Abstract
We have previously observed the spontaneous, pH-dependent insertion of a water-soluble peptide to form a helix across lipid bilayers [Hunt, J. F., Rath, P., Rothschild, K. J. & Engelman, D. M. (1997) Biochemistry 36, 15177–15192]. We now use a related peptide, pH (low) insertion peptide, to translocate cargo molecules attached to its C terminus across the plasma membranes of living cells. Translocation is selective for low pH, and various types of cargo molecules attached by disulfides can be released by reduction in the cytoplasm, including peptide nucleic acids, a cyclic peptide (phalloidin), and organic compounds. Because a high extracellular acidity is characteristic of a variety of pathological conditions (such as tumors, infarcts, stroke-afflicted tissue, atherosclerotic lesions, sites of inflammation or infection, or damaged tissue resulting from trauma) or might be created artificially, pH (low) insertion peptide may prove a useful tool for selective delivery of agents for drug therapy, diagnostic imaging, genetic control, or cell regulation.
Keywords: drug delivery, peptide nucleic acid delivery, tumors, membrane transport, helix formation
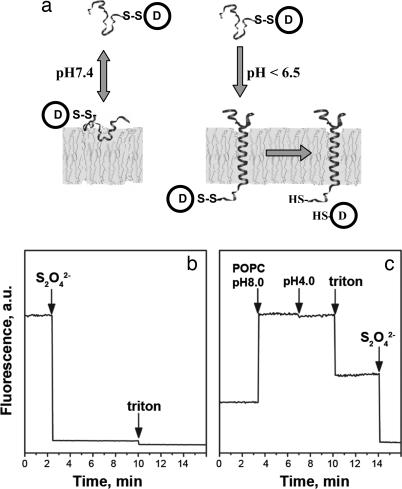
We suggest that spontaneous insertion and formation of transbilayer α-helices can be used as an approach to moving chemical reagents through the permeability barrier of biological membranes. Membrane proteins, which represent ≈30% of all ORFs in sequenced genomes (1, 2), play a key role in many fundamental cellular processes, such as signal and energy transduction, active transport, ion flow, cell–cell interactions, and nerve conduction. Based on the two-stage model of helical membrane protein folding, the formation of independently stable helices across the membrane lipid bilayer occurs first, and then the interaction of the helices with each other leads to the stabilization of higher-order structure (3, 4). Folding and insertion of large-membrane protein domains into targeted membranes require active participation of complex translocation machineries (5–7), whereas insertion of short (<50- to 60-residue) protein sequences can occur spontaneously (8–10). In recent work, some cases of apparent spontaneous insertion have been shown to require a membrane protein, YidC (11). We previously reported that a polypeptide derived from the bacteriorhodopsin C helix, consisting of the transmembrane sequence and two flanking sequences, is soluble in aqueous solution and spontaneously inserts across lipid bilayers in a pH-dependent manner (12, 13), with no requirement for additional proteins such as YidC. Here we report that a related peptide, pH (low) insertion peptide (pHLIP), which inserts across a cell membrane at low pH (<7.0), can translocate a drug or imaging molecule into the cell and release it in the cytoplasm (Fig. 1a).
Fig. 1.
pHLIP insertion and topology. (a) Schematic diagram of cargo molecule delivery into a cell. At physiological pH, the peptide–cargo conjugate interacts weakly with a membrane. At low pH, the peptide forms a transmembrane helix with its C terminus inserted in the cytoplasm. Reduction of the disulfide bond releases a drug. (b) The topology of the pHLIP peptide in a lipid bilayer was determined by using the NBD–dithionite quenching reaction. The fluorescence signal of NBD attached to the N terminus of peptide was monitored at 530 nm when excited at 470 nm. Sodium dithionite was added to the NBD peptide inserted into POPC large unilamellar vesicles. (c) POPC large unilamellar vesicles containing dithionite ion inside were added to the NBD peptide at pH 8.0, and then decreasing the pH triggered insertion of the peptide in liposomes. Triton X-100 was used for the disruption of liposomes. The concentration of the peptide used in the experiments was 7 μM.
Results
At neutral pH, a polypeptide derived from the bacteriorhodopsin C helix binds weakly to the surface of a liposome, whereas at acidic pH (<7.0) it inserts across a membrane and forms a transmembrane α-helix (12, 13). Our strategy is to conjugate a cargo molecule by means of a disulfide bond to the end of the transmembrane peptide that inserts into the cell, where the reducing environment of the cytoplasm will break the disulfide and release the cargo. Our first task was to identify the topology of the insertion of the peptide. We used the membrane-impermeable dithionite ion (S2O4−2), which can chemically modify the 4-chloro-7-nitrobenz-2-oxa-1,3 diazole (NBD) fluorophore and quench its fluorescence (14), to probe a label placed at one end of the peptide. The peptide, labeled with NBD at its N terminus, was inserted into 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) liposomes, which led to a shift of the maximum position of the NBD fluorescence spectrum from 555 (in the absence of liposomes) to 530 nm. The maximum position of the NBD fluorescence at 530 nm indicates that the fluorophore is located in an environment with a dielectric constant of ≈70 (15) on the border between water and membrane. Addition of the dithionite ion to the solution of peptide inserted into POPC led to the complete quenching of NBD fluorescence (Fig. 1b). We also prepared liposomes with dithionite trapped inside them. These liposomes were added to the labeled peptide at neutral pH, and then the pH was lowered to trigger peptide insertion. No change in fluorescence signal was seen (Fig. 1c), showing that dithionite trapped in the liposomes did not react with NBD, that there is no leakage of dithionite, and that the N terminus of the peptide is located outside of the liposome. Disruption of the liposomes by Triton X-100 led to the release of dithionite ions and the quenching of NBD fluorescence. Because we know that the peptide forms a transmembrane helix (12) and NBD labeling usually does not affect the process of protein or peptide insertion into lipid bilayers (16), we assume that the N terminus stays outside, whereas the C terminus of the peptide is translocated across the bilayer, which is the natural topology of the C-helix in bacteriorhodopsin. Our subsequent experiments (below) are consistent with this topology. We decided to use two strategies: to label the cell surface by attaching molecules to the N terminus or to insert molecules inside cells by attaching them to the C terminus.
