Abstract
The Arabidopsis ref2 mutant was identified in a screen for plants having altered fluorescence under UV light. Characterization of the ref2 mutants showed that they contained reduced levels of a number of phenylpropanoid pathway–derived products: sinapoylmalate in leaves, sinapoylcholine in seeds, and syringyl lignin in stems. Surprisingly, positional cloning of the REF2 locus revealed that it encodes CYP83A1, a cytochrome P450 sharing a high degree of similarity to CYP83B1, an enzyme involved in glucosinolate biosynthesis. Upon further investigation, ref2 mutants were found to have reduced levels of all aliphatic glucosinolates and increased levels of indole-derived glucosinolates in their leaves. These results show that CYP83A1 is involved in the biosynthesis of both short-chain and long-chain aliphatic glucosinolates and suggest a novel metabolic link between glucosinolate biosynthesis, a secondary biosynthetic pathway found only in plants in the order Capparales, and phenylpropanoid metabolism, a pathway found in all plants and considered essential to the survival of terrestrial plant species.
INTRODUCTION
The phenylpropanoid pathway converts Phe to a number of important secondary metabolites, including flavonoids, lignans, and lignin and hydroxycinnamic acid esters. Although the phenylpropanoid pathway is ubiquitous in vascular plants (Lewis and Yamamoto, 1990), downstream branches that lead to different classes of secondary metabolites often are unique to certain taxa, resulting in species-specific phenylpropanoid-derived products. Examples of these metabolites include the isoflavonoids found in members of the Fabaceae (Tahara and Ibrahim, 1995) and the sinapate esters found in members of the Brassicaceae (Strack, 1977; Chapple et al., 1992).
Sinapoylmalate and sinapoylcholine are two of the major sinapate esters of Arabidopsis (Chapple et al., 1992). Sinapoylmalate accumulates in leaves, where it acts as a UV light protectant (Landry et al., 1995; Booij-James et al., 2000), whereas sinapoylcholine accumulates in embryos and is thought to serve as a storage form of choline and sinapic acid for germinating seedlings (Strack, 1981; Ruegger et al., 1999). Under UV illumination, sinapoylmalate in the epidermis of wild-type plants causes their leaves to appear blue-green. By contrast, leaves of the sinapoylmalate-deficient fah1 mutant appear red under UV light as a result of the fluorescence of chlorophyll (Chapple et al., 1992). The fluorescent property of sinapate esters makes them convenient visible markers for the genetic dissection of phenylpropanoid metabolism (Chapple et al., 1992; Ruegger and Chapple, 2001). The cloning and characterization of genes defective in these mutants has improved our understanding of the functions of known phenylpropanoid enzymes (Humphreys et al., 1999; Franke et al., 2002a, 2002b) and identified a new class of enzymes not previously known to be involved in secondary metabolism (Lehfeldt et al., 2000; Shirley et al., 2001).
In contrast to phenylpropanoid metabolism, the glucosinolate biosynthetic pathway generally is restricted to members of the order Capparales (Halkier and Du, 1997). Glucosinolates are a heterogeneous family of compounds derived from a variety of protein and nonprotein amino acids. Similar biosynthetic mechanisms convert these amino acids to secondary metabolites that share a common glucosinolate backbone but differ in an R group derived from the original amino acid precursor (Chapple et al., 1994). These compounds are substrates for the enzyme myrosinase, which degrades them into biologically active products thought to play roles in the defense against herbivory (Halkier and Du, 1997; Lambrix et al., 2001).
Arabidopsis plants accumulate Met-, Phe-, and Trp-derived glucosinolates (Petersen et al., 2002). Despite decades of research, many gaps still exist in our understanding of the catalysts involved in glucosinolate biosynthesis (Bennett et al., 1995; Halkier and Du, 1997); however, the recent isolation and characterization of Arabidopsis glucosinolate mutants such as sur2/rnt1 and bus1/sps, plants reduced in their content of Trp- and short-chain Met-derived glucosinolates, respectively, has led to significant advances in our understanding of this metabolic pathway (Barlier et al., 2000; Bak et al., 2001; Chen and Andreassan, 2001; Reintanz et al., 2001; Tantikanjana et al., 2001). Many of these mutants show morphological and developmental defects, and there is increasing evidence that alterations in glucosinolate biosynthesis can lead to changes in plant hormone levels (Barlier et al., 2000; Bak and Feyereisen, 2001; Bak et al., 2001; Reintanz et al., 2001; Tantikanjana et al., 2001).
It is now known that a number of the reactions in glucosinolate biosynthesis are catalyzed by cytochrome P450-dependent monooxygenases (P450s). Members of the CYP79 family of P450s are responsible for the initial step in glucosinolate biosynthesis, in which the amino acid precursor is oxidized to its respective aldoxime. The CYP79B2 and CYP79B3 enzymes oxidize Trp to indole 3-acetaldoxime (Hull et al., 2000; Mikkelsen et al., 2000); whereas CYP79F1 oxidizes short-chain Met-derived homologs (Hansen et al., 2001; Reintanz et al., 2001). Recent work with the sur2 Arabidopsis mutants has shown that the next step in indole-derived glucosinolate biosynthesis is catalyzed by the cytochrome P450 CYP83B1 (Bak and Feyereisen, 2001; Bak et al., 2001; Hansen et al., 2001). These mutants accumulate less indole-derived glucosinolates than the wild type, and heterologously expressed CYP83B1 converts indole 3-acetaldoxime to a thiohydroximate adduct in the presence of a thiol donor (Bak and Feyereisen, 2001; Bak et al., 2001). It is becoming clear that the indole 3-acetaldoxime also is a precursor for indole 3-acetic acid, and as a result of defects in CYP83B1 activity, sur2 shows phenotypes thought to be associated with auxin overaccumulation (Delarue et al., 1998; Barlier et al., 2000; Bak et al., 2001).
A recent screen of ∼100,000 M2 seedlings for plants showing reduced fluorescence under UV light yielded multiple independent mutants disrupted in phenylpropanoid biosynthesis (Ruegger and Chapple, 2001). For all of these mutants, the reduced leaf fluorescence was found to be the result of significantly decreased levels of sinapoylmalate. Here, we report the further characterization of the ref2 mutant of Arabidopsis and the isolation of the REF2 gene. Our results show that REF2 encodes CYP83A1, an enzyme suggested previously to be involved in the synthesis of non-Trp-derived glucosinolates (Bak and Feyereisen, 2001). Our work provides genetic evidence of the role of CYP83A1 in alkylglucosinolate biosynthesis and suggests that a block in al-kylglucosinolate biosynthesis simultaneously perturbs phenylpropanoid metabolism in the ref2 mutant.
RESULTS
REF2 Maps to BAC F18A5
To understand how mutations in the REF2 gene affect phenylpropanoid biosynthesis, REF2 was isolated by positional cloning. Using 20 F2 plants from a ref2-1 (Columbia ecotype) × Landsberg erecta cross, a set of Arabidopsis restriction fragment length polymorphism markers spanning the Arabidopsis genome (Fabri and Schaffner, 1994; Schaffner, 1996) was used to identify an initial map position for the gene near the middle of chromosome 4. The position of REF2 was delineated further using PCR-based codominant cleaved amplified polymorphic sequence markers (Konieczny and Ausubel, 1993) and simple sequence length polymorphism markers (Bell and Ecker, 1994) available on the TAIR World Wide Web site (www.arabidopsis.org). Sequence information from both the Cereon database (www.arabidopsis.org/Cereon/index.html; Cereon Genomics, Cambridge, MA) as well as the Landsberg erecta database (www.tigr.org/tdb/e2k1/ath1/atgenome/Ler.shtml) was used to generate additional cleaved amplified polymorphic sequence markers to screen a mapping population of 850 plants, eventually narrowing the mapping interval to a 60-kb region on BAC F18A5. According to these results, REF2 is located between marker Cer466125 (31 kb from the centromeric end of BAC F18A5) and marker Cer466224 (89 kb) (Figure 1A).
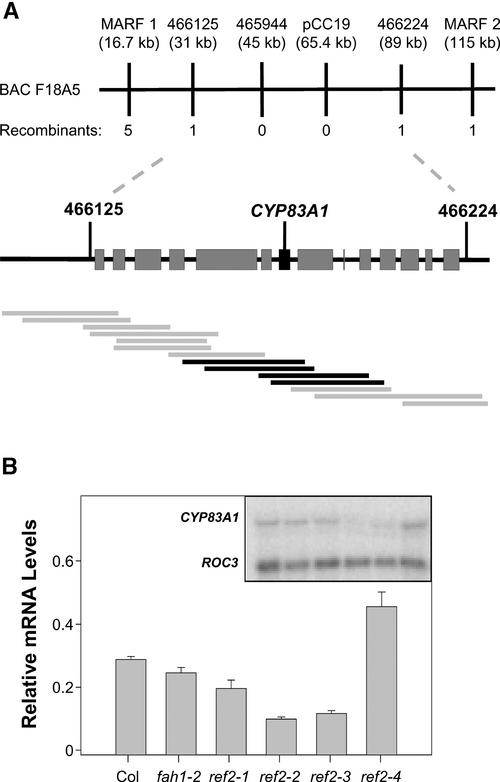
Figure 1.
