchange chromatography on DEAE-Sepharose and MonoQ and, finally, size-exclusion chromatography. After ion-exchange chromatography, the preparation contained the proteins recognized by the Stisa1 and Stisa2 antisera (Figures 7A to 7C). After size-exclusion chromatography, the isoamylase activity eluted with a molecular mass of 450 to 500 kD. The proteins recognized by the Stisa1 and Stisa2 antisera coeluted with the activity (Figure 7D). These data suggest that the isoamylase activity detected on native gels is attributable to a multi-meric protein containing approximately six isa proteins and that both Stisa1 and Stisa2 are present in this multimeric enzyme.
Figure 7.
Presence of Stisa1 and Stisa2 Proteins in Partially Purified Isoamylase.
Isoamylase activity was purified by an initial precipitation step (brought about by either ammonium sulfate or pH 5), followed by ion-exchange chromatography on DEAE-Sepharose and MonoQ and gel filtration on Superose 12.
(A) Fractions (lanes 11 to 20) from the MonoQ separation (after protein precipitation at pH 5) were subjected to electrophoresis on native, amylopectin-containing gels to show the isoamylase activity by staining with iodine solution.
(B) Coomassie blue staining of an SDS–polyacrylamide gel of protein from MonoQ fraction 16 (P). M indicates protein size markers with molecular mass in kD.
(C) Protein from MonoQ fraction 16 separated on SDS–polyacrylamide gels and transferred to nitrocellulose probed with antiserum to Stisa1 (lane 1) or Stisa2 (lane 2). M indicates protein size markers with molecular mass in kD.
(D) Fractions after Superose 12 chromatography, from a preparation using an ammonium sulfate precipitation. Isoamylase activity, as determined by native amylopectin gel separation followed by iodine staining, is compared with the presence of Stisa1 and Stisa2 peptides in the fractions, as determined by electrophoresis of the fractions (lanes 5 to 18) on SDS–polyacrylamide gels, transfer to nitrocellulose, and development with the anti-Stisa1 and anti-Stisa2 antisera. Arrows indicate molecular mass in kD of proteins eluting at particular points, as determined using size standards.
(E) Native, amylopectin-containing gel and immunoblots of SDS–polyacrylamide gels of supernatant fractions after incubation with antisera to Stisa1 and Stisa2. Partially purified isoamylase was incubated with protein A–Sepharose that had been preincubated as follows: lane 1, Stisa1 antiserum; lane 2, Stisa 2 antiserum; lane 3, Stisa1 preimmune serum; lane 4, Stisa2 preimmune serum; lane 5, BSA at 20 mg/mL in tuber extraction medium plus 0.15 M KCl. The left gel shows the isoamylase activity (arrow) remaining in the supernatants for these different treatments determined on amylopectin gels. Lane 2 shows the loss of the isoamylase band caused by the interaction of the Stisa2 antibody with the native isoamylase. The proteins from the antisera/protein A precipitations were recovered by centrifugation of the Sepharose and release by boiling in SDS sample buffer. These proteins were subjected to SDS-PAGE followed by immunoblotting with antiserum to Stisa1 (middle gel) or Stisa2 (right gel) as indicated. Both Stisa1 and Stisa2 peptides were recovered from the interaction between the Stisa2 antiserum and the isoamylase (lane 2 in both gels). Prestained markers are shown (M), with molecular masses indicated in kD.
Immunoprecipitation with the Stisa1 and Stisa2 antisera was performed on the partially purified enzyme (purified by an initial ammonium sulfate purification and the ion-exchange chromatography steps). In both preparations, Stisa2 antiserum inhibited isoamylase activity but the Stisa1 antiserum did not. Immunoblot analysis of proteins remaining in the soluble fractions of incubations after immunoprecipitation revealed that the Stisa1 antiserum did not immunoprecipitate the Stisa1 protein (Figure 7E). The anti-Stisa1 antiserum was raised to a peptide of 12 amino acids, and it seems likely that this motif is not accessible to the antiserum in the native enzyme. However, the Stisa2 antiserum immunoprecipitated both the Stisa1 and Stisa2 proteins (Figure 7E). The coprecipitation of Stisa1 and Stisa2 by the Stisa2 antiserum is consistent with the idea that both are associated in a single multimeric enzyme. An alternative explanation of the coprecipitation is that a component of the Stisa2 antiserum recognizes the native Stisa1 protein. We consider this to be highly unlikely. First, the preimmune serum immunoprecipitated neither Stisa2 nor Stisa1 (Figure 7E). Second, the peptide to which the Stisa2 antiserum was raised is absent from the Stisa1 protein. Third, the Stisa2 antiserum did not recognize the denatured Stisa1 protein (see supplemental data online).
Our data show that the relatively high molecular mass, multimeric enzyme separable from extracts of potato tubers analyzed on native gels comprises Stisa1 and Stisa2 subunits. These data fit very well the early isolation of isoamylase activity by Ishizaki et al. (1983), who identified a high molecular mass enzyme (estimated at 520 kD) that consisted of two polypeptides, one of 94 kD and one of 83 kD. Our data give mature Stisa2 a predicted molecular mass of 94 kD and mature Stisa1 a predicted molecular mass of 84 kD. There is no evidence, either from our analysis of this high molecular mass isoamylase or from the work of Ishizaki et al. (1983), for the inclusion of Stisa3 in this enzyme, even though the Stisa3 protein is present in the tubers and is targeted to the plastids.
