Abstract
The retinal G protein-coupled receptor (RGR) is a protein that structurally resembles visual pigments and other G protein-coupled receptors. RGR may play a role as a photoisomerase in the production of 11-cis-retinal, the chromophore of the visual pigments. As the proposed function of RGR, in a complex with 11-cis-retinol dehydrogenase (RDH5), is to regenerate 11-cis-retinal under light conditions and RDH5 is expected to function in the light-independent part of the retinoid cycle, we speculated that the simultaneous loss of function of both proteins should more severely affect the rhodopsin regeneration capacity. Here, we evaluated the role of RGR using rgr−/− single and rdh5−/− rgr−/− double knockout mice under a number of light conditions. The most striking phenotype of rgr−/− mice after a single flash of light includes light-dependent formation of 9-cis- and 13-cis-retinoid isomers. These isomers are not formed in wild-type mice because either all-trans-retinal is bound to RGR and protected from isomerization to 9-cis- or 13-cis-retinal or because RGR is able to eliminate these isomers directly or indirectly. After intense bleaching, a transient accumulation of all-trans-retinyl esters and an attenuated recovery of 11-cis-retinal were observed. Finally, even under conditions of prolonged light illumination, as investigated in vitro in biochemical assays or in vivo by electroretinogram (ERG) measurements, no evidence of catalytic-like photoisomerization-driven production of 11-cis-retinal could be attained. These and previous results suggest that RGR and RDH5 are likely to function in the retinoid cycle, although their role is not essential and regeneration of visual pigment is only mildly affected by the absence of both proteins in rod-dominated mice.
Keywords: photoreceptors, retina, retinal G protein-coupled receptor, retinal pigment epithelium, retinoid cycle, visual cycle
Abbreviations used: BTP, 1,3-bis[tris(hydroxymethyl)methylamino]propane; CRALBP, cellular retinaldehyde-binding protein; ERG, electroretinogram; GPCRs, G protein-coupled receptors; HPLC, high-performance liquid chromatography; LRAT, lecithin:retinol acyltransferase; RDH, retinol dehydrogenase; RGR, retinal G protein-coupled receptor; ROS, rod outer segments; RP, retinitis pigmentosa; RPE, retinal pigment epithelium; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; WT, wild type
Photochemical and enzymatic processing of retinoids in the eye is essential for perception of the light signal and for sustaining vision by regeneration of visual pigments (McBee et al. 2001a). In vertebrates, absorption of a photon results in photoisomerization of 11-cis-retinal, which is bound to opsin via a retinylidene linkage. This photoisomerization induces conformational changes in opsin, thereby triggering the phototransduction cascade that ultimately leads to visual sensation (Polans et al. 1996; Pugh et al. 1999; Okada et al. 2001). The photoisomerized chromophore is then converted back to 11-cis-retinal by an enzymatic pathway of chemical reactions termed the retinoid cycle. Recently, a second pathway was postulated that involves a photoisomerase called retinal G protein-coupled receptor (RGR), which would produce 11-cis-retinal by harnessing light energy (Chen et al. 2001a) (reviewed in McBee et al. 2001a; Pepperberg and Crouch 2001) (Fig. 1).
Fig. 1.
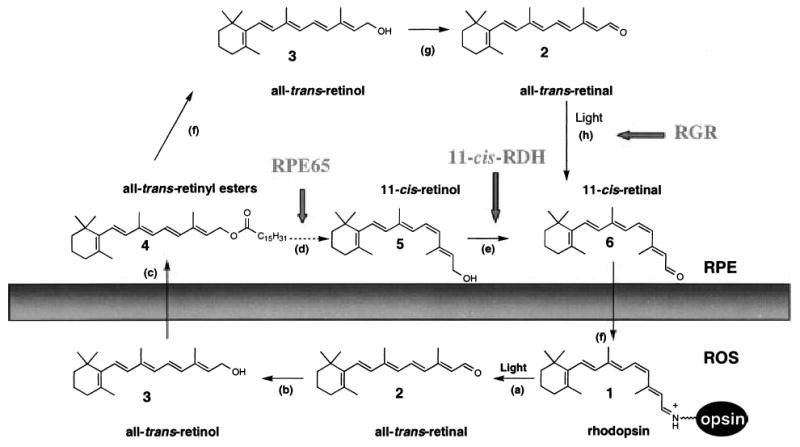
Chemistry of the retinoid cycle reactions in the vertebrate retina. The retinoid cycle reactions were reviewed recently (McBee et al. 2001a). In the rod outer segment (ROS), light causes the isomerization (reaction a) of rhodopsin chromophore, 11-cis-retinylidene (1), to all-trans-retinylidene. All-trans-retinal (2) is hydrolyzed and then reduced (reaction b) in the reaction catalyzed by all-trans-retinal-specific RDH(s). All-trans-retinol (3) diffuses to RPE where it is esterified by LRAT (reaction c) to retinyl esters (4). All-trans-retinyl esters can be hydrolyzed by a yet unidentified retinyl hydrolase (reaction f) generating all-trans-retinol. Next, the pathway branches and all-trans-retinol, or its derivative, is isomerized to 11-cis-retinol (5) in a reaction that involves an abundant RPE protein, termed RPE65 (poorly defined reactions d). 11-cis-retinol is then oxidized by 11-cis-RDH (RDH5) and other dehydrogenases (Haeseleer et al. 2002) to 11-cis-retinal (6) (reaction e) to complete the cycle (modified version from (Jang et al. 2000)). An alternative branch involves oxidation of all-trans-retinol to all-trans-retinal (reaction g) and photoisomerization by RGR of the aldehyde to 11-cis-retinal (reaction h). 11-cis-retinal diffuses back to ROS, where it recombines with opsin to reform rhodopsin.
Jiang et al. (1993) cloned RGR from a retinal pigment epithelium (RPE)-enriched cDNA library. RGR is an interesting protein because of its unique localization in specific intracellular compartments of RPE and Mu¨ller cells and its similarity to G protein-coupled receptors (GPCRs), in particular to visual pigments (Jiang et al. 1993). RGR is ~25% identical to rhodopsin and ~23% identical to a squid photoisomerase, an enzyme implicated in the photochemical production of the chromophore of the squid visual pigment (Hara-Nishimura et al. 1990). The homology to G protein-coupled receptors (GPCRs) and squid photoisomerase have led to the suggestion that RGR is either a signaling receptor coupled to a G protein, a vertebrate equivalent of the squid photoisomerase (Jiang et al. 1993), or is involved in cellular transport of 11-cis-retinal through RPE and Mu¨ller cells (Pandey et al. 1994). Further studies revealed that RGR preferably binds all-trans-retinal through a retinylidene linkage. The Lys residue of RGR involved in this linkage is in an equivalent position to Lys296 in rhodopsin (Shen et al. 1994). The λmax of the pigment formed by RGR and all-trans-retinal is 469 nm (at acidic pH values) or 370 nm (at basic pH values, with highly reduced absorption intensity at longer wavelengths), with a pKa-dependent conversion at pH 6.5 (Shen et al. 1994). Irradiation of the RGR sample had only a small effect on the spectrum (Hao and Fong 1996). These spectroscopic properties led to the hypothesis that RGR in vivo functions as a blue light and UV photoisomerase, although it is important to note that in the purified system, illumination with light of λ470nm and λ370 nm had similar effects on photoisomerization (Hao et al. 2000). Isolated RGR contains all-trans-retinal (85%) and small amounts of 11-cis- and 13-cis-retinal. After illumination, the bound chromophore was in the 11-cis-retinal configuration, a conversion that occurs with a quantum yield of 0.12 (Hao and Fong 1999). Unlike opsin, however, the chromophore did not dissociate from RGR upon photoisomerization. The proposed interaction with 11-cis-retinol dehydrogenase (11-cis-RDH, RDH5) was suggested to result in reduction of 11-cis-retinal and subsequent dissociation of 11-cis-retinol (Chen et al. 2001b). The reduction of 11-cis-retinal to 11-cis-retinol by 11-cis-RDH was found to enhance the net photoisomerization of all-trans-retinal bound to RGR (Chen et al. 2001b). All-trans-retinol dehydrogenase activity was detected in the RPE. The responsible dehydrogenase(s) may provide RGR with all-trans-retinal generated from all-trans-retinol (Yang and Fong 2002). Indeed, new retinol dehydrogenases with dual specificity for both all-trans- and cis-retinols/retinals (Haeseleer et al. 2002) or specific to all-trans-retinol (Wu et al. 2002) have recently been cloned from the retina/RPE.
