Abstract
CLOCK (CLK) is a core component of the transcriptional feedback loops that comprise the circadian timekeeping mechanism in Drosophila. As a heterodimer with CYCLE (CYC), CLK binds E-boxes to activate the transcription of rhythmically expressed genes within and downstream of the circadian clock, but this activation unexpectedly occurs at times when CLK is at its lowest levels on Western blots. Recent studies demonstrate that CLK also regulates nonrhythmic gene expression and behaviors. Despite the critical roles CLK plays within and outside the circadian clock, its spatial expression pattern has not been characterized. Using a newly developed CLK antibody, the authors show that CLK is coexpressed with PERIOD (PER) in canonical oscillator cells throughout the head and body. In contrast to PER, however, the levels of CLK immunoreactivity do not cycle in intensity, CLK is detected primarily in the nucleus throughout the circadian cycle, and CLK is expressed in nonoscillator cells within the lateral and dorsal brain, including Kenyon cells, which mediate various forms of learning and memory. These results indicate that constitutive levels of nuclear CLK regulate rhythmic transcription in circadian oscillator cells and suggest that CLK contributes to other behavioral processes by regulating gene expression in nonoscillator cells.
Keywords: CLOCK, Drosophila, circadian, oscillator cells, nonoscillator cells
The identification and analysis of clock genes in Drosophila have revealed that the circadian timekeeping mechanism is composed of 2 interlocked transcriptional feedback loops that operate in many neuronal and nonneuronal tissues (Hardin, 2004). Central to both loops are the basic helix-loop-helix (bHLH) transcription factors CLOCK (CLK) and CYCLE (CYC), which bind E-box regulatory elements as CLK-CYC heterodimers to activate genes that encode the circadian transcriptional feedback regulators PERIOD (PER), TIMELESS (TIM), VRILLE (VRI), and PAR DOMAIN PROTEIN 1ɛ (PDP1ɛ) (Allada et al., 1998; Blau and Young, 1999; Cyran et al., 2003; Darlington et al., 1998; Rutila et al., 1998). PER and TIM proteins bind to and inhibit CLK-CYC during the late night and early morning to control E-box-dependent clock gene transcription that peaks near dusk (Darlington et al., 1998; Lee et al., 1998; Lee et al., 1999), whereas VRI and PDP1ɛ proteins bind VRI/PDP1ɛ-boxes to control rhythms in Clk transcription that peak near dawn (Cyran et al., 2003; Glossop et al., 2003). Clk mRNA and protein levels both cycle with a peak phase when PER and TIM inhibit CLK-CYC activity around dawn (Lee et al., 1998), but CLK-dependent activation of circadian E-box-regulated genes around dusk suggests that CLK is always present. In addition to controlling rhythms in clock gene expression, these feedback loops also control rhythms in the expression of clock “output” genes, which mediate rhythms in metabolism, physiology, and behavior (Ceriani et al., 2002; Claridge-Chang et al., 2001; Lin et al., 2002; McDonald and Rosbash, 2001; Ueda et al., 2002).
A dominant negative form of CLK lacking most of the activation domain (ClkJrk) not only abolishes behavioral rhythms (Allada et al., 1998) but also eliminates molecular rhythms in fly heads (Ceriani et al., 2002; Claridge-Chang et al., 2001; McDonald and Rosbash, 2001; Ueda et al., 2002), which suggests that all rhythmic transcription is directly or indirectly CLK dependent. In addition, many noncycling transcripts are either up- or down-regulated in ClkJrk flies (McDonald and Rosbash, 2001). These noncycling transcripts could represent direct CLK targets (e.g., transcripts having long half-lives) or indirect CLK targets (e.g., transcripts controlled by CLK-dependent transcription factors having long half-lives) in oscillator cells (i.e., cells that show rhythms in PER and TIM abundance). Alternatively, these noncycling transcripts could represent direct CLK targets in cells that lack circadian oscillators. Several nonrhythmic behavioral phenomena such as the homeostatic control of sleep (Hendricks et al., 2003; Shaw et al., 2002) and sensitization to drugs of abuse (Andretic et al., 1999) are also affected by a subset of clock genes, including Clk, but the cells that mediate these phenomena have not been identified. Although PER-expressing cells have been well characterized in fly heads and are often thought of as canonical circadian oscillator cells, it has not been possible to determine whether CLK, as an activator of per transcription, is present only in oscillator cells—particularly in light of its regulation of noncycling transcripts and its affect on nonrhythmic behavioral phenomena.
To determine whether CLK expression is restricted to oscillator cells, we generated a new antibody that could be used for immunohistochemical and whole-mount immunofluorescent detection of CLK. We find that CLK is present primarily in the nuclei of canonical oscillator cells at all times of day and displays little if any cycling in signal intensity. CLK is found in many cells that lack PER expression (i.e., nonoscillator cells) in the brain, which may account for CLK regulation of nonrhythmic transcripts and phenomena.
MATERIALS AND METHODS
Generation of CLK Antibody
Full-length CLK antigen was produced in baculovirus using the Bac-N-Blue system (Invitrogen). The Clk coding region was amplified via PCR (5’ primer: 5’ GACCCGAAAATGGACGAC 3’; 3’ primer: 5’ TTGACTACTGCCTGGGGC 3’) and directly inserted into the pBlueBac4.5/V5-His-TOPO vector to generate an in-frame C-terminal fusion with 6xHIS and V-5 epitope tags. The resulting plasmid, pBlueBac-CLK, was then recombined into the Bac-N-Blue baculoviral DNA vector in Sf9 cells. Recombinant baculovirus containing the Clk coding region, referred to as baculo-CLK, were identified as blue plaques on X-gal-containing media. Purified baculo-CLK was grown on Sf9 cells to generate a high titer stock, and this stock was used for mass transfection of Sf9 cells (Orbigen, Inc.).
One liter of baculo-CLK-infected Sf9 cells were lysed in 6M guanidium hydrochloride. The cell lysate was adjusted to pH 8.0, and CLK was purified over a nickel column (Hi-Trap Chelating HP, Amersham Pharmacia Biotech AB). Elutions were tested for CLK using the previously characterized CLK antibody (Lee et al., 1998) and HIS antibody (Sigma). All fractions containing pure CLK protein were pooled, lyophilized, and resuspended in water. The amount of purified CLK antigen was then calculated, and 2.0 mg was sent for antibody production in guinea pigs (Cocalico Biologicals, Inc.).