DISCUSSION
The Three Isoamylase Isoforms Have Different Catalytic Specificities
cDNA clones encoding three isoforms of isoamylase-type starch-debranching enzyme have been identified from potato. The predicted products of these genes can be classified as isoamylase-like on the basis of their structural similarity to plant and bacterial isoamylases, and the proteins belong to the α-amylase superfamily. Genes that encode three isoamylases also can be identified within the complete genome sequences of Arabidopsis and rice (Arabidopsis Genome Initiative, 2000; Goff et al., 2002; Yu et al., 2002). From searches of these completed genome sequences and searches of EST collections from different higher plants, we believe that these three isoamylase isoforms are produced in most, if not all, monocots and dicots and that they represent the major types of isoamylase produced in angiosperms. From structural and functional analysis, Stisa1 appears to be most similar to the Su1 protein of maize and to other isoamylases from cereals that are associated with sugary phenotypes when mutated. Mutants of sugary1 of maize, sugary of rice, and notch2 of barley have reduced storage starch synthesis in endosperm and accumulate a highly branched, water-soluble polysaccharide, phytoglycogen (Pan and Nelson, 1984; Nakamura et al., 1996; Burton et al., 2002). They have reduced starch-debranching enzyme activity during endosperm development. Structurally, Stisa2 is related most closely to the product of the DBE1 locus of Arabidopsis.
Analysis of the predicted structure of Stisa2 suggests that it is unlikely to have catalytic activity, a prediction confirmed by our expression of Stisa2 in E. coli. Although Stisa2 shows no debranching catalytic activity in vitro and in vivo, it is known from the dbe1 mutant of Arabidopsis that Atisa2 is required for the isoamylase activity that can be detected as a slowly migrating protein that stains blue on amylopectin gels. In the dbe1 mutant, deletion of the Atisa2 gene results in the loss of this protein, reduced amylopectin synthesis, and the accumulation of phytoglycogen (Zeeman et al., 1998b). Our structural and functional analysis of Stisa2 suggests that the role of isa2 in starch synthesis is an indirect one: possibly, it regulates the activity of the isa1 isoform in the multimeric enzyme. This potential arrangement of catalytic and regulatory subunits in a multimeric enzyme is very similar to that of ADP-Glc pyrophosphorylase in higher plants. In bacteria, ADP-Glc pyrophosphorylase comprises a homotetramer. In plants, two subunits (the large and the small subunits), both with structural similarity to the bacterial enzyme subunit, are associated in a heteromeric enzyme. The small subunit has catalytic activity on its own, but the large subunit does not. The large subunit modifies the regulatory properties of the holoenzyme (Ballicora et al., 1995; Doan et al., 1999; Kavakli et al., 2001).
Stisa3 is a new form of isoamylase, and there are no known phenotypes associated with the loss of its function. The closest equivalent might be the product of the STA7 gene in Chlamydomonas, because available ESTs predict an isoamylase most similar to Stisa3. However, molecular analysis of the STA7 locus is required to confirm the structural and functional properties of the isoamylase that STA7 encodes or regulates. Biochemical evidence suggests that the isoamylase encoded by or regulated by STA7 assembles in a multimeric form (Dauvillée et al., 2000, 2001a, 2001b). Analysis of sta8 mutants of Chlamydomonas suggests that STA8 encodes a different component of the complex. The loss of STA8 activity causes a reduction, but not elimination, of isoamylase activity and the loss of a significant proportion of the large isoamylase complexes detected on native gels. By analogy to the situation in potato, STA7 might encode a catalytically active isoamylase and STA8, the regulatory component of the multimeric isoamylase (Dauvillée et al., 2001a, 2001b), although at present there is no evidence for isoamylase isoforms equivalent to Stisa1 or Stisa2 from the Chlamydomonas EST databases.
By comparing the primary amino acid sequences of the potato isoamylases and relating these to the known structure of isoamylase from P. amyloderamosa, we were able to make further predictions about the activities of the different potato isoforms. It has been suggested that differences in loop lengths between β-strands and α-helices provide specificity for substrate binding in different members of the (αβ)8 barrel starch hydrolase superfamily (MacGregor, 1993). The long loops between β-strand 2 and α-helix 2 and between β-strand 7 and α-helix 7 may allow for the binding of long branches within the substrate typical for isoamylases, and generally, the length of these loops is longer in isoamylases than in pullulanases, whether they are of plant or bacterial origin. Loop 2 and loop 7 of Stisa2 are significantly shorter than those of Stisa1 and Stisa3, suggesting that the Stisa2 isoform might preferentially bind a different glucan substrate to the other two isoforms from potato.
