Abstract
The abscisic acid (ABA) response promoter complexes (ABRCs) of the HVA1 and HVA22 genes have been shown to confer ABA-induced gene expression in cereals. A barley basic domain/Leu zipper (bZIP) transcription factor, HvABI5, is able to recognize ABRCs in vitro in a sequence-specific manner and to transactivate ABRC–β-glucuronidase reporter genes when introduced to barley aleurone cells via particle bombardment. This transactivation is dependent on the presence of another transcription factor, HvVP1, and cannot be blocked by the negative regulator abi1-1. Using the double-stranded RNA interference technique, we show that HvABI5 and HvVP1 are necessary for the ABA induction of gene expression but have no effect on another hormone-regulated process, the gibberellin-induced and ABA-suppressed expression of α-amylase. Our work indicates that although other typical plant bZIP transcription factors may bind ABRCs in vitro, HvABI5 is related to a subfamily of bZIPs responsible for the ABA induction of gene expression. Furthermore, HvABI5 and HvVP1 are not involved in the ABA suppression of gene expression.
INTRODUCTION
During seed development, the phytohormone abscisic acid (ABA) performs important roles in the onset of seed dormancy, the acquisition of desiccation tolerance, and the prevention of precocious germination to ensure the survival of the developing embryo. In vegetative growth, ABA also is involved in improving the plant's ability to adapt and endure adverse conditions such as drought and salinity (Leung and Giraudat, 1998). Such ABA-mediated adaptive responses include stomatal closure and the expression of a variety of genes involved in stress tolerance (Bray, 1997). Among these genes, the so-called late embryogenesis abundant (Lea) genes (Dure et al., 1989) are expressed during the desiccation process in late seed development and when germinating embryos are subjected to water or osmotic stress or as a result of treatment with exogenous ABA (Leung and Giraudat, 1998). LEA proteins are thought to play a role in the tolerance to desiccation by maintaining the structural integrity of membranes and proteins and controlling water exchange (Dure, 1993).
Although several components of ABA signal transduction pathways have been identified in plants, detailed mechanisms still are lacking. More substantial information has been obtained for the later steps of the signal transduction pathway, including the study of the cis- and trans-acting promoter elements involved in ABA responses (Rock, 2000). Regions in several promoters have revealed conserved DNA elements that are ABA responsive, named ABREs (for ABA response promoter complexes), G-boxes, and ACGT-boxes (Guiltinan et al., 1990; Izawa et al., 1993; Shen and Ho, 1997). Recently, a specific subfamily of plant basic domain/Leu zipper (bZIP) proteins that recognize cis elements present in ABA-responsive promoters was identified (Kim and Thomas, 1998). Unlike other bZIPs that also recognize ACGT-boxes of plant promoters, this new group shares some common features: (1) three unique N-terminal conserved motifs with potential phosphorylation sites; (2) a basic region partly different from that of other bZIPs; and (3) only four Leu residues in the Leu zipper. Some examples include TRAB1 (Hobo et al., 1999), DPBF1 to DPBF3 (Kim et al., 1997; Kim and Thomas, 1998), ABI5 (Finkelstein and Lynch, 2000), AREBs (Uno et al., 2000), and ABFs (Choi et al., 2000). Two of these bZIPs have been shown to regulate gene expression in seeds: TRAB1 can activate the Osem promoter in rice protoplasts (Hobo et al., 1999), and ABI5 affects the expression of embryo-specific genes in Arabidopsis (Finkelstein and Lynch, 2000). Moreover, both bZIPs were able to interact in a yeast two-hybrid system with their respective plant ortholog of the maize transcription factor VP1 (Hobo et al., 1999; Nakamura et al., 2001). It has been suggested that ABI5 plays a role in protecting germinating embryos from drought, and ABI5 accumulation, stability, and activity are regulated by ABA during germination (Lopez- Molina et al., 2001).
In barley seeds, ABA induces several genes, including HVA1 and HVA22, which also are induced by different stresses in vegetative tissues (Hong et al., 1992; Shen et al., 2001a). Although both genes encode proteins with unknown functions, it has been shown that constitutive expression of HVA1, which encodes a group 3 LEA protein, can confer drought resistance to transgenic plants (Xu et al., 1996). On the other hand, HVA22 probably is involved in intracellular vesicular trafficking (Brands and Ho, 2002). Promoter analyses of these two genes have demonstrated that two cis elements, an ACGT-box and a coupling element (together constituting ABA response complexes: ABRC3 in HVA1 and ABRC1 in HVA22), are necessary and sufficient for their induction by ABA (Shen and Ho, 1997). Nonetheless, no transcription factor has been demonstrated to mediate gene expression by binding to these cis elements in barley. To understand how ABA upregulates HVA1 and HVA22, we initiated the identification of the trans-acting factors that recognize their promoters. Here, we report the identification of two barley bZIP transcription factors, HvABI5 and HvZIP1. HvABI5 is related to the newly described subfamily of bZIPs responsible for ABA-dependent gene regulation, suggesting its role in the ABA induction of HVA1 and HVA22. Both gain-of-function (overexpression) and loss-of-function (double-stranded RNA interference [RNAi]) experiments suggest that HvABI5 and the barley ortholog of VP1 participate in the transactivation of HVA1 and HVA22 promoters. Furthermore, neither HvABI5 nor HvVP1 is involved in the ABA suppression of germination-specific genes.
RESULTS
Identification of Two New Barley bZIPs, HvABI5 and HvZIP1
Two cDNA libraries were screened for genes encoding bZIP transcription factors that may recognize the ACGT-boxes present in ABRC1 and ABRC3. Two isolates of the same gene were obtained by screening a cDNA library made with RNA from ABA-treated aleurones. This clone contained a cDNA of 1348 bp, and the deduced amino acid sequence encodes a protein of 353 amino acids, showing high similarity to previously described plant bZIP proteins (Figure 1). Because the probe used in the screening, AtDPBF1 cDNA, was described later to correspond to the Arabidopsis ABI5 gene (Finkelstein and Lynch, 2000), this barley bZIP was named HvABI5. A second library was screened with a cDNA fragment of OsZIP1a that constitutes the basic region and the Leu zipper (Nantel and Quatrano, 1996). Five isolates of only one gene were obtained, perhaps as a result of the high stringency used. This clone represents a cDNA of 1532 bp, encoding a protein of 368 amino acids, HvZIP1, that also contains a bZIP signature (Figure 1A).
Figure 1.
Comparison of Two New Barley bZIPs, HvABI5 and HvZIP1.
(A) Schemes of the HvABI5 and HvZIP1 proteins. Boxes represent the positions of conserved regions among ABI5-like bZIPs (C1 to C4), the Pro-rich region, the basic DNA binding domain (BR), and the Leu zipper dimerization domain.
(B) Comparison of the HvABI5 amino acid sequence with the sequences of other plant bZIPs. The amino acid sequence was deduced from the cDNA nucleotide sequence and was aligned with similar proteins found in the database: the rice TRAB1 protein and the Arabidopsis bZIPs AREB2 and ABI5. Identical residues are shaded. Conserved regions are underlined, and the basic region and the Leu zipper are double underlined. Asterisks denote potential phosphorylation sites.
HvABI5 Belongs to a Subclass of Plant bZIP Proteins
The amino acid sequence of HvABI5 was determined and compared with those of other proteins. It shows high similarity to the subfamily of plant bZIPs first described by Kim and Thomas (1998), which is now composed of at least 20 proteins. Figure 1B shows the alignment of HvABI5 with a few representative proteins of this class: its closest homolog, TRAB1 from rice, which is 53% identical (Hobo et al., 1999); AREB2 (Uno et al., 2000), its closest homolog from Arabidopsis; and the Arabidopsis protein ABI5 (Finkelstein and Lynch, 2000). Like HvABI5, members of this family of bZIPs share great similarity in the basic region and contain only four Leu residues forming the Leu zipper. In addition, four other regions of the protein, often containing putative phosphorylation sites, are highly conserved (Figures 1A and 1B). On the other hand, HvZIP1 shows more similarity to a different group of plant G-box binding factors, such as OsZIP1a (Nantel and Quatrano, 1996), CPRF4 (Kircher et al., 1998), HBP-1a (Tabata et al., 1991), and EmBP1 (Guiltinan et al., 1990), which are characterized by having a Pro-rich region near the N terminus (Figure 1A).