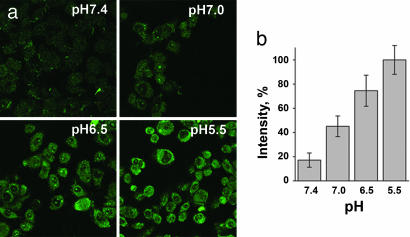
To evaluate the ability of the peptide to translocate molecules through a cell membrane, we synthesized a version of the peptide (pHLIP) with a single cysteine residue at its C terminus (AAEQNPIYWARYADWLFTTPLLLLDLALLVDADEGTCG), allowing it to be easily conjugated to a variety of molecules either via a disulfide bond, which should be cleaved in the reducing environment of the cytoplasm, or via a noncleavable covalent bond. To study pH-dependent translocation of molecules through the cell membrane, we first prepared pHLIP conjugated via a disulfide bond to dansyl dye (see chemical structure in Fig. 7a, which is published as supporting information on the PNAS web site) and tested the construct with live HeLa cells at different pHs (Fig. 2a). Our protocol was to add the fluorescently labeled peptide to cells and incubate for 15 min at pH 5.5, 6.5, 7.0, or 7.4 and then to wash the cells at pH 7.4 to remove any reversibly bound peptide. The uptake of dansyl was significantly higher at low pH (Fig. 2b). The relative uptakes were 18%, 48%, 78%, and 100% at pH 7.4, 7.0, 6.5, and 5.5, respectively.
Fig. 2.
pHLIP transport of dansyl into cells. (a) Fluorescence images of HeLa cells incubated (for 15 min) with cleavable pHLIP–S–S–dansyl construct (7 μM) at pH 5.5, 6.5, 7.0, and 7.4 and washed with PBS buffer at pH 7.4 are shown. (b) Quantification of fluorescence images. The fluorescence signal of cells at pH 5.5 was taken as 100%. The uptake of dansyl strongly decreases with an increase of extracellular incubation pH.
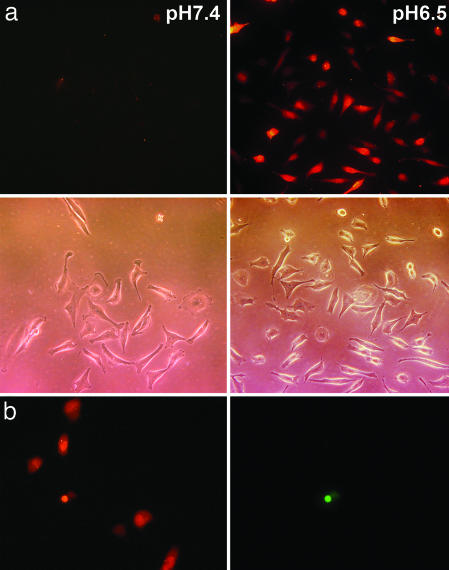
Because it is among the most useful functional cargo-molecules, we studied the translocation of peptide nucleic acid (PNA). Studies in vitro indicate that PNA can inhibit both transcription and translation of genes to which it has been targeted, which holds promise for its use for antigene and antisense therapy (17–19). However, the major obstacle is the delivery of PNA [as well as RNA or oligonucleotide (ODN)] across the membrane into a cell. We examined the ability of pHLIP to translocate a fluorescent-labeled 12-base PNA (see sequence of PNA in Fig. 7b) into the cells. Cells were incubated with pHLIP–S–S-PNA–carboxytetramethylrhodamine (TAMRA) and subsequently washed to remove reversibly bound peptide. No retention of fluorescence was seen in the cells at normal pH, but significant cytoplasmic fluorescent staining of cells was observed at low pH (Fig. 3a). The treatment of cells with PNA–TAMRA did not show fluorescent staining of cells at pH 6.5 or 7.4. We verified that the labeled cells were alive by using the dead cell marker SYTOX–Green. Fig. 3b shows the cells labeled with PNA–TAMRA translocated by pHLIP at pH 6.5 (Left) and the same cells treated with SYTOX–Green (Right). The majority of cells have only PNA–TAMRA, whereas only a few cells had both PNA–TAMRA and SYTOX–Green. We also tried to translocate a 20-base ODN; however, no uptake of a pHLIP–S–S–ODN–FITC (see sequence of ODN in Fig. 7c) construct at normal or low pH was seen (data not shown). The fact that pHLIP was unable to translocate ODN at any pH but can translocate other cargo molecules selectively at low pH argues against the involvement of endocytosis, as do the other data (see below). We conclude that relatively uncharged molecules such as PNAs can be translocated but that highly charged nucleic acids cannot.