Map-Based Cloning of the REF2 Gene.
(A) REF2 was mapped to a 60-kb mapping interval on BAC F18A5 containing 14 annotated genes. A contig of cosmids containing Landsberg genomic DNA was constructed that spanned the mapping interval (gray bars). Cosmids that complemented the sinapoylmalate and leaf glucosinolate phenotypes of ref2-1 overlap at CYP83A1 (black bars).
(B) RNA gel blot analysis of CYP83A1 expression in wild-type and ref2 rosettes. The graph shows levels of CYP83A1 expression normalized to cyclophilin (ROC3) expression. Error bars represent standard errors of triplicate assays. Col, Columbia wild type.
REF2 Encodes CYP83A1
To identify the location of REF2 on BAC F18A5, an overlapping contig was assembled from cosmid clones containing 6- to 14-kb fragments of Landsberg erecta genomic DNA. The clones were isolated by probing a Landsberg erecta genomic library constructed in the binary vector pBIC20 (Meyer et al., 1996) with PCR products of genomic DNA from the mapping interval. Thirty-five cosmid clones were isolated, with a minimum overlap of 10 kb between contiguous fragments. Using the floral dip method of Agrobacterium-mediated plant transformation (Clough and Bent, 1998), these binary cosmids were transformed into ref2-1 plants. All kanamycin-resistant T1 progeny were screened visually and by HPLC analysis for complementation of the ref2 phenotype. Four cosmids were found that complemented the ref2 sinapoylmalate deficiency phenotype (Figure 2A). The minimum sequence overlap of these fragments spanned a 7.4-kb region of F18A5 that contained one annotated gene, the P450 CYP83A1, a gene cloned previously based on a slight homology with ferulate 5-hydroxylase (Chapple, 1995).
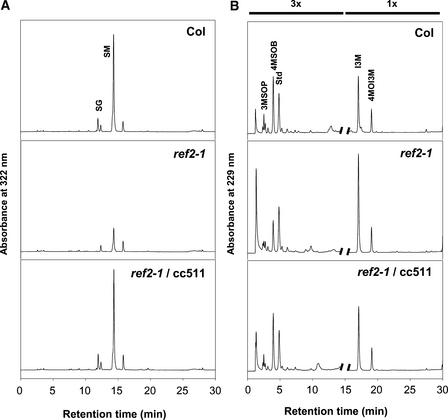
Figure 2.
HPLC Analysis of Soluble Secondary Metabolites Accumulated in Leaves of Wild-Type, ref2-1, and a Representative ref2-1 Plant Transformed with a Cosmid Containing CYP83A1.
(A) Methanolic leaf extracts analyzed by HPLC. The elution of UV light–absorbing compounds was monitored at 320 nm. Col, Columbia wild type; SG, sinapoylglucose; SM, sinapoylmalate.
(B) Leaf desulfoglucosinolate fractions analyzed by HPLC. The elution of UV light–absorbing compounds was monitored at 229 nm. Peaks are magnified threefold for the first 15 min of the chromatograms to more clearly show differences in peak areas. Glucosinolates are identified as follows: 3MSOP, 3-methylsulfinylpropyl; 4MSOB, 4-methylsulfinylbutyl; Std, internal standard; I3M, indol-3-ylmethyl; 4MOI3M, 4-methoxyindol-3-ylmethyl.
To examine the possibility that REF2 is an unannotated gene in the 7.4-kb region of complementing DNA rather than CYP83A1, the steady state level of CYP83A1 mRNA in the four ref2 mutant alleles was assayed by RNA gel blot hybridization. RNA isolated from 4-week-old ref2 and wild-type rosettes was probed with a CYP83A1 cDNA using a cyclophilin (ROC3) cDNA as a control (Chow and Gasser, 1997). The steady state level of CYP83A1 expression was decreased substantially in three of the four ref2 alleles (Figure 1B). Given that transcript levels of alleles containing frameshift or nonsense mutations frequently are decreased (Hilleren and Parker, 1999), these results strongly support the identity of REF2 as CYP83A1.
As a final confirmation of the identity of REF2, the genomic DNA corresponding to all four mutant alleles was sequenced. Each CYP83A1 allele from the ref2 mutants was found to contain a unique G-to-A base change. In ref2-1 and ref2-3 alleles, the codons for Trp-58 and Trp-406, respectively, were converted to stop codons. In ref2-2, the G-to-A change altered a 3′ intron-exon boundary recognition sequence (aaaaGGAT to aaaaaGAT; Simpson et al., 1998) such that it probably shifts the 3′ intron-exon border downstream by one base, causing a frameshift and termination of the polypeptide after another two amino acids. Finally, in the protein encoded by the ref2-4 allele, a conserved Gly in the proposed heme binding site at position 424 of the protein (Chapple, 1998) is converted to Glu. All four ref2 mutations would be expected to strongly reduce or eliminate CYP83A1 function (Graham and Peterson, 1999; Hilleren and Parker, 1999). These results confirm that REF2 is CYP83A1.
The ref2 Mutant Is Defective in Glucosinolate Biosynthesis
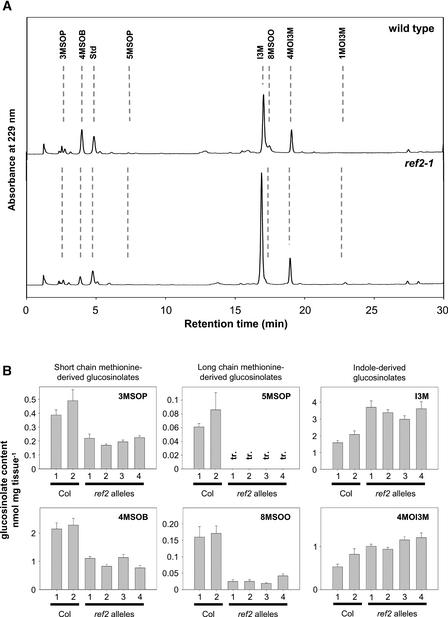
CYP83A1 shares 65% amino acid sequence identity with CYP83B1 (Bilodeau et al., 1999), a cytochrome P450 that catalyzes an early reaction in the indole glucosinolate biosynthetic pathway (Bak and Feyereisen, 2001). The proposed reaction catalyzed by CYP83B1 is the conversion of indole-3-acetaldoxime to 1-aci-nitro-2-indolyl-ethane, which reacts nonenzymatically to become an S-alkylthiohydroximate in the presence of a thiol donor (Bak et al., 2001; Hansen et al., 2001). To test the hypothesis that CYP83A1 also acts in glucosinolate biosynthesis, desulfoglucosinolate extracts were prepared from wild-type and ref2 tissues and analyzed by HPLC and liquid chromatography–mass spectrometry (LC-MS). HPLC analysis showed that ref2 leaf extracts contain less short-chain (3-methylsulfinylpropyl, 4-methylthiobutyl, and 4-methylsulfinylbutyl) and long-chain (5-methylsulfinylpropyl, 5-methylthiopropyl, 7-methylsulfinyl-heptyl, 7-methylthioheptyl, 8-methylsulfinyloctyl, and 8-meth-ylthio-octyl) Met-derived glucosinolates (Figure 3). Concomitant with the decreases in alkylglucosinolates, ref2 leaf extracts show increased levels of the indole glucosinolates indol-3-ylmethyl, 4-methoxyindol-3-ylmethyl, and 1-methoxyindol-3-ylmethyl glucosinolate (Figure 3). Because of the decrease in alkylglucosinolates and the increase in indole glucosinolates, the major glucosinolate accumulated in ref2 leaves is indol-3-ylglucosinolate. By contrast, the major glucosinolate present in wild-type leaves is 4-methylsulfinylbutyl glucosinolate.
Figure 3.
Analysis of Glucosinolate Levels in Wild-Type and ref2 Leaves.
(A) Leaf desulfoglucosinolate fractions analyzed by HPLC. The elution of UV light–absorbing compounds was monitored at 229 nm. Glucosinolates are identified as follows: 3MSOP, 3-methylsulfinylpropyl; 4MSOB, 4-methylsulfinylbutyl; Std, internal standard; 5MSOP, 5-methylsulfinylpropyl; I3M, indol-3-ylmethyl; 8MSOO, 8-methylsulfinyloctyl; 4MOI3M, 4-methoxyindol-3-ylmethyl; 1MOI3M, 1-methoxyindol-3-ylmethyl.
(B) Quantification of the levels of representative glucosinolates in wild-type and ref2 leaves. Error bars represent standard errors of at least triplicate assays. Col, Columbia wild type; tr., trace.