The role proposed for RGR in the retinoid cycle based on genetic findings closely follows the progress made on the elucidation of the biochemical properties of this protein. First, analysis of the RGR gene for mutations associated with recessive or dominant retinitis pigmentosa (RP) yielded two families with either a Ser66Arg mutation or a single base pair insertion (Gly2751-bpins), both most likely resulting in a non-functional, truncated protein (Morimura et al. 1999). The recessive inheritance of RP associated with a Ser66Arg mutation in RGR is as expected; however, the dominant inheritance of RP for the single base pair insertion was somewhat surprising. It is possible that the truncated RGR protein exhibits a dominant negative effect by interfering with ongoing biochemical processes possibly involving the retinoid metabolism. Alternatively, RGR may organize as a multimer, as was proposed recently for rhodopsin and other GPCRs (Overton and Blumer 2000; Angers et al. 2001; Dean et al. 2001; Rios et al. 2001; Fotiadis et al. 2003). Mutant mice with a disrupted rgr gene developed normal retina and RPE and, upon comparison with age-matched WT mice, did not show any morphological symptoms (Chen et al. 2001a). Constant illumination of rgr−/− mice led to depletion of rhodopsin, whereas after brief irradiation, the recovery of rhodopsin was comparable between RGR knockout and WT mice (Chen et al. 2001a).
The proposed role of RGR in vivo (Fig. 1) as a photoisomerase requires more experimental proof. First, the low quantum efficiency of RGR (approximately five times lower than rhodopsin), low abundance of RGR (~1/100 that of rhodopsin), and absorption of light through the photo-receptors before light reaches the RPE questions a high efficiency of 11-cis-retinal production by RGR in vivo. Second, mice lacking an abundant RPE protein (RPE65) involved in the enzymatic isomerization of all-trans-retinol to 11-cis-retinol (Fig. 1) showed only minute amounts of 11-cis-retinal produced by photochemical reaction, although RGR is present in these rpe65−/− mice (Van Hooser et al. 2002, see below). Therefore, in this study, we used rgr−/− single and rdh5−/− rgr−/− double knockout mice to further investigate the role of RGR and RDH5 in the retinoid cycle and to study possible additive aspects regarding retinal pathophysiology, as it was suggested that the enzymes are functionally linked (Chen et al. 2001b). Varying light intensities of different durations were used, followed by chemical analyses of retinoids and measurements of electro-retinograms (ERG).
Materials and methods
Animals
All animal experiments employed procedures approved by the University of Washington Animal Care Committee and conformed to recommendations of the American Veterinary Medical Association Panel on Euthanasia. All animals were maintained in complete darkness and all manipulations were carried out under dim red light employing a Kodak No. 1 safelight filter (transmittance > 560 nm). Six- to 12-week-old mice were used in all experiments. RPE65-deficient mice were obtained from Dr T.M. Redmond (National Eye Institute, National Institutes of Health, Bethesda, MD, USA) and genotyped as described previously (Redmond et al. 1998; Redmond and Hamel 2000). Rgr and rdh5 mice were generated and genotyped as described previously (Driessen et al. 2000; Chen et al. 2001a). Rdh5−/− rgr−/− mice were generated by crossing rgr−/− and rdh5−/− mice.
Analyses of retinoids and visual pigments
All procedures were performed under dim red light as described previously (Palczewski et al. 1999; Van Hooser et al. 2000b; Jang et al. 2001).
Retinoids
All reactions involving retinoids were carried out under dim red light. Retinoids were stored in N,N-dimethylformamide under argon at −80°C. All retinoids were purified by normal phase HPLC (Beckman, Ultrasphere-Si, 4.6 mm × 250 mm) with 10% ethyl acetate/90% hexane at a flow rate of 1.4 mL/min using an HP1100 HPLC with a diode-array detector and HP Chemstation A.03.03 software (Hewlett Packard, Palo Alto, CA, USA). Typical separation is shown in Fig. S1. The following extinction coefficients (Garwin and Saari 2000) were used for retinoids (in m/cm): all-trans-retinal, ɛ = 48 000 at 368 nm; 9-cis-retinal, ɛ = 36 100 at 373 nm; 11-cis-retinal, ɛ = 36 100 at 365 nm; 13-cis-retinal, ɛ = 38 770 at 363 nm; all-trans-retinol, ɛ = 51 770 at 325 nm; 9-cis-retinol, ɛ = 42 300 at 323 nm; 11-cis-retinol, ɛ = 34 320 at 318 nm; and 13-cis-retinol, ɛ = 48 305 at 328 nm.
Rhodopsin measurements
Typically, two mouse eyes were used per rhodopsin measurement. Mouse eyes were enucleated and rinsed with ddH2O. The lenses were removed and the eyes were cut into 3–4 pieces and frozen immediately on a dry ice/ethanol bath. Rhodopsin was extracted with 0.9 mL of 20 mm Bis-Tris propane {1,3-bis[tris(hydroxymethyl)amino]propane, BTP, pH 7.5} containing 10 mm dodecyl-α-maltoside and 5 mm freshly neutralized NH2OH·HCl. The sample was homogenized ~10 times with a Dounce tissue homogenizer and shaken for 5 min at room temperature (Eppendorf Mixer 5432). The sample was then centrifuged at 16 000 g for 5 min at room temperature (Eppendorf Centrifuge 5415C). The supernatant was collected, and the pellet was extracted a second time. The combined supernatants were centrifuged at 130 000 g for 10 min (Beckman Optima TLX centrifuge/TLA100.3 fixed angle rotor) and absorption spectra were recorded before and after a 12-min bleach (60 W incandescent bulb). The concentration of rhodopsin was determined by the decrease in absorption at 500 nm using the molar extinction coefficient ɛ = 42 000 m−1cm−1.
Photoisomerization experiments
Each of the 12 eyecups was cut into four pieces and placed in 492 μL 10 mm BTP, pH 7.5, containing 30 mm NaCl, 725 μm NAD, 725 μm NADH, and 100 μg cellular retinaldehyde-binding protein (CRALBP). The sample was kept for 30 min at in the dark, exposed to constant illumination (100 fc), or bleached using a flash lamp, all conditions at 37°C. apo-CRALBP was prepared as described previously (Stecher et al. 1999).
Electroretinograms (ERGs)
Mice were anesthetized by intraperitoneal injection using 15 μL/g body weight of 6 mg/mL ketamine and 0.44 mg/mL xylazine diluted with 10 mm phosphate buffer, pH 7.2, containing 100 mm NaCl. The pupils were dilated with 1% tropicamide. A contact lens electrode was placed on the eye with a drop of methylcellulose and a ground electrode was placed in the ear. ERGs were recorded with the universal testing and electrophysiologic system UTAS E-3000 (LKC Technologies, Inc.). The light intensity was calibrated and computer controlled. The mice were placed in a Ganzfield chamber and scotopic and photopic responses to flash stimuli were obtained from both eyes simultaneously. Flash stimuli had a range of intensities (0.00020–233 cd/s·m2), and white light flash duration was 10 ms. Between two and four recordings were made at intervals greater than 10 s, and for higher intensity intervals, intervals were 2 min or as indicated. There were no significant differences between the first and fourth flashes. Typically, 4–8 animals were used for the recording of each point in all conditions. Calculation of the rhodopsin bleaching rate was carried out as described previously (Lyubarsky et al. 2000). The bleaching level of rhodopsin was also measured by UV spectroscopy (Van Hooser et al. 2000a) and directly by HPLC retinoid analysis. The results were examined using the one-way anova test.
Immunoblot
Immunoblot blot analysis was performed as described previously (Maeda et al. 2001). Briefly, RPE and retinal extracts were prepared from mice by homogenizing the sample with the sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) sample buffer. The proteins (30 μg) were analyzed by SDS–PAGE, using a 12.5% polyacrylamide gel. Separated proteins in a gel were electrotransferred to polyvinylidene difluoride (PVDF) membranes in 10 mm BTP buffer, pH 8.4, containing 10% methanol. After blocking with 5% skim milk in PBS, the expression of proteins was probed with an appropriate primary antibody, followed by horse-radish peroxidase (HRP)-labeled secondary antibody (Amersham, NJ, USA). Immunoreactive bands were visualized by enhanced chemiluminescence (Amersham) according to the method described by the manufacturer.