Fig. 3.
The delivery of PNA into cells by pHLIP. (a) Fluorescence and phase-contrast images of HeLa cells incubated (for 30 min) with a pHLIP–S–S–PNA–TAMRA cleavable construct (1 μM) at pH 7.4 (Left) and 6.5 (Right) are shown. No translocation was observed of pHLIP–S–S–PNA–TAMRA at pH 7.4 or PNA–TAMRA at pH 7.4 or 6.5 (data not shown). (b) HeLa cells labeled with PNA–TAMRA translocated by pHLIP at pH 6.5 (Left) and the same cells treated with SYTOX–Green (Right). The majority of cells have only PNA–TAMRA, whereas only one cell had both PNA–TAMRA and SYTOX–Green.
We studied delivery of a cyclic peptide cargo using a toxin from the deadly Amanita phalloides mushroom, phalloidin (see chemical structure of phalloidin in Fig. 7d). Phalloidin is a water-soluble, cell-impermeable bicyclic peptide. If it enters a cell, it binds tightly to actin filaments at nanomolar concentration and strongly inhibits their depolymerization (20, 21). Fluorescent phalloidin is commonly used as a specific marker of actin filaments in permeabilized cells. Actin filaments stained with fluorescent phalloidin have an unmistakable filamentous pattern, distinct from the appearance of other cellular structures, organelles, or membrane staining. Therefore, the delivery of fluorescent phalloidin by our construct into live cells provides morphological evidence of the proposed molecular translocation mechanism. The biological effects of translocation of phalloidin into live cells should be an inhibition of cell contractility and division. In particular, we expect that attached cells would not contract and round up in response to EDTA/trypsin treatment. A long-term effect of phalloidin could be the formation of multinucleated cells, because nuclei might divide but treated cells cannot. Each of these expectations has been confirmed experimentally (see below).
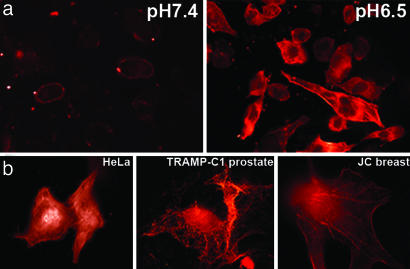
We conjugated a bifunctional, photoactivatable crosslinker via an S–S bond to the C terminus of pHLIP and coupled it to fluorescently labeled phalloidin [phalloidin–tetramethylrhodamine B isothiocyanate (Ph–TRITC)] by photoactivation. We observed staining of actin filaments of cells incubated with pHLIP–S–S–Ph–TRITC at low pH but not at normal pH (Fig. 4a). The treatment of cells with Ph–TRITC or crosslinker–Ph–TRITC in the presence or absence of peptide did not give fluorescent staining of actin filaments at any pH studied. We tested delivery in human (HeLa) and mouse (prostate, TRAMP-C1; breast, JC) cancer cell lines. In each case, we observed the characteristic staining of actin filaments in the target cells at pH < 7.0 (Fig. 4b).
Fig. 4.
The delivery of phalloidin into cells by pHLIP. (a) Fluorescence images of HeLa cells incubated (for 1 h) with a pHLIP–S–S–Ph–TRITC cleavable construct (2 μM) at pH 7.4 (Left) and 6.5 (Right) are shown. The fluorescence was extremely weak after pH 7.4 incubation and localized to the plasma membrane. Strong fluorescence of actin filaments was observed after pH 6.5 incubation. (b) Images of HeLa (Left), breast JC (Center), and prostate TRAMP-C1 (Right) cancer cells with fluorescent actin filaments are shown. Cells were incubated for 1 h with the cleavable pHLIP–S–S–Ph–TRITC (0.5–1 μM) at pH 6.5 followed by washing with PBS at pH 7.4.
To demonstrate that pHLIP itself does not enter the cell, we exploited the fact (reported in ref. 11) that a pH increase leads to the reversal of insertion and the release of the peptide from lipid bilayers. We conjugated Ph–TRITC to the C terminus of the peptide via a noncleavable covalent bond incubated with cells at pH 6.5 and washed with buffer at the same pH. As expected, we observed fluorescence mostly from the plasma membranes; however, when cells were washed with the buffer at pH 7.4, the pHLIP–Ph–TRITC constructs were removed, and only traces of residual fluorescence were seen (Fig. 8a, which is published as supporting information on the PNAS web site). These procedures were repeated with the peptide conjugated to Texas red and rhodamine (data not shown), with similar results. These experiments demonstrate that cells do not internalize the peptide with covalently attached cargo and argues against any involvement of endocytosis or receptor-mediated uptake.