Immunoblotting
CLK antisera were tested on Western blots containing head extracts from wild-type (Canton-S) and ClkJrk flies. Flies were entrained for at least 3 days in 12-h LD cycles, then collected during LD at different ZTs, where ZT0 is lights-on and ZT12 is lights-off. Fly head isolation, head extract preparation, and Western blot analysis were carried out as described (Edery et al., 1994). The CLK antibody that gave the highest specific signal and the fewest number of cross-reacting bands (GP47) was used for all experiments. GP47 antiserum was used at a 1:3500 dilution in blocking solution, and incubation with the membrane was carried out overnight at room temperature. Horseradish peroxidase (HRP) conjugated anti–guinea pig secondary antibody was used at a dilution of 1:3000 (Abcam, Inc.). All Western blot results were repeated independently at least 3 times using freshly collected fly heads. In experiments to verify that CLK was recognized by GP47 on Western blots, 30 ng/mL of purified CLK was incubated with GP47 in 10 mL of blocking solution for 30 min before this solution was incubated with the membrane. PVDF membrane was used according to the manufacturer’s instructions to detect CLKJrk (Millipore).
Immunohistochemistry
Wild-type and ClkJrk flies were entrained and collected as described for immunoblotting. Consecutive 12-μm cryosections were collected and air-dried for 30 to 60 min. Horseradish peroxidase immunochemistry was performed as described (Hao et al., 1999), except that sections were incubated with GP47 at a dilution of 1:7500 (horizontal head sections), 1:3500 (frontal head sections), or 1:1500 (body sections) for 1 h at room temperature. Mutant and wild-type flies were sectioned, collected, and treated on the same slide. For each genotype, CLK immunostaining was repeated independently at least 3 times at every time point. In experiments to verify that immunostaining was due to CLK, 30 ng/mL of purified CLK (final concentration) was added to blocking buffer containing CLK antibody (1:1500 dilution) for 30 min before this buffer was incubated with samples.
Whole-Mount Immunofluorescence
Wild-type flies were entrained and collected as described for immunoblotting. Brains were dissected at ZT21 for colocalization of CLK with PER, CLK with EYELESS (EY), and PER with EY. Brains were also processed for colocalization of CLK with PER at ZT9 and CLK with PIGMENT DISPERSING FACTOR (PDF) at ZT9 and ZT21. PER was used as a marker for oscillator cells (Helfrich-Forster, 2003), and EY was used as a marker for Kenyon cells (KCs) (Callaerts et al., 2001). Brains were prepared for immunofluorescent detection as described (Mardon et al., 1994), except that 3% normal goat serum was added during the blocking step to reduce nonspecific signal. CLK antiserum was diluted in PAXD to a final concentration of 1:15,000. Polyclonal PER antiserum from rabbit was preabsorbed with per01 embryos as described (Cheng and Hardin, 1998), then diluted to a final concentration of 1:15,000 in PAXD. Polyclonal PDF antiserum from rabbit was diluted to a final concentration of 1:500 in PAXD. Polyclonal EY antiserum from rat was diluted to a final concentration of 1:400 in PAXD. Cy-3 (FITC) conjugated anti–guinea pig and anti-rabbit secondary antibodies (Jackson ImmunoResearch) and Alexafluor conjugated anti-rabbit and anti-rat secondary antibodies (Molecular Probes) were diluted to a final concentration of 1:200 in PAXD. After antibody incubations were complete, brains were mounted with Vectashield mounting media (Vector Laboratories). Brains were examined via confocal microscopy (Zeiss LSM 310) using a 488-nm laser to detect Alexafluor immunofluorescence and a 586-nm laser to detect Cy-3 immunofluorescence. Confocal images were scanned at a 1-μm thickness and acquired using LSM software. For each set of experiments, 6 or more brains were analyzed with similar results. In experiments to verify that CLK immunofluorescence was specific, 30 ng/mL of purified CLK (final concentration) was added to PAXD containing CLK antibody for 30 min before this solution was incubated with samples.
RESULTS
CLK Accumulates in Oscillator and Nonoscillator Cells
To confirm that newly generated antisera detected CLK, we probed fly head extracts from wild-type and clock mutant flies. In wild-type flies, previously characterized CLK antisera detect high levels of CLK between late night and early morning and low levels of CLK between midday and early evening on Western blots (Lee et al., 1998). Newly generated CLK antiserum detects a similar rhythm in CLK abundance on Western blots as well as a previously described rhythm of phosphorylation, in which the apparent molecular weight of CLK is highest when it is most abundant around dawn and lowest when CLK is least abundant around dusk (Kim et al., 2002; Lee et al., 1998) (Fig. 1A). As expected, the new CLK antiserum does not detect truncated CLKJrk protein in the ~115- to 150-kD range of wild-type CLK (Fig. 1A), but low levels of CLKJrk are detected at the predicted molecular weight of ~87 kD (Fig. 1B). To further test the specificity of our CLK antiserum, we incubated purified CLK antigen with CLK antiserum to block any CLK-specific signal and found that the CLK band was eliminated (Fig. 1C). Taken as a whole, these Western results demonstrate that our newly generated CLK antiserum readily detects CLK.
Figure 1.
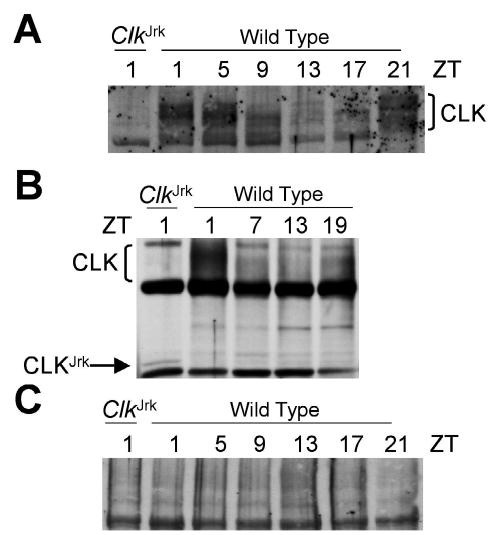
Western analysis of CLOCK (CLK) in wild-type and ClkJrk mutant flies. (A) Total head extracts were prepared from wild-type and ClkJrk flies collected at the ZTs indicated and probed using GP47 antiserum to reveal CLK levels. (B) Experiment performed identically to A, except the gel was run for a shorter time to retain the ~87-kD CLKJrk band. (C) Experiment performed identically to A except that purified CLK was added to specifically block CLK detection.
