Abstract
Identifying the genes regulated by the floral homeotic genes APETALA3 (AP3) and PISTILLATA (PI) is crucial for understanding the molecular mechanisms that lead to petal and stamen formation. We have used microarray analysis to conduct a broad survey of genes whose expression is affected by AP3 and PI activity. DNA microarrays consisting of 9216 Arabidopsis ESTs were screened with probes corresponding to mRNAs from different mutant and transgenic lines that misexpress AP3 and/or PI. The microarray results were further confirmed by RNA gel blot analyses. Our results suggest that AP3 and PI regulate a relatively small number of genes, implying that many genes used in petal and stamen development are not tissue specific and likely have roles in other processes as well. We recovered genes similar to previously identified petal- and stamen-expressed genes as well as genes that were not implicated previously in petal and stamen development. A very low percentage of the genes recovered encoded transcription factors. This finding suggests that AP3 and PI act relatively directly to regulate the genes required for the basic cellular processes responsible for petal and stamen morphogenesis.
INTRODUCTION
Arabidopsis flowers are composed of four distinct organ types arranged in concentric whorls. Each of these organ types has a characteristic ontogeny and morphology. The sterile organs, or the perianth, consist of four sepals in the first or outer whorl and four petals in the second whorl. Sepals are leaf-like organs in which the abaxial surface contains many stomata interspersed among irregularly shaped epidermal cells. The petals have a simple laminar structure, and the petal blade is covered with specialized, dome-shaped cells whose surface is finely ridged in a radial pattern. The two inner whorls contain the reproductive organs: the third whorl comprises six (four long and two short) stamens, and the fourth whorl consists of a gynoecium formed by two fused carpels. The stamens, the male reproductive organs, consist of a filament bearing an anther containing several differentiated tissues that are involved in producing and releasing the pollen. The gynoecium, the female reproductive structure, is composed of a basal ovary, in which the ovules develop, and a distal style capped by stigmatic papillae.
How do these distinct organ types develop from the morphologically homogeneous cells of the floral meristem? Genetic and molecular studies of floral homeotic mutants, in which one floral organ type is transformed into another, have led to the identification of several master regulatory genes that specify floral organ identity, known as the ABC genes (reviewed by Theissen and Saedler, 2001; Lohmann and Weigel, 2002). These genes act in a combinatorial manner in overlapping domains to regulate the specification of organ identity in the flower. The activity of A-class genes leads to the formation of sepals in the first whorl, whereas the activity of A-class genes in conjunction with that of B-class genes promotes the formation of petals in the second whorl. Similarly, the combination of B- and C-class activity is required for stamen formation in the third whorl, whereas C-class genes specify carpel identity in the fourth whorl. More recently, additional functions have been shown to be required in combination with the ABC genes in the specification of petals, stamens, and carpels (Honma and Goto, 2001; Pelaz et al., 2001). The organ identity genes all encode transcription factors and thus presumably do not act directly to specify morphological differences. Rather, it is likely that they regulate the expression of suites of subordinate genes that encode cellular functions required directly in differentiation processes. Identifying the genes regulated by these master transcription factors is crucial for understanding the molecular mechanisms that lead to organ-specific cell and tissue differentiation.
APETALA3 (AP3) and PISTILLATA (PI) genes together confer B-class function in Arabidopsis. Loss-of-function mutations in either of these genes result in the conversion of petals to sepals and stamens to carpeloid organs (Bowman et al., 1989; Hill and Lord, 1989; Jack et al., 1992). Furthermore, in the presence of intact A and C function, their ectopic coexpression is sufficient to transform sepals to petals and carpels to stamens (Krizek and Meyerowitz, 1996a). In accordance with their roles in specifying petal and stamen identity, AP3 and PI expression coincides largely with their functional domains and is maintained throughout petal and stamen development (Jack et al., 1992; Goto and Meyerowitz, 1994).
AP3 and PI encode MADS domain–containing transcription factors (Jack et al., 1992; Goto and Meyerowitz, 1994). Their gene products form heterodimers that recognize and bind in vitro to a conserved DNA sequence, CC(A/T)6GG, called the CArG box (Schwarz-Sommer et al., 1992; Riechmann et al., 1996; Hill et al., 1998).
Only a few genes have been shown to be regulated directly by the AP3/PI heterodimer. The expression of AP3 or PI is reduced in ap3 or pi mutant backgrounds, suggesting that they are required to maintain their own expression in a positive feedback loop (Jack et al., 1992; Goto and Meyerowitz, 1994). In the case of AP3, this autoregulation is direct, because the AP3 promoter can be activated by a steroid-inducible form of AP3 even in the presence of the protein synthesis inhibitor cycloheximide (Honma and Goto, 2001). Furthermore, this activation is likely to be mediated by at least two CArG boxes in the AP3 promoter that are required for promoter function and are bound by AP3/PI heterodimers (Riechmann et al., 1996; Hill et al., 1998; Tilly et al., 1998). However, PI autoregulation probably is indirect, because it requires de novo protein synthesis and PI regulatory sequences do not contain CArG boxes (Chen et al., 2000; Honma and Goto, 2000). To date, other than AP3, only the NAP (NAC-LIKE, ACTIVATED BY AP3/PI) gene has been shown to be a direct target of AP3/PI. NAP was identified in a differential display–based screen for genes expressed in the presence of cycloheximide in response to the activation of a steroid-inducible form of AP3 in the presence of PI protein (Sablowski and Meyerowitz, 1998). NAP is expressed relatively late in floral development in the developing petals and stamens and is thought to play a role in the transition from the cell division to the cell expansion phase of growth of these organs (Sablowski and Meyerowitz, 1998). SUPERMAN, a gene that is required to maintain the boundary between the third and fourth whorls of the flower, also has been postulated to be a target of AP3/PI action, because its expression is reduced in ap3 and pi mutants; however, direct regulation by AP3/PI has not been demonstrated (Sakai et al., 2000).
A number of other candidate AP3/PI targets have been isolated using differential display–based screens to identify genes expressed predominantly in petals and stamens (Rubinelli et al., 1998; Irish, 1999; Kotilainen et al., 1999). These studies yielded a relatively small number of genes that likely play a role in petal and stamen development. Two of these studies demonstrated reduced expression of candidate genes in an ap3 loss-of-function mutant (Rubinelli et al., 1998) or in a loss-of-function mutant of the Antirrhinum AP3 ortholog DEFICIENS (Nacken et al., 1991), suggesting that their expression is regulated by the B-class genes. Based on sequence similarity to known genes, the putative function of several of the genes recovered in these studies was postulated. These genes encode proteins that are likely involved in cell wall formation (polygalacturonase, pectate lyase, and cell wall structural proteins) as well as a lipid transfer protein, β-glucosidase, and a Leu-rich repeat protein (Nacken et al., 1991; Rubinelli et al., 1998).
To conduct a broader survey for genes regulated by AP3/PI that are involved in petal and stamen development, we took advantage of high-throughput DNA microarray technology. DNA microarray analysis allows the systematic monitoring of expression profiles of thousands of genes (Schena et al., 1995). It has been used to study genes involved in developmental processes in animals (White et al., 1999; Reinke et al., 2000), such as identifying potential downstream targets of a homeodomain transcription factor (Leemans et al., 2001). In plants, increasing numbers of studies have applied DNA microarray technology to investigate the regulation of environmental responses and developmental processes. These genome-scale analyses have revealed the coordinated regulation of different metabolic pathways (Harmer et al., 2000; Schenk et al., 2000; Wang et al., 2000; Ma et al., 2001; Schaffer et al., 2001). Furthermore, the ability to assess the regulation of expression of a large number of genes has allowed the identification of novel genes acting in a specific pathway (Wang et al., 2000; Perez-Amador et al., 2001; Schaffer et al., 2001) and has suggested novel functions for previously characterized genes (Ruan et al., 1998; Chen et al., 2002). In addition, such analyses have enabled the recovery of cis-acting DNA elements required for the regulation of specific patterns of gene expression (Girke et al., 2000; Harmer et al., 2000).
In this study, we have compared the gene expression profiles of flowers from either mutant or transgenic lines that misexpress AP3/PI, resulting in altered petal and stamen development. In addition to identifying potential downstream targets of AP3/PI, our results provide new insights regarding the nature of the developmental pathways that underlie petal and stamen formation.
RESULTS
Brief Description of the Microarray and the Experimental Protocol
We used an Arabidopsis EST clone–based microarray to perform these studies (Ma et al., 2001). This array contained 9216 Arabidopsis ESTs from all stages of the Arabidopsis life cycle spotted as two duplicate arrays on individual glass slides. These ESTs correspond to ∼6120 unique genes (Ma et al., 2001). Four microarray slides (eight replicates) were used to analyze the relative mRNA abundance of each sample. Two slides were probed with cDNA synthesized from one sample RNA labeled with Cy-3 and the other sample RNA labeled with Cy-5 deoxy-UTP, and the labeling was reversed in the other two slides. Two independent RNA preparations were made for each biological sample and used to prepare labeled probes. The hybridization signals from the eight replicate experiments were averaged and used for analysis. The cutoff for significant difference in expression between the sample pair was >1.8. Although the commonly used threshold value is twofold (DeRisi et al., 1997; Wildsmith and Elcock, 2001), several studies have shown that a lower cutoff ranging from 1.4 to 1.74 can be used reliably (Wang et al., 2000; Perez-Amador et al., 2001; Yue et al., 2001), particularly if the results are reproducible in seven or more replicates (Perez-Amador et al., 2001). A BLAST (Basic Local Alignment Search Tool) search against the Arabidopsis genome sequence database identified the gene corresponding to each EST and also revealed multiple ESTs representing the same gene. In most cases, ESTs representing the same gene showed similar changes in expression, verifying the reproducibility of the microarray results.
Strategy to Identify Genes That Act Downstream of AP3/PI
The AP3 and PI proteins bind to DNA as an obligate heterodimer, so their activity requires the function of both proteins (Goto and Meyerowitz, 1994; Jack et al., 1994; McGonigle et al., 1996). Therefore, loss of function of either of these genes results in a similar mutant phenotype (Figure 1) (Jack et al., 1992; Goto and Meyerowitz, 1994). This finding suggests that the regulation of AP3 and PI downstream targets is affected similarly in both mutants. Hence, similar genes are expected to be recovered when comparing the expression profiles in either ap3 or pi mutants with those of their wild-type counterparts. ap3-3 and pi-1 are strong mutant alleles of AP3 and PI, respectively (Jack et al., 1992; Goto and Meyerowitz, 1994). In both mutants, a single nucleotide change results in the conversion of an individual amino acid residue (Gln at amino acid 18 in ap3-3 and Trp at amino acid 80 in pi-1) to a stop codon, resulting in an early termination of the encoded protein.