HvABI5 Binds to ABRCs in a Sequence-Specific Manner
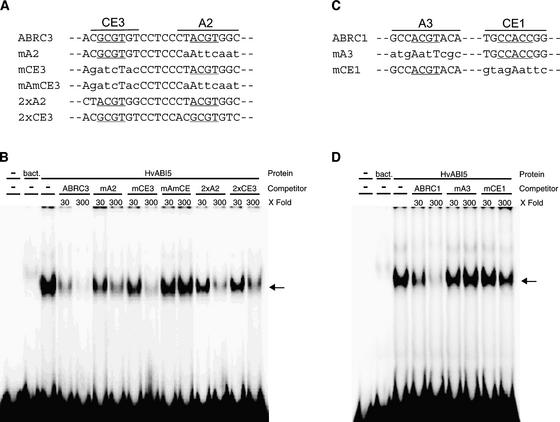
By the time HvABI5 was isolated, similar bZIP proteins had been described to mediate ABA signaling (Hobo et al., 1999; Finkelstein and Lynch, 2000; Uno et al., 2000). Thus, we explored the possibility that HvABI5 could recognize the ABRCs present in the promoters of HVA1 and HVA22. Electrophoretic mobility shift assays with recombinant HvABI5 were conducted to establish whether it can recognize ABRCs in vitro. Figure 2B shows that HvABI5 was able to bind ABRC3. The binding activity was abolished when an excess amount of unlabeled ABRC3 was added. The binding also was diminished if the competitor had only one of the cis elements, either the ACGT-box or the coupling element, mutated (designated mA2 or mCE3, respectively). However, a competitor with both elements mutated (mAmCE) could not abolish the binding of the factor to ABRC3. Because both cis elements seemed to affect the binding, competitions using two synthetic versions of ABRC3, consisting of two copies of each of the cis elements, also were performed (Figure 2B). Two copies of the ACGT-box (2xA2) affected the binding more than two copies of CE3 (2xCE3). This finding correlates with the observations using competitors with one mutated element, suggesting that HvABI5 may have more affinity for the ACGT-box than for CE3.
Figure 2.
HvABI5 Binds ABRCs in Vitro in a Sequence-Specific Manner.
(A) and (C) Partial nucleotide sequences of ABRC3 (A) and ABRC1 (C) mutant and synthetic versions used as competitors in electrophoretic mobility shift assays.
(B) and (D) Electrophoretic mobility shift assays with recombinant HvABI5. A 124-bp fragment probe containing ABRC3 (B) or a 90-bp fragment probe containing ABRC1 (D) and 2 μg of HvABI5 protein were used in each assay. bact indicates the control binding reaction with bacterial protein from Escherichia coli that carried an empty expression vector. The molar excess of each competitor used (30- and 300-fold) is indicated at the top of each lane. Arrows indicate the DNA-protein complexes.
To determine whether HvABI5 also can interact directly with ABRC1, we performed similar assays with an ABRC1 probe and recombinant HvABI5. HvABI5 also was able to bind ABRC1 in vitro (Figure 2D), and an excess of unlabeled ABRC1 abolished the binding. In this case, a competitor with the ACGT-box mutated (mA3) did not compete against the wild type ABRC1, suggesting that CE1 may not be the target of HvABI5. We were expecting that the competitor with the CE mutated (mCE1), with an intact ACGT-box, would affect the binding, but we found only a slight competition.
In vitro binding of HvZIP1 also was tested with both ABRCs (data not shown). HvZIP1 required the presence of both A2 and CE3 of ABRC3 for complete binding. This bZIP also recognized A3 and CE1 of ABRC1. With both ABRCs, the binding of HvZIP1 was stronger when the ACGT-box was present. Together, these data indicate that both HvABI5 and HvZIP1 can recognize both ABRCs in vitro and have a preference for the ACGT-boxes.
Ectopic Expression of HvABI5 and VP1 Is Sufficient to Transactivate ABA-Upregulated Genes but Has No Effect on the GA Induction of α-Amylase
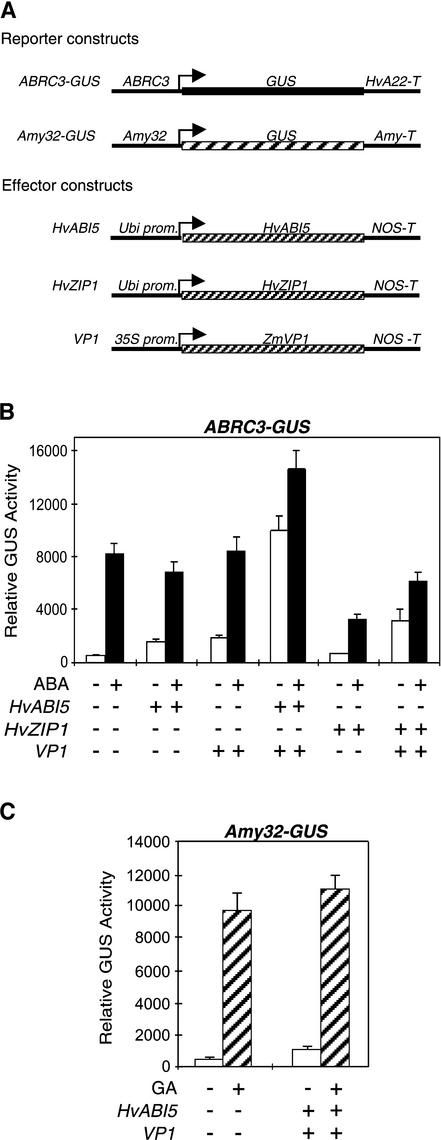
To determine whether HvABI5 can recognize the ABRCs in vivo and whether it plays a role in HVA1 and HVA22 expression, biolistic transformation of barley embryoless half-seeds was performed. Different effector constructs containing a cDNA driven by a constitutive promoter along with an ABRC–β-glucuronidase (ABRC-GUS) reporter construct were cobombarded into barley half-seeds treated with or without ABA. We first tested the effect of HvABI5 by itself and observed a 3-fold activation of ABRC3-GUS expression (Figure 3B), whereas the effect of ABA alone was a 19-fold enhancement. The activation of ABRC3 has been shown to be affected positively by the maize transactivator VP1 (Shen et al., 1996); thus, we also tested the effector construct 35S-VP1. VP1 alone also gave a small activation of the reporter construct (threefold increase). However, when both HvABI5 and VP1 were cobombarded into the aleurone cells in the absence of ABA, they transactivated ABRC3 to the levels observed with ABA treatment or even higher (22-fold in this assay).
Figure 3.
HvABI5 and VP1 Are Sufficient to Transactivate ABRC-GUS.
(A) Schemes of the reporter and effector constructs used in the transient expression assays.
(B) The reporter construct ABRC3-GUS was cobombarded into barley embryoless half-seeds with (+) or without (−) the effector constructs Ubi-HvABI5, Ubi-HvZIP1, and 35S-VP1 using 1 μg of each construct. Bars indicate GUS activities ± se after 24 h of incubation of the bombarded seeds with (closed bars) or without (open bars) 20 μM ABA.
(C) The reporter construct Amy32-GUS was cobombarded into barley embryoless half-seeds with (+) or without (−) the effector constructs Ubi-HvABI5 and 35S-VP1 using 1 μg of each construct. Bars indicate GUS activities ± se after 24 h of incubation of the bombarded seeds with (hatched bars) or without (open bars) 1 μM GA3.
Because another barley bZIP isolated from aleurones, HvZIP1, also was able to bind both ABRCs in vitro (data not shown), an effector construct that constitutively expresses HvZIP1 was tested. Constitutive expression of HvZIP1 did not affect the activation of the ABRC3-GUS construct (Figure 3B); however, it had a negative effect on ABA induction, perhaps because it competes against the endogenous HvABI5. When cobombarded along with VP1, HvZIP1 showed only a fivefold activation in the absence of ABA, much less than the 22-fold enhancement observed with HvABI5 and VP1. The same experiments were conducted with the ABRC1-GUS reporter construct, and similar patterns of GUS activity were observed with the same combination of effector constructs (data not shown), suggesting that HvABI5 plays a role in the expression of HVA1 and HVA22. The effect of HvABI5 and VP1 also was tested on the expression of α-amylase (Amy). Cobombardment of these two factors showed an approximately threefold activation of the Amy-GUS reporter construct in the absence of gibberellic acid (GA) and no significant effect on α-amylase expression in the presence of GA (Figure 3C). These data suggest that HvABI5 and VP1 are sufficient to activate ABA-induced genes but not GA-induced α-amylase.