To determine the specificity of our CLK antiserum in situ, immunostaining was compared between wild-type flies expressing high levels of CLK at ZT21 and ClkJrk mutants that express very low levels of truncated CLK. For this comparison, sectioned wild-type and ClkJrk flies were placed on the same slides and processed in parallel. Since per and tim expression is virtually eliminated in ClkJrk flies (Allada et al., 1998), CLK is expected to be expressed in all circadian oscillator cells. In wild-type heads, CLK is detected in retinal photoreceptors; in the lateral protocerebrum (LP), which contains the lateral neurons (LNs) that control locomotor activity rhythms (Helfrich-Forster, 2003); and in the dorsal brain (Fig. 2A,C). However, immunostaining in the LP could not definitively be attributed to LNs since CLK immunoreactivity is detected in more cells than those previously characterized as oscillator cells (i.e., PER-expressing cells) in the lateral protocerebrum and dorsal brain based on the number of cells staining in serial sections (data not shown) and on cells that express CLK but not PER or PDF in brains (see Figs. 5, 6). We refer to these CLK-positive and PER- or PDF-negative cells as nonoscillator cells. In ClkJrk flies, CLK immunoreactivity is sharply reduced or eliminated in almost all cells that were CLK immunoreactive in wild-type flies (Fig. 2B,D). CLK immunoreactivity is essentially eliminated in photoreceptors and the lateral protocerebrum in ClkJrk flies (Fig. 2B) but can be detected at lower levels in cells corresponding to dorsal neurons (DNs) and nonoscillator cells in the dorsomedial portion of the brain (Fig. 2D). The presence of CLK immunoreactivity in characterized cells and brain regions known to harbor clock cells in wild-type flies, along with the reduction or elimination of CLK immunoreactivity in ClkJrk flies, demonstrates that our CLK antibody specifically detects CLK in situ. The low levels of truncated CLKJrk protein can be detected in oscillator cells if higher concentrations of CLK antiserum (1:1500) are used (data not shown). As an independent verification of CLK immunostaining specificity, sections were incubated with CLK antibody that had been preabsorbed with purified CLK antigen. This treatment eliminated all CLK immunoreactivity, further indicating that our antibody indeed detects CLK (Fig. 2E).
Figure 2.
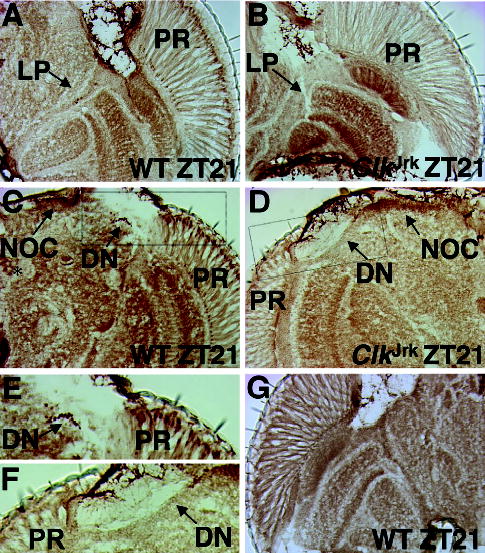
Immunohistochemical detection of CLOCK (CLK) in wild-type and ClkJrk heads. (A) Wild-type fly collected at ZT21, cryosectioned in the horizontal orientation, and analyzed for CLK immunostaining with GP47 antiserum. CLK immunostaining in the head (right hemisphere) is shown, where anterior is at the top. PR, photoreceptor cells; LP, lateral protocerebrum. (B) Identical to A except that a ClkJrk fly was analyzed. Flies shown in panels A and B were processed for immunohistochemistry on the same slide. Abbreviations are as in panel A. (C) Identical to A except that the fly was sectioned in a frontal orientation, where dorsal is at the top. A left brain hemisphere is shown. DN, dorsal neurons; NOC, nonoscillator cells. The asterisk denotes CLK staining not seen in other wild-type flies. Dashed box represents the region magnified is panel E. (D) Identical to C except that a ClkJrk fly was analyzed and a right hemisphere is shown. Flies shown in panels C and D were processed for immunohistochemistry on the same slide. Dashed box represents the region magnified in panel F. (E) Boxed region in panel C has been magnified. Abbreviations are as described in panels A and C. (F) Boxed region in panel D has been magnified. Abbreviations are as described in panels A and C. (G) Experiment performed identically to that in panel A except that purified CLK was added to specifically block CLK immunostaining.
Figure 5.