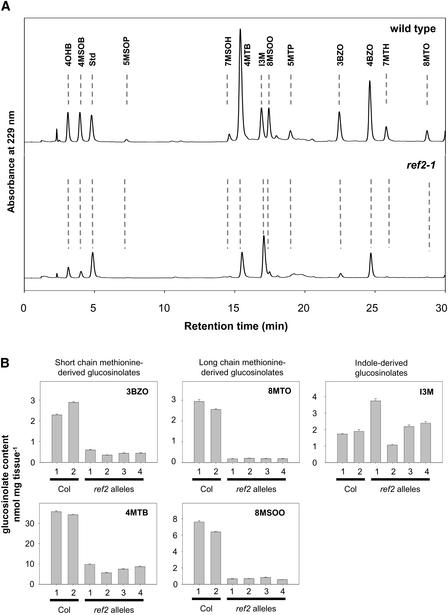
Comparable reductions in alkylglucosinolate profiles were observed when samples of ref2 seeds were analyzed (Figure 4); however, no consistent increases in indole glucosinolates were seen. In addition to decreases in the glucosinolates listed above, seeds of the mutants also contained significantly less benzoyloxyalkyl glucosinolates (Figure 4). Because the glucosinolate core structure of benzoyloxyalkyl glucosinolates is synthesized from Met in Arabidopsis (Graser et al., 2001), a decrease in alkylglucosinolate precursors probably is responsible for the decreased levels of ben-zoylalkyl glucosinolates in ref2 seeds. These results demonstrate that CYP83A1 is required for the accumulation of wild-type levels of Met-derived glucosinolates in Arabidopsis (Figure 5).
Figure 4.
Analysis of Glucosinolate Levels in Wild-Type and ref2 Seeds.
(A) Seed desulfoglucosinolate fractions analyzed by HPLC. The elution of UV light–absorbing compounds was monitored at 320 nm. Glucosinolates are identified as follows: 4OHB, 4-hydroxybutyl; 4MSOB, 4-methylsulfinylbutyl; Std, internal standard; 5MSOP, 5-methylsulfinylpropyl; 7MSOH, 7-methylsulfinylheptyl; 4MTB, 4-methylthiobutyl; I3M, indol-3-ylmethyl; 8MSOO, 8-methylsulfinyloctyl; 5MTP, 5-methylthiopropyl; 3BZO, 3-benzoyloxypropyl; 4BZO, 4-benzoyloxybutyl; 7MTH, 7-methylthioheptyl; 8MTO, 8-methylthiooctyl.
(B) Quantification of levels of representative glucosinolates in wild-type and ref2 seeds. Error bars represent standard errors of at least triplicate assays. Col, Columbia wild type.
Figure 5.
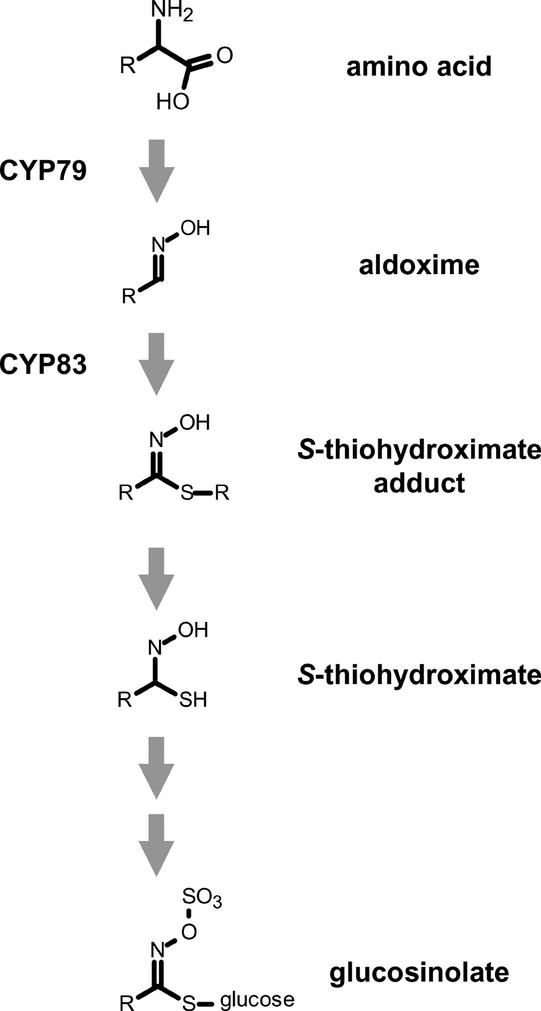
Scheme of Glucosinolate Biosynthesis.
The oxidation of amino acids to their respective aldoximes is catalyzed by cytochrome P450 monooxygenases in the CYP79 family. Members of the CYP83 family subsequently oxidize these aldoximes to a product that is thought to react spontaneously to give an S-thiohydroximate. These S-thiohydroximates are further glycosylated and sulfated to yield the final glucosinolates.
As stated above, REF2 was identified originally as CYP83A1 based on the complementation of the ref2-1 sinapoylmalate deficiency by cosmids containing the Landsberg CYP83A1 genomic sequence. After determining that the ref2 mutations affected glucosinolate accumulation as well, glucosinolate levels also were examined in ref2-1 plants transformed with the CYP83A1-containing cosmids. HPLC analysis of desulfoglucosinolate extracts from leaves of wild-type, ref2-1, and transgenic ref2-1 plants indicated that the Landsberg CYP83A1 sequence complemented the ref2-1 alkylglucosinolate phenotypes as well as its phenylpropanoid defects (Figure 2B).
ref2 Lacks the Morphological and Developmental Phenotypes Seen in Other Arabidopsis Glucosinolate Mutants
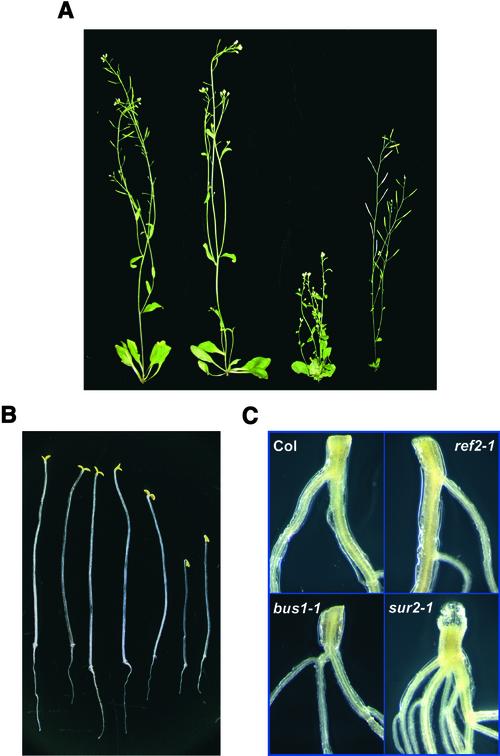
Several previously identified Arabidopsis glucosinolate mutants show developmental abnormalities. These include bus1, which lacks wild-type levels of CYP79F1 function and is blocked in the first step of short-chain alkylglucosinolate biosynthesis (Hansen et al., 2001; Reintanz et al., 2001), and sur2, which eliminates CYP83B1 function and blocks the aldoxime-oxidizing reaction in indole glucosinolate biosynthesis (Barlier et al., 2000; Bak et al., 2001; Hansen et al., 2001). Other than a slight delay in average time to bolt (data not shown), analysis of a range of developmental characteristics in ref2 plants, including germination time and frequency, leaf size, internode length, and silique shape, showed no obvious differences between wild-type and ref2 plants. ref2 inflorescences reached wild-type heights and displayed none of the obvious defects of bus1 and sur2 plants (Figure 6A). Similarly, light-grown ref2 seedlings did not develop the adventitious roots seen in sur2 plants (Figure 6C) as a result of auxin overproduction. Finally, dark-grown ref2 seedlings exhibited a normal etiolated seedling phenotype compared with the reduced hypocotyl and root length phenotypes seen in sur2 dark-grown seedlings (Figure 6B). These results show that a block at CYP83A1 in ref2 plants does not lead to changes in hormone levels or metabolic pools capable of causing the phenotypes seen in related glucosinolate mutants.
Figure 6.
Morphology of ref2 Plants Compared with Wild-Type Plants and with the Glucosinolate Biosynthetic Mutants bus1 and sur2.
(A) Appearance of Columbia wild-type, ref2-1, bus1, and sur2 plants (left to right).
(B) Appearance of 10-day-old etiolated seedlings of Columbia wild-type, ref2-1, ref2-2, bus1, Wassilewskija wild-type, and two seedlings of sur2-1 (left to right).
(C) Hypocotyl-root junction of 2-week-old Columbia (Col) wild-type, ref2-1, bus1, and sur2-1 light-grown seedlings.
Detailed Analysis Confirms the Lignin Phenotypes of ref2
A preliminary analysis of lignin composition had shown that ref2 stems have wild-type levels of total lignin but that its lignin contains significantly reduced levels of syringyl monomers (Ruegger and Chapple, 2001). Because the chemically resistant nature of lignin makes unambiguous analysis difficult, multiple methods of analysis often are used to achieve a more accurate evaluation of the lignin composition of plant tissue. Considering the unexpected role of REF2 in glucosinolate biosynthesis, the impact of the ref2 mutation on lignin content was revisited using the Klason method (Kaar and Brink, 1991), and ref2 lignin composition was assayed by the derivatization followed by reductive cleavage (DFRC) method (Lu and Ralph, 1997).