We investigated pHLIP’s cytotoxicity and its ability to induce membrane leakage. The incubation of HeLa cells with peptide at concentrations up to 16 μM for 24 h under physiological conditions does not affect cell viability (Fig. 9a, which is published as supporting information on the PNAS web site). Membrane leakage was tested by incubation of HeLa cells at various concentrations of peptide (up to 10 μM) at pH 7.4 and 6.5 together with cell-impermeable agents: Ph–TRITC and nuclear-staining SYTOX–Orange. Then, cells were washed and fluorescence was measured (Fig. 9b). The fluorescence values obtained in absence or presence of various concentrations of pHLIP were all about the same and ≈10% of those obtained after cell membrane disruption by Triton X-100. The data indicated that pHLIP did not induce the uptake of free Ph–TRITC or SYTOX–Orange, establishing that the cells are viable and that the phalloidin uptake depends on linkage to the pHLIP.
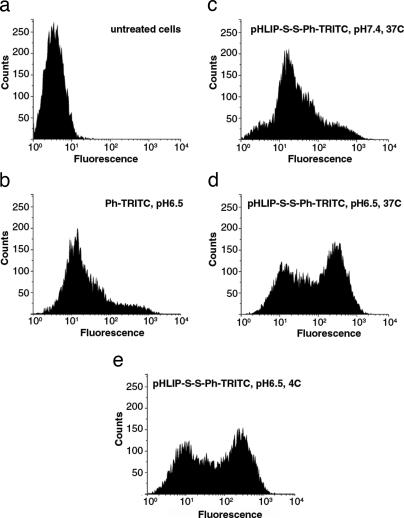
We used FACS of HeLa cells in suspension to improve the quantification of the pH-dependent translocation of fluorescent phalloidin by pHLIP through the membranes (Fig. 5). Cells treated with pHLIP–S–S–Ph–TRITC exhibited a very low level of fluorescence because of limited membrane attachment at pH 7.4. However, at low pH, a population of highly fluorescent cells was observed. The efficiency of Ph–TRITC delivery into cells at low temperature was the same as at 37°C. The appearance of discrete populations of fluorescent cells might reflect a difference in the delivery of Ph–TRITC to cells in different phases of the cell cycle. It seems unlikely that the insertion of pHLIP–Ph–TRITC would depend on the cell cycle, so the difference might arise from a variation in the rate of reduction of the disulfide bonds and/or the variation of the surface area of cell membrane. If phalloidin were not released during the incubation time, it would be removed together with the peptide during cell washing at pH 7.4, because the insertion of peptide is reversible. The release of Ph–TRITC inside the cell might depend on the glutathione concentration, which is known to vary significantly during the cell cycle, reaching a maximum at G2/M and a minimum at G1 (22). Because the incubation time (1 h) is much less than the HeLa cell cycle time (18 h), the cells that happen to be in G2/M might be expected to take up more phalloidin than the rest. On the other hand, the surface area of cells in G2/M phase increases by almost a factor of two, which might lead to double the number of inserted peptides. Indeed, we observed that bigger cells (cell size can be estimated from forward light scattering intensity) emitted more light.
Fig. 5.
Cytofluorometry of HeLa cells. (a and b) The untreated cells (a) and cells treated with Ph–TRITC (b) at pH 6.5 and 37°C are shown. (c–e) Cells treated (for 1 h) with 6 μM pHLIP–S–S–Ph–TRITC at pH 7.4 and 37°C (c), pH 6.5 and 37°C (d), and pH 6.5 and 4°C (e) are shown.
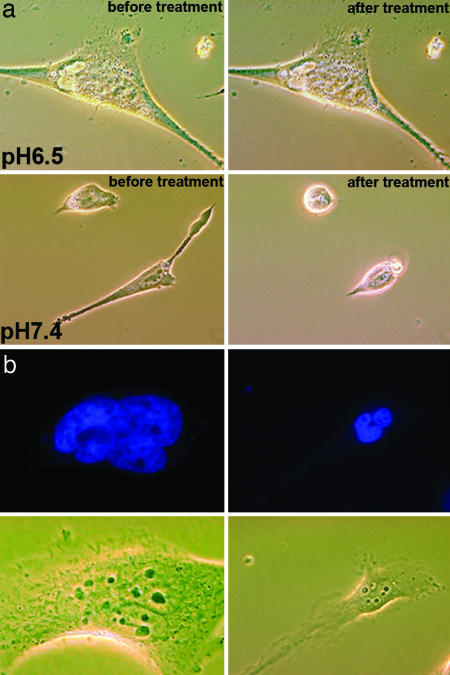
The translocation of phalloidin into cells led to inhibition of cytoskeleton dynamics and consequent loss of the ability of cells to contract and round up in response to treatment by EDTA/trypsin dissociation solution (Fig. 6a and Fig. 10, which is published as supporting information on the PNAS web site). However, untreated cells (data not shown) or cells treated with the construct at normal pH were able to round up and dissociate from the surface. We observed formation of multinucleate cells after 48 h of treatment with pHLIP–S–S–Ph–TRITC (Fig. 6b). The appearance of multinucleated cells has been also reported after treatment of cells with jasplakinolide, which is a cell-permeable analog of phalloidin (23).
Fig. 6.