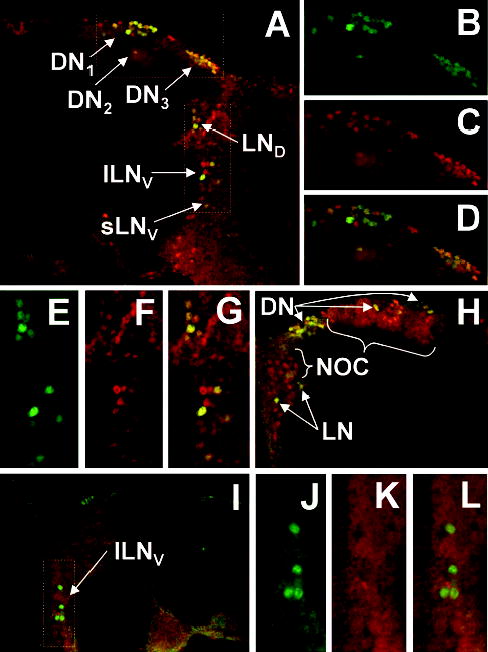
Colocalization CLOCK (CLK) and PERIOD (PER) immunofluorescence in the brain. (A) Brains from wild-type flies collected at ZT21 were dissected, prepared for CLK and PER whole-mount immunofluorescence, and imaged via confocal microscopy. A 1-μm optical section through the right hemisphere of a brain is shown, where dorsal is at the top. Colocalization of CLK (red) and PER (green) immunofluorescence in this superimposed dual laser image is seen as yellow. Arrows point to the different groups of oscillator neurons that coexpress PER and CLK. LND, dorsal lateral neurons; lLNV, large ventral lateral neurons; sLNV, small ventral lateral neurons; DN1, dorsal neuron 1; DN2, dorsal neuron 2; DN3, dorsal neuron 3. (B) Enlarged version of PER immunofluorescence in the dorsal region of the brain shown in A. (C) CLK immunofluorescence in the same region as panel B. (D) Superimposed dual laser image of PER and CLK immunofluorescence in the same region as panels B and C. (E) Enlarged version of PER immunofluorescence in the lateral region of the brain shown in A. (F) CLK immunofluorescence in the same region as panel E. (G) Superimposed dual laser image of PER and CLK immunofluorescence in the same region as panels E and F. (H) Superimposed dual laser image of a 1-μm optical section through the left hemisphere of a brain, where dorsal is at the top. CLK is colocalized with PER in dorsal neurons (DN) and lateral neurons (LN) but is also detected in cells that do not express PER (NOC). (I) Superimposed dual laser image of a plane through the left hemisphere of a brain (oriented dorsal to the top) in which CLK immunofluorescence has been blocked by the addition of purified CLK. (J) Enlarged version of PER immunofluorescence in the lateral region of the brain shown in I. (K) CLK immunofluorescence in the same region as panel J. (L) Superimposed dual laser image of PER and CLK immunofluorescence in the same region as panels J and K.
Figure 6.
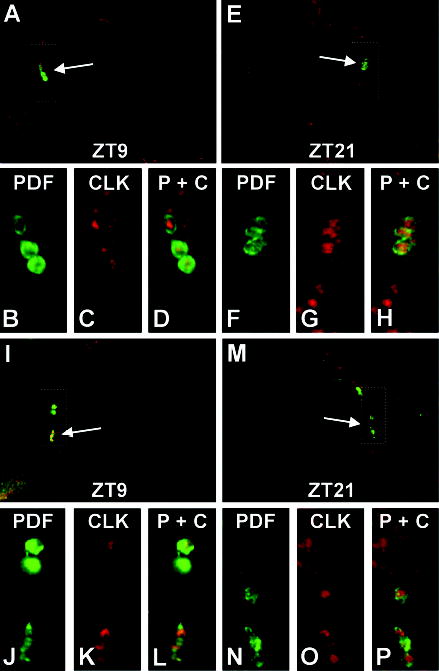
Immunolocalization of CLOCK (CLK) in PIGMENT DISPERSING FACTOR (PDF)–expressing cells. Brains from wild-type flies collected at ZT9 (panels A and I) or ZT21 (panels E and M) were dissected, prepared for CLK and PDF whole-mount immunofluorescence, and imaged via confocal microscopy. The 1-μm optical sections through left (panels A and I) or right (panels E and M) brain hemispheres are shown, where dorsal is at the top. CLK immunofluorescence is shown in red and PDF immunofluorescence is shown in green. (A) Arrow points to large ventral lateral neurons (lLNVs) that coexpress PDF and CLK. Box represents the area enlarged in panels B, C, and D. (B) Enlarged version of PDF immunofluorescence in the brain area shown in A. (C) CLK immunofluorescence in the same region as panel B. (D) Superimposed dual laser image of PDF and CLK immunofluorescence in the same region as panels B and C. (E) Arrow points to lLNVs that coexpress PDF and CLK. Box represents the area enlarged in panels F, G, and H. (F) Enlarged version of PDF immunofluorescence in the brain area shown in E. (G) CLK immunofluorescence in the same region as panel F. (H) Superimposed dual laser image of PDF and CLK immunofluorescence in the same region as panels F and G. (I) Arrow points to small ventral lateral neurons (sLNVs) that coexpress PDF and CLK. Box represents the area enlarged in panels J, K, and L. (J) Enlarged version of PDF immunofluorescence in the brain area shown in I. (K) CLK immunofluorescence in the same region as panel J. (L) Superimposed dual laser image of PDF and CLK immunofluorescence in the same region as panels J and K. (M) Arrow points to sLNVs that coexpress PDF and CLK. Box represents the area enlarged in panels N, O, and P. (N) Enlarged version of PDF immunofluorescence in the brain area shown in M. (O) CLK immunofluorescence in the same region as panel N. (P) Superimposed dual laser image of PDF and CLK immunofluorescence in the same region as panels N and P.
Circadian oscillator cells are not only present in the head but also reside in many peripheral tissues in Drosophila (Bell-Pedersen et al., 2005). Sectioned wild-type animals collected at ZT21 were immunostained with CLK antiserum to determine if oscillators in peripheral tissues express CLK. CLK immunoreactivity was detected in the cardia, gut, Malpighian tubules, and fat body (Fig. 3A–D), each of which are known oscillator tissues (Bell-Pedersen et al., 2005). Importantly, the punctate immunostaining seen in these body oscillator cells is lacking flight muscles (Fig. 3C,D), which are not known to harbor circadian oscillators. Localization of CLK immunoreactivity to peripheral tissues that contain circadian oscillators further demonstrates the specificity of this CLK antiserum and strengthens the linkage between CLK expression and circadian oscillator function.
Figure 3.
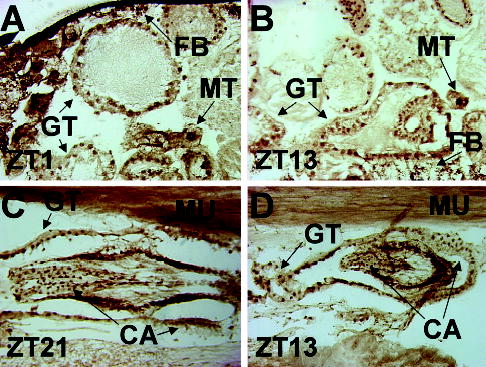
Immunohistochemical detection of CLOCK (CLK) in wild-type fly bodies. Wild-type flies were collected at the times indicated, cryosectioned in the sagittal orientation, and analyzed for CLK immunostaining using GP47 antiserum. (A, B) CLK immunostaining in the abdomen is shown where dorsal is at the top and anterior is to the right. GT, gut; FB, fat body; MT, Malpighian tubule. (C, D) CLK immunostaining in the thorax is shown where dorsal is at the top and anterior is to the right. CA, cardia; MU, flight muscle.