Klason analysis of ref2 stems indicated that ref2 plants accumulate wild-type levels of total lignin (Table 1). DFRC analysis of the same samples also confirmed the previous nitrobenzene oxidation results showing that the syringyl monomer content of ref2 lignin was 25 to 50% of that found in wild-type lignin (Table 1). ref2 cell walls contained, on average, 8% syringyl lignin, whereas wild-type cell walls contained ∼18% syringyl lignin. Together, these assays suggest that the ref2 mutation does not alter total carbon flux into the phenylpropanoid pathway but blocks phenylpropanoid biosynthesis downstream of guaiacyl lignin biosynthesis and upstream of sinapate ester–specific and syringyl lignin–specific reactions.
Table 1.
Klason Lignin Content and DFRC Lignin Monomer Composition of Mutant and Wild-Type Cell Walls (n = 3)
| Klason lignin (n = 3) (mg/g cell wall) |
Monomer Composition (μmol/g dry wt) |
Monomer Composition (μmol/g Klason lignin) |
Subunit mol %
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample | G | S | Total | G | S | Total | G | S | |
| Wild type | 185 ± 5 | 189 ± 19 | 43 ± 3 | 232 ± 28 | 671 ± 50 | 148 ± 11 | 820 ± 49 | 81 ± 1 | 18 ± 1 |
| ref2-1 | 196 ± 4 | 215 ± 26 | 22 ± 1 | 238 ± 32 | 694 ± 158 | 75 ± 29 | 769 ± 188 | 91 ± 1 | 9 ± 1 |
| ref2-2 | 190 ± 4 | 192 ± 20 | 20 ± 2 | 213 ± 20 | 499 ± 73 | 51 ± 4 | 551 ± 74 | 90 ± 1 | 10 ± 1 |
| ref2-3 | 189 ± 3 | 134 ± 14 | 10 ± 1 | 146 ± 16 | 500 ± 86 | 38 ± 8 | 539 ± 93 | 93 ± 1 | 7 ± 1 |
| ref2-4 | 198 ± 10 | 191 ± 42 | 15 ± 2 | 206 ± 46 | 456 ± 93 | 32 ± 10 | 489 ± 103 | 94 ± 0.1 | 6 ± 0.1 |
Values shown are for the units indicated ± se. G, guaiacyl; S, syringyl.
Perturbation of Phenylpropanoid O-Methylation Reactions May Be the Cause of the ref2 Phenotype
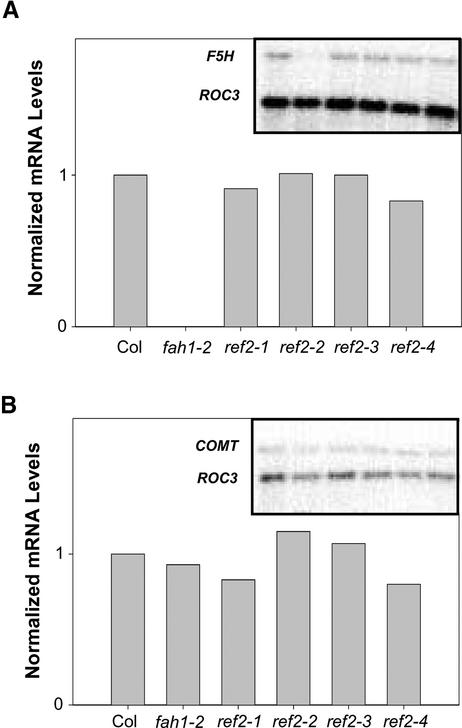
Characterization of the ref2 mutant's phenylpropanoid phenotypes strongly indicated some type of connection between alkylglucosinolate and phenylpropanoid biosynthesis, at least in the context of defective ref2 alleles. Based on the current model of phenylpropanoid biosynthesis (Humphreys and Chapple, 2002), a block in either ferulate 5-hydroxylase (F5H) or caffeic acid O-methyltransferase (COMT) could lead to the ref2 sinapate ester and syringyl lignin phenotypes. To investigate the possibility that the expression of these genes was reduced in the ref2 background, steady state levels of F5H and COMT mRNA were assayed in 4-week- old ref2 rosettes by RNA gel blot analysis (Figure 7). Quantification of expression levels using cyclophilin (ROC3) as a loading control showed that neither F5H nor COMT expression was decreased in ref2 rosettes.
Figure 7.
RNA Gel Blot Analysis of Phenylpropanoid Gene Expression in Wild-Type and ref2 Rosettes.
(A) F5H gene expression in wild-type, fah1-2, and ref2 rosettes normalized to cyclophilin (ROC3). Col, Columbia wild type.
(B) COMT gene expression in wild-type, fah1-2, and ref2 rosettes normalized to cyclophilin (ROC3).
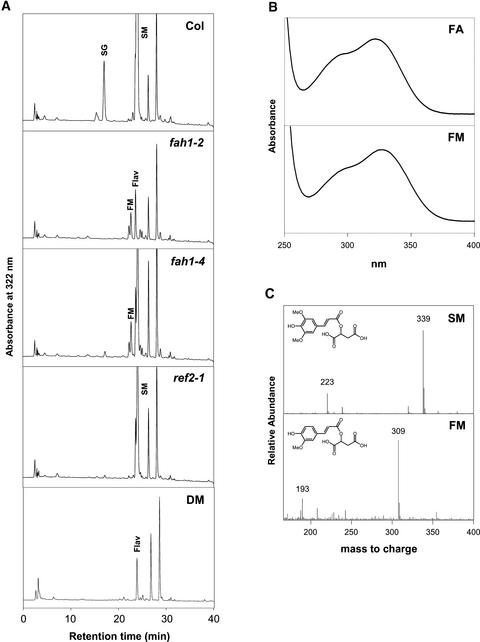
To determine if decreased F5H enzyme activity is responsible for the ref2 phenylpropanoid phenotypes, attempts were made to assay F5H activity in plant microsomes prepared from wild-type and ref2 rosettes. Unfortunately, activity could be detected in neither wild-type nor ref2 microsomes, even using the enzyme's preferred substrates, coniferaldehyde and coniferyl alcohol (data not shown) (Humphreys et al., 1999; Osakabe et al., 1999). To circumvent the inability to assay F5H in vitro, a genetic approach was taken to investigate F5H activity in the ref2 mutant. Although it has been stated frequently that fah1 leaves lack hydroxycinnamic acid esters, they accumulate a novel compound that is undetectable in wild-type extracts and that displays a UV light spectrum identical to that of ferulic acid (Figures 8A and 8B). LC-MS analysis of the peak showed that it has a mass consistent with that of feruloylmalate (negative ion mass-to-charge ratio of 309) (Figure 8C). This compound was detected in all null and leaky fah1 alleles tested (Figure 8A) and was absent in wild-type extracts analyzed by HPLC and LC-MS. These findings suggest that feruloylmalate accumulation is a common phenotype shared by Arabidopsis mutants with decreased levels of F5H. Thus, as a measure of in vivo F5H activity, plants homozygous for each ref2 allele were analyzed by HPLC for the presence of feruloylmalate; however, this compound was not found in any of the mutants (Figure 8A). The absence of detectable levels of feruloylmalate in ref2 extracts suggests that ref2 rosettes contain wild-type levels of F5H activity. Finally, to determine if a decrease in COMT activity was responsible for the ref2 phenylpropanoid phenotypes, COMT activity was assayed in wild-type and ref2 protein extracts. HPLC analysis of in vitro enzyme assays showed that ref2 extracts have wild-type levels of COMT (Table 2).
Figure 8.
Soluble Secondary Metabolites Accumulated in the Leaves of the Wild Type, fah1-2, fah1-4, ref2-1, and a fah1-2 ref2-1 Double Mutant.
(A) HPLC analysis of phenylpropanoids monitored at 320 nm. fah1-2 is a null allele of F5H, whereas fah1-4 is a leaky fah1 allele. Col, Columbia wild type; DM, double mutant; Flav, flavonoid; FM, feruloylmalate; SG, sinapoylglucose; SM, sinapoylmalate.
(B) Absorbance spectrum of the novel ferulate ester peak accumulated in fah1 mutants, and the spectrum of an authentic standard of ferulic acid (FA) for comparison.
(C) Mass spectra of sinapoylmalate from wild-type leaf extracts and feruloylmalate in fah1-2 leaf extracts.
Table 2.
COMT Activity and Ethylene Production in Wild-Type and ref2 Arabidopsis (n = 3)
| Sample | COMT Activity (pkat mg−1 protein) |
Ethylene Production (ppb·mg−1 fresh wt·min−1) |
|---|---|---|
| Wild type | 8.39 ± 0.78 | 9.4 ± 1.8 |
| Wild type (2) | 11.97 ± 0.33 | – |
| ref2-1 | 9.37 ± 0.07 | 16.3 ± 1.4 |
| ref2-2 | 8.25 ± 0.13 | 15.7 ± 3.0 |
| ref2-3 | 10.46 ± 0.14 | – |
| ref2-4 | 9.74 ± 0.36 | – |
Values shown are for the units indicated ± se.