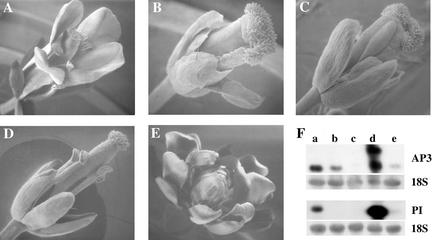
Figure 1.
Characterization of Flowers from the Different Genotypes Used in This Study.
(A) to (E) Scanning electron micrographs of mature flowers of each genotype treated with dexamethasone. Genotypes are as follows: wild type (A), pi-1 (B), ap3-3 (C), D6::DTA (D), and ap3-3; ag-3; 35S::PI; 35S::AP3-GR (abbreviated as AP3-GR) treated with dexamethosone (E).
(F) RNA gel blot analysis of AP3 (top gels) and PI (bottom gels) expression in the different genetic backgrounds: wild type (lane a), pi-1 (lane b), ap3-3 (lane c), AP3-GR (lane d), and D6::DTA (lane e). Lanes contained 20 μg of total RNA extracted from flowers. The AP3 higher molecular mass band in lane d corresponds to the transcript of the transgene encoding the AP3-GR fusion protein. The lower molecular mass band corresponds to the transcript of the endogenous mutant ap3 gene, whose expression also was regulated positively by AP3-GR. Hybridization to 18S rRNA shows the RNA loading in each lane.
To corroborate the results obtained from studying gene expression in the ap3/pi loss-of-function mutants, as well as to differentiate between AP3/PI downstream targets acting specifically in the formation of either petals or stamens, we also examined two transgenic lines that differentially affect the development of these organs. Plants of the genetic constitution ap3-3; ag-3; 35S::PI; 35S::AP3-GR (abbreviated as AP3-GR) have been used previously to identify direct downstream targets of AP3/PI (Sablowski and Meyerowitz, 1998). When the AP3 fusion protein, in this genetic background, is activated post-translationally by the application of the steroid dexamethasone, these plants produce flowers containing all petals (Figure 1). Genes regulated by AP3/PI in petals should show opposite expression patterns in the AP3-GR line compared with the ap3 and pi mutants (i.e., such positively regulated genes should show increased expression in AP3-GR and reduced expression in ap3/pi); conversely, negatively regulated genes should show a reduction of expression in AP3-GR and increased expression levels in ap3/pi.
We also examined the expression of ESTs in a second transgenic line, D6::DTA, in which flowers fail to produce petals but still produce stamens (Figure 1) (Hill et al., 1998). D6::DTA plants express the diphtheria toxin A chain (DTA) gene under the control of a fragment of the AP3 promoter driving significant expression only in the second whorl of the flower. DTA gene expression results in cell ablation; thus, the second-whorl petals do not develop, whereas the rest of the flower develops almost normally, except for occasional loss of stamens (Hill et al., 1998). In this line, the expression of genes regulated by AP3/PI specifically in petals should be similar to that in the pi-1 and ap3-3 loss-of-function mutants, because petals are lacking in both. On the other hand, genes showing an expression pattern similar in the wild-type and D6::DTA backgrounds, both of which have stamens, but having reduced expression in pi-1, ap3-3, and AP3-GR, all of which lack stamens, are likely to be genes regulated by AP3/PI specifically in stamens.
The mRNA levels of PI and AP3 themselves in the different genetic backgrounds were analyzed by gel blot analysis (Figure 1F). PI mRNA levels are almost undetectable in both ap3 and pi loss-of-function mutants, reflecting the autoregulation of PI expression by AP3/PI. Similarly, AP3 mRNA levels are reduced to background in ap3. However, AP3 expression is maintained in second-whorl organs of pi (Jack et al., 1992); thus, there is only a partial reduction in its mRNA level in this mutant background. Both AP3 and PI mRNA levels are increased highly in AP3-GR, because their ectopic expression is driven by the strong viral 35S promoter. The mRNA levels of both AP3 and PI are reduced significantly in D6::DTA, because these flowers lack petals, where these genes are normally highly expressed.
To be able to compare all of the mutant and transgenic lines, the microarray experiments were performed by comparing the gene expression profiles of each genotype with the wild type. Total RNA was extracted from flowers at all stages of development from each of the different genotypes and used to synthesize probes for the microarray experiments. The genes that showed significant changes in expression in the different lines compared with the wild type then were compared with each other, and eight different groups based on relative levels of expression were compiled. These data are shown in Table 1 as the ratio of hybridization signals between each mutant or transgenic line to the wild type. The table includes the list of genes (and the corresponding EST clones) that were recovered as well as the probable functions of these genes (based on annotation by the Munich Information Center for Protein Sequences [MIPS] Arabidopsis database).
Table 1.
| Gene Identifier |
EST Clone Identifier |
pi | ap3 | AP3GR | D6::DTA | Putative Functiona |
|---|---|---|---|---|---|---|
| Group A: genes expressed in both petals and stamens | ||||||
| At5g49360 | H5A10 | 0.479 ± 0.178 | 0.838 ± 0.301 | 3.474 ± 0.382 | 1.300 ± 0.364 | Xylosidase |
| At5g11090 | 123M11 | 0.492 ± 0.049 | 0.451 ± 0.294 | 3.186 ± 0.397 | 1.602 ± 0.326 | Putative protein |
| At1g32100 | 192P14 | 0.508 ± 0.054 | 0.269 ± 0.379 | 2.957 ± 0.289 | 1.666 ± 0.278 | Putative pinoresinol-lariciresinol reductase |
| At4g33150 | 206I4 | 0.483 ± 0.109 | 0.698 ± 0.256 | 2.015 ± 0.304 | 0.858 ± 0.135 | Lys-ketoglutarate reductase |
| G12C4 | 0.500 ± 0.167 | 0.682 ± 0.257 | 1.938 ± 0.385 | 0.942 ± 0.195 | ||
| Group B: genes expressed in petals and highly expressed in stamens | ||||||
| At1g20440 | 242B8 | 0.180 ± 0.057 | 0.290 ± 0.195 | 1.942 ± 0.390 | 1.971 ± 0.774 | COR47, dehydrin |
| 137A7 | 0.357 ± 0.076 | 0.444 ± 0.172 | 1.776 ± 0.348 | 2.270 ± 0.456 | ||
| At1g20450 | 240L5 | 0.211 ± 0.075 | 0.357 ± 0.235 | 1.607 ± 0.396 | 2.373 ± 0.845 | ERD10/dehydrin |
| 242A18 | 0.346 ±0.123 | 0.388 ± 0.244 | 1.591 ± 0.381 | 2.482 ± 0.859 | ||
| At1g07600 | 162J19 | 0.448 ± 0.059 | 0.269 ± 0.355 | 3.502 ± 0.454 | 1.826 ± 0.351 | Metallothionein-like |
| 222N3 | 0.482 ± 0.045 | 0.297 ± 0.355 | 3.181 ± 0.361 | 1.829 ± 0.333 | ||
| 104H3 | 0.433 ± 0.053 | 0.201 ± 0.372 | 3.213 ± 0.321 | 1.805 ± 0.278 | ||
| 201I2 | 0.437 ± 0.059 | 0.205 ± 0.329 | 3.531 ± 0.444 | 1.942 ± 0.316 | ||
| 167G14 | 0.461 ± 0.039 | 0.273 ± 0.382 | 3.223 ± 0.313 | 1.097 ± 0.191 | ||
| 200L16 | 0.507 ± 0.081 | 0.359 ± 0.278 | 2.522 ± 0.367 | 1.538 ± 0.417 | ||
| At5g02160 | 125K7 | 0.410 ± 0.085 | 0.261 ± 0.349 | 3.153 ± 0.492 | 1.817 ± 0.335 | Unknown protein |
| At4g35770 | 224A3 | 0.385 ± 0.152 | 0.442 ± 0.340 | 1.998 ± 0.428 | NDb | Senescence-associated protein |
| 242N24 | 0.374 ± 0.091 | 0.494 ± 0.256 | 1.176 ± 0.593 | 1.835 ± 0.619 | ||
| 212B17 | 0.508 ± 0.881 | 0.545 ± 0.272 | 1.497 ± 0.272 | 1.442 ± 0.437 | ||
| At4g39090 | 240K3 | 0.509 ± 0.168 | 0.825 ± 0.392 | 2.400 ± 0.548 | 1.805 ± 0.547 | Drought-inducible thiol protease |
| 240O6 | 0.480 ± 0.170 | 0.755 ± 0.426 | 2.309 ± 0.522 | 1.737 ± 0.545 | ||
| At4g36040 | 242O4 | 0.541 ± 0.098 | 0.873 ± 0.544 | 2.763 ± 0.990 | 2.617 ± 0.997 | DNAJ-11 chaperone |
| Group C: genes expressed in petals | ||||||
| At3g11930 | 151O22 | 0.368 ± 0.037 | 0.407 ± 0.095 | 1.523 ± 0.346 | 0.502 ± 0.092 | Unknown protein (receptor-like kinase) |
| At1g25230 | F9B4 | 0.387 ± 0.051 | 0.524 ± 0.093 | 1.100 ± 0.346 | 0.342 ± 01.4 | Hypothetical protein (phosphatase) |
| At5g45950 | F4D7 | 0.398 ± 0.067 | 0.312 ± 0.054 | 3.373 ± 0.340 | 0.410 ± 0.152 | GDSL-motif lipase/hydrolase |
| At3g47340 | G3F8 | 0.475 ± 0.227 | 0.439 ± 0.139 | 0.860 ± 0.161 | 0.603 ± 0.151 | Glutamine-dependent Asn synthetase |
| At1g62480 | 223M24 | 0.696 ± 0.133 | 0.408 ± 0.054 | 0.799 ± 0.140 | 0.464 ± 0.113 | T3P18.4/hypothetical protein |
| At4g04460 | 148M12 | 0.264 ± 0.079 | 0.361 ± 0.109 | 0.558 ± 0.201 | 0.124 ± 0.047 | Putative Asp protease |
| Group D: genes expressed in stamens | ||||||
| At5g14380 | 171N22 | 0.237 ± 0.047 | 0.282 ± 0.137 | 0.194 ± 0.174 | ND | AGP6 |
| At3g15400 | 188H17 | 0.085 ± 0.029 | 0.072 ± 0.016 | 0.122 ± 0.055 | 0.982 ± 0.178 | ATA20; anther development protein |
| At3g28670 | 190D9 | 0.185 ± 0.053 | 0.247 ± 0.039 | 0.122 ± 0.054 | 1.055 ± 0.180 | Unknown protein (Meth adenotransferase) |
| Group E: genes expressed in petals and stamens at late stages of development | ||||||
| At5g57660 | F3A10 | 0.594 ± 0.168 | 0.808 ± 0.065 | 1.900 ± 0.473 | 3.064 ± 0.944 | Constans-like |
| At2g33380 | 158N17 | 0.766 ± 0.192 | 0.946 ± 0.378 | 1.975 ± 0.391 | 1.861 ± 0.607 | Putative calcium-binding EF-hand protein |
| At1g62060 | 140P1 | 0.791 ± 0.096 | 0.878 ± 0.229 | 2.027 ± 0.276 | 1.801 ± 0.675 | Hypothetical protein (ribosomal) |
| At1g64370 | 105C1 | 0.91 ± 0.081 | 0.912 ± 0.276 | 2.619 ± 0.641 | 2.167 ± 0.846 | Unknown protein |
| Group F: genes downregulated in petals and stamens at late stages of development | ||||||