Cell phenotypes induced by phalodin transport. (a) Phase-contrast images of HeLa cells incubated (for 1 h) with pHLIP–S–S–Ph–TRITC (1 μM) at pH 6.5 and 7.4 followed by washing with PBS (pH 7.4) before (Left) and 5 min after adding of the dissociation solution (Right). Cells treated with the peptide–phalloidin at low pH remained unchanged, consistent with stabilization of the cytoskeleton by Ph–TRITC delivered by the pHLIP. (b) Fluorescence images of nuclei stained with DAPI (0.5 μM) and corresponding phase-contrast images of the multinucleated HeLa cells are presented. Multinucleation was observed at 48 h after treatment of cells with of pHLIP–S–S–Ph–TRITC (1 μM) at pH 6.5 for 1 h.
Discussion
We have used a peptide (pHLIP) that predominantly inserts across a cell membrane at low pH (<7.0) but not at normal physiological pH. By translocating a molecule into a cell and releasing it in the cytoplasm, pHLIP functions, in effect, as a nanosyringe. The peptide does not exhibit any elements of helical secondary structure in solution or on the cell membrane at neutral pH; however, it becomes rigid (as a syringe needle) when it inserts into a lipid bilayer, and it forms a transmembrane helix and injects molecules into cells. We demonstrated that pHLIP inserts across the membrane at low pH and can release disulfide-linked membrane-impermeable molecules, such as PNA and phalloidin, inside cells. Based on all our data, we conclude that the pathway of peptide entry into the membrane and the translocation of molecules into cells are not mediated by endocytosis, by interactions with cell receptors, or by formation of pores in cell membranes; rather, it is the formation of a helix across the lipid bilayer, triggered by the increase of the peptide hydrophobicity due to the protonation of Asp residues induced by low pH. This insertion is consistent with the recent scale of Hessa et al. (24), where the membrane insertion scale is shown to be more favorable for the protonated form of Asp. Our estimation of the free energy of insertion showed that the transbilayer configuration of the protonated pHLIP peptide is favored compared with the most stable peripheral configuration by at least 3 kcal/mol (ref. 11 and Y.K.R. and D.M.E., unpublished results). We hypothesize that this energy might be used to bias the translocation of cargo molecules by pHLIP through the cell membrane. ODN, with high negative charge, might have too high a barrier for its translocation across a membrane on the time scale of our experiments. In contrast, PNA has a charge neutral backbone [N-(2-aminoethyl)glycine units] and thus a lower energy barrier, which makes it suitable for pHLIP translocation. The size range and polarity of cargo molecules that pHLIP can translocate through the membrane remain to be studied.
pHLIP offers a new technology for the fast and efficient delivery of drugs, imaging probes, or cell and gene regulation agents into living cells. The method allows translocation of certain cell-impermeable molecules into a cell or the attachment of a variety of functional moieties and particles to the cell surface at low pH. The pH-selective delivery properties of pHLIP may provide a new approach for diagnosis and treatment of diseases with naturally occurring (or artificially created) low-pH extracellular environments, such as tumors, infarcts, stroke-afflicted tissue, atherosclerotic lesions, sites of inflammation or infection, or damaged tissue resulting from trauma (25–28).
Methods
Synthesis of Peptide and Peptide–Cargo Constructs.
pHLIP was prepared by solid-phase peptide synthesis by using standard 9-fluorenylmethyloxycarbonyl chemistry and purified by reverse-phase chromatography (on a C18 column) at the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University. In a typical preparation of the soluble form of the peptide, the lyophilized powder was dissolved in a solution containing 6 M urea and was transferred to working buffer by using a G-10 size-exclusion spin column. The concentration of the peptide was determined by absorbance (ε280 = 13,940 M−1cm−1) and was confirmed by quantitative amino acid analysis. The peptide was labeled at its single C-terminal cysteine residue with dansyl, Texas red, or rhodamine by incubation with didansyl (Sigma), Texas red maleimide (Molecular Probes), or tetramethylrhodamine maleimide (Molecular Probes) in 10 mM Tris·HCl/20 mM NaCl/6 M urea, pH 8.0, in dark at 4°C for 24 h. The conjugated peptides were purified on a G-10 size-exclusion column and transferred to PBS buffer (pH 7.4). The concentration of labeled peptide was determined by absorbance (dansyl, ε340 = 4,300 M−1cm−1; Texas red, ε582 = 112,000 M−1cm−1; rhodamine, ε542 = 65,000 M−1cm−1).
pHLIP was conjugated to Ph–TRITC (Sigma) by using bifunctional photocrosslinkers: S-[2-(4-azidosalicylamido)ethylthio]-2-thiopyridine (Molecular Probes) or benzophenone-4-iodoacetamide (Molecular Probes). The first crosslinker {S-[2-(4-azidosalicylamido)ethylthio]-2-thiopyridine} makes an S–S bond with the C terminus of the peptide and binds to Ph–TRITC under UV irradiation (pHLIP–S–S–Ph–TRITC). The second crosslinker was used for the synthesis of the noncleavable construct (pHLIP–Ph–TRITC). pHLIP was incubated with the crosslinker in PBS in the dark at 4°C for 24 h. Excess crosslinker was removed by using a G-10 size-exclusion spin column. A 5× molar excess of Ph–TRITC was added to the pHLIP crosslinker and illuminated at 340 nm for 30 min. The unreacted Ph–TRITC was removed by using a G-10 size-exclusion column. The concentration of rhodamine was determined by absorption at 542 nm (ε542 = 65,000 M−1cm−1). Separately, the Ph–TRITC was conjugated to S-[2-(4-azidosalicylamido)ethylthio]-2-thiopyridine without peptide. The conjugation of Ph–TRITC to crosslinker or pHLIP did not affect its ability to bind to F-actin (data not shown).