A Dominant-Negative Form of HvABI5 Represses the ABA Activation of ABRC-GUS
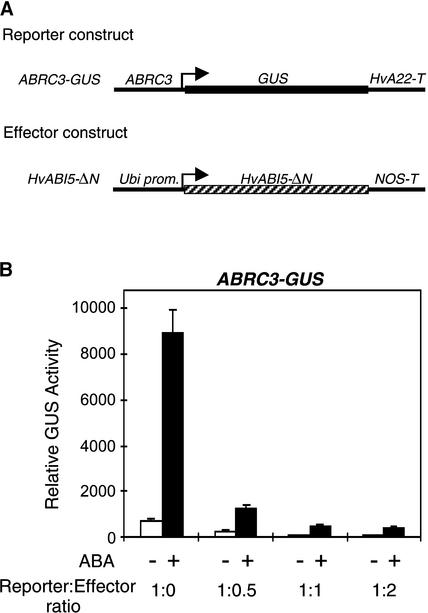
The role of transcription factors in plants often has been analyzed using gain-of-function approaches; few loss-of function experiments have been performed, especially when the desired mutants have been available. It has been shown that introducing a dominant-negative form of a bZIP protein can suppress the activity of the endogenous transcription factor in stable and transient expression studies (Fukazawa et al., 2000; Sprenger-Haussels and Weisshaar, 2000). To investigate the role of HvABI5 in the ABA induction of HVA1, we tried to repress HvABI5 activity using a construct made of the C-terminal region of the protein that contains only the basic and Leu zipper regions lacking the first 279 amino acids. When the HvABI5-ΔN construct was cobombarded into aleurone layers, it repressed the ABA induction of the ABRC3-GUS reporter construct in a dose-dependent manner (Figure 4). Using a reporter:effector ratio of 1:0.5, the ABA activation decreased to 14% of that obtained with no effector construct; it decreased to ∼5% with a 1:1 reporter:effector ratio, showing that the ABA induction of ABRC-GUS can be overcome effectively by a dominant-negative form of HvABI5.
Figure 4.
A Dominant-Negative Form of HvABI5 Represses the ABA Induction of ABRC-GUS.
(A) Schemes of the reporter and effector constructs used in the transient expression assays.
(B) One microgram of the reporter construct ABRC3-GUS was cobombarded into barley embryoless half-seeds with (+) or without (−) increasing amounts of the effector construct Ubi-HvABI5-ΔN as indicated in the reporter:effector ratios. Bars indicate GUS activities ± se after 24 h of incubation of the bombarded seeds with (closed bars) or without (open bars) 20 μM ABA.
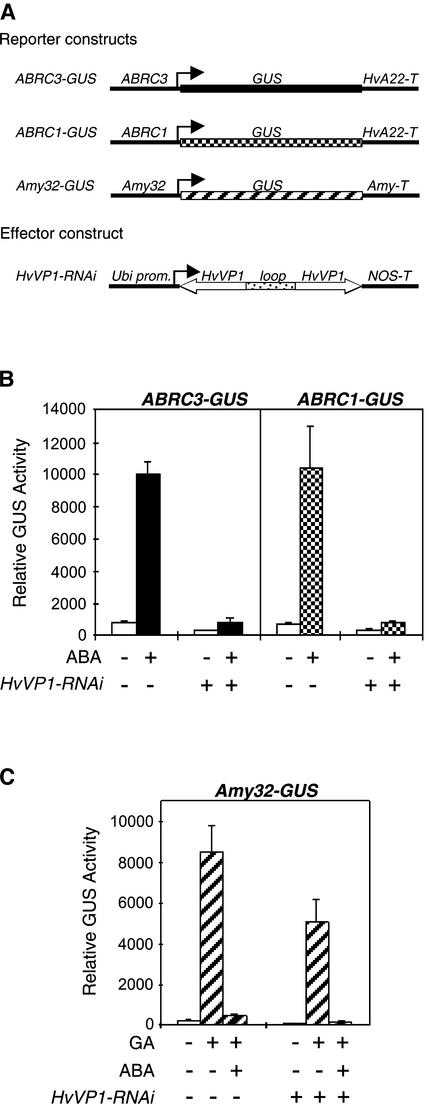
HvABI5 RNAi Inhibits the ABA Activation of ABRC-GUS but Has No Effect on the ABA Suppression of α-Amylase Expression
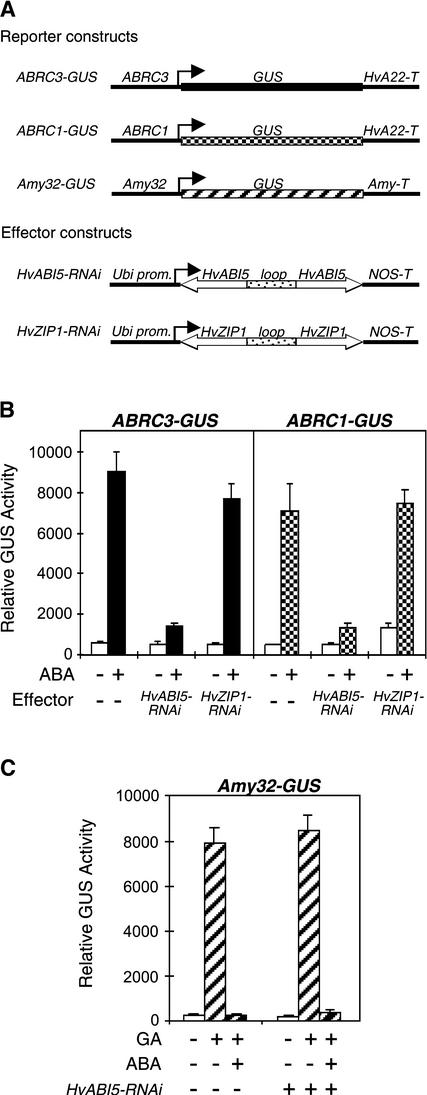
To exclude the possibility that the HvABI5-ΔN construct blocked the binding of other bZIP proteins, we used the double-stranded RNAi system as a more specific loss-of-function approach. RNAi technology has been used successfully in transient expression experiments in barley aleurone tissue (Zentella et al., 2002). To determine whether HvABI5 is necessary to induce the ABA-regulated genes HVA1 and HVA22, HvABI5 transcripts were targeted specifically and the ABA-mediated induction of ABRC-GUS reporter constructs was examined. The effector construct Ubi1-HvABI5-RNAi was generated using the coding region for the first 144 amino acids of HvABI5, excluding the basic region and the Leu zipper to decrease any possible cross-interference with other bZIP factors. Figure 5B shows that HvABI5-RNAi suppressed the ABA induction of ABRC3 and ABRC1 to ∼10% of the GUS activity obtained with ABA treatment alone. This reduction of HvABI5 expression by RNAi was observed when the inducing hormone was added 6 h after the bombardment of the effector and reporter constructs, allowing any preexisting HvABI5 to turn over. When the seeds were incubated with ABA immediately after particle bombardment, the effect of HvABI5-RNAi already was evident, with the activity reduced to 25% (data not shown). HvABI5-RNAi also overcame the transactivation activity observed when HvABI5 and VP1 were overexpressed transiently in aleurones (data not shown).
Figure 5.
HvABI5-RNAi Specifically Inhibits the ABA Induction of ABRC-GUS but Has No Effect on the ABA Suppression of α-Amylase Expression.
(A) Schemes of the reporter and effector constructs used in the transient expression assays.
(B) The reporter constructs ABRC3-GUS and ABRC1-GUS were cobombarded into barley embryoless half-seeds with (+) or without (−) the effector constructs Ubi-HvABI5-RNAi or Ubi-HvZIP1-RNAi using 1 μg of each construct. Bars indicate GUS activities ± se after 24 h of incubation of the bombarded seeds with (closed or checked bars) or without (open bars) 20 μM ABA. Incubation with the hormone was initiated 6 h after bombardment.