Results
Mice with disrupted retinoid cycle genes
Five genetic backgrounds were used in our study to investigate the role of RGR in vivo, including four strains of pigmented mice with a targeted elimination of proteins playing a role in the retinoid cycle and WT pigmented mice as a reference model (Saari et al. 1998; Palczewski et al. 1999; Driessen et al. 2000; Jang et al. 2001; McBee et al. 2001b). Rgr−/− mice, characterized initially by Chen and coworkers (Chen et al. 2001b), were employed to identify how disruption of the rgr gene affects the production of 11-cis-retinal in the retina. In this study, we also crossed rgr−/− mice and rdh5−/− mice to obtain mutant mice with disruptions in both of these genes. The RDH5 gene product, 11-cis-retinol dehydrogenase (11-cis-RDH), has been implicated to form a complex with RGR (Chen et al. 2001b). Therefore, elimination of both RGR and 11-cis-RDH in rdh5−/−rgr−/− mice could affect the production of 11-cis-retinal more significantly than in single knockouts. A number of the visual functions of Rdh5−/− and rpe65−/− mice were also characterized previously (Redmond et al. 1998; Driessen et al. 2000; Jang et al. 2001; Seeliger et al. 2001; Ablonczy et al. 2002; Znoiko et al. 2002). To eliminate any interference from background light on the isomeric composition of retinoids in the eye, all mice were raised in the dark. All retinoids were identified by co-elution with authentic retinoids, on-line UV spectroscopy, and, in some cases, by mass spectrometry. Retinoid analyses of rgr−/− mice showed 530 ± 97 pmol/eye of 11-cis-retinal, an amount comparable to mice of other genetic backgrounds used in our studies, including WT mice (Fig. 2, Table 1). This observation along with histological analyses (Driessen et al. 2000; Chen et al. 2001a) revealed that the amount of rhodopsin and formation of rod outer segments (ROS) is unaffected in rgr−/− and rdh5−/− mice. The mice differed in their level of retinyl esters and their isomeric composition. Rgr−/− mice had elevated amounts of all-trans-retinyl esters (103 ± 29 pmol/eye versus WT 28.5 ± 13) and 13-cis-retinyl esters (30 ± 13 pmol/eye versus WT almost undetectable) (Table 1). Other non-polar retinoids were present in comparable low amounts. Rdh5−/− mice accumulated high levels of 11-cis-retinyl esters (18 ± 4.5 pmol/eye) and 13-cis-retinyl esters (82 ± 19 pmol/eye) and only small amounts of all-trans-retinyl esters (32 ± 15 pmol/eye). Rdh5−/− rgr−/− displayed a phenotype similar to that of rdh5−/−, with a high accumulation of cis-retinyl esters (Fig. 2). In all cases, the majority (> 95%) of retinyl esters were present in RPE (data not shown).
Fig. 2.
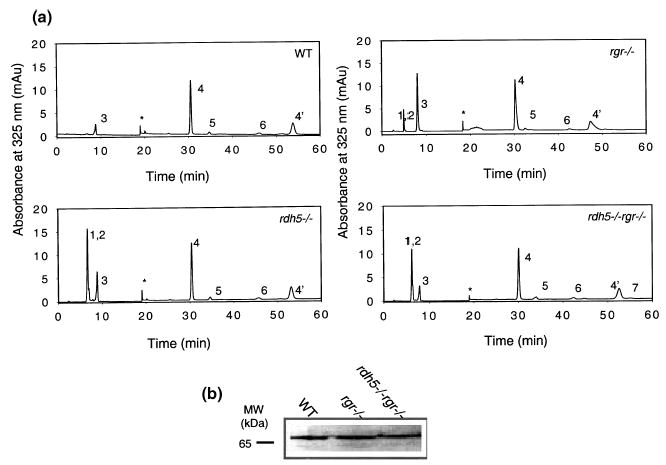
(a) Chromatographic separation of non-polar retinoids from mice of different genetic backgrounds used in this study. Retinoids were extracted from the eye and separated on normal-phase HPLC. The peaks correspond to the following retinoids: 1, 13-cis-retinyl esters; 2, 11-cis-retinyl esters; 3, all-trans-retinyl esters; 4, 4′ syn-and anti-11-cis-retinal oximes; 5, syn-all-trans-retinal oximes; 6, 11-cis-retinol; 7, all-trans-retinol. *Artifact related to a change in the solvent composition. (b) Immunoblot analysis of RPE65 in RPE extracts obtained from WT, rgr−/−, or rdh5−/−rgr−/− mice, as detected by anti-RPE65 polyclonal antibody (1 : 5000 dilution).
Table 1.
Retinoid contents in mice from different genetic backgrounds*
| Retinoid | rdh5−/− (pmol/eye) | rgr−/− (pmol/eye) | rdh5−/− rgr−/− (pmol/eye) | WT (pmol/eye) |
|---|---|---|---|---|
| 13-cis-Retinyl esters/ | 82 ± 19 | 30 ± 13 (together) | 62 ± 29 | Trace |
| 11-cis-Retinyl esters | 18 ± 4.5 | – | 8 ± 3.5 | – |
| all-trans-Retinyl esters | 32 ± 15 | 103 ± 29 | 25 ± 12 | 28.5 ± 13 |
| 11-cis-Retinal | 490 ± 35 | 530 ± 97 | 505 ± 25 | 535 ± 32 |
| all-trans-Retinal | 19.7 ± 4.2 | 22 ± 7 | 15.6 ± 8.2 | 10.2 ± 3 |
| 11-cis-Retinol | 9 ± 5 | 11 ± 5 | 7 ± 2 | 6 ± 3 |
| all-trans-Retinol | ND | ND | 6.1 ± 2.0 | 10.1 ± 2.1 |
Rhodopsin was measured from the light-dependent changes at 500 nm as described previously (Saari et al. 1998; Palczewski et al. 1999). The levels of rhodopsin are comparable to those of 11-cis-retinal, as measured by HPLC analysis (Saari et al. 1998). ND, not detected (below detection level). The results are presented with standard deviation, and n was between 5 and 10.
Using immunoblotting, the protein levels of lecithin/retinol acyltransferase (LRAT), RDH5, RDH11, CRALBP (data not shown), or RPE65 (Fig. 2b) in rgr−/− mice appeared comparable to WT mice.
Flow of retinoids in rgr−/− mice
To understand how the flow of retinoids in the eye was affected by disruption of the rgr gene, dark-adapted rgr−/− mice were exposed to an intense bleach (bleaching ~40% rhodopsin), transferred to the dark, and analyzed at the indicated time points (Fig. 3). As expected, bleaching caused formation of all-trans-retinal, which was reduced to all-trans-retinol by photoreceptor all-trans-retinol dehydrogenases (McBee et al. 2001a) with kinetics comparable to WT (Fig. 3a, panel i). Consistently, we observed elevated levels of all-trans-retinol in rgr−/− mice (28 ± 8 pmol/eye at 0–40 min) compared to WT mice (7.8 ± 2 pmol/eye). Note that these amounts of retinol are only 1.5–5.2% of the total amount of 11-cis-retinal. All-trans-retinol in rgr−/− mice leveled out at WT amounts at 60 min postbleaching (Fig. 3a, panel ii). The total amount of retinyl esters also showed a clear accumulation in rgr−/− mice (133 ± 29 pmol/eye versus WT 29 ± 12), even further elevated after bleaching (for example 260 ± 26 pmol/eye at 45 min) and sustained at higher levels until 120 min postbleaching (Fig. 3a, panel iii). This accumulation of retinyl esters might result from a slower utilization of esters in the regeneration of 11-cis-retinal and might also cause slower removal of all-trans-retinol. 11-cis-Retinol kinetics appeared to be unaffected in rgr−/− mice (Fig. 3a, panel iv), suggesting that oxidation to 11-cis-retinal was still faster than other rate-limiting processes in the retinoid cycle. Finally, the accumulation and subsequent decrease of retinyl esters paralleled the rate of 11-cis-retinal decrease and formation. The decrease in esters formed after the bleach roughly paralleled synthesis of 11-cis-retinal. The increase in the ester levels mostly affected formation of all-trans-retinyl esters (Fig. 3b). In summary, disruption of the rgr gene in mice leads to significant accumulation of all-trans-retinol and retinyl esters that results in slower formation of 11-cis-retinal. These properties are inconsistent with the proposed role for RGR as only a photoisomerase.