Mutagenesis experiments on bacterial isoamylase from Flavobacterium odoratum KU have shown that the distances between the active site carboxylic acid residues are important to substrate specificity (Abe et al., 1999). These distances are defined by the lengths of loops 4 and 5 in the barrel structure. Loop 4 in Stisa1 is predicted to be 55 amino acids long, very similar to the Su1 protein of maize. In both Stisa2 and Stisa3, loop 4 is 36 amino acids long, which is similar to the length of loop 4 in the glgX isoamylases from E. coli and Chlamydia, which preferentially hydrolyze β-limit dextrins (Jeanningros et al., 1976; Abe et al., 1999). This difference in the lengths of loop 4 suggests that Stisa2 and Stisa3 isoforms may preferentially bind glucan chains that are relatively short on the nonreducing side of the α-1,6 branch point and show relatively higher affinity for β-limit dextrins than for amylopectin (like the glgX isoamylase of E. coli [Jeanningros et al., 1976]). The plant isoamylases that are most similar to Stisa1 are likely to have higher affinity for longer branches, such as those found in amylopectin (Rahman et al., 1998). Therefore, Stisa3 is predicted to encode an isoamylase with a preference for shorter branches than Stisa1. We gathered experimental support for these predictions by demonstrating that the Stisa3 enzyme (when expressed in E. coli) shows a strong substrate preference for β-limit dextrin over amylopectin, whereas Stisa1 prefers amylopectin over β-limit dextrin as a substrate.
Roles of Stisa1, Stisa2, and Stisa3 in Starch Synthesis
We have shown that all three isoforms are targeted to plastids and so have the potential to be involved in starch synthesis as well as mobilization. All three genes are expressed during storage and transitory starch synthesis, consistent with the view that all of them may influence starch synthesis. Our biochemical analysis of debranching enzymes in potato tubers shows that Stisa1 and Stisa2 interact in a multimeric enzyme (probably a hexamer), whereas Stisa3 is not associated with this complex. Many groups have used the high molecular mass protein that can be observed as a blue band on native amylopectin gels as a measure of isoamylase activity. It is clear from our analysis that this protein includes Stisa1 and Stisa2 but not Stisa3. This means that assays based on the single band of isoamylase activity on native amylopectin gels do not measure all of the isoamylase activity present in a plant cell, just that associated with the isa1 and isa2 isoforms. However, given that it is this high molecular mass enzyme that disappears in every mutant that accumulates phytoglycogen, it seems likely that it is Stisa1 and Stisa2 (and their functional homologs in other plant species) that play the central role in starch synthesis (James et al., 1995; Nakamura et al., 1996; Zeeman et al., 1998b; Kubo et al., 1999; Burton et al., 2002).
Our biochemical analysis has shown that the specificities of Stisa1 (or Stisa1 and Stisa2 together) and Stisa3 differ significantly for different glucan substrates. These differences are likely to be very important in determining the role that each isoform plays in starch synthesis or in starch mobilization. Although in vitro, the preferred substrate for Stisa1 (or Stisa1 and Stisa2 together) is soluble amylopectin, the starch-debranching enzymes active in the plastid stroma during starch synthesis are unlikely to encounter much glucan in this form. We believe that the activity of Stisa1 on phytoglycogen is significant, particularly because it is increased by association with Stisa2, a situation that mirrors their interaction in a multimer in the starch-synthesizing plastid. The low activity of Stisa1 or Stisa1/Stisa2 on whole starch suggests that in vivo the primary substrate for the Stisa1/Stisa2 complex is soluble branched glucan rather than any highly branched regions on the outside of starch granules. Similar conclusions were drawn for the activity of an isoamylase characterized recently in wheat (Genschel et al., 2002). The Stisa1 and Stisa3 isoforms also show interactions in their ability to debranch some substrates. This is probably different from the interaction between Stisa1 and Stisa2 and likely results from the sequential activity of isoforms with different specificities on a complex substrate. The activity of one isoform may expose α-1,6 branches that are better substrates for the second isoform, thus allowing more extensive debranching of a complex substrate when the two isoforms work together. This type of interactive activity was suggested recently for the activities of starch-branching enzyme isoforms from maize (Seo et al., 2002).
By analogy to the equivalent isoamylase isoforms in cereals and Arabidopsis and their mutant phenotypes, we conclude that the multimeric enzyme formed by isa1 and isa2 exists in many different plant species and that it is the activity of this multimer that plays a central role in amylopectin biosynthesis. However, in rice, although proteins homologous with both Stisa1 and Stisa2 are encoded by the genome (Goff et al., 2002; Yu et al., 2002), biochemical analysis of the enzyme active in endosperm has shown it to be a monomeric multimer (Fujita et al., 1999). Perhaps the isa2 isoform is not expressed in developing rice endosperm. In maize endosperm, Doehlert and Knutson (1991) identified and separated two isoamylase activities. One peak of activity (type II) was of high molecular mass and possibly represented a multimeric enzyme, although the authors did not determine whether it was monomeric or multimeric. The generality of the association between isa1 and isa2 isoforms and the effects of this association on isoamylase activity await further biochemical characterization of isoamylases in other plant species.
Based on our evidence of Stisa1/Stisa2 substrate specificity, the primary function of this complex in potato starch synthesis is likely to be the removal of branched, soluble glucan. The removal of branched, soluble glucan may be important for effective granule synthesis either because it competes for ADP-Glc substrate with starch synthesis or because it serves to prime ectopic starch granule initiation if allowed to form unchecked (Burton et al., 2002). We believe that the fact that the inhibition of isoamylase activity results in increased numbers of starch granules (crystallization-competent units) (Boyer et al., 1977; Yeh et al., 1981; Shannon and Garwood, 1984; Zeeman et al., 1998b; Kubo et al., 1999; Burton et al., 2002) argues against the glucan-trimming model as an explanation of the role of debranching enzymes in starch synthesis, and the substrate preference of Stisa1/Stisa2 supports this interpretation. The physiological role of isa3 awaits definition through mutant analysis, although its substrate preference for β-limit dextrins suggests that it may play a more significant role in starch mobilization than in starch synthesis.