(C) The reporter construct Amy32-GUS was cobombarded into barley embryoless half-seeds with (+) or without (−) the effector construct Ubi-HvABI5-RNAi using 1 μg of each construct. Bars indicate GUS activities ± se after 24 h of incubation of the bombarded seeds with (hatched bars) or without (open bars) 20 μM ABA or 1 μM GA3. Incubation with the hormone was initiated 6 h after bombardment.
To demonstrate that HvABI5 was the bZIP involved in the ABA regulation of HVA1 and HVA22, an RNAi construct for the other barley bZIP, HvZIP1, also was tested. The Ubi1-HvZIP1-RNAi construct was generated using the coding region for the last 194 amino acids of HvZIP1, including the basic region and the Leu zipper. No effect on the ABA induction of both reporter constructs was observed with the HvZIP1-RNAi construct (Figure 5B), in agreement with the low transactivation activity of HvZIP1 on both ABRCs (Figure 3). In aleurone cells, ABA also suppresses the GA induction of α-amylase and proteases (Gómez-Cadenas et al., 2001); thus, we examined whether HvABI5 also mediates the ABA suppression of gene expression. The reporter construct Amy32-GUS, which is induced by GA and downregulated by ABA, was used. HvABI5-RNAi did not affect the GA induction of Amy32-GUS or the ABA suppression of the GA induction of the same reporter construct (Figure 5C). Together, these data indicate that the expression of HvABI5 is required for the ABA induction of HVA1 and HVA22 and that the Ubi1-HvABI5-RNAi construct specifically affects the ABA upregulatory pathway.
HvVP1 RNAi Inhibits the ABA Activation of ABRC-GUS but Has Little Effect on the ABA Suppression of α-Amylase Expression
The observation that HvABI5 along with VP1 can transactivate ABRC-containing promoters (Figure 3) is comparable with the findings of other studies (Hobo et al., 1999). However, the requirement of VP1 for such activity has never been established. Thus, RNAi also was used to verify whether the endogenous VP1 was necessary for the activation of ABRC-containing promoters. To perform gain-of-function assays, we used the maize VP1 cDNA under the control of a constitutive promoter because a complete barley VP1 cDNA has not been obtained. For the RNAi experiment, a PCR approach was designed to clone part of the barley VP1 cDNA sufficient to produce an HvVP1-RNAi construct. The PCR product was obtained from a barley cDNA library and comprises 187 amino acids that span from basic region 1 to basic region 2 of the VP1 protein (McCarty et al., 1991). When the HvVP1-RNAi construct was tested in transient expression, it blocked the ABA induction of ABRC3 and ABRC1 (Figure 6B), suggesting that VP1 is required for a positive ABA regulation of HVA1 and HVA22. As observed with HvABI5-RNAi, HvVP1-RNAi did not have any effect on the ABA suppression of GA-induced α-amylase but showed a 40% reduction of the GA induction of α-amylase (Figure 6C).
Figure 6.
ABA Induction of ABRC-GUS also Is Inhibited by HvVP1-RNAi.
(A) Schemes of the reporter and effector constructs used in the transient expression assays.
(B) The reporter constructs ABRC3-GUS and ABRC1-GUS were cobombarded into barley embryoless half-seeds with (+) or without (−) the effector construct Ubi-HvVP1-RNAi using 1 μg of each construct. Bars indicate GUS activities ± se after 24 h of incubation of the bombarded seeds with (closed or checked bars) or without (open bars) 20 μM ABA. Incubation with the hormone was initiated 8 h after bombardment.
(C) The reporter construct Amy32-GUS was cobombarded into barley embryoless half-seeds with (+) or without (−) the effector construct Ubi-HvVP1-RNAi using 1 μg of each construct. Bars indicate GUS activities ± se after 24 h of incubation of the bombarded seeds with (hatched bars) or without (open bars) 20 μM ABA or 1 μM GA3. Incubation with the hormone was initiated 8 h after bombardment.
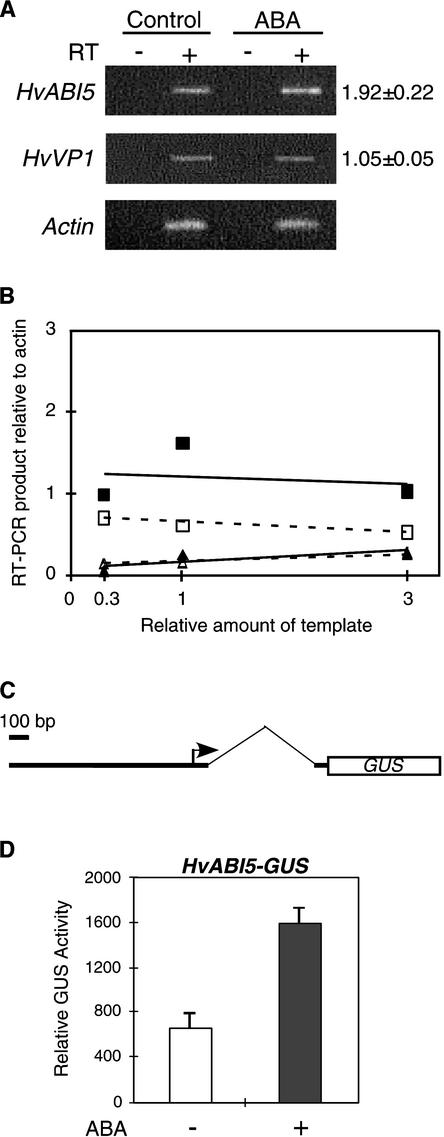
Expression of HvABI5 but Not HvVP1 Is Upregulated by ABA
The expression patterns of HvABI5 and HvVP1 in aleurones were analyzed by reverse transcriptase–mediated PCR because detection of HvABI5 was difficult by RNA gel blot hybridization, probably as a result of its low abundance. Total RNA isolated from aleurones treated or not treated with 100 μM ABA for 12 h was used to generate first-strand cDNAs. Expression of both HvABI5 and HvVP1 was detected in nontreated aleurones. Using a semiquantitative method, we determined that HvABI5 was upregulated by ABA by almost twofold, whereas HvVP1 was practically unaffected (Figures 7A and 7B). In addition, a genomic clone of HvABI5 was isolated and a transcriptional GUS fusion with the HvABI5 promoter was used to analyze its expression in aleurone cells. Incubation with ABA for 12 h enhanced the expression by approximately twofold of a construct containing up to position −757 of the HvABI5 promoter (Figure 7D). Both reverse transcriptase–mediated PCR and transient expression assays indicated that the expression of HvABI5 is upregulated by ABA.
Figure 7.
Expression of HvABI5 but Not HvVP1 Is Upregulated by ABA.
(A) Reverse transcriptase–mediated PCR analysis of HvABI5 and HvVP1 expression in barley aleurones. Reactions with (+) or without (−) reverse transcriptase (RT) and with total RNA isolated from aleurone layers incubated with (ABA) or without (Control) 100 μM ABA for 12 h are shown. PCR products were separated on a 2% agarose gel and stained with ethidium bromide. Numbers at right represent fold of induction by ABA ± se (n = 7).
(B) Abundance of HvABI5 (squares) and HvVP1 (triangles) from aleurones incubated with (closed symbols) or without (open symbols) ABA was examined within a linear range of 10-fold dilution of the starting template and normalized with respect to the actin products.
(C) Scheme of a transcriptional GUS fusion with the promoter and first intron (thin line) of HvABI5 (HvABI5-GUS) used in transient expression assays.
(D) The reporter construct HvABI5-GUS (2 μg) was cobombarded into barley embryoless half-seeds with 1 μg of the Ubi-LUC internal control construct. Bars indicate GUS activities ± se after 12 h of incubation of the bombarded seeds with (+) or without (−) 20 μM ABA.