CLK Is Present at Constant Levels and Is Predominantly Nuclear
The rhythms in CLK abundance seen on Western blots would predict that CLK immunoreactivity in oscillator cells also cycles in situ. To test this prediction, wild-type flies were collected every 4 h during an LD cycle, cryosectioned, and immunostained with CLK antiserum. On these horizontal sections, CLK immunostaining was detected in photoreceptors and cells in the lateral protocerebrum, but the intensity of CLK immunostaining in these cells did not vary appreciably between time points (Fig. 4). The lack of rhythms in CLK immunostaining intensity is particularly surprising in photoreceptors given that these cells account for ~80% of oscillator cells in the head (Glossop and Hardin, 2002), and CLK levels cycle • 5-fold in head extracts (Lee et al., 1998) (Fig. 1A). These immunostaining results contrast with the rhythms in CLK abundance on Western blots and suggest that similar levels of CLK are present whether E-box-dependent transcription is being activated (i.e., mid to late morning and early evening) or repressed (i.e., mid to late evening and early morning).
Figure 4.
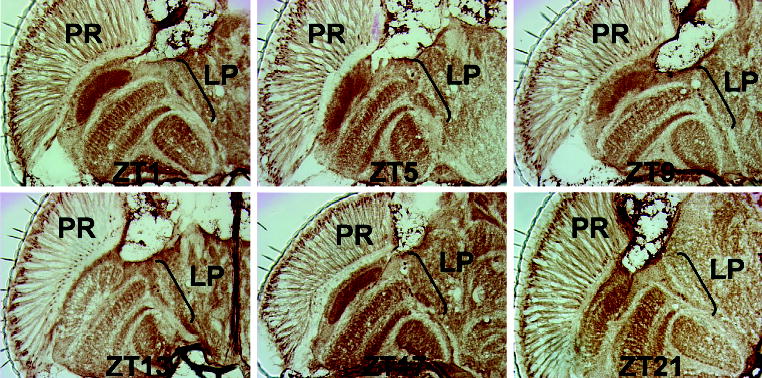
Time course of CLOCK (CLK) immunohistochemistry in wild-type fly heads. Wild-type flies were collected at the times indicated, cryosectioned in the horizontal orientation, and analyzed for CLK immunostaining using GP47 antiserum. For each time point, CLK immunostaining in the head (left hemisphere) is shown, where anterior is at the top. Abbreviations are as described in Figure 2A.
At each time point, CLK immunostaining is detected primarily in photoreceptor nuclei, which lie near the apical surface of the compound eye (Figs. 2A, 4). Whether CLK is primarily localized to nuclei in LNs could not be determined definitively via immunohistochemical staining, but this question will be addressed further below using coimmunofluorescent localization. In fly bodies, CLK is detected primarily in nuclei of cuboidal and columnar epithelia from the cardia and gut, adipose cells that comprise the fat body, and large ovoid Malpighian tubule epithelia around dawn (Fig. 3A,C) or dusk (Fig. 3B,D). Detection of CLK in the nucleus at different circadian times is consistent with E-box binding and transcriptional activation during the mid to late day and early evening and PER-dependent repression of CLK-CYC in the nucleus during the mid to late evening and early morning (Hardin, 2004).
CLK Is Present in Nuclei of Oscillator and Nonoscillator Cells in Brains
Immunohistochemical analysis of CLK spatial expression indicates that CLK is present in oscillator cells within the head and body. However, the large number of neurons and glia in the brain precludes the positive identification of oscillator cells based solely on location. To determine whether CLK is expressed in brain oscillator cells, brains were dissected from flies collected at ZT21 and prepared for whole-mount coimmunofluorescent detection using PER and CLK antisera. At ZT21, PER is bound to CLK-CYC in the nucleus (Lee et al., 1998), thus enhancing our ability to detect colocalization with CLK.
There are 6 groups of neurons in the brain that contain circadian oscillators based on PER expression: 3 groups of neurons in the lateral protocerebrum (dorsal lateral, LND; large ventral lateral, lLNV; small ventral lateral, sLNV) and 3 groups of neurons in the dorsal brain (dorsal 1, DN1; dorsal 2, DN2; dorsal 3, DN3) (Helfrich-Forster, 2003). Confocal images of wild-type brains show that CLK immunofluorescence is present in all 6 groups of PER-expressing neurons (Fig. 5A–H), confirming that CLK is expressed in all canonical brain oscillator cells. Within these oscillator cells, the overlap in PER and CLK immunofluorescence at ZT21 indicates that CLK is primarily nuclear, which is consistent with the punctate immunohistochemical staining in the lateral protocerebrum (Figs. 2A, 4). Since PER falls to low levels in brain neurons during the mid to late day and early night (Zerr et al., 1990), it could not be used to mark oscillator cells during this phase of the circadian cycle. As an alternative, we employed the neuropeptide PDF, which constitutively expressed in the cell bodies of lLNVs and 4 of the 5 sLNVs (Park et al., 2000). Confocal images of brains from wild-type flies collected at ZT9 and ZT21 show that the majority of CLK immunofluorescence is surrounded by cytoplasmic PDF immunofluorescence in both lLNVs (Fig. 6A–H) and sLNVs (Fig. 6I–P). These 1-μm optical sections contain different portions of LNV nuclei, thus leading to variations in CLK staining intensity within and between time points. These results demonstrate that CLK is primarily nuclear in these cells, whether E-box-dependent transcription is being activated (ZT9) or repressed (ZT21). These results, along with those showing that CLK is primarily localized to photoreceptor and body oscillator cell nuclei, suggest that CLK is continuously present in nuclei of all oscillator cells.