| At5g42530/At2g25510 | 103C7 | 0.917 ± 0.065 | 1.120 ± 0.445 | 0.491 ± 0.038 | 0.410 ± 0.306 | Unknown protein |
| At4g11320 | 203N14 | 1.477 ± 0.264 | 0.819 ± 0.194 | 0.533 ± 0.100 | 0.313 ± 0.116 | Drought-inducible Cys proteinase |
| Group G: genes expressed in petals at late stages of development | ||||||
| At1g15260 | 229L4 | 0.745 ± 0.087 | 0.752 ± 0.086 | 2.359 ± 0.287 | 0.338 ± 0.187 | Hypothetical protein |
| At1g74670 | 149E11 | 0.904 ± 0.112 | 0.819 ± 0.396 | 2.924 ± 0.395 | 0.320 ± 0.086 | GAST-1 |
| At5g33370 | 93M23 | 0.960 ± 0.133 | 0.937 ± 0.155 | 2.290 ± 1.075 | 0.231 ± 0.050 | Putative protein (lipase) |
| At2g10940 | 137N1 | 0.992 ± 0.111 | 1.153 ± 0.360 | 2.486 ± 0.781 | 0.514 ± 0.101 | Unknown protein (cell wall) |
| At5g22430 | 143D1 | 1.003 ± 0.154 | 0.856 ± 0.087 | 1.975 ± 1.018 | 0.108 ± 0.061 | Unknown protein |
| At1g12090 | F10D9 | 1.110 ± 0.172 | 1.302 ± 0.423 | 2.268 ± 0.439 | 0.426 ± 0.100 | pEARLI 1-like (cell wall) |
| At4g24510 | 154C7 | 1.346 ± 0.191 | 0.999 ± 0.075 | 1.918 ± 0.479 | 0.549 ± 0.297 | CER2/fatty acid elongation |
| Group H: genes expressed in petals and/or stamens at early stages of development | ||||||
| At2g32150 | F2C7 | 0.282 ± 0.059 | 0.459 ± 0.122 | 1.016 ± 0.216 | 1.023 ± 0.093 | Putative hydrolase |
| At1g80920 | 241N24 | 0.286 ± 0.080 | 0.318 ± 0.341 | 1.348 ± 0.319 | 1.342 ± 0.392 | J8 (DNA-J like) |
| At4g30270 | 182G21 | 0.315 ± 0.065 | 0.439 ± 0.057 | 1.338 ± 0.113 | 0.692 ± 0.091 | Endo-xyloglucan transferase (Meri-5) |
| At5g19120 | 231A9 | 0.326 ± 0.732 | 0.413 ± 0.189 | 1.124 ± 0.336 | 0.971 ± 0.223 | Conglutin-gamma like |
| 220B19 | 0.373 ± 0.102 | 0.487 ± 0.192 | 1.102 ± 0.32 | 1.066 ± 0.168 | ||
| At5g62460 | F4H11 | 0.361 ± 0.085 | 0.475 ± 0.224 | 0.931 ± 0.413 | 0.960 ± 0.181 | Unknown protein |
| At3g15450 | 105P15 | 0.211 ± 0.075 | 0.333 ± 0.266 | 1.338 ± 0.528 | 1.207 ± 0.311 | Asn synthetase (ASN3) |
| At1g29930 | 130E5 | 0.299 ± 0.092 | 0.415 ± 0.206 | 1.129 ± 0.354 | 1.316 ± 0.533 | Photosystem II type I chlorophyll a/b binding protein |
| At2g37520 | 131D1 | 0.342 ± 0.083 | 0.438 ± 0.065 | 1.286 ± 0.135 | 0.889 ± 0.241 | Unknown protein (PHD finger) |
| 209L20 | 0.348 ± 0.093 | 0.438 ± 0.202 | 1.307 ± 0.407 | 1.078 ± 0.189 | Unknown protein | |
| 104E3 | 0.387 ± 0.076 | 0.416 ± 0.241 | 1.104 ± 0.307 | 1.233 ± 0.287 | Unknown protein | |
| At2g15890 | 232D20 | 0.422 ± 0.089 | 0.519 ± 0.404 | 1.062 ± 0.455 | 1.357 ± 0.316 | Unknown protein |
| 232C20 | 0.475 ± 0.104 | 0.499 ± 0.424 | 1.176 ± 0.338 | 1.150 ± 0.314 | ||
| At1g65970 | 93A21 | 0.447 ± 0.064 | 0.533 ± 0.079 | 1.174 ± 0.138 | ND | Peroxiredoxin |
| At5g40730 | 109G13 | 0.460 ± 0.109 | 0.460 ± 0.081 | 0.754 ± 0.162 | 0.703 ± 0.246 | AGP24 |
| At5g54770 | 158H10 | 0.519 ± 0.120 | 0.546 ± 0.455 | 1.116 ± 0.275 | 1.430 ± 0.352 | Thiazole biosynthetic enzyme (targeted to chloroplast) |
The results are presented as averages of the ratio of signal between each mutant or transgenic line to the wild type in eight replicates ±sd. Gene identifier numbers shown in boldface indicate EST sequences that showed 100% identity with the corresponding gene. Lightface type is used for genes that gave the highest Expect value in BLAST searches against genes from the Arabidopsis Genome Initiative but that did not show 100% identity with the corresponding EST.
According to the MIPS database or based on conserved motifs identified using NCBI Protein BLAST.
ND, not determined.
These genes were grouped into categories based on different expression profiles. Group A consists of genes whose expression was reduced in pi-1 and ap3-3, increased in AP3-GR, and not altered significantly in D6::DTA compared with the wild type. These genes are likely to be positively regulated by AP3/PI in both petals and stamens. Group B includes genes whose expression was reduced in pi-1 and ap3-3 and increased in AP3-GR and D6::DTA. The observation that these genes are more highly expressed in D6::DTA likely reflects the fact that stamen RNA is a larger proportion of the florally derived RNAs in these transgenic lines than in the wild type; thus, this group presumably consists of genes that are upregulated in stamens (and positively regulated in petals) in response to AP3/PI action. Group C includes genes whose expression was reduced in pi-1 and ap3-3 as well as in D6::DTA but not in AP3-GR. Expression of these genes is reduced in all lines lacking petals, suggesting that these genes are positively regulated by AP3/PI specifically in petals. Group D is composed of genes that show lower levels of expression in pi-1, ap3-3, and AP3-GR but not in D6::DTA; thus, these genes are likely to be positively regulated by AP3/PI in stamens.
Three additional groups include genes that did not show significant changes in their expression in pi-1 and ap3-3 mutants. However, their expression was either increased (group E) or reduced (group F) in both AP3-GR and D6::DTA or increased in AP3-GR but reduced in D6::DTA (group G). The recovery of these groups of genes might reflect a bias in the collection of floral samples. The more noticeable phenotype of AP3-GR and D6::DTA at later stages of development might have led to a higher percentage of older flowers in these samples compared with the wild-type, pi-1, and ap3-3 samples. Thus, the genes in groups E, F, and G may represent genes that are regulated positively or negatively at later stages of petal and stamen development. In addition, genes in group G, whose expression is unchanged in pi-1 and ap3-3, increased in AP3-GR, and decreased in D6::DTA, might represent genes expressed in both petals and sepals. Thus, change in their mRNA abundance is apparent only when significantly more petals are produced (AP3-GR) or when the organs are missing completely (D6::DTA).
The final group, group H, consists of genes whose expression is reduced in pi-1 and ap3-3 but is not changed significantly in AP3-GR and D6::DTA relative to the wild type. In accordance with a possible bias in sampling, these genes likely represent AP3/PI-regulated genes that act early in petal and/or stamen development. Alternatively, this group might correspond to genes that are expressed weakly in petals and strongly in stamens. Thus, the lack of petals in D6::DTA does not significantly alter the mRNA levels of these genes compared with the wild type, whereas the large number of petals in AP3-GR compensates for the lack of stamens. The validity of these hypotheses can be tested by monitoring the precise spatial and temporal expression of these genes using in situ hybridization.
The Expression of a Relatively Small Number of Genes Is Altered in Response to Changes in AP3/PI Activity
In this study, 61 ESTs corresponding to 47 genes were recovered as genes likely to be regulated by AP3/PI in petal and/or stamen development. The array used contains ∼6120 unique genes, which represents ∼25% of the genes in the Arabidopsis genome (Arabidopsis Genome Initiative, 2000). Therefore, our results suggest that the expression of a relatively small number of genes, on the order of 200, is affected in response to changes in AP3/PI activity. In turn, this implies that many genes used in petal and stamen development are not tissue specific and are likely to have roles in other tissues as well.
Assessing the Microarray Results by RNA Gel Blot Analysis
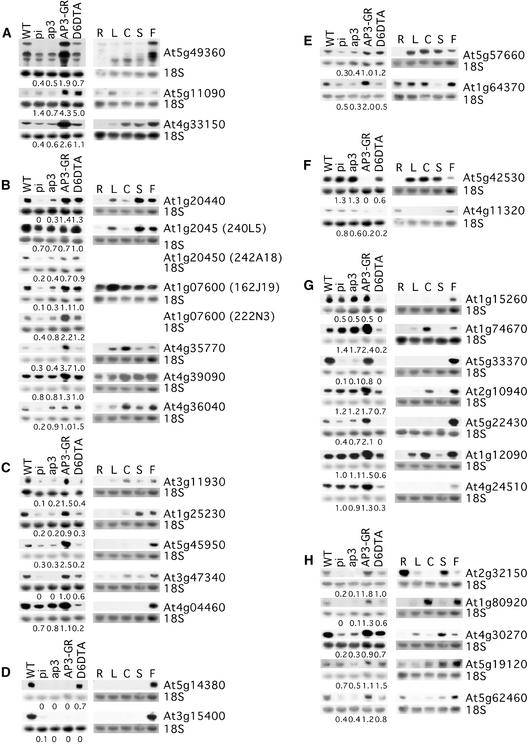
To test the reliability of the microarray results, the expression patterns of 32 of 47 candidate genes were examined by RNA gel blot analysis using total RNA prepared from the same lines that were used for the microarray experiments (Figure 2, left). The levels of the hybridization signals in the different samples were normalized relative to 18S RNA. For >90% of the genes, the differences in gene expression determined by RNA gel blot analysis correlated with the differences detected using microarrays, although values for the relative levels of mRNA abundance often were lower in the blot analysis (Figure 2, At5g57660; group E). In some of these cases, the hybridization signal overall was low, making it more difficult to accurately measure the signal over background (Figure 2, At4g11320; group F). In all 32 cases, the genes recovered by microarray analysis were shown to be regulated by AP3/PI. However, the pattern of upregulation and downregulation occasionally was somewhat different in the blot analysis versus the microarray analysis. For example, according to the microarray results, the expression of At5g33370 (group G) is unchanged in pi-1 and ap3-3 compared with the wild type; however, by gel blot analysis, lower mRNA levels of this gene are detected in these mutants. Two genes (At1g74670 and At2g10940; group G) were identified that according to the microarray results showed no change in expression in pi-1 and ap-3, an increase in AP3-GR, and a reduction in D6::DTA. These genes actually showed a small increase in RNA levels in pi-1 and ap3-3 by RNA gel blot analysis, suggesting that in the wild type, these genes are downregulated specifically by AP3/PI in stamens.