METHODS
Isolation of the Stisa cDNA Clones
Two cDNA libraries were prepared in λgt10 from RNA isolated from potato (Solanum tuberosum) mini tubers grown in vitro on stem explants (Visser et al., 1989) and from developing tubers from greenhouse-grown plants (cv Desiree) according to the manufacturer's instructions (Amersham). A total of 120,000 plaque-forming units from the unamplified libraries were screened using a 1-kb fragment from EST At69012 as a probe. From screening both libraries, 18 positive plaques were isolated and purified. DNA was isolated from the λ clones and digested with EcoRI. The EcoRI-cut cDNA fragments were cloned into pBluescript SK+ (Stratagene). cDNA clones encoding the three different isoamylase isoforms were identified by sequencing and designated Stisa1, Stisa2, and Stisa3.
Construction of Plasmids for Stisa Expression in Escherichia coli
All of the plasmids for Stisa expression in E. coli were constructed as in-frame fusions of the mature isoamylase proteins to the S-TAG in the vector pSTAG (Edwards et al., 1999). The DNA fragments encoding the mature proteins were generated by PCR mutagenesis to introduce suitable restriction sites.
The cDNA fragment encoding mature Stisa1 was generated by PCR with the primers 5′-CCCGGGGCTGTTGATAGTGGACGTGGA-GGTG-3′ and 5′-AAAATTCACCCTTAGGAGCTAGCG-3′ to generate a 1-kb PCR product. The sequence encoding the fusion protein was assembled using triple ligation of a 1-kb SmaI-NheI fragment from this PCR product and a 1.6-kb NheI-EcoRI fragment from the Stisa1 cDNA between the EcoRV and EcoRI sites in pSTAG.
A cDNA fragment encoding mature Stisa2 was generated by PCR with the primers 5′-CCATGGGTCTAAGGAGGCTGGAATTGGAAGA-3′ and 5′-CCATATCCTTCATCGATTTAATGG-3′ to generate a 1.2-kb PCR product. The sequence encoding the fusion protein was assembled by triple ligation of a 1.2-kb NcoI-ClaI fragment from the PCR product and a 1.5-kb ClaI-NcoI fragment from the Stisa2 cDNA into the NcoI site in pSTAG.
A cDNA clone encoding mature Stisa3 was generated by PCR with the primers 5′-GATATGGCTAAACTTCAGGAAGAAGC-3′ and 5′-CGA-CATGATACTCGGTGACCC-3′ to generate a 1-kb fragment. The sequence encoding the fusion protein was assembled by a triple ligation of a 200-bp EcoRV-BamHI fragment from the PCR product and a 2-kb BamHI-EcoRI fragment from the Stisa3 cDNA between the EcoRV and EcoRI sites of pSTAG.
Chloroplast Import Assays
Intact chloroplasts were isolated from pea (Pisum sativum var Kelvedon Wonder) (Brock et al., 1993). After optimization, a wheat germ cell-free lysate was used to translate mRNA derived by T3 RNA polymerase–driven transcription of the Stisa3 cDNA clone, whereas Stisa1 and Stisa2 cDNA clones were transcribed by T3 and T7 RNA polymerases, respectively, and translated in a rabbit reticulocyte lysate system. Chloroplasts (50 μg of chlorophyll in a final volume of 125 μL) in HS (50 mM Hepes-KOH, pH 8.0, and 330 mM sorbitol) were preincubated with 8 mM MgATP for 5 min at 25°C in the light (100 μmol·m−2·s−1). A 35S-Met–labeled in vitro translation mixture (12.5 μL) was mixed with an equal volume of unlabeled Met (final concentration of 5 mM) and added to the chloroplast suspension.
Incubation was for 30 min in the light. To remove unbound proteins, chloroplasts were washed in ice-cold HS and treated with 0.2 mg/mL thermolysin for 40 min on ice. After the protease treatment, the chloroplasts were lysed in 10 mM Hepes-KOH, pH 8.0, 5 mM MgCl2, and 10 mM EDTA for 5 min on ice. The envelope and thylakoid membranes then were separated from the stromal fraction by centrifugation at 20,000g at 4°C for 10 min in a microcentrifuge. Chloroplast, stromal, and membrane fractions were analyzed by SDS-PAGE and fluorography.
Expression of Debranching Enzymes in E. coli BL21 (DE3) Rosetta
The plasmids for Stisa expression were transformed into E. coli BL21 (DE3) Rosetta (Novagen, Madison, WI). Transformed cells were inoculated into 10 mL of Luria broth and incubated overnight at 27°C with shaking at 200 rpm. A 2-mL aliquot from the overnight culture was inoculated into 50 mL of Luria broth containing 100 μg/mL ampicillin and 34 μg/mL chloramphenicol and was incubated at 30°C with shaking at 350 rpm. After 5 to 6 h, when the OD600 reached 0.6 to 0.8, expression was induced by the addition of 10 mM isopropylthio-β-galactoside followed by incubation at 20°C with shaking at 350 rpm overnight. Cells were collected by centrifugation at 10,000g and resuspended in Mes buffer (50 mM Mes, pH 6.0, 5 mM DTT, and 50 mL/L ethanediol). Resuspended cells were lysed by two passages through a French press at 20,000 p.s.i. Cell debris were removed by centrifugation at 14,000g for 5 min. The crude extract was dialyzed on a NAP-25 column (Amersham Pharmacia) to remove excess sugar and contaminants from the growth medium before being used for the S-TAG protein gel blot and protein assays.