If inhibitors of methylation reactions, including that catalyzed by COMT, accumulate in ref2 leaves, in vitro analysis of COMT activity in desalted protein extracts in the presence of saturating levels of substrates may not accurately reflect the enzyme's in vivo activity. For this reason, and as a parallel to the genetic approach used to evaluate F5H action in vivo, the phenylpropanoid ester content of a ref2-1 fah1-2 double mutant was evaluated. HPLC analysis of ref2-1 fah1-2 leaf extracts showed that the double mutant lacked the sinapoylmalate peak present in ref2-1 extracts and the feruloylmalate peak identified in fah1-2 extracts (Figure 8A). Thus, fah1 is epistatic to ref2 for sinapoylmalate accumulation, whereas ref2 is epistatic to fah1 for the presence of feruloylmalate. These data reveal that at least in leaves, the ref2 mutation can have an impact on phenylpropanoid metabolism upstream of F5H despite the fact that the ref2 mutant deposits normal levels of guaiacyl lignin in stems. Given that the phenylpropanoid ring O-meth-ylation steps catalyzed by caffeoyl-CoA O-methyltransferase and COMT are required for the synthesis of ferulic acid and sinapic acid, respectively, these two enzymes represent possible sites at which phenylpropanoid biosynthesis could be perturbed in the ref2 mutant. In this context, it is notable that ref2 rosettes produce normal levels of ethylene (Table 2), suggesting that if phenylpropanoid O-methylation reactions are perturbed in the mutant, it is probably not the result of the general poisoning of S-adenosylmethionine pools.
sur2 Exhibits a ref Phenotype
To further investigate the link between glucosinolate and phenylpropanoid biosynthesis in the ref2 mutant, the sinapate ester levels in leaves of two other glucosinolate biosynthetic mutants were analyzed by HPLC. Analysis of bus1 leaf extracts showed that bus1 has wild-type levels of leaf sinapate esters (Figure 9). Given that the bus1 mutation blocks alkylglucosinolate biosynthesis immediately preceding the reaction catalyzed by REF2, this result suggests that the ref2 phenylpropanoid phenotypes are not attributable to a lack of alkylglucosinolate intermediates downstream of both REF2 and BUS1 but instead probably are the result of an accumulation of the REF2 substrate, Met-derived aldoximes.
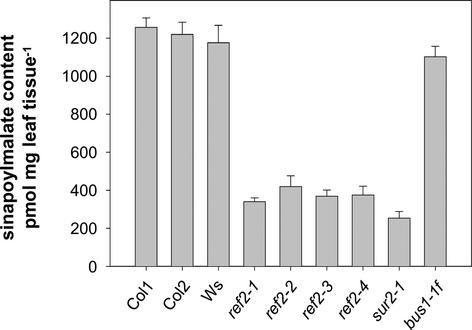
Figure 9.
Sinapoylmalate Levels in Columbia Wild-Type, Wassilewskija Wild-Type, ref2, sur2-1, and bus1 4-Week-Old Rosettes.
Quantification of sinapate esters by HPLC was performed by comparison with a sinapate standard at 320 nm. Error bars represent standard errors of triplicate assays. Col, Columbia; Ws, Wassilewskija.
HPLC analysis of sur2 leaf extracts showed that they contain significantly less sinapoylmalate than Wassilewskija, the parent ecotype of sur2-1 (Figure 9). The reduced sinapate ester content of sur2 rosettes shows that mutations in other glucosinolate biosynthetic genes lead to alterations in phenylpropanoid biosynthesis. Furthermore, given that CYP83B1 mutants may accumulate increased levels of indole-3-acetaldoxime, it also suggests that that perturbation of phenylpropanoid biosynthesis may be an effect common to multiple aldoximes.
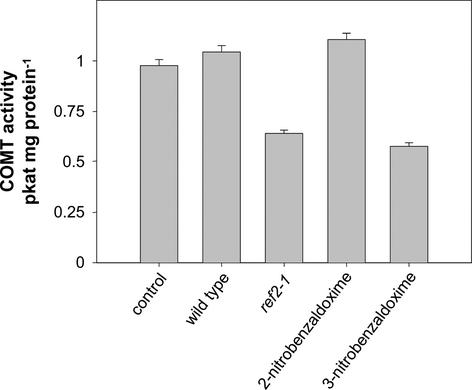
COMT Assayed in Vitro Is Inhibited by ref2 Leaf Extracts and Synthetic Aldoximes
To further test the hypothesis that aldoximes accumulate in ref2 tissues and inhibit phenylpropanoid O-methylation reactions, the activity of heterologously expressed COMT was assayed after preincubation with two commercially available aldoximes and with methanolic leaf extracts from wild-type and ref2 plants (Figure 10). Based on the assumption that aldoximes may be competitive inhibitors of COMT, caffeic acid and S-adenosylmethionine were added to assays at concentrations close to the reported Km values of COMT for these substrates (Edwards and Dixon, 1991). Compared with assays containing wild-type extracts, COMT activity by ref2 extracts was between 25 and 38% in three independent experiments (Figure 10). These results indicate that ref2 leaves may contain one or more inhibitors of COMT activity, including Met-derived aldoximes or their degradation products. Similarly, 3-nitrobenzaldoxime, but not its 2-nitro isomer, inhibited COMT activity by 40%, further suggesting that aldoximes may play a role in the ref2 phenylpropanoid phenotypes.
Figure 10.
Activity of Heterologously Expressed COMT in the Presence of Wild-Type and Mutant Leaf Extracts and Synthetic Aldoximes.
Quantification of the ferulic acid product by HPLC was performed by comparison with a ferulic acid standard at 320 nm. Error bars represent standard errors for quadruplicate assays.
DISCUSSION
Genetic approaches can elucidate the functions of known proteins in plant metabolism but also have the potential to identify novel factors that regulate or otherwise affect biosynthetic pathways. The ref2 mutant was identified in a screen for plants showing altered phenylpropanoid phenotypes, but further analysis of ref2 showed that mutations in CYP83A1 also lead to decreased levels of glucosinolates, a family of secondary compounds thought to be metabolized independently from phenylpropanoids. With this fact in mind, a more detailed investigation into the phenylpropanoid, glucosinolate, and morphological phenotypes of ref2 was conducted to determine if these phenotypes were the result of changes in phenylpropanoid pathway gene expression, a specific metabolic interaction between these two pathways, or a broad metabolic disruption in the mutant.
CYP83A1 Is an Enzyme in Met-Derived Glucosinolate Biosynthesis
Analysis of glucosinolates in ref2 leaf and seed extracts shows that ref2 plants accumulate significantly lower amounts of all of the alkylglucosinolates found in wild-type plants. Given that heterologously expressed CYP83A1 has been shown to be an aldoxime-oxidizing enzyme (Bak and Feyereisen, 2001), this genetic analysis shows that in vivo, CYP83A1 oxidizes Met-derived aldoximes in alkylglucosinolate biosynthesis (Figures 5 and 11). These results are in contrast to similar analyses conducted with bus1 tissues, which showed that the bus1 mutation leads to reductions in only short-chain Met-derived glucosinolates. Thus, although the oxidation of chain-elongated Met homologs is shared in a nonredundant manner by CYP79F1 and CYP79F2, CYP83A1 oxidizes all Met-derived aldoximes.
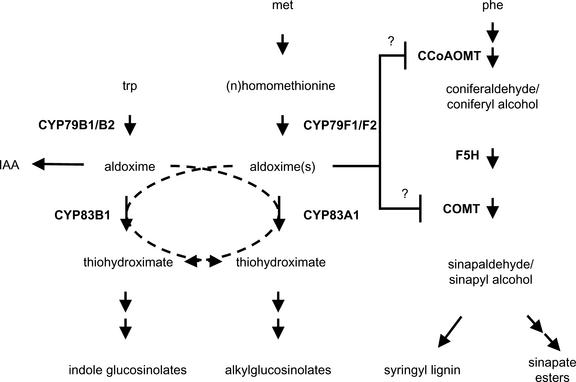
Figure 11.
Proposed Model for Alkylglucosinolate and Phenylpropanoid Biosynthesis and Their Interaction in ref2.
The model illustrates the role of the six P450s known to function in alkylglucosinolate and indole glucosinolate biosynthesis. The model also indicates that CYP83A1 and CYP83B1 may have partially overlapping functions, at least in the context of certain mutants. Finally, the model illustrates the hypothesis that a block at CYP83A1 may lead directly or indirectly to decreased COMT and caffeoyl-CoA O-methyltransferase (CCoAOMT) activity as a result of the accumulation of Met-derived aldoximes. IAA, indole 3-acetic acid.