Figure 2.
RNA Gel Blot Analysis of Genes Recovered in the Microarray Analysis.
Total RNA (20 μg) extracted from flowers of the different genotypes used in this study or from different tissues of wild-type (WT) plants (R, roots; L, leaves; C, cauline leaves; S, inflorescence stems; and F, flowers) were used for gel blot analysis. Blots were hybridized with 32P-labeled probes corresponding to the indicated genes and then rehybridized with a probe corresponding to 18S rRNA as a loading control. The values given under the blots are the ratio of signal in each mutant or transgenic lines to the wild type, normalized relative to the 18S rRNA signal. In cases in which probes from two ESTs corresponding to the same gene were used, the EST clone identifier is given in parentheses. Blots are arranged in groups corresponding to the divisions in Table 1.
In addition to comparing mRNA levels in flowers of the different genotypes used in the microarray studies against wild-type floral mRNA, the expression levels in different parts of the wild-type plant (roots, leaves, cauline leaves, inflorescence stems, and flowers) also were measured by RNA gel blot analysis (Figure 2, right). All genes tested showed expression in flowers, and 11 of the 30 genes analyzed showed expression predominantly or exclusively in flowers, further supporting a role for these genes specifically in flower development. Overall, the RNA gel blot analysis confirmed the utility of our microarray-based strategy to identify genes whose expression is affected by AP3/PI activity. Possible roles for these genes are addressed below.
Stress-Related Genes
In groups A and B, which consist of genes putatively positively regulated by AP3/PI in both petals and stamens, several genes identified previously as stress-related genes were recovered. Both petal and stamen development can be divided into two main phases. The first phase involves cell division and differentiation, and the second phase is characterized by rapid growth of the organs predominantly through cell expansion (Goldberg et al., 1993; Pyke and Page, 1998; Irish, 1999). The induction of stress-related genes during petal and stamen development possibly reflects a requirement for similar functions during rapid organ expansion.
COR47 (At1g20440; group B) and ERD10 (At1g20450; group B) are dehydrin-encoding genes. Dehydrin genes are induced by stresses such as low temperature, drought, salts, and treatment with abscisic acid (Rouse et al., 1996; Nylander et al., 2001). In addition, several Arabidopsis dehydrin genes show tissue and cell type specificity in unstressed plants, which suggests a possible function under nonstress conditions (Nylander et al., 2001). Metallothioneins are small heavy metal binding proteins that can be activated by certain metals as well as a variety of other stimuli, including abscisic acid treatment, heat shock, cold shock, wounding, viral infection, senescence, and salt stress (Garcia- Hernandez et al., 1998). In Arabidopsis, three METALLOTHIO-NEIN gene families have been identified, MT1, MT2, and MT3, which exhibit characteristic temporal and tissue-specific expression patterns (Zhou and Goldsbrough, 1995; Garcia-Hernandez et al., 1998). Our data suggest a change only in the expression of the MT1-like genes (At1g07600/At1g076100; group B) in response to changes in AP3/PI activity. Another gene (At1g32100; group A) encodes a putative pinoresinol-lariciresinol reductase (PLR). PLRs are enzymes involved in the synthesis of plant defense lignans and isoflavonoids, and PLR-like genes are induced by various stresses (Gang et al., 1999). In addition to these more general stress-related genes, SENESCENCE-ASSOCIATED1 (SEN1; At4g35770; group B), a gene that has senescence-correlated expression in leaves (Oh et al., 1996), also was recovered as positively regulated by AP3/PI in petals and stamens.
The rapid growth of the petals and stamens also is likely to require increases in the rate of cellular metabolism. In accordance with this idea, several genes involved in amino acid metabolism were recovered in this study, such as Lys-ketoglutarate reductase (At4g33150; group A) and Gln-dependent Asn synthetase (At3g47340; group C). A gene that, according to our results, is putatively expressed early in petal and stamen development (At3g15450; group H) also is predicted to encode an Asn synthetase isoform. In addition, a gene (At4g04460; group C) putatively expressed in petals but not in stamens, as assessed by gel blot analysis, encodes a protein similar to Asp protease.
SEN1 and proteases are associated with senescence (Oh et al., 1996; Wagstaff et al., 2002). The reduction in the expression of the genes that encode these proteins in pi-1 and ap3-3 loss-of-function mutants correlates with the delayed senescence observed in these infertile mutants (Vivian-Smith et al., 2001; our unpublished observation). An early and seminal feature of plant senescence is the deterioration of the membrane, involving the deesterification of membrane fatty acids mediated by lipases. Reduction of the levels of a senescence-induced lipase protein in transgenic Arabidopsis plants leads to delayed leaf senescence (Thompson et al., 2000). Interestingly, two genes encoding a lipase (At5g45950; group C) and a putative protein with similarity to lipase (At5g33370; group G) were recovered among the genes putatively positively regulated in petals. However, these lipase-encoding genes are expressed exclusively in floral tissue (Figure 2), which implies a specific role in petals rather than a general role in senescence.
Cellular Signaling
Hydrolases, like lipases, also can play a role in cellular signaling, because lipids are involved in signal transduction pathways and a lipase-like gene has been shown to be important for salicylic acid signaling in plants (Jirage et al., 1999). A gene putatively expressed early in petal and/or stamen development encodes a hydrolase-like protein (At2g32150; group H) similar to a ripening-related protein from grape (Grip21) (Davies and Robinson, 2000). In addition to the hydrolase domain, the protein encoded by At2g32150 also contains a TIR domain, which is found in proteins such as the Drosophila melanogaster and human TOLL and Interleukin1 receptors as well as in some plant resistance gene products (van der Biezen and Jones, 1998). In these proteins, the TIR domain is involved in cellular signaling (van der Biezen and Jones, 1998). Two other genes that are likely to play a role in signaling cascades were identified as putative petal-expressed genes. One gene encodes an unknown protein that shows similarity to a receptor-like kinase (At3g11930; group C). The second gene encodes a protein with similarity to protein phosphatase (At1g25230; group C). Recent studies suggest roles for receptor-like kinases and phosphatases in plant morphogenesis (Lease et al., 1998; Takahashi et al., 1998).
Cell Wall–Associated Gene Products
Several genes that encode cell wall proteins and enzymes that modify cell wall components were recovered as being expressed differentially in the different mutant and transgenic lines. The cell wall has a profound effect on plant morphogenesis, both because its rigid structure constrains cell expansion and cell division and because, as increasing evidence suggests, the cell wall may be a direct source of factors that specify cell fate (reviewed by Fowler and Quatrano, 1997; Brownlee et al., 1998). Two genes expressed specifically in stamens that encode putative cell wall–associated proteins were recovered. The anther development gene ATA20 (At3g15400; group D) was identified previously as stamen specific (Rubinelli et al., 1998). It encodes a protein containing a Gly-rich region that is characteristic of one of the groups of cell wall proteins (Sachetto-Martins et al., 2000; Ringli et al., 2001). The second gene encodes an arabinogalactan protein, AGP6 (At5g14380; group D). AGPs are highly glycosylated Hyp-rich proteins that are found throughout the plant kingdom and are expressed mainly at cell surfaces, where they are thought to play important roles in plant growth and development (Majewska-Sawka and Nothnagel, 2000; Showalter, 2001). We also recovered another gene encoding an AGP, AGP24 (At5g40730; group H), which appears to be expressed in early petal and/or stamen development. Three genes that appear to be expressed late in petal development encode putative cell wall proteins (At1g74670, At2g10940, and At1g12090; group G). In addition to these cell wall proteins, two genes that encode enzymes that modify xyloglucan, a major structural component of the plant cell wall (Campbell and Braam, 1999), were recovered as petal- and stamen-expressed genes. One of these genes encodes an endoxyloglucan transferase (At4g30270; group H) commonly known as Meri-5 (Medford et al., 1991), whereas the other gene encodes a xylosidase (At5g49360; group A).
Transcriptional Regulation
Most of the genes recovered in this study encode products that appear to play roles in basic cellular functions. Although several of the recovered genes encode proteins that could play a role in cellular signaling, only two genes encoding products that might function as transcription factors were identified. One of these genes appears to be expressed early in petal and stamen development and encodes an unknown protein (At2g37520; group H) that contains a PHD (plant homeodomain) finger that is present in one class of plant homeodomain proteins (Schindler et al., 1993). The other gene encodes a CONSTANS-like protein (At5g57660; group E). The Arabidopsis CONSTANS gene accelerates flowering in response to long days, and its expression is modulated by the circadian clock and daylength (Suarez-Lopez et al., 2001). CONSTANS and CONSTANS-like genes encode proteins with an N-terminal zinc finger, a B-box domain predicted to mediate protein–protein interactions, and a C-terminal domain containing a putative nuclear localization signal. Because they lack any evident DNA binding motifs, other DNA binding proteins may recruit such factors to promoters (Ledger et al., 2001; Samach and Gover, 2001).
The extremely low recovery of transcription factors as targets of AP3/PI action might be the result of either the low representation of such factors on the arrays or the fact that the expression levels of such factors is below the limits of detection. The first possibility is unlikely, because these arrays are estimated to contain ESTs corresponding to 330 transcription factor–encoding genes (Ma et al., 2002), representing ∼20% of the transcription factors in the Arabidopsis genome (Riechmann and Ratcliffe, 2000). To determine whether such genes escaped recovery because their expression levels are under the threshold of signal detection with the microarray protocol used in our experiments, we performed two control microarray analyses of known genes. First, we examined whether two known floral transcription factors represented on the array, APETALA1 (Mandel et al., 1992) and SEPALLATA3 (Mandel and Yanofsky, 1998; Pelaz et al., 2000), showed detectable and significant changes in expression in comparisons of flower versus leaf tissue. These proteins showed greater than threefold increases in signal ratios in flowers versus leaves (3.000 ± 1.489 and 3.257 ± 0.668, respectively). We then examined the expression of 14 genes chosen at random that encode putative transcription factors on the arrays comparing the mutant and transgenic lines misexpressing AP3/PI (Table 2). Reliable hybridization signals were obtained for all of the genes tested; however, there was no significant difference in the expression of these genes in the various genotypes. These results indicate that the expression of these genes can be detected easily using our protocol and suggest that any changes in the expression of genes encoding transcription factors would have been detected.