S-TAG Protein Assay
Fusion proteins were assayed using the S-TAG protein assay kit (Novagen) according to the method of Kim and Raines (1993). Enzyme concentrations were determined by plotting the absorption of supernatant at OD280 against the S-TAG standard.
Assay of Isoamylase Activity Using the Bicinchoninic Acid Reducing Sugar Assay
Reducing end formation released by the debranching enzyme activity on different glucan substrates was quantified by the methods of Fox and Robyt (1991) and Meeuwsen et al. (2000) with a few modifications. Isoamylases expressed in E. coli were purified partially from crude extracts by binding to S-agarose beads according to the manufacturer's instructions (Novagen). Aliquots of 100 μg/mL (Stisa1 and Stisa2) and 50 μg/mL (Stisa3) were used in assays. The assays contained 5 mg/mL glucan substrate in 50 mM Mes, pH 6.0, incubated at 30°C. After 3 h, 100-μL aliquots were taken out and mixed with 50 μL of 1 M Na2CO3 to stop the reaction. A 100-μL aliquot also was taken immediately after the addition of substrate and stopped with 1 M Na2CO3 (time 0). Samples were derived from two independent preparations of each protein, and every sample was assayed in triplicate. Samples were diluted with water as required to get the absorption in the linear range, and 500 μL of the diluted reaction was added to 500 μL of BCA reagent (0.5 M Na2CO3, 0.288 M NaHCO3, and 5 mM sodium bicinchoninic acid [Sigma] mixed with 12 mM l-Ser and 5 mM CuSO4·5H2O at a ratio of 1:1). The mixture was incubated at 80°C for 1 h and then cooled to room temperature. Three aliquots of 200 μL from each assay were ordered on a microtiter plate, and samples were read at OD562. The amount of reducing sugar was determined by plotting the absorption from the reaction against the maltose standard curve. Assays were checked to establish that they were linear with respect to enzyme concentration and time and that substrate concentration was saturating. The pH optimum for Stisa1 was sharp at 6.0, whereas that for Stisa3 was broad but maximal at 7.0. The temperature curves for both Stisa1 and Stisa3 were broad: the optimum for Stisa1 was 30°C, and the optimum for Stisa3 was 40°C.
Purification of Isoamylase from Tubers
Potatoes were purchased locally and were freshly harvested tubers of 60 to 90 g fresh weight of cv Cara or cv Carlingford. All steps were performed at 0 to 4°C. Approximately 1 kg of tuber was homogenized in 2 volumes of extraction medium (50 mM Mes, pH 6.0, 10 mM Ca-acetate, 5 mM DTT, and 50 mL/L ethanediol) at 4°C. The extract was filtered through two layers of muslin and centrifuged at 20,000g for 30 min. The supernatant was subjected either to ammonium sulfate precipitation or precipitation with acetic acid. In the former case, the fraction of protein precipitating between 0 and 40% ammonium sulfate was collected by centrifugation. In the latter case, the pH of the supernatant was adjusted to 5.0 by the slow addition of 1.2 M acetic acid. After stirring overnight, the precipitate was collected by centrifugation. Precipitates were resuspended in ∼150 mL of extraction medium, centrifuged to remove undissolved material, and mixed with ∼150 mL of DEAE-Sepharose Fast Flow resin equilibrated in extraction medium. After incubation with rotation for 30 min, the liquid was removed and the resin was washed several times with extraction medium. The resin was resuspended and incubated further with 150 mL of extraction medium containing 0.3 M KCl and washed in this medium as described above. The resin then was suspended in 150 mL of extraction medium containing 0.45 M KCl, and after incubation, the liquid was collected and dialyzed against 2.5 L of extraction medium without KCl for 16 h. The dialysate was applied to a MonoQ column (Amersham Pharmacia) equilibrated with extraction medium. After washing with extraction medium, the column was eluted with a gradient of 0 to 0.6 M KCl at a flow rate of 0.7 mL/min. Fractions of 1 mL were collected. Fractions containing isoamylase activity were identified by native gel electrophoresis. A sample of 0.2 mL of the fraction of highest activity was applied to a Superose 12 gel-filtration column (Pharmacia Amersham) equilibrated with extraction medium containing 0.15 M KCl. The column was eluted with this medium at a flow rate of 0.2 mL/min, and 0.5-mL fractions were collected.
Preparation of Peptides and Antisera
Synthetic peptides were used to produce antibodies specific to the three isoforms of isoamylase from potato: peptide 1, (C)DVPERETA-AKQY, which is specific to Stisa1; peptide 2, (C)IDSSKRKKQIR-LSSKRQ, which is specific to Stisa2; and peptide 3, (C)NEADDENP-YTTS, which is specific to Stisa3. Polyclonal antisera were produced in rabbits and analyzed as 98 term bleeds.