CLK is expressed in cells that do not express PER, which are consequently defined as nonoscillator cells. These nonoscillator cells are present in both the lateral and dorsal brain near known oscillator cells (Fig. 5A–H). When CLK immunostaining in the brains of flies collected at ZT9 and ZT21 was compared, no differences in intensity were detected (data not shown), indicating that CLK levels are constant in oscillator and nonoscillator cells alike. CLK immunofluorescence in oscillator and nonoscillator cells is blocked when CLK antiserum is preabsorbed with purified CLK antigen, thus demonstrating that the immunofluorescent signal is due to CLK (Fig. 5I–L). Preabsorbing CLK antisera with a similar amount of purified bovine serum albumin (BSA) did not diminish the CLK signal (data not shown). These results demonstrate that CLK is present in both oscillator and nonoscillator cells in the brain and that CLK is nuclear at least in oscillator cells.
Kenyon Cells Are CLK-Expressing Nonoscillator Cells
The presence of CLK in nonoscillator cells suggests that it is involved in regulating clock-independent processes. One large group of nonoscillator cells that express CLK is situated in the dorsal brain near the DNs (Fig. 5H). Based on their location, these nonoscillator cells could correspond to KCs, whose axonal and dendritic projections form the mushroom bodies (MBs) that are necessary for olfactory learning and memory (Heisenberg, 2003). Although several genes are preferentially expressed in mushroom bodies, we chose to use EY because it is localized to KC nuclei (Callaerts et al., 2001).
Brains were dissected from adults collected at ZT21 and prepared for whole-mount coimmunofluorescent detection using EY and CLK antisera. CLK is present in essentially all EY-expressing KCs (Fig. 7A–D). In this focal plane, it is also apparent that CLK is expressed in several clusters of cells situated near the KCs that do not express EY, and EY is expressed in 2 clusters of cells medial and lateral to KCs that do not express CLK (Fig. 7A–D). Colocalization of CLK with EY indicates that CLK is present in Kenyon cell nuclei (Fig. 7D). Although PER feeds back to inhibit CLK in oscillator cells, PER is not colocalized with EY in a focal plane containing both PER-expressing DNs and EY-expressing KCs (Fig. 7E–H), thus indicating that KCs do not harbor circadian oscillators. These results demonstrate that KCs represent 1 nonoscillator cell type that expresses CLK and that CLK is present in KC nuclei.
Figure 7.
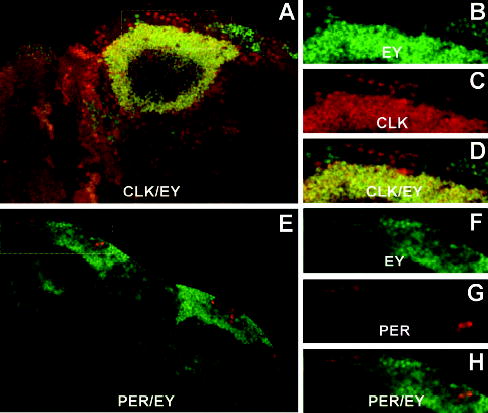
CLOCK (CLK) is expressed in Kenyon cells independent of PERIOD (PER). (A) Brains from wild-type flies collected at ZT21 were dissected, prepared for CLK and EYELESS (EY) immunofluorescence, and imaged via confocal microscopy. A 1-μm optical section through the left hemisphere of a brain is shown, where dorsal is at the top. Colocalization of CLK (red) and EY (green) immunofluorescence in this superimposed dual laser image is seen as yellow. Box represents the area enlarged in panels B, C, and D. (B) Enlarged version of EY immunofluorescence in the boxed area shown in A. (C) CLK immunofluorescence in the same region as panel B. (D) Superimposed dual laser image of EY and CLK immunofluorescence in the same region as panels B and C. (E) Superimposed dual laser image of a plane through the dorsal region of a brain (dorsal is to the top left) showing PER (red) and EY (green) immunofluorescence. Box represents the area enlarged in panels F, G, and H. (F) Enlarged version of EY immunofluorescence in the boxed is shown in E. (G) PER immunofluorescence in the same region as panel F. (H) Superimposed dual laser image of EY and PER immunofluorescence in the same region as panels F and G.
DISCUSSION
CLK Expression in Oscillator Cells
We generated a novel antibody against Drosophila CLK that is useful for immunohistochemistry to determine the spatial expression pattern of CLK in adults. This antibody detects CLK in peripheral tissues known to contain circadian oscillators and in PER-expressing brain neurons, including those that mediate locomotor activity rhythms. These results suggest that CLK is expressed in all circadian oscillator cells throughout the head and body. Expression of CLK in all oscillator cells is expected since the ClkJrk mutant virtually eliminates transcription of per and other rhythmically expressed E-box-dependent clock genes (Allada et al., 1998; Blau and Young, 1999; Cyran et al., 2003). However, per promoter-driven transgenes continue to be expressed in lLNVs and other brain neurons of ClkJrk mutant flies, which might suggest that CLK is not expressed in these cells (Kaneko and Hall, 2000). Our results confirm that CLK is indeed expressed in lLNVs and the other 5 clusters of brain oscillator neurons, which, together with the transgene expression results in ClkJrk flies, implies that clock gene expression may not be dependent on CLK in all oscillator cells. The reduction or elimination of CLK immunoreactivity in ClkJrk flies and the lack of CLK immunoreactivity after blocking with purified CLK demonstrates that our newly generated antibody specifically detects CLK in situ.
The intensity of CLK immunoreactivity in wild-type heads, including the most abundant oscillator cells—photoreceptors (Glossop and Hardin, 2002)—is essentially constant over a diurnal cycle. This result contrasts with previous Western blot results showing that CLK abundance cycles with a peak during late night/early morning and a trough during the late morning and early night (Lee et al., 1998). This discrepancy is not due to the inability of our CLK antibody to detect differences in CLK abundance since CLK immunoreactivity is low or undetectable in ClkJrk flies but is readily detectable in wild-type flies. CLK can nevertheless be detected in ClkJrk flies if higher concentrations of CLK antibody are used. Moreover, rhythms in PER abundance are readily observed using HRP immunohistochemistry (Zerr et al., 1990), indicating that this detection system is capable of distinguishing ~5-fold changes in CLK abundance. Rhythms in CLK abundance on Western blots might also be due to a time-specific masking of CLK epitopes or an inability to extract CLK from cells at certain times of day. Given that the CLK antibodies are polyclonal and were raised against the full-length protein, masking would have to occur at multiple epitopes to yield a ~5-fold reduction in signal intensity. On Western blots, the lowest levels of CLK are detected when CLK-CYC binds E-boxes to activate transcription, which may imply that binding is so strong that CLK cannot be readily released during the extraction procedure. This possibility can be tested in the future by determining whether more transcriptionally active CLK (i.e., CLK that is bound to E-boxes) is released by extraction buffers having higher detergent concentrations and/or ionic strengths.