TAMRA-o-o-CATAGTATAAGT-o-Cys-NH2–PNA with Cys and fluorescent dye, TAMRA, was synthesized by Applied Biosystems. This PNA targets MDM2 mRNA (29). PNA–TAMRA was incubated with a 4× molar excess of pHLIP for 24 h in PBS (pH 7.4) at 4°C. pHLIP–S–S–PNA–TAMRA construct was purified on G-10 column. The concentration was determined by measuring absorption at 546 nm (ε542 = 65,000 M−1cm−1).
FITC-(5′-GTTCTCCCAGCGTGCGCCAT-3′)-SH was synthesized by Eurogentec. FITC–ODN targets Bcl-2 protooncogene mRNA (30). It was conjugated to pHLIP by the same method as described above for PNA conjugation. Concentration of pHLIP–S–S–ODN–FITC was measured by the absorption at 492 nm (ε492 = 56,000 M−1cm−1).
Liposome Preparation.
Large unilamellar vesicles were prepared by sonication. The POPC (Avanti Polar Lipids) phospholipid was dissolved in chloroform. After removal of the solvent using a rotary evaporator, the phospholipid film was dried overnight and then rehydrated in 10 mM Tris·HCl/20 mM NaCl, pH 8.0, and vortexed. The suspension was sonicated by using a Branson titanium tip ultrasonicator until the solution became transparent. The liposomes distribution was evaluated by dynamic light scattering. The vesicles radius was 72.6 ± 4.6 nm (polydispersity was 13.6 ± 6.7%). To prepare liposomes with dithionite inside them, the phospholipid film was rehydrated in 1 M Tris buffer (pH 8.0) containing 1 M dithionite. Untrapped dithionite was removed from the solution of liposomes by using a dialysis cassette with 10-kDa cutoff pores. Dialysis was performed against 10 mM Tris·HCl/20 mM NaCl, pH 8.0, for 4 h with hourly solution changes.
Detection of Peptide Topology.
External addition of dithionite (Na2S2O4) to large unilamellar vesicles chemically quenches the NBD fluorescence in the outer leaflets of the bilayers. The peptide was labeled at N terminus with NBD by incubation with NBD-Cl (Molecular Probes) in 10 mM Tris·HCl/20 mM NaCl, pH 7.0, in dark at 4°C for 24 h (the peptide:NBD ratio was 1:20). The conjugated peptide was purified in a G-10 size-exclusion column. The concentration of labeled peptide was determined by absorbance (ε480 = 25,000 M−1cm−1; ε336 = 9,800 M−1cm−1). The labeled peptide was incubated with liposomes at neutral pH, and insertion was triggered by the reducing of pH to 4.0. The fluorescence spectra of the peptide labeled with NBD in solution and inserted into lipid bilayer were recorded on a SLM8000 spectrofluorometer. Changes in the fluorescence signal of NBD were monitored at 530 nm (excited at 470 nm) after addition of the dithionite and disruption of the liposomes by Triton X-100. In other experiments, liposomes containing dithionite were added to the labeled peptide in solution at pH 8.0, the pH was then decreased to 4.0, Triton X-100 was added to disrupt the liposomes, and more dithionite was added for the complete quenching of the NBD fluorescence.
Cell Lines.
Both human and mouse cancer cell lines were used in our study. HeLa cells were provided by the Cancer Center of Yale University Medical School. Prostate, TRAMP-C1 (CRL-2730), and breast adenocarcinoma (CRL-2116) were from the American Type Culture Collection. Cells were cultured in DMEM supplemented with 10% FBS, 100 units/ml penicillin, 0.1 mg/ml streptomycin, and 2 mM glutamine in a humidified atmosphere of 5% CO2 and 95% air at 37°C.
Cytotoxicity Assay.
Cytotoxicity was tested by using a standard colorimetric assay according to an established protocol provided by Promega. HeLa cells were loaded in the wells of 96-well plates (20,000 cells per well) and incubated for 24 h in DMEM supplemented with 10% FBS, 100 units/ml penicillin, 0.1 mg/ml streptomycin, and 2 mM glutamine in a humidified atmosphere of 5% CO2 and 95% air at 37°C. The growth medium was then replaced with the same medium but containing 1% of FBS and increasing amounts of pHLIP (0.5, 1, 2, 4, 8, and 16 μM). After 24 h of incubation, the solution was replaced with DMEM and a colorimetric reagent (CellTiter 96 AQueous One Solution Assay) was added for 1 h followed by measuring absorbance at 490 nm in the plate reader. All samples were prepared in triplicate.
Membrane Leakage Assay.