Table 2.
Microarray Results of Several Transcription Factors Represented on the Array
| Gene Identifier |
EST Clone Identifier |
pi | ap3 | AP3GR | D6::DTA | Putative Functiona |
|---|---|---|---|---|---|---|
| At3g14230 | 196J8T7 | 0.869 ± 0.149 | 0.996 ± 0.067 | 1.107 ± 0.133 | 0.887 ± 0.171 | AP2 domain–containing protein |
| At3g54990 | 222A13T7 | 0.947 ± 0.141 | 0.894 ± 0.254 | 1.083 ± 0.202 | 1.065 ± 0.183 | APETALA2-like protein |
| At5g14750 | F2G6T7 | 0.961 ± 0.323 | 0.970 ± 0.222 | 1.004 ± 0.085 | 1.125 ± 0.248 | Myb transcription factor werewolf (WER)/ MYB66 |
| At3g13810 | G11B6T7 | 1.066 ± 0.125 | 1.029 ± 0.230 | 1.847 ± 0.238 | 0.928 ± 0.098 | Zinc finger protein, similar to finger protein pcp1 |
| At4g08150 | G1B1T7 | 1.265 ± 0.042 | 1.203 ± 0.265 | 0.926 ± 0.073 | 0.960 ± 0.163 | KNAT1 homeobox-like protein |
| At3g01470 | 208D10T7 | 0.754 ± 0.139 | 0.634 ± 0.102 | 1.300 ± 0.334 | 0.926 ± 0.263 | Homeobox-Leu zipper protein HAT5 |
| At3g57390 | 171E11T7 | 0.849 ± 0.099 | 0.804 ± 0.147 | 0.956 ± 0.060 | 0.889 ± 0.142 | MADS transcription factor–like agamous-like protein 15 |
| At1g24260 | 182B3T7 | 1.050 ± 0.055 | 0.955 ± 0.109 | 1.769 ± 0.209 | 0.916 ± 0.188 | Putative floral homeotic protein, AGL9 MADS box |
| At4g36990 | 190J12T7 | 1.037 ± 0.051 | 1.045 ± 0.058 | 1.068 ± 0.028 | 1.561 ± 0.212 | Heat shock transcription factor HSF4 |
| At5g24800 | 205M1T7 | 0.919 ± 0.209 | 0.979 ± 0.147 | 0.971 ± 0.074 | 1.077 ± 0.193 | Transcription factor–like protein, light-induced protein CPRF-2 |
| At5g52510 | G4D1T7 | 1.015 ± 0.176 | 0.978 ± 0.104 | 1.193 ± 0.282 | 1.051 ± 0.228 | SCARECROW transcriptional regulator-like |
| At5g12840 | 92H23T7 | 1.039 ± 0.088 | 1.019 ± 0.072 | 0.922 ± 0.036 | 0.872 ± 0.162 | CCAAT box binding factor/transcription factor Hap2a |
| At3g24050 | 146K2T7 | 1.041 ± 0.159 | 0.975 ± 0.095 | 0.980 ± 0.152 | 0.834 ± 0.153 | GATA transcription factor |
| At5g43700 | G5C3T7 | 1.206 ± 0.170 | 0.991 ± 0.112 | 0.984 ± 0.169 | 1.270 ± 0.283 | Auxin-induced protein AUX2-11 |
The results are presented as averages of the ratio of signal between each mutant or transgenic line to the wild type in eight replicates ±sd.
According to the MIPS database.
DISCUSSION
Identifying Downstream Targets of AP3/PI Using a Microarray-Based Strategy
More than a decade of research has led to significant progress in understanding the mechanisms of floral patterning, specifically the identification and characterization of the floral homeotic genes (for a recent review, see Lohmann and Weigel, 2002). However, one of the main questions that remains largely unanswered is how the regulatory activity of the floral homeotic genes is translated into changes in cell division, cell shape, size, and differentiation underlying specific organ formation. A first step in addressing this question is to identify the genes that act downstream of these master regulatory genes. To date, though, only a few downstream targets of the floral homeotic genes have been identified.
In the present study, we used microarrays to conduct a broad survey for genes whose expression is affected by the activity of AP3/PI, which together specify petal and stamen identity. We further tested the microarray results for most of the candidate genes by RNA gel blot analysis. For all of the genes tested, RNA gel blot analysis confirmed that their RNA levels were changed in response to changes in AP3/PI activity. However, in some cases, the pattern of RNA levels detected by gel blot analysis in the different genotypes used in our study did not accurately follow the pattern detected using microarray analysis. The incomplete correlation between the microarray and the RNA gel blot analysis results emphasizes the importance of comparing the gene expression profiles of several populations and the need to validate microarray results by other methods.
Using microarray analyses, we identified 47 genes that display differential expression in the various lines with altered AP3/PI activity versus the wild type, and we estimate that the number of genes presumably regulated by AP3/PI activity is on the order of 200. This relatively small number of differentially expressed genes suggests that, to a large extent, similar suites of genes are used in other developmental processes. Because the arrays used in this study represent an EST population obtained from all stages of Arabidopsis development, it is likely that floral tissues are underrepresented. As a result, we likely underestimated the number of genes required specifically in petal and stamen development. Nonetheless, many of the genes identified in our study have putative functions similar to those of previously identified petal- and/or stamen-expressed genes (see below). Although many of the genes have the expected characteristics of AP3/PI-regulated genes, others have been implicated previously in seemingly unrelated processes.
Many of the Genes Recovered May Play a Role in the Rapid-Growth Phase of Petals and Stamens
A prominent feature in both petal and stamen development is a phase of rapid organ growth, mainly through cell expansion (Goldberg et al., 1993; Pyke and Page, 1998; Irish, 1999). Many of the genes recovered in our study have functions that could be required for this rapid cell expansion.
In plants, cell expansion is dependent largely on changes in the cell wall. Thus, it is very likely that the expression of genes that encode proteins involved in the structure of the cell wall will be regulated during petal and stamen development. In our study, we identified several genes that encode proteins that modify cell wall components or that are likely to be cell wall proteins. One of these genes, which appears to be expressed late in petal development, is a GAST-1–like gene. Genes that encode proteins with high similarity to GAST-1 appear to play a specific role in corolla cell elongation (Shi et al., 1992; Ben-Nissan and Weiss, 1996; Kotilainen et al., 1999). Screens for stamen- and pollen-expressed genes also have yielded many cell wall–associated genes, including polygalacturonases, pectate lyases, and Gly-rich proteins (McCormick et al., 1987; Wing et al., 1989; Brown and Crouch, 1990; Nacken et al., 1991; Chen et al., 1994; Rubinelli et al., 1998). However, several of these genes probably have a specific function in pollen development or pollen tube growth.
Rapid cell expansion likely requires the acceleration of basic metabolic processes. In agreement with such a requirement, we recovered several genes that encode proteins involved in amino acid metabolism. Among these genes is a gene that encodes Lys-ketoglutarate, which was shown previously in Arabidopsis to have much higher expression levels in flowers than in vegetative tissue (Tang et al., 1997). Furthermore, a gene that encodes 5-enolypyruvylshikimate-3-phosphate, which catalyzes a step in the biosynthesis of aromatic amino acids, was shown to have a very high level of expression in petunia petals concomitant with the cell expansion phase (Benfey and Chua, 1989).
One of the surprising findings of our study was the recovery of several classes of stress-related genes that were positively regulated in petals and stamens. Upregulation of these genes in the transgenic lines used here might be a bona fide stress response. However, the reduced expression of these genes in ap3 and pi loss-of-function mutants suggests that they play an intrinsic role in petal and stamen development. Because stress conditions require a rapid cellular response, we speculate that similar gene functions might be required in processes that lead to rapid cell division and expansion in normal developmental pathways. Other studies also indicate possible roles for similar stress- and defense-related genes in development. In animals, the programmed expression of metallothioneins during development and their response to endogenous factors such as hormones and growth factors has suggested a possible role in cellular regulation (Robinson et al., 1993). Also, there is evidence for the regulation of transcription factor activity by metallothioneins with high affinity for Zn2+ (Robinson et al., 1993). In plants, different metallothioneins have distinct temporal and tissue-specific expression patterns (Garcia-Hernandez et al., 1998). Possible roles for these gene products in plant development also are suggested by the high expression of metallothioneins in the fruit of strawberry and grape (Nam et al., 1999; Davies and Robinson, 2000) and in the early stages of embryogenesis in wheat, soon after the onset of rapid cell division and differentiation (Kawashima et al., 1992). Similarly, dehydrins have been shown to display tissue- and cell type–specific expression in unstressed Arabidopsis plants (Nylander et al., 2001) and show enhanced expression during fruit ripening in strawberry (Nam et al., 1999). A function for defense response genes in normal flower development was suggested after the detection of developmentally regulated expression of pathogenesis-related genes in tobacco flowers (Lotan et al., 1989).
Genes That Encode Proteins Involved in Cellular Signaling Are Likely Candidates for AP3/PI Downstream Targets
AP3 has been shown to act in a non-cell-autonomous manner to regulate various aspects of petal and stamen development (Jenik and Irish, 2001). These nonautonomous effects of AP3 are not attributable to AP3 protein movement, implying that downstream targets of AP3 are required to mediate these intercellular interactions (Jenik and Irish, 2001). Therefore, the identification of genes encoding proteins that play a role in signaling cascades, specifically those that have been implicated in cell–cell interactions, is a strong expectation from our study. Indeed, we identified several classes of genes that might perform such functions.
In our study, we recovered a gene that encodes a protein similar to receptor-like kinase and a gene that encodes a putative phosphatase. Genes that encode proteins similar to receptor-like kinases and phosphatases are good candidates to function in these signaling pathways, particularly because similar gene products have been implicated in the regulation of morphogenesis. The best characterized receptor-like kinase is CLAVATA1, which is required to maintain the proper balance of cell proliferation and differentiation in the shoot and floral meristems (Clark et al., 1997). Other receptor-like kinases that are involved in morphogenesis include the Arabidopsis BRl1 and ERECTA gene products as well as the maize CRINKLY4 gene product and its homolog ACR4 in Arabidopsis (Becraft et al., 1996; Torii et al., 1996; Li and Chory, 1997; Tanaka et al., 2002). Studies of the CLAVATA1 signaling pathway have suggested a role for a protein phosphatase in the negative regulation of CLAVATA1 (Williams et al., 1997; Stone et al., 1998). A possible role for phosphatases in floral development is suggested by the finding that the carpel-specific protein VSP1, which has phosphatase activity, interacts in vitro with AGAMOUS, a MADS box protein that specifies stamen and carpel identity in Arabidopsis (Gamboa et al., 2001).