CLK immunohistochemical staining is primarily nuclear in photoreceptors and peripheral oscillator tissues (e.g., gut, cardia, Malpighian tubules) in the thorax and abdomen, whether CLK-CYC is activating transcription during the mid to late day and early night or being inhibited by PER or PER-TIM during the mid to late night and early morning. In fly brains, CLK is colocalized with PER in the nucleus of LNVs, LNDs, and DNs at ZT21 and is present in the nucleus of PDF-expressing LNVs at both ZT9 and ZT21. Thus, in contrast to the rhythmic nuclear localization of PER and TIM, these results suggest that CLK is always present in oscillator cell nuclei, consistent with its role as a transcriptional activator when nuclear PER and/or PER-TIM are absent. Constant nuclear localization of CLK in flies also contrasts with rhythmic CLOCK nuclear localization in mammals (Kondratov et al., 2003). Paradoxically, low levels of CLOCK are present in the nucleus when CLOCK-BMAL1 target gene transcription is high, suggesting that post-translational modifications are important regulators of CLOCK transcriptional activity (Kondratov et al., 2003; Lee et al., 2001).
CLK Expression in Nonoscillator Cells
When confirming that CLK was expressed in brain oscillator cells via PER coimmunofluorescence, we noticed that CLK is also present in cells that lack PER. Given that PER is a required component of all characterized animal circadian clocks, cells that are PER negative and CLK positive presumably lack circadian oscillators. The presence of CLK-positive/PER-negative cells could be due to the lack of CYC, which is a necessary for per transcription in clock cells (Rutila et al., 1998). Alternatively, CYC may be expressed in these cells, but it may form higher affinity heterodimers with other bHLH proteins. Neither of these possibilities can currently be evaluated since the spatial expression pattern of CYC is not known. Expression of CLK in nonoscillator cells strongly suggests that some CLK-dependent regulation of nonrhythmically expressed genes in fly heads occurs in these cells. If this nonrhythmic expression is CYC independent, CLK may activate transcription in nonoscillator cells as a homodimer or a heterodimer with other bHLH transcription factors.
Most nonoscillator cells that express CLK are located in the dorsal and lateral regions of the brain. The largest group of nonoscillator cells in the brain that express CLK are KCs. This result suggests that CLK regulates some aspect of KC function in adults, which includes several forms of learning and memory and the termination of locomotor activity (Zars, 2000). A strong connection between circadian clock function and learning and memory has been established in several organisms. Rats that learned to associate a conditioned stimulus, a puff of air, with light were able to reset the circadian clock in response to a puff of air during the dark phase (Amir and Stewart, 1996). In mice, a close relative of CLOCK, called NPAS2, is rhythmically expressed in the forebrain (Reick et al., 2001), and mice lacking NPAS2 function show deficits in certain forms of long-term memory (Garcia et al., 2000). A rhythm in long-term sensitization of the siphon withdrawal reflex is also seen in Aplysia and is due to a rhythm in learning rather than recall (Fernandez et al., 2003). In Drosophila, an analysis of clock mutants revealed that per01 flies, but not ClkJrk, cyc01, or tim01 flies, are defective in long-term memory (LTM) formation after courtship conditioning (Sakai et al., 2004). Since LTM formation resulting from courtship conditioning requires MBs (McBride et al., 1999), expression of CLK in MBs is unlikely to be involved in courtship conditioning–dependent LTM. It is, nevertheless, possible that CLK expression in MBs mediates memory formation or recall arising from other associative learning paradigms.
Even though KCs do not support circadian oscillator function, CLK may nevertheless function within these cells to regulate locomotor activity rhythms. Evidence for such a role includes the following: MBs are required for the cessation of locomotor activity (Zars, 2000), projections from the sLNVs that control locomotor activity rhythms terminate close to the MB calyces (Helfrich-Forster, 1995), and cyclic AMP response element binding protein (CREB), which is involved in the consolidation of memory in MBs (Yin and Tully, 1996), mediates rhythms in cyclic AMP response element (CRE)–dependent transcription (Belvin et al., 1999). Flies bearing chemically ablated or structurally mutant MBs had relatively normal locomotor activity rhythms, however, indicating that MBs are not involved in regulating circadian rhythms in locomotor activity (Helfrich-Forster et al., 2002).
In addition to being involved in mediating MB-dependent behaviors, CLK may also contribute to the development of MBs. Defects in the axonal projections from sLNVs in ClkJrk flies suggest that CLK is required for the normal development of these clock neurons (Blanchardon et al., 2001; Park et al., 2000). ClkJrk mutants nevertheless show no gross defects in MB structure (data not shown), although more subtle defects in axonal routing or connectivity may be present. Ultimately, understanding how CLK functions in KCs will require a detailed analysis of MB anatomy and MB-dependent behavior.
Acknowledgments
We thank Mary Estes and Sue Crawford for assistance with baculovirus propagation, purification, and titering; Glen Legge for use of his lyophilizer; Michael Rea for the use of his confocal microscope; Patrick Callaerts for EY antibody; Jeff Hall for PER antibody; and Francois Rouyer for comments on the manuscript.