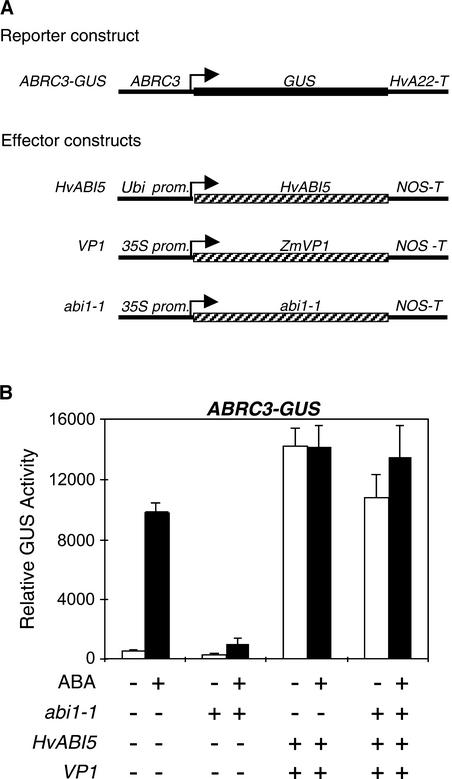
HvABI5-Mediated Transactivation of ABRC Is Not Inhibited by abi1-1
A protein phosphatase 2C encoded by the Arabidopsis gene ABI1 has been shown to negatively regulate responses to ABA. Its dominant-negative mutant form, abi1-1, is capable of blocking ABA responses in Arabidopsis (Leung et al., 1994). In barley aleurone cells, the gene product of abi1-1 was shown to be much more effective than wild-type ABI1 in blocking the ABA induction of ABRC-containing reporter constructs (Shen et al., 2001b). Therefore, the effect of abi1-1 on the HvABI5 transactivation of ABRC3-GUS was studied. As shown in Figure 8, coexpression of 35S–abi1-1 had a negative effect on the ABA induction of ABRC3-GUS; however, it did not decrease the transactivation of ABRC3-GUS by HvABI5 and VP1. This finding indicates that HvABI5 acts downstream of abi1 in the ABA upregulatory pathway.
Figure 8.
The HvABI5/VP1 Transactivation of ABRC-GUS Is Not Inhibited by abi1-1.
(A) Schemes of the reporter and effector constructs used in the transient expression assays.
(B) The reporter construct ABRC3-GUS was cobombarded into barley embryoless half-seeds with (+) or without (−) the effector constructs 35S-abi1-1, Ubi-HvABI5, and 35S-VP1 using 1 μg of each construct. Bars indicate GUS activities ± se after 24 h of incubation of the bombarded seeds with (closed bars) or without (open bars) 20 μM ABA.
DISCUSSION
We have described the isolation of a barley gene, HvABI5, that belongs to a subfamily of bZIPs involved in ABA-dependent signaling. Combining gain-of-function and loss-of-function approaches, we demonstrated that HvABI5 and the barley ortholog of VP1 are necessary for the ABA upregulation of the expression of HVA1 and HVA22. In addition, the ectopic expression of these two factors combined is sufficient to transactivate HVA1 and HVA22 promoters. However, neither of these transcription factors is involved in the ABA suppression of germination-specific α-amylase expression.
Several studies have shown bZIP factors binding to ABA-responsive promoter elements, but there is no functional evidence indicating their role in ABA or stress response (Busk and Pages, 1998). However, a group of bZIP factors was identified recently as mediating ABA-regulated gene expression (Hobo et al., 1999; Choi et al., 2000; Finkelstein and Lynch, 2000; Uno et al., 2000). Here, we refer to this subclass of bZIPs as ABI5-like bZIP factors, because of their similarity to the Arabidopsis ABI5 gene product. ABI5-like bZIP factors are unique because they possess very conserved regions that include putative phosphorylation sites (Figure 1) that may be important in regulating their activity. The possible phosphorylation of ABI5-like factors has been explored (Uno et al., 2000; Lopez-Molina et al., 2001), yet there is no direct evidence that phosphorylation is required for their activity. Currently, ABI5-like factors are described as mediating ABA-regulated gene expression in seeds and vegetative tissues in both monocots and dicots; however, an important functional feature of these factors could be the tissue specificity that they exhibit. Two of them, TRAB1 and ABI5, are known to be expressed mainly in seeds and to regulate the expression of embryo-specific genes (Hobo et al., 1999; Finkelstein and Lynch, 2000); others, such as ABFs and AREBs, are expressed in green tissues and roots (Uno et al., 2000; Kang et al., 2002). All of them have been shown to be involved in ABA signaling. The existence of multiple factors of this class able to recognize similar promoter elements suggests that the signaling mediated by ABA may be controlled by a complex battery of transcription factors. Moreover, not every seed-specific gene that is regulated by ABA is affected in the Arabidopsis abi5 mutant (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000), indicating that other bZIPs or another class of transcription factors may be involved in the regulation of stress- or ABA-induced genes.
ABI5-like bZIP factors also are distinct from other bZIPs because they can recognize a broader spectrum of ABREs, including a nonperfect ACGT-box, CGCGTG (Choi et al., 2000), which also is present in CE3 of the HVA1 promoter (Figure 2A). Our in vitro binding assays indicate that HvABI5 can recognize both imperfect palindromic ACGT-boxes present in ABRC1 and ABRC3 and that the binding activity is specific to the ACGT-boxes and CE3 but not to CE1 (Figures 2B and 2D). Because CE3 resembles an ACGT-box with the A replaced by G (Q. Shen, J. Casaretto, and T.-h.D. Ho, unpublished data), and because ABI5-like bZIPs were shown to recognize such target sequences (Kim et al., 1997), we tested two copies of CE3 in a competition assay. 2xCE3 did not render the same result as two copies of the ACGT-box A2 (Figure 2B). Therefore, we postulate that the protein-ABRC3 complex is composed of a homodimer of HvABI5 that recognizes the ACGT-box and another homodimer or heterodimer that binds CE3. In ABRC1, an HvABI5 homodimer binding to the ACGT-box could work with another factor that recognizes CE1. Further studies will be needed to determine the actual affinity of this bZIP to different cis elements.
Interestingly, we found that a partially purified nuclear extract from barley embryos has specific binding activity for the class of ACGT-boxes present in ABRC3 (Q. Shen, J. Casaretto, and T.-h.D. Ho, unpublished data). It recognizes the wild-type version of the ABRC3 and two copies of the ACGT-box but possesses low affinity for two copies of the coupling element CE3. Similar to HvABI5, the binding activity of the protein extract was dependent on the presence of both the ACGT-box and CE3 in the HVA1 promoter. It will be interesting to determine if the binding activity is attributable to the presence of HvABI5 in the nuclear extract. Furthermore, the binding activity of HvABI5 and that of the nuclear extract correlates with the in vivo activity of the promoter-GUS constructs used in transient expression studies (Q. Shen, J. Casaretto, T.-h.D. Ho, unpublished data). Binding specificity studies of several plant bZIPs have been reported (Izawa et al., 1993). However, it is important to note that although other plant bZIPs, such as HvZIP1, may be able to recognize ABREs or ACGT-boxes, only in vivo assays, like those presented in this work, would allow us to identify the actual transcription factors that operate on specific promoters. To date, only ABI5-like bZIPs have been shown to mediate ABA responses.
It has been shown that overexpression of ABI5-like factors alone cannot render the same level of activation of an ABA-responsive promoter as treatment with ABA. However, a synergistic effect has been observed with the bZIP and ABA, or with the bZIP, VP1, and ABA (Hobo et al., 1999; Gampala et al., 2002). Our data also agree with these observations. HvABI5 was unable to fully activate an ABRC-GUS construct. However, when HvABI5 was expressed along with VP1, we were able to mimic the activation effect of ABA (Figure 3B). We also observed a slight further increment in the activation of the reporter construct when ABA was present along with HvABI5 and VP1, suggesting that ABA may perform an additional task in transactivating these promoters.
To date, the bZIPs TRAB1 and ABI5 have been shown to regulate ABA-induced seed-specific genes in vivo (Hobo et al., 1999; Finkelstein and Lynch, 2000). In addition, genetic evidence for the role of a bZIP in ABA signaling exists only for ABI5. The abi5 mutation has been identified in several independent genetic screens (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Carles et al., 2002). In the case of barley, for which mutants of HvABI5 are not available, one way to study the effect of a transcription factor is through the expression of a dominant-negative form of the protein to repress the function of the wild-type gene. This has been particularly suitable for bZIP factors (Unger et al., 1993; Fukazawa et al., 2000; Sprenger-Haussels and Weisshaar, 2000). We constructed a dominant-negative form of HvABI5, and when introduced into aleurones, it efficiently repressed the ABA induction of the ABRC3-GUS reporter construct (Figure 4). However, to avoid the possibility of affecting other bZIPs, we also implemented RNAi as another loss-of-function approach to this problem. The RNAi technology has been shown to cause sequence-specific repression of several target genes in barley aleurones. The activity of the endogenous factors GAMyb and SLN1, as well as the expression of transiently transformed LUC, GUS, and the Ser/Thr protein kinase PKABA genes, were abolished using RNAi (Zentella et al., 2002). Overexpression studies cannot always reflect the in vivo function of a transcription factor. For example, overexpression of HvZIP1 showed fivefold activation of ABRC3 (this would be regarded as significant activity in other transient expression systems; Figure 3B), but that does not necessarily indicate that this factor activates the ABRC-containing promoter. Using RNAi against HvABI5 and HvZIP1, we identified HvABI5 as the transcription factor responsible for specifically mediating the ABA induction of ABRC-GUS. Furthermore, we demonstrated that HvABI5 is not involved in the ABA suppression of GA-induced α-amylase (Figure 5C).