Fig. 3.
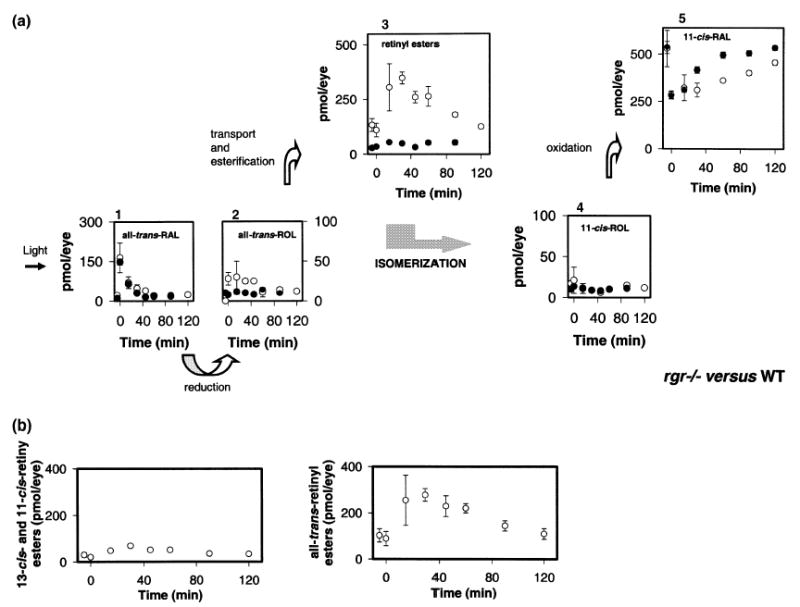
Kinetics of retinoid recovery in rgr−/− and WT mice. (a) Mice were reared in the dark. HPLC retinoid analysis was performed either before or after a flash that bleached ~40% of the visual pigment. Photoactivated rhodopsin releases all-trans-retinal (1), which is reduced to all-trans-retinol (2), transported to the RPE, and then esterified to retinyl esters (3). All-trans-retinol, or its derivative, is isomerized to 11-cis-retinol (4), which in turn is oxidized to 11-cis-retinal (5). ○, rgr−/− mice; •, data obtained from WT mice. Error bars indicate the standard error of the mean (n = 3 or 4). (b) Changes in the amounts of 13-cis-, 11-cis-, and all-trans-retinyl esters in rgr−/−mice.
Consistently, we observed the presence of 13-cis-retinal (12 ± 2 pmol/eye) and 9-cis-retinal (26 ± 6 pmol/eye) in rgr−/− mice formed as a result of illumination (Fig. 4a). These aldehydes, detected in the form of oximes, remained constant during dark adaptation (Fig. 4b). Under similar conditions, these aldehydes were below detection levels in WT mice (data not shown). To understand the conditions under which these aldehydes are formed, three groups of mice were continuously reared in the dark (Fig. 5a) and either exposed to 12 flashes (each bleaching ~40% of rhodopsin) followed by overnight dark adaptation (Fig. 5b) or exposed for 10 h of constant light from a 100-W bulb followed by overnight dark adaptation (Fig. 5c). Significant levels of 9-cis-retinal and 13-cis-retinal were formed only when mice were exposed to light, suggesting that these aldehydes were produced by photochemical reactions. This may suggest that uncomplexed all-trans-retinal (normally bound to RGR) readily undergoes photo-interconversion in vivo. The cis-aldehydes were present in RPE membranes (data not shown).
Fig. 4.

Kinetics of changes in amounts of retinal isomers in rgr−/− mice. Conditions of this experiment are described in Fig. 2. (a) Typical separation profile and spectra of retinal oximes from rgr−/− mice. 4, 11-cis-retinal; 5, all-trans-retinal; 8, 9-cis-retinal; and 9, 13-cis-retinal. (b) Changes in the amounts of retinals.
Fig. 5.

Formation of 9-cis- and 13-cis-retinals after bleaching. Rgr−/−mice were reared in the dark (a), exposed to 12 flashes (~40% bleach) with 10 min intervals followed by overnight dark adaptation (different intervals from 10 to 90 min did not change the results) (b) or 10 h constant light from a 100-W bulb followed by overnight dark adaptation (c). The elution regions of 9-cis- and 13-cis-retinal oximes are marked by lines and arrows. 4,4′-syn- and anti-11-cis-retinal oximes; 5, all-trans-retinal; 6, 11-cis-retinol; 8, 9-cis-retinal; and 9, 13-cis-retinal.
Double knockout mice for rdh5 and rgr genes
RGR appears to form a complex with 11-cis-RDH (Chen et al. 2001b). In addition, RGR has been suggested to play a role in 11-cis-retinal formation in the light (photopic cycle; Chen et al. 2001a), whereas RDH5 has been shown to be important for dark adaptation (Yamamoto et al. 1999; Cideciyan et al. 2000) and, hence, 11-cis-retinal formation in the dark. Therefore, we reasoned that inactivation of both genes may amplify the phenotypes observed following inactivation of each individual gene. During the course of this study rdh5−/− rgr−/− mice were generated and analyzed for retinoid composition (Fig. 2) and ERG (below). Although rdh5−/− rgr−/− mice had normal amounts of 11-cis-retinal (and rhodopsin), the rdh5−/− rgr−/− retinoid profile is identical to that of rdh5−/− mice. The flow of retinoids was, within experimental error, similar to the flow observed in rgr−/− mice (data not shown). Therefore, in summary, it appears that the still mild phenotype of rdh5−/− rgr−/− double knockout mice displays a sum of abnormalities observed for mice with either rdh5 or rgr inactivated. Below, we will evaluate the visual properties of rdh5−/− rgr−/− mice using ERG.
Photoisomerization in vivo and in the eyecup
The next set of experiments was set to detect photoisomerization activity in rgr−/− mice. First, dark-reared mice were exposed to 12 intense flashes (bleaching ~40% of the visual pigment) with short intervals. Unexpectedly, multiple bleaches caused more pigment to be bleached in rgr−/− mice (rhodopsin content after bleaching 87 ± 20 pmol per eye) compared to WT (115 ± 18 pmol/eye) after the last flash. Slower regeneration of the visual pigment was not expected based on the postulated role of RGR as a photoisomerase. However, the rate of pigment formation in the dark was higher in rgr−/− mice (~3.2 pmol/min) compared to WT mice (~2 pmol/min) (Fig. 6). These results demonstrate that the enzymatic isomerization reaction in rgr−/− mice in the dark is capable of generating 11-cis-retinal at higher, or at least comparable, rates compared to WT mice. Based on our analysis, it is likely that the increased ester hydrolysis (Fig. 7a versus 7b) correlates with the increased 11-cis-retinal formation rate in rgr−/− mice.
Fig. 6.

Recovery of 11-cis-retinal and changes in all-trans-retinal and all-trans-retinyl esters in rgr−/− mice after multiple bleaches. Mice, WT (a) and rgr−/− (b) were reared in the dark and exposed to 12 flashes with 10 min intervals and dark adaptation for 0, 15, 45 and 60 min. The first flash bleached ~40% of the visual pigment. HPLC retinoid and rhodopsin analyses were performed as described in Materials and methods. •, all-trans-retinyl ester; ○, 11-cis-retinal; all-trans-retinal.
Fig. 7.
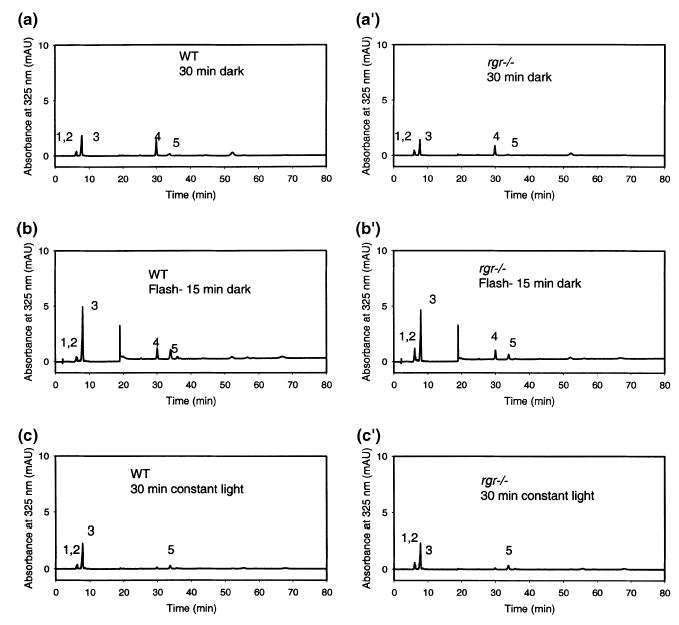
Lack of photoisomerization of retinoids to 11-cis-retinal in eyecups derived from rgr−/− and WT mice. Separation chromatograms of retinoids from eyecups incubated in the dark (a and a′); after an intense bleach (b and b′); or after constant white light illumination at 100 fc (c and c′). 1, 13-cis-retinyl esters; 2, 11-cis-retinyl esters; 3, all-trans-retinyl esters; 4, syn-11-cis-retinal oximes; 5, syn-all-trans-retinal oximes. Omission of CRALBP or NAD/NADH did not affect production of 11-cis-retinals.