There May Be Distinct Functions for, but Compensatory Interplay between, CYP83A1 and CYP83B1
Residual levels of alkylglucosinolates in the four null ref2 alleles examined in this study indicate that one or more other enzymes, most likely CYP83B1, can oxidize Met-derived aldoximes in the absence of CYP83A1 function (Figure 11). Similarly, indole glucosinolate levels are decreased by only ∼50% in a null sur2 allele (Bak et al., 2001). These data could be interpreted as evidence to support overlapping roles for CYP83A1 and CYP83B1 in glucosinolate biosynthesis, particularly in light of the recent demonstration that these enzymes oxidize a range of aldoximes (Bak and Feyereisen, 2001). By contrast, the accumulation of wild-type or greater levels of indole glucosinolates in ref2 argues against a significant role for CYP83A1 in their biosynthesis, at least in the presence of wild-type CYP83B1 function. The lack of sur2-like developmental phenotypes in ref2 plants also is consistent with this hypothesis, because a loss of CYP83A1 activity did not lead to indole 3-acetic acid–related growth perturbations. Together, these results suggest that in wild-type plants, the primary functions of CYP83A1 and CYP83B1 are in alkylglucosinolate and indole glucosinolate biosynthesis, respectively; however, in the sur2 and ref2 mutants, the loss of one enzyme is mitigated by the activity of its homolog.
Interestingly, ref2 leaf extracts were found consistently to contain higher levels of indole glucosinolates, a phenotype shared by bus1 plants (Reintanz et al., 2001). These observations indicate that a block in alkylglucosinolate biosynthesis, regardless of whether it is at the amino acid– or the aldoxime-oxidizing step, leads to an increase in indole glucosinolates. Similarly, 3-methylsulfinylpropyl glucosinolate, the most abundant glucosinolate in Wassilewskija leaves, is hyperaccumulated in sur2 (M. Hemm and C. Chapple, unpublished results). These results suggest that glucosinolate biosynthesis may be regulated in Arabidopsis such that a block in the biosynthesis of one class of glucosinolates leads to an increase in the synthesis of others, resulting in some degree of glucosinolate homeostasis.
Phenylpropanoid Biosynthesis Is Altered in ref2
Given that ref2 mutants are morphologically indistinguishable from wild-type plants, it seems unlikely that their phenylpropanoid phenotypes are caused by broad-scale perturbation of hormone levels or plant development. Instead, biochemical analysis of ref2, and the inhibition of COMT by ref2 extracts in vitro, suggest that the phenylpropanoid phenotypes of the mutant may be attributable to the inhibition of O-methyltransferase activities by one or more aldoximes accumulated in the mutant (Figure 11). Furthermore, the ref phenotype displayed by sur2 plants, and the inhibition of COMT by 3-nitrobenzaldoxime, suggest that the perturbation of phenylpropanoid metabolism may result from the accumulation of a wide variety of aldoximes. Alternatively, because aldoximes are relatively reactive compounds (Dawson et al., 1993), it might be their degradation products that inhibit COMT.
Whether the inhibition of O-methylation reactions alone leads to the ref2 phenylpropanoid phenotypes remains to be determined. Another possible explanation for the ref2 phenylpropanoid phenotypes is that CYP83A1 plays an enzymatic role in sinapate ester and/or syringyl lignin biosynthesis. Although it is difficult to explain where another P450 could act in the phenylpropanoid pathway, two other members of the CYP83 P450 family, CYP83C1 (Bilodeau et al., 1999) and CYP83D1 (Siminszky et al., 1999), have been identified in parsley and soybean, respectively, two species that do not accumulate glucosinolates. The presence of CYP83 family members in these taxa suggest that these enzymes may be involved in pathways other than glucosinolate biosynthesis, potentially in phenylpropanoid metabolism.
The Impact of ref2 Mutations on Phenylpropanoid Metabolism May Constrain Glucosinolate Variability in Arabidopsis Populations
Recently, areas of the Arabidopsis genome have been identified as quantitative trait loci for glucosinolate accumulation. For example, quantitative trait loci have been identified that lead to differences in the levels of glucosinolates between the Landsberg and Cape Verdi ecotypes (Kliebenstein et al., 2001). Neither BUS1 nor SUR2 is located in significant quantitative trait loci, consistent with the selective disadvantage that mutations at these loci would provide in natural populations because of their impact on overall plant vigor (Delarue et al., 1998; Bak et al., 2001; Reintanz et al., 2001). By contrast, REF2 is contained within one of the quantitative trait loci responsible for a significant increase in indole glucosinolates in the Landsberg ecotype, suggesting that allelic variation at the REF2 locus may exist among Arabidopsis ecotypes. In this context, however, it is interesting that plants lacking wild-type levels of sinapoylmalate show increased susceptibility to damage from UV light (Landry et al., 1995; Booij-James et al., 2000). Thus, even though null CYP83A1 mutants develop normally under laboratory conditions, changes in glucosinolate biosynthesis through allelic variation at the REF2 locus may be constrained in natural populations by the effects of decreased CYP83A1 activity on the synthesis of UV light–protective sinapate esters. This illustrates the potential for natural variation in one group of secondary metabolites to be linked directly to the accumulation of a group of biochemically unrelated molecules and for selection for variation in metabolites involved in plant–insect interactions to be constrained by relative plant fitness in the context of UV light stress.
METHODS
Plant Growth and Genetic Mapping
Arabidopsis thaliana plants were cultivated at a light intensity of 100 μE·m−2·s−1 at 23°C under a 16-h-light/8-h-dark photoperiod in Redi-Earth potting mix (Scotts-Sierra Horticulture Products, Marysville, OH). Unless noted otherwise, all Arabidopsis plants used in the experiments were ecotype Columbia. As described previously, the four ref2 alleles were isolated from a F2 screen of ethyl methanesulfonate–mutagenized plants (Ruegger and Chapple, 2001). ref2-1 and ref2-2 were backcrossed three times, and the ref2-3 and ref2-4 alleles were backcrossed once to Columbia before analysis. Isolation and backcross information on the sur2-1 and bus1-1 mutants has been described elsewhere (Delarue et al., 1998; Reintanz et al., 2001).
Cloning and Plant Transformation
The ref2 mutant (Columbia background) was used in a cross to the Landsberg erecta ecotype. F1 individuals were allowed to self-pollinate, and F2 plants were screened for the ref2 phenotype. DNA was extracted from homozygous mutant lines for Arabidopsis restriction fragment length polymorphism marker mapping (Schaffner, 1996) to determine an initial map position for the REF2 gene. Subsequently, DNA was extracted from additional F2 plants and F3 families for use in PCR-based genotyping experiments. Individuals carrying recombinant chromosomes in the region of the REF2 locus were used to determine a mapping interval for the gene and were analyzed further.
Cosmids containing Landsberg genomic fragments spanning the REF2 locus were identified from a transformation-competent Landsberg genomic library (Meyer et al., 1996). These binary cosmid vectors then were introduced into Agrobacterium tumefaciens C58 pGV3850 (Zambrisky et al., 1983) by electroporation. Plant transformation was performed by the floral dip method (Clough and Bent, 1998).
To screen for transformants, ref2-1 T0 seeds were surface-sterilized for 10 min in a 2:1 mixture of 0.1% Triton X-100 and bleach. Seeds were rinsed thoroughly with sterile water and plated on modified Murashige and Skoog (1962) medium (ammonia-free medium to which an additional 20.6 mM potassium nitrate was added in place of ammonium nitrate) containing 1% Suc, 0.7% agar, 50 mg/L kanamycin, and 200 mg/L Timentin. After 2 weeks, kanamycin-resistant progeny were transferred to soil. Two to 3 weeks after transplanting, plants were screened visually and by HPLC analysis for complementation of the ref2 fluorescence and sinapoylmalate phenotypes.
RNA Gel Blot Analysis
RNA was isolated from 4-week old Arabidopsis rosettes as described previously (Goldsbrough and Cullis, 1981). RNA samples were separated electrophoretically, transferred to Hybond N+ membranes (Amersham), and hybridized at 65°C with a DNA probe (DECAprime II system; Amersham) made from PCR amplification of the gene of interest as well as with a probe made from the cDNA of the Arabidopsis cyclophilin ROC3. Gene expression was quantified using a Typhoon Imager (Molecular Dynamics, Sunnyvale, CA).
Glucosinolate Analysis
For glucosinolate analysis of plant tissues, desulfoglucosinolate extracts were prepared according to the protocol described by Glover et al. (1988) with minor modifications. Leaves were harvested and extracted twice in 1.5 mL of 50% methanol heated to 70°C for 15 min. Before heating, 40 μL of 500 μM allylglucosinolate (sinigrin) (Indofine Chemical Company, Somerville, NJ) was added to the first methanol extraction of each sample as an internal standard. Extracts were combined and dried completely under vacuum using an RC1010 Speed-Vac (Jouan, Winchester, VA). Samples were redissolved in 1.5 mL of water and passed over an SP-50 anion-exchange column (500-μL bed volume suspended in water). Columns then were washed with 3 mL of water. A total of 200 μL of a 1:3 dilution of type-I sulfatase solution (Helix pomatia; Sigma) in 26 mM sodium acetate buffer, pH 5.5, was added to each column. Sulfatase was prepared as described by Kiddle et al. (2001). Columns were capped and incubated for at least 12 h at 37°C. After incubation, 1.5 mL of water was passed through the column and collected as the desulfoglucosinolate-containing fraction. For seed sam-ples, the same protocol was used with the following alterations: 40 μL of 5 mM allylglucosinolate was added to each sample as a standard, and samples weighing ∼40 mg were incubated in 1 mL of 50% methanol for 10 min at 70°C to soften the seeds followed by grinding in the methanol and further incubation at 70°C for 20 min.