Recently, it was shown that TRAB1 and ABI5 can interact with the rice and Arabidopsis orthologs of VP1 (OsVP1 and ABI3, respectively) in yeast (Hobo et al., 1999; Nakamura et al., 2001). Interestingly, the portion of the ABI5 protein that seems to interact with ABI3 in the yeast two-hybrid system includes two of the N-terminal conserved regions containing putative phosphorylation residues (Nakamura et al., 2001). These regions are present only in the subfamily of bZIPs involved in ABA signaling. VP1 is the gene product of the maize Viviparous1 (Vp1) locus, with its expression restricted to seed tissue (McCarty et al., 1991). Molecular analysis of the vp1 mutant has shown that the seeds do not accumulate ABA-regulated Lea genes such as rab17 (Pla et al., 1989) and the maize ortholog of the wheat Em gene (McCarty et al., 1991), suggesting its involvement in ABA-regulated gene expression. The expression of some ABA response genes, such as Em, Osem, and HVA1, has been reported to be activated by VP1 (Hattori et al., 1995; Hill et al., 1996; Shen et al., 1996). In this study, using an HvVP1-RNAi construct, we demonstrated that VP1 also is required for the activation of HVA1 and HVA22 promoters by ABA in aleurones (Figure 6). Notably, HVA1 and HVA22 have been shown to be expressed in young vegetative tissues specifically upon ABA treatment (Hong et al., 1992; Shen et al., 2001a). If VP1 is required for their response to ABA, how can these genes be expressed if VP1 is embryo specific? One explanation is that the promoters of HVA1 and HVA22 respond to ABA in young vegetative tissues as a result of the continuous existence of embryo-specific ABI5-like bZIPs in young tissues and later as a result of other ABI5-like bZIPs that also can activate them. As with AREBs and ABFs, they may not need VP1 for their expression but perhaps another equivalent transactivator present in vegetative tissues. In the case of TRAB1, it has been shown that this bZIP is expressed not only in seed tissues but also in leaves and roots of rice seedlings (Hobo et al., 1999).
The effect of HvVP1-RNAi on the GA induction of α-amylase is puzzling. VP1 has been described to play dual roles as a positive regulator of the ABA induction of maturation-specific genes in late seed development and as a repressor of the induction of germination-specific α-amylase genes (Hoecker et al., 1995). If VP1 has inhibitory activity on the GA induction of α-amylase in seeds, we would expect an increase of GUS activity when the HvVP1-RNAi construct is used. By contrast, we observed a 40% reduction of the GA activation of α-amylase (Figure 6C). Moreover, in our system, overexpression of VP1 alone showed only a small repression (∼30%) of GA-induced α-amylase (data not shown). As expected, the HvVP1-RNAi construct had no effect on the ABA suppression of GA-induced α-amylase.
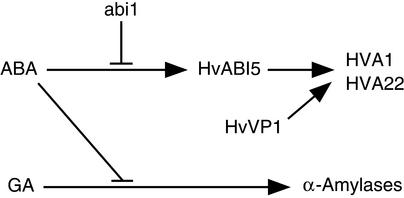
Several lines of evidence suggest that similar transcription complexes may be found in several ABA- or stress-induced promoters, in particular promoters of seed-specific genes such as Lea genes: (1) the similarity in the two-cis-element configuration of the promoters; (2) their ABREs being recognized by ABI5-like bZIPs; and (3) the ability of VP1 to activate some of them. In the case of HVA1 and HVA22, little is known about how ABA controls their expression. In this study, which integrates overexpression and RNAi experiments, we have demonstrated that HvABI5 and VP1 are sufficient and necessary for the transactivation of ABRC-containing promoters. In our system, this transactivation was not impaired by the dominant-negative ABA repressor abi1-1 (Figure 8), suggesting that abi1 acts upstream of HvABI5 in the ABA signal transduction pathway. We also showed that ABA slightly upregulates the transcription of HvABI5 (Figure 7), as happens with other ABI5-like bZIPs (Hobo et al., 1999; Choi et al., 2000; Finkelstein and Lynch, 2000; Uno et al., 2000). It is difficult to say how such minimal induction of a transcription factor could account for the ABA activation of our promoters, especially because the overexpression of HvABI5 cannot fully transactivate them. Therefore, we postulate that ABA also can activate HvABI5 via post-transcriptional modifications such as phosphorylation and possibly promote the interaction with other factor(s) such as VP1 (Figure 9). It will be crucial to determine how ABA affects the activity or formation of the DNA binding protein complex to understand how gene expression is regulated.
Figure 9.
Model for the ABA Regulation of HVA1 and HVA22.
ABA could regulate the expression of the stress-induced genes HVA1 and HVA22 by activating a protein complex composed of HvABI5 and HvVP1 gathered at the ABRCs. These two transcription factors are involved specifically in the ABA upregulatory pathway and act downstream of abi1.
METHODS
Plant Materials
Barley seeds (Hordeum vulgare cv Himalaya) from the 1991 and 1998 harvests at Washington State University in Pullman were used in all experiments.
Cloning of Barley bZIP and VP1 cDNAs
A cDNA library made with mRNA obtained from abscisic acid (ABA)–treated aleurone layers and another library from nontreated aleurones (Stratagene, La Jolla, CA) were used to screen for barley bZIPs. The Arabidopsis AtDPBF1 and the rice OsZIP1a (Nantel and Quatrano, 1996) cDNAs were used as probes to screen 1 × 106 colony-forming units. All positive clones were cloned into the EcoRI or EcoRI-XhoI sites in the pBluescript SK− vector (Stratagene). A partial sequence of the barley VP1 gene was amplified by PCR using the primers 5′-GCGCCAGGGCACCATGCA-3′ and 5′-CTGCTGGCT-CCGCTGCTGCTG-3′ and a barley cDNA library (Stratagene). A 613-bp fragment was cloned into the EcoRV site in pBluescript KS+.
Isolation of an HvABI5 Genomic Clone
The entire coding region of HvABI5 was used as a probe to screen 1 × 106 colony-forming units of a barley (cv Igri) genomic library (Stratagene). One clone of ∼7.5 kb containing the coding region and 3.2 kb upstream of the open reading frame was isolated. A 3.5-kb SalI fragment containing the region −2503 to +1031 of the HvABI5 gene was sequenced and subcloned into the SalI site of pBluescript KS+.
DNA and Amino Acid Sequence Analysis
Plasmid DNA containing cDNA clones was used as a DNA template for sequencing. The DNA sequence was determined using the T7 and T3 promoter primers and the BigDye Terminator mix (PE Applied Biosystems, Foster City, CA). Amino acid sequences were deduced and sequence alignment was performed using the DNASTAR analysis software package (Madison, WI).
Preparation of the DNA Constructs
The reporter constructs used in transient expression assays were prepared as follows. (1) ABRC1-GUS consisted of the 49-bp ABRC1 fragment of the HVA22 promoter fused to the progenitor MP64, which has a minimal (−60) promoter of the Amy64 gene (Khursheed and Rogers, 1988) and its 5′ untranslated region (+57 relative to the transcription start site), the HVA22 intron1-exon2-intron2 segment, the Escherichia coli β-glucuronidase (GUS) coding region, and the HVA22 3′ region (Shen and Ho, 1995). (2) ABRC3-GUS was made by linking a 68-bp promoter fragment of the HVA1 gene to the SmaI site of the MP64 progenitor (Shen and Ho, 1995). (3) Amy32-GUS consisted of the promoter (−331), the 5′ untranslated region and the first intron of a low-pI α-amylase gene, Amy32, fused to the GUS coding sequence, and the 3′ untranslated region of the same α-amylase gene (Lanahan et al., 1992). (4) HvABI5-GUS was made by digesting the SalI genomic fragment of HvABI5 in pBluescript SK− with SnaBI and EcoRV and religated to generate a construct containing 1459 bp upstream of the open reading frame. A fragment corresponding to nucleotides 1747 to 3226 of the gene then was amplified by PCR using the primers 5′-TGCTCTAGAGCTCCTGAAGTCCATGACC-3′ and T7 and cloned in front of the GUS coding region. This construct contains 757 bp of the promoter, a 643-bp intron, and the sequence encoding for the first seven amino acids of the HvABI5 protein fused in frame to the β-glucuronidase coding region and the 3′ untranslated region of the HVA22 barley gene.