Because short pulses of intense light result in two opposite effects, bleaching of pigments and the induction of photoisomerization, another set of experiments was designed in which we used freshly isolated eyecups from WT and rgr−/− mice with lysed cells in a low ionic strength buffer and from which the neural retinas were removed. Eyecups were exposed to three different light conditions: (i) dark (Figs 7a and a′), (ii) an intense bleach (Figs 7b and b′), or (iii) constant white light illumination at 100 fc (Figs 7c and c′).
We did not observe production of 11-cis-retinol in eyecup experiments when performed without the addition of an acceptor for cis-retinoids (data not shown). These experiments were performed under constant illumination or under flash experiments. This could be the result of esterification by LRAT. Therefore, we included an acceptor, CRALBP, to promote production of 11-cis-retinal and dinucleotides to support redox reactions of retinoids and to prevent esterification. Under the condition of no light, 11-cis-retinal was not present above the small amounts present as a result of contamination by the retina (rhodopsin). Neither intense bleaching nor constant light induced production of the chromophore 11-cis-retinal in eyecups derived from WT mice. Furthermore, we did not notice any differences among any of the three illumination conditions in eyecups from WT versus rgr−/− animals.
Electroretinogram analyses (ERG)
To evaluate the light response, rgr−/− and rdh5−/−rgr−/− mice were studied using ERG under multiple conditions (Hetling and Pepperberg 1999). Analysis of a- and b-waves of dark-adapted mice statistically showed no significant differences (one-way anova) in the amplitudes for WT, rgr−/−, and rdh5−/− rgr−/− mice. Light-adapted amplitudes also showed only minor variations. Furthermore, flicker responses at a fixed frequency of 10 Hz were attenuated for rdh5−/− rgr−/− mice at low intensities (data now shown). The recovery rate from a bleach that photoactivated ~1% of the visual pigment (Fig. S2) was within experimental error for all genetic backgrounds tested. However, dark adaptation in rdh5−/− rgr−/− and rgr−/− mice was significantly lower than WT after bleaching at 75 cd/s·m2 for 1 or 2 min, whereas single flash responses immediately after bleach for the both periods showed no significant differences between genetic backgrounds (Fig. 8). ERG data suggest that rod function of mice was only minimally affected by the deficiency of only rgr or both rgr and rdh5 genes. Abnormalities could be revealed only under intense illumination.
Fig. 8.

Dark adaptation after bleaching with 75 cd/s·m2 for 1 min or 2 min as measured by ERG. Mice were dark adapted overnight and bleached for 1 min (upper panels) or 2 min (lower panels) using 75 cd/s·m2 (steady state 15% bleaching level). Scotopic single flash EGRs were recorded immediately after bleaching at different intensities (a). In addition, scotopic ERGs were recorded every minute during a 30-min period using a flash intensity of 0.1 cd/s/m2. Recovery percentages of a-wave amplitudes were plotted (mean ± SE) (b). In both conditions, recovery rates from rgr−/− and rdh5−/−rgr−/− these two knockout mice were significantly lower when compared with WT (p < 0.02; one-way anova). The calculation of the rhodopsin bleaching rate was carried out as described previously (Lyubarsky et al. 2000).
Discussion
Deletion of rgr, rdh5 and rpe65 genes cause changes in retinoid content
Deletion of rgr, rdh5, and rpe65 genes (their putative roles in the retinoid cycle are presented in Fig. 1) results in changes in retinoid content in the dark (Fig. 2). Rgr−/− mice raised in complete darkness show elevated amounts of all-trans-retinyl and cis-retinyl esters. The reason for this accumulation is not yet clear, however, similar observations were made for rdh5−/− and rdh5−/− rgr−/− mice. In those mice strains, an even larger accumulation of cis-retinyl esters levels was observed. 13-cis-Retinyl esters in these mice strains could be formed because of two independent mechanisms. In rgr−/− mice, lack of RGR results in the presence of a higher concentration of free all-trans-retinal that is in an equilibrium with 13-cis-retinal. The latter compound could be reduced to 13-cis-retinol by RDH5 and subsequently esterified, generating 13-cis-retinyl esters. In addition, over-accumulation of 13-cis-retinol resulting from disruption of the RDH5 gene might be due to very low residual 13-cis-retinol dehydrogenase activity (Jang et al. 2001). Accumulation of 13-cis-retinol could result in formation of high concentrations of 13-cis-retinyl esters. RDH11 is a candidate enzyme that could be responsible for the redox reaction in the absence of 11-cis-RDH and may not be able to convert 13-cis-isomers (Haeseleer et al. 2002). In contrast to the level of all-trans-retinyl esters in rgr−/− mice, the level of 13-cis-retinyl esters appears to be unaffected by light.
Attenuation of the retinoid flow in rgr−/− mice after an intense bleach
The recovery rate of 11-cis-retinal after an intense bleach appears to be affected in rgr−/− mice. This is probably accompanied by a transient over-accumulation of all-trans-retinyl esters. RGR in the RPE is part of a multicomplex protein structure. Hence, like RDH5 and RPE65, lack of RGR may perturb the normal flow of retinoids. Regeneration after multiple flashes occurs in rgr−/− mice at rates at least comparable to those of WT mice, and probably at rates even greater because more all-trans-retinoid may be transferred to the RPE in rgr−/− mice. Thus, the esters serve as a pool for biosynthesis of 11-cis-retinal in the dark part of the retinoid cycle (Fig. 6).
Isomeric compositions of rgr−/− mice exposed to light
Upon strong bleaching of rgr−/− mice, 9-cis- and 13-cis-retinal are formed. Both derivatives are not detected in samples from WT and dark-adapted rgr−/− mice. These aldehydes may be products of a photochemical reaction and appear to be trapped in the retina and uninvolved in the flow of retinoids that leads to 11-cis-retinal production. By inference, it appears that the presence of RGR significantly prevents the formation of 9-cis- and 13-cis-isomers. This property of RGR seems to play a more important role under intense light conditions.
Lack of photoisomerization in RPE membranes containing RGR
No 11-cis-retinal production by direct photoisomerization could be detected in the RPE of rgr−/− and WT mice under various conditions. Admittedly, not all conditions can be reproduced in vitro. However, under the most obvious conditions of prolonged high intensity light and the presence of both the acceptor molecule CRALBP and dinucleotides, no significant production of 11-cis-retinal or 11-cis-retinol could be detected. Unfortunately, these results still do not provide us with clear evidence that RGR functions as an efficient photoisomerase.
Interpretation of the RPE65 phenotype in light of RGR function
We and others detected no significant production of 11-cis-retinal in rpe65−/− mice due to a disabled retinoid cycle under both dark and light conditions (Redmond et al. 1998; Van Hooser et al. 2000b; Kuksa et al. 2002; Van Hooser et al. 2002). Strong bleaches and prolonged illumination do not change this status. If RGR is involved in the production of 11-cis-retinoids, it would be able to produce 11-cis-retinal upon illumination and the observed phenotype of rpe65−/− mice would be expected to be different. As a cautionary note, it is important to emphasize that RGR could be more important in the production of 11-cis-retinoids as a supply for cones than for rods and therefore the scarcity of cones in the murine retina may obscure the role of RGR in the retina of species with a higher abundance of these photoreceptors, as is the case in humans and other primates.