Another class of cell surface macromolecules that play multiple roles in plant development are the arabinogalactan proteins (AGPs) (Majewska-Sawka and Nothnagel, 2000; Gaspar et al., 2001; Showalter, 2001). The localization of AGPs on the plasmalemma, bound to the cell wall or soluble in the cell wall space, and their complex molecular structure could serve in signaling to neighbor cells (Majewska-Sawka and Nothnagel, 2000). We recovered two genes that encode arabinogalactan proteins that belong to two different subfamilies. AGP6 belongs to the “classic” AGPs, whereas AGP24 is an “AG peptide” (Borner et al., 2002).
The Low Percentage of Transcription Factors Recovered Suggests That AP3/PI Act Relatively Directly in Regulating Downstream Targets
A key question regarding the function of homeotic genes is whether they effect their function by regulating the onset of a gene activity cascade or, alternatively, by directly controlling a large subset of genes involved in the different processes required for organ formation. To date, the best studied group of homeotic genes are the Drosophila Hox genes. Among the Hox gene downstream targets, more than half of the identified genes encode other transcription factors and proteins involved in cellular signaling (Graba et al., 1997; Leemans et al., 2001). These findings suggest that many other intermediary regulators delegate many of the aspects of Hox gene regulation. By contrast, among the genes we recovered whose expression is affected by AP3/PI activity, only two genes encode putative transcription factors. This finding suggests that AP3/PI act fairly directly in regulating the basic cellular functions required for morphogenesis.
Studies using ap3/pi temperature-sensitive mutants or unstable transposon insertion lines demonstrated that the expression of AP3/PI (or their orthologs in Antirrhinum) is required throughout petal and stamen development (Bowman et al., 1989; Carpenter and Coen, 1990; Zachgo et al., 1995). Furthermore, the NAP gene, which is a direct target of AP3/PI, is expressed at late stages of petal and stamen development (Sablowski and Meyerowitz, 1998). These observations are consistent with the idea that AP3/PI must function throughout floral development and likely regulate many different suites of downstream genes, although this does not exclude the possibility that AP3/PI directly regulate only a small set of genes. Distinguishing between these possibilities will require the identification of the direct targets of AP3/PI.
The AP3 and PI genes encode MADS domain–containing proteins, and like other MADS domain proteins, they recognize and bind to a DNA motif known as the CArG box (Shore and Sharrocks, 1995). To try to identify possible direct targets of AP3/PI from the genes recovered in our study, we searched for the presence of a CArG box sequence in the upstream region of the candidate genes. Twenty-eight of 47 genes had at least one CArG box–like sequence in the 1-kb region upstream of the first ATG (data not shown). However, the significance of the presence of a CArG box–like motif is questionable. The presence of a CArG box does not necessarily imply the binding of a particular factor, because different MADS proteins are capable of binding to the same CArG box DNA binding sites; conversely, MADS domain proteins have been shown to bind to non-CArG box sequences in vitro (Riechmann et al., 1996; Hill et al., 1998). In addition, swapping the DNA binding domains of different MADS proteins does not affect their function in vivo (Krizek and Meyerowitz, 1996b; Riechmann and Meyerowitz, 1997). Therefore, the presence of a CArG box in a regulatory sequence does not necessarily indicate that such a gene is regulated directly by a particular MADS domain–containing transcription factor.
The multiple genes that we have identified as differentially regulated by AP3/PI have been implicated in a wide range of basic cellular processes. Identifying those genes that are direct targets of AP3/PI will be valuable in defining the regulatory cascades controlled by these master floral homeotic regulatory gene products.
METHODS
Plant Material
Arabidopsis thaliana plants of the ecotype Landsberg erecta were grown on a 12:3:1 mix of vermiculite:soil:sand at 22°C under 16-h-light/ 8-h-dark conditions. All of the mutant lines (pi-1 and ap3-3) and transgenic lines (D6::DTA and ap3-3; ag-3; 35S::PI; 35S::AP3-GR [AP3-GR]) are in the Landsberg erecta background (Bowman et al., 1989; Hill et al., 1998; Sablowski and Meyerowitz, 1998). The AP3-GR line was a gift from Robert W.M. Sablowski (John Innes Centre, Norwich, UK) (Sablowski and Meyerowitz, 1998). Starting at 7 days after planting, AP3-GR plants were watered every 2 days with 5 μM dexamethasone dissolved in ethanol.
Microscopy
Flowers were fixed in 3.7% formaldehyde, 50% ethanol, and 5% acetic acid for 8 h and dehydrated in a graded ethanol series. Dehydrated flowers were critical point dried in liquid CO2 and sputter-coated with gold palladium. Specimens were analyzed and photographed with an ISI SS40 scanning electron microscope (ISI Systems, Santa Barbara, CA).
RNA Preparation and Fluorescent Labeling of Probe
Total RNA was extracted from plant tissue using Trizol (Gibco BRL) according to the manufacturer's instructions. The RNA was labeled by direct incorporation of fluorescent deoxy-UTP or by labeling with aminoallyl deoxy-UTP followed by conjugation of fluorescent dye. Cy-3– or Cy-5–conjugated deoxy-UTP (Amersham Pharmacia Biotech, Piscataway, NJ) or 5-(3-aminoallyl)-2-deoxy-UTP (Sigma, St. Louis, MO) was incorporated into the cDNA probe during reverse transcription as follows: 50 μg (for direct labeling) or 25 μg (for aminoallyl labeling) of total RNA was combined with 4 μg of anchored oligo(dT) 19-mer and 5 μg of random hexamers, heated to 65°C for 5 min, and then cooled on ice. To this mixture, the remaining components were added to obtain the following reaction mixture in a total volume of 42 μL: 1 × Superscript II reverse transcriptase buffer (Invitrogen, Carlsbad, CA), 2 μL of Superscript II reverse transcriptase, 1 μL of RNaseOUT (Invitrogen), 100 μM Cy-3 or Cy-5 deoxy-UTP, 500 μM each deoxy-ATP, deoxy-CTP, and deoxy-GTP, and 200 μM deoxy-thymine triphosphate. After a 60-min incubation at 42°C, another 2 μL of Superscript II reverse transcriptase was added to the mixture and incubated at 42°C for another 120 min.
The reaction was stopped by adding 5 μL of 0.5 M EDTA and incubating at 94°C for 3 min, and RNA was hydrolyzed by adding 10 μL of 1 M NaOH and incubating at 65°C for 20 min. The cDNA was cleaned by one phenol/chloroform extraction. The labeled cDNA then was purified from the unincorporated dye molecules by spinning through a Microcon YM-30 filter (Millipore, Bedford, MA). The labeled cDNA was purified from the unincorporated dye molecules by adding 400 μL of water and spinning through a Microcon YM-30 filter for 7 min at 11,000g, after which it was washed again two times. The purified, labeled probe was concentrated to a final volume of 7 μL. For aminoallyl labeling, an additional conjugation step was added. One vial (0.2 to 0.3 mg of dye) of Cy-3 or Cy-5 dye (Amersham Pharmacia Biotech) was resuspended in 10 μL of DMSO, and 1.25 μL was added to the concentrated probe. After a 90-min incubation at room temperature in the dark, the reaction was stopped by adding 4.5 μL of 4 M hydroxylamine (Sigma) and incubating at room temperature for 15 min. The labeled cDNA was purified from the unincorporated dye molecules by spinning through a Microcon YM-30 filter and concentrated to a final volume of 7 μL.
Hybridization to the Microarray, Washing, and Scanning
The Arabidopsis EST 9.2K array was produced at the W.M. Keck Biotechnology Resource Laboratory, DNA Microarray Resource (Yale University School of Medicine, New Haven, CT) (Ma et al., 2001). For hybridization to the Arabidopsis array, the labeled probe was combined with 1 μL of 20× SSPE (1× SSPE is 0.115 M NaCl, 10 mM sodium phosphate, and 1 mM EDTA, pH 7.4), 1 μL of blocking solution (2 mg/mL poly[dA] and 4 mg/mL tRNA), and 16 μL of hybridization solution (62.8% formamide, 0.8% SDS, 4× Denhardt's solution [1× Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA], and 5× SSPE) to a final volume of 25 μL. This mixture was incubated at 90°C for 2 min, cooled on ice, applied to the prehybridized microarray, covered with a cover slip, and incubated in a GeneMachines hybridization chamber (San Carlos, CA) at 42°C (air or water bath) for 16 to 20 h. After incubation, the array was washed with 2× SSC [1× SSC is 0.15 M NaCl and 0.015 M sodium citrate] and 0.1% SDS, 0.2× SSC and 0.1% SDS, 0.2× SSC, and 0.02× SSC for 10 min each at room temperature. After washing, the slides were dried by spinning at 1000g for 5 min.
Hybridized microarray slides were scanned at 532-nm (Cy3) and 635-nm (Cy5) wavelengths with an Axon GenePix 4000A scanner (Foster City, CA) at 10-nm resolution, generating two separate TIFF images. Photomultiplier tube voltages were adjusted manually to minimize background and reduce the percentage of spots on the array with saturated signal values. The normalization of the two channels with respect to signal intensity also was achieved by adjusting the photomultiplier tube voltage settings. We chose photomultiplier tube voltages that resulted in a Cy3:Cy5 signal ratio for the majority of control genes as close to 1.0 as possible.
Data Analysis
Spot intensities were quantified using Axon GenePix Pro 3 image-analysis software. The channel ratios were measured using the GenePix Pro 3 median-of-ratio method, and they were normalized using the corresponding GenePix default normalization factor. To merge the replicated GenePix Pro 3 output data files (.gpr files), the GPMERGE computer program (Ma et al., 2001) was used. With this program, eight replicated data sets from each experiment were pooled. A number of quality control procedures were conducted before data points from the eight replicates were averaged. First, all spots flagged Bad or Not Found by GenePix software were removed from the final data analysis. Second, GPMERGE contains an outlier-searching algorithm that defined as outliers and eliminated from the analysis those spots that exhibited a large difference between the ratio mean and the ratio median. ESTs corresponding to specific spots, which were analyzed further, were sequenced to confirm their identity.