The effector constructs Ubi-HvABI5 and Ubi-HvZIP1 consisted of the maize Ubi1 promoter (Christensen and Quail, 1996) linked to the coding sequence of HvABI5 and HvZIP1, respectively, and to the 3′ untranslated region of the nopaline synthase gene (NOS-T). The HvABI5-ΔN construct was made by amplifying by PCR the coding sequence that corresponds to amino acids 280 to 353, using the primers 5′-GGCAGAGGAGGATGATCAAG-3′ and T3, and the HvABI5 cDNA cloned in the pBluescript SK− vector. The amplified sequence then was inserted between the Ubi1 promoter and NOS-T. For the generation of double-stranded RNA interference constructs, two identical fragments of a region of the transcribed sequence of HvABI5 (corresponding to nucleotides 1 to 455 of the cDNA), HvZIP1 (nucleotides 677 to 1515), or HvVP1 (nucleotides 87 to 561) were cloned in opposite directions, separated by a DNA fragment of ∼700 bp of plasmid origin, and cloned between the maize Ubi1 promoter and the NOS terminator. The 35S-VP1 and 35S-abi1-1 constructs have been described (McCarty et al., 1991; Armstrong et al., 1995).
Transient Expression Assays
Barley embryoless half-seeds were prepared and transformed transiently by particle bombardment as described previously (Shen et al., 1993). Briefly, 1 μg of each reporter and effector construct was cobombarded in a molar ratio of 1:1 unless indicated otherwise. After particle bombardment, the half-seeds were incubated for 24 h in the presence or absence of 20 μM ABA or 1 μM GA3. Four bombarded seeds were processed and assayed for luciferase and GUS activities. An internal control of transformation (Ubi1-LUC) also was included at a ratio of 1:1 with the reporter plasmid for the purpose of normalizing GUS activities. The expression of this Ubi1-LUC construct was not affected by any of the treatments (Shen et al., 1993). All experiments consisted of four replicates. The values shown in the figures represent relative GUS activities ± se.
Electrophoretic Mobility Shift Assays
HvABI5 and HvZIP1 coding sequences were cloned in TrcHis bacterial expression vectors (Invitrogen, Carlsbad, CA) to produced 6xHis-tagged fusion proteins, and overexpressed proteins were purified using nickel–nitrilotriacetic acid agarose columns (Qiagen, Valencia, CA). Recombinant proteins were used in electrophoretic mobility shift assays along with radiolabeled fragments of the HVA1 and HVA22 promoters containing the ABRC3 and ABRC1 sequences, respectively. Briefly, double-stranded oligonucleotide probes were obtained by digesting the ABRC3-GUS and ABRC1-GUS constructs used in transient expression assays with NotI and XbaI and labeling them with 32P-dCTP by the Klenow fill-in reaction. Binding reactions (20 μL) contained 1 ng of radiolabeled probe, 1 μg of poly(dIdC), 10 mM Tris-HCl, pH 7.6, 50 mM KCl, 0.5 mM EDTA, 10% (v/v) glycerol, 1 mM DTT, and 2 μg of recombinant protein and were incubated at 4°C for 30 min. Competitors were added in a 30- or 300-fold molar excess. All reaction mixtures were resolved by electrophoresis on a 4% polyacrylamide gel in 0.5× TBE buffer (45 mM Tris, 45 mM boric acid, and 1 mM EDTA).
Reverse Transcriptase–Mediated PCR Analysis
Total RNA was isolated from aleurone layers of 2-day imbibed embryoless half-seeds treated or not treated with 100 μM ABA for 12 h according to a procedure described elsewhere (Logemann et al., 1987). First-strand cDNA was made from 5 μg of total RNA using 50 units of SuperScript II reverse transcriptase (Invitrogen) and 0.5 μg of oligo(dT) according to the manufacturer's instructions. Reactions were performed at 42°C for 50 min. The resulting single-stranded cDNAs were amplified with KlentaqLA (Clontech, Palo Alto, CA) using the primers 5′-ATGCCGTACTCGTTCGAGG-3′ and 5′-CCAGGG-CCCGGTCAGC-3′ for HvABI5 and 5′-CAGAAGGTGCTGAAGCAG-AG-3′ and 5′-TCAGATGCTCACCGCCATCTGG-3′ for HvVP1. The barley actin gene was chosen as a constitutive control RNA and was amplified with the primers 5-CAGCATTGTAGGAAGGCCAC-3′ and 5′-CCAGTTGTTGACAATGCCA-3′. PCR was performed for 30 cycles within a linear range of amplification of the actin, HvABI5, and HvVP1 genes. Each cycle consisted of 97°C for 50 s, 58°C for 1 min, and 68°C for 30 s, and the procedure terminated at 68°C for 4 min. PCR products were separated on a 2% agarose gel and stained with ethidium bromide for photography. Analysis and quantification of cDNA abundance were performed by comparing each cDNA product with a quantitative standard DNA ladder and using the NIH Image program (http://rsb.info.nih.gov/nih-image/).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
GenBank accession numbers for the genes described in this work are as follows: HvABI5 (AY150676), HvZIP1 (AY150677), partial HvVP1 (AY150678), and partial HvABI5 genomic sequence (AY156992). Accession numbers for the protein sequences shown in Figure 1 are as follows: TRAB1 (BAA83740), AREB2 (BAB12405), and ABI5 (AAD21438). Accession numbers for the genes used as probes are AtDPBF1 (AF334206) and OsZIP1a (U04295).
Acknowledgments
We thank Terry Thomas (Texas A&M University) for providing the Arabidopsis DPBF1 cDNA, Ralph Quatrano (Washington University) for providing the OsZIP1a cDNA, Donald McCarty (University of Florida) for the 35S-VP1 construct, and Daisuke Yamaguchi (Tokyo Metropolitan University) for providing a barley cDNA library.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.007096.