HeLa cells were loaded in 96-well plates (2,000 cells per well) and incubated for 24 h in DMEM supplemented with 10% FBS, 100 units/ml penicillin, 0.1 mg/ml streptomycin, and 2 mM glutamine in a humidified atmosphere of 5% CO2 and 95% air at 37°C. The growth medium was then replaced with PBS buffer containing 1 mM CaCl2 and 1 mM MgCl2 at pH 7.4 or 6.5 and increasing amounts of the peptide (0.5, 2, 5, and 10 μM). After 1 h, 2 μM Ph–TRITC and 0.5 μM SYTOX–Orange (Molecular Probes) were added for 10 min in the presence of peptide and then washed with PBS buffer (pH 7.4). The rhodamine fluorescence was measured at 580 nm with excitation at 544 nm by plate reader. The cell membrane was subsequently disrupted by adding 0.5% Triton X-100 and a new portion of the 2 μM Ph–TRITC and 0.5 μM SYTOX–Orange followed by washing with PBS. The fluorescence signal was detected before and after washing, respectively. All samples were done in triplicate.
Fluorescence Microscopy.
For the fluorescence microscopy studies, the cells were grown in 35-mm dishes with 10-mm glass-bottom windows coated with collagen. Cells were washed with PBS buffer containing 1 mM CaCl2 and 1 mM MgCl2 with pH 5.5, 6.5, 7.0, or 7.4 and then incubated in PBS at the experimental pH in the absence or presence of varied concentrations (0.1–7 μM) of the pHLIP–S–S–dansyl, pHLIP–dye (Texas red or rhodamine), pHLIP–S–S–Ph–TRITC, pHLIP–Ph–TRITC, pHLIP–S–S–PNA–TAMRA, or pHLIP–S–S–ODN–FITC constructs. The medium pH was measured before and after incubation. The time of incubation was varied from 15 to 60 min. The cancer cells can acidify the medium (PBS buffer) in a few minutes if the incubation volume is small, creating the problem of maintaining constant pH 7.4 in the PBS incubation buffer. We therefore preferred to use a low density of cells in the chamber, a larger volume, and a higher phosphate concentration (up to 50 mM instead of the standard 10 mM PBS), and we routinely checked the pH before and after the experiments. The incubation was followed by the replacement of the PBS buffer with Leibovitz’s L-15 phenol free medium (supplemented with 5% FBS, 100 units/ml penicillin, 0.1 mg/ml streptomycin, and 2 mM glutamine) at the experimental pH for 1 h and then (if not indicated otherwise) at pH 7.4. To monitor multinucleated cells we used nuclear-staining dye DAPI (Sigma–Aldrich). HeLa cells were treated with pHLIP–S–S–Ph–TRITC in PBS at pH 6.5 for 1 h followed by changing solution with DMEM supplemented with 10% FBS, 100 units/ml penicillin, 0.1 mg/ml streptomycin, and 2 mM glutamine in a humidified atmosphere of 5% CO2 and 95% air at 37°C. After 48 h 0.5 μM DAPI was added followed by washing. Fluorescent images were taken by using an inverted epifluorescence microscope (Olympus IX71). Some images of cells stained with pHLIP–phalloidin–TRITC noncleavable construct were taken on the Zeiss Axioplan 2 light microscope with Zeiss LSM 5 PASCAL laser scanning module with excitation at 543 nm of He/Ne laser. The images of cells stained with dansyl were taken on a Bio-Rad MRC-1024 two-photon confocal microscope with excitation at 740 nm. Each time we performed fluorescence microscopy experiments and observed the translocation of cargo by pHLIP, we verified that labeled cells were alive by using the dead cell marker SYTOX–Green (Molecular Probes).
Flow Cytometry.
Analytic flow cytometric measurements were performed by using a FACS instrument. Ten thousand cells were analyzed in each sample. The cells were suspended in PBS at pH 6.5 or 7.4 in presence and absence of (6 μM) pHLIP–S–S–Ph–TRITC at 4°C or 37°C for 1 h. Then they were washed twice with PBS buffer (pH 7.4), resuspended in PBS (pH 7.4), and analyzed on the FACS instrument.
Supplementary Material
Acknowledgments
We thank Drs. Mira Krendel and Mark Mooseker (Yale University) for providing microscopy and cell culture facilities for some initial experiments; Rocco Carbone (Flow Cytometry Shared Resource at the Yale Cancer Center) for assistance with flow cytometric measurements; the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University; the Tissue Culture and Media Preparation shared resource of the Yale Cancer Center; the Center for Cell Imaging at Yale University; the Rhode Island Idea Network of Biomedical Research Excellence (RI-INBRE) core facility at the University of Rhode Island; and the Genomics and Sequencing Center at the University of Rhode Island. This work was supported in part by National Institutes of Health Grant GM054160 (to D.M.E.), Department of Defense Grant PCRP CDMRP PC050351 (to Y.K.R. and O.A.A.), National Center for Research Resources/National Institutes of Health RI-INBRE Grant P20 RR016457, and a Research Development Grant from the Council for Research, University of Rhode Island (to Y.K.R.).