Gel Electrophoresis and Immunoblot Analysis
Native, amylopectin-containing PAGE was performed according to Zhu et al. (1998). SDS-PAGE and immunoblot analysis were performed according to Edwards et al. (1995).
Immunoprecipitation
Immunoprecipitation experiments were performed according to the method described by Marshall et al. (1996) for rabbit antisera. Samples of 30 mg of protein A–Sepharose were preincubated with 50-μL samples of serum, preimmune serum, or BSA solution. After five washes in tuber extraction medium plus 0.15 M KCl, protein A–Sepharose that had been preincubated with Stisa1 antiserum, Stisa 2 antiserum, Stisa1 preimmune serum, Stisa2 preimmune serum, or BSA at 20 mg/mL in tuber extraction medium plus 0.15 M KCl was incubated with 100-μL samples of partially purified isoamylase. Isoamylase was analyzed after incubation on native amylopectin-containing gels. The supernatants from the incubations after centrifugation were subjected to SDS-PAGE followed by immunoblot analysis with antiserum to Stisa1 or Stisa2.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The accession numbers for Stisa1, Stisa2, and Stisa3 are AY132996, AY132997, and AY132998, respectively. The accession number for EST At69012 from Arabidopsis is H36690.
Supplementary Material
Acknowledgments
We dedicate this article to the memory of Oliver E. Nelson, Jr. (1920-2002), the father of plant biochemical genetics and an inspiration for this work. Many thanks are due to all participants of the European Union FAIR program Tailoring of Novel Starches for lively and stimulating debate on starch-debranching enzymes. Part of the work described in this article was funded by Zeneca (Jealot's Hill, UK), and part was funded by the Core Strategic Grant to the John Innes Centre from the Biotechnology and Biological Science Research Council. H.H. was supported by a scholarship from the Universiti Malaysia (Sarawak), and A.E. was supported by a Biotechnology and Biological Science Research Council–Wealth Creating Products of Plants grant (WCP 11512). A.M. is the recipient of a long-term European Molecular Biology Organization fellowship.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006635.
Footnotes
Online version contains Web-only data.
References
- Abe, J.-I., Ushijima, C., and Hizukuri, S. (1999). Expression of the isoamylase gene of Flavobacterium odoratum KU in Escherichia coli and identification of essential residues in the enzyme by site-directed mutagenesis. Appl. Environ. Microbiol. 65, 4163–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Ball, S., Guan, H.-P., James, M., Myers, A., Keeling, P., Mouille, G., Buleon, A., Colonna, P., and Preiss, J. (1996). From glycogen to amylopectin: A model for the biogenesis of the plant starch granule. Cell 86, 349–352. [DOI] [PubMed] [Google Scholar]
- Ballicora, M.A., Laughlin, M.J., Fu, Y.B., Okita, T.W., Barry, G.F., and Preiss, J. (1995). Adenosine 5′-diphosphate-glucose pyrophosphorylase from potato tuber: Significance of the N terminus of the small subunit for catalytic properties and heat stability. Plant Physiol. 109, 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty, M.K., Rahman, A., Cao, H.P., Woodman, W., Lee, M., Myers, A.M., and James, M.G. (1999). Purification and molecular genetic characterization of ZPU1, a pullulanase-type starch-debranching enzyme from maize. Plant Physiol. 119, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, R.C., Loerch, J.D., McArdle, F.J., and Creech, R.G. (1966). Genetic interactions affecting maize phytoglycogen and the phytoglycogen-forming branching enzyme. Genetics 53, 661–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer, C., Daniels, R.R., and Shannon, J.C. (1977). Starch granule (amyloplast) development in endosperm of several Zea mays L. genotypes affecting kernel polysaccharides. Am. J. Bot. 64, 50–56. [Google Scholar]
- Brock, I.W., Hazell, L., Michl, D., Nielsen, V.S., Møller, B.L., Herrmann, R.G., Klösgen, R.B., and Robinson, C. (1993). Precursors of one integral and five lumenal thylakoid proteins are imported by isolated pea and barley thylakoids: Optimisation of in vitro assays. Plant Mol. Biol. 23, 717–725. [DOI] [PubMed] [Google Scholar]
- Buisson, G., Duée, E., Haser, R., and Payan, F. (1987). Three dimensional structure of porcine pancreatic alpha-amylase at 2.9 Å resolution: Role of calcium in structure and activity. EMBO J. 6, 3909–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton, R., Jenner, H., Carrangis, L., Fahy, B., Fincher, G., Hylton, C., Laurie, D., Parker, M., Waite, D., van Wegen, S., Verhoeven, T., and Denyer, K. (2002). Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity. Plant J. 31, 97–112. [DOI] [PubMed] [Google Scholar]
- Correns, C. (1901). Bastarde zwichen maisrassen, mit besonder Berucksichtung der Xenien. Bibl. Bot. 53, 1–161. [Google Scholar]