References
- Armstrong, F., Leung, J., Grabov, A., Brearly, J., Giraudat, J., and Blatt, M.R. (1995). Sensitivity to abscisic acid of guard-cell K+ channels is suppressed by abi1-1, a mutant Arabidopsis gene encoding a putative protein phosphatase. Proc. Natl. Acad. Sci. USA 86, 9692–9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brands, A., and Ho, T.-H.D. (2002). Function of a plant stress-induced gene, HVA22: Synthetic enhancement screen with its yeast homolog reveals its role in vesicular traffic. Plant Physiol. 130, 1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, E.A. (1997). Plant responses to water deficit. Trends Plant Sci. 2, 48–54. [Google Scholar]
- Busk, P.K., and Pages, M. (1998). Regulation of abscisic acid-induced transcription. Plant Mol. Biol. 37, 425–435. [DOI] [PubMed] [Google Scholar]
- Carles, C., Bies-Etheve, N., Aspart, L., Leon-Kloosterziel, K.M., Koornneef, M., Echeverria, M., and Delseny, M. (2002). Regulation of Arabidopsis thaliana Em genes: Role of ABI5. Plant J. 30, 373–383. [DOI] [PubMed] [Google Scholar]
- Choi, H., Hong, J., Ha, J., Kang, J., and Kim, S.Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730. [DOI] [PubMed] [Google Scholar]
- Christensen, A.H., and Quail, P.H. (1996). Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 5, 213–218. [DOI] [PubMed] [Google Scholar]
- Dure, L., III (1993). A repeating 11-mer amino acid motif and plant desiccation. Plant J. 3, 363–369. [DOI] [PubMed] [Google Scholar]
- Dure, L., III, Crouch, M., Harada, J., Ho, T.-H.D., Mundy, J., Quatrano, R.S., Thomas, T., and Sung, Z.R. (1989). Common amino acid sequence domains among the LEA proteins of higher plants. Plant Mol. Biol. 12, 475–486. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R., and Lynch, T. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa, J., Sakai, T., Ishida, S., Yamaguchi, I., Kamiya, Y., and Takahashi, Y. (2000). REPRESSION OF SHOOT GROWTH, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12, 901–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala, S.S., Finkelstein, R., Sun, S.S., and Rock, C.D. (2002). ABI5 interacts with abscisic acid signaling effectors in rice protoplasts. J. Biol. Chem. 277, 1689–1694. [DOI] [PubMed] [Google Scholar]
- Gómez-Cadenas, A., Zentella, R., Walker-Simmons, M.K., and Ho, T.-H.D. (2001). Gibberellin/abscisic acid antagonism in barley aleurone cells: Site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13, 667–679. [PMC free article] [PubMed] [Google Scholar]
- Guiltinan, M.J., Marcotte, W.R., Jr., and Quatrano, R.S. (1990). A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250, 267–271. [DOI] [PubMed] [Google Scholar]
- Hattori, T., Terada, T., and Hamasuna, S. (1995). Regulation of the Osem gene by abscisic acid and the transcriptional activator VP1: Analysis of cis-acting promoter elements required for regulation by abscisic acid and VP1. Plant J. 7, 913–925. [DOI] [PubMed] [Google Scholar]
- Hill, A., Nantel, A., Rock, C.D., and Quatrano, R.S. (1996). A conserved domain of the viviparous-1 gene product enhances the DNA binding activity of the bZIP protein EmBP-1 and other transcription factors. J. Biol. Chem. 271, 3366–3374. [DOI] [PubMed] [Google Scholar]
- Hobo, T., Kowyama, Y., and Hattori, T. (1999). A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. USA 96, 15348–15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker, U., Vasil, I.K., and McCarty, D.R. (1995). Integrated control of seed maturation and germination programs by activator and repressor functions of Viviparous-1 of maize. Genes Dev. 9, 2459–2469. [DOI] [PubMed] [Google Scholar]
- Hong, B., Barg, R., and Ho, T.-H.D. (1992). Developmental and organ-specific expression of an ABA- and stress-induced protein in barley. Plant Mol. Biol. 18, 663–674. [DOI] [PubMed] [Google Scholar]
- Izawa, T., Foster, R., and Chua, N.-H. (1993). Plant bZIP protein DNA binding specificity. J. Mol. Biol. 230, 1131–1144. [DOI] [PubMed] [Google Scholar]
- Kang, J., Choi, H., Im, M., and Kim, S.Y. (2002). Arabidopsis basic leucine zipper proteins mediate stress-responsive abscisic acid signaling. Plant Cell 14, 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khursheed, B., and Rogers, J.C. (1988). Barley α-amylase genes: Quantitative comparison of steady-state mRNA levels from individual members of the two different families expressed in aleurone cells. J. Biol. Chem. 263, 18953–18960. [PubMed] [Google Scholar]
- Kim, S.Y., Chung, H.-J., and Thomas, T.L. (1997). Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant J. 11, 1237–1251. [DOI] [PubMed] [Google Scholar]
- Kim, S.Y., and Thomas, T.L. (1998). A family of novel basic leucine zipper proteins binds to seed-specification elements in the carrot Dc3 gene promoter. J. Plant Physiol. 152, 607–613. [Google Scholar]
- Kircher, S., Ledger, S., Hagashi, H., Weisshaar, B., Schafer, E., and Frohnmeyer, H. (1998). CPRF4a, a novel plant bZIP protein of the CPRF family: Cooperative analyses of light-dependent expression, post-transcriptional regulation, nuclear import and heterodimerisation. Mol. Gen. Genet. 257, 595–605. [DOI] [PubMed] [Google Scholar]
- Lanahan, M.B., Ho, T.-H.D., Rogers, S.W., and Rogers, J.C. (1992). A gibberellin response complex in cereal α-amylase gene promoters. Plant Cell 4, 202–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, J., Bouvier-Durand, M., Morris, P.C., Guerrier, D., Chefdor, F., and Giraudat, J. (1994). Arabidopsis ABA response gene ABI1: Features of a calcium-modulated protein phosphatase. Science 264, 1448–1452. [DOI] [PubMed] [Google Scholar]
- Leung, J., and Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 199–222. [DOI] [PubMed] [Google Scholar]
- Logemann, J., Schell, J., and Willmitzer, L. (1987). Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 163, 16–20. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina, L., and Chua, N.-H. (2000). A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 41, 541–547. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina, L., Mongrand, S., and Chua, N.-H. (2001). A post-germination developmental arrest checkpoint is mediated by ABA and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98, 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty, D.R., Hattori, T., Carson, C.B., Vasil, V., Lazar, M., and Vasil, I.K. (1991). The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66, 895–905. [DOI] [PubMed] [Google Scholar]
- Nakamura, S., Lynch, T.J., and Finkelstein, R.R. (2001). Physical interactions between ABA response loci of Arabidopsis. Plant J. 26, 627–635. [DOI] [PubMed] [Google Scholar]
- Nantel, A., and Quatrano, R.S. (1996). Characterization of three rice basic/leucine zipper factors, including two inhibitors of EmBP-1 DNA binding activity. J. Biol. Chem. 271, 31296–31305. [DOI] [PubMed] [Google Scholar]
- Pla, M., Goday, A., Vilardell, J., Gómez, J., and Pagès, M. (1989). Differential regulation of ABA-induced 23–25 kDa proteins in embryo and vegetative tissues of the viviparous mutants of maize. Plant Mol. Biol. 13, 385–394. [DOI] [PubMed] [Google Scholar]
- Rock, C. (2000). Pathways to abscisic acid-regulated gene expression. New Phytol. 148, 357–396. [DOI] [PubMed] [Google Scholar]
- Shen, Q., Chen, C.-N., Brands, A., Pan, S.-M., and Ho, T.-H.D. (2001. a). The stress- and abscisic acid-induced barley gene HVA22: Developmental regulation and homologues in diverse organisms. Plant Mol. Biol. 45, 327–340. [DOI] [PubMed] [Google Scholar]
- Shen, Q., Gomez-Cadenas, A., Zhang, P., Walker-Simmons, M.K., Sheen, J., and Ho, T.-H.D. (2001. b). Dissection of abscisic acid signal transduction pathways in barley aleurone layers. Plant Mol. Biol. 47, 437–448. [DOI] [PubMed] [Google Scholar]
- Shen, Q., and Ho, T.-H.D. (1995). Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. Plant Cell 7, 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Q., and Ho, T.-H.D. (1997). Promoter switches specific for abscisic acid (ABA)-induced gene expression in cereals. Physiol. Plant. 101, 653–664. [Google Scholar]
- Shen, Q., Uknes, S.J., and Ho, T.-H.D. (1993). Hormone response complex of a novel abscisic acid and cycloheximide inducible barley gene. J. Biol. Chem. 268, 23652–23660. [PubMed] [Google Scholar]
- Shen, Q., Zhang, P., and Ho, T.-H.D. (1996). Modular nature of abscisic acid (ABA) response complexes: Composite promoter units which are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell 8, 1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger-Haussels, M., and Weisshaar, B. (2000). Transactivation properties of parsley proline-rich bZIP transcription factors. Plant J. 22, 1–8. [DOI] [PubMed] [Google Scholar]
- Tabata, T., Nakayama, T., Mikami, K., and Iwabuchi, M. (1991). HBP-1a and HBP-1b: Leucine zipper-type transcription factors of wheat. EMBO J. 10, 1459–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger, E., Parsons, R.L., Schmidt, R.J., Bowen, B., and Roth, B.A. (1993). Dominant-negative mutants of opaque2 suppress transactivation of a 22-kD zein promoter by opaque2 in maize endosperm cells. Plant Cell 5, 831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno, Y., Furuhata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 97, 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, D., Duan, X., Wang, B., Hong, B., Ho, T.-H.D., and Wu, R. (1996). Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 110, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella, R., Yamauchi, D., and Ho, T.-H.D. (2002). Molecular dissection of the gibberellin/abscisic acid signaling pathways by transiently expressed RNA interference in barley aleurone cells. Plant Cell 14, 2289–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]