Abbreviations
- NBD
4-chloro-7-nitrobenz-2-oxa-1,3 diazole
- PNA
peptide nucleic acid
- pHLIP
pH (low) insertion peptide
- ODN
oligonucleotide
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- Ph–TRITC
phalloidin–tetramethylrhodamine B isothiocyanate
- TAMRA
carboxytetramethylrhodamine.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 2.Lehnert U., Xia Y., Royce T. E., Goh C. S., Liu Y., Senes A., Yu H., Zhang Z. L., Engelman D. M., Gerstein M. Q. Rev. Biophys. 2004;37:121–146. doi: 10.1017/s003358350400397x. [DOI] [PubMed] [Google Scholar]
- 3.Popot J.-L., Engelman D. M. Biochemistry. 1990;29:4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- 4.Engelman D. M., Chen Y., Chin C. N., Curran A. R., Dixon A. M., Dupuy A. D., Lee A. S., Lehnert U., Matthews E. E., Reshetnyak Y. K., et al. FEBS Lett. 2003;555:122–125. doi: 10.1016/s0014-5793(03)01106-2. [DOI] [PubMed] [Google Scholar]
- 5.Van den Berg B., Clemons W. M., Jr., Collinson I., Modis Y., Hartmann E., Harrison S. C., Rapoport T. A. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 6.Osborne A. R., Rapoport T. A., van den Berg B. Annu. Rev. Cell Dev. Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- 7.White S. H., von Heijne G. Curr. Opin. Struct. Biol. 2005;15:378–386. doi: 10.1016/j.sbi.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 8.von Heijne G. FEBS Lett. 1994;346:69–72. doi: 10.1016/0014-5793(94)00296-7. [DOI] [PubMed] [Google Scholar]
- 9.Whitley P., Zander T., Ehrmann M., Haardt M., Bremer E., von Heijne G. EMBO J. 1994;13:4653–4661. doi: 10.1002/j.1460-2075.1994.tb06788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wimley W. C., White S. H. Biochemistry. 2000;39:4432–4442. doi: 10.1021/bi992746j. [DOI] [PubMed] [Google Scholar]
- 11.Dalbey R. E., Kuhn A. J. Cell Biol. 2004;166:769–774. doi: 10.1083/jcb.200405161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunt J. F., Rath P., Rothschild K. J., Engelman D. M. Biochemistry. 1997;36:15177–15192. doi: 10.1021/bi970147b. [DOI] [PubMed] [Google Scholar]
- 13.Engelman D. M., Hunt J. F. 5,739,273. U.S. Patent. 1998
- 14.McIntyre J. C., Sleight R. G. Biochemistry. 1991;30:11819–11827. doi: 10.1021/bi00115a012. [DOI] [PubMed] [Google Scholar]
- 15.Yano Y., Takemoto T., Kobayashi S., Yasui H., Sakurai H., Ohashi W., Niwa M., Futaki S., Sugiura Y., Matsuzaki K. Biochemistry. 2002;41:3073–3080. doi: 10.1021/bi011161y. [DOI] [PubMed] [Google Scholar]
- 16.Ladokhin A. S., Isas J. M., Haigler H. T., White S. H. Biochemistry. 2002;41:13617–13626. doi: 10.1021/bi0264418. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen P. E., Egholm M., Berg R. H., Buchardt O. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 18.Ray A., Nordén B. FASEB J. 2000;14:1041–1060. doi: 10.1096/fasebj.14.9.1041. [DOI] [PubMed] [Google Scholar]
- 19.Kaihatsu K., Janowski A., Corey D. R. Chem. Biol. 2004;11:749–758. doi: 10.1016/j.chembiol.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 20.Weiland J., Osborn M., Weber K. Curr. Probl. Clin. Biochem. 1977;7:11–14. [PubMed] [Google Scholar]
- 21.Wehland J., Osborn M., Weber K. Proc. Natl. Acad. Sci. USA. 1977;74:5613–5617. doi: 10.1073/pnas.74.12.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conour J. E., Graham W. V., Gaskins H. R. Physiol. Genomics. 2004;18:196–205. doi: 10.1152/physiolgenomics.00058.2004. [DOI] [PubMed] [Google Scholar]
- 23.Senderowicz A. M., Kaur G., Sainz E., Laing C., Inman W. D., Rodriguez J., Crews P., Malspeis L., Grever M. R., Sausville E. A., et al. J. Natl. Cancer Inst. 1995;87:46–51. doi: 10.1093/jnci/87.1.46. [DOI] [PubMed] [Google Scholar]
- 24.Hessa T., Kim H., Bihlmaier K., Lundin C., Boekel J., Andersson H., Nilsson I., White S. H., von Heijne G. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 25.Stubbs M., McSheehy P. M. J., Griffiths J. R., Bashford C. L. Mol. Med. Today. 2000;6:15–19. doi: 10.1016/s1357-4310(99)01615-9. [DOI] [PubMed] [Google Scholar]
- 26.Leake D. S. Atherosclerosis. 1997;129:149–157. doi: 10.1016/s0021-9150(96)06035-2. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto S., Ehara T. Am J. Physiol. 2006 in press. [Google Scholar]
- 28.Ying W., Shan-Kuo H., Miller J. W., Swanson R .A. J. Neurochem. 1999;73:1549–1556. doi: 10.1046/j.1471-4159.1999.0731549.x. [DOI] [PubMed] [Google Scholar]
- 29.Shiraishi T., Nielsen P. E. Nucleic Acids Res. 2004;32:4893. doi: 10.1093/nar/gkh820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buck A. C., Shen C., Schirrmeister H., Schmid-Kotsas A., Munzert G., Guhlmann A., Mehrke G., Klug N., Gross H. J., Bachem M., Reske S. N. Cancer Biother. Radiopharm. 2002;17:281–289. doi: 10.1089/10849780260179242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.