Desulfoglucosinolates were analyzed by diode array HPLC and liquid chromatography–mass spectrometry (LC-MS). A total of 100 μL of the desulfoglucosinolate fractions was injected onto a Varian C18 reverse-phase column and run using the HPLC method described by Reintanz et al. (2001). Peaks were identified based on retention times from published results as well as on UV light spectra (Reintanz et al., 2001). Peak identity was confirmed by LC-MS (LCMS-2010 single-quadrupole mass spectrometer detector with an atmospheric pressure chemical ionization source; Shimadzu, Kyoto, Japan). The LCMS-2010 was operated under the following conditions: curved desolvation line temperature of 250°C, atmospheric pressure chemical ionization probe temperature of 400°C, nitrogen nebulizer gas flow of 2.5 L/min, detector voltage of 1.5 kV, and curved desolvation line voltage of −35 kV. Major peaks were identified by scanning ions with mass-to-charge ratios of 150 to 400 in positive mode. Quantification of glucosinolates was performed by comparison with the allylglucosinolate internal standard using HPLC response factors for desulfoglucosinolates at 229 nm (J. Gershenzon, personal communication).
Sinapate Ester Analysis
For analysis of sinapate esters, tissue was extracted in 50% methanol and analyzed by reverse-phase HPLC. Leaf and seed extracts were separated on a Microsorb-MV C18 column (Rainin Instruments, Woburn, MA) using a gradient from 1.5% acetic acid to 35% acetonitrile in 1.5% acetic acid at a flow rate of 1 mL/min. To better separate the novel ferulic acid ester found in fah1-2 plants from the sinapoylmalate found in wild-type and ref2 plants, samples were run on the same column using a gradient from 5% acetic acid to 25% acetonitrile in 5% acetic acid at a flow rate of 1 mL/min. Compounds were identified based on their retention times and UV light spectra. The identity of the feruloylmalate present in fah1 leaf extracts was confirmed by LC-MS using the same system and method described for glucosinolate analysis. Samples were run on a Microsorb-MV C18 column using a gradient from 0 to 35% acetonitrile at a flow rate of 1 mL/min. Major peaks were identified by scanning ions with mass-to-charge ratios of 150 to 400 in negative and positive modes. Sinapate and ferulate esters were quantified based on comparison with standards of their respective free acids.
Lignin Analysis
For cell wall preparation, Arabidopsis rachis tissue was ground to a fine powder and extracted with neutral phosphate buffer, 80% ethanol, and acetone (Meyer et al., 1998). To measure lignin content, cell wall samples were analyzed using the microscale Klason method (Kaar and Brink, 1991). Lignin monomer composition was determined by the derivatization followed by reductive cleavage method (Lu and Ralph, 1997) modified as described previously (Franke et al., 2000). Derivatization followed by reductive cleavage products were quantified by comparison with authentic standards using gas chromatography–mass spectrometry and selective ion monitoring.
Ethylene Measurements
Rosettes of 4-week-old plants were harvested, weighed, and incubated under ambient laboratory lighting in a sealed 5-mL scintillation vial for 90 min. Ethylene content of the gas phase was measured as described previously (Jones and Woodson, 1999).
O-Methyltransferase Assays
Protein extracts were prepared and assayed for caffeic acid O-methyltransferase activity as described previously (Inoue et al., 1998) except that the product was measured by HPLC. Samples were separated on a Microsorb-MV C18 column (Rainin Instruments) using a gradient from 1.5% acetic acid to 35% acetonitrile in 1.5% acetic acid at a flow rate of 1 mL/min. Formation of the ferulic acid product was measured at 322 nm and quantified by comparison with standards. Total protein content was measured using the Pierce bicinchoninic acid assay using BSA as a standard.
Protein extracts from Escherichia coli expressing caffeic acid O-methyltransferase were prepared as described previously (Humphreys et al., 1999). Methanolic leaf extracts were prepared by incubating ground rosettes in 50% methanol for 15 min at 4°C. After samples were centrifuged, the supernatants were removed, stored at −70°C, and used in the add-back assays. In each assay, 10 μL of plant extracts was incubated in a total volume of 100 μL. Control assays contained 10 μL of 50% methanol. In reactions incubated with 2-nitrobenzaldoxime or 3-nitrobenzaldoxime, 5 μL of 10 mM aldoxime was incubated in a total volume of 100 μL, making the final aldoxime concentration 500 μM. Enzyme and extracts were incubated at 30°C for 30 min before reactions were started by the addition of S-adenosylmethionine and caffeic acid to final concentrations of 5 μM S-adenosylmethionine and 60 μM caffeic acid. After 1 h at 30°C, the reactions were stopped by the addition of 100 μL of methanol followed by vortexing. Supernatants of the reaction were analyzed by HPLC.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
The authors are grateful to Daryl J. Murry for access to his LC-MS facilities. We also thank Catherine Bellini and Klaus Palme for the generous donation of sur2-1 and bus1-1 seeds, respectively. This work was supported by a grant from the Division of Energy Biosciences, U.S. Department of Energy, and also by a fellowship from the Innovation Realization Laboratory (IRL) at Purdue University's Krannert School of Management. The IRL is funded through the National Science Foundation's Integrative Graduate Engineering Research and Training grant program. This is journal paper number 16939 of the Purdue University Agricultural Experiment Station.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006544.
References
- Bak, S., and Feyereisen, R. (2001). The involvement of two P450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol. 127, 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak, S., Tax, F.E., Feldmann, K.A., Galbraith, D.W., and Feyereisen, R. (2001). CYP83B1, a cytochrome P450 at the metabolic branch paint in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13, 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlier, I., Kowalczyk, M., Marchant, A., Ljung, K., Bhalerao, R., Bennett, M., Sandberg, G., and Bellini, C. (2000). The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl. Acad. Sci. USA 97, 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Bennett, R.N., Hick, A.J., Dawson, G.W., and Wallsgrove, R.M. (1995). Glucosinolate biosynthesis: Further characterization of the aldoxime-forming microsomal monooxygenases in oilseed rape leaves. Plant Physiol. 109, 299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau, P., Udvardi, M.K., Peacock, W.J., and Dennis, E.S. (1999). A prolonged cold treatment-induced cytochrome P450 gene from Arabidopsis thaliana. Plant Cell Environ. 22, 791–800. [Google Scholar]
- Booij-James, I.S., Dube, S.K., Jansen, M.A.K., Edelman, M., and Mattoo, A.K. (2000). Ultraviolet-B radiation impacts light-mediated turnover of the photosystem II reaction center heterodimer in Arabidopsis mutants altered in phenolic metabolism. Plant Physiol. 124, 1275–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple, C. (1998). Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 311–343. [DOI] [PubMed] [Google Scholar]
- Chapple, C.C.S. (1995). A cDNA encoding a novel cytochrome P450-dependent monooxygenase from Arabidopsis thaliana. Plant Physiol. 108, 875–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple, C.C.S., Shirley, B.W., Zook, M., Hammerschmidt, R., and Somerville, S.C. (1994). Secondary metabolism in Arabidopsis. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 989–1030.