RNA Gel Blot Hybridization
RNA (20 μg of total RNA per sample) was separated by electrophoresis on 1.2% agarose gels containing 6% formaldehyde and blotted onto nylon membranes (Hybond-N; Amersham Pharmacia Biotech). DNA probes were labeled with α-32P-dCTP by the random primer method (Feinberg and Vogelstein, 1983). EST-specific cDNAs were derived from the corresponding EST clones, which were obtained from the ABRC (Columbus, OH). PI and AP3 full-length cDNAs were derived from the plasmids pSPI (McGonigle et al., 1996) and pIa (Irish and Yamamoto, 1995), respectively. The RNA gel blots were hybridized using the indicated 32P-labeled probes. For loading controls, RNA gel blots were hybridized with a 32P-labeled cDNA probe corresponding to 18S rRNA. Hybridizations were performed at 60°C in a solution containing 5× SSPE, 2% SDS, and 100 μg/mL herring sperm DNA. After hybridization, membranes were washed once with 2× SSC and 0.1% SDS for 20 min at room temperature, then twice with 1× SSC and 0.1% SDS for 15 min at 65°C, and then a final wash with 0.1× SSC and 0.1% SDS for 15 min at 65°C. Blots were stripped between hybridizations with boiled 0.5% SDS followed by a 20-min incubation at room temperature. Quantification of hybridization signals was performed using NIH Image software (http://rsb.info.nih.gov/nih-image).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank Janet Hager and the Keck Microarray Facility at Yale University for fabricating the microarrays and for help with analyses. We also appreciate the help of Ligeng Ma in analyzing the microarray data. We thank Robert Sablowski for the gift of the AP3-GR line and the ABRC for DNA samples. We thank our colleagues at the Osborn Memorial Laboratories and an anonymous reviewer for comments on the manuscript. This work was supported by National Science Foundation Grants 9904876 and 0212222 to V.F.I.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006353.
References
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Becraft, P.W., Stinard, P.S., and McCarty, D.R. (1996). CRINKLY4: A TNFR-like receptor kinase involved in maize epidermal differentiation. Science 273, 1406–1409. [DOI] [PubMed] [Google Scholar]
- Benfey, P., and Chua, N. (1989). Regulated genes in transgenic plants. Science 244, 174–181. [DOI] [PubMed] [Google Scholar]
- Ben-Nissan, G., and Weiss, D. (1996). The petunia homologue of tomato gast1: Transcript accumulation coincides with gibberellin-induced corolla cell elongation. Plant Mol. Biol. 32, 1067–1074. [DOI] [PubMed] [Google Scholar]
- Borner, G.H., Sherrier, D.J., Stevens, T.J., Arkin, I.T., and Dupree, P. (2002). Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A genomic analysis. Plant Physiol. 129, 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, S.M., and Crouch, M.L. (1990). Characterization of a gene family abundantly expressed in Oenothera organensis pollen that shows sequence similarity to polygalacturonase. Plant Cell 2, 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee, C., Berger, F., and Bouget, F.Y. (1998). Signals involved in control of polarity, cell fate and developmental pattern in plants. Symp. Soc. Exp. Biol. 51, 33–41. [PubMed] [Google Scholar]
- Campbell, P., and Braam, J. (1999). Xyloglucan endotransglycosylases: Diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci. 4, 361–366. [DOI] [PubMed] [Google Scholar]
- Carpenter, R., and Coen, E.S. (1990). Floral homeotic mutations produced by transposon-mutagenesis in Antirrhinum majus. Genes Dev. 4, 1483–1493. [DOI] [PubMed] [Google Scholar]
- Chen, R., Aguirre, P., and Smith, A. (1994). Characterization of an anther- and tapetum-specific gene encoding a glycine-rich protein from tomato. J. Plant Physiol. 143, 651–658. [Google Scholar]
- Chen, W., et al. (2002). Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14, 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X.M., Riechmann, J.L., Jia, D.X., and Meyerowitz, E. (2000). Minimal regions in the Arabidopsis PISTILLATA promoter responsive to the APETALA3/PISTILLATA feed back control do not contain a CArG box. Sex. Plant Reprod. 13, 85–94. [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. [DOI] [PubMed] [Google Scholar]
- Davies, C., and Robinson, S.P. (2000). Differential screening indicates a dramatic change in mRNA profiles during grape berry ripening: Cloning and characterization of cDNAs encoding putative cell wall and stress response proteins. Plant Physiol. 122, 803–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi, J.L., Iyer, V.R., and Brown, P.O. (1997). Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278, 680–686. [DOI] [PubMed] [Google Scholar]
- Feinberg, A.P., and Vogelstein, B. (1983). A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132, 6–13. [DOI] [PubMed] [Google Scholar]
- Fowler, J.E., and Quatrano, R.S. (1997). Plant cell morphogenesis: Plasma membrane interactions with the cytoskeleton and cell wall. Annu. Rev. Cell Dev. Biol. 13, 697–743. [DOI] [PubMed] [Google Scholar]
- Gamboa, A., Paez-Valencia, J., Acevedo, G.F., Vazquez-Moreno, L., and Alvarez-Buylla, R.E. (2001). Floral transcription factor AGAMOUS interacts in vitro with a leucine-rich repeat and an acid phosphatase protein complex. Biochem. Biophys. Res. Commun. 288, 1018–1026. [DOI] [PubMed] [Google Scholar]
- Gang, D.R., Kasahara, H., Xia, Z.Q., Vander Mijnsbrugge, K., Bauw, G., Boerjan, W., Van Montagu, M., Davin, L.B., and Lewis, N.G. (1999). Evolution of plant defense mechanisms: Relationships of phenylcoumaran benzylic ether reductases to pinoresinol-lariciresinol and isoflavone reductases. J. Biol. Chem. 274, 7516–7527. [DOI] [PubMed] [Google Scholar]
- Garcia-Hernandez, M., Murphy, A., and Taiz, L. (1998). Metallothioneins 1 and 2 have distinct but overlapping expression patterns in Arabidopsis. Plant Physiol. 118, 387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar, Y., Johnson, K.L., McKenna, J.A., Bacic, A., and Schultz, C.J. (2001). The complex structures of arabinogalactan-proteins and the journey towards understanding function. Plant Mol. Biol. 47, 161–176. [PubMed] [Google Scholar]
- Girke, T., Todd, J., Ruuska, S., White, J., Benning, C., and Ohlrogge, J. (2000). Microarray analysis of developing Arabidopsis seeds. Plant Physiol. 124, 1570–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, R.B., Beals, T.P., and Sanders, P.M. (1993). Anther development: Basic principles and practical applications. Plant Cell 5, 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, K., and Meyerowitz, E.M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8, 1548–1560. [DOI] [PubMed] [Google Scholar]
- Graba, Y., Aragnol, D., and Pradel, J. (1997). Drosophila Hox complex downstream targets and the function of homeotic genes. Bioessays 19, 379–388. [DOI] [PubMed] [Google Scholar]
- Harmer, S.L., Hogenesch, J.B., Straume, M., Chang, H.S., Han, B., Zhu, T., Wang, X., Kreps, J.A., and Kay, S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113. [DOI] [PubMed] [Google Scholar]
- Hill, J.P., and Lord, E.M. (1989). Floral development in Arabidopsis thaliana: A comparison of the wild type and the homeotic pistillata mutant. Can. J. Bot. 67, 2922–2936. [Google Scholar]
- Hill, T.A., Day, C.D., Zondlo, S.C., Thackeray, A.G., and Irish, V.F. (1998). Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development 125, 1711–1721. [DOI] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2000). The Arabidopsis floral homeotic gene PISTILLATA is regulated by discrete cis-elements responsive to induction and maintenance signals. Development 127, 2021–2030. [DOI] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 469–471. [DOI] [PubMed] [Google Scholar]
- Irish, V.F. (1999). Petal and stamen development. Curr. Top. Dev. Biol. 41, 133–161. [DOI] [PubMed] [Google Scholar]
- Irish, V.F., and Yamamoto, Y.T. (1995). Conservation of floral homeotic gene function between Arabidopsis and Antirrhinum. Plant Cell 7, 1635–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack, T., Brockman, L.L., and Meyerowitz, E.M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. [DOI] [PubMed] [Google Scholar]
- Jack, T., Fox, G.L., and Meyerowitz, E.M. (1994). Arabidopsis homeotic gene APETALA3 ectopic expression: Transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76, 703–716. [DOI] [PubMed] [Google Scholar]
- Jenik, P.D., and Irish, V.F. (2001). The Arabidopsis floral homeotic gene APETALA3 differentially regulates intercellular signaling required for petal and stamen development. Development 128, 13–23. [DOI] [PubMed] [Google Scholar]
- Jirage, D., Tootle, T.L., Reuber, T.L., Frost, L.N., Feys, B.J., Parker, J.E., Ausubel, F.M., and Glazebrook, J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96, 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima, I., Kennedy, T.D., Chino, M., and Lane, B.G. (1992). Wheat Ec metallothionein genes: Like mammalian Zn2+ metallothionein genes, wheat Zn2+ metallothionein genes are conspicuously expressed during embryogenesis. Eur. J. Biochem. 209, 971–976. [DOI] [PubMed] [Google Scholar]
- Kotilainen, M., Helariutta, Y., Mehto, M., Pollanen, E., Albert, V.A., Elomaa, P., and Teeri, T.H. (1999). GEG participates in the regulation of cell and organ shape during corolla and carpel development in Gerbera hybrida. Plant Cell 11, 1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek, B.A., and Meyerowitz, E.M. (1996. a). The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122, 11–22. [DOI] [PubMed] [Google Scholar]
- Krizek, B.A., and Meyerowitz, E.M. (1996. b). Mapping the protein regions responsible for the functional specificities of the Arabidopsis MADS domain organ-identity proteins. Proc. Natl. Acad. Sci. USA 93, 4063–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease, K., Ingham, E., and Walker, J.C. (1998). Challenges in understanding RLK function. Curr. Opin. Plant Biol. 1, 388–392. [DOI] [PubMed] [Google Scholar]
- Ledger, S., Strayer, C., Ashton, F., Kay, S.A., and Putterill, J. (2001). Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 26, 15–22. [DOI] [PubMed] [Google Scholar]
- Leemans, R., Loop, T., Egger, B., He, H., Kammermeier, L., Hartmann, B., Certa, U., Reichert, H., and Hirth, F. (2001). Identification of candidate downstream genes for the homeodomain transcription factor Labial in Drosophila through oligonucleotide-array transcript imaging. Genome Biol. 2, B0015.1–B0015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- Lohmann, J.U., and Weigel, D. (2002). Building beauty: The genetic control of floral patterning. Dev. Cell 2, 135–142. [DOI] [PubMed] [Google Scholar]
- Lotan, T., Ori, N., and Fluhr, R. (1989). Pathogenesis-related proteins are developmentally regulated in tobacco flowers. Plant Cell 1, 881–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Gao, Y., Qu, L., Chen, Z., Li, J., Zhao, H., and Deng, X.W. (2002). Genomic evidence for COP1 as a repressor of light-regulated gene expression and development in Arabidopsis. Plant Cell 14, 2383–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Li, J., Qu, L., Hager, J., Chen, Z., Zhao, H., and Deng, X.W. (2001). Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13, 2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska-Sawka, A., and Nothnagel, E.A. (2000). The multiple roles of arabinogalactan proteins in plant development. Plant Physiol. 122, 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel, M.A., Gustafson-Brown, C., Savidge, B., and Yanofsky, M.F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273–277. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., and Yanofsky, M.F. (1998). The Arabidopsis AGL9 MADS box gene is expressed in young flower primordia. Sex. Plant Reprod. 11, 22–28. [Google Scholar]
- McCormick, S., et al. (1987). Identification of genes specifically expressed in reproductive organs of tomato. In Tomato Biotechnology, D.J. Nevins and R.A. Jones, eds (New York: Alan R. Liss), pp. 255–265.