Molecular model of RGR based on the high-resolution crystal structure of rhodopsin
To gain molecular insight into the arrangement of the chromophore in RGR, molecular models based on the crystal structure of bovine rhodopsin (Palczewski et al. 2000; Teller et al. 2001) were generated for RGR, Drosophila rhodopsin, and melanopsin (Fig. S3). In contrast to bovine rhodopsin, RGR, Drosophila rhodopsin and melanopsin appear to have the chromophore stably linked via retinylidene linkage in the active site. Bovine rhodopsin eventually loses its chromophore as Meta II, the signaling form of rhodopsin, decays (Okada et al. 2001). In all cases, position 113, corresponding to Glu113 in bovine rhodopsin, is substituted by a residue that cannot support counter-ion function. RGR, Drosophila rhodopsin and melanopsin may utilize for this role a Glu that corresponds to Glu181 of bovine rhodopsin as an alternative counter-ion (Terakita et al. 2000) (Fig. S4a). For example, water-polarizing Glu113 is replaced by His113 in RGR (Fig. S4). This switch in the chromophore counter-ion could be one of the reasons why the chromophore does not dissociate spontaneously, as was confirmed for Drosophila rhodopsin and RGR (reviewed in Shichida and Imai 1998; Ebrey and Koutalos 2001). Importantly, in such case, RGR would switch reversibly from all-trans-retinal to 11-cis-retinal, rather than release the chromophore as does bovine rhodopsin. Close structural homology of these pigments suggests that they are all most likely involved in signaling functions.
Recently, we performed a comprehensive sequence analysis of 277 human GPCRs from GPCR family A (Mirzadegan et al. 2003). We found that the ligand binding extracellular domains are the least conserved, while GPCRs display considerable conservation toward the cytoplasmic side, suggesting a common mechanism by which GPCRs activate G proteins. Among the highly conserved residues (60–100% frequency) within the GPCR family (Fig. S4b), 50% are present in RGR. Moreover, additional ~25% of residues in RGR are identical to rhodopsin, the remaining ~25% are sporadically present among GPCRs, and only Gly in helix VI of RGR is not present in any GPCRs from family A (Fig. 4b). The presence of modified ‘DRY’ and ‘NPXXY’ regions found in other GPCRs (GRY, and NAXXY, respectively) that are critical to G protein interaction (Okada et al. 2001; Filipek et al. 2003) further strengthen the argument that RGR is a signaling molecule. Peropsin (Sun et al. 1997), a homologous RPE receptor, has features similar to RGR, including similar gene structure (Bellingham et al. 2003). Interestingly, the chromophore of peropsin also appears to undergo 11-cis-retinal isomerization to all-trans-retinal (Koyanagi et al. 2002).
ERG analysis of rgr−/− and rdh5−/− rgr−/− mice
Retinitis pigmentosa (RP) patients with mutations in the RGR gene show severe ERG abnormalities of rods and cones accompanied by retinal and RPE degeneration (Morimura et al. 1999), whereas rgr−/− mice develop a normal retina and RPE, without any morphological symptoms or severe abnormality of ERG (Chen et al. 2001a and this study). In this study, ERG analysis of dark-adapted mice showed comparable responses for WT mice, rgr−/−, and rdh5−/− rgr−/− mice. Furthermore, flicker responses at a fixed frequency of 10 Hz were attenuated for rdh5−/− rgr−/− mice at low intensities. Dark adaptation after 1 min or 2 min of an intense (75 cd/s/m2) illumination (Fig. 8) showed significant differences only between rdh5−/− rgr−/− and rgr−/− mice.
The differences in ERG responses between humans and mice with a disrupted RGR (rgr) gene could result from a different number of cone photoreceptor cells in the two species. Cones, very infrequent in mice, differ from rods in phototransduction processes and the kinetics of regeneration (McBee et al. 2001a). Furthermore, responses of flicker ERG to weak stimuli or b-waves in the dark conditions, which reflect the function of neural retina, were attenuated in rgr−/− and rdh5−/− rgr−/− mice compared to WT mice, respectively. The function of RGR expressed in Mu¨ller cells is unclear, but the absence of RGR may have some effect on the responses generated in the neural retina.
RGR function and properties
Based on several biochemical experiments, Fong and colleagues convincingly demonstrated that RGR has the capability of converting bound all-trans-retinylidene to the 11-cis-isomer (more recent publication Chen et al. 2001b). This demonstration was carried out using a stoichiometrical amount of chromophore. However, for RGR to be an efficient photoisomerase, a catalytic-like property needs to be demonstrated, where RGR would continuously produce 11-cis-retinoids upon exposure to light (like in the conditions of experiment 6). Using transgenic mice and various techniques, we were unable to observe robust photoisomerization in the presence of RGR and/or RDH5 compared to knockout mice. The original characterization of rgr−/− mice (Chen et al. 2001a) leaves us with a few unexplained observations that can be interpreted differently based on the original and our results. A decrease in rhodopsin concentration upon exposure of mice for 8 h to 4000 lux light (Figs 2 and 3 in Chen et al. 2001a) could simply be a result of shifting retinoid to the ester pool, as shown in Fig. 6 in this paper. RGR has many characteristics that are not compatible with a high throughput photoisomerase, such as a stable arrangement of the chromophore in the active site (Figs S3 and S4), low quantum efficiency below that of rhodopsin (~1/5), and lower abundance compared to rhodopsin (~1/100). These properties would create situation where rhodopsin bleaching exceeds formation of 11-cis-retinal by RGR. Finally, perhaps the strongest evidence that questions the function of RGR in vivo as a photoisomerase comes from genetics. Lack of RPE65 results in severely diminished visual function due to a very low level of 11-cis-retinal production (reviewed in Rattner et al. 1999; McBee et al. 2001a). Double knockout mice with disrupted rgr and rpe65 genes are not different in residual visual activities from rpe65−/− mice (Van Hooser et al. 2002), although expression of RGR in rpe65−/− mice was likely unaffected (Fig. 2b). These results point to a minor role of RGR in the production of the chromophore. Whether RGR is a signaling molecule is an open question, but it is justifiable to seek further answers on its function. Possibly, the first step would be to test coupling of this receptor to different G proteins and other proteins in order to identify the pathway in which RGR is involved.
Acknowledgments
We would like to thank Dr Henry Fong for providing the rgr mice, anti-RGR antibodies, and many comments during the course of this work and on the manuscript and Dr Michael Redmond (National Eye Institute) for rpe65−/− and anti-RPE65 antibody. We are grateful to Yunie Kim and Matthew Batten for help with the manuscript preparation, Drs Volker Gerke and Alex Moise for their comments on the manuscript. This research was supported by NIH grants EY09339 and EY13385, a grant from Research to Prevent Blindness, Inc. (RPB) to the Department of Ophthalmology at the University of Washington, and a grant from the E.K. Bishop Foundation. KP is a RPB Senior Investigator. Modeling tasks were partly done in the ICM Computer Center, University of Warsaw, Poland.
Footnotes
Supplementary material
The following material is available from http://www.blackwellpublishing.com/products/journals/suppmat/JNC/JNC1741/JNC1741sm.htm
Fig. S1 Chromatogram for separation of retinyl palmitate, retinal oxime and retinol standards. Peaks were identified by retention time and UV spectra and compared to standards. The spike around 19 min resulted from changes in the solvent composition. Retinoid analysis was performed on an HP1100 HPLC equipped with a diode array detector and HP Chemstation (A.07.01) software, allowing identification of retinoid isomers according to their specific retention time and absorption maxima. A normal-phase column (Beckman Ultrasphere Si 5μ, 4.6 × 250 mm) and an isocratic solvent system of 0.5% ethyl acetate in hexane (v/v) for 15 min, followed by 4% ethyl acetate in hexane for 60 min at a flow rate of 1.4 mL/min (total 80 min), with detection at 325 nm allowed the partial separation of 11-cis-retinyl esters, 13-cis-retinyl esters, and all-trans-retinyl esters at 20°C.
Fig. S2 Recovery of ERG responses for rgr−/−, rdh5−/−rgr−/− and WT mice exposed to prior > 1% bleach. (a) ERG responses for dark-adapted mice. Traces below represent responses after 1, 120 and 300 s. (b) The plots below represent quantification of a-wave versus time after bleaching.