- Chapple, C.C.S., Vogt, T., Ellis, B.E., and Somerville, C.R. (1992). An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4, 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., and Andreassan, E. (2001). Update on glucosinolate metabolism and transport. Plant Physiol. Biochem. 39, 743–759. [Google Scholar]
- Chow, I.T., and Gasser, C.S. (1997). Characterization of the cyclophilin gene family of Arabidopsis thaliana and phylogenetic analysis of known cyclophilin proteins. Plant Mol. Biol. 35, 873–892. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dawson, G.W., Hick, A.J., Bennett, R.N., Donald, A., Pickett, J.A., and Wallsgrove, R.M. (1993). Synthesis of glucosinolate precursors and investigations into the biosynthesis of phenylalkyl- and methylthioalkylglucosinolates. J. Biol. Chem. 268, 27154–27159. [PubMed] [Google Scholar]
- Delarue, M., Prinsen, E., Onckelen, H.V., Caboche, M., and Bellini, C. (1998). sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 14, 603–611. [DOI] [PubMed] [Google Scholar]
- Edwards, R., and Dixon, R.A. (1991). Purification and characterization of S-adenosyl-l-methionine:caffeic acid 3-O-methyltransferase from suspension cultures of alfalfa (Medicago sativa L.). Arch. Biochem. Biophys. 287, 372–379. [DOI] [PubMed] [Google Scholar]
- Fabri, C.O., and Schaffner, A.R. (1994). An Arabidopsis thaliana RFLP mapping set to localize mutations to chromosomal regions. Plant J. 5, 149–156. [Google Scholar]
- Franke, R., Hemm, M.R., Denault, J.W., Ruegger, M.O., Humphreys, J.M., and Chapple, C. (2002. a). Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J. 30, 47–59. [DOI] [PubMed] [Google Scholar]
- Franke, R., Humphreys, J.M., Hemm, M.R., Denault, J.W., Ruegger, M.O., Cusumano, J.C., and Chapple, C. (2002. b). The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 30, 33–45. [DOI] [PubMed] [Google Scholar]
- Franke, R., McMichael, C.M., Meyer, K., Shirley, A.M., Cusumano, J.C., and Chapple, C. (2000). Modified lignin in tobacco and poplar plants over-expressing the Arabidopsis gene encoding ferulate 5-hydroxylase. Plant J. 22, 223–234. [DOI] [PubMed] [Google Scholar]
- Glover, J.R., Chapple, C.C.S., Rothwell, S., Tober, I., and Ellis, B.E. (1988). Allylglucosinolate biosynthesis in Brassica carinata. Phytochemistry 27, 1345–1348. [Google Scholar]
- Goldsbrough, P.B., and Cullis, C.A. (1981). Characterization of the genes for ribosomal RNA in flax. Nucleic Acids Res. 9, 1301–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, S.E., and Peterson, J.A. (1999). How similar are P450s and what can their differences teach us? Arch. Biochem. Biophys. 369, 24–29. [DOI] [PubMed] [Google Scholar]
- Graser, G., Oldham, N.J., Brown, P.D., Temp, U., and Gershenzon, J. (2001). The biosynthesis of benzoic acid glucosinolate esters in Arabidopsis thaliana. Phytochemistry 57, 23–32. [DOI] [PubMed] [Google Scholar]
- Halkier, B.A., and Du, L. (1997). The biosynthesis of glucosinolates. Trends Plant Sci. 2, 425–431. [DOI] [PubMed] [Google Scholar]
- Hansen, C.H., Du, L.C., Naur, P., Olsen, C.E., Axelsen, K.B., Hick, A.J., Pickett, J.A., and Halkier, B.A. (2001). CYP83B1 is the oxime-metabolizing enzyme in the glucosinolate pathway in Arabidopsis. J. Biol. Chem. 276, 24790–24796. [DOI] [PubMed] [Google Scholar]
- Hilleren, P., and Parker, R. (1999). mRNA surveillance in eukaryotes: Kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA 5, 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull, A.K., Vij, R., and Celenza, J.L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA 97, 2379–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys, J.M., and Chapple, C. (2002). Rewriting the lignin roadmap. Curr. Opin. Plant Biol. 5, 224–229. [DOI] [PubMed] [Google Scholar]
- Humphreys, J.M., Hemm, M.R., and Chapple, C. (1999). New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc. Natl. Acad. Sci. USA 96, 10045–10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, K., Sewalt, V.J.H., Balance, G.M., Ni, W., Stürzer, C., and Dixon, R. (1998). Developmental expression and substrate specificities of alfalfa caffeic acid 3-O-methyltransferase and caffeoyl coenzyme A 3-O-methyltransferase in relation to lignification. Plant Physiol. 117, 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M.L., and Woodson, W.R. (1999). Differential expression of three members of the 1-aminocyclopropane-1-carboxylate synthase gene family in carnation. Plant Physiol. 119, 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaar, W.E., and Brink, D.L. (1991). Simplified analysis of acid-soluble lignin. J. Wood Chem. Technol. 11, 465–477. [Google Scholar]
- Kiddle, G., Bennett, R.N., Botting, N.P., Davidson, N.E., Robertson, A.A.B., and Wallsgrove, R.M. (2001). High-performance liquid chromatographic separation of natural and synthetic desulphoglucosinolates and their chemical validation by UV, NMR, and chemical ionisation-MS methods. Phytochem. Anal. 12, 226–242. [DOI] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Gershenzon, J., and Mitchell-Olds, T. (2001). Comparative quantitative trait loci mapping of aliphatic, indolic and benzylic glucosinolate production in Arabidopsis thaliana leaves and seeds. Genetics 159, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Lambrix, V., Reichelt, M., Mitchell-Olds, T., Kliebenstein, D.J., and Gershenzon, J. (2001). The Arabidopsis epithiospecifier protein promotes the hydrolysis of glucosinolates to nitriles and influences Trichoplusia ni herbivory. Plant Cell 13, 2793–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry, L.G., Chapple, C.C.S., and Last, R.L. (1995). Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol. 109, 1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehfeldt, C., Shirley, A.M., Meyer, K., Ruegger, M.O., Cusumano, J.C., Viitanen, P.V., Strack, D., and Chapple, C. (2000). Cloning of the SNG1 gene of Arabidopsis reveals a role for serine carboxy-peptidase-like protein as an acyltransferase in secondary metabolism. Plant Cell 12, 1295–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, N.G., and Yamamoto, E. (1990). Lignin: Occurrence, biogenesis and biodegradation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41, 455–496. [DOI] [PubMed] [Google Scholar]
- Lu, F.C., and Ralph, J. (1997). Derivatization followed by reductive cleavage (DFRC method), a new method for lignin analysis: Protocol for analysis of DFRC monomers. J. Agric. Food Chem. 45, 2590–2592. [Google Scholar]
- Meyer, K., Benning, G., and Grill, E. (1996). Cloning of plant genes based on genetic map location. In Genome Mapping in Plants, A.H. Paterson, ed (New York: Academic Press/Austin, TX: Landes Bioscience Publishers), pp. 137–154.
- Meyer, K., Shirley, A.M., Cusumano, J.C., Bell-Lelong, D.A., and Chapple, C. (1998). Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 6619–6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen, M.D., Hansen, C.H., Wittstock, U., and Halkier, B.A. (2000). Cytochrome P450CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 275, 33712–33717. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Osakabe, K., Tsao, C.C., Li, L.G., Popko, J.L., Umezawa, T., Carraway, D.T., Smetzer, R.H., Joshi, C.P., and Chiang, V.L. (1999). Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc. Natl. Acad. Sci. USA 96, 8955–8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, B.L., Chen, S.X., Hansen, C.H., Olsen, C.E., and Halkier, B.A. (2002). Composition and content of glucosinolates in developing Arabidopsis thaliana. Planta 214, 562–571. [DOI] [PubMed] [Google Scholar]
- Reintanz, B., Lehnen, M., Reichelt, M., Gershenzon, J., Kowalczyk, M., Sandberg, G., Godde, M., Uhl, R., and Palme, K. (2001). bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell 13, 351–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger, M., and Chapple, C. (2001). Mutations that reduce sinapoylmalate accumulation in Arabidopsis thaliana define loci with diverse roles in phenylpropanoid metabolism. Genetics 159, 1741–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger, M., Meyer, K., Cusumano, J.C., and Chapple, C. (1999). Regulation of ferulate-5-hydroxylase expression in Arabidopsis in the context of sinapate ester biosynthesis. Plant Physiol. 119, 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner, A. (1996). pARMS, multiple marker-containing plasmids for easy RFLP analysis in Arabidopsis thaliana. Plant Mol. Biol. Rep. 14, 11–16. [Google Scholar]
- Shirley, A.M., McMichael, C.M., and Chapple, C. (2001). The sng2 mutant of Arabidopsis is defective in the gene encoding the serine carboxypeptidase-like protein sinapoylglucose:choline sinapoyltransferase. Plant J. 28, 83–94. [DOI] [PubMed] [Google Scholar]
- Siminszky, B., Corbin, F.T., Ward, E.R., Fleischmann, T.J., and Dewey, R.E. (1999). Expression of a soybean cytochrome P450 monooxygenase cDNA in yeast and tobacco enhances the metabolism of phenylurea herbicides. Proc. Natl. Acad. Sci. USA 96, 1750–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, G.S., McQuade, C., Lyon, J., and Brown, J.W.S. (1998). Characterization of exon skipping mutants of the COP1 gene from Arabidopsis. Plant J. 15, 125–131. [DOI] [PubMed] [Google Scholar]
- Strack, D. (1977). Sinapic acid ester fluctuations in cotyledons of Raphanus sativus. Z. Pflanzenphysiol. 84, 139–145. [Google Scholar]
- Strack, D. (1981). Sinapine as a supply of choline for the biosynthesis of phosphatidylcholine in Raphanus sativus seedlings. Z. Naturforsch. 36c, 215–221. [Google Scholar]
- Tahara, S., and Ibrahim, R.K. (1995). Prenylated isoflavonoids: An update. Phytochemistry 38, 1073–1094. [Google Scholar]
- Tantikanjana, T., Yong, J.W.H., Letham, D.S., Griffith, M., Ljung, K., Sandberg, G., and Sundaresan, V. (2001). Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes Dev. 15, 1577–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrisky, P., Joos, H., Genetello, C., Leemans, J., van Montagu, M., and Schell, J. (1983). Ti plasmid vector for the introduction of DNA into plant cells without alteration of their normal regeneration capacity. EMBO J. 2, 2143–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]