- Dauvillée, D., Colleoni, C., Mouille, G., Buleon, A., Gallant, D.J., Bouchet, B., Morell, M.K., d'Huilst, C., Myers, A.M., and Ball, S.G. (2001. a). Two loci control phytoglycogen production in the monocellular green alga Chlamydomonas reinhardtii. Plant Physiol. 125, 1710–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillée, D., Colleoni, C., Mouille, G., Morell, M.K., d'Huilst, C., Wattebled, F., Liénard, L., Delvallé, D., Ral, J.-P., Myers, A.M., and Ball, S.G. (2001. b). Biochemical characterization of wild-type and mutant isoamylases of Chlamydomonas reinhardtii supports a function of a multimeric enzyme organization in amylopectin maturation. Plant Physiol. 125, 1723–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillée, D., Mestre, V., Colleoni, C., Slomianny, M.-C., Mouille, G., Delrue, B., d'Huilst, C., Bliard, C., Nuzillard, J.-M., and Ball, S. (2000). The debranching enzyme complex missing in glycogen-accumulating mutants of Chlamydomonas reinhardtii displays an isoamylase-type specificity. Plant Sci. 157, 145–156. [DOI] [PubMed] [Google Scholar]
- Doan, D.N.P., Rudi, H., and Olsen, O.A. (1999). The allosterically unregulated isoform of ADP-glucose pyrophosphorylase from barley endosperm is the most likely source of ADP-glucose incorporated into endosperm starch. Plant Physiol. 121, 965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlert, D.C., and Knutson, C.A. (1991). Two classes of starch debranching enzymes from developing maize kernels. J. Plant Physiol. 138, 566–572. [Google Scholar]
- Edwards, A., Borthakur, A., Bornemann, S., Venail, J., Denyer, K., Waite, D., Fulton, D., Smith, A., and Martin, C. (1999). Specificity of starch synthase isoforms from potato. Eur. J. Biochem. 266, 724–736. [DOI] [PubMed] [Google Scholar]
- Edwards, A., Marshall, J., Sidebottom, C., Visser, R.G.F., Smith, A.M., and Martin, C. (1995). Biochemical and molecular characterization of a novel starch synthase from potato tubers. Plant J. 8, 283–294. [DOI] [PubMed] [Google Scholar]
- Fox, J.D., and Robyt, J.F. (1991). Miniaturization of three carbohydrate analyses using a microsample plate reader. Anal. Biochem. 195, 93–96. [DOI] [PubMed] [Google Scholar]
- Fujita, N., Kubo, A., Francisco, P.B., Nakakita, M., Harada, K., Minaka, N., and Nakamura, Y. (1999). Purification, characterisation and cDNA structure of isoamylase from developing endosperm of rice. Planta 208, 283–293. [DOI] [PubMed] [Google Scholar]
- Gavel, Y., and von Heijne, G. (1990). A conserved cleavage-site motif in chloroplast transit peptides. FEBS Lett. 261, 455–458. [DOI] [PubMed] [Google Scholar]
- Genschel, U., Abel, G., Lorz, H., and Lutticke, S. (2002). The sugary-type isoamylase in wheat: Tissue distribution and subcellular localisation. Planta 214, 813–820. [DOI] [PubMed] [Google Scholar]
- Goff, S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp japonica). Science 296, 92–100. [DOI] [PubMed] [Google Scholar]
- Ishizaki, Y., Taniguchi, H., Maruyama, Y., and Nakamura, M. (1983). Debranching enzymes of potato tubers (Solanum tuberosum L.). I. Purification and some properties of potato isoamylase. Agric. Biol. Chem. 47, 771–779. [Google Scholar]
- James, M.G., Robertson, D.S., and Myers, A.M. (1995). Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell 7, 417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanningros, R., Creuzet-Sigal, N., Frixon, C., and Cattaneo, J. (1976). Purification and properties of a debranching enzyme from Escherichia coli. Biochim. Biophys. Acta 438, 186–199. [DOI] [PubMed] [Google Scholar]
- Jespersen, H.M., MacGregor, E.A., Henrissat, B., Sierks, M.R., and Svenson, B. (1993). Starch- and glycogen-debranching and branching enzymes: Prediction of structural features of the catalytic (βα8)-barrel domain and evolutionary relationship to other amylolytic enzymes. J. Protein Chem. 12, 791–805. [DOI] [PubMed] [Google Scholar]
- Jespersen, H.M., MacGregor, E.A., Sierks, M.R., and Svensson, B. (1991). Comparison of the domain-level organization of starch hydrolases and related enzymes. Biochem. J. 280, 51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuya, Y., Mezaki, Y., Kubota, M., and Matsuura, Y. (1998). Three-dimensional structure of Pseudomonas isoamylase at 2.2 Å resolution. J. Mol. Biol. 281, 885–897. [DOI] [PubMed] [Google Scholar]
- Kavakli, I.H., Greene, T.W., Salamone, P.R., Choi, S.B., and Okita, T.W. (2001). Investigation of subunit function in ADP-glucose pyrophosphorylase. Biochem. Biophys. Res. Commun. 281, 783–787. [DOI] [PubMed] [Google Scholar]
- Kim, J.S., and Raines, R.T. (1993). Ribonuclease S-peptide as a carrier in fusion proteins. Protein Sci. 2, 348–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, C., Hollender, J., Bender, H., and Schulz, G.E. (1992). Catalytic center of cyclodextrin glycosyltransferase derived from X-ray structure-analysis combined with site-directed mutagenesis. Biochemistry 31, 8740–8746. [DOI] [PubMed] [Google Scholar]
- Kubo, A., Fujita, N., Harada, K., Matsuda, T., Satoh, H., and Nakamura, Y. (1999). The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiol. 121, 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor, E.A. (1993). Relationships between structure and activity in the α-amylase family of starch-metabolising enzymes. Starch 45, 232–237. [Google Scholar]