Fig. S3 Molecular models of the 11-cis-retinal-binding pocket of bovine and Drosophila rhodopsins, melanopsin, and RGR. Bovine rhodopsin (a, green), Drosophila (b, yellow), melanopsin (c, blue), and RGR (d, red). Homology modeling was done with InsightII v. 2000 (Accelrys, Inc.) using the 1HZX coordinates (Teller et al. 2001) as a starting structure. In (a–d), 11-cis-retinylidene-Lys296 is shown in green for amino acid residues and violet for retinyl residues within the binding sites. The numbering follows that of bovine rhodopsin.
Fig. S4 (a) Molecular models of the retinoid-binding pocket of RGR, bovine and Drosophila rhodopsin, and melanopsin in the vicinity of the counter-ion. Bovine rhodopsin (green), Drosophila (yellow), melanopsin (blue), and RGR (red). (b) Sequence alignment between bovine rhodopsin and RGR. For rhodopsin, helices are in red, and β-strands are in yellow. For RGR, residues that are 60–100% conserved among 277 identified human GPCRs are on a red background, and residues that are 80–100% conserved are on a brown background.
References
- Ablonczy Z, Crouch RK, Goletz PW, Redmond TM, Knapp DR, Ma JX, Rohrer B. 11-cis-Retinal reduces constitutive opsin phosphorylation and improves quantum catch in retinoid-deficient mouse rod photoreceptors. J Biol Chem. 2002;277:40491–40498. doi: 10.1074/jbc.M205507200. [DOI] [PubMed] [Google Scholar]
- Angers S, Salahpour A, Bouvier M. Biochemical and biophysical demonstration of GPCR oligomerization in mammalian cells. Life Sci. 2001;68:2243–2250. doi: 10.1016/s0024-3205(01)01012-8. [DOI] [PubMed] [Google Scholar]
- Bellingham J, Wells DJ, Foster RG. In silico characterisation and chromosomal localisation of human RRH (peropsin) –implications for opsin evolution. BMC Genomics. 2003;4:3. doi: 10.1186/1471-2164-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Hao W, Rife L, Wang XP, Shen D, Chen J, Ogden T, Van Boemel GB, Wu L, Yang M, Fong HK. A photic visual cycle of rhodopsin regeneration is dependent on Rgr. Nat Genet. 2001a;28:256–260. doi: 10.1038/90089. [DOI] [PubMed] [Google Scholar]
- Chen P, Lee TD, Fong HK. Interaction of 11-cis-retinol dehydrogenase with the chromophore of retinal G protein-coupled receptor opsin. J Biol Chem. 2001b;276:21098–21104. doi: 10.1074/jbc.M010441200. [DOI] [PubMed] [Google Scholar]
- Cideciyan AV, Haeseleer F, Fariss RN, Aleman TS, Jang GF, Verlinde CL, Marmor MF, Jacobson SG, Palczewski K. Rod and cone visual cycle consequences of a null mutation in the 11-cis-retinol dehydrogenase gene in man. Vis Neurosci. 2000;17:667–678. doi: 10.1017/s0952523800175029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean MK, Higgs C, Smith RE, Bywater RP, Snell CR, Scott PD, Upton GJ, Howe TJ, Reynolds CA. Dimerization of G-protein-coupled receptors. J Med Chem. 2001;44:4595–4614. doi: 10.1021/jm010290+. [DOI] [PubMed] [Google Scholar]
- Driessen CA, Winkens HJ, Hoffmann K, Kuhlmann LD, Janssen BP, Van Vugt AH, Van Hooser JP, Wieringa BE, Deutman AF, Palczewski K, Ruether K, Janssen JJ. Disruption of the 11-cis-retinol dehydrogenase gene leads to accumulation of cis-retinols and cis-retinyl esters. Mol Cell Biol. 2000;20:4275–4287. doi: 10.1128/mcb.20.12.4275-4287.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrey T, Koutalos Y. Vertebrate photoreceptors. Prog Retina Eye Res. 2001;20:49–94. doi: 10.1016/s1350-9462(00)00014-8. [DOI] [PubMed] [Google Scholar]
- Filipek S, Teller DC, Palczewski K, Stenkamp RE. The cyrstallographic model of rhodopsin and its use in studies of other G-protein coupled receptors. Annu Rev Biophys Biomol Struct. 2003;32:375–397. doi: 10.1146/annurev.biophys.32.110601.142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Atomic-force microscopy: rhodopsin dimers in native disc membranes. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- Garwin GG, Saari JC. High-performance liquid chromatography analysis of visual cycle retinoids. Methods Enzymol. 2000;316:313–324. doi: 10.1016/s0076-6879(00)16731-x. [DOI] [PubMed] [Google Scholar]
- Haeseleer F, Jang GF, Imanishi Y, Driessen CA, Matsumura M, Nelson PS, Palczewski K. Dual-substrate specificity short chain retinol dehydrogenases from the vertebrate retina. J Biol Chem. 2002;277:45537–45546. doi: 10.1074/jbc.M208882200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W, Fong HK. Blue and ultraviolet light-absorbing opsin from the retinal pigment epithelium. Biochemistry. 1996;35:6251–6256. doi: 10.1021/bi952420k. [DOI] [PubMed] [Google Scholar]
- Hao W, Fong HK. The endogenous chromophore of retinal G protein-coupled receptor opsin from the pigment epithelium. J Biol Chem. 1999;274:6085–6090. doi: 10.1074/jbc.274.10.6085. [DOI] [PubMed] [Google Scholar]
- Hao W, Chen P, Fong HK. Analysis of chromophore of RGR: retinal G-protein-coupled receptor from pigment epithelium. Methods Enzymol. 2000;316:413–422. doi: 10.1016/s0076-6879(00)16739-4. [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I, Matsumoto T, Mori H, Nishimura M, Hara R, Hara T. Cloning and nucleotide sequence of cDNA for retinochrome, retinal. FEBS Lett. 1990;271:106–110. doi: 10.1016/0014-5793(90)80383-t. [DOI] [PubMed] [Google Scholar]
- Hetling JR, Pepperberg DR. Sensitivity and kinetics of mouse rod flash responses determined in vivo. J Physiol Lond. 1999;516:593–609. doi: 10.1111/j.1469-7793.1999.0593v.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang GF, McBee JK, Alekseev AM, Haeseleer F, Palczewski K. Stereoisomeric specificity of the retinoid cycle in the vertebrate retina. J Biol Chem. 2000;275:28128–28138. doi: 10.1074/jbc.M004488200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang GF, Van Hooser JP, Kuksa V, McBee JK, He YG, Janssen JJ, Driessen CA, Palczewski K. Characterization of a dehydrogenase activity responsible for oxidation of 11-cis-retinol in the retinal pigment epithelium of mice with a disrupted rdh5 gene. A model for the human hereditary disease fundus albi-punctatus. J Biol Chem. 2001;276:32456–32465. doi: 10.1074/jbc.M104949200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Pandey S, Fong HK. An opsin homologue in the retina and pigment epithelium. Invest Ophthalmol Vis Sci. 1993;34:3669–3678. [PubMed] [Google Scholar]
- Koyanagi M, Terakita A, Kubokawa K, Shichida Y. Amphioxus homologs of Go-coupled rhodopsin and peropsin having 11-cis- and all-trans-retinals as their chromophores. FEBS Lett. 2002;531:525–528. doi: 10.1016/s0014-5793(02)03616-5. [DOI] [PubMed] [Google Scholar]
- Kuksa V, Bartl F, Maeda T, Jang GF, Ritter E, Heck M, Van Hooser JP, Liang Y, Filipek S, Gelb MH, Hofmann KP, Palczewski K. Biochemical and physiological properties of rhodopsin regenerated with 11-cis-6-ring- and 7-ring-retinals. J Biol Chem. 2002;277:42315–42324. doi: 10.1074/jbc.M206014200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubarsky AL, Chen C, Simon MI, Pugh EN., Jr Mice lacking G-protein receptor kinase 1 have profoundly slowed recovery of cone-driven retinal responses. J Neurosci. 2000;20:2209–2217. doi: 10.1523/JNEUROSCI.20-06-02209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Maeda A, Maruyama I, Ogawa KI, Kuroki Y, Sahara H, Sato N, Ohguro H. Mechanisms of photoreceptor cell death in cancer-associated retinopathy. Invest Ophthalmol Vis Sci. 2001;42:705–712. [PubMed] [Google Scholar]