References
- Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- Amir S, Stewart J. Resetting of the circadian clock by a conditioned stimulus. Nature. 1996;379:542–545. doi: 10.1038/379542a0. [DOI] [PubMed] [Google Scholar]
- Andretic R, Chaney S, Hirsh J. Requirement of circadian genes for cocaine sensitization in Drosophila. Science. 1999;285:1066–1068. doi: 10.1126/science.285.5430.1066. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: Lessons from diverse species. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belvin MP, Zhou H, Yin JC. The Drosophila dCREB2 gene affects the circadian clock. Neuron. 1999;22:777–787. doi: 10.1016/s0896-6273(00)80736-9. [DOI] [PubMed] [Google Scholar]
- Blanchardon E, Grima B, Klarsfeld A, Chelot E, Hardin PE, Preat T, Rouyer F. Defining the role of Drosophila lateral neurons in the control of circadian rhythms in motor activity and eclosion by targeted genetic ablation and PERIOD protein overexpression. Eur J Neurosci. 2001;13:871–888. doi: 10.1046/j.0953-816x.2000.01450.x. [DOI] [PubMed] [Google Scholar]
- Blau J, Young MW. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/s0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- Callaerts P, Leng S, Clements J, Benassayag C, Cribbs D, Kang YY, Walldorf U, Fischbach KF, Strauss R. Drosophila Pax-6/eyeless is essential for normal adult brain structure and function. J Neurobiol. 2001;46:73–88. doi: 10.1002/1097-4695(20010205)46:2<73::aid-neu10>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Ceriani MF, Hogenesch JB, Yanovsky M, Panda S, Straume M, Kay SA. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosci. 2002;22:9305–9319. doi: 10.1523/JNEUROSCI.22-21-09305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Hardin PE. Drosophila photoreceptors contain an autonomous circadian oscillator that can function without period mRNA cycling. J Neurosci. 1998;18:741–750. doi: 10.1523/JNEUROSCI.18-02-00741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, Young MW. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci USA. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez RI, Lyons LC, Levenson J, Khabour O, Eskin A. Circadian modulation of long-term sensitization in Aplysia. Proc Natl Acad Sci USA. 2003;100:14415–14420. doi: 10.1073/pnas.2336172100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JA, Zhang D, Estill SJ, Michnoff C, Rutter J, Reick M, Scott K, Diaz-Arrastia R, McKnight SL. Impaired cued and contextual memory in NPAS2-deficient mice. Science. 2000;288:2226–2230. doi: 10.1126/science.288.5474.2226. [DOI] [PubMed] [Google Scholar]
- Glossop NR, Hardin PE. Central and peripheral circadian oscillator mechanisms in flies and mammals. J Cell Sci. 2002;115:3369–3377. doi: 10.1242/jcs.115.17.3369. [DOI] [PubMed] [Google Scholar]
- Glossop NR, Houl JH, Zheng H, Ng FS, Dudek SM, Hardin PE. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron. 2003;37:249–261. doi: 10.1016/s0896-6273(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Hao H, Glossop NR, Lyons L, Qiu J, Morrish B, Cheng Y, Helfrich-Forster C, Hardin P. The 69 bp circadian regulatory sequence (CRS) mediates per-like developmental, spatial, and circadian expression and behavioral rescue in Drosophila. J Neurosci. 1999;19:987–994. doi: 10.1523/JNEUROSCI.19-03-00987.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE. Transcription regulation within the circadian clock: The E-box and beyond. J Biol Rhythms. 2004;19:348–360. doi: 10.1177/0748730404268052. [DOI] [PubMed] [Google Scholar]
- Heisenberg M. Mushroom body memoir: From maps to models. Nat Rev Neurosci. 2003;4:266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C. The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proc Natl Acad Sci USA. 1995;92:612–616. doi: 10.1073/pnas.92.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C. The neuroarchitecture of the circadian clock in the brain of Drosophila melanogaster. Microsc Res Tech. 2003;62:94–102. doi: 10.1002/jemt.10357. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C, Wulf J, de Belle JS. Mushroom body influence on locomotor activity and circadian rhythms in Drosophila melanogaster. J Neuro-genet. 2002;16:73–109. doi: 10.1080/01677060213158. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Lu S, Kume K, Yin JC, Yang Z, Sehgal A. Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J Biol Rhythms. 2003;18:12–25. doi: 10.1177/0748730402239673. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: Transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Kim EY, Bae K, Ng FS, Glossop NR, Hardin PE, Edery I. Drosophila CLOCK protein is under posttranscriptional control and influences light-induced activity. Neuron. 2002;34:69–81. doi: 10.1016/s0896-6273(02)00639-6. [DOI] [PubMed] [Google Scholar]
- Kondratov RV, Chernov MV, Kondratova AA, Gorbacheva VY, Gudkov AV, Antoch MP. BMAL1-dependent circadian oscillation of nuclear CLOCK: Posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev. 2003;17:1921–1932. doi: 10.1101/gad.1099503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Bae K, Edery I. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron. 1998;21:857–867. doi: 10.1016/s0896-6273(00)80601-7. [DOI] [PubMed] [Google Scholar]
- Lee C, Bae K, Edery I. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: A basis for circadian transcription. Mol Cell Biol. 1999;19:5316–5325. doi: 10.1128/mcb.19.8.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Lin Y, Han M, Shimada B, Wang L, Gibler TM, Amarakone A, Awad TA, Stormo GD, Van Gelder RN, Taghert PH. Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc Natl Acad Sci USA. 2002;99:9562–9567. doi: 10.1073/pnas.132269699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardon G, Solomon NM, Rubin GM. dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development. 1994;120:3473–3486. doi: 10.1242/dev.120.12.3473. [DOI] [PubMed] [Google Scholar]
- McBride SM, Giuliani G, Choi C, Krause P, Correale D, Watson K, Baker G, Siwicki KK. Mushroom body ablation impairs short-term memory and long-term memory of courtship conditioning in Drosophila melanogaster. Neuron. 1999;24:967–977. doi: 10.1016/s0896-6273(00)81043-0. [DOI] [PubMed] [Google Scholar]
- McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- Park JH, Helfrich-Forster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci USA. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: An analog of clock operative in the mammalian forebrain. Science. 2001;293:506–509. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- Sakai T, Tamura T, Kitamoto T, Kidokoro Y. A clock gene, period, plays a key role in long-term memory formation in Drosophila. Proc Natl Acad Sci USA. 2004;101:16058–16063. doi: 10.1073/pnas.0401472101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–291. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, Hashimoto S. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem. 2002;277:14048–14052. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- Yin JC, Tully T. CREB and the formation of long-term memory. Curr Opin Neurobiol. 1996;6:264–268. doi: 10.1016/s0959-4388(96)80082-1. [DOI] [PubMed] [Google Scholar]
- Zars T. Behavioral functions of the insect mushroom bodies. Curr Opin Neurobiol. 2000;10:790–795. doi: 10.1016/s0959-4388(00)00147-1. [DOI] [PubMed] [Google Scholar]
- Zerr DM, Hall JC, Rosbash M, Siwicki KK. Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J Neurosci. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]