- Marshall, J., Sidebottom, C., Debet, M., Martin, C., Smith, A.M., and Edwards, A. (1996). Identification of the major starch synthase in the soluble fraction of potato tubers. Plant Cell 8, 1121–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura, Y., Kusunoki, M., Harada, W., and Kakudo, M. (1984). Structure and possible catalytic residues of taka-amylase A. J. Biochem. 95, 697–702. [DOI] [PubMed] [Google Scholar]
- Meeuwsen, P.J.A., Vincken, J.P., Beldman, G., and Voragen, A.G.J. (2000). A universal assay for screening expression libraries for carbohydrases. J. Biosci. Bioeng. 89, 107–109. [DOI] [PubMed] [Google Scholar]
- Mizuguchi, K., Deane, C.M., Blundell, T.L., and Overington, J.P. (1998). HOMSTRAD: A database of protein structure alignments for homologous families. Protein Sci. 7, 2469–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouille, G., Maddelein, M.-L., Libessart, N., Talaga, P., Decq, A., Delrue, B., and Ball, S.G. (1996). Preamylopectin processing: A mandatory step for starch biosynthesis in plants. Plant Cell 8, 1353–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, A.M., Morell, M.K., James, M.G., and Ball, S.G. (2000). Recent progress toward understanding the biosynthesis of the amylopectin crystal. Plant Physiol. 122, 989–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, Y., Umemoto, T., Ogata, N., Kuboki, Y., Yano, M., and Sasaki, T. (1996). Starch debranching enzyme (R-enzyme or pullulanase) from developing rice endosperm: Purification, cDNA and chromosomal localisation of the gene. Planta 199, 209–218. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., Umemoto, T., Takahata, Y., Komae, K., Amano, E., and Satoh, H. (1997). Changes in the structure of starch and enzyme activities affected by sugary mutations in developing rice endosperm: Possible role of starch debranching enzyme (R-enzyme) in amylopectin biosynthesis. Physiol. Plant. 97, 491–498. [Google Scholar]
- Pan, D., and Nelson, O.E. (1984). A debranching enzyme deficiency in endosperms of the sugary1 mutants of maize. Plant Physiol. 74, 324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, A., Wong, K.S., Jane, J.L., Myers, A.M., and James, M.G. (1998). Characterization of SU1 isoamylase, a determinant of storage starch structure in maize. Plant Physiol. 117, 425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, B.-S., Kim, S., Scott, M.P., Singletary, G.W., Wong, K.-S., James, M.G., and Myers, A.M. (2002). Functional interactions between heterologously-expressed starch-branching enzymes of maize and the glycogen synthases of brewer's yeast. Plant Physiol. 128, 1189–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, J.C., and Garwood, D.L. (1984). Genetics and physiology of starch development. In Starch: Chemistry and Technology, R.L. Whistler, J.N. BeMiller, and E.F. Paschall, eds (San Diego, CA: Academic Press), pp. 25–86.
- Shi, J., Blundell, T.L., and Mizuguchi, K. (2001). FUGUE: Sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310, 243–257. [DOI] [PubMed] [Google Scholar]
- Sun, C.X., Sathish, P., Ahlandsberg, S., and Jansson, C. (1999). Analyses of isoamylase gene activity in wild-type barley indicate its involvement in starch synthesis. Plant Mol. Biol. 40, 431–443. [DOI] [PubMed] [Google Scholar]
- Visser, R.G.F., Jacobsen, E., Hesseling-Meinders, A., Schans, M.J., Witholt, B., and Feenstra, W.J. (1989). Transformation of homozygous diploid potato with an Agrobacterium tumefaciens binary vector system by adventitious shoot regeneration on leaf and stem segments. Plant Mol. Biol. 12, 329–337. [DOI] [PubMed] [Google Scholar]
- Yeh, J.Y., Garwood, D.L., and Shannon, J.C. (1981). Characterisation of starch from maize endosperm mutants. Starch 33, 222–230. [Google Scholar]
- Yu, J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp indica). Science 296, 79–92. [DOI] [PubMed] [Google Scholar]
- Yu, Y., Mu, H.H., Mu-Forster, C., and Wasserman, B.P. (1998). Polypeptides of the maize amyloplast stroma: Stromal localization of starch-biosynthetic enzymes and identification of an 81-kilodalton amyloplast stromal heat-shock cognate. Plant Physiol. 116, 1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman, S.C., Northrop, F., Smith, A.M., and ap Rees, T. (1998. a). A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolysing enzyme. Plant J., 15, 357–365. [DOI] [PubMed] [Google Scholar]
- Zeeman, S.C., Umemoto, T., Lue, W.-L., Au-Yeung, P., Martin, C., Smith, A.M., and Chen, J. (1998. b). A mutant of Arabidopsis lacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. Plant Cell 10, 1699–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Z.-P., Hylton, C.M., Rossner, U., and Smith, A.M. (1998). Characterization of starch-debranching enzymes in pea embryos. Plant Physiol. 118, 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.