- McGonigle, B., Bouhidel, K., and Irish, V.F. (1996). Nuclear localization of the Arabidopsis APETALA3 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes Dev. 10, 1812–1821. [DOI] [PubMed] [Google Scholar]
- Medford, J.I., Elmer, J.S., and Klee, H.J. (1991). Molecular cloning and characterization of genes expressed in shoot apical meristems. Plant Cell 3, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacken, W.K.F., Huijser, P., Beltran, J.-P., Saedler, H., and Sommer, H. (1991). Molecular characterization of two stamen-specific genes, tap1 and fil1, that are expressed in the wild type, but not in the deficiens mutant of Antirrhinum majus. Mol. Gen. Genet. 229, 129–136. [DOI] [PubMed] [Google Scholar]
- Nam, Y.W., Tichit, L., Leperlier, M., Cuerq, B., Marty, I., and Lelievre, J.M. (1999). Isolation and characterization of mRNAs differentially expressed during ripening of wild strawberry (Fra-garia vesca L.) fruits. Plant Mol. Biol. 39, 629–636. [DOI] [PubMed] [Google Scholar]
- Nylander, M., Svensson, J., Palva, E.T., and Welin, B.V. (2001). Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol. Biol. 45, 263–279. [DOI] [PubMed] [Google Scholar]
- Oh, S.A., Lee, S.Y., Chung, I.K., Lee, C.H., and Nam, H.G. (1996). A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol. Biol. 30, 739–754. [DOI] [PubMed] [Google Scholar]
- Pelaz, S., Ditta, G.S., Baumann, E., Wisman, E., and Yanofsky, M.F. (2000). B and C floral organ identity functions require SEPAL-LATA MADS-box genes. Nature 405, 200–203. [DOI] [PubMed] [Google Scholar]
- Pelaz, S., Gustafson-Brown, C., Kohalmi, S.E., Crosby, W.L., and Yanofsky, M.F. (2001). APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 26, 385–394. [DOI] [PubMed] [Google Scholar]
- Perez-Amador, M.A., Lidder, P., Johnson, M.A., Landgraf, J., Wisman, E., and Green, P.J. (2001). New molecular phenotypes in the dst mutants of Arabidopsis revealed by DNA microarray analysis. Plant Cell 13, 2703–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke, K.A., and Page, A.M. (1998). Plastid ontogeny during petal development in Arabidopsis. Plant Physiol. 116, 797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke, V., Smith, H.E., Nance, J., Wang, J., Van Doren, C., Begley, R., Jones, S.J., Davis, E.B., Scherer, S., Ward, S., and Kim, S.K. (2000). A global profile of germline gene expression in C. elegans. Mol. Cell 6, 605–616. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., and Meyerowitz, E.M. (1997). Determination of floral organ identity by Arabidopsis MADS domain homeotic proteins AP1, AP3, PI, and AG is independent of their DNA-binding specificity. Mol. Biol. Cell 8, 1243–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., and Ratcliffe, O.J. (2000). A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 3, 423–434. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., Wang, M., and Meyerowitz, E.M. (1996). DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 24, 3134–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringli, C., Keller, B., and Ryser, U. (2001). Glycine-rich proteins as structural components of plant cell walls. Cell. Mol. Life Sci. 58, 1430–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, N.J., Tommey, A.M., Kuske, C., and Jackson, P.J. (1993). Plant metallothioneins. Biochem. J. 295, 1.–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse, D.T., Marotta, R., and Parish, R.W. (1996). Promoter and expression studies on an Arabidopsis thaliana dehydrin gene. FEBS Lett. 381, 252–256. [DOI] [PubMed] [Google Scholar]
- Ruan, Y., Gilmore, J., and Conner, T. (1998). Towards Arabidopsis genome analysis: Monitoring expression profiles of 1400 genes using cDNA microarrays. Plant J. 15, 821–833. [DOI] [PubMed] [Google Scholar]
- Rubinelli, P., Hu, Y., and Ma, H. (1998). Identification, sequence analysis and expression studies of novel anther-specific genes of Arabidopsis thaliana. Plant Mol. Biol. 37, 607–619. [DOI] [PubMed] [Google Scholar]
- Sablowski, R.W.M., and Meyerowitz, E.M. (1998). A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92, 93–103. [DOI] [PubMed] [Google Scholar]
- Sachetto-Martins, G., Franco, L.O., and de Oliveira, D.E. (2000). Plant glycine-rich proteins: A family or just proteins with a common motif? Biochim. Biophys. Acta 1492, 1–14. [DOI] [PubMed] [Google Scholar]
- Sakai, H., Krizek, B.A., Jacobsen, S.E., and Meyerowitz, E.M. (2000). Regulation of SUP expression identifies multiple regulators involved in Arabidopsis floral meristem development. Plant Cell 12, 1607–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., and Gover, A. (2001). Photoperiodism: The consistent use of CONSTANS. Curr. Biol. 11, R651–R654. [DOI] [PubMed] [Google Scholar]
- Schaffer, R., Landgraf, J., Accerbi, M., Simon, V.V., Larson, M., and Wisman, E. (2001). Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13, 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena, M., Shalon, D., Davis, R.W., and Brown, P.O. (1995). Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470. [DOI] [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Wilson, I., Anderson, J.P., Richmond, T., Somerville, S.C., and Manners, J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97, 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler, U., Beckmann, H., and Cashmore, A.R. (1993). HAT3.1, a novel Arabidopsis homeodomain protein containing a conserved cysteine-rich region. Plant J. 4, 137–150. [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer, Z., Hue, I., Huijser, P., Flor, P.J., Hansen, R., Tetens, F., Lonnig, W.-E., Saedler, H., and Sommer, H. (1992). Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: Evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 11, 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, L., Gast, R.T., Gopalraj, M., and Olszewski, N.E. (1992). Characterization of a shoot-specific, GA3- and ABA-regulated gene from tomato. Plant J. 2, 153–159. [PubMed] [Google Scholar]
- Shore, P., and Sharrocks, A.D. (1995). The MADS-box family of transcription factors. Eur. J. Biochem. 229, 1–13. [DOI] [PubMed] [Google Scholar]
- Showalter, A.M. (2001). Arabinogalactan-proteins: Structure, expression and function. Cell. Mol. Life Sci. 58, 1399–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, J.M., Trotochaud, A.E., Walker, J.C., and Clark, S.E. (1998). Control of meristem development by CLAVATA1 receptor kinase and kinase-associated protein phosphatase interactions. Plant Physiol. 117, 1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Lopez, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120. [DOI] [PubMed] [Google Scholar]
- Takahashi, T., Mu, J.H., Gasch, A., and Chua, N.H. (1998). Identification by PCR of receptor-like protein kinases from Arabidopsis flowers. Plant Mol. Biol. 37, 587–596. [DOI] [PubMed] [Google Scholar]
- Tanaka, H., Watanabe, M., Watanabe, D., Tanaka, T., Machida, C., and Machida, Y. (2002). ACR4, a putative receptor kinase gene of Arabidopsis thaliana, that is expressed in the outer cell layers of embryos and plants, is involved in proper embryogenesis. Plant Cell Physiol. 43, 419–428. [DOI] [PubMed] [Google Scholar]
- Tang, G., Miron, D., Zhu-Shimoni, J.X., and Galili, G. (1997). Regulation of lysine catabolism through lysine-ketoglutarate reductase and saccharopine dehydrogenase in Arabidopsis. Plant Cell 9, 1305–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen, G., and Saedler, H. (2001). Plant biology: Floral quartets. Nature 409, 469–471. [DOI] [PubMed] [Google Scholar]
- Thompson, J., Taylor, C., and Wang, T.W. (2000). Altered membrane lipase expression delays leaf senescence. Biochem. Soc. Trans. 28, 775–777. [PubMed] [Google Scholar]
- Tilly, J., Allen, D.W., and Jack, T. (1998). The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development 125, 1647–1657. [DOI] [PubMed] [Google Scholar]
- Torii, K.U., Mitsukawa, N., Oosumi, T., Matsuura, Y., Yokoyama, R., Whittier, R.F., and Komeda, Y. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8, 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen, E.A., and Jones, J.D. (1998). The NB-ARC domain: A novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 8, R226–R227. [DOI] [PubMed] [Google Scholar]
- Vivian-Smith, A., Luo, M., Chaudhury, A., and Koltunow, A. (2001). Fruit development is actively restricted in the absence of fertilization in Arabidopsis. Development 128, 2321–2331. [DOI] [PubMed] [Google Scholar]
- Wagstaff, C., Leverentz, M.K., Griffiths, G., Thomas, B., Chanasut, U., Stead, A.D., and Rogers, H.J. (2002). Cysteine protease gene expression and proteolytic activity during senescence of Alstroemeria petals. J. Exp. Bot. 53, 233–240. [DOI] [PubMed] [Google Scholar]
- Wang, R., Guegler, K., LaBrie, S.T., and Crawford, N.M. (2000). Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12, 1491–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, K.P., Rifkin, S.A., Hurban, P., and Hogness, D.S. (1999). Microarray analysis of Drosophila development during metamorphosis. Science 286, 2179–2184. [DOI] [PubMed] [Google Scholar]
- Wildsmith, S.E., and Elcock, F.J. (2001). Microarrays under the microscope. Mol. Pathol. 54, 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, R.W., Wilson, J.M., and Meyerowitz, E.M. (1997). A possible role for kinase-associated protein phosphatase in the Arabidopsis CLAVATA1 signaling pathway. Proc. Natl. Acad. Sci. USA 94, 10467–10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing, R., Yamaguchi, J., Larabell, S., Ursin, V., and McCormick, S. (1989). Molecular and genetic characterization of two pollen-expressed genes that have sequence similarity to pectate lyases of the plant pathogen Erwinia. Plant Mol. Biol. 14, 17–28. [DOI] [PubMed] [Google Scholar]
- Yue, H., Eastman, P.S., Wang, B.B., Minor, J., Doctolero, M.H., Nuttall, R.L., Stack, R., Becker, J.W., Montgomery, J.R., Vainer, M., and Johnston, R. (2001). An evaluation of the performance of cDNA microarrays for detecting changes in global mRNA expression. Nucleic Acids Res. 29, E41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachgo, S., de Andrade Silva, E., Motte, P., Trobner, W., Saedler, H., and Schwarz-Sommer, Z. (1995). Functional analysis of the Antirrhinum floral homeotic Deficiens gene in vivo and in vitro by using a temperature-sensitive mutant. Development 121, 2861–2875. [DOI] [PubMed] [Google Scholar]
- Zhou, J., and Goldsbrough, P.B. (1995). Structure, organization and expression of the metallothionein gene family in Arabidopsis. Mol. Gen. Genet. 248, 318–328. [DOI] [PubMed] [Google Scholar]