- McBee JK, Palczewski K, Baehr W, Pepperberg DR. Confronting complexity: the interlink of phototransduction and retinoid metabolism in the vertebrate retina. Prog Retin Eye Res. 2001a;20:469–529. doi: 10.1016/s1350-9462(01)00002-7. [DOI] [PubMed] [Google Scholar]
- McBee JK, Van Hooser JP, Jang GF, Palczewski K. Isomerization of 11-cis-retinoids to all-trans-retinoids in vitro and in vivo. J Biol Chem. 2001b;276:48483–48493. doi: 10.1074/jbc.M105840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadegan T, Benk G, Filipek S, Palczewski K. Sequence analyses of G-protein-coupled receptors: Similarities to rhodopsin. Biochemistry. 2003;42:2759–2767. doi: 10.1021/bi027224+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimura H, Saindelle Ribeaudeau F, Berson EL, Dryja TP. Mutations in RGR, encoding a light-sensitive opsin homologue, in patients with retinitis pigmentosa. Nat Genet. 1999;23:393–394. doi: 10.1038/70496. [DOI] [PubMed] [Google Scholar]
- Okada T, Ernst OP, Palczewski K, Hofmann KP. Activation of rhodopsin: new insights from structural and biochemical studies. Trends Biochem Sci. 2001;26:318–324. doi: 10.1016/s0968-0004(01)01799-6. [DOI] [PubMed] [Google Scholar]
- Overton MC, Blumer KJ. G-protein-coupled receptors function as oligomers in vivo. Curr Biol. 2000;10:341–344. doi: 10.1016/s0960-9822(00)00386-9. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Van Hooser JP, Garwin GG, Chen J, Liou GI, Saari JC. Kinetics of visual pigment regeneration in excised mouse eyes and in mice with a targeted disruption of the gene encoding interphotoreceptor retinoid-binding protein or arrestin. Biochemistry. 1999;38:12012–12019. doi: 10.1021/bi990504d. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Pandey S, Blanks JC, Spee C, Jiang M, Fong HK. Cytoplasmic retinal localization of an evolutionary homolog of the visual pigments. Exp Eye Res. 1994;58:605–613. doi: 10.1006/exer.1994.1055. [DOI] [PubMed] [Google Scholar]
- Pepperberg DR, Crouch RK. An illuminating new step in visual-pigment regeneration. Lancet. 2001;358:2098–2099. doi: 10.1016/S0140-6736(01)07231-2. [DOI] [PubMed] [Google Scholar]
- Polans A, Baehr W, Palczewski K. Turned on by Ca2+! The physiology and pathology of Ca2+-binding proteins in the retina. Trends Neurosci. 1996;19:547–554. doi: 10.1016/s0166-2236(96)10059-x. [DOI] [PubMed] [Google Scholar]
- Pugh EN, Jr, Nikonov S, Lamb TD. Molecular mechanisms of vertebrate photoreceptor light adaptation. Curr Opin Neurobiol. 1999;9:410–418. doi: 10.1016/S0959-4388(99)80062-2. [DOI] [PubMed] [Google Scholar]
- Rattner A, Sun H, Nathans J. Molecular genetics of human retinal disease. Annu Rev Genet. 1999;33:89–131. doi: 10.1146/annurev.genet.33.1.89. [DOI] [PubMed] [Google Scholar]
- Redmond TM, Hamel CP. Genetic analysis of RPE65: from human disease to mouse model. Methods Enzymol. 2000;316:705–724. doi: 10.1016/s0076-6879(00)16758-8. [DOI] [PubMed] [Google Scholar]
- Redmond TM, Yu S, Lee E, Bok D, Hamasaki D, Chen N, Goletz P, Ma JX, Crouch RK, Pfeifer K. Rpe65 is necessary for production of 11-cis-vitamin A in the retinal visual cycle. Nat Genet. 1998;20:344–351. doi: 10.1038/3813. [DOI] [PubMed] [Google Scholar]
- Rios CD, Jordan BA, Gomes I, Devi LA. G-protein-coupled receptor dimerization: modulation of receptor function. Pharmacol Ther. 2001;92:71–87. doi: 10.1016/s0163-7258(01)00160-7. [DOI] [PubMed] [Google Scholar]
- Saari JC, Garwin GG, Van Hooser JP, Palczewski K. Reduction of all-trans-retinal limits regeneration of visual pigment in mice. Vis Res. 1998;38:1325–1333. doi: 10.1016/s0042-6989(97)00198-3. [DOI] [PubMed] [Google Scholar]
- Seeliger MW, Grimm C, Stahlberg F, Friedburg C, Jaissle G, Zrenner E, Guo H, Reme CE, Humphries P, Hofmann F, Biel M, Fariss RN, Redmond TM, Wenzel A. New views on RPE65 deficiency: the rod system is the source of vision in a mouse model of Leber congenital amaurosis. Nat Genet. 2001;29:70–74. doi: 10.1038/ng712. [DOI] [PubMed] [Google Scholar]
- Shen D, Jiang M, Hao W, Tao L, Salazar M, Fong HK. A human opsin-related gene that encodes a retinaldehyde-binding protein. Biochemistry. 1994;33:13117–13125. doi: 10.1021/bi00248a022. [DOI] [PubMed] [Google Scholar]
- Shichida Y, Imai H. Visual pigment: G-protein-coupled receptor for light signals. Cell Mol Life Sci. 1998;54:1299–1315. doi: 10.1007/s000180050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher H, Gelb MH, Saari JC, Palczewski K. Preferential release of 11-cis-retinol from retinal pigment epithelial cells in the presence of cellular retinaldehyde-binding protein. J Biol Chem. 1999;274:8577–8585. doi: 10.1074/jbc.274.13.8577. [DOI] [PubMed] [Google Scholar]
- Sun H, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J. Peropsin, a novel visual pigment-like protein located in the apical microvilli of the retinal pigment epithelium. Proc Natl Acad Sci USA. 1997;94:9893–9898. doi: 10.1073/pnas.94.18.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teller DC, Okada T, Behnke CA, Palczewski K, Stenkamp RE. Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs) Biochemistry. 2001;40:7761–7772. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakita A, Yamashita T, Shichida Y. Highly conserved glutamic acid in the extracellular IV–V loop in rhodopsins acts as the counterion in retinochrome, a member of the rhodopsin family [in process citation] Proc Natl Acad Sci USA. 2000;97:14263–14267. doi: 10.1073/pnas.260349597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser JP, Garwin GG, Saari JC. Analysis of visual cycle in normal and transgenic mice. Methods Enzymol. 2000a;316:565–575. doi: 10.1016/s0076-6879(00)16750-3. [DOI] [PubMed] [Google Scholar]
- Van Hooser JP, Aleman TS, He YG, Cideciyan AV, Kuksa V, Pittler SJ, Stone EM, Jacobson SG, Palczewski K. Rapid restoration of visual pigment and function with oral retinoid in a mouse model of childhood blindness. Proc Natl Acad Sci USA. 2000b;97:8623–8628. doi: 10.1073/pnas.150236297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser JP, Liang Y, Maeda T, Kuksa V, Jang GF, He YG, Rieke F, Fong HK, Detwiler PB, Palczewski K. Recovery of visual functions in a mouse model of Leber congenital amaurosis. J Biol Chem. 2002;277:19173–19182. doi: 10.1074/jbc.M112384200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu BX, Chen Y, Fan J, Rohrer B, Crouch RK, Ma JX. Cloning and characterization of a novel all-trans retinol short-chain dehydrogenase/reductase from the RPE. Invest Ophthalmol Vis Sci. 2002;43:3365–3372. [PubMed] [Google Scholar]
- Yamamoto H, Simon A, Eriksson U, Harris E, Berson EL, Dryja TP. Mutations in the gene encoding 11-cis retinol dehydrogenase cause delayed dark adaptation and fundus albipunctatus. Nat Genet. 1999;22:188–191. doi: 10.1038/9707. [DOI] [PubMed] [Google Scholar]
- Yang M, Fong HK. Synthesis of the all-trans-retinal chromophore of retinal G protein-coupled receptor opsin in cultured pigment epithelial cells. J Biol Chem. 2002;277:3318–3324. doi: 10.1074/jbc.M108946200. [DOI] [PubMed] [Google Scholar]
- Znoiko SL, Crouch RK, Moiseyev G, Ma JX. Identification of the RPE65 protein in mammalian cone photoreceptors. Invest Ophthalmol Vis Sci. 2002;43:1604–1609. [PubMed] [Google Scholar]


